Abstract
Exhausted CD8 T cells during chronic inflammatory responses against viral infections and cancer are phenotypically and functionally heterogeneous. In particular, CD8 T cells with cytolytic effector function have been recently identified among the exhausted CD8 T cell subsets. However, the regulation of their differentiation and function remains largely unknown. Here, we report that Krüppel-like factor 4 (KLF4) is a critical regulator of the exhaustion process, promoting the cytolytic effector function of exhausted CD8 T cells. KLF4-expressing CD8 T cells in exhaustion contexts showed the features of transitory effector CD8 T cells. Enforced KLF4 expression increased CD8 T cell differentiation into transitory effector subsets and enhanced their antitumor immunity. We further demonstrated that KLF4 also showed a capacity of reinvigorating exhausted CD8 T cells. Last, high KLF4 expression was positively correlated with a favorable prognosis in human patients with cancer. Our study highlights the potential impacts of KLF4 on CD8 T cell exhaustion and antitumor immune therapy.
KLF4 regulates the exhaustion process by promoting CD8 T cell differentiation into cytolytic transitory effector subsets.
INTRODUCTION
Antigen-specific CD8 T cells reach a severely dysfunctional state during chronic inflammatory responses against viral infections and cancer, which is described as exhaustion (1–3). Exhausted CD8 T cells are phenotypically and functionally heterogeneous (4–11). Recent studies have identified progenitor exhausted CD8 T cells and terminally exhausted CD8 T cells and their characteristics (4, 7, 10, 11). Progenitor exhausted CD8 T cells highly express T cell factor 1 (TCF1) and CXCR5 and show a stem-like phenotype with an increased proliferative capacity. In contrast, terminally exhausted CD8 T cells down-regulate TCF1 and CXCR5 and up-regulate PD1 (Programmed cell death protein 1), TIM3 (T cell immunoglobulin and mucin domain containing protein 3), and other inhibitory receptors. Notably, the number of progenitor exhausted CD8 T cells increases in response to anti-PD1 therapy, which appears to be a critical factor for the successful control of chronic viral infection and cancer (5–7, 10, 12).
Effective immune responses exerted by the progenitor exhausted CD8 T cells imply that cells with effector functions may develop from the progenitor cells. CD8 T cells with effector characteristics in the exhaustion contexts have recently been identified (13–15). In chronic viral infections, stem-like TCF1+ CD8 T cells initially differentiated into transitory CD101−CX3CR1+TIM3+ cells that exert effector-like transcriptional signatures and later converted into CD101+CX3CR1−TIM3+ cells (13). CX3CR1+ CD8 T cells are found not only in chronic infections but also in the tumor microenvironment and show effector-like functions (14). Moreover, four different subsets of exhausted CD8 T cells are identified in chronic viral infections: Texprog1 (CD69+Ly108+), Texprog2 (CD69−Ly108+), Texint (CD69−Ly108−), and Texterm (CD69+Ly108−) (15). Texint cells display effector-like signatures including the expression of CX3CR1, proinflammatory cytokines, and granzyme B (GzmB), and they eventually convert into Texterm. Thus, the functional shift from the effector-like CD8 T cells to the terminally exhausted CD8 T cells may be the true nature of exhaustion, suggesting that increasing the number and/or function of effector-like CD8 T cells may be an ideal strategy for immune therapy of viral infections and cancer. However, little is known about the characteristics of the effector-like CD8 T cells in the context of exhaustion, especially in terms of their genetic regulation.
Here, we report that Krüppel-like factor 4 (KLF4) is a hallmark of cytolytic effector–like CD8 T cells during the exhaustion process, which we term as transitory effector CD8 T cells in this study. We found that KLF4 promotes the CD8 T cell differentiation into transitory effector subsets. In consequence, KLF4 expression in CD8 T cells increased antitumor immunity and provided a great advantage in controlling tumor growth. Moreover, we demonstrated that KLF4 expression could reinvigorate the effector function of exhausted CD8 T cells. Last, we showed that high KLF4 expression also correlated with increased survival of human patients with cancer. Thus, the potential impact of KLF4 on CD8 T cell exhaustion can be underscored in terms of antitumor immune therapy.
RESULTS
KLF4 is a potential regulator of CD8 T cell effector function during the exhaustion process and is specifically increased by chronic TCR stimulation
To identify the factors that regulate CD8 T cell effector function during the exhaustion process, we first examined consensus motifs and their expected binding factors within the promoter regions of inhibitory receptors including Pdcd1 (PD1), Ctla4 [CTLA4 (Cytotoxic T lymphocyte associated protein 4)], Lag3 [LAG3 (Lymphocyte activation gene 3)], and Havcr2 (TIM3), which can be regarded as hallmarks of exhaustion (fig. S1, A to C). The list of identified factors was overlapped with potential binding factors on differentially methylated regions (DMRs) that are highly enriched in effector CD8 T cells (16). We identified EGR1 (Early growth response 1), EGR2, and SP1 (Sp1 transcription factor) as candidate factors binding to demethylated DMR-enriched motifs, while FLI1 (Friend leukemia integration 1 transcription factor), CTCFL (CCCTC-binding factor like), SMAD3 (SMAD family member 3), NEUROD1 (Neuronal differentiation 1), KLF4, and ZNF263 (Zinc finger protein 263) as factors binding to methylated DMR-enriched motifs (fig. S1D). Among these, it has been recently shown that KLF4 is positively correlated with T-bet, a well-known regulator of CD8 T cell effector function, during chronic viral infections (15).
To check the relatedness between KLF4 and an exhaustion process, we adopted an in vitro system to induce exhaustion by repetitive peptide stimulation on OT-I CD8 T cells, as previously described (fig. S2A) (17). Compared to CD8 T cells treated with cytokines [cytokine only (C cells)] or stimulated by a single treatment with ovalbumin (OVA) peptide for 2 days [single stimulation (SS cells)], cells repeatedly stimulated with OVA peptide [repeated stimulation (RS cells)] up-regulated PD1 and TOX (Tox) expression (fig. S2, B to D). Klf4 expression significantly increased in RS cells compared to that in C and SS cells (fig. S2E). In addition, we sought to validate our in vitro exhaustion model whether in vitro generated CD8 T cells show known features of reported in vivo exhausted CD8 T cells. After day 3, we observed a gradual increase of PD1 and TOX with a decrease of tumor necrosis factor–α (TNF-α), which are typical phenotypes of exhaustion (fig. S2, F to H). Moreover, we found that most of the cells were Texterm (CD69+Ly108−) on day 5, while Texint (CD69−Ly108−) cells appeared on day 3 and their proportion was gradually decreased (fig. S2I). Thus, our in vitro exhaustion model resembles in vivo exhaustion, suggesting that KLF4, which is specifically increased by chronic T cell receptor (TCR) stimulation, is a potential regulator of CD8 T cell effector function during the exhaustion process.
KLF4 is highly expressed in the transitory effector subset of exhausted CD8 T cells with cytolytic effector function
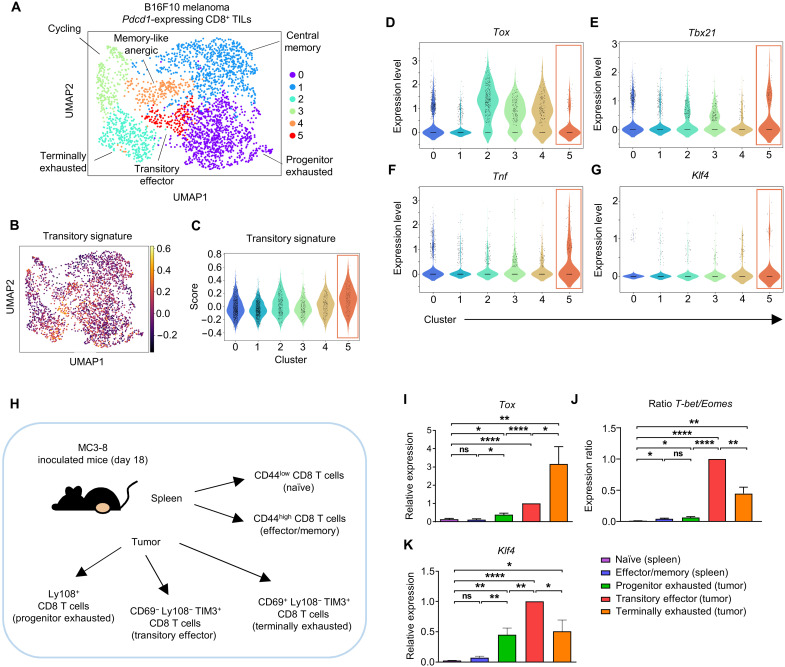
To further investigate Klf4 expression in tumor-infiltrating CD8 T cells, we analyzed single-cell RNA sequencing (scRNA-seq) data of CD8+ tumor-infiltrating lymphocytes (TILs) isolated from B16F10 melanoma–bearing C57BL/6 mice (18). Pdcd1-expressing CD8+ TILs were filtered and grouped into six clusters on the basis of their expression of distinct markers (progenitor exhausted: Slamf6 and Gzmk; central memory: Tcf7, Lef1, and Sell; terminally exhausted: Tox, Tigit, and Pdcd1; cycling: Stmn1, Cks1b, and Mki67; memory-like anergic: Ccr7, Xcl1, and Nrn1; and transitory effector: Junb, Fosb, and Nr4a1; Fig. 1A and fig. S3). To validate whether cluster 5 shows known features of transitory/intermediate exhausted subsets, we scored the expression level of their signature genes (Tbx21, Tnf, Cx3cr1, Ets1, Klrk1, Ifng, Klrd1, Klrg1, S1pr1, Klf3, Foxj3, Hoxb4, Nfyb, Atf2, Sp3, and Maz) and compared it with other clusters. We found that the score was the highest in cluster 5 (transitory effector) cells (Fig. 1, B and C). In addition, we further validated phenotypes of transitory effector cells by gene set enrichment analysis (GSEA) using Texint and Texterm marker genes described by Beltra et al. (15). When GSEA was performed between cluster 2 (terminally exhausted) and cluster 5 (transitory effector) cells, Texint marker genes were significantly enriched in cluster 5 cells, whereas Texterm marker genes were significantly enriched in cluster 2 cells (fig. S4, A and B). Moreover, cluster 5 cells showed high expression of Cx3cr1 and Havcr2 (fig. S4, C and D), which are also known phenotypes of transitory/intermediate exhausted subsets (13–15). Moreover, cluster 5 cells showed a low Tox expression with the highest level of Tbx21 and Tnf expression among the clusters (Fig. 1, D to F). Thus, we defined the cells in cluster 5 as transitory effector subset of tumor-infiltrating CD8 T cells. The expression level of Klf4 was the highest in cluster 5 cells (Fig. 1G).
Fig. 1. KLF4 is highly expressed in the transitory effector subset of exhausted CD8 T cells with cytolytic effector function.
(A to G) scRNA-seq data from B16F10 melanoma–infiltrating CD8+ TILs (GSE116390) were analyzed. Pdcd1-expressing CD8+ TILs were filtered. (A) Clustering and uniform manifold approximation and projection (UMAP) visualization of Pdcd1-expressing CD8 T cells from B16F10 melanoma tumor (n = 2652 cells) on day 15. Colors denote transcriptional clusters, labeled with functional annotations. (B) UMAP visualization of transitory signature score within Pdcd1-expressing CD8+ TILs. (C) Violin plot of transitory signature score within clusters. Cluster 5 denotes transitory effector cells. (D to G) Violin plot of the expression level of (D) Tox, (E) Tbx21, (F) Tnf, and (G) Klf4 within clusters. Cluster 5 denotes transitory effector cells. (H to K) Five different CD8 T cell subpopulations were isolated from MC38 tumor–bearing C57BL/6 mice on day 18. (H) Schematic design of the experiment. (I) Histogram of Tox mRNA expression on five different CD8 T cell subpopulations (n = 5 per group). (J) Histogram of ratio T-bet/Eomes (mRNA) on five different CD8 T cell subpopulations (n = 4 per group). (K) Histogram of Klf4 mRNA expression on five different CD8 T cell subpopulations (n = 5 per group). All data are means ± SEM. Statistical analysis was performed using Student’s t test. ns (nonsignificant), P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001.
To validate the results of the single-cell analysis, we verified Klf4 expression among different subsets of exhausted CD8 T cells obtained from tumor tissue of MC38 tumor–bearing mice. According to previous reports (13–15) and our data, we grouped CD8+ TILs into three subsets: progenitor (Ly108+), TIM3-expressing transitory effector (CD69−Ly108−TIM3+), and terminally exhausted cells (CD69+Ly108−TIM3+; fig. S5, A to E). Then, we isolated CD44low (naïve) and CD44high (effector/memory) CD8 T cells from the spleen and each subset of the exhausted CD8 T cells from the tumor tissue and compared their gene expression patterns (Fig. 1H). The expression of Tox gradually increased during the exhaustion process (Fig. 1I). In contrast, the Tbet/Eomes ratio, a signature of CD8 T cell effector function (15, 19), was the highest in transitory effector cells and decreased in terminally exhausted cells (Fig. 1J). In addition, while gene expression of Tcf7, Havcr2, and Gzmb was comparable between the two subsets (fig. S5, F to H), gene expression of Ifng, Tnf, and Cx3cr1 was the highest in transitory effector cells and decreased in terminally exhausted cells (fig. S5, I to K). Klf4 expression was the highest in the transitory effector cells, showing similar kinetics to the effector signature genes (Fig. 1K). Together, our data demonstrated that Klf4 is highly expressed in the transitory effector subset and its expression decreased in the terminally exhausted subset of tumor-infiltrating CD8 T cells.
KLF4 enhances effector function and characteristics of transitory effector CD8 T cells
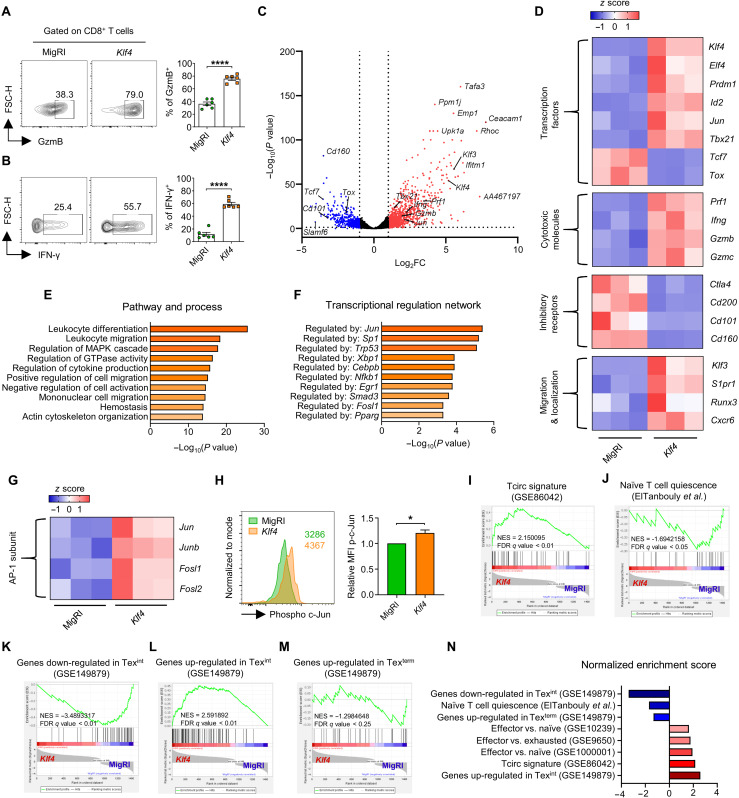
To identify the role of KLF4 on exhausted CD8 T cells, we retrovirally transduced Klf4 into OT-I CD8 T cells on day 1 of in vitro exhaustion model and repeatedly treated OVA peptide to induce exhaustion (fig. S6A). We found that CD8 T cells transduced with Klf4 showed a significantly increased proportion of GzmB+ and interferon-γ–positive (IFN-γ+) cells compared to that of cells transduced with the MigRI control (Fig. 2, A and B). We further analyzed the gene expression profiles of these transduced cells by RNA-seq. Among the 1435 differentially expressed genes (DEGs), genes related to the effector program (Tbx21, Prdm1, and Jun), cytotoxic molecules (Prf1, Gzmb, Gzmc, and Ifng), and T cell migration and localization (Klf3, S1pr1, Runx3, and Cxcr6) were up-regulated by Klf4 expression. In contrast, Tcf7 and Slamf6, which are known to be down-regulated during effector CD8 T cell differentiation (20–22), were decreased by Klf4 expression. In addition, the expression of genes encoding inhibitory receptors (Ctla4, Cd200, Cd101, and Cd160) and the exhaustion state (Tox) decreased (Fig. 2, C and D). Gene Ontology (GO) enrichment analysis showed that genes involved in leukocyte differentiation, leukocyte migration, and regulation of cytokine production were highly enriched within the DEGs (Fig. 2E). In addition, the result showed that the genes involved in the mitogen-activated protein kinase cascade pathway and the guanosine triphosphatase (GTPase) activity, both of which are closely related to c-Jun (23–25), were significantly enriched within DEGs. GO analysis also showed that the transcriptional network regulated by Klf4 shared prominently with Jun-regulated processes (Fig. 2F). CD8 T cells transduced with Klf4 highly increased the expression of Jun and AP-1 (Activator protein 1) subunit factors Junb, Fosl1, and Fosl2 (Fig. 2G). Moreover, CD8 T cells transduced with Klf4 showed an increased phospho–c-Jun level, which is critical for its activity (Fig. 2H) (26). In line with a previous report that the expression of c-Jun enhances effector function and counteracts exhaustion in CAR T cells (27), our results suggest that c-Jun activity increased by Klf4 expression may contribute to enhancing CD8 T cell effector function.
Fig. 2. KLF4 enhances effector function and characteristics of transitory effector CD8 T cells.
(A to N) OT-I CD8 T cells were retrovirally transduced with MigRI/Klf4 on day 1 during in vitro exhaustion process and were harvested on day 5. GFP+ cells were gated for all analyses. (A) Representative flow cytometry plot of GzmB expression and the proportion of GzmB+ cells in CD8 T cells transduced with MigRI/Klf4 (n = 6 per group). (B) Representative flow cytometry plot of IFN-γ expression and the proportion of IFN-γ+ cells in CD8 T cells transduced with MigRI/Klf4 (n = 6 per group). (C to N) CD8 T cells transduced with MigRI/Klf4 were isolated and RNA-seq was performed. (C) Volcano plot highlighting differential transcripts between CD8 T cells transduced with MigRI/Klf4 (n = 3 per group). (D) RNA expression heatmap of DEGs from CD8 T cells transduced with MigRI/Klf4 (n = 3 per group). (E) Gene Ontology (GO) (pathway and process) analysis of DEGs. (F) GO (transcriptional regulation network) analysis of DEGs. (G) RNA expression heatmap of AP-1 subunit genes from CD8 T cells transduced with MigRI/Klf4 (n = 3 per group). (H) Representative flow cytometry plot and histogram of relative MFI (mean fluorescence intensity) level of phospho–c-Jun in CD8 T cells transduced with MigRI/Klf4 (n = 4 per group). (I to M) GSEA between CD8 T cells transduced with MigRI/Klf4 using gene sets of (I) Tcirc signature, (J) naïve T cell quiescence, (K) genes down-regulated in Texint, (L) genes up-regulated in Texint, and (M) genes up-regulated in Texterm. (N) Histogram of normalized enrichment score from GSEA using various gene sets. (A, B, and H) Data are means ± SEM. Statistical analysis was performed using Student’s t test. *P < 0.05; ****P < 0.0001. Log2FC, log2 fold change; FDR, false discovery rate; NES, normalized enrichment score; FSC-H, forward scatter height.
To consolidate the role of KLF4 during the exhaustion process, we compared the RNA-seq data with published data through GSEA. We found that signature genes for circulating T cells (28) were highly enriched in CD8 T cells transduced with Klf4 (Fig. 2I), whereas signature genes for naïve T cell quiescence (29) were highly enriched in cells transduced with MigRI (Fig. 2J). Detailed analysis of four subsets (Texprog1, Texprog2, Texint, and Texterm) of exhausted CD8 T cells (15) revealed that genes down-regulated in Texint were highly enriched in the cells transduced with MigRI (Fig. 2K), while genes up-regulated in Texint were highly enriched in cells transduced with Klf4 (Fig. 2L). Furthermore, genes up-regulated in Texterm were significantly enriched in the cells transduced with MigRI (Fig. 2M). In addition, genes related to the effector function of CD8 T cells in various contexts (30–32) were significantly enriched in cells transduced with Klf4 (Fig. 2N). These data indicated that Klf4-expressing CD8 T cells show transcriptional parallelism with transitory effector cells, as well as resistance to quiescence and terminal exhaustion. Collectively, all these data support that Klf4-expressing CD8 T cells in the exhaustion contexts resemble transitory effector CD8 T cells with increased effector function.
CEACAM1 is highly expressed in transitory effector CD8 T cells during the exhaustion process
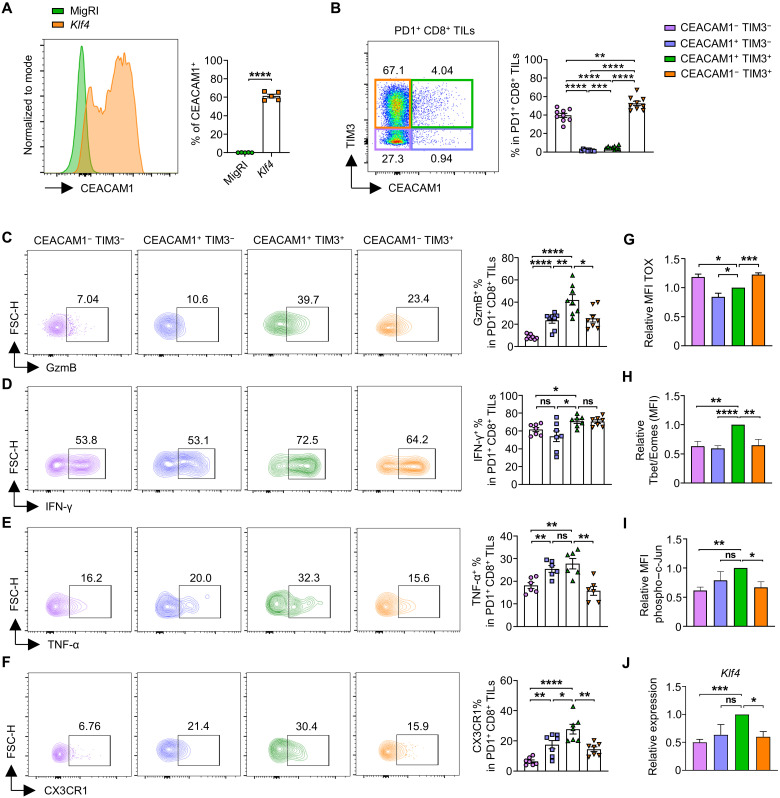
Our RNA-seq analysis revealed that Ceacam1 was the gene with the highest fold change among 1435 DEGs in CD8 T cells transduced with Klf4 compared to the control (Fig. 2C). The majority (61.3 ± 0.9%) of CD8 T cells transduced with Klf4 were CEACAM1 (CEA cell adhesion molecule 1)+ (Fig. 3A). In the CD8+ TILs, Ceacam1 was expressed at the highest level in the transitory effector subset but down-regulated in the terminally exhausted subset (fig. S5L), which is similar to the expression pattern of Klf4 (Fig. 1K). To check CEACAM1 protein level in exhausted CD8 T cells, we analyzed the PD1+CD8+ TILs from MC38 tumor–bearing C57BL/6 mice. The cells were grouped into four different subsets depending on the expression of CEACAM1 and TIM3: CEACAM1−TIM3−, CEACAM1+TIM3−, CEACAM1+TIM3+, and CEACAM1−TIM3+. The majority (92.2 ± 0.3%) of PD1+CD8+ TILs were CEACAM1− (with TIM3+ and TIM3−), and the proportion of CEACAM1+TIM3+ cells was low (4.6 ± 0.2%; Fig. 3B). When the effector features were compared among the four groups, the highest proportion of GzmB+ and TNF-α+ cells was observed in CEACAM1+TIM3+ cells, and the proportion was decreased in CEACAM1−TIM3+ cells (Fig. 3, C and E). The proportion of IFN-γ+ cells was also the highest in CEACAM1+TIM3+ cells; however, it did not show a significantly decreased level in CEACAM1−TIM3+ cells (Fig. 3D). In addition, CEACAM1+TIM3+ cells contained the highest proportion of CX3CR1+ cells, which are known as effector-like CD8 T cells during exhaustion, while the proportion decreased in CEACAM1−TIM3+ cells (Fig. 3F). Furthermore, CEACAM1+TIM3+ cells showed relatively lower level of TOX (Fig. 3G), whereas the levels of Tbet/Eomes ratio and phospho–c-Jun were the highest among the subpopulations (Fig. 3, H and I). Similarly, the profiles of the effector signatures shown by the subpopulations matched well with Klf4 expression (Fig. 3J). Together, we concluded that CEACAM1+TIM3+ cells, which show the highest effector features among the exhausted subsets, are in the transitory effector stage, whereas CEACAM1−TIM3+ cells are in the terminally exhausted stage.
Fig. 3. CEACAM1 is highly expressed in transitory effector CD8 T cells during the exhaustion process.
(A) Representative flow cytometry plot of CEACAM1 expression and the proportion of CEACAM1+ cells in CD8 T cells transduced with MigRI/Klf4 (n = 5 per group) from in vitro exhaustion experiment. GFP+ cells were gated for the analysis. (B to J) CD8+ TILs were analyzed from MC38 tumor–bearing C57BL/6 mice on day 18. (B) Representative flow cytometry plot of CEACAM1 and TIM3 expression, and the proportion of each subpopulation in PD1+CD8+ TILs (n = 9 per group). (C to F) Representative flow cytometry plot and the proportion of (C) GzmB+ (n = 8 per group), (D) IFN-γ + (n = 7 per group), (E) TNF-α+ (n = 6 per group), and (F) CX3CR1+ (n = 7 per group) cells in CEACAM1-TIM3–defined subsets. (G to J) Histogram of (G) relative TOX (MFI) ratio (n = 4 per group), (H) relative T-bet/Eomes (MFI) ratio (n = 5 per group), (I) relative phospho–c-Jun (n = 3 per group), and (J) relative expression of Klf4 (n = 3 per group) in CEACAM1-TIM3–defined subsets. Relative values were calculated on the basis of the level of CEACAM1+TIM3+ subset. All data are means ± SEM. Statistical analysis was performed using Student’s t test. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
KLF4 enhances CD8 T cell differentiation into transitory effector subsets and the antitumor function
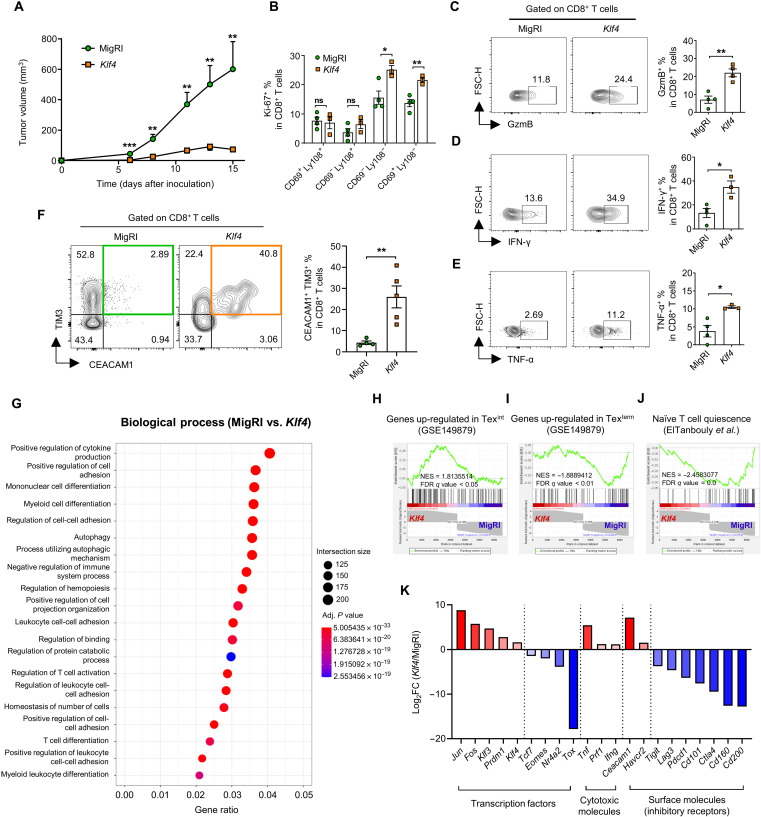
To investigate the function of KLF4 during CD8 T cell exhaustion in vivo, we induced a tumor in Rag2 knockout (KO) mice by injecting MC38-gp100 cells, which could be recognized by CD8 T cells derived from PmelI TCR transgenic mice (33). PmelI CD8 T cells were transduced with MigRI control or Klf4 and then adoptively transferred into mice bearing the tumor (fig. S6B). Mice that received CD8 T cells transduced with Klf4 showed a decreased tumor volume compared to that in control mice (Fig. 4A). The proportion of proliferative Ki-67+ cells within the CD69−Ly108− and CD69+Ly108− subsets also significantly increased in CD8 T cells transduced with Klf4 (Fig. 4B). In addition, the proportion of GzmB+, IFN-γ +, and TNF-α+ cells significantly increased in CD8 T cells transduced with Klf4 (Fig. 4, C to E). Notably, CEACAM1+TIM3+ cells, which showed the highest effector function among the tumor-infiltrating CD8 T cells, were highly enriched in CD8 T cells transduced with Klf4 (Fig. 4F).
Fig. 4. KLF4 enhances CD8 T cell differentiation into transitory effector subsets and the antitumor function.
(A to F) PmelI CD8 T cells transduced with MigRI/Klf4 were adoptively transferred into MC38-gp100–inoculated Rag2 KO mice on day 1. The tumor-infiltrating CD8 T cells were analyzed on day 15. GFP+ cells were gated for all analyses. (A) Tumor growth curve of mice received CD8 T cells transduced with MigRI (n = 6)/Klf4 (n = 7). (B) The proportion of Ki-67+ cells in CD8 T cells transduced with MigRI/Klf4 (n = 4 per group). (C to E) Representative flow cytometry plot and the proportion of (C) GzmB+, (D) IFN-γ+, and (E) TNF-α+ cells in CD8 T cells transduced with MigRI (n = 4)/Klf4 (n = 3 to 4). (F) Representative flow cytometry plot and the proportion of CEACAM1+TIM3+ cells in CD8 T cells transduced with MigRI (n = 4)/Klf4 (n = 5). (G to K) PmelI CD8 T cells transduced with MigRI/Klf4 were adoptively transferred into MC38-gp100–inoculated Rag2 KO mice on day 7. The tumor-infiltrating CD8 T cells transduced with MigRI (from four mice) and Klf4 (from four mice) were pooled (GFP+ cells were sorted) on day 15, and SMART-seq was performed. (G) GO (GO biological process) analysis of DEGs. (H to J) GSEA between CD8 T cells transduced with MigRI/Klf4 using gene sets of (H) genes up-regulated in Texint, (I) genes up-regulated in Texterm, and (J) naïve T cell quiescence. (K) Histogram of fold change of genes between CD8 T cells transduced with MigRI/Klf4. (A to F) Data are means ± SEM. Statistical analysis was performed using Student’s t test. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
To further investigate the in vivo function of KLF4, we analyzed gene expression profiles of tumor-infiltrating CD8 T cells transduced with MigRI or Klf4 by SMART (Switching mechanism at 5' end of RNA template) sequencing (SMART-seq). CD8 T cells transduced with MigRI or Klf4 were adoptively transferred on day 7 (fig. S7A), instead of day 1, to increase overall tumor sizes, as well as the total number of CD8 T cells from tumor tissues, against to the model described in fig. S6B. GO enrichment analysis showed that genes involved in regulation of cytokine production, cell adhesion, and immune cell differentiations were highly enriched within DEGs (Fig. 4G), similarly to the results from in vitro experiments (Fig. 2E). In addition, GSEA showed that genes up-regulated in Texint were highly enriched in CD8 T cells transduced with Klf4, whereas genes up-regulated in Texterm and signature genes for naïve T cell quiescence were highly enriched in CD8 T cells transduced with MigRI (Fig. 4, H to J). These results indicated that CD8 T cells transduced with Klf4 displayed similar transcription profiles to transitory effector subsets. We found that the proportion of TIM3+CX3CR1+, CD69−Ly108−, and CEACAM1+TIM3+ cells, which are considered as transitory effector subsets, were all significantly increased in CD8 T cells transduced with Klf4 (fig. S7, B to F). In particular, the expression of AP-1 family factor Jun and Fos, as well as Klf3 and Prdm1, was highly increased in CD8 T cells transduced with Klf4, while the expression of Tcf7, Eomes, Nr4a2, and Tox was decreased. The expression of cytotoxic molecules (Tnf, Prf1, and Ifng) was also increased by Klf4 overexpression. Moreover, the expression of Ceacam1 and Havcr2 was increased by Klf4 overexpression, whereas the expression of most inhibitory receptors (Tigit, Lag3, Pdcd1, Cd101, Ctla4, Cd160, and Cd200) was down-regulated (Fig. 4K). Consistently, Klf4 overexpression increased the proportion of IFN-γ+ cells in all CEACAM1-TIM3–defined subsets (fig. S7G). Increased functionality even in terminally exhausted subsets (CEACAM1−TIM3+) suggests that Klf4 may increase the effector function of CD8 T cells and/or counteracts the exhaustion status in these subsets. Together, these results demonstrated that overexpression of Klf4 in tumor-specific CD8 T cells increases their functionality, by up-regulating effector-related genes and down-regulating exhaustion-related genes, which, in turn, promotes differentiation into the transitory effector subsets and counteracts terminal exhaustion.
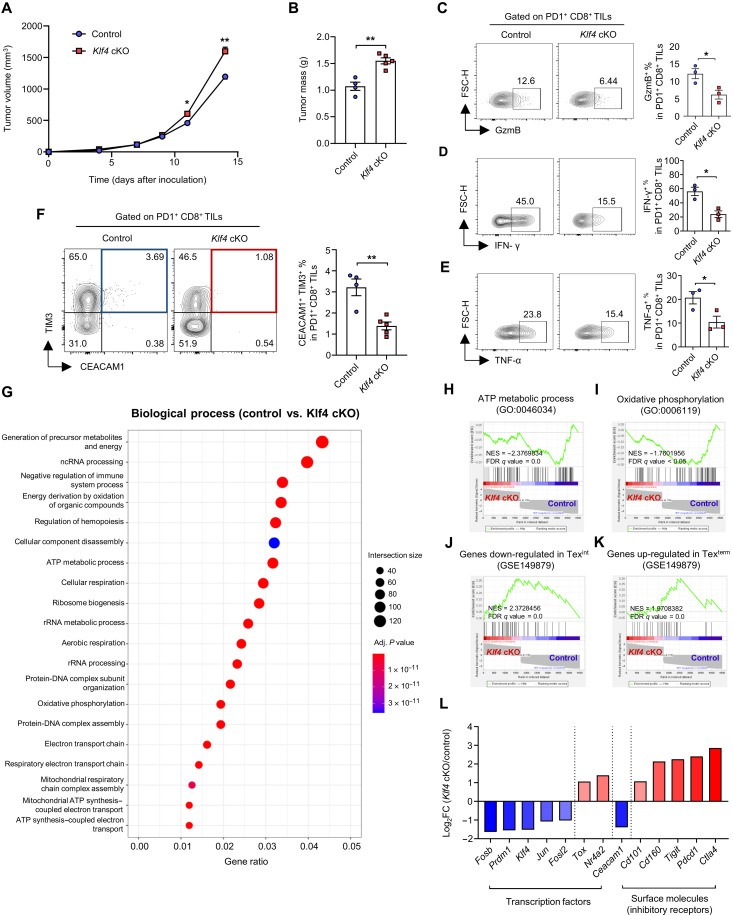
KLF4 deficiency impairs CD8 T cell differentiation into transitory effector subsets and the antitumor function
Next, we further investigated the effects of the loss of Klf4 in CD8 T cells on their antitumor response using Klf4fl/fl; E8i-cre [Klf4 conditional KO (cKO)] mice. A tumor was induced by subcutaneous injection of MC38 cells into control or Klf4 cKO mice. The tumor volume and mass increased slightly but significantly in Klf4 cKO mice compared to that in control mice (Fig. 5, A and B). The proportion of GzmB+, IFN-γ +, and TNF-α+ cells also decreased in Klf4 cKO PD1+CD8+ TILs (Fig. 5, C to E). Note that although the proportion of CEACAM1+TIM3+ cells decreased significantly in Klf4 cKO PD1+CD8+ TILs (Fig. 5F), the degree of the decrease was minor when compared to that of the increase observed after Klf4 overexpression (Fig. 4F). Thus, there appears to be a correlation between a high increase in CEACAM1+TIM3+ cells and effective control of tumor growth. Similarly, the proportion of transitory effector subsets, defined by various combination of markers, appeared to be decreased by Klf4 deficiency (fig. S8, A to E). Although the proportion of progenitor exhausted subsets appeared to increase and that of terminally exhausted subsets was largely unchanged, Klf4 deficiency greatly reduced the absolute cell number of transitory effector subsets, as well as terminally exhausted subsets, while the number of progenitor exhausted subsets did not change significantly (fig. S8, F to H).
Fig. 5. KLF4 deficiency impairs CD8 T cell differentiation into transitory effector subsets and the antitumor function.
(A to L) MC38 cells were subcutaneously injected into the right flank of control/Klf4 cKO mice. CD8+ TILs were analyzed on day 14. (A) Tumor growth curve of control (n = 4)/Klf4 cKO (n = 5) mice. (B) Histogram of tumor mass of control (n = 4)/Klf4 cKO (n = 5) mice. (C to E) Representative flow cytometry plot and the proportion of (C) GzmB+, (D) IFN-γ+, and (E) TNF-α+ cells in PD1+CD8+ TILs from control/Klf4 cKO mice (n = 3 per group). (F) Representative flow cytometry plot and the proportion of CEACAM1+TIM3+ cells in PD1+CD8+ TILs from control (n = 4)/Klf4 cKO (n = 5) mice. (G to L) PD1+CD8+ TILs from tumor tissues of four control mice and six Klf4 cKO mice were pooled on day 14, and SMART-seq was performed. (G) GO (GO biological process) analysis of DEGs. rRNA, ribosomal RNA. (H to K) GSEA between control and Klf4 cKO PD1+CD8+ TILs using gene sets of (H) adenosine triphosphate (ATP) metabolic process, (I) oxidative phosphorylation, (J) genes down-regulated in Texint, and (K) genes up-regulated in Texterm. (L) Histogram of fold change of genes between control and Klf4 cKO PD1+CD8+ TILs. (A to F) Data are means ± SEM. Statistical analysis was performed using Student’s t test. *P < 0.05; **P < 0.01.
To better understand the role of KLF4, we analyzed gene expression profile of control and Klf4 cKO PD1+CD8+ TILs by SMART-seq. GO enrichment analysis showed that genes involved in energy production, such as adenosine triphosphate (ATP) metabolic process and oxidative phosphorylation, were highly enriched within DEGs (Fig. 5G). GSEA analysis also showed that genes involved in ATP metabolic process and oxidative phosphorylation were significantly enriched in control PD1+CD8+ TILs (Fig. 5, H and I). It has been known that dysfunctional and terminally exhausted CD8 T cells from tumors display defective oxidative phosphorylation and ATP synthesis (34). Genes down-regulated in Texint and up-regulated in Texterm were highly enriched in Klf4 cKO PD1+CD8+ TILs (Fig. 5, J and K), indicating that Klf4 deficiency increases the severity of exhaustion. Consistent with this, the expression of AP-1 family (Jun, Fosb, and Fosl2), as well as Prdm1, was decreased in Klf4 cKO PD1+CD8+ TILs, while the expression of exhaustion genes (Nr4a2 and Tox) was increased. Furthermore, the expression of inhibitory receptors such as Cd101, Cd160, Tigit, Pdcd1, and Ctla4 was up-regulated, whereas the expression of Ceacam1 was down-regulated in Klf4 cKO PD1+ CD8+ TILs (Fig. 5L).
To integrate the results from the analyses of both Klf4 overexpression and Klf4 deficiency, genes up-regulated by Klf4 overexpression and those down-regulated by Klf4 deficiency were overlapped. GO enrichment analysis showed that genes involved in GTPase-mediated signal transductions and regulation of phosphatidylinositol 3-kinase (PI3K) activity were highly enriched within these overlapping genes (fig. S9A). Among the genes that were decreased by Klf4 deficiency, genes involved in small GTPase-mediated signal transduction were highly enriched in CD8 T cells transduced with Klf4 compared to the MigRI control. In contrast, genes that were increased by Klf4 overexpression were decreased in Klf4 cKO PD1+ CD8+ TILs and highly enriched in the control (fig. S9B). Genes involved in PI3K activity showed a similar tendency (fig. S9C). It was reported that signal transduction by PI3K and Ras small GTPase induces the expression of AP-1 family factors such as c-Jun (35). Thus, there is a possibility that decreased signaling in small GTPase and PI3K pathway by Klf4 deficiency may down-regulate the expression of Jun. To sum, the results revealed that Klf4 deficiency impairs the generation of transitory effector subsets and increases the severity of exhaustion—such as impaired energy metabolism, small GTPase, and PI3K signaling pathways—which, in turn, decreases the antitumor function of CD8 T cells.
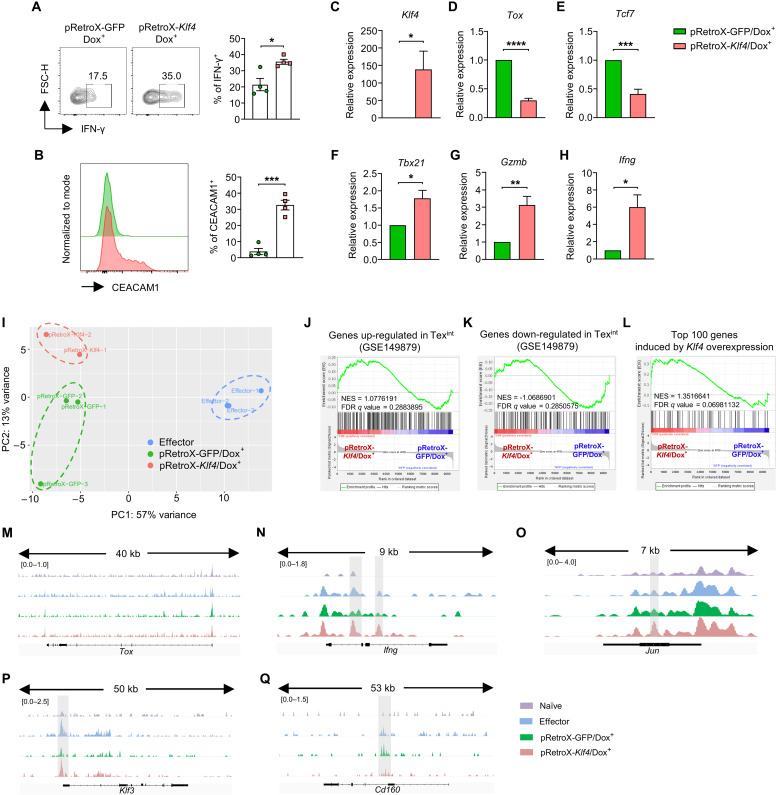
KLF4 reinvigorates the effector function of exhausted CD8 T cells
Next, we tested whether Klf4 can reinvigorate the effector function of exhausted CD8 T cells. For this, Retro-X Tet-One system was used to induce Klf4 expression in exhausted CD8 T cells through doxycycline treatment. The pRetroX vector was inserted with internal ribosomal entry site (IRES)–green fluorescent protein (GFP; pRetroX-GFP) or Klf4-IRES-GFP (pRetroX-Klf4), and cells transduced with the constructs were identified by GFP expression (fig. S10A). CD8 T cells were first transduced with pRetroX-GFP or pRetroX-Klf4 and then induced to be exhausted by repeated antigen stimulation in vitro. The exhausted CD8 T cells transduced with the constructs were then cultured for additional 2 days with or without doxycycline (fig. S10B), and GFP+ cells appeared only after the doxycycline treatment (fig. S10C). GFP- or Klf4-induced exhausted CD8 T cells (pRetroX-GFP/Dox+ or pRetroX-Klf4/Dox+ cells) were isolated, and their phenotypes were analyzed. The proportion of IFN-γ+ cells significantly increased in pRetroX-Klf4/Dox+ cells compared to that in pRetroX-GFP/Dox+ cells (Fig. 6A). The proportion of CEACAM1+ cells also markedly increased in pRetroX-Klf4/Dox+ cells (Fig. 6B). Moreover, pRetroX-Klf4/Dox+ cells, which showed increased expression of Klf4 (Fig. 6C), expressed Tox and Tcf7 at a considerably reduced level (Fig. 6, D and E). Conversely, they showed significantly increased expression of genes related to the effector functions including Tbx21, Gzmb, and Ifng (Fig. 6, F to H). Thus, it appears that CD8 T cells exhausted by repeated antigen stimulation were reinvigorated by the induction of Klf4 expression.
Fig. 6. KLF4 reinvigorates the effector function of exhausted CD8 T cells.
(A to H) OT-I CD8 T cells were transduced with pRetroX-GFP/pRetroX-Klf4 on day 1 during in vitro exhaustion process. Exhausted CD8 T cells were harvested on day 5 and cultured with doxycycline for additional 2 days. GFP+ cells were gated for all analyses. (A) Representative flow cytometry plot and the proportion of IFN-γ+ cells in pRetro-GFP/Dox+ and pRetro-Klf4/Dox+ cells (n = 4 per group). (B) Representative flow cytometry plot of CEACAM1 expression and the proportion of CEACAM1+ cells in pRetro-GFP/Dox+ and pRetro-Klf4/Dox+ cells (n = 4 per group). (C to H) Histogram of relative mRNA expression of (C) Klf4 (n = 4 per group), (D) Tox (n = 4 per group), (E) Tcf7 (n = 4 per group), (F) Tbx21 (n = 3 per group), (G) Gzmb (n = 4 per group), and (H) Ifng (n = 4 per group) in pRetro-GFP/Dox+ and pRetro-Klf4/Dox+ cells. (I to Q) ATAC-seq was performed on naïve, effector, pRetro-GFP/Dox+, and pRetro-Klf4/Dox+ cells. Effector cells were in vitro generated by peptide stimulation for 2 days. (I) Principal components analysis plot of ATAC-seq data from effector CD8 T cells (n = 3), pRetro-GFP/Dox+ (n = 3), and pRetro-Klf4/Dox+ (n = 2) cells. (J to L) GSEA on chromatin accessibility of genes adjacent to peak regions between pRetro-GFP/Dox+ and pRetro-Klf4/Dox+ cells using gene sets of (J) genes up-regulated in Texint, (K) genes down-regulated in Texint, and (L) top 100 genes increased DEGs by Klf4 overexpression (RNA-seq from in vitro exhaustion model). (M to Q) Representative ATAC-seq genome track of (M) Tox, (N) Ifng, (O) Jun, (P) Klf3, (Q) Cd160 in naïve, effector, pRetro-GFP/Dox+, and pRetro-Klf4/Dox+ cells. (A to H) Data are means ± SEM. Statistical analysis was performed using Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Because exhausted CD8 T cells are known to have a strong epigenetic imprint, which is known as a major obstacle for reinvigoration (36–38), we investigated whether the induced Klf4 expression could change the epigenetic status of the exhausted CD8 T cells. For this, we performed assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) and compared chromatin landscapes between in vitro generated effector, pRetroX-GFP/Dox+, and pRetroX-Klf4/Dox+ CD8 T cells. The principal components analysis (PCA) plot showed that the cluster of pRetroX-Klf4/Dox+ cells was closer to the cluster of pRetroX-GFP/Dox+ cells than that of the effector cells (Fig. 6I). In addition, the proportion of overlapping differentially accessible regions (DARs) between effector CD8 T cells and pRetroX-GFP/Dox+ cells or pRetroX-Klf4/Dox+ cells was comparable (59 and 61%, respectively; fig. S10D). When DARs were clustered according to their chromatin accessibility, most were clusters 2 and 5, showing similar patterns between pRetroX-GFP/Dox+ cells and pRetroX-Klf4/Dox+ cells (fig. S10E). These data suggest that Klf4 expression could partly change the epigenetic status of the already exhausted CD8 T cells. Nevertheless, the PCA plot suggests that effector cells, pRetroX-GFP/Dox+ cells, and pRetroX-Klf4/Dox+ cells could be segregated into distinct phenotypic groups (Fig. 6I). The GSEA analysis showed that regions adjacent to the genes up-regulated in Texint appeared to be more accessible in pRetroX-Klf4/Dox+ cells than that in pRetroX-GFP/Dox+ cells. In contrast, regions adjacent to the genes down-regulated in Texint appeared to be more accessible in pRetroX-GFP/Dox+ cells; however, the differences were statistically insignificant (Fig. 6, J and K). We also noticed that regions adjacent to the top 100 up-regulated DEGs from RNA-seq (Fig. 2C) were more accessible in pRetroX-Klf4/Dox+ cells (Fig. 6L). In addition, most of the opened DARs were located at promoter regions, and potential binding targets of enriched motifs in the DARs were majorly SP and KLF families (fig. S10, F to I). Although the chromatin accessibility of Tox was comparable between pRetroX-GFP/Dox+ cells and pRetroX-Klf4/Dox+ cells (Fig. 6M), pRetroX-Klf4/Dox+ cells showed increased chromatin accessibility of genes related to cytotoxicity (Ifng), effector program (Jun), and T cell migration (Klf3), with decreased chromatin accessibility of inhibitory receptor Cd160 (Fig. 6, N to Q). Together, our data suggest that the exhausted CD8 T cells could be reinvigorated by Klf4 expression through transcriptional and partial epigenetic changes, recapitulating the features of a transitory effector CD8 T cell differentiation.
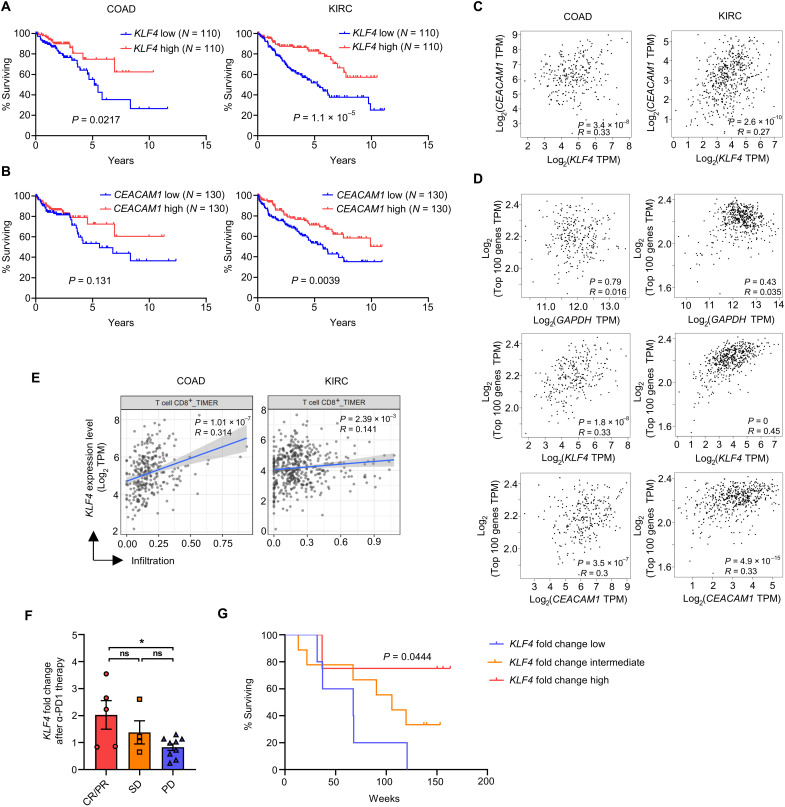
High KLF4 expression correlates with a favorable tumor prognosis
To check the clinical relevance between KLF4 expression and tumor prognosis in patients with cancer, we analyzed data from The Cancer Genome Atlas (TCGA) using various analytical tools (39–41). Among the cancer types, the survival rate of patients with colon adenocarcinoma (COAD) and kidney renal clear cell carcinoma (KIRC) was positively correlated with KLF4 expression (Fig. 7A). Similarly, CEACAM1 expression was also positively correlated with the survival rate of the patients (Fig. 7B). In line with the results of mouse experiments, the expression of KLF4 and CEACAM1 was positively correlated in patients with COAD and KIRC (Fig. 7C). Analysis on the average expression of human orthologs of top 100 up-regulated DEGs by Klf4 expression (Fig. 2C) revealed a positive correlation with KLF4 and CEACAM1 expression in these patients, whereas there was no such correlation with the control gene GAPDH (Fig. 7D). The average expression of Texint, Tcirc, and Teff signature genes was also positively correlated with KLF4 and CEACAM1 expression (fig. S11, A to C). Furthermore, CD8 T cell infiltration was positively correlated with KLF4 expression, in concordance with the survival rates of patients with COAD and KIRC (Fig. 7E).
Fig. 7. High KLF4 expression correlates with a favorable tumor prognosis.
(A and B) Kaplan-Meier curve of patients with COAD and KIRC based on their (A) KLF4 and (B) CEACAM1 expression (upper, 25%; and lower, 25%). Analyzed by OncoLnc. (C) Pearson correlation between KLF4 and CEACAM1 expression on COAD (n = 275) and KIRC (n = 523) tumor tissue. Analyzed by GEPIA2. (D) Pearson correlation between the expression of GAPDH/KLF4/CEACAM1 and average expression of effector signature genes (top 100 genes induced by Klf4 overexpression from RNA-seq data) on COAD (n = 275) and KIRC (n = 523) tumor tissue. Analyzed by GEPIA2. (E) Correlation of KLF4 expression and CD8 T cell infiltration score on COAD (n = 458) and KIRC (n = 533) tumor tissue. Analyzed by TIMER2.0. (F) KLF4 fold change after anti-PD1 therapy in CR (complete response)/PR (partial response; n = 5), SD (stable disease; n = 4), and PD (progressive disease; n = 9) group from patients with melanoma. Data are means ± SEM. Statistical analysis was performed using Student’s t test. ns, P > 0.05; *P < 0.05. (G) Kaplan-Meier curve of melanoma patient groups distinguished by KLF4 fold change. P values less than 0.05 were considered to be significant for the log-rank test and Pearson correlation test.
To further investigate a possibility that CD8 T cells expressing KLF4 actually play a major anticancer function in human, the cell type and their gene expression profiles were analyzed by scRNA-seq analysis. Then, a signature matrix was generated, and the abundance of the cell type within human cancer was calculated by CIBERSORTx (42). The calculated abundance was used for survival analysis of human patients with cancer (fig. S12A). scRNA-seq data of high-grade serous ovarian cancer (HGSOC) have been deposited (43) and used for the subsequent analysis. CD8 T cells were filtered from the datasets and grouped into six subpopulations (LAYN-CD8, GZMK-CD8, STMN1-CD8, FOSB-CD8, KLF2-CD8, and FOXP3-CD8) on the basis of their highly or exclusively expressed genes (fig. S12, B and C). Among the subpopulations, cluster 4 (FOSB-CD8) cells highly expressed TRANSITORY gene sets that are human orthologs of top 10 marker genes in transitory effector subsets of the mouse CD8 T cells (fig. S12D). When we compared KLF4 expression between clusters, the expression was the highest in cluster 4 (FOSB-CD8) cells (fig. S12E). The results suggest that cluster 4 (FOSB-CD8) cells are human counterparts of mouse transitory effector subsets. Then, the signature matrix of 10 cell types in cancer, including six CD8 subpopulations, was generated using CIBERSORTx (fig. S12F). Using the signature matrix, we calculated the abundances of each cell types from expression data of 532 patients with KIRC and analyzed their survival rates. We found that the abundance of FOSB-CD8 T cells showed a positive correlation with better prognosis in patients with cancer (fig. S13, A to F). In line with that, when the patients were grouped by their prognostic stage (stages I to IV), the abundance of FOSB-CD8 T cells decreased as cancer progressed (fig. S13G). These results indicated that a high abundance of FOSB-CD8 T cells expressing high level of KLF4 correlates positively with better outcomes for patients with KIRC.
Because immune checkpoint blockade therapy enhances the antitumor immunity of CD8 T cells (44), we investigated whether immune checkpoint blockade increases KLF4 expression in patients with cancer. We analyzed the reported RNA-seq data obtained from patients with melanoma both before and after anti-PD1 therapy (45). Notably, the fold change in KLF4 expression was the greatest in the CR (complete response)/PR (partial response) group, followed by the SD (stable disease) and PD (progressive disease) groups (Fig. 7F). We also found that the higher fold change in KLF4 expression correlated well with the survival rate of the patients (Fig. 7G). All these data strongly suggest that higher KLF4 expression confers a significant advantage in controlling tumor growth and survival rates of patients with cancer.
DISCUSSION
KLF4 is well known as a reprogramming factor that is highly expressed in embryonic stem cells. Ectopic expression of reprogramming factors (Oct4, Sox2, c-Myc, and Klf4) is known to reprogram mouse somatic cells into pluripotent stem cells (46). In T cells, it has been reported that KLF4 negatively controls their proliferation and is rapidly down-regulated after TCR activation (47–49). However, these studies focused on T cell activities for a short period after TCR stimulation, and the role of KLF4 in the context of exhaustion has been unknown. In this study, we showed that Klf4 expression of CD8 T cells specifically increased during the process of exhaustion induced by chronic antigen stimulation. Klf4 is expressed at its highest level in the transitory effector CD8 T cells during the exhaustion process, while it is decreased in the terminally exhausted CD8 T cells. Klf4 overexpression study revealed that high expression of Klf4 in tumor-specific CD8 T cells up-regulates effector program genes such as AP-1 family factor Jun to acquire the properties of transitory effector subsets. c-Jun is known to enhance the effector function of CD8 T cells (50–52). Recent studies have also demonstrated that the expression of c-Jun or BATF (Basic leucine zipper ATF-like transcription factor), the binding partner of c-Jun, enhances effector function and counteracts exhaustion in CAR T cells (27, 53). In line with that, Klf4 expression down-regulated genes associated with terminal exhaustion and T cell quiescence, highlighting the role of Klf4 in counteracting terminal exhaustion. In contrast, Klf4 deficiency impairs CD8 T cell differentiation into transitory effector subsets and increases the severity of exhaustion, such as impaired energy metabolism, small GTPase, and PI3K signaling pathways, which might, in turn, weaken the effector functions. Thus, our data indicate that high Klf4 expression is important for maintaining functions of transitory effector CD8 T cells, while down-regulation of Klf4 is related to terminal exhaustion.
Several surface molecules have been discovered as signatures of effector CD8 T cells (31, 54). CD69 and CD25 are the most typical markers of activated effector CD8 T cells (55, 56). However, in chronic infections and cancers, these molecules are also up-regulated in exhausted CD8 T cells despite their low-responsive state (15, 57). Thus, identifying previously unknown surface markers for effector CD8 T cells, specifically in the exhaustion contexts, is crucial for understanding their exhaustion process. We found that CEACAM1 was highly induced by Klf4 expression. CEACAM1 is known to mediate immune synapse formation between CD8 T cells and dendritic cells, thereby enhancing the effector function of CD8 T cells during chronic viral infections (58). In addition, it has been reported that CEACAM1 signaling enhances Ras small GTPase and PI3K pathways (59), both of which induce the expression of AP-1 family factors such as c-Jun (35). Thus, CEACAM1 may play a critical role in the antitumor function of CD8 T cells as well. Our results show that Ceacam1 is highly expressed in the transitory effector CD8+ TILs. Moreover, CEACAM1+TIM3+ cells displayed the highest level of effector signatures among CD8+ TIL subsets, while the level decreased in CEACAM1−TIM3+ cells, suggesting that CEACAM1+TIM3+ cells are in the transitory effector stage and CEACAM1−TIM3+ cells are in the terminally exhausted stage. We also noticed that the proportion of transitory effector (CEACAM1+TIM3+) cells among the CD8+ TILs from normal conditioned mice was even very low, whereas the proportion of terminally exhausted (CEACAM1−TIM3+) cells was high, implying insufficient control of overwhelming tumor growth. We discovered that Klf4 expression greatly increased the proportion of CEACAM1+TIM3+ cells and markedly increased their antitumor immunity, suggesting that maintaining a high number and/or proportion of CEACAM1+TIM3+ cells is effective for the control of tumor growth. Our study provides a new window of analyzing exhausted CD8 T cell subsets; however, further studies on detailed characteristics of CEACAM1-TIM3–defined subsets are needed.
Recent studies have shown that terminal exhaustion inherits strong epigenetic scars in CD8 T cells, which could not be cured by “passive” methods such as removal of chronic antigen stimulation (36–38). Rather, more “active” methods are needed to reinvigorate terminally exhausted CD8 T cells. Several studies have shown that CD8 T cells from tumor environment can be reinvigorated by immune checkpoint blockades, gene manipulations, and exogenous cytokine treatments (27, 44, 53, 60, 61). However, most of these studies investigated at the scope of overall antitumor immunity, overlooking the heterogeneity of CD8+ TILs. Our data, combining in vitro exhaustion and Retro-X Tet-one system, showed that Klf4 expression could reinvigorate the effector function of exhausted CD8 T cells, in part, by restoring the transcriptional and epigenetic status of genes related to effector functions such as Ifng, Jun, and Klf3. Although we did not dissect whether Klf4 expression causes dedifferentiation and rejuvenation of terminally exhausted CD8 T cells or just strengthens the effector function of the cells, our study suggests that KLF4 may serve as an effective reinvigorating factor for cancer immune therapy. Further studies will be required to understand the mechanistic role of KLF4 in reinvigorating exhausted CD8 T cells and to improve the efficacy of the process.
Last, our TCGA data analyses suggested that KLF4 expression positively correlates with the prognosis of patients with cancer. The expression of KLF4 and CEACAM1 was positively correlated with the expression of effector signature genes and survival rates of patients with COAD and KIRC. Similarly, the abundance of KLF4-expressing human counterparts of transitory effector subsets was also positively correlated with favorable prognosis among patients with KIRC. Although these correlations were observed only in certain types of cancer, our results support that KLF4 plays an important role also in regulating human cancer. Moreover, we found that up-regulation of KLF4 expression by anti-PD1 increased the survival rates of patients with melanoma. The increase of KLF4 by anti-PD1 suggests two possibilities: anti-PD1 directly increases the expression of KLF4 on CD8+ TILs or anti-PD1 increases the number of KLF4-expressing transitory effector CD8 T cells. Up-regulated KLF4 expression might increase CD8 T cell differentiation into transitory effector subsets and the effector function. Meanwhile, because anti-PD1 is known to increase the number of progenitor exhausted CD8 T cells, the number of transitory effector CD8 T cells is also likely to increase upon the treatment. In either case, anti-PD1 therapy may increase the function and/or the number of transitory effector CD8 T cells, thereby effectively controlling human cancer. We also noticed that the increase of KLF4 was detected only within responders, while the expression did not change within nonresponders. The results imply that the responsiveness of anti-PD1 therapy may positively correlate with increased KLF4 expression, suggesting that a combinatorial therapy using anti-PD1 with KLF4 inducers would be an efficient way of treating cancer.
In conclusion, our study demonstrates that KLF4 promotes CD8 T cell differentiation into cytolytic transitory effector CD8 T cells, enhances their effector function during the exhaustion process, blocks reaching into terminal exhaustion, and reinvigorates the exhausted cells (fig. S14). Thus, the potential effect of KLF4 on CD8 T cell exhaustion can be highlighted in terms of antitumor immune therapy. A major limitation of the study is a lack of understanding on the molecular mechanism of how KLF4 expression is regulated during the exhaustion process. Further studies are required to discover any signal and/or factor(s) regulating KLF4 expression during the exhaustion process.
MATERIALS AND METHODS
Study design
The aim of this study was to understand the role of KLF4 on CD8 T cells specifically in the context of exhaustion. To examine the expression of KLF4 on exhausted CD8 T cells, we used in vitro CD8 T cell exhaustion model and mouse tumor model, as well as scRNA-seq analyses. In addition, we combined retroviral transduction of KLF4 on CD8 T cells with the models and used a cKO mice model to figure out the role of KLF4 on antitumor immunity. To investigate whether KLF4 could reinvigorate exhausted CD8 T cells, we used a modified in vitro exhaustion model in which KLF4 could be induced after the exhaustion process, not during the activation stage. Last, to apply the results to human patients with cancer, we analyzed TCGA data by various analytical tools to examine the correlation between gene expression and tumor prognosis.
Mice
Klf4fl/fl mice were purchased from Mutant Mouse Resource & Research Centers. C57BL/6, Rag2 KO, OT-I, and E8i-cre were purchased from the Jackson Laboratory. PmelI mice were gifted by E. J. Park (Korea National Cancer Center). Klf4 cKO mice were generated by crossing Klf4fl/fl mice with E8i-cre mice for deletion of Klf4 on CD8 T cells. Klf4fl/fl or Klf4fl/+ mice were used as control mice for comparison with Klf4 cKO mice (Klf4fl/fl; E8i-cre). All mice were bred and maintained in specific pathogen–free barrier facilities at Seoul National University and were used according to protocols approved by Institutional Animal Care and Use Committees of Seoul National University.
Cell culture
Tumor cell lines (MC38, MC38-gp100) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco, HyClone), streptomycin (100 U/ml), and penicillin. Platinum-E (Plat-E) cells were cultured in DMEM supplemented with 10% FBS (Gibco, HyClone), streptomycin (100 U/ml), and penicillin, and with blasticidin (10 μg/ml) and puromycin (1 μg/ml) for selection. All primary cells (CD8 T cells) were cultured in RPMI supplemented with 10% FBS (Gibco, HyClone), streptomycin and penicillin (100 U/ml), and 2-ME (2-mercaptoethanol).
In vitro CD8 T cell exhaustion experiment
In vitro CD8 T cell exhaustion was performed as described (17) with little modification. Briefly, magnetic-activated cell sorting (MACS)–purified naïve CD44low CD8 T cells from OT-I mice were cultured with RPMI medium supplemented with mouse interleukin-7 (mIL-7) (5 ng/ml), mIL-15 (5 ng/ml), and OVA peptide (257 to 264; 10 ng/ml). After 24 hours, cells were washed twice with RPMI medium and stimulated with the same condition as stated before. The daily stimulation process was repeated for additional 4 days, and cells were harvested at day 5 for further experiments. To generate cytokine only samples, cells were cultured without OVA peptide; and for single stimulated samples, cells were stimulated with OVA peptide for 2 days and cultured without OVA peptide for another 3 days. In case of retroviral transduction of CD8 T cells, the cells from day 1 (24 hours stimulated) were retrovirally transduced with MigRI, Klf4, pRetroX-GFP, and pRetroX-Klf4 and repeatedly stimulated until day 5. For reinvigoration experiments using Retro-X Tet-one system (Takara Bio), the cells were treated with OVA peptides and doxycycline (500 ng/ml) for additional 2 days.
Transfection and production of retrovirus
A total of 2.5 × 106 Plat E cells were seeded on 60-mm plates in DMEM without antibiotics; and after 6 hours, attached cells were transfected with MigRI, MigRI-Klf4, pRetroX-GFP, and pRetroX-Klf4 (10 μg) according to calcium phosphate transfection methods. Briefly, DNA–calcium chloride mix was added onto 2× HBS (HEPES buffured saline) along with weak vortexing. After 30 min of a resting period, mix was gently pipetted and added to the plate dropwisely. Cells were cultured overnight at 37°C. On the next day, plates were gently washed twice with warm medium or phosphate-buffered saline (PBS) and added 3 ml of fresh DMEM without antibiotics. After 48 hours, viral supernatants were harvested and filtered through 0.45-μm filter (ADVANTEC). Prepared viral supernatants were used directly for transduction or snap-frozen and stored at −70°C.
Retroviral transduction of CD8 T cells
For efficient retroviral transduction of CD8 T cells, we isolated blasting CD8 T cells using Percoll enrichment method as described (62). Briefly, OT-I or PmelI CD44low naïve CD8 T cells were MACS-enriched and activated with OVA peptide (10 ng/ml) or gp100 peptide (1 μg/ml), soluble anti-CD28 (2 μg/ml), and mIL-2 (100 U/ml) for 18 to 24 hours. Then, cells were density gradient–centrifuged with 30%/60% Percoll for 20 min at 2000 rpm, 25°C. Cells on the interface layer were harvested and spin-infected for 90 min at 2000 rpm, 30°C in the presence of polybrene (8 μg/ml). After spin infection, cells were incubated at 37°C for 30 min and harvested for further experiments.
Tumor transplantation and CD8 T cell adoptive transfer experiment
A total of 3 × 105 MC38/MC38-gp100 tumor cells (less cultured than 3 weeks from thawed cell stock) were subcutaneously injected into the shaved-right flank of mice. For CD8 T cell adoptive transfer experiments, 5 × 105 retrovirally transduced PmelI CD8 T cells were transferred into MC38-gp100 tumor–inoculated Rag2 KO mice through tail vein. Tumor size was monitored every 2 to 3 days, and tumor cells were harvested on days 14 to 18 from inoculation for further analyses.
Isolating TILs from tumor tissue
Tumor tissues were minced using scissors, and digestion solution [RPMI medium supplemented with collagenase IV (1 mg/ml), deoxyribonuclease I (100 μg/ml), 1% FBS, antibiotics, and 2-ME] was added. Samples were incubated at 37°C for 30 min and stirred with a magnetic stirrer. After that, samples were passed through 70-μm strainers, and RPMI medium (10% FBS, antibiotics, and 2-ME) was added. Cells were washed and density gradient–centrifuged using 40%/70% Percoll for 20 min at 2000 rpm, 25°C. Interfaced cells were harvested and washed twice with 1× PBS. Then, prepared samples were used for further experiments.
Flow cytometry and cell sorting
Single-cell suspensions were prepared by passing through a strainer to get rid of cell debris. ACK (Ammonium-Chloride-Potassium) lysing buffer was added to the cells and incubated at room temperature for 3 min to remove red blood cells. After that, PBS was added up to 10 ml and washed. In case of using MACS for an enrichment of naïve CD8 T cells, experiments were performed according to the manufacturer’s protocol (Miltenyi Biotec). Cells were treated with Fc Block (BD Biosciences) for at least 20 min and stained with monoclonal antibodies in various combinations in 1× PBS for 15 to 30 min. Flow cytometry analyses were performed using FACSCanto II (BD Biosciences), and Sony Sorter SH-800 (Sony) and FACSAria II (BD Biosciences) were used for cell sorting. Data were analyzed using FlowJo v10 software. The antibodies used are as follows: The following antibody conjugates were purchased from BD Biosciences: Biotin-conjugated mouse lineage (Lin) panel that contains anti-B220 (RA3-6B2), anti-CD3e (145-2C11), anti–Gr-1 (RB6-8C5), anti-CD11b (Mac1, M1/70), and anti–Ter-119 antibodies; CD62L (MEL-14)–PerCP/Cy5.5; IFN-γ (XMG1.2)–allophycocyanin (APC); Ly108 (13G3)–Alexa Fluor 647; and Fc Block (2.4G2). The following antibody conjugates were purchased from BioLegend: LAG3 (C9B7W)–PE-Cy7, TIGIT (1G9)–phycoerythrin (PE)–Cy7, TIM3 (RMT3-23)–PE, TIM3 (RMT3-23)–APC, TNF-α (MP6-XT22)–APC-Cy7, CEACAM1 (Mab-CC1)–PerCP/Cy5.5, and CX3CR1 (SA011F11)–PerCP/Cy5.5. The following antibody conjugates were purchased from Miltenyi Biotec: GzmB (REA226)–PE, TOX (REA473)–PE, and TOX (REA473)–APC. The following antibody conjugates were purchased from Invitrogen: CD44 (IM7)–APC, CD45 (HI30)–PerCP-Cy5.5, CD45.1 (A20)–APC eFluor780, CD45.2 (104)–PE-Cy7, CD8 (53-6.7)–fluorescein isothiocyanate (FITC), CD8 (53-6.7)–APC-Cy7, CD8 (53-6.7)–PE; CTLA-4 (UC10-4B9)–Biotin, Eomes (Dan11mag)–PE-Cy7, Ki-67 (SolA15)–Biotin, PD-1 (J43)–FITC, T-bet (4B10)–PE, CD69 (H1.2F3)–PerCP-Cy5.5, and TOX (TXRX10)–PE. TCF1/TCF7 (C63D9)–Alexa Fluor 647 and Phospho–c-Jun (Ser63, 54B3)–Biotin were purchased from Cell Signaling Technology.
Intracellular staining of cytokine and transcription factors
For intracellular staining of cytokine, cells were treated with phorbol 12-myristate 13-acetate (50 ng/ml), ionomycin (1 μg/ml), and brefeldin A for 4 hours. In case of OT-I CD8 T cells, the cells were treated with OVA peptide (10 ng/ml) and brefeldin A for 6 hours. Then, intracellular cytokine staining was performed according to BD Biosciences protocol. For intracellular staining of transcription factors, staining was performed according to the eBioscience protocol. In case of detecting GFP signal, because Fix/Perm from BD Biosciences and eBioscience causes loss of GFP signal, cells were prefixed with 4% paraformaldehyde for 20 min in ice. After that, cells were washed with 1× PBS and added Fix/Perm from BD Biosciences/eBioscience.
Quantitative reverse transcription PCR
Total RNA was extracted from cells using TRI Reagent according to the manufacturer’s instructions (Molecular Research Center Inc.). Equivalent quantities of total RNA were reverse-transcribed with the Quantitect Reverse Transcription Kit (QIAGEN). Complementary DNAs (cDNAs) were diluted and were analyzed by quantitative real-time polymerase chain reaction (PCR) analysis (Applied Biosystems, StepOnePlus). The expression of each gene was normalized to Actb expression. The primer sets used in experiments are listed in table S1.
Motif analysis
Motif analysis for searching consensus motifs in inhibitory receptor gene promoters was performed using the MEME Suite (63). For comparing motifs against a database of known motifs, TomTom analysis (64) was performed. For discovering motifs from the ATAC-seq peak regions, MEME-ChIP was used (65).
scRNA-seq analysis
Mouse scRNA-seq analysis was performed using data from GSE116390. All data processing and analysis were performed using galaxy (66). Filtered 3574 cells expressing Cd8a, Cd8b1, and Cd2 but not Cd4 were kept for further analysis (processed data available in Gene Expression Omnibus (GEO) entry). Among the cells, Pdcd1-expressing 2652 cells were filtered. Cell size normalization was performed using Scanpy as target sum 10,000, and then, the data matrix was logarithmized. For dimensionality reduction, we identified highly variable genes using Scanpy as Seurat computing method: minimal mean cutoff, 0.0125; maximal mean cutoff, 3; and minimal normalized dispersion cutoff, 0.5. Then, data were scaled to unit variance and zero mean. A total of 1987 highly variable genes were used for dimensionality reduction using PCA. We chose 10 neighbors for a K-nearest neighbor (KNN) graph, the Euclidean distance metrics, and the uniform manifold approximation and projection (UMAP) method to compute the connectivities. Louvain graph clustering method was used to cluster neighborhood graph: vtraag method, resolution 0.3. The expression of Tbx21, Tnf, Cx3cr1, Ets1, Klrk1, Ifng, Klrd1, Klrg1, S1pr1, Klf3, Foxj3, Hoxb4, Nfyb, Atf2, Sp3, and Maz was scored for the signature genes of transitory/intermediate exhausted CD8 T cells, which are known to positively correlate with Tbx21 expression and CD8 T cell effector functions (15). Marker genes for each cluster were analyzed by comparing each cluster to the union of the rest of the cluster using Wilcoxon rank sum method. UMAP visualization and violin plots were plotted using Scanpy.
Human scRNA-seq analysis was performed using data from GSE184880. Datasets from seven patients with HGSOC were merged and analyzed. Among the cells, CD14-expressing monocytes, CD79A-expressing B cells, and EPCAM-expressing tumor cells were filtered out, and CD8A-expressing cells were kept for CD8+ TIL analysis (n = 3137). Cell size normalization was performed using Scanpy as target sum 10,000, and then, the data matrix was logarithmized. For dimensionality reduction, we identified highly variable genes using Scanpy as Seurat computing method: minimal mean cutoff, 0.0125; maximal mean cutoff, 3; and minimal normalized dispersion cutoff, 0.5. Then, data were scaled to unit variance and zero mean. A total of 2338 highly variable genes were used for dimensionality reduction using PCA. We chose 22 neighbors for a KNN graph, the Euclidean distance metrics, and the UMAP method to compute the connectivities. Louvain graph clustering method was used to cluster neighborhood graph: vtraag method, resolution 0.45. The expression of TRANSITORY gene sets (JUNB, BTG1, TNFAIP3, BTG2, DUSP1, PNRC1, NR4A1, NFKBIA, DUSP2, and ZFP36) was scored for signature genes of transitory effector human counterparts. UMAP visualization and violin plots were plotted using Scanpy.
Bulk RNA-seq library preparation and analysis
OT-I CD8 T cells transduced with MigRI (n = 3)/Klf4 (n = 3) were repeatedly stimulated with OVA peptide in vitro and then harvested. Total RNA was extracted using TRI Reagent according to the manufacturer’s instructions (Molecular Research Center Inc.). cDNA libraries were prepared using the TruSeq stranded mRNA Sample Preparation Kit (Illumina, CA, USA). cDNA libraries were quantified with the KAPA library quantification kit (Kapa Biosystems, MA, USA) according to the manufacturer’s library quantification protocol. Agilent 2100 BioAnalyzer (Agilent, CA, USA) was used to evaluate the quality of these cDNA libraries. Sequencing was performed as paired-end [2 × 150 base pairs (bp)] using an Illumina NovaSeq6000 (Illumina, CA, USA) by Theragen Bio sequencing service (Theragen Bio Co. Ltd., Seongnam, Gyeonggi, Korea). The ends of the reads less than Phred quality score of 20 and the adapter sequences were trimmed. In addition, the reads shorter than 50 bp were removed by using cutadapt (v2.8) (67). Aligner STAR (v.2.7.1.a) (68) was used to map filtered reads to the reference genome (mm10), and RSEM (v.1.3.1) (69) was used to estimate gene expression. FPKM (Fragments per kilobase of transcript per million mapped fragments), RPKM (Reads per kilobase of transcript per million mapped reads), and TPM (Transcripts per million) values were calculated to normalize the sequencing depth among the samples. The TCC (Tag count comparison) R package (v.1.26.0) (70) was used to normalize and compare the tag count data. To identify physiologically meaningful gene expression changes, genes with read counts lower than 10 and/or RPKM lower than 2 were filtered out. Normalization factors were calculated using the iterative DEseq2 method (71). The DEGs were identified on the basis of the P value less than 0.05 and an absolute value of fold change over 2. GO analysis was performed using Metascape (72). For GSEA, GSEA 4.1.0 software was used.
RNA-seq of tumor-infiltrating CD8 T cells using SMART-seq
PmelI CD8 T cells transduced with MigRI/Klf4 were adoptively transferred into MC38-gp100–inoculated Rag2 KO mice on day 7. The tumor-infiltrating CD8 T cells transduced with MigRI (from four mice) and Klf4 (from four mice) were pooled (GFP+ cells were sorted) on day 15. For cKO study, PD1+CD8+ TILs from tumor tissues of four control mice and six Klf4 cKO mice were pooled on day 14. Total RNA was extracted from the isolated cells using TRI Reagent according to the manufacturer’s instructions (Molecular Research Center Inc.). The RNA isolated from each sample was used to construct sequencing libraries with the SMART-Seq v4 Ultra Low Input RNA Kit for Illumina, following the manufacturer’s protocol. cDNA libraries were quantified with the KAPA library quantification kit (Kapa Biosystems, MA, USA) according to the manufacturer’s library quantification protocol. Agilent 4200 TapeStation (Agilent, CA, USA) was used to evaluate the quality of these cDNA libraries. Sequencing was performed as single-end (100 bp) using an Illumina NovaSeq6000 (Illumina, CA, USA) by Macrogen sequencing service (Macrogen Inc., Seoul, Korea). Low-quality reads were filtered out, and adaptor sequences were trimmed. HISAT (Hierarchical indexing for spliced alignment of transcripts) (v.2.1.0) (73) was used to map filtered reads to the reference genome (mm10), and StringTie (v.2.1.3b) (74) was used to estimate gene expression. Size factor for read count data was calculated using edgeR (calcNormFactors), and counts were TMM (trimmed mean of M values)–normalized. DEG analysis was performed using edgeR (75). The DEGs were identified on the basis of the P value less than 0.05 and an absolute value of fold change over 2. GO analysis was performed using Metascape (72) and g:Profiler (76). For GSEA, GSEA 4.1.0 software was used.
ATAC-seq library preparation and analysis
A total of 50,000 cells from naïve OT-I CD8 T cells (n = 3), effector OT-I CD8 T cells (n = 3), pRetroX-GFP/Dox+ (n = 3), and pRetroX-Klf4/Dox+ CD8 T cells (n = 2) were prepared for ATAC-seq samples. Naïve OT-I CD8 T cells were treated with OVA peptide (10 ng/ml), anti-CD28 (2 μg/ml), and mIL-2 (100 U/ml) for 2 days to generate effector OT-I CD8 T cells. For generation of gene-induced exhausted CD8 T cells, OT-I CD8 T cells transduced with pRetroX-GFP/pRetroX-Klf4 were repeatedly stimulated with OVA peptide in vitro. After that, cells were cultured with OVA peptide (10 ng/ml), anti-CD28 (2 μg/ml), mIL-2 (100 U/ml), and doxycycline (500 ng/ml) for 2 days. Gene-induced cells (GFP+) were isolated using flow cytometry. DNA libraries of each group were prepared using an ATAC-Seq kit (Cell Biologics) according to the manufacturer’s instructions. DNA libraries were quantified with the KAPA library quantification kit (Kapa Biosystems, MA, USA) according to the manufacturer’s library quantification protocol. Agilent High Sensitivity D1000 ScreenTape (Agilent, CA, USA) was used to evaluate the quality of these DNA libraries. Sequencing was performed as paired-end (2 × 150 bp) using an Illumina NovaSeq6000 (Illumina, CA, USA) by Theragen Bio sequencing service (Theragen Bio Co. Ltd., Seongnam, Gyeonggi, Korea). All data processing was performed using galaxy (66). Raw ATAC-seq FASTQ files were trimmed with cutadapt. Samples were aligned to the mm10 reference genome using Bowtie2. Unmapped, unpaired, and mitochondrial reads were removed using BamTools filter. PCR duplicates were removed using Picard MarkDuplicates. Peak calling was performed using MACS2. Then, we combined the peaks of all samples to create union peaks and merged overlapping peaks using bedtools mergeBed. The number of reads in each peak was calculated using BedCov. DARs were identified using DEseq2 and P value cutoff < 0.05. bigWig files were generated from Bam files using bamCoverage with BPM (bins per million) normalization, and genome tracks were visualized using pyGenomeTracks and Integrative Genomics Viewer (v2.11.2). We used MEME-ChIP to search for enriched motifs from the DARs. We used ChIPseeker for peak region annotation.
Cancer patient data analysis
Data from TCGA were analyzed using various analytic tools (39–41). Kaplan-Meier survival analysis was performed using OncoLnc (39) and Prism8 (GraphPad software). Gene expression correlation analysis was performed using GEPIA2 (40). Scoring of CD8 T cell infiltration and correlation with gene expression were calculated using TIMER2.0 (41). RNA-seq data and clinical information of patients with melanoma were acquired from the published report (45).
For analyzing abundance of the cell types and survival rates of patients with cancer, count matrix of 10 cell types was generated from the single-cell analysis, and the signature matrix was generated by CIBERSORTx (42). scRNA-seq mode was used with quantile normalization option and min. Expression was set to 0. Then, TPM expression data of 532 patients with KIRC were downloaded from TCGA and were deconvoluted using the signature matrix. The analysis was performed using S-mode batch correction with quantile normalization and absolute mode options. Calculated abundances were used for survival analysis of patients with cancer. All other options in CIBERSORTx were conducted as in default option unless stated otherwise.
Statistical analysis
For calculations of statistical significance, Prism 8 (GraphPad software) was used. Data are presented as means ± SEM and were analyzed using two-tailed Student’s t test unless stated otherwise. P values less than 0.05 were considered to be significant.
Acknowledgments
Funding: This work was supported by National Research Foundation of Korea (NRF) grant NRF-2021R1A2B5B03002202 (R.H.S.) and MedGene Therapeutics Inc. grant (R.H.S.).
Author contributions: Conceptualization: J.N. and R.H.S. Methodology: J.N. and R.H.S. Investigation: J.N. Visualization: J.N. Funding acquisition: R.H.S. Project administration: R.H.S. Supervision: R.H.S. Writing—original draft: J.N. Writing—review and editing: R.H.S.
Competing interests: J.N. and R.H.S. disclose a submitted patent application (PCT/KR2022/003482) related to this research on 11 March 2022. R.H.S. is a cofounder of MedGene Therapeutics Inc. The authors declare they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Accession number to access sequencing data (RNA-seq, SMART-seq, and ATAC-seq) at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE212806 is GSE212806.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Table S1
REFERENCES AND NOTES
- 1.A. J. Zajac, J. N. Blattman, K. Murali-Krishna, D. J. D. Sourdive, M. Suresh, J. D. Altman, R. Ahmed,Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188,2205–2213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. Gallimore, A. Glithero, A. Godkin, A. C. Tissot, A. Plückthun, T. Elliott, H. Hengartner, R. Zinkernagel,Induction and exhaustion of lymphocytic choriomeningitis virus–specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I–peptide complexes. J. Exp. Med. 187,1383–1393 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. Schietinger, M. Philip, V. E. Krisnawan, E. Y. Chiu, J. J. Delrow, R. S. Basom, P. Lauer, D. G. Brockstedt, S. E. Knoblaugh, G. J. Hämmerling, T. D. Schell, N. Garbi, P. D. Greenberg,Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity 45,389–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. A. Paley, D. C. Kroy, P. M. Odorizzi, J. B. Johnnidis, D. V. Dolfi, B. E. Barnett, E. K. Bikoff, E. J. Robertson, G. M. Lauer, S. L. Reiner, E. J. Wherry,Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338,1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D. T. Utzschneider, M. Charmoy, V. Chennupati, L. Pousse, D. P. Ferreira, S. Calderon-Copete, M. Danilo, F. Alfei, M. Hofmann, D. Wieland, S. Pradervand, R. Thimme, D. Zehn, W. Held,T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45,415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 6.S. J. Im, M. Hashimoto, M. Y. Gerner, J. Lee, H. T. Kissick, M. C. Burger, Q. Shan, J. S. Hale, J. Lee, T. H. Nasti, A. H. Sharpe, G. J. Freeman, R. N. Germain, H. I. Nakaya, H. H. Xue, R. Ahmed,Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537,417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R. He, S. Hou, C. Liu, A. Zhang, Q. Bai, M. Han, Y. Yang, G. Wei, T. Shen, X. Yang, L. Xu, X. Chen, Y. Hao, P. Wang, C. Zhu, J. Ou, H. Liang, T. Ni, X. Zhang, X. Zhou, K. Deng, Y. Chen, Y. Luo, J. Xu, H. Qi, Y. Wu, L. Ye,Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature 537,412–416 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Y. A. Leong, Y. Chen, H. S. Ong, D. Wu, K. Man, C. Deleage, M. Minnich, B. J. Meckiff, Y. Wei, Z. Hou, D. Zotos, K. A. Fenix, A. Atnerkar, S. Preston, J. G. Chipman, G. J. Beilman, C. C. Allison, L. Sun, P. Wang, J. Xu, J. G. Toe, H. K. Lu, Y. Tao, U. Palendira, A. L. Dent, A. L. Landay, M. Pellegrini, I. Comerford, S. R. McColl, T. W. Schacker, H. M. Long, J. D. Estes, M. Busslinger, G. T. Belz, S. R. Lewin, A. Kallies, D. Yu,CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17,1187–1196 (2016). [DOI] [PubMed] [Google Scholar]
- 9.T. Wu, Y. Ji, E. A. Moseman, H. C. Xu, M. Manglani, M. Kirby, S. M. Anderson, R. Handon, E. Kenyon, A. Elkahloun, W. Wu, P. A. Lang, L. Gattinoni, D. B. McGavern, P. L. Schwartzberg,The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.I. Siddiqui, K. Schaeuble, V. Chennupati, S. A. Fuertes Marraco, S. Calderon-Copete, D. P. Ferreira, S. J. Carmona, L. Scarpellino, D. Gfeller, S. Pradervand, S. A. Luther, D. E. Speiser, W. Held,Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50,195–211.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 11.B. C. Miller, D. R. Sen, R. al Abosy, K. Bi, Y. V. Virkud, M. W. LaFleur, K. B. Yates, A. Lako, K. Felt, G. S. Naik, M. Manos, E. Gjini, J. R. Kuchroo, J. J. Ishizuka, J. L. Collier, G. K. Griffin, S. Maleri, D. E. Comstock, S. A. Weiss, F. D. Brown, A. Panda, M. D. Zimmer, R. T. Manguso, F. S. Hodi, S. J. Rodig, A. H. Sharpe, W. N. Haining,Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20,326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. D. Blackburn, H. Shin, G. J. Freeman, E. J. Wherry,Selective expansion of a subset of exhausted CD8 T cells by αPD-L1 blockade. Proc. Natl. Acad. Sci. U.S.A. 105,15016–15021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.W. H. Hudson, J. Gensheimer, M. Hashimoto, A. Wieland, R. M. Valanparambil, P. Li, J.-X. Lin, B. T. Konieczny, S. J. Im, G. J. Freeman, W. J. Leonard, H. T. Kissick, R. Ahmed,Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51,1043–1058.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. Zander, D. Schauder, G. Xin, C. Nguyen, X. Wu, A. Zajac, W. Cui,CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity 51,1028–1042.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.J.-C. Beltra, S. Manne, M. S. Abdel-Hakeem, M. Kurachi, J. R. Giles, Z. Chen, V. Casella, S. F. Ngiow, O. Khan, Y. J. Huang, P. Yan, K. Nzingha, W. Xu, R. K. Amaravadi, X. Xu, G. C. Karakousis, T. C. Mitchell, L. M. Schuchter, A. C. Huang,Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52,825–841.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.C. D. Scharer, B. G. Barwick, B. A. Youngblood, R. Ahmed, J. M. Boss,Global DNA methylation remodeling accompanies CD8 T cell effector function. J. Immunol. 191,3419–3429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Zhao, C. H. Kiernan, C. J. Stairiker, J. L. Hope, L. G. Leon, M. van Meurs, I. Brouwers-Haspels, R. Boers, J. Boers, J. Gribnau, W. F. J. van IJcken, E. M. Bindels, R. M. Hoogenboezem, S. J. Erkeland, Y. M. Mueller, P. D. Katsikis,Rapid in vitro generation of bona fide exhausted CD8+ T cells is accompanied by Tcf7 promotor methylation. PLOS Pathog. 16,e1008555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. J. Carmona, I. Siddiqui, M. Bilous, W. Held, D. Gfeller,Deciphering the transcriptomic landscape of tumor-infiltrating CD8 lymphocytes in B16 melanoma tumors with single-cell RNA-seq. Onco. Targets. Ther. 9,1737369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. M. McLane, S. F. Ngiow, Z. Chen, J. Attanasio, S. Manne, G. Ruthel, J. E. Wu, R. P. Staupe, W. Xu, R. K. Amaravadi, X. Xu, G. C. Karakousis, T. C. Mitchell, L. M. Schuchter, A. C. Huang, B. D. Freedman, M. R. Betts, E. J. Wherry,Role of nuclear localization in the regulation and function of T-bet and Eomes in exhausted CD8 T cells. Cell Rep. 35,109120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.S. Xing, F. Li, Z. Zeng, Y. Zhao, S. Yu, Q. Shan, Y. Li, F. C. Phillips, P. K. Maina, H. H. Qi, C. Liu, J. Zhu, R. M. Pope, C. A. Musselman, C. Zeng, W. Peng, H. H. Xue,Tcf1 and Lef1 transcription factors establish CD8+ T cell identity through intrinsic HDAC activity. Nat. Immunol. 17,695–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M. Danilo, V. Chennupati, J. G. Silva, S. Siegert, W. Held,Suppression of Tcf1 by inflammatory cytokines facilitates effector CD8 T cell differentiation. Cell Rep. 22,2107–2117 (2018). [DOI] [PubMed] [Google Scholar]
- 22.E. Hajaj, G. Eisenberg, S. Klein, S. Frankenburg, S. Merims, I. Ben David, T. Eisenhaure, S. E. Henrickson, A. C. Villani, N. Hacohen, N. Abudi, R. Abramovich, J. E. Cohen, T. Peretz, A. Veillette, M. Lotem,SLAMF6 deficiency augments tumor killing and skews toward an effector phenotype revealing it as a novel T cell checkpoint. eLife 9,e52539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Y. Zhang, M. Xu, X. Zhang, F. Chu, T. Zhou,MAPK/c-Jun signaling pathway contributes to the upregulation of the anti-apoptotic proteins Bcl-2 and Bcl-xL induced by Epstein-Barr virus-encoded BARF1 in gastric carcinoma cells. Oncol. Lett. 15,7537–7544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. Batarseh, J. Li, V. Papadopoulos,Protein kinase Cε regulation of translocator protein (18 kDa) Tspo gene expression is mediated through a MAPK pathway targeting STAT3 and c-Jun transcription factors. Biochemistry 49,4766–4778 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. J. Marinissen, M. Chiariello, T. Tanos, O. Bernard, S. Narumiya, J. S. Gutkind,The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol. Cell 14,29–41 (2004). [DOI] [PubMed] [Google Scholar]
- 26.T. Smeal, B. Binetruy, D. Mercola, A. Grover-Bardwick, G. Heidecker, U. R. Rapp, M. Karin,Oncoprotein-mediated signalling cascade stimulates c-Jun activity by phosphorylation of serines 63 and 73. Mol. Cell Biol. 12,3507–3513 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. C. Lynn, E. W. Weber, E. Sotillo, D. Gennert, P. Xu, Z. Good, H. Anbunathan, J. Lattin, R. Jones, V. Tieu, S. Nagaraja, J. Granja, C. F. A. de Bourcy, R. Majzner, A. T. Satpathy, S. R. Quake, M. Monje, H. Y. Chang, C. L. Mackall,c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576,293–300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. J. Milner, C. Toma, B. Yu, K. Zhang, K. Omilusik, A. T. Phan, D. Wang, A. J. Getzler, T. Nguyen, S. Crotty, W. Wang, M. E. Pipkin, A. W. Goldrath,Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552,253–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. A. ElTanbouly, Y. Zhao, E. Nowak, J. Li, E. Schaafsma, I. le Mercier, S. Ceeraz, J. L. Lines, C. Peng, C. Carriere, X. Huang, M. Day, B. Koehn, S. W. Lee, M. Silva Morales, K. A. Hogquist, S. C. Jameson, D. Mueller, J. Rothstein, B. R. Blazar, C. Cheng, R. J. Noelle,VISTA is a checkpoint regulator for naïve T cell quiescence and peripheral tolerance. Science 367, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.E. J. Wherry, S. J. Ha, S. M. Kaech, W. N. Haining, S. Sarkar, V. Kalia, S. Subramaniam, J. N. Blattman, D. L. Barber, R. Ahmed,Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27,670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 31.S. Sarkar, V. Kalia, W. N. Haining, B. T. Konieczny, S. Subramaniam, R. Ahmed,Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205,625–640 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. M. Kaech, S. Hemby, E. Kersh, R. Ahmed,Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111,837–851 (2002). [DOI] [PubMed] [Google Scholar]
- 33.W. W. Overwijk, M. R. Theoret, S. E. Finkelstein, D. R. Surman, L. A. de Jong, F. A. Vyth-Dreese, T. A. Dellemijn, P. A. Antony, P. J. Spiess, D. C. Palmer, D. M. Heimann, C. A. Klebanoff, Z. Yu, L. N. Hwang, L. Feigenbaum, A. M. Kruisbeek, S. A. Rosenberg, N. P. Restifo,Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J. Exp. Med. 198,569–580 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. A. Vardhana, M. A. Hwee, M. Berisa, D. K. Wells, K. E. Yost, B. King, M. Smith, P. S. Herrera, H. Y. Chang, A. T. Satpathy, M. R. M. van den Brink, J. R. Cross, C. B. Thompson,Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 21,1022–1033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. Zhao, Y. Qiao, P. Jonsson, J. Wang, L. Xu, P. Rouhi, I. Sinha, Y. Cao, C. Williams, K. Dahlman-Wright,Genome-wide profiling of AP-1–regulated transcription provides insights into the invasiveness of triple-negative breast cancer. Cancer Res. 74,3983–3994 (2014). [DOI] [PubMed] [Google Scholar]
- 36.M. S. Abdel-Hakeem, S. Manne, J. C. Beltra, E. Stelekati, Z. Chen, K. Nzingha, M. A. Ali, J. L. Johnson, J. R. Giles, D. Mathew, A. R. Greenplate, G. Vahedi, E. J. Wherry,Epigenetic scarring of exhausted T cells hinders memory differentiation upon eliminating chronic antigenic stimulation. Nat. Immunol. 22,1008–1019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.P. Tonnerre, D. Wolski, S. Subudhi, J. Aljabban, R. C. Hoogeveen, M. Damasio, H. K. Drescher, L. M. Bartsch, D. C. Tully, D. R. Sen, D. J. Bean, J. Brown, A. Torres-Cornejo, M. Robidoux, D. Kvistad, N. Alatrakchi, A. Cui, D. Lieb, J. A. Cheney, J. Gustafson, L. L. Lewis-Ximenez, L. Massenet-Regad, T. Eisenhaure, J. Aneja, W. N. Haining, R. T. Chung, N. Hacohen, T. M. Allen, A. Y. Kim, G. M. Lauer,Differentiation of exhausted CD8 T cells after termination of chronic antigen stimulation stops short of achieving functional T cell memory. Nat. Immunol. 22,1030–1041 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.K. B. Yates, P. Tonnerre, G. E. Martin, U. Gerdemann, R. al Abosy, D. E. Comstock, S. A. Weiss, D. Wolski, D. C. Tully, R. T. Chung, T. M. Allen, A. Y. Kim, S. Fidler, J. Fox, J. Frater, G. M. Lauer, W. N. Haining, D. R. Sen,Epigenetic scars of CD8+ T cell exhaustion persist after cure of chronic infection in humans. Nat. Immunol. 22,1020–1029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Anaya,OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2,e67 (2016). [Google Scholar]
- 40.Z. Tang, B. Kang, C. Li, T. Chen, Z. Zhang,GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47,W556–W560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.T. Li, J. Fu, Z. Zeng, D. Cohen, J. Li, Q. Chen, B. Li, X. S. Liu,TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48,W509–W514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. M. Newman, C. B. Steen, C. L. Liu, A. J. Gentles, A. A. Chaudhuri, F. Scherer, M. S. Khodadoust, M. S. Esfahani, B. A. Luca, D. Steiner, M. Diehn, A. A. Alizadeh,Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37,773–782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. Xu, Y. Fang, K. Chen, S. Li, S. Tang, Y. Ren, Y. Cen, W. Fei, B. Zhang, Y. Shen, W. Lu,Single-cell RNA sequencing reveals the tissue architecture in human high-grade serous ovarian cancer. Clin. Cancer Res. 28,3590–3602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. A. Postow, M. K. Callahan, J. D. Wolchok,Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 33,1974–1982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.N. Riaz, J. J. Havel, V. Makarov, A. Desrichard, W. J. Urba, J. S. Sims, F. S. Hodi, S. Martín-Algarra, R. Mandal, W. H. Sharfman, S. Bhatia, W.-J. Hwu, T. F. Gajewski, C. L. Slingluff Jr., D. Chowell, S. M. Kendall, H. Chang, R. Shah, F. Kuo, L. G. T. Morris, J.-W. Sidhom, J. P. Schneck, C. E. Horak, N. Weinhold, T. A. Chan,Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171,934–949.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.K. Takahashi, S. Yamanaka,Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126,663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Z. Hao, Y. Sheng, G. S. Duncan, W. Y. Li, C. Dominguez, J. Sylvester, Y.-W. Su, G. H. Y. Lin, B. E. Snow, D. Brenner, A. You-Ten, J. Haight, S. Inoue, A. Wakeham, A. Elford, S. Hamilton, Y. Liang, J. C. Zúñiga-Pflücker, H. H. He, P. S. Ohashi, T. W. Mak,K48-linked KLF4 ubiquitination by E3 ligase Mule controls T-cell proliferation and cell cycle progression. Nat. Commun. 8,14003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.T. Yamada, C. S. Park, M. Mamonkin, H. D. Lacorazza,Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Krüppel-like factors KLF4 and KLF2. Nat. Immunol. 10,618–626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. An, S. Golech, J. Klaewsongkram, Y. Zhang, K. Subedi, G. E. Huston, W. H. Wood III, R. P. Wersto, K. G. Becker, S. L. Swain, N. Weng,Krüppel-like factor 4 (KLF4) directly regulates proliferation in thymocyte development and IL-17 expression during Th17 differentiation. FASEB J. 25,3634–3645 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.F. Macián, C. García-Rodríguez, A. Rao,Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 19,4783–4795 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.J. Jain, P. G. McCafffrey, Z. Miner, T. K. Kerppola, J. N. Lambert, G. L. Verdine, T. Curran, A. Rao,The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature 365,352–355 (1993). [DOI] [PubMed] [Google Scholar]
- 52.A. Kel, O. Kel-Margoulis, V. Babenko, E. Wingender,Recognition of NFATp/AP-1 composite elements within genes induced upon the activation of immune cells. J. Mol. Biol. 288,353–376 (1999). [DOI] [PubMed] [Google Scholar]
- 53.H. Seo, E. González-Avalos, W. Zhang, P. Ramchandani, C. Yang, C. W. J. Lio, A. Rao, P. G. Hogan,BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 22,983–995 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Shipkova, E. Wieland,Surface markers of lymphocyte activation and markers of cell proliferation. Clin. Chim. Acta 413,1338–1349 (2012). [DOI] [PubMed] [Google Scholar]
- 55.R. Marzio, J. Mauel, S. Betz-Corradin,CD69 and regulation of the immune function. Immunopharmacol. Immunotoxicol. 21,565–582 (1999). [DOI] [PubMed] [Google Scholar]
- 56.M. Reddy, E. Eirikis, C. Davis, H. M. Davis, U. Prabhakar,Comparative analysis of lymphocyte activation marker expression and cytokine secretion profile in stimulated human peripheral blood mononuclear cell cultures: An in vitro model to monitor cellular immune function. J. Immunol. Methods 293,127–142 (2004). [DOI] [PubMed] [Google Scholar]
- 57.A. Diederichsen, J. Zeuthen, P. B. Christensen, T. Kristensen,Characterisation of tumour infiltrating lymphocytes and correlations with immunological surface molecules in colorectal cancer. Eur. J. Cancer 35,721–726 (1999). [DOI] [PubMed] [Google Scholar]
- 58.V. Khairnar, V. Duhan, A. M. Patil, F. Zhou, H. Bhat, C. Thoens, P. Sharma, T. Adomati, S.-K. Friendrich, J. Bezgovsek, J. D. Dreesen, G. Wennemuth, A. M. Westendorf, G. Zelinskyy, U. Dittmer, C. Hardt, J. Timm, J. R. Göthert, P. A. Lang, B. B. Singer, K. S. Lang,CEACAM1 promotes CD8+ T cell responses and improves control of a chronic viral infection. Nat. Commun. 9,2561 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Y. Ling, J. Wang, L. Wang, J. Hou, P. Qian, W. Xiang-dong,Roles of CEACAM1 in cell communication and signaling of lung cancer and other diseases. Cancer Metastasis Rev. 34,347–357 (2015). [DOI] [PubMed] [Google Scholar]
- 60.E. E. West, H. T. Jin, A. U. Rasheed, P. Penaloza-MacMaster, S. J. Ha, W. G. Tan, B. Youngblood, G. J. Freeman, K. A. Smith, R. Ahmed,PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J. Clin. Invest. 123,2604–2615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Y. Li, Y. Cong, M. Jia, Q. He, H. Zhong, Y. Zhao, H. Li, M. Yan, J. You, J. Liu, L. Chen, H. Hang, S. Wang,Targeting IL-21 to tumor-reactive T cells enhances memory T cell responses and anti-PD-1 antibody therapy. Nat. Commun. 12,951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. Kurachi, J. Kurachi, Z. Chen, J. Johnson, O. Khan, B. Bengsch, E. Stelekati, J. Attanasio, L. M. McLane, M. Tomura, S. Ueha, E. J. Wherry,Optimized retroviral transduction of mouse T cells for in vivo assessment of gene function. Nat. Protoc. 12,1980–1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.T. L. Bailey, J. Johnson, C. E. Grant, W. S. Noble,The MEME Suite. Nucleic Acids Res. 43,W39–W49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.E. Tanaka, T. Bailey, C. E. Grant, W. S. Noble, U. Keich,Improved similarity scores for comparing motifs. Bioinformatics 27,1603–1609 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.P. Machanick, T. L. Bailey,MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27,1696–1697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.E. Afgan, D. Baker, B. Batut, M. van den Beek, D. Bouvier, M. Čech, J. Chilton, D. Clements, N. Coraor, B. A. Grüning, A. Guerler, J. Hillman-Jackson, S. Hiltemann, V. Jalili, H. Rasche, N. Soranzo, J. Goecks, J. Taylor, A. Nekrutenko, D. Blankenberg,The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 Update. Nucleic Acids Res. 46,W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.M. Martin,Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17,10–12 (2011). [Google Scholar]
- 68.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras,STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29,15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.B. Li, C. N. Dewey,RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12,323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.J. Sun, T. Nishiyama, K. Shimizu, K. Kadota,TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14,219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.M. I. Love, W. Huber, S. Anders,Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15,550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Y. Zhou, B. Zhou, L. Pache, M. Chang, A. H. Khodabakhshi, O. Tanaseichuk, C. Benner, S. K. Chanda,Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10,1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D. Kim, B. Langmead, S. L. Salzberg,HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12,357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.M. Pertea, G. M. Pertea, C. M. Antonescu, T. C. Chang, J. T. Mendell, S. L. Salzberg,StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33,290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.M. D. Robinson, D. J. McCarthy, G. K. Smyth,edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26,139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.U. Raudvere, L. Kolberg, I. Kuzmin, T. Arak, P. Adler, H. Peterson, J. Vilo,g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47,W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Table S1