Abstract
Rickettsia typhi, the causative agent of murine typhus, grows directly within the host cell cytoplasm, accumulating a large number of progeny, and eventually lyses the cells. Typhus group rickettsiae (R. typhi and R. prowazekii) adhere to and lyse human and sheep erythrocytes. However, the molecular mechanism underlying erythrocyte lysis by R. typhi has not been defined. Here we describe the cloning and nucleotide sequence analysis of the gene (tlyC) encoding a hemolysin from R. typhi. DNA sequence analysis of R. typhi tlyC revealed an open reading frame of 912 bp, which encodes a protein of 304 amino acids with a predicted molecular mass of 38 kDa. To associate the R. typhi tlyC gene product with hemolytic activity, we performed complementation studies with hemolysin-negative Proteus mirabilis WPM111 (a HpmA− mutant of BA6163) transformed with R. typhi tlyC or R. typhi GFPuv-tlyC constructs. We demonstrated that the cloned tlyC gene conferred a hemolytic phenotype on an otherwise nonhemolytic mutant of P. mirabilis. The availability of the cloned R. typhi tlyC will permit further characterization and definition of its role in rickettsial virulence.
The genus Rickettsia contains several well-known human pathogens, such as R. prowazekii, R. typhi, R. rickettsii, and R. conorii, the etiologic agents of epidemic and endemic typhus, Rocky Mountain spotted fever, and Boutonneuse fever, respectively (6).
As obligate intracellular parasites, typhus group (TG) rickettsiae (R. prowazekii and R. typhi) enter nonprofessional phagocytes (e.g., epithelial and endothelial cells and fibroblasts) by a process of induced phagocytosis, multiply intracellularly, and lyse the cells only after the accumulation of a large number of rickettsial progeny (reviewed in reference 6). Little is known about rickettsial pathogenesis and virulence factors. Previously, the pathogenesis of R. typhi has been attributed to several putative virulence factors such as phospholipases A2 and C, increased concentration of intracellular peroxide, the presence of trypsin-like proteases, and iron metabolism (6, 9, 14).
Hemolysins have often been implicated as virulence factors for variety of human pathogens such as Escherichia coli, Streptococcus pneumoniae, Listeria monocytogenes, Pasteurella haemolytica, Proteus mirabilis, and Actinobacillus pleuropneumoniae (3–5). Despite the similarity in attachment, entry, and cytoplasmic location, members of the TG and spotted fever group (SFG) (R. rickettsii and R. akari) rickettsiae differ in several features. R. typhi and R. prowazekii adhere to and lyse erythrocytes at mildly acidic pH, in contrast to SFG rickettsiae, whose members are incapable of hemolysis (6, 15). For R. prowazekii, hemolysis at acidic pH but not at neutral pH is enhanced by Ca2+, raising the possibility that more than one membranolytic factor is produced by the rickettsiae (9). The evidence suggests that the rickettsial membranolytic activity may contribute to the escape of the bacteria from the early phagosome during infection.
Recently Andersson et al. (2) described the sequence of the 1.11-Mbp genome of R. prowazekii. Two genes (tlyA and tlyC) encoding hemolysins have been identified in the genome of R. prowazekii (2). Using PCR primers based on the R. prowazekii tlyC sequence, we identified the R. typhi homologue of tlyC. R. typhi tlyC exhibits hemolytic activity when expressed in E. coli and P. mirabilis.
Here we describe the cloning, nucleotide sequence analysis, and molecular characterization of the R. typhi gene encoding a hemolysin. Expression of R. typhi tlyC in nonhemolytic mutants of P. mirabilis WPM111 (HpmA−) confers a hemolytic phenotype to these otherwise nonhemolytic bacteria.
MATERIALS AND METHODS
Rickettsial strains.
Plaque-purified R. typhi (Wilmington, 42E/3TC/2E0), R. prowazekii (Breinl, 155E/3TC/2E), R. rickettsii (Sheila Smith, 10E/1TC), and R. akari (Kaplan, ATCC/VR-148) were used in this study. R. australis (JC) was kindly provided by D. H. Walker (University of Texas Medical Branch at Galveston).
Isolation of genomic DNA.
Rickettsiae were cultivated in Vero cells in minimal essential medium supplemented with 4% fetal calf serum (Atlanta Biological, Atlanta, Ga.). The infected monolayers were harvested, and rickettsiae were purified by discontinuous Renografin gradient ultracentrifugation. Genomic DNAs were isolated as previously described (12). Briefly, 200 μl of Renografin-purified rickettsiae were suspended overnight at 37°C in 400 μl of TE buffer (10 mM Tris-HCl, [pH 8.0], 1 mM EDTA) supplemented with 0.1 mg of proteinase K per ml and 1.0% sodium dodecyl sulfate, and an equal volume of phenol-chloroform-isoamyl alcohol was added. Samples were vortexed for 15 s and then centrifuged at 12,000 × g for 10 min. The upper (aqueous) phase was collected and subjected to ethanol precipitation. The resultant pellet was washed with cold 70% ethanol, resuspended in 20 μl of TE buffer, and quantified spectrophotometrically.
PCR amplification.
Primers used in the PCRs were designed based on the nucleotide sequences of the R. prowazekii tlyC gene (GenBank accession no. Y11778): forward primer (nucleotides [nt] 70 to 88) (5′-GTA CGA CAA TTA TTC TAT-3′) and reverse primer (nt 825 to 843) (5′-GTT GCA TCT GTT ACT TCA-3′). For PCR, 2 μg of purified R. typhi genomic DNA was used as the template. The thermal cycling conditions consisted of 35 cycles of denaturation at 94°C for 1 min, annealing at 40°C for 2 min, and extension at 72°C for 3 min. Following PCR, 15 μl of each reaction product was electrophoresed on 1% agarose gel and visualized by ethidium bromide staining.
Cloning and sequencing of R. typhi tlyC.
PCR products were subcloned into PCRII vector as described by the manufacturer (Invitrogen, San Diego, Calif.). Cells were plated onto Luria-Bertani (LB) agar plates containing 50 μg of ampicillin per ml and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 20 mg/ml), and white colonies were selected for further analysis. Plasmid DNA was purified with the Nucleospin kit (The Nest Group Inc., Southboro, Mass.). The insert was sequenced by the dye terminator method on a model 373 automated fluorescence sequencing system (Applied Biosystems, Foster City, Calif.). Sequence analysis was carried out with the Sequence editor 675 software package (Applied Biosystems), and the BLAST program (1) was used for sequence comparisons (National Center for Biotechnology Information, Bethesda, Md.). An internal forward primer (5′-CTC CTC GAA GAG ATT ATC-3′) was designed on the basis of the R. typhi tlyC sequence (nt 660 to 678) and the 3′ end of tlyC was amplified from the R. typhi Lambda Zap genomic library with T7 (5′-GTA ATA CGA CTC ACT ATA GGG C-3′) as the reverse primer. The 5′ end of the gene was obtained from the R. typhi Lambda Zap genomic library with T3 as the forward primer (5′-AAT TAA CCC TCA CTA AAG GG-3′), and the reverse primer was based on the internal sequence of R. typhi tlyC (nt 70 to 88; 5′-GTA CGA CAA TTA TTC TAT-3′). The tlyC gene was sequenced three times in both directions to ensure sequence fidelity.
Dot blot detection of tlyC in rickettsiae.
A 1.5-μg sample of rickettsial DNA was applied to a nylon membrane (Hybond-N+; Amersham) and probed with 32P-labelled tlyC (230 bp, 311 to 541 nt) (nick translation kit; Promega, Madison, Wis.). For overnight oligonucleotide probing, hybridization and washing were carried out at 42°C.
Cloning of R. typhi tlyC in E. coli.
To determine the role of tlyC in hemolysis, nonhemolytic E. coli was used in our complementation studies (10). The 1.3-kb R. typhi tlyC was amplified by PCR with primers containing an EcoRI site (G′AATTC) and ligated into the pGEX-2TK expression vector (Pharmacia Biotech, Inc.). Ligation products (2 μl) were used to transform nonhemolytic E. coli K-12.
SDS-PAGE and immunoblotting.
To determine the size of the TlyC protein, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on a slab gel by a modification of the method described by Laemmli (7) with a 15.0% separating gel. The purified nonhemolytic E. coli, E. coli containing plasmid pGEX-2TK, and E. coli containing pGEX-2TK-tlyC were solubilized at room temperature in sample buffer (0.25 M Tris-HCl [pH 6.8], 10% SDS, 0.5 M dithiothreitol, 0.5% bromophenol blue, 50% glycerol [5′-3′, Inc., Boulder, Colo.]) at 2.0 mg of protein per ml. A 10-μl sample was loaded into each lane, and a 10-kDa protein ladder (Gibco BRL, Gaithersburg, Md.) was used as a molecular standard. Electrophoretically separated polypeptides were transferred from the gels to a 0.2-μm-pore-size nitrocellulose membrane (Gibco) by electrophoresis at 365 mA for 1 h at 4°C. After nonspecific protein binding was blocked with 5% nonfat dry milk in 25 mM Tris-buffered saline (TBS), the blot was incubated with antibody to glutathione S-transferase (GST) (Pharmacia) for 1 h at room temperature. Next, the blot was incubated with anti-goat immunoglobulin G (IgG)-alkaline phosphatase conjugate (Sigma, St. Louis, Mo.) for 1 h at room temperature and then developed with BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) as the substrate.
Hemolytic assay.
The hemolytic activities of the culture supernatant of nonhemolytic E. coli K-12, E. coli expressing GST-tlyC, and E. coli containing pGEX-2TK were assayed as follows. Bacteria were cultured in LB broth in a shaking incubator for 24 h at 37°C. The cells were removed by centrifugation (3,000 × g at 4°C for 10 min), and the supernatants were filtered through 0.22-μm-pore-size nitrocellulose. Samples of filtrate (0.1 ml) were serially diluted twofold in 10 mM Tris-HCl (pH 7.4) containing 0.9% NaCl and were then added to an equal volume of a 2% suspension of sheep erythrocytes (Cleveland Scientific, Bath, Ohio). After incubation at 25°C for 90 min, the titer was expressed as the highest dilution causing complete hemolysis.
GFP ligation to R. typhi tlyC.
A green fluorescence protein (GFP)-tlyC fusion was constructed in the following manner. The 800-bp tlyC fragment was amplified by PCR with primers containing a SacI site (forward primer, nt 72 to 88 [5′-GGA GCT CGT ACG ACA ATT ATT CTA T-3′]; reverse primer, nt 800 to 816 [5′-GGA GCT CGT TGC ATC TGT TAC TTC A-3′]). For PCR, 2 μg of purified R. typhi genomic DNA was used as the template. The thermal cycling conditions consisted of 30 cycles of denaturation at 94°C for 1 min, annealing at 37°C for 1 min, and extension at 72°C for 1 min. The PCR product was subcloned into TA vector for propagation. Plasmid preparation was performed with a Wizard miniprep kit (Promega). Purified plasmid (10 μg) was digested with 5 U of SacI to excise the tlyC insert. The 800-bp SacI digestion product was electrophoresed on a 1% agarose gel and eluted from the gel with GeneClean (Bio 101, Vista, Calif.). pGFPuv (Clontech, Palo Alto, Calif.) was linearized by SacI digestion (2 μg of pGFPuv and 5 U of SacI in 1× buffer) at 37°C for 1 h, treated with 1 μl of calf intestinal alkaline phosphatase in 1× buffer (New England BioLabs, Beverly, Mass.) at 37°C for 1 h, and subjected to heat inactivation at 75°C for 10 min. Following phenol-chloroform-isoamyl extraction, the linearized GFPuv was electrophoresed on a 1% agarose gel, excised, and eluted as previously described (12). The vector (linearized GFPuv) and insert (tlyC) at a 1:3 ratio were ligated at 16°C overnight with 10 U of the T4 DNA ligase and 1× buffer (New England BioLabs). Ligation products (2 μl) were used to transform E. coli (One Shot competent cells; TA cloning kit [Invitrogen, San Diego, Calif.]). Transformants were grown on LB plates containing 100 μg of ampicillin per ml. Green fluorescent colonies were screened for incorporation of the tlyC by PCR amplification with the protocol and program described above and by SacI restriction digestion. The proper orientation of the tlyC in the pGFPuv vector was confirmed by sequencing.
Creation of the frameshift GFP-tlyC construct.
A frameshift GFP construct was created by deleting 7 nt from the 3′ end of the GFP coding region by the following method. A 10-μg portion of the GFP-tlyC plasmid was linearized by digestion with 10 U of XhoI (Gibco BRL) in 1× buffer for 2 h at 37°C. Completion of total digestion was checked by agarose gel. The XhoI site is located at the 3′ end of GFP, before the tlyC coding region (nt 710 of pGFPuv). The linearized vector was treated with 80 U of mung bean nuclease (Gibco) in 1× buffer in a total volume of 500 μl. The reaction mixture was heated to 37°C for 10 min and then extracted twice with phenol-chloroform-isoamyl alcohol. DNA was ethanol precipitated and resuspended in 8 μl of water; 400 U of T4 DNA ligase and 1 μl of 10× buffer were added at 16°C overnight to religate the plasmid. Then 2 μl of the ligation reaction mixture was used to transform E. coli (One Shot competent cells; TA cloning kit). The colonies were screened for loss of the XhoI site, and positive colonies were sequenced to confirm the frameshift of GFP. The frameshift in the GFP-tlyC produces a premature stop codon in the GFPuv coding region. This results in a truncated GFP, which does not fluoresce, and the tlyC is not expressed.
Transformation of nonhemolytic P. mirabilis with R. typhi tlyC.
P. mirabilis WPM111 hpmA (a deletion mutant of the complete hemolysin gene) lacks hemolytic activity and is unable to lyse human renal proximal tubular epithelial cells (3, 8). P. mirabilis WPM111 hpmA and wild-type P. mirabilis BA6163 were inoculated into LB broth and grown for 18 h with aeration at 37°C. These cultures were used to inoculate 50-ml portions of LB broth and incubated for 3 h at 37°C. The cells were centrifuged at 3,000 × g at 4°C for 10 min and then washed twice with cold phosphate-buffered saline and once with cold ultrapure H2O. They were then resuspended in 100 μl of H2O and immediately electroporated with 3 to 6 μg of DNA of GFP-R. typhi tlyC or frameshift GFP-tlyC construct.
Western blot analysis.
The bacterial cells were removed from the cultures by centrifugation at 12,000 × g for 30 min at 4°C. Cell lysates were subjected to polyacrylamide gel electrophoresis with 15% acrylamide (7). For Western blot analysis, proteins were transferred to nitrocelulose membrane. The membrane was then blocked with 5% nonfat milk in TBS and incubated with GFP monoclonal antibody (Clontech) for 1 h at room temperature. Next, the blot was incubated with anti-mouse IgG-alkaline phosphatase Conjugate for 1 h at room temperature and then developed with BCIP/NBT as a chromogenic substrate for alkaline phosphatase.
Hemolytic titers.
The hemolytic titers of the wild-type P. mirabilis BA6163, nonhemolytic P. mirabilis WPM111 hpmA, P. mirabilis WPM111 containing GFP-tlyC, and P. mirabilis WPM111 containing frameshift GFP-tlyC were determined as follows. The optical density of the bacterial overnight cultures was adjusted to 106 CFU/ml in phosphate-buffered saline (pH 7.2). Bacterial suspensions were serially diluted twofold. Samples (100 μl) were mixed with a 1% suspension of sheep erythrocytes and incubated at 37°C, and the results were recorded macroscopically after 1, 2, 3, and 4 h. The hemolytic titer was defined as the reciprocal of the last dilution to give complete hemolysis of erythrocytes.
Nucleotide sequence accession number.
The nucleotide sequence of R. typhi hemolysin has been deposited in GenBank under accession no. U97482.
RESULTS
PCR, cloning, and sequencing of R. typhi tlyC.
We have cloned and sequenced the R. typhi hemolysin gene (tlyC) with primers based on the sequence of R. prowazekii tlyC (GenBank Y11778). DNA sequence analysis of R. typhi tlyC revealed an open reading frame of 912 bp, which encodes a protein of 304 amino acids with a predicted molecular mass of 38 kDa. Comparison of the 912 bp of R. typhi tlyC with the equivalent region of R. prowazekii tlyC revealed 97% homology at the amino acid level (Fig. 1). We have noted 15-amino-acid substitutions in R. typhi tlyC. The R. typhi TlyC amino acid sequences revealed 68% homology to the putative hemolysin (HlyX) of Streptococcus mutants (AF051356) and 63% homology to Mycoplasma genitalium TlyC (U39694).
FIG. 1.
(A). Amino acid alignment of TlyC between R. prowazekii and R. typhi. Boldface letters represent amino acid differences. The TlyC amino acid sequence revealed 68% homology to HlyX of S. mutans and 63% homology to TlyC of M. genitalium.
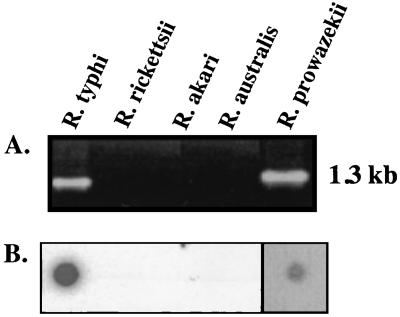
The PCR primer pair designed for the entire R. typhi tlyC yielded a 1.3-kb product in TG rickettsiae (R. typhi and R. prowazekii) but not in SFG rickettsiae (R. rickettsii, R. australis, and R. akari) (Fig. 2A). The presence of the tlyC gene in TG rickettsiae was confirmed by dot blot analysis (Fig. 2B). Successful PCR amplifications were also obtained with genomic DNA from several R. typhi strains isolated from humans and rats from various geographic regions (data not shown).
FIG. 2.
(A) PCR demonstrating the presence of the tlyC gene in TG (R. typhi and R. prowazekii) but not in SFG group (R. rickettsii, R. akari, and R. australis) rickettsiae. (B) Results showing dot blot hybridization on the five rickettsial DNA types in panel A, using a tlyC-specific probe. (The darker background for R. prowazekii is due to a longer exposure of the film to blot. Only one blot is represented here).
R. typhi tlyC expression in nonhemolytic E. coli.
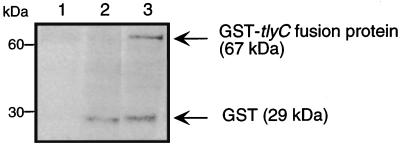
PCR was used to amplify R. typhi tlyC and to subclone the gene into pGEX-2TK expression vector. The ligation products were used to transform nonhemolytic E. coli K-12 (10). Western analysis of nonhemolytic E. coli expressing the GST-tlyC fusion protein revealed a band of 67 kDa (Fig. 3). In contrast, controls including nonhemolytic E. coli as well as E. coli carrying only the vector pGEX-2TK lack the 67-kDa band. The size of the R. typhi tlyC product did not change when it was analyzed on a nondenaturing gel. Introducing the tlyC gene into nonhemolytic E. coli K-12 resulted in a hemolytic phenotype. E. coli expressing the GST-tlyC fusion protein led to hemolysis of sheep erythrocytes (data not shown).
FIG. 3.
Western analysis of 10 μg of proteins extracted from E. coli. Nonhemolytic E. coli K-12 (lane 1), E. coli containing vector only (lane 2), and E. coli expressing R. typhi tlyC (lane 3) were incubated with the anti-GST antibody in conjunction with anti-goat IgG conjugate. GST-tlyC fusion protein was detected at 67 kDa.
Transformation of nonhemolytic P. mirabilis WPM111 hpmA with R. typhi GFPuv-tlyC and frameshifted GFPuv-tlyC.
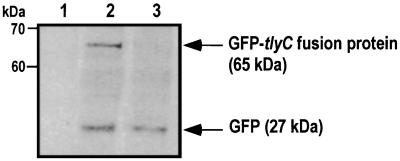
Lack of rickettsial mutants has restricted our ability to carry complementation studies to determine the function of the tlyC gene. Initially, we used nonhemolytic E. coli K-12, and the introduction of the tlyC into bacteria resulted in a hemolytic phenotype. However, we could not rule out the possibility that rickettsial tlyC could act as an activator of the cryptic cytolysin A (clyA) expression in transformed E. coli. Therefore, we selected the well-described and well-characterized hemolysin-negative P. mirabilis WPM111 hpmA, which is incapable of lysing sheep erythrocytes (3, 8). We transformed R. typhi GFPuv-tlyC and also frameshifted the GFPuv-tlyC construct into the P. mirabilis nonhemolytic mutant WPM111 hpmA. Expression of the GFP-tlyC fusion protein was confirmed by SDS-PAGE and Western analysis. The 65-kDa band was present in the supernatant of the cell lysate of P. mirabilis WPM111 hpmA carrying the R. typhi tlyC construct (Fig. 4). In addition, P. mirabilis WPM111 hpmA complemented with R. typhi GFPuv-tlyC formed colonies expressing GFP grown on LB plates, and the colonies fluoresced brightly when viewed on a UV light box. The P. mirabilis GFPuv-tlyC frameshifted mutant was clearly visible as pale green colonies.
FIG. 4.
Detection of a GFP-tlyC fusion protein by immunoblotting with the anti-GFP monoclonal antibody. Lanes 1 to 3 contain nonhemolytic P. mirabilis WPM111/hpmA, P. mirabilis WPM111/hpmA complemented with GFP-tlyC, and P. mirabilis WPM111/hpmA transformed with GFPuv only, respectively. GFP-tlyC fusion protein was detected at 65 kDa.
Analysis of hemolytic activity.
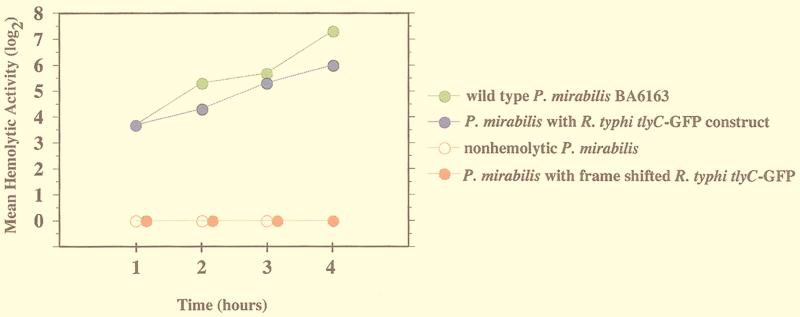
P. mirabilis expressing the GFP-tlyC fusion protein readily lysed sheep erythrocytes. The hemolytic activity of wild-type P. mirabilis BA6163 peaked at 1:128 4 h after the bacteria were mixed with a 1% suspension of sheep erythrocyte (Fig. 5). The WPM111 hpmA P. mirabilis mutant demonstrated no lysis of sheep erythrocytes at any time point. When this mutant was complemented with GFPuv-tlyC, the hemolytic titer peaked at 1:64 at the 4-h time point. The hpmA mutant transformed only with the GFPuv vector and the mutant with the frameshift deletion of GFPuv-tlyC demonstrated no hemolysis at any time point.
FIG. 5.
Hemolytic activity of P. mirabilis BA6163 (wild type) and nonhemolytic WPM111/hpmA complemented with R. typhi tlyC. Each point is the geometric mean of three separate experiments. The hemolytic activity is defined as the reciprocal of the last dilution that results in complete hemolysis of the sheep erythrocytes. Nonhemolytic strain (P. mirabilis WPM111/hpmA) and P. mirabilis (WPM111) containing frameshift GFP-tlyC had no hemolytic activity.
DISCUSSION
Hemolysins or cytolysins are membrane-damaging agents which serve as important virulence factors not only for gram-positive human pathogens but also for a variety of gram-negative bacteria such as Proteus, Bordetella, Morganella, Pasteurella, and Actinobacillus (4, 5, 8).
Members of the TG and SFG rickettsiae cause important human diseases by invading endothelial cells in widely disseminated sites throughout the body. Although they are genetically closely related, they differ antigenically and behaviorally. While the TG rickettsiae adhere to and lyse sheep and human erythrocytes, the SFG rickettsiae cannot lyse erythrocytes. The hemolytic activity of R. prowazekii has been extensively studied for four decades (6, 12, 14), but the DNA sequences of two genes (tlyA and tlyC) encoding hemolysins of R. prowazekii have been identified recently (2). Rickettsial entry and exit from phagosomes have also been indirectly associated with two phospholipases, A2 and C (14, 15), but the molecular events that lead to rickettsial intracellular placement, cell-to-cell spread, and eventual lysis of the host cells have yet to be elucidated.
In the present study, we have identified, cloned, sequenced, and expressed the R. typhi tlyC gene. tlyC was detected by PCR in several R. typhi isolates obtained from humans and rats from various geographic regions. The identification of R. typhi tlyC prompted us to look for related genes encoding hemolysin in other rickettsiae. The PCR primer pair targeting the entire tlyC of R. typhi yielded no PCR amplicons in SFG rickettsiae. The failure to detect the tlyC gene in SFG rickettsiae could have been caused by substantial differences between the SFG tlyC sequence and that of R. typhi. Another possibility is the presence of a nonfunctional fragment of tlyC in SFG rickettsiae, as is well known for most nonhemolytic isolates of S. pneumoniae (13).
When R. typhi tlyC was subcloned into the pGEX-2TK expression vector and transformed into nonhemolytic E. coli K-12, the resulting GST-tlyC fusion protein was expressed as a 67-kDa protein and conferred a hemolytic phenotype to E. coli. Although R. typhi tlyC shows no homology to the cytolysin A gene (clyA), which encodes the only cytolytic factor found in nonpathogenic strain of E. coli including the laboratory strain K-12, we could not rule out the possibility that tlyC was activating a cryptic E. coli K-12 hemolysin.
To associate the R. typhi tlyC gene product with hemolytic activity, we have complemented a nonhemolytic mutant of P. mirabilis WPM111, the hpmA mutant, with the GFPuv-tlyC construct. We have demonstrated that the GFP-tlyC fusion protein was detected in supernatants of cell lysates and transferred the hemolytic phenotype to the otherwise nonhemolytic mutant of P. mirabilis. Nonhemolytic P. mirabilis strains transformed only with vector (pGFPuv) were unable to lyse sheep erythrocytes.
We are aware that all of our complementation studies of tlyC of R. typhi were done with nonhemolytic bacteria such as E. coli and P. mirabilis. The major methodological constraint to studying gene function in rickettsiae has been the lack of mutants. Although our results associate the R. typhi tlyC gene product with lysis of human and sheep erythrocytes, complementation experiments with R. typhi mutants lacking tlyC would have been helpful. Fortunately, work in progress in our laboratory and a few others is closing this gap. Transformation of rickettsiae has been performed by homologous recombination (11, 12). Rickettsial transformation, availability of selectable markers such as GFPuv-tlyC, and molecular manipulation will allow us to study the function of the tlyC gene product in R. typhi and to shed light on the role of hemolysin in host cell entry, exit from the phagosome, and host cell lysis.
ACKNOWLEDGMENTS
We thank Harry Mobley for the generous gift of wild-type and nonhemolytic mutant P. mirabilis.
This research was supported by grants from the National Institutes of Health (R37AI 17828 and R01 AI 43006).
REFERENCES
- 1.Altschul S F, Gisch W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark C M U, Podowski R M, Naslund A K, Eriksson A-C, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 3.Chippendale G R, Warren J W, Trifillis A L, Mobley H L T. Internalization of Proteus mirabilis by human renal epithelial cells. Infect Immun. 1994;62:3115–3121. doi: 10.1128/iai.62.8.3115-3121.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkow S. The evolution of pathogenicity in Escherichia, Shigella, and Salmonella. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 85–96. [Google Scholar]
- 5.Galan J E. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 6.Hackstadt T. The biology of rickettsiae. Infect Agents Dis. 1996;5:127–143. [PubMed] [Google Scholar]
- 7.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 8.Mobley H L T, Chippendale G R, Swihart K G, Welch R A. Cytotoxicity of the HpmA hemolysin and urease of Proteus mirabilis and Proteus vulgaris against cultured human renal proximal tubular epithelial cells. Infect Immun. 1991;59:2036–2042. doi: 10.1128/iai.59.6.2036-2042.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojcius D M, Thibon M, Mounier C, Dautry-Varsat A D. pH and calcium dependence of hemolysis due to Rickettsia prowazekii: comparison with phospholipase activity. Infect Immun. 1995;63:3069–3072. doi: 10.1128/iai.63.8.3069-3072.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oscarsson J, Mizunoe Y, Uhlin B E, Haydon D J. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol Microbiol. 1996;20:191–199. doi: 10.1111/j.1365-2958.1996.tb02500.x. [DOI] [PubMed] [Google Scholar]
- 11.Rachek L I, Tucker A M, Winkler H H, Wood D O. Transformation of Rickettsia prowazekii to rifampin resistance. J Bacteriol. 1998;180:2118–2124. doi: 10.1128/jb.180.8.2118-2124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troyer M J, Radulovic S, Azad A F. Green fluorescent protein as a marker in Rickettsia typhi transformation. Infect Immun. 1999;67:3308–3311. doi: 10.1128/iai.67.7.3308-3311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whatmore A M, King S J, Doherty N C, Sturgeon D, Chanter N, Dowson C G. Molecular characterization of equine isolates of Streptococcus pneumoniae: natural disruption of genes encoding the virulence factors pneumolysin and autolysin. Infect Immun. 1999;67:2776–2782. doi: 10.1128/iai.67.6.2776-2782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkler H H, Miller E T. Phospholipase A and the interaction of Rickettsia prowazekii and mouse fibroblasts (L-929 cells) Infect Immun. 1982;38:109–113. doi: 10.1128/iai.38.1.109-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler H H, Miller E T. Phospholipase A activity in the hemolysis of sheep and human erythrocytes by Rickettsia prowazekii. Infect Immun. 1980;29:316–321. doi: 10.1128/iai.29.2.316-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]