Abstract
Rapid and accurate classification of the etiology for acute respiratory illness not only helps establish timely therapeutic plans but also prevents inappropriate use of antibiotics. Host gene expression patterns in peripheral blood can discriminate bacterial from viral causes of acute respiratory infection (ARI) but suffer from long turnaround time, as well as high cost resulting from the measurement methods of microarrays and next-generation sequencing. Here, we developed an automated DNA computing–based platform that can implement an in silico trained classification model at the molecular level with seven different mRNA expression patterns for accurate diagnosis of ARI etiology in 4 hours. By integrating sample loading, marker amplification, classifier implementation, and results reporting into one platform, we obtained a diagnostic accuracy of 87% in 80 clinical samples without the aid of computer and laboratory technicians. This platform creates opportunities toward an accurate, rapid, low-cost, and automated diagnosis of disease etiology in emergency departments or point-of-care clinics.
A DNA computation platform enables automated, rapid, and accurate classification of acute respiratory illness etiology.
INTRODUCTION
Acute respiratory infections (ARIs) caused by bacterial or viral pathogens are the leading causes for seeking medical attention. Difficulties in distinguishing between bacterial and viral infections can lead to the overuse of antibiotics. For instance, some clinical and emergency room (ER) visits with suspected respiratory infections still lead to prescribing antibiotics, although most of these infections still originate from viruses. Driven by the overuse of antibiotics, antibiotic resistance is progressing at an alarming rate, outpacing the development of new antibiotics and contributing to the rise of health care costs. Diagnostic tests that rapidly and accurately identify the causes of ARI can provide more personalized care and reduce the inappropriate use of antibiotics.
Traditional etiological diagnostic methods rely on the identification of pathogenic antigen or genetic molecules in the host (1). However, these methods may be limited when the concentration of a pathogen is too low for reliable detection or the pathogenic role of a detected microbial agent derives from normal colonization instead of infection. Measuring host response induced by ARI provides an alternative diagnostic strategy. For instance, complement-reactive protein (CRP) (2), procalcitonin (3, 4), and human neutrophilic apolipoprotein (5, 6) normally increase in bacterial infections. However, these biomarkers have only shown moderate accuracy (65 to 75%) in distinguishing between bacterial and viral infections in the clinic. Analysis of the transcriptional profile in host peripheral blood offers a novel strategy to diagnose ARI, as circulating blood leukocytes react differently to bacterial and viral pathogens and lead to variations of the expression of host genes related to immune functions (7). In previous studies, host transcriptional profiles have been characterized for viral and bacterial ARI with high-throughput assays, and a classifier with 104 host genes could diagnose ARI with an overall accuracy of 87% in a cohort of 273 individuals (8–14). Existing high-throughput RNA assays, such as microarrays (15) and RNA sequencing (16), can be used to quantitatively observe changes in massive host gene expressions. However, they are difficult to implement in the ER or point-of-care clinics where fast data turnaround time and low cost are prerequisites for diagnosing the cause of ARI (17–19). To overcome these potential limitations, it is essential to develop rapid and practical diagnostic tests that can be used to detect the transcriptional profile for classifying ARI in a clinical setting.
DNA-based molecular computation relies on DNA as hardware and biochemical reactions as software and has been experimentally proven to be a powerful tool for implementing complex computation on different substrates toward various applications, such as solving mathematical problems, recognizing complicated patterns, and intelligent delivery of drugs (20–27). With the development of DNA programming methods, different logical connectives (e.g., AND, OR, and NOT) and algorithms, such as weighting, summing, and subtraction, can be implemented at the molecular level without human intervention (28–30). In principle, DNA molecular computing can extend the applications of diagnostic methods from single to multiple markers, as well as integrate the information encoded in a multimarker signature with logical analysis for more intelligent diagnostics. These features also enable the direct reporting of diagnostic results in a “sensing-thinking” mode of implementing complicated classifier models at molecular levels. So far, well-designed DNA computation systems have been used to generate diagnostic information from computational models for cancer screening without the aid of computers, but human intervention is still required, such as sample preparation and loadings that prevent the implementation of complete automation of DNA computing–based diagnostics (31). Therefore, we here sought to explore a fully automated DNA computational system to diagnose the etiology of ARIs by integrating sample loading, marker amplification, classifier implementation, and results reporting into one platform to achieve a “sample in and result out” flow without the aid of any computer or laboratory specialists. We expect this automated DNA computing–based platform to offer a more rapid and accurate way of identifying ARI etiology in ERs or point-of-care clinics, thereby guiding appropriate antibiotic use and combating emerging antibiotic resistance.
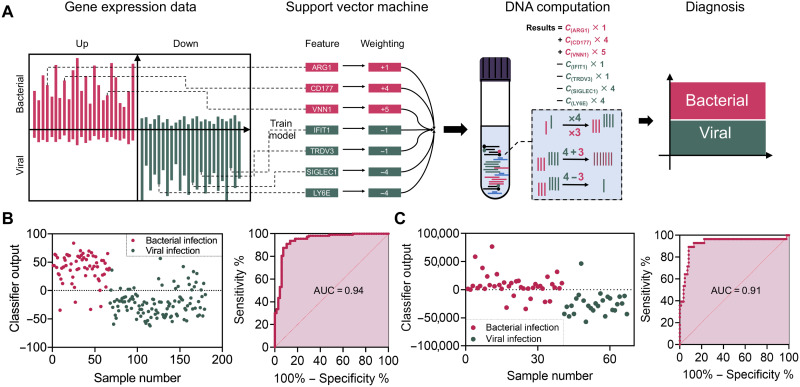
As shown in Fig. 1 (A and B), the workflow of the DNA-based molecular classifier includes three main steps. First, an in silico classifier model trained with a support-vector machine (SVM) is constructed using publicly available transcription of viral and bacterial ARI from the National Center for Biotechnology Information (NCBI) database. This step aims to identify classifiers that contain minimum varieties (i.e., genes) with no redundancy and high diagnostic accuracy. Second, a DNA-based molecular classifier is designed to implement the in silico trained model at the molecular level. Specifically, it can perform arithmetic operations, such as weighting, summing, and subtraction, in response to the transcriptional signals in solution and report results as either bacterial or viral in nature. Last, the molecular classifier is used to diagnose the cause of ARI in clinical samples. We automated the diagnostic flow of this system by integrating a mechanical sample loading machine, consisting of a thermocycler and a fluorescence reader, thereby achieving fully automated diagnostics of ARI. Using this method, we obtained a diagnostic accuracy of classifying bacterial and viral ARI of 87% in 80 human blood samples within 4 hours, compared to the accuracy of most clinically used CRP testing and complete blood count (CBC) of 74% and 54 to 62%, respectively, with the same cohort of blood samples.
Fig. 1. DNA computational platform for ARI diagnosis.
(A) Workflow for the classification of ARI etiology with the DNA computation platform. (B) Performance of the molecular classifier with data from the training set. The training dataset contains transcriptome sequencing data from samples containing 113 viral and 67 bacterial ARIs, in which 159 of samples were classified correctly with area under the curve (AUC) = 0.94 (95% CI: 0.9040 to 0.9803). Data are from the NCBI database (GSE63990). (C) The validation dataset contains transcriptome sequencing data from 27 viral ARI and 57 bacterial ARI samples with AUC = 0.91 (95% CI: 0.8270 to 0.9915). Data are from the NCBI database (GSE6269).
RESULTS
Model construction
To build an effective model classifier to discriminate between bacterial and viral ARI, we chose the SVM, a supervised learning model with specific algorithms, to train the data according to the following conditions and steps (32). First, publicly available transcriptional profiles of host peripheral blood corresponding to 113 viral ARI and 67 bacterial ARI from the NCBI database (GSE63990) were selected as the training set. These data were used for differential expression analysis with the goal of classifying the samples into bacterial and viral infection groups with an anticipated accuracy >80%. We successfully identified 30 up-regulated and 30 down-regulated mRNA candidates with stable expression differences that are four times larger, given that larger differences typically have higher classification sensitivity and specificity (fig. S1). Next, more constraints were applied to the 60 selected mRNA candidates to obtain minimal input sets while maintaining the discriminatory accuracy of the classifier (>80%). For example, we artificially limited weights to integers less than 10 and mathematical operations to addition, multiplication, and subtraction. This was done to identify the most practical models with the smallest number of mRNA inputs for subsequent molecular computation. Among the multiple in silico trained models, we selected a classifier with the highest area under the curve (AUC) value as well as reasonable numbers of mRNAs, the components of which included seven mRNAs, namely, SIGLEC1, LY6E, IFIT1, TRDV3, VNN1, CD177, and ARG1, associated with negative and positive weights, ranging from integer values of −4 to +5. This model could discriminate between bacterial and viral ARI with an AUC of 0.94 [95% confidence interval (CI): 0.9040 to 0.9803] with the training database (Fig. 1C). We also validated the model using another dataset of NCBI (GSE6269) with an obtained AUC of 0.91 (95% CI: 0.8270 to 0.9915; Fig. 1D). In addition, of the seven noted candidates, individual mRNAs had limited diagnostic accuracy by either low sensitivity or insufficient specificity (fig. S2 and table S1). These results highlight the importance of using multiple markers for more accurate diagnostics. Therefore, once having constructed a molecular classifier that displayed high discriminatory sensitivity and specificity using simplified mathematical operations, we proceeded to explore the implementation of molecular computation using a set of well-designed DNA probes for diagnosing the causes of ARI (33).
RNA amplification
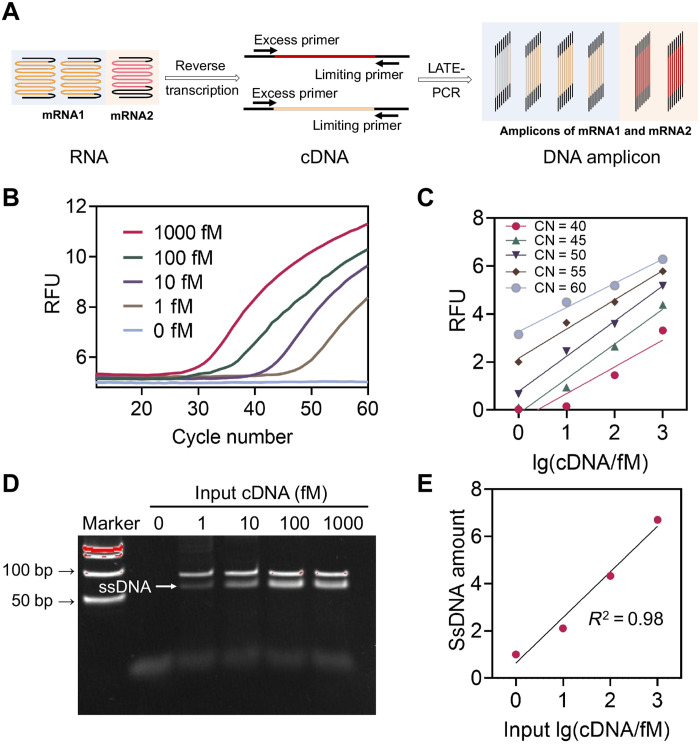
Low concentration of analytes, such as microRNA and mRNA in biological samples, poses a barrier limiting the clinical applications of DNA-based molecular computation and hence the reporting accuracy of DNA-based interactions. Our previously developed amplification method used an improved asymmetric polymerase chain reaction [Linear-After-The-Exponential (LATE)-PCR] and achieved near-linear amplification of complementary DNA (cDNA) targets from low concentrations in clinical samples (≤pM; figs. S3 and S4) to a higher detectable range (>nM) (34). In this way, the amplified sample still has data associated with the quantity of initial mRNA, a key index for accurate classification. As shown in Fig. 2, we first extracted total RNA from fresh human whole blood using the QIAamp RNA Blood Mini Kit. Total RNAs were then reversely transcribed into cDNA with Moloney murine leukemia virus (M-MLV) reverse transcriptase. Then, the acquired cDNAs were amplified by LATE-PCR with specific primer pairs. By adjusting the nucleotide composition and numbers of the excess and limiting primers, as well as their ratios (figs. S5 and S6), the exponential amplification stage of the PCR could be quickly switched to the linear stage when the limiting primers are exhausted (Fig. 2, B and D) (35, 36). Meanwhile, we can maintain the amplified single-stranded DNA (ssDNA) products in an appropriate concentration range by controlling the number of amplification cycles, thus providing optimal conditions for subsequent DNA computing. Figure 2C shows the relative fluorescence units of ssDNA products amplified from initial mRNA concentrations ranging from 1 fM to 1 pM using LATE-PCR. The results show the linear relationship between the initial concentrations of mRNAs and the final concentrations of cDNAs. In general, the LATE-PCR can help reduce the bias caused by traditional exponential amplification and, at the same time, convert the low concentrations of mRNA inputs (≤pM) to higher concentrations of ssDNAs (≥nM; fig. S7) without interfering with their original ratios. The accuracy and efficiency of each primer set for the seven mRNA targets were verified through qPCR (SYBR Green) amplification. The fluorescence melting curve and polyacrylamide gel electrophoresis (PAGE) of the PCR products are shown in figs. S8 and S9, respectively.
Fig. 2. RNA amplification for molecular computation.
(A) Schematic illustration of mRNA amplification in clinical samples. (B) Detection of LATE-PCR products with different initial concentrations of cDNA (reverse transcription template of mRNA IFIT1, as an example) using a FAM-labeled TaqMan probe. (C) Plot of endpoint PCR fluorescence at different cycle numbers versus initial IFIT1 cDNA concentrations, proving the linear amplification behavior of LATE-PCR with the target concentrations from 1 fM up to 1 pM [cycle number (CN) = 40, R2 = 0.89; CN = 45, R2 = 0.98; CN = 50, R2 = 0.99; CN = 55, R2 = 0.98; CN = 60, R2 = 0.98]. (D) Native PAGE analysis of products after LATE-PCR. (E) Plot of band intensities of the amplified ssDNA products versus initial IFIT1 cDNA concentrations. R2 = 0.98. RFU, relative fluorescence units.
Implementation of molecular classifier
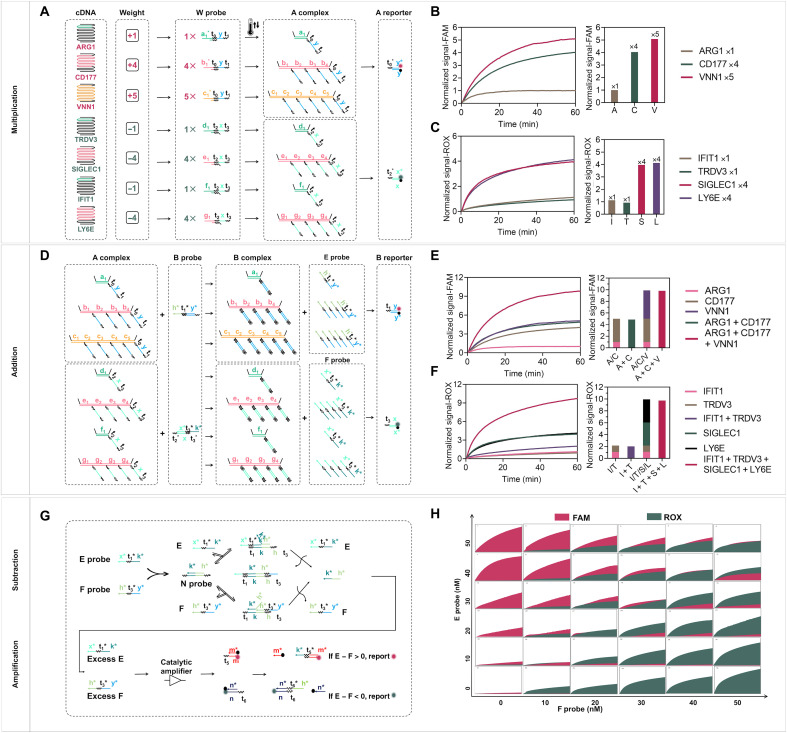
Performing the classifier at molecular levels can directly report the calculated results and reduce experimental errors caused by parallel measurement of multiple mRNA inputs with traditional PCR-based methods. Here, we adapted a simplified winner-take-all computing scheme initially reported by Cherry and Qian (37) to implement the trained molecular classifier systematically. Specifically, the mathematical weighted sums of bacterial infection–related mRNA inputs are compared with those of viral infection mRNA inputs, and only the larger value is reported with the corresponding fluorescence signals for clinical diagnostics. The computing scheme of the trained classifier is abstracted into three substeps, including multiplication, addition, and subtraction at a molecular level, as implemented by cascading DNA strand displacement reactions (38–40).
The detailed DNA computing process is shown in Fig. 3. First, different cDNA amplicons linearly amplified from original mRNA inputs undergo weighted multiplication [Wn × c(cDNA)n = An], where Wn is the weight predetermined by the in silico model, c(cDNA)n is the concentration of cDNA, and An is the weighted multiplication, respectively. In particular, mRNA VNN1, CD177, and ARG1 were found to be positively correlated to bacterial infection with weights of +5, +4, and +1, respectively. Meanwhile, SIGLEC1, LY6E, IFIT1, and TRDV3 were found to be negatively associated with bacterial infection with weights of −4, −4, −1, and −1, respectively. The numerical weights associated with these seven mRNAs actually capture their degrees of importance in the diagnostic determination for ARI. To take advantage of this, we designed multiplication probes (W probe) that could be weighted for different target sequence regions of each cDNA amplicon (Fig. 3A). For instance, weights (Wn) of 5, 4, or 1 are realized by having 5, 4, or 1 distinct sequence region that can be located by W probes in the same cDNA amplicon (33). We used a fluorescence reporting scheme based on a one-step DNA strand displacement reaction to confirm the correctness of multiplication calculation [Wn × c(cDNA)n = An] by examining the concentrations of resulting A complexes (An). To facilitate strand displacement reaction between W probe and the cDNA inputs with extensive secondary structures, we implemented a thermal annealing strategy in which W probes were first annealed with the cDNAs reversely transcribed from the original mRNA before the addition of the subsequent computing probes. As expected, we observed a better response of the cDNA target to the reporting probes (based on strand displacement reaction; fig. S10) after thermal annealing. As shown in Fig. 3 (B and C), the steady-state signals accurately reflect the weighted signals of different cDNA inputs with equal initial concentration (10 nM), indicating that the molecules effectively perform the multiplication operations as designed. Next, the different weights of mRNA inputs are summed for comparison (A1 + A2 + A3 = E; A4 + A5 + A6 + A7 = F). This step is done by using a summation probe (B probe) that reacts with the A complex generated in the previous multiplication step. In particular, this B probe can bind to the same sequence regions in the A complex and convert the same category of inputs into a common weighted sum species (E or F probe) (Fig. 3D). We also detected the summed signals of An (i.e., E and F probes) using a DNA displacement reaction–based fluorescence reporter probe (B reporter) and obtained the matched signals reflecting the corresponding correct total concentrations (Fig. 3, E and F). Therefore, this summation scheme can precisely determine the weighted sum of the same type of inputs at the molecular level. Following this step, the classification result is reported after subtracting the sums of two different types of summed inputs, i.e., diagnostic result = E − F (Fig. 3G). It is realized by using an N probe able to annihilate the different types of weighted sums (E and F probes). Specifically, E and F probes bind to the N probe in a reversible annihilation manner such that only the remaining one can be reported by FAM (E) or ROX (F) fluorescence with a catalytic entropy-driven amplification step for improving reporting sensitivity (fig. S11) (41). Details of N probe optimization are shown in fig. S12. Last, we validated this process in Fig. 3H using 36 different combinations of weighted sums (E and F probes) with concentrations ranging from 0 to 50 nM. The results demonstrated that higher concentrations of the weighted sums could report higher signals, which is consistent with the proposed scheme.
Fig. 3. Validation of the DNA molecular classifier.
(A) Scheme for multiplication [Wn × c(cDNA)n = An]. A reporter is used to verify the multiplication results. (B) Fluorescence kinetics of 10 nM cDNA of ARG1, CD177, and VNN1 genes in the FAM channel (Wn = 1, 4, and 5). (C) Fluorescence kinetics of 10 nM cDNA of IFIT1, TRDV3, SIGLEC1, and LY6E genes with different weights in the ROX channel (Wn = 1, 1, 4, and 4). (D) Scheme for summation (E = A1 + A2 + A3; F = A4 + A5 + A6 + A7). (E) Fluorescence kinetics of A1, A2, and A3 alone and their sums (A1 + A2, A1 + A2 + A3) in the presence of equivalent B probes and B reporter in the FAM channel (i.e., 1, 4, 5, 1 + 4, 1 + 5, and 4 + 5). The normalized FAM fluorescence at 60 min was used to compare the individual and summation signals. (F) Fluorescence kinetics of A4, A5, A6, and A7 alone and their sums (A4 + A5 and A4 + A5 + A6 + A7) in the presence of equivalent B probes and B reporter (i.e., 1, 4, 5, 1 + 4, 1 + 5, and 4 + 5). The normalized ROX fluorescence at 60 min was used to compare the individual and summation signals. (G) Scheme for subtraction (diagnostic result = E − F) and amplification step for sensitivity improvement. Excess E and F probes can be reported by FAM and ROX, respectively. (H) Winner-take-all behavior for different ratios of E and F. The x axis of each small graph is time (from 0 to 60 min), and the y axis is the relative fluorescence units. Samples resulting in a normalized signal of [FAM] − [ROX] > 0 belong to the bacterial infection category, and samples with [FAM] − [ROX] < 0 belong to the viral infection category.
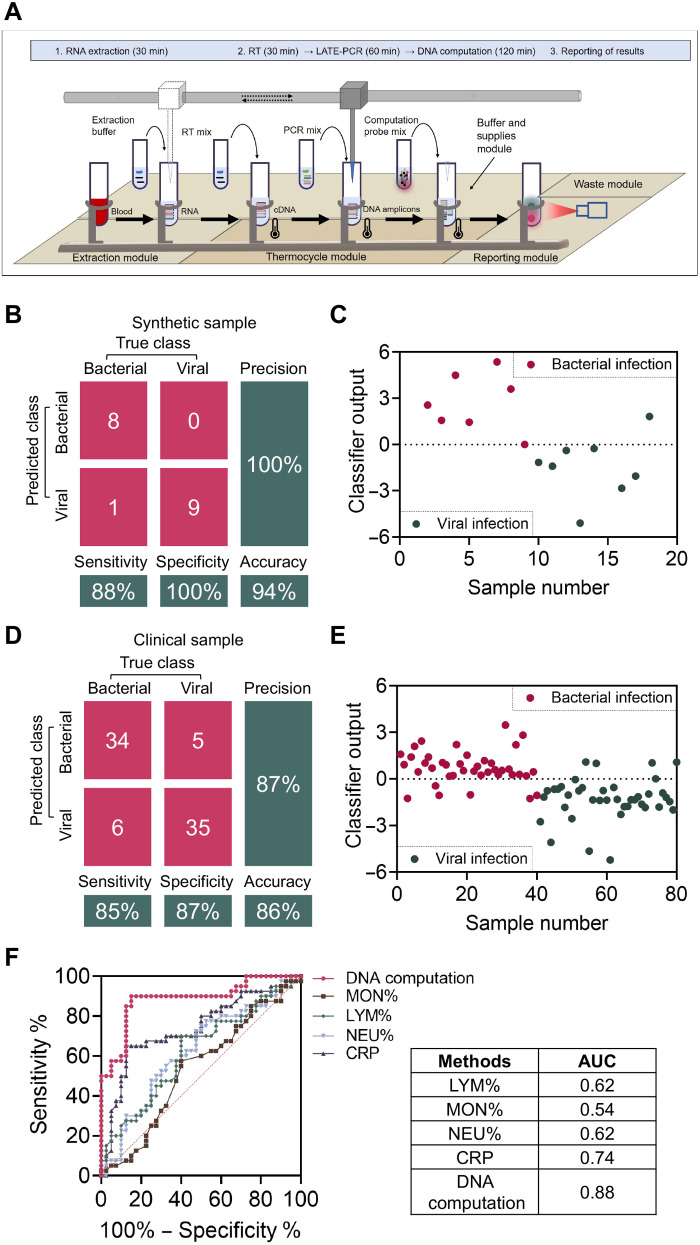
We next integrate the entire workflow of the DNA computing–based platform to classify bacterial and viral ARI from “input” to “output.” We replicated 18 synthetic samples containing 9 bacterial and 9 viral ARI and used each cDNA target at calculated concentrations from table S2 as inputs. After LATE-PCR–based amplification and DNA computation (multiplication, summation, and subtraction), fluorescent signals were directly detected to obtain the diagnostic results of these synthetic samples. As shown in Fig. 4B and fig. S13, only one viral infection sample was misdiagnosed as bacterial infection, and the remaining eight viral infection samples and nine bacterial infection samples were classified correctly with a sensitivity of 88.9% (95% CI: 0.5067 to 0.9942), a specificity of 100% (95% CI: 0.6288 to 1), and an accuracy of 94.4% (95% CI: 0.5067 to 0.9942). These data verified that the DNA-based molecular classifier can intelligently perform computation as designed for synthetic inputs (Fig. 4C).
Fig. 4. Validation of the DNA computation–based molecular classifier.
(A) Automated operational flow using DNA computation–based molecular classifier for the classification of ARI with clinical whole-blood samples automatically. Specifically, we integrated an RNA extractor (ZJ Bio EX3600 Plus), a thermocycler [PCR thermal module by a qPCR machine (Roche Z480)], and a fluorescence reader [reporting module by a qPCR machine (Roche Z480)] to a complete workflow. A mechanical sample loading machine (Hamilton & Microlab STARlet 8) was used to transfer sample tubes and add reactants. (B) Confusion matrix analysis of the 18 synthetic samples. (C) Classification results of the DNA computation–based diagnostic system with 18 synthetic samples. (D) Confusion matrix analysis of the 80 clinical samples. (E) Classification results of the DNA computation–based diagnostic system with 80 clinical samples. (F) Performance of this method in diagnosing bacterial infection. The clinical samples were correctly classified with AUC = 0.88 (95% CI: 0.8097 to 0.9615).
Automated diagnosis of ARI etiology with the DNA computing platform
To prove that our method can be a reliable and convenient platform for the clinical diagnosis of ARI, two key factors need to be satisfied. The first is high accuracy of classifying ARI in clinical samples. The second is high throughput and automation level to fit the requirements of a clinical setting. Correspondingly, we studied the classification accuracy of our system using whole-blood samples (1 ml) from 80 patients with ARI. In particular, these samples were confirmed with ARI by successfully identifying the pathogens with either antibody tests or bacterial cultures for viral and bacterial infections. Table S3 summarizes the clinical features of the samples. Here, we assembled the diagnostic platform into a completely automated flow by integrating an RNA extractor, a mechanical sample loading machine, a thermocycler, and a fluorescence reader. After direct reading of the output fluorescence of these samples using the described automated workflow (Fig. 4A), it can be seen that 34 of 40 bacterial infection samples were identified correctly with a sensitivity of 85.0% (95% CI: 0.6947 to 0.9375), while 6 of 40 viral infection samples were misdiagnosed as bacterial with a specificity of 87.5% (95% CI: 0.7239 to 0.9530; Fig. 4, D and E, and fig. S14). The total accuracy of this method in clinical samples was 86.2% (95% CI: 0.7630 to 0.9260) with an AUC of 0.88 (95% CI: 0.8097 to 0.9615). We also classified these samples with other clinically available methods, including CBC based on neutrophilic granulocyte, lymphocyte, and monocyte percentage (NEU%, LYM%, and MON%), as well as CRP test for the same cohort. The obtained AUCs of these two methods are between 0.54 and 0.74, indicating less accuracy than that of our DNA computation method, as shown in Fig. 4F. In addition, with the help of complete automation flow, the entire testing procedure only took 3.5 to 4 hours from raw blood sample to the final diagnostic results, which is fully compatible with the need for fast turnaround time for ARI diagnostics in ERs and point-of-care settings.
Compared with the existing methods (table S4) for acute respiratory illness etiology identification, our automated DNA computation method shows some advantages such as high accuracy, fast turnaround time, and high automation level. Specifically, the power of data training in silico, coupled with the molecular implementation of a winner-take-all computing scheme, enables us to perform disease diagnosis in an automated manner. More fundamentally, the workload required to classify gene expression with our system is independent of the number of genes used in the analysis, as DNA computation allows the parallel introduction of multiple inputs. In contrast, RT-qPCR is currently the gold standard for quantitation of gene expression profiles in clinics, and its complexity is proportionally correlated with the number of genes being analyzed, as more genes require more human resources on the tedious and step-by-step quantitation of individual RNA. At the same time, RNA sequencing and microarray also allow for multigene expression analysis in a single reaction, but they require expensive instrumentation and consumables and have slow data turnaround time. Second, an automated system is established in the workflow to couple with DNA computation, providing a feasible path for realizing automatic and rapid diagnostics of ARI in clinics. If provided with suitable consumables, such as a 384-well plate, the entire system could produce up to 600 tests per day per machine. Last, the diagnostic accuracy of our method is higher than that of existing clinical methods in a head-to-head comparison. This further highlights the potential of using the transcriptional profile in host peripheral blood for diagnostics of diseases in clinical applications.
DISCUSSION
Accurate and rapid identification of bacterial and viral causes in ARI with only a droplet of blood has been a clinical challenge. Our method integrated an effective diagnostic model and a powerful DNA computation scheme into a completely automated classification workflow for ARI etiology diagnostics. Using this system, we can realize accurate and automated identification of the etiology in acute infections using 1 ml of blood sample in 4 hours with a much higher accuracy of 86% compared to that of other currently available methods. In general, there are multiple factors that may have an impact on the accuracy of clinical diagnostics with this DNA computing–based method. For instance, the technical variables affecting transcriptome analysis include blood collection and storage conditions, RNA extraction kits (42), and PCR amplification process (43). Previous studies have shown that the efficiency of various extraction kits is different, but little impact has been found on the quantification of relative amounts of multiple RNA targets in the same sample (34). In addition, evaluation of primer design and amplification cycles must be made for the PCR amplification process to exclude nonspecific and nonlinear amplification that will introduce bias in the relative ratios of multiple RNA targets. Overall, we believe that the potential of DNA computation to analyze and classify complex input profiles is impressive, especially toward the future applications of intelligent and automated diagnostics in clinics (42, 43). Furthermore, development of the DNA computing scheme with simpler nucleic acid extraction and preamplification steps (e.g., isothermal amplification), as well as user-friendly output (color change recognizable by the naked eye), will continue to propel the platform to wider applications, such as at-home self-inspection. We envision that the power of DNA computation will shed light on more clinical applications with its beneficial features: low cost, automated, and convenient.
MATERIALS AND METHODS
DNA oligonucleotides
All DNA oligonucleotides used in the present study were synthesized and purified by Shanghai Sangon Biological Engineering Technology & Services Co. Ltd. (China). Individual DNA oligonucleotides were suspended to deionized water to a concentration of 100 μM. All oligonucleotides were high-performance liquid chromatography–purified.
Blood RNA extraction
Total RNA was extracted from human whole blood (1 ml) using the Qiagen PAXgene Blood RNA Kit according to the manufacturer’s instructions. Specifically, a lysis reagent is first added to each blood sample. Then, the lysate is separated by adding chloroform and centrifugating. The sample is then applied to a spin column where the total RNAs, including mRNAs, can be eluted in a small volume of nuclease-free water. The extracted total RNA was stored at −80°C in nuclease-free water until needed.
RT-qPCR quantification
Total RNA was extracted from each blood sample with Qiagen Research kits. The isolated RNA was first reversely transcribed to cDNA using procedures described below. Then, the acquired cDNAs were amplified by using the qPCR Mix (Takara) according to the procedures in the instructions.
Reverse transcription
We used a reverse transcription kit (Takara) to convert RNAs into cDNAs under the conditions and protocol shown in table S5.
SVM training and validation
To train the SVM algorithm, microarray data of 273 patients diagnosed with ARI were obtained from NCBI GSE63990. We processed the dataset by first selecting samples labeled only as bacterial or viral infections (70 and 115 samples, respectively) and then converting the microarray gene expression ratios by logarithm (base 2) to estimate biological expression levels. We trained a linear SVM algorithm on this dataset to distinguish between viral and bacterial infections. We evaluated these classifiers using another microarray dataset (NCBI GSE6269) where they still performed well (AUC > 0.90). Last, the classifier with the highest AUC value as well as reasonable numbers of mRNA inputs were selected for experimental implementation.
Design of DNA sequences
All DNA strands used in the DNA computation were composed of short toehold domains and long-branch migration domains with a three-letter code (A, T, and C) to diminish secondary structures and undesired strand interactions. The annihilator complexes had 8-nucleotide toeholds compared to 6-nucleotide toeholds in the signal restoration complexes. This increased the strand displacement reaction rate between the annihilator complexes and E or F, compared to that between the signal restoration complexes and E or F. Last, the candidate sequences were validated by Nucleic Acid Package (NUPACK) for binding energy and specificity inspection. All DNA sequence can be found in table S6.
Preparation of hybridization probes
The hybridization probe is an annealed complex consisting of two DNA oligonucleotides: a bottom strand and a top strand. The top strands were stoichiometrically mixed with 20% excess of the bottom strand and then thermally annealed by heating to 95°C for 10 s, followed by cooling from 95° to 25°C over the course of 70 min.
Purification of hybridization probes
The prepared hybridization probes were purified with 12% nondenaturing PAGE gel. Subsequently, gel bands were visualized using ultraviolet light with a fluorescent backplate and then cut out and eluted into 1 ml of 1× tris-acetate-EDTA and 12.5 mM Mg2+ overnight. The concentrations of purified hybridization probes were determined by detecting their absorbance at 260 nm (Bio-Rad GelDoc EZ) with an IDT-specified extinction coefficients.
Integration of the automated platform
First, we used an RNA extractor [extraction module composed of an automated RNA extractor (ZJ Bio EX3600 Plus)] for the RNA extraction from whole-blood samples in 30 min. Second, the RT mix (Takara RR047A) was added in the extracted total RNA for the production of cDNAs in 30 min by the mechanical sample loading machine. Third, seven kinds of PCR mix were added by the mechanical sample loading machine and reacted in the thermocycler for 60 min [PCR thermal module by a qPCR machine (Roche Z480)]. Last, the DNA computation probe mix was added by the mechanical sample loading machine and reacted in the fluorescence reader for 120 min for results reporting [reporting module by a qPCR machine (Roche Z480)].
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (2021YFA0909400), the National Natural Science Foundation of China (22225402, 22104085, and 21974087), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (2018709), the Shanghai Rising-Star Program (20QA1405800), the Shanghai Sailing Program (21YF1424700), the Innovative Research Team of High-Level Local University in Shanghai, the Faculty Start-up Funding Support from the Institute of Molecular Medicine of Shanghai Jiao Tong University, and the Recruitment Program of Global Youth Experts of China.
Author contributions: Q.M., D.H., and C.Z. designed the research. Q.M., M.Z., L.Y., X.T., Y.T., and J.W. performed the research. Q.M. and C.Z. analyzed data. Q.M., D.H., and W.T. wrote the manuscript. All authors reviewed the manuscript and approved the final version.
Competing interests: Q.M. and C.Z. are employees of Intellinosis Biotechnologies Co. Ltd. with equity in the company. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Supplementary information is available for this paper at the publisher’s website. Correspondence and requests for materials should be addressed to D.H., W.T., or C.Z.
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Tables S1 to S6
REFERENCES AND NOTES
- 1.Zaas A. K., Burke T., Chen M., Clain M. M., Nicholson B., Veldman T., Tsalik E. L., Fowler V., Rivers E. P., Otero R., Kingsmore S. F., Voora D., Lucas J., Hero A. O., Carin L., Woods C. W., Ginsburg G. S., A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci. Transl. Med. 5, 203ra126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sproston N. R., Ashworth J. J., Role of C-reactive protein at sites of inflammation and infection. Front. Immunol. 9, 754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyriazopoulou E., Liaskou-Antoniou L., Adamis G., Panagaki A., Melachroinopoulos N., Drakou E., Marousis K., Chrysos G., Spyrou A., Alexiou N., Symbardi S., Alexiou Z., Lagou S., Kolonia V., Gkavogianni T., Kyprianou M., Anagnostopoulos I., Poulakou G., Lada M., Makina A., Roulia E., Koupetori M., Apostolopoulos V., Petrou D., Nitsotolis T., Antoniadou A., Giamarellos-Bourboulis E. J., Procalcitonin to reduce long-term infection-associated adverse events in sepsis. A randomized trial. Am. J. Respir. Crit. Care Med. 203, 202–210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon L., Gauvin F., Amre D. K., Saint-Louis P., Lacroix J., Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: A systematic review and meta-analysis. Clin. Infect. Dis. 39, 206–217 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Honda T., Uehara T., Matsumoto G., Arai S., Sugano M., Neutrophil left shift and white blood cell count as markers of bacterial infection. Clin. Chim. Acta 457, 46–53 (2016). [DOI] [PubMed] [Google Scholar]
- 6.van der Does Y., Rood P. P. M., Ramakers C., Schuit S. C. E., Patka P., van Gorp E. C. M., Limper M., Identifying patients with bacterial infections using a combination of C-reactive protein, procalcitonin, TRAIL, and IP-10 in the emergency department: A prospective observational cohort study. Clin. Microbiol. Infect. 24, 1297–1304 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Lei H., Wang C., Wang Y., Wang C., Single-cell RNA-seq revealed profound immune alteration in the peripheral blood of patients with bacterial infection. Int. J. Infect. Dis. 103, 527–535 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Ramilo O., Allman W., Chung W., Mejias A., Ardura M., Glaser C., Wittkowski K. M., Piqueras B., Banchereau J., Palucka A. K., Chaussabel D., Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 109, 2066–2077 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaas A. K., Chen M., Varkey J., Veldman T., Hero A. O. III, Lucas J., Huang Y., Turner R., Gilbert A., Lambkin-Williams R., Øien N. C., Nicholson B., Kingsmore S., Carin L., Woods C. W., Ginsburg G. S., Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 6, 207–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parnell G. P., McLean A. S., Booth D. R., Armstrong N. J., Nalos M., Huang S. J., Manak J., Tang W., Tam O. Y., Chan S., Tang B. M., A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit. Care 16, R157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu X., Yu J., Crosby S. D., Storch G. A., Gene expression profiles in febrile children with defined viral and bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 110, 12792–12797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahajan P., Kuppermann N., Mejias A., Suarez N., Chaussabel D., Casper T. C., Smith B., Alpern E. R., Anders J., Atabaki S. M., Bennett J. E., Blumberg S., Bonsu B., Borgialli D., Brayer A., Browne L., Cohen D. M., Crain E. F., Cruz A. T., Dayan P. S., Gattu R., Greenberg R., Hoyle J. D. Jr., Jaffe D. M., Levine D. A., Lillis K., Linakis J. G., Muenzer J., Nigrovic L. E., Powell E. C., Rogers A. J., Roosevelt G., Ruddy R. M., Saunders M., Tunik M. G., Tzimenatos L., Vitale, Dean J. M., Ramilo O.; for the Pediatric Emergency Care Applied Research Network (PECARN) , Association of RNA biosignatures with bacterial infections in febrile infants aged 60 days or younger. JAMA 316, 846–857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharya S., Rosenberg A. F., Peterson D. R., Grzesik K., Baran A. M., Ashton J. M., Gill S. R., Corbett A. M., Holden-Wiltse J., Topham D. J., Walsh E. E., Mariani T. J., Falsey A. R., Transcriptomic biomarkers to discriminate bacterial from nonbacterial infection in adults hospitalized with respiratory illness. Sci. Rep. 7, 6548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suarez N. M., Bunsow E., Falsey A. R., Walsh E. E., Mejias A., Ramilo O., Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J Infect Dis 212, 213–222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sealfon S. C., Chu T. T., RNA and DNA microarrays. Methods Mol. Biol. 671, 3–34 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Stark R., Grzelak M., Hadfield J., RNA sequencing: The teenage years. Nat. Rev. Genet. 20, 631–656 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Tsalik E. L., Henao R., Nichols M., Burke T., Ko E. R., McClain M. T., Hudson L. L., Mazur A., Freeman D. H., Veldman T., Langley R. J., Quackenbush E. B., Glickman S. W., Cairns C. B., Jaehne A. K., Rivers E. P., Otero R. M., Zaas A. K., Kingsmore S. F., Lucas J., Fowler V. G. Jr., Carin L., Ginsburg G. S., Woods C. W., Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 8, 322ra311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herberg J. A., Kaforou M., Wright V. J., Shailes H., Eleftherohorinou H., Hoggart C. J., Cebey-López M., Carter M. J., Janes V. A., Gormley S., Shimizu C., Tremoulet A. H., Barendregt A. M., Salas A., Kanegaye J., Pollard A. J., Faust S. N., Patel S., Kuijpers T., Martinón-Torres F., Burns J. C., Coin L. J. M., Levin M.; for the IRIS Consortium , Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA 316, 835–845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lydon E. C., Henao R., Burke T. W., Aydin M., Nicholson B. P., Glickman S. W., Fowler V. G., Quackenbush E. B., Cairns C. B., Kingsmore S. F., Jaehne A. K., Rivers E. P., Langley R. J., Petzold E., Ko E. R., McClain M. T., Ginsburg G. S., Woods C. W., Tsalik E. L., Validation of a host response test to distinguish bacterial and viral respiratory infection. EBioMedicine 48, 453–461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adleman L. M., Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Liu Q., Wang L., Frutos A. G., Condon A. E., Corn R. M., Smith L. M., DNA computing on surfaces. Nature 403, 175–179 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Han D., Wu C., You M., Zhang T., Wan S., Chen T., Qiu L., Zheng Z., Liang H., Tan W., A cascade reaction network mimicking the basic functional steps of adaptive immune response. Nat. Chem. 7, 835–841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braich R. S., Chelyapov N., Johnson C., Rothemund P. W., Adleman L., Solution of a 20-variable 3-SAT problem on a DNA computer. Science 296, 499–502 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Han D., Zhu G., Wu C., Zhu Z., Chen T., Zhang X., Tan W., Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy. ACS Nano 7, 2312–2319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas S. M., Bachelet I., Church G. M., A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Yang L., Zhao Y., Xu X., Xu K., Zhang M., Huang K., Kang H., Lin H. C., Yang Y., Han D., An intelligent DNA nanorobot for autonomous anticoagulation. Angew. Chemie 59, 17697–17704 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Bai M., Chen F., Cao X., Zhao Y., Xue J., Yu X., Fan C., Zhao Y., Intracellular entropy-driven multi-bit DNA computing for tumor progression discrimination. Angew. Chemie 59, 13267–13272 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Fu D., Shah S., Song T., Reif J., DNA-based analog computing. Methods Mol. Biol. 1772, 411–417 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Chang X., Zhang C., Lv C., Sun Y., Zhang M., Zhao Y., Yang L., Han D., Tan W., Construction of a multiple-aptamer-based DNA logic device on live cell membranes via associative toehold activation for accurate cancer cell identification. J. Am. Chem. Soc. 141, 12738–12743 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Gao Q., Zhao Y., Xu K., Zhang C., Ma Q., Qi L., Chao D., Zheng T., Yang L., Miao Y., Han D., Highly specific, single-step cancer cell isolation with multi-aptamer-mediated proximity ligation on live cell membranes. Angew. Chemie 59, 23564–23568 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Moerman P. G., Schulman R., DNA computation improves diagnostic workflows. Nat. Nanotechnol. 15, 626–627 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Brown M. P., Grundy W. N., Lin D., Cristianini N., Sugnet C. W., Furey T. S., Ares M. Jr., Haussler D., Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. U.S.A. 97, 262–267 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez R., Wang R., Seelig G., A molecular multi-gene classifier for disease diagnostics. Nat. Chem. 10, 746–754 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Zhang C., Zhao Y., Xu X., Xu R., Li H., Teng X., du Y., Miao Y., Lin H. C., Han D., Cancer diagnosis with DNA molecular computation. Nat. Nanotechnol. 15, 709–715 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Pierce K. E., Sanchez J. A., Rice J. E., Wangh L. J., Linear-After-The-Exponential (LATE)-PCR: Primer design criteria for high yields of specific single-stranded DNA and improved real-time detection. Proc. Natl. Acad. Sci. U.S.A. 102, 8609–8614 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice J. E., Sanchez J. A., Pierce K. E., Reis A. H. Jr., Osborne A., Wangh L. J., Monoplex/multiplex linear-after-the-exponential-PCR assays combined with PrimeSafe and dilute-‘N’-go sequencing. Nat. Protoc. 2, 2429–2438 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Cherry K. M., Qian L., Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature 559, 370–376 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Tian R., Zou H., Wang L., Liu L., Song M., Zhang H., Analysis of differentially expressed genes in bacterial and fungal keratitis. Indian J. Ophthalmol. 68, 39–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian L., Winfree E., Bruck J., Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Qian L., Winfree E., Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Zhang D. Y., Cooperative hybridization of oligonucleotides. J. Am. Chem. Soc. 133, 1077–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 42.exRNAQC Consortium, Anckaert J., Cobos F. A., Decock A., Deleu J., De Wever O., De Wilde J., Dhondt B., D’huyvetter T., Everaert C., Fierro C., Helsmoortel H. H., Hendrix A., Hulstaert E., Kuersten S., Mestdagh P., Morlion A., Nijs N., Nuytens J., Philippron A., Piofczyk T., Schoofs K., Schroth G. P., Eynde E. V., Vandesompele J., Van Maerken T., Van Paemel R., Verniers K., Yigit N., Performance of RNA purification kits and blood collection tubes in the Extracellular RNA Quality Control (exRNAQC) study. Cold Spring Harbor Laboratory 10.1101/2021.05.11.442610 (2021). [Google Scholar]
- 43.Tan G. W., Khoo A. S. B., Tan L. P., Evaluation of extraction kits and RT-qPCR systems adapted to high-throughput platform for circulating miRNAs. Sci. Rep. 5, 9430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Tables S1 to S6