Abstract
Background: N6-methyladenosine (m6A) is the most abundant epigenetic modification. Although the dysregulation of m6A regulators has been associated with cancer progression in several studies, its relationship with cancer prognosis and clinicopathology is still controversial. Therefore, we evaluated the prognostic and clinicopathological value of m6A regulators in cancers by performing a comprehensive meta-analysis.
Methods: The PubMed, Cochrane Library, Web of Science, and Embase databases were searched up to April 2022. Hazard ratios were used to analyze the association between m6A with prognosis. We also analyze the relationship between m6A and clinicopathology using odds ratios.
Results: METTL3 overexpression predicted poor overall survival and disease-free survival in cancer patients (p < 0.001) such as gastric cancer (p < 0.001), esophageal squamous cell carcinoma (p < 0.001), oral squamous cell carcinoma (p = 0.002) and so on. Additionally, METTL3 overexpression was associated with poor pT stage (p < 0.001), pN stage (p < 0.001), TNM stage (p < 0.001), tumor size >5 cm (p < 0.001) and vascular invasion (p = 0.024). Conversely, METTL14 overexpression was positively associated with better OS (p < 0.001), negatively with poor pT stage (p = 0.001), pM stage (p = 0.002), pN stage (p = 0.011) and TNM stage (p < 0.001). Moreover, KIAA1429 overexpression was associated with poor OS (p = 0.001). YTHDF1 overexpression was also associated with advanced pM stage (p < 0.001) and tumor size >5 cm (p < 0.001). However, ALKBH5 overexpression was negatively associated with vascular invasion (p = 0.032).
Conclusions: High expression of METTL3 predicted poor outcome. In contrast, high expression of METTL14 was associated with better outcome. Thus, we suggest that among all the m6A regulators, METTL3 and METTL14 could be potential prognostic markers in cancers.
Keywords: m6A regulators, cancers, prognosis, clinicopathology, meta-analysis
INTRODUCTION
According to world cancer report 2020, there will be an estimated 60% increase in cancer cases over the next two decades and they will cause about one sixth of deaths worldwide [1]. Although certain progresses have been made in cancer treatment in the past decades, the overall survival of cancer patients is still unsatisfactory. Therefore, biomarkers which can function as prognosticators for the survival time in cancer are necessarily needed. N6-methyladenosine (m6A) modification, an epigenetic modification found in eukaryotes, has been a hot topic in recent years. As the most abundant epigenetic modification in eukaryotes [2], m6A modification is associated with RNA splicing [3–5], maturation [5], stabilization [6] and translation initiation [7]. As a result, m6A modification participates in several biological processes: neural development [8], disease occurrence [9, 10] and tumorigenesis [11–13]. This reversible modification can be added or removed by writers and erasers [14]. Writers are known as m6A methyltransferases, such as METTL3, METTL14, WTAP, KIAA1429 and RBM15/RBM15B. The two major erasers, FTO and ALKBH5, function as m6A demethylases. Furthermore, there are binding proteins called readers [14], represented by YTHDC, IGF2BP and HNRNPC, which recognize specifically modified RNA to exercise different subsequent reactions, including translation and degradation. Recently, emerging studies reported that the above mentioned m6A regulators were of great significance in tumorigenesis [15–17], tumor progression [18, 19] and metastasis [20]. For example, the writer METTL14 could suppress UVB-induced skin tumorigenesis and act as a critical epitranscriptomic mechanism to facilitate global genome repair which is essential for preventing mutagenesis and skin cancer [17]. Moreover, Bo Tang and his colleagues revealed that the eraser ALKBH5 suppressed pancreatic cancer tumorigenesis through promoting transcription of WIF-1 mRNA and inhibiting Wnt signaling pathway in a m6A dependent manner [21]. Additionally, the reader YTHDF1 could promote translation of autophagy-related genes ATG2A and ATG14 by binding to m6A-modified ATG2A and ATG14 mRNA, which facilitated autophagy and autophagy-related human hepatocellular carcinoma progression [15]. YTHDF1 could also enhance ferroptosis by promoting the activation of autophagy and BECN1 mRNA stability in hepatic stellate cells [22]. Overall, there are high-complexity links between m6A and different types of programmed cell death, which are closely related with the initiation, progression and resistance of cancer [23]. Furthermore, there is increasing evidence suggesting that dysregulated expression of m6A regulators exists in major types of cancers and correlates with poor prognosis. However, these survival data were contradictory among different cancer types and regulators, suggesting that a meta-analysis is required to identify prognostic markers. Therefore, in this study, we conducted a systematic review and meta-analysis to assess the prognostic and clinicopathological value of m6A regulators in cancer patients.
RESULTS
Study characteristics
The literature selection is presented in Figure 1, and the characteristics of eligible studies are shown in Table 1. A total of 3069 relevant studies were retrieved through an initial search. Among them, 915 duplicated records and 1944 unrelated records were excluded based on title or abstract. We subjected 210 studies to full-text screening, of which 159 studies were excluded because they did not meet the inclusion criteria. The remaining 51 articles were further assessed for quality by the Newcastle-Ottawa Scale (NOS) system, and only high-quality studies (NOS ≥ 6) were included in the meta-analysis. Finally, we included 49 cohort studies [6, 15, 24–70] comprising 7006 patients. All studies were published between 2017 and 2022. Forty-eight studies were conducted in Asia and one was conducted in Europe. Sample size ranged from 31 to 603 patients per study. In 49 included studies, 27 studies involved m6A writers, 15 studies referred to erasers and 9 studies were related to readers. The included studies totally reported 20 types of cancers, including digestive system cancer (n = 33), respiratory system cancer (n = 6), urinary system cancer (n = 4), female reproductive system cancer (n = 2) and others (n = 4). With respect to survival data, 48 studies reported overall survival (OS), 9 studies presented disease-free survival (DFS), and 4 studies showed relapse-free survival (RFS).
Figure 1.
Flow diagram of reviewing and selecting studies.
Table 1. The main characteristics of included studies.
| Study | m6A regulators | Country | Ethnicity | Cancer types | Follow-up (months) | Sample size (M/F) | TMN stage | Cut-off value | Outcome | HR and 95% CI | NOS score | Status |
| Yang 2020 (3) | METTL14 | China | Asian | Colorectal cancer | NA | 37 (27/10) | I–IV | score > 6 (0–12) | RFS | Reported | 6 | Included |
| Chen 2020 | METTL14 | China | Asian | Colorectal cancer | NA | 112 (74/38) | I–IV | > median | OS | Reported | 7 | Included |
| Wang 2022 | METTL14 | China | Asian | Colorectal cancer | 60 | 72 (44/28) | I–IV | NA | 0S | Reported | 7 | Included |
| Deng 2019 | METTL3 | China | Asian | Colorectal cancer | 72–108 | 181 (97/84) | I–IV | NA | OS | Reported | 7 | Included |
| Li 2019 (1) | METTL3 | China | Asian | Colorectal carcinoma | 80 | OS:432 (257/175) DFS:389 | NA | > median | OS DFS | OS: Reported DFS: Calculated | 6 | Included |
| Shengli 2022 | METTL3 | China | Asian | Colorectal cancer | 60 | 111 (51/60) | I–IV | score ≥ 4 (0–12) | OS | Calculated | 7 | Included |
| Ma 2022 | KIAA1429 | China | Asian | Colorectal cancer | 100 | 111 (75/36) | I–IV | NA | OS | Calculated | 7 | Included |
| Yang 2020 (1) | ALKBH5 | China | Asian | Colon cancer | 80 | 60 (25/35) | I–IV | score ≥ 4 (0–12) | OS DFS | Reported | 7 | Included |
| Ruan 2021 | FTO | China | Asian | Colorectal cancer | 140 | 369 (209/160) | I–III | > median | OS RFS | Reported | 6 | Included |
| Nishizawa 2018 | YTHDF1 | Japan | Asian | Colorectal cancer | NA | 63 (41/22) | I–IV | score = 2+ or 3+ (0–3) | OS | Reported | 7 | Included |
| Yue 2019 | METTL3 | China | Asian | Gastric cancer | NA | 120 (79/41) | I–IV | NA | OS DFS | Reported | 7 | Included |
| Wang 2020 | METTL3 | China | Asian | Gastric cancer | 60 | 83 (61/22) | I–IV | score > 7 (0-–12) | OS | Reported | 6 | Included |
| Yang 2020 (2) | METTL3 | China | Asian | Gastric cancer | 21-84 | OS:196 (131/65) DFS:156 | I–IV | score > 145 (0-300) | OS DFS | Reported | 8 | Included |
| Sun 2020 | METTL3 | China | Asian | Gastric cancer | NA | OS:80 RFS:58 (NA) | I–IV | score = 2+ or 3+ (0–3) | OS RFS | Reported | 7 | Included |
| Wang 2021 (1) | METTL16 | China | Asian | Gastric cancer | 49.1 | 231 (155/76) | I–IV | > median | OS | Reported | 8 | Included |
| Liu 2021 | METTL14 | China | Asian | Gastric cancer | 100 | 248 (183/65) | I–IV | score > 6 (0–12) | OS | Reported | 7 | Included |
| Li 2019 (2) | FTO ALKBH5 | China | Asian | Gastric cancer | 100 | 450 (308/142) | I–IV | score ≥ 6 (0–12) | OS | Reported | 6 | Included |
| Xu 2017 | FTO | China | Asian | Gastric cancer | 60 | 128 (68/60) | I–IV | NA | OS | Reported | 7 | Included |
| Yuan 2022 | YTHDC2 | China | Asian | Gastric cancer | 80 | 120 (86/34) | I–IV | NA | OS | Reported | 6 | Included |
| Xia 2019 | METTL3 | China | Asian | Pancreatic cancer | 15-26 | 40 (35/5) | I–III | > median | OS | Calculated | 6 | Included |
| Guo 2020 | ALKBH5 | China | Asian | Pancreatic cancer | 60 | 42 (19/23) | I–III | median | OS | Calculated | 7 | Included |
| Zeng 2021 | FTO | China | Asian | Pancreatic cancer | NA | 50 (27/23) | I–IV | > average | OS | Calculated | 8 | Included |
| Tan 2022 | FTO | China | Asian | Pancreatic cancer | NA | 209 (NA) | I–IV | score > 6 (0–12) | OS | Reported | 8 | Included |
| Li 2021 | YTHDF1 | China | Asian | Hepatocellular carcinoma | 60 | 120 (32/88) | I–III | NA | OS DFS | Reported | 7 | Included |
| Ma 2017 | METTL14 | China | Asian | Hepatocellular carcinoma | NA | 220 (193/27) | I–IV | > median | OS RFS | Calculated | 3 | Not included |
| Xu 2022 (1) | METTL3 | China | Asian | Intrahepatic cholangiocarcinoma | NA | 96 (53/43) | I–IV | > median | OS DFS | OS: Reported DFS: Calculated | 6 | Included |
| Ye 2020 | FTO | China | Asian | Liver cancer | 60 | 309 (NA) | I–III | score ≥ 6 (0–12) | OS | Reported | 7 | Included |
| Wang 2021 (2) | METTL3 | China | Asian | Oesophageal squamous cell carcinoma | NA | 81 (64/17) | I–IV | score > 300 (0–400) | OS | Calculated | 7 | Included |
| Xia 2020 | METTL3 | China | Asian | Esophageal squamous cell carcinoma | 108 | 207 (151/56) | I–IV | score > 8 (0–12) | OS DFS | Reported | 7 | Included |
| Nagaki 2020 | FTO ALKBH5 | Japan | Asian | Esophageal squamous cell carcinoma | 41.5–60 | 177 (153/24) | NA | score = 2+ or 3+ (0–3) | OS | ALKBH5: Reported FTO: Calculated | 6 | Included |
| Liu 2020 | METTL3 | China | Asian | Oral squamous cell carcinoma | 3–106 | 101 (68/33) | I–IV | Youden index | OS | Reported | 7 | Included |
| Xu 2021 | METTL3 | China | Asian | Oral squamous cell carcinoma | 80 | 94 (51/43) | I–IV | score ≥ 4 (0–12) | OS | Calculated | 7 | Included |
| Guo 2022 | METTL3 | China | Asian | Head and neck squamous cell carcinoma | 80 | 100 (99/1) | I–IV | score ≥ 8 (0–12) | OS | Reported | 7 | Included |
| Chen 2021 | METTL3 | China | Asian | Gallbladder-cancer | NA | 120 (57/63) | I–IV | > median | OS DFS | Reported | 6 | Included |
| Yang 2021 | FTO | China | Asian | Lung adenocarcinoma | 120 | 83 (55/28) | I–IV | score ≥ 6 (0–8) | OS | Calculated | 8 | Included |
| Huang 2018 | ALKBH5 | China | Asian | Lung adenocarcinoma | 3–125 | 88 (47/41) | I–IV | HSCORE > 100% | OS | Reported | 7 | Included |
| Xu 2022 (2) | YTHDF2 | China | Asian | Lung squamous cell carcinoma | 60 | 73 (66/7) | I–III | > median | OS | Reported | 7 | Included |
| Tsuchiya 2021 | YTHDF1 and YTHDF2 | Japan | Asian | Non–small-cell lung cancer | NA | 603 (414/189) | I–IV | YTHDF1: score > 118(0-300) YTHDF2: score > 118 (0–300) | OS RFS | Reported | 6 | Included |
| Lu 2020 | METTL3 | China | Asian | Nasopharyngeal carcinoma | 10.33–91.67 | 55 (30/25) | I–IV | score > 3 (0–9) | OS DFS | OS: Reported DFS: Calculated | 7 | Included |
| Du 2022 | IGF2BP3 | China | Asian | Nasopharyngeal carcinoma | 150 | 70 (56/14) | I–IV | NA | OS | Reported | 7 | Included |
| Gu 2019 | METTL14 | China | Asian | Bladder cancer | NA | 98 (NA) | NA | NA | OS | Calculated | 3 | Not included |
| Han 2019 | METTL3 | China | Asian | Bladder cancer | 60–96 | 180 (141/39) | I–IV | score > 3 (0–9) | OS | Calculated | 7 | Included |
| Yu 2021 | ALKBH5 | China | Asian | Bladder cancer | 60 | 161 (124/37) | I–IV | score ≥ 8 (0–12) | OS | Calculated | 7 | Included |
| Li 2017 | METTL3 | China | Asian | Renal cell carcinoma | 100 | 145 (89/56) | I–IV | NA | OS | Reported | 7 | Included |
| Zhang 2020 | ALKBH5 | China | Asian | Renal cell carcinoma | 100 | 96 (60/36) | I–IV | score ≥ 8 (0–12) | OS | Calculated | 7 | Included |
| Niu 2019 | FTO | China | Asian | Breast tumor | 96 | 53 (0/53) | NA | NA | OS | Calculated | 7 | Included |
| Hua 2018 | METTL3 | China | Asian | Ovarian carcinoma | NA | 162 (0/162) | I–IV | > median | OS | Reported | 8 | Included |
| Lin 2022 | METTL3 | China | Asian | Thyroid carcinoma | 36 | 80 (25/55) | I–IV | > median | OS | Calculated | 6 | Included |
| Orouji E 2020 | YTHDF1 | Germany | European | Merkel cell carcinoma | NA | 31 (NA) | NA | score > 8 | OS | Calculated | 7 | Included |
| Li 2020 | WTAP, KIAA1429, RBM15, RBM15B, METTL3, METTL14, METTL16, HNRNPC, HNRNPA2B1, YTHDF1, YTHDF2, YTHDF3, YTHDC1, FTO, ALKBH5 | China | Asian | Osteosarcoma | 60 | 120 (NA) | NA | score > 6 | OS | Reported | 6 | Included |
| Wei 2022 | YTHDF1 | China | Asian | Osteosarcoma | 60 | 56 (NA) | I–IV | > median | OS | Calculated | 6 | Included |
Abbreviations: CI: confidence interval; HR: hazard ratio; OS: overall survival; DFS: disease-free survival; RFS: relapse-free survival; NA: not available; F: female; M: male.
Expression of m6A regulators and prognosis of cancer patients
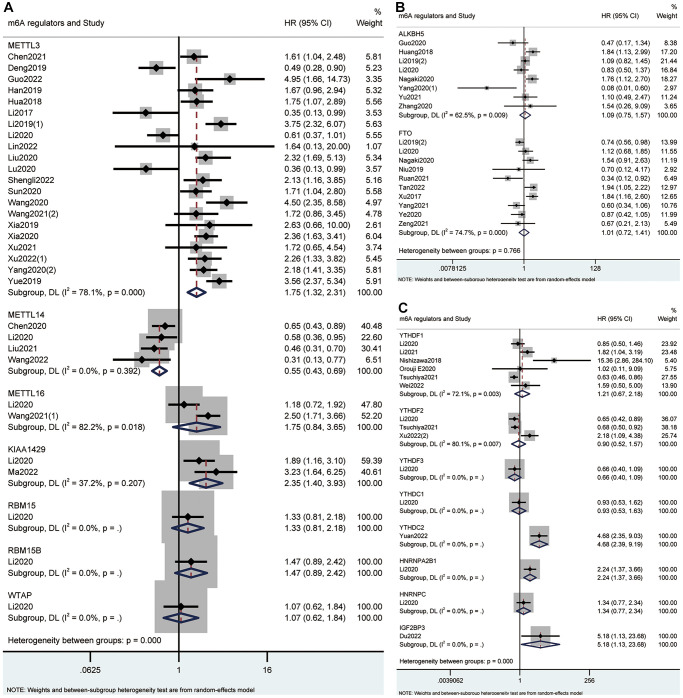
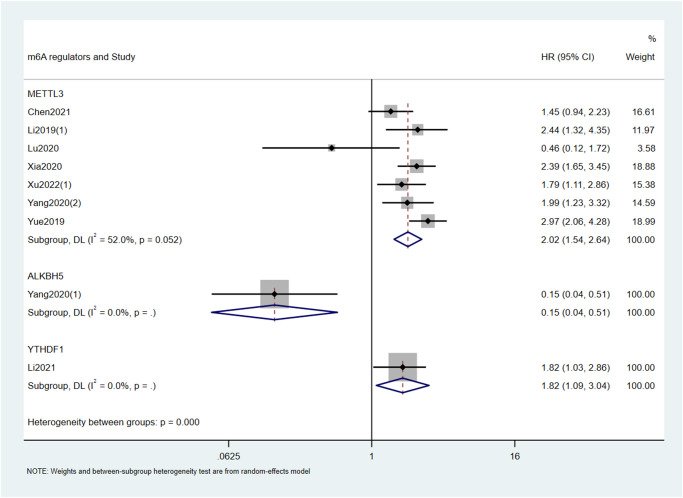
Based on the type of m6A writers, we carried out meta-analysis and found that high expression of METTL3 had an unfavorable effect on OS (HR = 1.75; 95% CI: 1.32–2.31, p < 0.001; I2 = 78.1%, p < 0.001; Figure 2, Table 2) and DFS (HR = 2.02; 95% CI: 1.54–2.64, p < 0.001; I2 = 52%, p = 0.052; Figure 3, Table 2) in cancer patients. Similarly, high expression of KIAA1429 was associated with poor OS (HR = 2.35; 95% CI: 1.40–3.93, p = 0.001; I2 = 37.2%, p = 0.207; Figure 2, Table 2). On the contrary, high expression of METTL14 had a favorable effect on OS (HR = 0.55; 95% CI: 0.43–0.69, p < 0.001; I2 = 0.0%, p = 0.392; Figure 2, Table 2). Furthermore, the expression of METTL16 was not significantly associated with OS in cancer patients (Figure 2, Table 2). Similarly, neither erasers nor readers were significantly associated with OS in cancer patients. (Figure 2, Table 2). We did not perform a meta-analysis of m6A regulators and RFS because there were not enough studies.
Figure 2.
Forest plots for the association of m6A writers (A), erasers (B) and readers (C) with OS in cancer patients.
Table 2. Summary of the meta-analysis of m6A regulators and prognosis in cancer patients.
| Regulators | Outcome | Studies | HR | 95% Cl | P-value | Heterogeneity | Effects model | |
| I2 | P-value | |||||||
| METTL3 | OS | 21 | 1.75 | 1.32–2.31 | 0 | 78.10% | 0 | Random |
| DFS | 7 | 2.02 | 1.54–2.64 | 0 | 52% | 0.052 | Random | |
| METTL14 | OS | 4 | 0.55 | 0.43–0.69 | 0 | 0.00% | 0.392 | Random |
| KIAA1429 | OS | 2 | 2.35 | 1.40–3.93 | 0.001 | 37.20% | 0.207 | Random |
| METTL16 | OS | 2 | 1.75 | 0.84–3.65 | 0.137 | 82.20% | 0.018 | Random |
| ALKBH5 | OS | 8 | 1.09 | 0.75–1.57 | 0.657 | 62.50% | 0.009 | Random |
| FTO | OS | 10 | 1.01 | 0.72–1.41 | 0.966 | 74.70% | 0 | Random |
| YTHDF1 | OS | 6 | 1.21 | 0.67–2.18 | 0.532 | 72.10% | 0.003 | Random |
| YTHDF2 | OS | 3 | 0.9 | 0.52–1.57 | 0.715 | 80.10% | 0.007 | Random |
Abbreviations: CI: confidence interval; HR: hazard ratio; OS: overall survival; DFS: disease-free survival.
Figure 3.
Forest plots for the association of m6A regulators with DFS in cancer patients.
Subgroup analysis for different m6A regulators and cancer types
For further exploration, subgroup analyses were conducted according to cancer types. As shown in Table 3, high expression of METTL3 was correlated with poor OS (HR = 2.72; 95% CI: 1.81–4.07, p < 0.001; I2 = 64.2%, p = 0.039) and DFS (HR = 2.58; 95% CI: 1.92–3.47, p < 0.001; I2 = 37.9%, p = 0.205) in gastric cancer. Moreover, high expression of METTL3 was significantly associated with poor OS in esophageal squamous cell carcinoma (HR = 2.20; 95% CI: 1.59–3.05, p < 0.001; I2 = 0.0%, p = 0.436) and oral squamous cell carcinoma (HR = 2.16; 95% CI: 1.33–3.49, p = 0.002; I2 = 0.0%, p = 0.602). However, the expression of METTL3 or METTL14 was not significantly associated with OS in colorectal cancer. The expression of FTO was also not significantly associated with OS in gastric cancer and pancreatic cancer. Furthermore, we did not find a significant association between YTHDF1 and OS in osteosarcoma.
Table 3. Subgroup analysis of the correlation between m6A regulators and cancer prognosis based on cancer types.
| Regulators | Cancer types | Outcome | Studies | HR | 95% Cl | P-value | Heterogeneity | Effects model | |
| I2 | P-value | ||||||||
| METTL3 | oral squamous cell carcinoma | OS | 2 | 2.16 | 1.33–3.49 | 0.002 | 0.00% | 0.602 | Fix |
| esophageal squamous cell carcinoma | OS | 2 | 2.2 | 1.59–3.05 | 0 | 0.00% | 0.436 | Fix | |
| gastric cancer | OS | 4 | 2.72 | 1.81–4.07 | 0 | 64.20% | 0.039 | Random | |
| gastric cancer | DFS | 2 | 2.58 | 1.92–3.47 | 0 | 37.90% | 0.205 | Fix | |
| colorectal cancer | OS | 3 | 1.59 | 0.48–5.26 | 0.448 | 92.9%, | 0 | Random | |
| METTL14 | colorectal cancer | OS | 2 | 0.51 | 0.26–1.00 | 0.051 | 53.50% | 0.142 | Random |
| FTO | pancreatic cancer | OS | 2 | 1.32 | 0.48–3.60 | 0.586 | 65.90% | 0.087 | Random |
| gastric cancer | OS | 2 | 1.15 | 0.47–2.81 | 0.756 | 92.40% | 0 | Random | |
| YTHDF1 | osteosarcoma | OS | 2 | 0.95 | 0.58–1.54 | 0.833 | 0.00% | 0.337 | Fix |
Abbreviations: CI: confidence interval; HR: hazard ratio; OS: overall survival; DFS: disease-free survival.
Expression of m6A regulators and the clinicopathological parameters
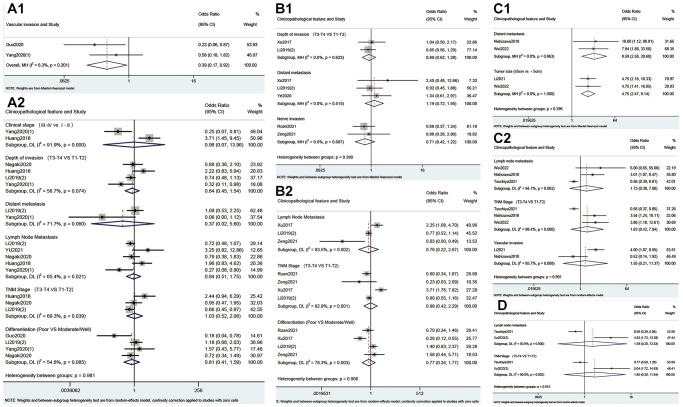
As shown in Figure 4 and Table 4, high expression of METTL3 was associated with advanced pT stage (OR = 1.85; 95% CI: 1.40–2.45, p < 0.001; I2 = 47.4%, p = 0.055), pN stage (OR = 2.37; 95% CI: 1.58–3.56, p < 0.001; I2 = 63.7%, p = 0.001), TNM stage (OR = 2.61; 95% CI: 2.03–3.36, p < 0.001; I2 = 12.7%, p = 0.323), tumor size >5 cm (OR = 2.33; 95% CI: 1.51–3.61, p < 0.001; I2 = 0.0%, p = 0.886) and vascular invasion (OR = 1.47; 95% CI: 1.05–2.05, p = 0.024; I2 = 0.0%, p = 0.508). Conversely, high expression of METTL14 correlated negatively with pT stage (OR = 0.27; 95% CI: 0.13–0.58, p = 0.001; I2 = 0.0%, p = 0.739), pM stage (OR = 0.12; 95% CI: 0.03–0.46, p = 0.002; I2 = 0.0%, p = 0.497), pN stage (OR = 0.26; 95% CI: 0.09–0.73, p = 0.011; I2 = 60.6%, p = 0.079) and TNM stage (OR = 0.21; 95% CI: 0.13–0.34, p < 0.001; I2 = 0.0%, p = 0.575). Meanwhile, there was a statistical association between overexpression of ALKBH5 and negative vascular invasion (OR=0.39; 95%CI: 0.17-0.92, p = 0.032; I2 = 6.3%, p = 0.301, Figure 5). Furthermore, overexpression of YTHDF1 was associated with advanced pM stage (OR = 8.59; 95% CI: 2.58–28.60, p < 0.001; I2 = 0.0%, p = 0.863, Figure 5) and tumor size >5 cm (OR = 4.75; 95% CI: 2.47–9.14, p < 0.001; I2 = 0.0%, p = 1.000, Figure 5).
Figure 4.
Forest plots for the association of METTL3 (A) and METTL14 (B) with clinicopathological parameters in cancer patients.
Table 4. The correlations between m6A regulators with clinicopathological characteristics in cancer patients.
| m6A regulator | Clinicopathological feature | Studies (n) | Patients (n) | References | OR (95% CI) | P value | Heterogeneity | Effects model | |
| I² (%) | P value | ||||||||
| METTL3 | Depth of invasion (T3–T4 vs. T1–T2) | 9 | 1057 | Hua 2018; Lu 2020; Wang 2020; Xia 2020; Xu 2021; Yang 2020 (2); Xia 2019; Chen 2021; Guo 2022 | 1.85 (1.40–2.45) | 0.000 | 47.4 | 0.055 | Fix |
| Lymph Node Metastasis | 13 | 1421 | Liu 2020; Chen 2021; Guo 2022; Sun 2020; Lin 2022; Xia 2020; Xu 2022 (1); Xia 2019; Hua 2018; Lu 2020; Wang 2020; Yang 2020 (2); Xu 2021 | 2.37 (1.58–3.56) | 0.000 | 63.7 | 0.001 | Random | |
| TNM Stage (T3–T4 vs. T1–T2) | 11 | 1303 | Xia 2019; Sun 2020; Xia 2020; Chen 2021; Lin 2022; Yang 2020 (2); Xu 2022 (1); Shengli 2022; Wang 2020; Deng 2019; Liu 2020 | 2.61 (2.03–3.36) | 0.000 | 12.7 | 0.323 | Fix | |
| Tumor size (>5 cm vs ≤ 5 cm) | 3 | 375 | Wang 2020; Xu 2022 (1); Yang 2020 (2) | 2.33 (1.51–3.61) | 0.000 | 0.0 | 0.886 | Fix | |
| Vascular invasion | 4 | 781 | Sun 2020; Li 2019 (1); Xu 2022 (1); Yue 2019 | 1.47 (1.05–2.05) | 0.024 | 0.0 | 0.508 | Fix | |
| Distant metastasis | 9 | 1091 | Chen 2021; Shengli 2022; Xu 2022 (1); Deng 2019; Sun 2020; Hua 2018; Lu 2020; Wang 2020; Yang 2020 (2) | 1.93 (0.99–3.78) | 0.054 | 67.5 | 0.002 | Random | |
| Clinical stage III–IV vs. II–II | 4 | 688 | Guo 2022; Li 2019 (1); Liu 2020; Lu 2020 | 1.05 (0.28–3.91) | 0.936 | 89.7 | 0.000 | Random | |
| Differentiation (Poor vs. Moderate/Well) | 4 | 997 | Li 2019 (1); Xia 2020; Yang 2020 (2); Hua 2018 | 1.22 (0.65–2.30) | 0.529 | 73.3 | 0.011 | Random | |
| Nerve invasion | 3 | 724 | Xu 2022 (1); Yue 2019; Li 2019 (1) | 1.26 (0.92–1.74) | 0.150 | 0.0 | 0.666 | Fix | |
| METTL14 | Depth of invasion (T3–T4 vs. T1–T2) | 2 | 285 | Liu 2021; Yang 2020 (3) | 0.27 (0.13–0.58) | 0.001 | 0.0 | 0.739 | Fix |
| Lymph Node Metastasis | 3 | 357 | Wang 2022; Yang 2020 (3); Liu 2021 | 0.26 (0.09–0.73) | 0.011 | 60.6 | 0.079 | Random | |
| Distant metastasis | 2 | 285 | Liu 2021; Yang 2020 (3) | 0.12 (0.03–0.46) | 0.002 | 0.0 | 0.497 | Fix | |
| TNM Stage (T3–T4 vs. T1–T2) | 4 | 466 | Chen 2020; Liu 2021; Wang 2022; Yang 2020 (3) | 0.21 (0.13–0.34) | 0.000 | 0.0 | 0.575 | Fix | |
| Tumor size (>5 cm vs. ≤5 cm) | 2 | 285 | Liu 2021; Yang 2020 (3) | 0.32 (0.05–2.14) | 0.241 | 79.6 | 0.027 | Random | |
| ALKBH5 | Vascular invasion | 2 | 102 | Guo 2020; Yang 2020 (1) | 0.39 (0.17–0.92) | 0.032 | 6.3 | 0.301 | Fix |
| Clinical stage (III–IV vs. I– II) | 2 | 148 | Yang 2020 (1); Huang 2018 | 0.98 (0.07–13.96) | 0.988 | 91.9 | 0.000 | Random | |
| Depth of invasion (T3–T4 vs. T1–T2) | 4 | 775 | Nagaki 2020; Huang 2018; Li 2019 (2); Yang 2020 (1) | 0.84 (0.45–1.54) | 0.564 | 56.7 | 0.074 | Random | |
| Differentiation (Poor vs. Moderate/Well) | 4 | 729 | Guo 2020; Li 2019 (2); Yang 2020 (1); Nagaki 2020 | 0.81 (0.41–1.59) | 0.532 | 54.8 | 0.085 | Random | |
| Distant metastasis | 2 | 510 | Li 2019 (2); Yang 2020 (1) | 0.37 (0.02–5.60) | 0.475 | 71.7 | 0.060 | Random | |
| Lymph Node Metastasis | 5 | 936 | Li 2019 (2); Yu 2021; Nagaki 2020; Huang 2018; Yang 2020 (1) | 0.94 (0.51–1.75) | 0.851 | 65.4 | 0.021 | Random | |
| TNM Stage (T3–T4 vs. T1–T2) | 3 | 715 | Huang 2018; Nagaki 2020; Li 2019 (2) | 1.03 (0.52–2.06) | 0.925 | 69.3 | 0.039 | Random | |
| FTO | Depth of invasion (T3-T4 vs. T1–T2) | 2 | 578 | Xu 2017; Li 2019 (2) | 0.89 (0.62–1.28) | 0.533 | 0 | 0.623 | Fix |
| Differentiation (Poor vs. Moderate/Well) | 4 | 997 | Ruan 2021; Xu 2017; Li 2019 (2); Zeng 2021 | 0.77 (0.34–1.77) | 0.537 | 78.3 | 0.003 | Random | |
| Distant metastasis | 3 | 902 | Xu 2017; Li 2019 (2); Ye 2020 | 1.19 (0.72–1.95) | 0.502 | 0 | 0.515 | Fix | |
| Lymph Node Metastasis | 3 | 628 | Xu 2017; Li 2019 (2); Zeng 2021 | 0.76 (0.22–2.67) | 0.671 | 83.5 | 0.002 | Random | |
| Nerve invasion | 2 | 419 | Ruan 2021; Zeng 2021 | 0.71 (0.42–1.22) | 0.218 | 0 | 0.687 | Fix | |
| TNM Stage (T3–T4 vs. T1–T2) | 4 | 997 | Ruan 2021; Zeng 2021; Xu 2017; Li 2019 (2) | 0.98 (0.42–2.29) | 0.969 | 82.8 | 0.001 | Random | |
| YTHDF1 | Distant metastasis | 2 | 113 | Nishizawa 2018; Wei 2022 | 8.59 (2.58–28.60) | 0.000 | 0.0 | 0.863 | Fix |
| Tumor size (>5 cm vs ≤5 cm) | 2 | 170 | Li 2021; Wei 2022 | 4.75 (2.47–9.14) | 0.000 | 0.0 | 1.000 | Fix | |
| Lymph Node Metastasis | 3 | 716 | Wei 2022; Nishizawa 2018; Tsuchiya 2021 | 1.73 (0.38–7.80) | 0.476 | 84.1 | 0.002 | Random | |
| TNM Stage (T3–T4 vs. T1–T2) | 3 | 716 | Wei 2022; Nishizawa 2018; Tsuchiya 2021 | 1.83 (0.42–7.94) | 0.418 | 88.4 | 0.000 | Random | |
| Vascular invasion | 2 | 183 | Li 2021; Nishizawa 2018 | 1.55 (0.21–11.37) | 0.665 | 85.7 | 0.008 | Random | |
| YTHDF2 | Lymph Node Metastasis | 2 | 676 | Tsuchiya 2021; Xu 2022 (2) | 1.59 (0.20–12.53) | 0.660 | 92.9 | 0.000 | Random |
| TNM Stage (T3–T4 vs. T1–T2) | 2 | 676 | Tsuchiya 2021; Xu 2022 (2) | 1.85 (0.30–11.54) | 0.512 | 90.0 | 0.002 | Random | |
Abbreviations: CI: confidence interval; OR: odds ratio.
Figure 5.
Forest plots for the association of ALKBH5 (A), FTO (B), YTHDF1 (C) and YTHDF2 (D) with clinicopathological parameters in cancer patients.
Sensitivity analysis
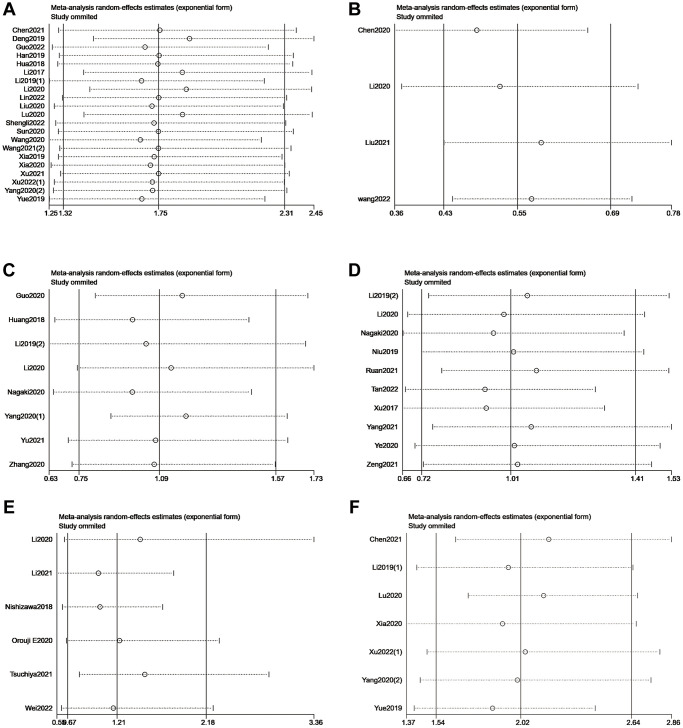
We omitted individual studies successively to estimate the impact of each study in our meta-analysis. No individual study modified the pooled HR of included studies reporting OS or DFS significantly, which proved that the results were stable (Figure 6).
Figure 6.
Sensitivity analysis of METTL3 (A), METTL14 (B), ALKBH5 (C), FTO (D), and YTHDF1 (E) for OS. Sensitivity analysis of METTL3 (F) for DFS.
Publication bias
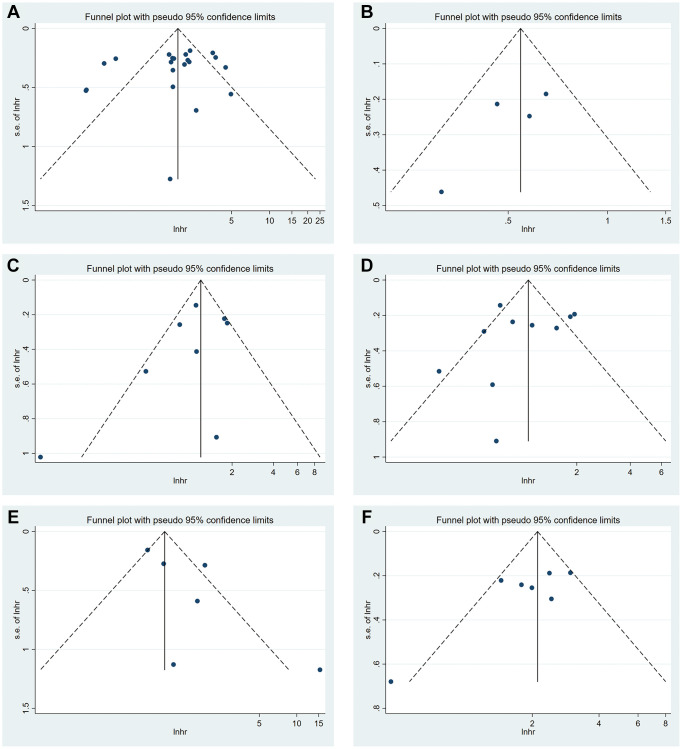
Funnel plots were generated to detect publication bias (Figure 7). The studies were distributed uniformly around the axis, indicating no obvious publication bias. Meanwhile, no obvious publication bias was found according to Begg’s test and Egger’s test (Table 5).
Figure 7.
Funnel plot of METTL3 (A), METTL14 (B), ALKBH5 (C), FTO (D) and YTHDF1 (E) for OS. Funnel plot of METTL3 (F) for DFS.
Table 5. Publication bias test of included studies in our meta-analysis.
| Regulators | Outcome | Begg’s test (P value) | Egger’s test (P value) |
| METTL3 | OS | 0.415 | 0.319 |
| METTL4 | OS | 0.308 | 0.229 |
| ALKBH5 | OS | 0.174 | 0.290 |
| FTO | OS | 0.592 | 0.571 |
| YTHDF1 | OS | 0.260 | 0.117 |
| METTL3 | DFS | 0.230 | 0.083 |
Abbreviations: OS: overall survival; DFS: disease-free survival.
DISCUSSION
m6A modification, a reversible epigenetic modification regulated by three types of proteins (writers, erasers and readers), plays a complicated role in cancer initiation and development [14, 71, 72]. Recent studies have explored how m6A regulators influenced the prognosis of cancer patients. However, results were frequently inconsistent among different cancer types. Therefore, a comprehensive study to summarize the results from current publications is necessary. To report prognostic value of m6A regulators in cancer patients, we analyzed the survival time and clinicopathological features of 7006 patients from 49 studies who expressed different levels of m6A regulators. Results showed that expression level of m6A writers was related to cancer prognosis. In addition, different m6A writers had opposite associations with the prognosis and clinicopathological features in cancer patients. According to the results, there was a possible trend for poor OS and DFS in patients with the high expression of METTL3. Similarly, previous bioinformatic analysis from databases like TCGA, GEO and HPA, supported that high expression of METTL3 was correlated with unfavorable prognosis in various cancers, including gastric cancer [73], colorectal cancer [74], liver cancer [75], prostate cancer [76] and glioma [77]. In most of these databases, RNA-seq was used to detect the level of METTL3. Moreover, a previous meta-analysis including 9 studies showed that high METTL3 expression was associated with poor prognosis in cancer patients, and the expression of METTL3 in included 9 studies were all detected by qRT-PCR. While in the studies included in our analysis, METTL3 was detected only by IHC staining. Combining our studies with the results from databases, we can conclude that METTL3 is related to cancer prognosis at protein level, which strongly suggests that it could be a prognostic predictor. Additionally, this tendency was more prominent in gastric cancer. Previous studies indicated that in human gastric cancer cells, high expression of METTL3 stimulates the expression of GLUT4 and ENO2 via the METTL3/HDGF axis, thereby promoting tumor angiogenesis and glycolysis [6]. Moreover, Ben Yue et al. unveiled that METTL3 stabilized ZMYM1 mRNA in gastric cancer cells, which facilitated EMT and metastasis by repressing E-cadherin promoter [26]. These might account, at least to some extent, for the poor survival of patients with gastric cancer. Furthermore, aberrant expression of METTL3 was involved in the dysfunction of cellular signaling pathways, such as MAPK [74], JAK/STAT [78], PI3K/AKT [79, 80] and Wnt/β-catenin [81] cascades, which are involved in tumor progression, metastasis, migration and stemness. We also found that high expression of METTL3 was associated with advanced TNM stage and pT stage, pN stage, tumor size >5 cm and vascular invasion respectively. Therefore, based on these current results, we believe that METTL3 plays an important role in multiple stages of cancer progression and ultimately affects prognosis. Interestingly, in contrast to METTL3, METTL14, another m6A methylation writer, might be a positive prognosticator. Previous studies have shown that METTL14 might have various functions that have not been fully identified yet, thus its role in cancer remained controversial [82]. In this study, our result confirmed that high level of METTL14 was associated with better OS. Different studies have reported that METTL14 suppressed progression and metastasis in several cancers, such as colorectal cancer [83] and hepatocellular carcinoma [84]. Besides, Panneerdoss et al. found that in METTL14-silenced breast cancer cells, RhoA and PI3K-AKT signaling pathways were highly enriched, which are well-known to be mediators of cancer progression and angiogenesis [85]. Moreover, our study showed that high expression of METTL14 was inversely associated with poor TNM stage, pT stage, pN stage and pM stage. Combining the results of other studies and ours, we inferred that METTL14 plays a role in cancer suppression and could be a favorable index of cancer progression and prognosis. Moreover, METTL3 and METTL14 show completely contrary effects on cancer progression, indicating that METTL3 and METTL14 may have some biological functions that are independent of m6A modification, which deserves further study.
Besides, from the analysis results of clinicopathological features, high expression of YTHDF1 was associated with advanced pM stage and tumor size >5 cm, while high expression of ALKBH5 was negatively associated with vascular invasion. Consistently, a recent study reported that YTHDF1 regulates CRC tumorigenesis and metastasis by promoting ARHGEF2 translation and RhoA signaling in colorectal cancer [20]. High YTHDF1 level is significantly associated with metastatic gene signature in colorectal cancer, while YTHDF1-knockout mice inhibited tumor growth in vivo [20]. Therefore, targeting the YTHDF1-m 6 A-ARHGEF2 axis may be a promising therapeutic strategy to inhibit tumor growth, invasion, and metastasis. In addition, ALKBH5, as the second m6A demethylated enzyme discovered after FTO, was reported to promote tumor stem formation in gliomas and promote tumor progression in breast cancer, colon cancer and hepatocellular carcinoma [85, 86]. Conversely, ALKBH5 could inhibit tumor growth in bladder cancer and pancreatic cancer. These findings suggest the complexity of the action of ALKBH5 in cancers. However, no significant relationship was found between high expressions of m6A erasers or readers and poor prognosis. Limitation of sample size and a certain degree of heterogeneity may partly account for this. Additionally, the mechanisms of m6A modification and cancers are complicated [87]. Therefore, more studies are needed to provide further mechanistic insights.
To the best of our knowledge, this is the first study to conduct a meta-analysis of the association between m6A regulators and the prognosis and clinicopathology in cancer patients systematically. Nonetheless, there are still several limitations in our meta-analysis. First, several original data were not available, therefore we had to extract data from the Kaplan-Meier survival curves and this might increase the inaccuracy in our study. Secondly, the ethnicity of included patients was mostly Asian, which may increase the population selection bias. Thirdly, IHC was adopted to detect the expression of m6A regulators in all studies, but the IHC protocols, antibodies and cut-off values were not consistent across the included studies, which may have led to significant heterogeneity between included studies. Therefore, future research should standardize the cut-off values for the expression of m6A regulators, detection antibodies used and IHC staining protocols to better compare the results of different studies. In summary, our meta-analysis provides evidence that the expression level of m6A writers is related to cancer progression and prognosis. Different m6A writer proteins play different roles in patients’ outcome: high expression level of METTL3 is significantly associated with poor prognosis, while high expression of METTL14 leads to better survival rate. Both m6A regulators possess a great potential to become practicable prognosticators in various cancers. Meanwhile, future studies with more complete and representative datasets are required for further exploration.
METHODS
Literature search
Relevant articles published up to April 2022 were obtained from PubMed, Embase, Web of Science and the Cochrane library. There were no restrictions on language or date of publication. “N (6)-methyladenosine” and “cancers” were the two main key words we used. The comprehensive search strategy for each database is provided in Supplementary Table 1. All references were managed using EndNote X9. Three reviewers independently analyzed search results. Any disagreements between reviewers were resolved by discussion.
Inclusion and exclusion criteria
The process of selecting eligible studies was conducted by three reviewers independently. Articles were included when they met the following inclusion criteria: (1) the text evaluated the relation between m6A regulators expression and cancer prognosis; (2) HR and 95% CI were reported or could be calculated from the text; (3) original research; (4) the expression of m6A regulators in tissues was detected by immunohistochemistry; (5) patients were confirmed cancers definitively. The exclusion criteria were: (1) reviews, letters, meeting abstracts; (2) nonhuman studies; (3) sample cases were from databases; (4) duplicate data; (5) studies did not provide necessary and complete data.
Data extraction and quality assessment
The following information were extracted from eligible studies independently by three researchers: author, published year, country, m6A regulators, cancer types, cancer stage, sample size, gender, cut-off value of m6A regulators and survival data including OS, DFS and RFS. The HR with its 95% CI were extracted from the text directly or calculated from Kaplan-Meier survival curve using Engauge Digitizer. The quality of the included studies was evaluated using the Newcastle Ottawa Scale (NOS) criteria. NOS scores range from 0 to 9. It would be considered as high-quality study if score was more than 5; otherwise, it would be considered as low-quality study. Only studies with NOS ≥ 6 were finally selected for inclusion in meta-analysis. Disagreements were resolved by discussion.
Statistical analysis
The pooled HR and 95% CI were used to evaluate the relation between m6A regulators and cancer prognosis (OS, DFS and RFS). The pooled odds ratio (OR) and 95%CI were used to evaluate the relationship between m6A regulators and clinicopathological parameters. HRs or ORs >1 represented a poor prognosis in cancer. Heterogeneity among the studies was evaluated by Coltrane’s Q statistic and the I2 statistic. If a p < 0.1 or I2 > 50%, we applied a random-effect model. Otherwise, a fixed-effect model was applied. Subgroup analysis was conducted according to cancer types. In the sensitivity analysis, we omitted individual studies successively to estimate the impact of each study in our meta-analysis. Begg’s test and Egger’s test were used to evaluate publication bias. A two-tailed p value < 0.05 was considered statistically significant in all statistical tests. All data analyses were performed using StataSE15.1 (Stata Corporation, College Station, TX, USA).
Supplementary Materials
ACKNOWLEDGMENTS
We would like to thank all researchers for their contributions.
Abbreviations
- METTL3
Methyltransferase Like 3
- METTL14
Methyltransferase Like 14
- METTL16
Methyltransferase Like 16
- RBM15
RNA-binding protein 15
- RBM15B
Putative RNA-binding protein 15B
- HNRNPC
Heterogeneous nuclear ribonucleoproteins
- HNRNPA2B1
Heterogeneous nuclear ribonucleoproteins A2/B1
- YTHDF1
YTH domain-containing family protein 1
- YTHDF2
YTH domain-containing family protein 2
- YTHDF3
YTH domain-containing family protein 3
- YTHDC1
YTH domain-containing protein 1
- FTO
Alpha-ketoglutarate-dependent dioxygenase FTO
- ALKBH5
RNA demethylase ALKBH5
- OS
overall survival
- DFS
disease-free survival
- RFS
recurrence-free survival
- HR
hazard ratio
- OR
odds ratio
- M/F
male/female
- NA
not available
- cut-off value
the value that can be diagnosed as positive/high expression of a m6A regulator
- IHC
immunohistochemistry
- IF
immunofluorescence
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- P
prospective
- CI
confidence interval
AUTHOR CONTRIBUTIONS: Zhangci Su, Leyao Xu, Xinning Dai and Yun Wang conceived and designed the study. Zhangci Su, Leyao Xu and Xinning Dai analyzed the data, prepared the figures and tables, and wrote the paper. Mengyao Zhu validated the data. Xiaodan Chen, Yuanyuan Li, Jie Li and Ruihan Ge contributed analysis tools and materials. Yun Wang and Bin Cheng reviewed drafts of the paper and participated in its coordination. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST: The authors declare no conflicts of interest related to this study.
FUNDING: This work was supported by Guangzhou Science and Technology Project (Grant Number: 201804010040); Guangdong Basic and Applied Basic Research Foundation (Grant Number: 2019A1515011203); Sun Yat-sen University Young Teacher Cultivation Project (Grant Number: 18ykpy29); Science and Technology Planning Project of Guangzhou, China (Grant Number: 201704020063).
REFERENCES
- 1.World Cancer Report: Cancer Research for Cancer Prevention. https://publications.iarc.fr/586.
- 2.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013; 29:108–15. 10.1016/j.tig.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017; 45:6051–63. 10.1093/nar/gkx141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–89. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015; 518:560–4. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, Zhou J, Sun B, Zou X, Wang S. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020; 69:1193–205. 10.1136/gutjnl-2019-319639 [DOI] [PubMed] [Google Scholar]
- 7.Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, Mori M, Sahara H. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016; 376:34–42. 10.1016/j.canlet.2016.02.022 [DOI] [PubMed] [Google Scholar]
- 8.Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020; 21:36–51. 10.1038/s41583-019-0244-z [DOI] [PubMed] [Google Scholar]
- 9.Maity A, Das B. N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J. 2016; 283:1607–30. 10.1111/febs.13614 [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Shen F, Huang W, Qin S, Huang JT, Sergi C, Yuan BF, Liu SM. Glucose Is Involved in the Dynamic Regulation of m6A in Patients With Type 2 Diabetes. J Clin Endocrinol Metab. 2019; 104:665–73. 10.1210/jc.2018-00619 [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y, Huang Y, Zheng R, Yu H, Wang J, Hu M, Miao J, Li J. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m6A-dependent manner. Cell Death Dis. 2020; 11:659. 10.1038/s41419-020-02847-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell. 2017; 31:127–41. 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, Chen H, Su R, Yin Z, Li W, Deng X, Chen Y, Hu YC, et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell. 2020; 27:64–80.e9. 10.1016/j.stem.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao G, Li HB, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol. 2016; 6:160003. 10.1098/rsob.160003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, Hu Y, Qiu J, Pu L, Tang J, Wang X. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021; 6:76. 10.1038/s41392-020-00453-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui YH, Yang S, Wei J, Shea CR, Zhong W, Wang F, Shah P, Kibriya MG, Cui X, Ahsan H, He C, He YY. Autophagy of the m6A mRNA demethylase FTO is impaired by low-level arsenic exposure to promote tumorigenesis. Nat Commun. 2021; 12:2183. 10.1038/s41467-021-22469-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Yang S, Cui YH, Wei J, Shah P, Park G, Cui X, He C, He YY. METTL14 facilitates global genome repair and suppresses skin tumorigenesis. Proc Natl Acad Sci U S A. 2021; 118:e2025948118. 10.1073/pnas.2025948118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao L, Mu X, Chen H, Jin D, Zhang R, Zhao Y, Fan J, Cao M, Zhou Z. FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin Transl Med. 2021; 11:e310. 10.1002/ctm2.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma L, Xue X, Zhang X, Yu K, Xu X, Tian X, Miao Y, Meng F, Liu X, Guo S, Qiu S, Wang Y, Cui J, et al. The essential roles of m6A RNA modification to stimulate ENO1-dependent glycolysis and tumorigenesis in lung adenocarcinoma. J Exp Clin Cancer Res. 2022; 41:36. 10.1186/s13046-021-02200-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Gao S, Zeng Y, Zhu L, Mo Y, Wong CC, Bao Y, Su P, Zhai J, Wang L, Soares F, Xu X, Chen H, et al. N6-Methyladenosine Reader YTHDF1 Promotes ARHGEF2 Translation and RhoA Signaling in Colorectal Cancer. Gastroenterology. 2022; 162:1183–96. 10.1053/j.gastro.2021.12.269 [DOI] [PubMed] [Google Scholar]
- 21.Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, He S, Shimamoto F. m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020; 19:3. 10.1186/s12943-019-1128-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Shen M, Li Y, Wang Y, Shao J, Zhang F, Yin G, Chen A, Zhang Z, Zheng S. N6-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021; 47:102151. 10.1016/j.redox.2021.102151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Li H, Hu D, Wang Y, Shao W, Zhong J, Yang S, Liu J, Zhang J. Insights into N6-methyladenosine and programmed cell death in cancer. Mol Cancer. 2022; 21:32. 10.1186/s12943-022-01508-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Wang F, Wang Z, Yang X, Yu H, Si S, Lu J, Zhou Z, Lu Q, Wang Z, Yang H. ALKBH5 promotes the proliferation of renal cell carcinoma by regulating AURKB expression in an m6A-dependent manner. Ann Transl Med. 2020; 8:646. 10.21037/atm-20-3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng J, Zhang H, Tan Y, Wang Z, Li Y, Yang X. m6A demethylase FTO suppresses pancreatic cancer tumorigenesis by demethylating PJA2 and inhibiting Wnt signaling. Mol Ther Nucleic Acids. 2021; 25:277–92. 10.1016/j.omtn.2021.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019; 18:142. 10.1186/s12943-019-1065-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan W, Chen S, Li B, Han X, Meng B, Zou Y, Chang S. The N6-methyladenosine reader protein YTHDC2 promotes gastric cancer progression via enhancing YAP mRNA translation. Transl Oncol. 2022; 16:101308. 10.1016/j.tranon.2021.101308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Yang X, Tang J, Si S, Zhou Z, Lu J, Han J, Yuan B, Wu Q, Lu Q, Yang H. ALKBH5 Inhibited Cell Proliferation and Sensitized Bladder Cancer Cells to Cisplatin by m6A-CK2α-Mediated Glycolysis. Mol Ther Nucleic Acids. 2020; 23:27–41. 10.1016/j.omtn.2020.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Z, Wang S, Chen W, Zhang X, Chen J, Jiang J, Wang M, Zhang L, Xuan Z. Fat mass and obesity-associated protein promotes the tumorigenesis and development of liver cancer. Oncol Lett. 2020; 20:1409–17. 10.3892/ol.2020.11673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Zhang S, He C, Xue P, Zhang L, He Z, Zang L, Feng B, Sun J, Zheng M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020; 19:46. 10.1186/s12943-020-1146-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Shao F, Guo D, Wang W, Wang J, Zhu R, Gao Y, He J, Lu Z. WNT/β-catenin-suppressed FTO expression increases m6A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis. Cell Death Dis. 2021; 12:462. 10.1038/s41419-021-03739-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang P, Wang Q, Liu A, Zhu J, Feng J. ALKBH5 Holds Prognostic Values and Inhibits the Metastasis of Colon Cancer. Pathol Oncol Res. 2020; 26:1615–23. 10.1007/s12253-019-00737-7 [DOI] [PubMed] [Google Scholar]
- 33.Yang DD, Chen ZH, Yu K, Lu JH, Wu QN, Wang Y, Ju HQ, Xu RH, Liu ZX, Zeng ZL. METTL3 Promotes the Progression of Gastric Cancer via Targeting the MYC Pathway. Front Oncol. 2020; 10:115. 10.3389/fonc.2020.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu QC, Tien YC, Shi YH, Chen S, Zhu YQ, Huang XT, Huang CS, Zhao W, Yin XY. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m6A-YTHDF2-dependent manner. Oncogene. 2022; 41:1622–33. 10.1038/s41388-022-02185-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu P, Hu K, Zhang P, Sun ZG, Zhang N. Hypoxia-mediated YTHDF2 overexpression promotes lung squamous cell carcinoma progression by activation of the mTOR/AKT axis. Cancer Cell Int. 2022; 22:13. 10.1186/s12935-021-02368-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, Li Q, Wang Y, Wang L, Guo Y, Yang R, Zhao N, Ge N, Wang Y, Guo C. m6A methyltransferase METTL3 promotes oral squamous cell carcinoma progression through enhancement of IGF2BP2-mediated SLC7A11 mRNA stability. Am J Cancer Res. 2021; 11:5282–98. [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017; 38:2285–92. 10.3892/or.2017.5904 [DOI] [PubMed] [Google Scholar]
- 38.Xia TL, Yan SM, Yuan L, Zeng MS. Upregulation of METTL3 Expression Predicts Poor Prognosis in Patients with Esophageal Squamous Cell Carcinoma. Cancer Manag Res. 2020; 12:5729–37. 10.2147/CMAR.S245019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang J, Lu Z, Wu P, Cai B, Miao Y, Jiang K. The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol Res Pract. 2019; 215:152666. 10.1016/j.prp.2019.152666 [DOI] [PubMed] [Google Scholar]
- 40.Wei K, Gao Y, Wang B, Qu YX. Methylation recognition protein YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) regulates the proliferation, migration and invasion of osteosarcoma by regulating m6A level of CCR4-NOT transcription complex subunit 7 (CNOT7). Bioengineered. 2022; 13:5236–50. 10.1080/21655979.2022.2037381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang XK, Zhang YW, Wang CM, Li B, Zhang TZ, Zhou WJ, Cheng LJ, Huo MY, Zhang CH, He YL. METTL16 promotes cell proliferation by up-regulating cyclin D1 expression in gastric cancer. J Cell Mol Med. 2021; 25:6602–17. 10.1111/jcmm.16664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Shao F, Yang X, Wang J, Zhu R, Yang Y, Zhao G, Guo D, Sun Y, Wang J, Xue Q, Gao S, Gao Y, et al. METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N6-methyladenosine-dependent YTHDF binding. Nat Commun. 2021; 12:3803. 10.1038/s41467-021-23501-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Wei W, Zhang ZY, Liu Y, Shi B, Zhong W, Zhang HS, Fang X, Sun CL, Wang JB, Liu LX. TCF4 and HuR mediated-METTL14 suppresses dissemination of colorectal cancer via N6-methyladenosine-dependent silencing of ARRDC4. Cell Death Dis. 2021; 13:3. 10.1038/s41419-021-04459-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuchiya K, Yoshimura K, Inoue Y, Iwashita Y, Yamada H, Kawase A, Watanabe T, Tanahashi M, Ogawa H, Funai K, Shinmura K, Suda T, Sugimura H. YTHDF1 and YTHDF2 are associated with better patient survival and an inflamed tumor-immune microenvironment in non-small-cell lung cancer. Oncoimmunology. 2021; 10:1962656. 10.1080/2162402X.2021.1962656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan Z, Shi S, Xu J, Liu X, Lei Y, Zhang B, Hua J, Meng Q, Wang W, Yu X, Liang C. RNA N6-methyladenosine demethylase FTO promotes pancreatic cancer progression by inducing the autocrine activity of PDGFC in an m6A-YTHDF2-dependent manner. Oncogene. 2022; 41:2860–72. 10.1038/s41388-022-02306-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y, Li S, Yu W, Zhao Z, Gao J, Chen C, Wei M, Liu T, Li L, Liu L. N6-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis. 2020; 11:836. 10.1038/s41419-020-03049-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan S, Deng Y, Fu J, Zhang Y, Zhang Z, Qin X. N6-methyladenosine upregulates miR-181d-5p in exosomes derived from cancer-associated fibroblasts to inhibit 5-FU sensitivity by targeting NCALD in colorectal cancer. Int J Oncol. 2022; 60:14. 10.3892/ijo.2022.5304 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Ruan DY, Li T, Wang YN, Meng Q, Li Y, Yu K, Wang M, Lin JF, Luo LZ, Wang DS, Lin JZ, Bai L, Liu ZX, et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene. 2021; 40:5168–81. 10.1038/s41388-021-01916-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orouji E, Peitsch WK, Orouji A, Houben R, Utikal J. Oncogenic Role of an Epigenetic Reader of m6A RNA Modification: YTHDF1 in Merkel Cell Carcinoma. Cancers (Basel). 2020; 12:202. 10.3390/cancers12010202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, Wu X, Wan G. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019; 18:46. 10.1186/s12943-019-1004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai D, Kudo T, Hata T, Matsuda C, et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2017; 9:7476–86. 10.18632/oncotarget.23554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagaki Y, Motoyama S, Yamaguchi T, Hoshizaki M, Sato Y, Sato T, Koizumi Y, Wakita A, Kawakita Y, Imai K, Nanjo H, Watanabe H, Imai Y, et al. m6 A demethylase ALKBH5 promotes proliferation of esophageal squamous cell carcinoma associated with poor prognosis. Genes Cells. 2020; 25:547–61. 10.1111/gtc.12792 [DOI] [PubMed] [Google Scholar]
- 53.Ma L, Lin Y, Sun SW, Xu J, Yu T, Chen WL, Zhang LH, Guo YC, Wang YW, Chen T, Wei JF, Zhu LJ. KIAA1429 is a potential prognostic marker in colorectal cancer by promoting the proliferation via downregulating WEE1 expression in an m6A-independent manner. Oncogene. 2022; 41:692–703. 10.1038/s41388-021-02066-z [DOI] [PubMed] [Google Scholar]
- 54.Lu S, Yu Z, Xiao Z, Zhang Y. Gene Signatures and Prognostic Values of m6A Genes in Nasopharyngeal Carcinoma. Front Oncol. 2020; 10:875. 10.3389/fonc.2020.00875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Xiao M, Zhang L, Li L, Zhu G, Shen E, Lv M, Lu X, Sun Z. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal. 2021; 35:e23655. 10.1002/jcla.23655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L, Wu Y, Li Q, Liang J, He Q, Zhao L, Chen J, Cheng M, Huang Z, Ren H, Chen J, Peng L, Gao F, et al. METTL3 Promotes Tumorigenesis and Metastasis through BMI1 m6A Methylation in Oral Squamous Cell Carcinoma. Mol Ther. 2020; 28:2177–90. 10.1016/j.ymthe.2020.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin S, Zhu Y, Ji C, Yu W, Zhang C, Tan L, Long M, Luo D, Peng X. METTL3-Induced miR-222-3p Upregulation Inhibits STK4 and Promotes the Malignant Behaviors of Thyroid Carcinoma Cells. J Clin Endocrinol Metab. 2022; 107:474–90. 10.1210/clinem/dgab480 [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of Demethylase Genes, FTO and ALKBH1, Is Associated with Prognosis of Gastric Cancer. Dig Dis Sci. 2019; 64:1503–13. 10.1007/s10620-018-5452-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X, Tang J, Huang W, Wang F, Li P, Qin C, Qin Z, Zou Q, Wei J, Hua L, Yang H, Wang Z. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget. 2017; 8:96103–16. 10.18632/oncotarget.21726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, Chen D, Li B, Kang TB, et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019; 18:112. 10.1186/s12943-019-1038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J, Rao B, Yang J, Liu L, Huang M, Liu X, Cui G, Li C, Han Q, Yang H, Cui X, Sun R. Dysregulated m6A-Related Regulators Are Associated With Tumor Metastasis and Poor Prognosis in Osteosarcoma. Front Oncol. 2020; 10:769. 10.3389/fonc.2020.00769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang H, Li L, Chen S, Lü W, Hu J. Expression of demethylase ALKBH5 in lung adenocarcinoma and its relationship with cell proliferation. Tumor. 2018; 38:572–80. 10.3781/j.issn.1000-7431.2018.33.009 [DOI] [Google Scholar]
- 63.Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, Duan P. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018; 151:356–65. 10.1016/j.ygyno.2018.09.015 [DOI] [PubMed] [Google Scholar]
- 64.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, Wei JF, Yang H. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019; 18:110. 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo YQ, Wang Q, Wang JG, Gu YJ, Song PP, Wang SY, Qian XY, Gao X. METTL3 modulates m6A modification of CDC25B and promotes head and neck squamous cell carcinoma malignant progression. Exp Hematol Oncol. 2022; 11:14. 10.1186/s40164-022-00256-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, Feng Y, Pan Q, Wan R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 2020; 19:91. 10.1186/s12943-020-01158-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du M, Peng Y, Li Y, Sun W, Zhu H, Wu J, Zong D, Wu L, He X. MYC-activated RNA N6-methyladenosine reader IGF2BP3 promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cell Death Discov. 2022; 8:53. 10.1038/s41420-022-00844-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng R, Cheng Y, Ye S, Zhang J, Huang R, Li P, Liu H, Deng Q, Wu X, Lan P, Deng Y. m6A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019; 12:4391–402. 10.2147/OTT.S201052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, Pan B, He B, Pan Y, Sun H, Xia X, Wang S. RETRACTED: METTL14 Suppresses CRC Progression via Regulating N6-Methyladenosine-Dependent Primary miR-375 Processing. Mol Ther. 2020; 28:599–612. 10.1016/j.ymthe.2019.11.016. Retraction in: Mol Ther. 2022; 30: 2640. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Chen HD, Li F, Chen S, Zhong ZH, Gao PF, Gao WZ. METTL3-mediated N6-methyladenosine modification of DUSP5 mRNA promotes gallbladder-cancer progression. Cancer Gene Ther. 2022; 29:1012–20. 10.1038/s41417-021-00406-5 [DOI] [PubMed] [Google Scholar]
- 71.Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer. 2019; 18:103. 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu ZX, Li LM, Sun HL, Liu SM. Link Between m6A Modification and Cancers. Front Bioeng Biotechnol. 2018; 6:89. 10.3389/fbioe.2018.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu T, Yang S, Sui J, Xu SY, Cheng YP, Shen B, Zhang Y, Zhang XM, Yin LH, Pu YP, Liang GY. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol. 2020; 235:548–62. 10.1002/jcp.28994 [DOI] [PubMed] [Google Scholar]
- 74.Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019; 38:393. 10.1186/s13046-019-1408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu J, Sun G, Pan S, Qin M, Ouyang R, Li Z, Huang J. The Cancer Genome Atlas (TCGA) based m6A methylation-related genes predict prognosis in hepatocellular carcinoma. Bioengineered. 2020; 11:759–68. 10.1080/21655979.2020.1787764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan Y, Du Y, Wang L, Liu X. The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J Cancer. 2020; 11:3588–95. 10.7150/jca.42338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018; 37:522–33. 10.1038/onc.2017.351 [DOI] [PubMed] [Google Scholar]
- 78.Yao Y, Bi Z, Wu R, Zhao Y, Liu Y, Liu Q, Wang Y, Wang X. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2-dependent manner. FASEB J. 2019; 33:7529–44. 10.1096/fj.201802644R [DOI] [PubMed] [Google Scholar]
- 79.Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, Gong J, Shen L. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019; 8:4766–81. 10.1002/cam4.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, Liang X, Yang Y. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021; 28:335–49. 10.1038/s41417-020-00222-3 [DOI] [PubMed] [Google Scholar]
- 81.Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, Zhao W, Fan Y, Zhang D, Sun R. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020; 53:e12768. 10.1111/cpr.12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang BH, Yan LN, Yang JY. Pending role of METTL14 in liver cancer. Hepatobiliary Surg Nutr. 2019; 8:669–70. 10.21037/hbsn.2019.10.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, Li C, Sun L, Qin J, Xu T, He B, Pan Y, Sun H, Wang S. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020; 19:106. 10.1186/s12943-020-01220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Z, Li F, Peng Y, Fang J, Zhou J. Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma. Cancer Med. 2020; 9:1877–89. 10.1002/cam4.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, Abdelfattah N, Onyeagucha BC, Cui X, Lai Z, Mohammad TA, Gupta YK, Huang TH, et al. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Sci Adv. 2018; 4:eaar8263. 10.1126/sciadv.aar8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.You Y, Wen D, Zeng L, Lu J, Xiao X, Chen Y, Song H, Liu Z. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int J Biol Sci. 2022; 18:5001–18. 10.7150/ijbs.70149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019; 112:108613. 10.1016/j.biopha.2019.108613 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.