Abstract
Secondary cell walls (SCWs) in stem cells provide mechanical strength and structural support for growth. SCW thickening varies under different light conditions. Our previous study revealed that blue light enhances SCW thickening through the redundant function of MYC2 and MYC4 directed by CRYPTOCHROME1 (CRY1) signaling in fiber cells of the Arabidopsis inflorescence stem. In this study, we find that the Arabidopsis PHYTOCHROME B mutant phyB displays thinner SCWs in stem fibers, but thicker SCWs are deposited in the PHYTOCHROME INTERACTING FACTOR (PIF) quadruple mutant pif1pif3pif4pif5 (pifq). The shaded light condition with a low ratio of red to far-red light inhibits stem SCW thickening. PIF4 interacts with MYC2 and MYC4 to affect their localization in nuclei, and this interaction results in inhibition of the MYCs’ transactivation activity on the NST1 promoter. Genetic evidence shows that regulation of SCW thickening by PIFs is dependent on MYC2/MYC4 function. Together, the results of this study reveal a PHYB-PIF4-MYC2/MYC4 module that inhibits SCW thickening in fiber cells of the Arabidopsis stem.
Key words: far-red light, fiber cell, MYC2, secondary cell wall, xylem
Light plays a critical role in the regulation of plant cell wall formation. This study reveals a PHYB-PIF4-MYC2/MYC4 module that inhibits secondary cell wall thickening in stem fiber cells via NST1-directed transcriptional regulatory networks, providing new insights into plant cell wall formation in response to shaded conditions.
Introduction
In higher plants, all cells are encased in a primary cell wall laid down during cell elongation that is flexible (to allow growth) yet possesses sufficient tensile strength to withstand the turgor pressure that drives growth. The primary cell wall defines the shape of a plant cell and is important for communication between plants and their environments (Doblin et al., 2010). Some types of specialized cells, such as fiber and vessel cells in stem xylem, deposit a rigid secondary cell wall (SCW) inside the primary cell wall after cell elongation has ceased, which provides plants with the mechanical strength to withstand enormous compressive forces and the capacity to transport water to aerial organs (Zhong and Ye, 2015). The main components of lignified SCWs include cellulose, hemicelluloses, and lignin, with deposition of lignin being a sign of SCW thickening. Expression of SCW biosynthesis genes is controlled by a hierarchy of transcriptional regulatory networks. SND1/NST1 and VND6/VND7 are key regulators at the top tier of the regulatory network that specifically control SCW formation in fiber and vessel cells, respectively, in Arabidopsis (Kubo et al., 2005; Zhong et al., 2006; Mitsuda et al., 2007; Yamaguchi et al., 2010; Zhu and Li, 2021).
In addition to developmental signals, various external environmental cues, including light, water, and temperature, affect SCW formation (Le Gall et al., 2015). Light induces a range of effects on plant cell wall formation (Le Gall et al., 2015). For example, when grown under blue light, Arabidopsis produces an inflorescence stem that is mechanically strengthened due to thickening of the SCWs of fiber cells. The blue light signal has been shown to induce MYC2/MYC4 expression, which activates NST1 expression by binding to its promoter, leading to an enhancement of SCW thickening (Zhang et al., 2018a). However, under shaded light conditions with a lower ratio of red to far-red (R:FR) light, plants exhibit an increase in cell elongation and a subsequent reduction in SCW thickening (Sasidharan et al., 2008, 2010; Casal, 2012; Wu et al., 2017). The molecular mechanism underlying this reduction in SCW thickening caused by shaded light conditions is not well defined. The R:FR photoreceptor, PHYTOCHROME B (PHYB), exists in two forms that are reversibly interconvertible through perception of red and far-red light (Quail, 1991). The Pr form of PHYB absorbs red light and rapidly reverts to the Pfr form, which absorbs far-red light to return to the Pr form (Franklin, 2008). In response to the R:FR ratio signal, interconversion of the PHYB Pr-Pfr forms activates downstream molecular pathways to regulate induction of seed germination, seedling de-etiolation, shade avoidance, and floral initiation (Somers et al., 1991; Poppe and Schafer, 1997; Franklin and Quail, 2010; Strasser et al., 2010).
Red light activates PHYB to interact with PHYTOCHROME INTERACTING FACTORs (PIFs; mainly a quartet of members: PIF1, PIF3, PIF4, and PIF5), leading to their degradation (Bauer et al., 2004; Monte et al., 2004; Al-Sady et al., 2006; Shen et al., 2007; Lorrain et al., 2008), whereas far-red light inactivates PHYB and stabilizes PIFs, inducing stem elongation and other morphogenesis processes (Hornitschek et al., 2012; Leivar et al., 2012).
The transcription factor (TF) MYC2 is considered to be a transcriptional regulatory hub that interconnects a variety of biological processes (Kazan and Manners, 2013). MYC2 interacts with different TFs to integrate the crosstalk among different signaling pathways, including jasmonate (JA)-mediated pathogen defenses and abscisic acid (ABA), ethylene, gibberellic acid (GA), and light signals (Chen et al., 2012; Hong et al., 2012; Song et al., 2014). The blue light signal upregulates expression of MYC2/MYC4, which then activates NST1-mediated SCW thickening by directly binding to the NST1 promoter (Zhang et al., 2018a). Here, we show that the low R:FR ratio under shaded light conditions inhibits SCW thickening in Arabidopsis inflorescence stems. Our analyses indicate that R:FR light is perceived by PHYB to alter the status of PIF4, which acts as a direct regulator of MYC2/MYC4 to modulate SCW thickening. This study reveals a molecular pathway that controls SCW thickening in response to the R:FR light ratio.
Results
Light R:FR ratio affects SCW thickening in the inflorescence stem of Arabidopsis
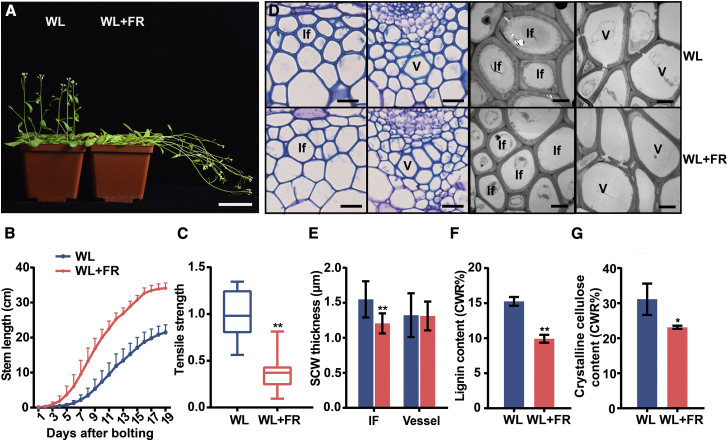
As our previous study showed that a blue light signal enhances SCW thickening in Arabidopsis inflorescence stems (Zhang et al., 2018a), we were interested in further examining the effect of light with a lower R:FR ratio, which imitates shaded light conditions, on SCW thickening during inflorescence stem growth. First, wild-type (WT) plants were grown to bolting in normal white light (WL) conditions. When the inflorescence stem started to grow, plants were transferred to light conditions with different R:FR ratios to monitor the growth of the inflorescence stem. The inflorescence stem grew faster (Figures 1A and 1B) and had a significantly lower tensile strength (Figure 1C) under WL supplemented with far-red light compared with WL. This result indicated that a low R:FR ratio affected both inflorescence stem growth and mechanical strength. Anatomical analyses of stem structure revealed that the SCW thickness in fiber cells was significantly decreased, but vessel cell SCW thickness showed little difference (Figures 1D and 1E). Expression of SCW thickening marker genes (NST1, SND1, 4CL1, and IRX8) (Lee et al., 1997; Zhong et al., 2006; Mitsuda et al., 2007; Hao et al., 2014) was downregulated under low R:FR conditions (Supplemental Figure 1). Contents of lignin and crystalline cellulose, typical SCW components, were significantly lower under low R:FR conditions (Figures 1F and 1G). These data suggest that the additional far-red light promotes stem elongation and inhibits SCW thickening in stem fiber cells.
Figure 1.
Shaded light inhibits SCW thickening in the inflorescence stem.
(A) Growth of Arabidopsis inflorescence stems in white-light (WL) and WL + far-red conditions. FR, far red. Scale bar, 5 cm.
(B) Elongation of inflorescence stems in WL and WL + far-red conditions during growth. n = 9, mean ± SD.
(C) Tensile strength of inflorescence stems; the WT stem tensile strength was set to 1. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, n = 18, mean ± SD.
(D) Cross sections of the inflorescence stem grown under different light conditions (WL and WL + far red) visualized under a light microscope (after toluidine blue staining; left panels) and a transmission electron microscope (right panels). If, interfascicular fiber cell; V, vessel cell. Left: scale bar, 20 μm; right: scale bar, 5 μm.
(E) Measurements of SCW thickness in the interfascicular fiber cells in (D). There were three biological replicates, and more than 10 cells were measured per biological replicate. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, mean ± SD.
(F) Lignin content in inflorescence stems of plants grown in different R:FR conditions. Student’s t-test (∗P < 0.05) was used for statistical analyses, n = 3, mean ± SD.
(G) Crystalline cellulose content in inflorescence stems of plants grown in different R:FR conditions. Student’s t-test (∗P < 0.05) was used for statistical analyses, n = 3, mean ± SD.
PHYB and PIFs are involved in regulating cell elongation and SCW thickening in the Arabidopsis inflorescence stem
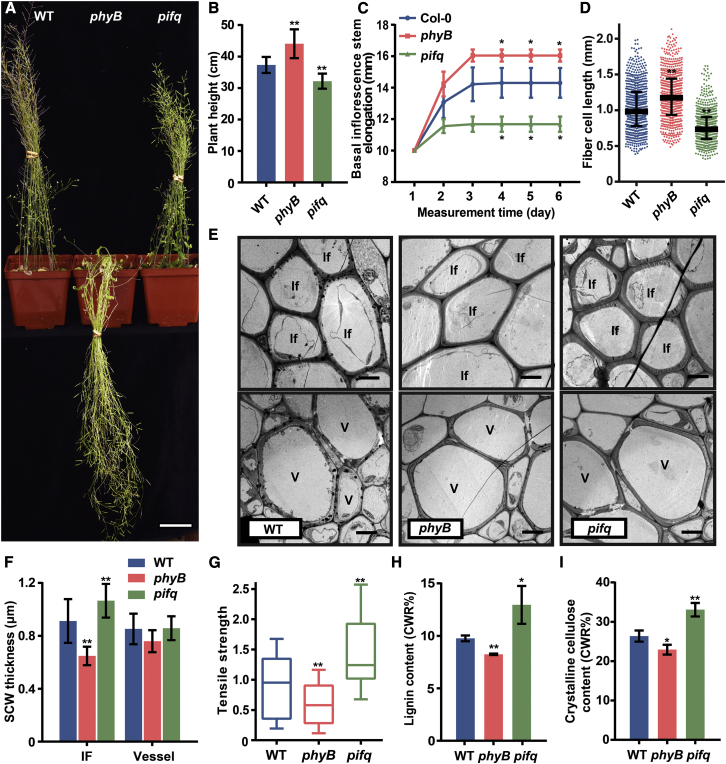
Red/far-red light is perceived by the photoreceptor PHYB, which induces a series of responses through PIF proteins (Reed et al., 1993; Pham et al., 2018). To dissect the genetic basis of the R:FR effect on SCW thickening, we analyzed the inflorescence stem growth of phyB and quadruple pif1pif3pif4pif5 (pifq) mutants. phyB mutants displayed a lodging phenotype and grew longer inflorescence stems than the WT, whereas pifq mutants showed erect growth with shorter inflorescence stems compared with the WT (Figures 2A and 2B and Supplemental Figure 2A). Stem elongation growth was determined by measuring the distance between two markers on the inflorescence stem during its growth. Elongation growth was increased in phyB but decreased in pifq mutant plants (Figure 2C). Mechanical properties of the inflorescence stem, measured as tensile strength, were also significantly affected: phyB inflorescence stems had decreased tensile strength, whereas the tensile strength of pifq mutant inflorescence stems was increased (Figure 2G). To analyze the cell length and cell wall structure of the inflorescence stem, stems were disintegrated to measure the length of xylem fibers, which are the predominant cell type in mature inflorescence stems. The fiber cells of phyB plants were longer, whereas those of pifq plants were shorter, relative to those in the WT (Figure 2D and Supplemental Figure 2B).
Figure 2.
PHYB and PIFs regulate SCW thickening in fiber cells.
(A) Plants were grown in WL at 8 weeks of age. Scale bar, 5 cm.
(B) Measurements of plant height in (A). Student’s t-test (∗∗P < 0.01) was used for statistical analyses, n = 20, mean ± SD.
(C) Inflorescence stem elongation. The stem was marked with two points at the basal region, and the distance between the two points was measured every day during growth. Student’s t-test (∗P < 0.05) was used for statistical analyses, n = 3, mean ± SD.
(D) Fiber cell length measured in disaggregated fiber cells. Data were collected from three biological replicates, and more than 200 cells were measured per biological replicate. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, mean ± SD.
(E) Transmission electron micrographs of inflorescence stem cross sections. If, interfascicular fiber cell; V, vessel cell. Scale bar, 5 μm.
(F) Measurements of SCW thickness in the interfascicular fiber cells and vessel cells in (E). Data were collected from three biological replicates, and more than five cells were measured per biological replicate. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, mean ± SD.
(G) Tensile strength measurements of the inflorescence stem; the WT stem tensile strength was set to 1. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, n = 16, mean ± SD.
(H) Lignin content in inflorescence stems. Student’s t-test (∗∗P < 0.01, ∗P < 0.05) was used for statistical analyses, n = 3, mean ± SD.
(I) Crystalline cellulose content in inflorescence stems. Student’s t-test (∗∗P < 0.01, ∗P < 0.05) was used for statistical analyses, n = 3, mean ± SD.
Cross sections of the stem showed a difference in the cell wall thickness of interfascicular fiber cells (Figures 2E and 2F and Supplemental Figure 2C). Compared with that in the WT, cell wall thickness in the interfascicular fiber cells was decreased in phyB but increased in pifq, whereas thickness of the vessel cells was unaltered (Figures 2E and 2F). Furthermore, both lignin and crystalline cellulose contents were decreased in phyB but increased in pifq (Figures 2H and 2I, respectively).
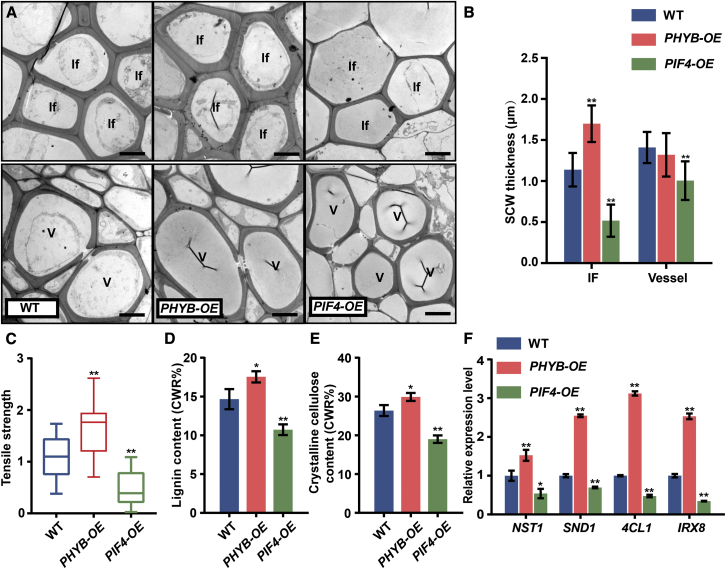
In parallel with the knockout mutant analyses, Arabidopsis plants overexpressing PHYB (PHYB-OE) and PIF4 (PIF4-OE) were also generated and their inflorescence stem properties analyzed. PHYB-OE transgenics had shorter and stronger (increased tensile strength) inflorescence stems, whereas those of PIF4-OE plants were thinner and weaker (Supplemental Figure 3 and Figure 3C). Examination of stem cross sections indicated that PHYB overexpression resulted in thicker cell walls of interfascicular fiber cells, whereas PIF4 overexpression led to thinner fiber cell walls relative to the WT (Figures 3A and 3B). Lignin and crystalline cellulose contents were increased in the PHYB-OE inflorescence stems but decreased in PIF4-OE stems (Figures 3D and 3E). By contrast, the cell wall thickness of vessel cells was largely unaffected in PHYB-OE plants but was decreased in PIF4-OE plants (Figures 3A and 3B). Transcriptional analyses of SCW thickening genes (NST1, SND1, 4CL1, and IRX8) demonstrated that their expression was upregulated in PHYB-OE plants but suppressed in PIF4-OE plants (Figure 3F). These results indicate that SCW thickening is positively regulated by PHYB but negatively regulated by PIFs in interfascicular fibers of inflorescence stems.
Figure 3.
SCW phenotypes of PHYB-OE and PIF4-OE plants.
(A) Transmission electron micrographs of inflorescence stem cross sections of plants grown in WL. If, interfascicular fiber cell; V, vessel cells. Scale bar, 5 μm.
(B) Measurements of SCW thickness of cells in (A). Data were collected from three biological replicates, and more than 10 cells were measured per biological replicate. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, mean ± SD.
(C) Tensile strength of inflorescence stems; the WT stem tensile strength was set to 1. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, n = 15, mean ± SD.
(D) Lignin content in inflorescence stems. Student’s t-test (∗P < 0.05) was used for statistical analyses, n = 3, mean ± SD.
(E) Cellulose content in inflorescence stems. Student’s t-test (∗∗P < 0.01, ∗P < 0.05) was used for statistical analyses, n = 3, mean ± SD.
(F) Expression of SCW regulatory (NST1 and SND1) and biosynthesis-related (4CL1 and IRX8) genes in different genotypes grown under WL. Three biological replicates were performed. Student’s t-test (∗∗P < 0.01, ∗P < 0.05) was used for statistical analysis, mean ± SD.
The PHYB-PIFs signal module links to MYC2 for SCW thickening
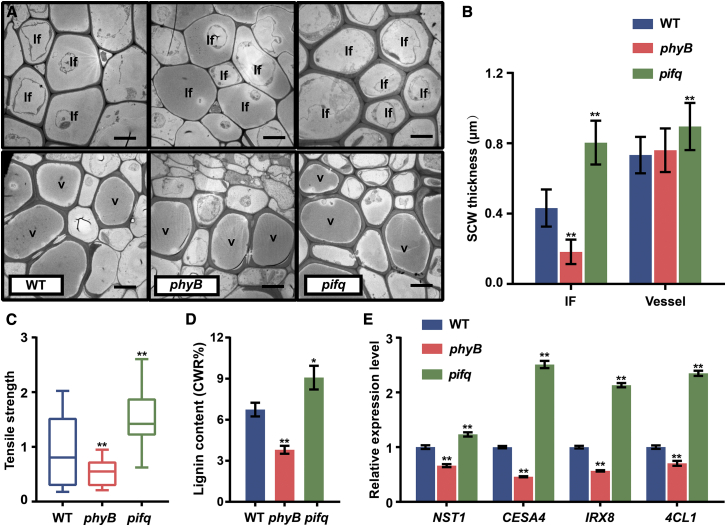
As shown above, SCW thickening is genetically regulated by PHYB and PIFs and also conditionally modified by light R:FR ratio. Next, we examined the effect of red light on SCW thickening. phyB and pifq mutants were grown under red light (high R:FR). The inflorescence stem of phyB mutant plants showed a significant decrease in stem tensile strength; SCW thickening was severely affected in fiber cells but was essentially unchanged in vessel cells (Figures 4A–4C). Conversely, the pifq mutants displayed increased stem tensile strength, accumulated much thicker SCWs in interfascicular fiber cells, and had a modest increase in vessel cells, compared with those in WT plants (Figures 4A–4C). Lignin content was decreased in the phyB mutant but increased in the pifq mutant (Figure 4D). Consistent with these findings, the expression of SCW thickening genes (NST1, SND1, 4CL1, and IRX8) was downregulated in the phyB mutant but upregulated in the pifq mutant (Figure 4E). These data suggest that high R:FR facilitates SCW thickening through PHYB-enhanced SCW gene expression, whereas PIFs inhibit this process. Analysis of gene expression showed that PHYB and PIF quadruple members are both expressed in inflorescence stems, and among them, PIF4 and PIF5 had the highest abundance (Supplemental Figure 4A). Also, PIF4 promoter-β-glucuronidase (GUS) analysis indicated its expression in interfascicular fiber cells (Supplemental Figure 4B).
Figure 4.
Red-light signaling regulates SCW thickening, dependent on PHYB and PIFs.
(A) The inflorescence stems of phyB and pifq mutants were grown under red light (high R:FR) and anatomically analyzed. Transmission electron micrographs of the inflorescence stem cross sections are shown. If, interfascicular fiber cell; V, vessel. Scale bar, 5 μm.
(B) Statistics of SCW thickness in (A). Data were collected from three biological replicates, and more than 10 cells were measured per biological replicate. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, mean ± SD.
(C) Tensile strength of the inflorescence stem; the WT stem tensile strength was set to 1. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, n = 20, mean ± SD.
(D) Lignin content in the inflorescence stem. Student’s t-test (∗∗P < 0.01, ∗P < 0.05) was used for statistical analyses, n = 3, mean ± SD.
(E) Expression of SCW regulatory (NST1) and biosynthesis-related (CESA4, 4CL1, and IRX8) genes was measured by qRT-PCR analysis. Analysis was performed on three biological replicates. Student’s t-test (∗∗P < 0.01) was used for statistical analysis, mean ± SD.
Next, we dissected the molecular pathway that connects the PHYB-PIFs signal to SCW thickening. Five-week-old WT plants at the stage of inflorescence stem elongation were moved from normal light to the dark for 24 h in order to shut down light-induced gene expression, then transferred to red-light (high R:FR) conditions for 2 h. Stem samples were collected for RNA sequencing (see Supplemental Figure 5A). Compared with 24 h of darkness, a group of 2203 differentially expressed genes (DEGs) were detected after red-light treatment, among which two-thirds were upregulated and one-third were downregulated (Supplemental Table 1; Supplemental Figure 4B). The regulated genes included red-light-responsive photopigment genes (PSY, PORC, GUN5) and target genes of PIFs (PIL1, ATHB2, BBX28) (Zhang et al., 2013; Toledo-Ortiz et al., 2014), indicative of the red-light signaling effectiveness. Expression of cell expansion genes (XTH22, XTH27, XTH30, EXPA1) was inhibited, but SCW-thickening-related regulatory genes, including NST1 and MYC2, were upregulated in response to red light (Supplemental Figure 5C) (Matsui et al., 2005; Claisse et al., 2007; Mitsuda et al., 2007; Goh et al., 2012; Zhang et al., 2018a). Interestingly, expression of VND6/VND7, which control vessel SCW thickening, was not induced by red light (Supplemental Table 1) (Kubo et al., 2005). The red-light induction of NST1, MYC2, and other key genes for SCW thickening was further confirmed by RT-qPCR analysis (Supplemental Figure 6). Conversely, after exposure to far-red light conditions, MYC2 and NST1 expression was inhibited in WT plants; however, this inhibition was reduced in phyB and pifq mutants (Supplemental Figure 7). These results suggest that expression of MYC2 is induced by red light, inhibited by far-red light, and regulated through PHYB and PIFs.
PIF4 affects MYC2 stability and inhibits its transcriptional activity
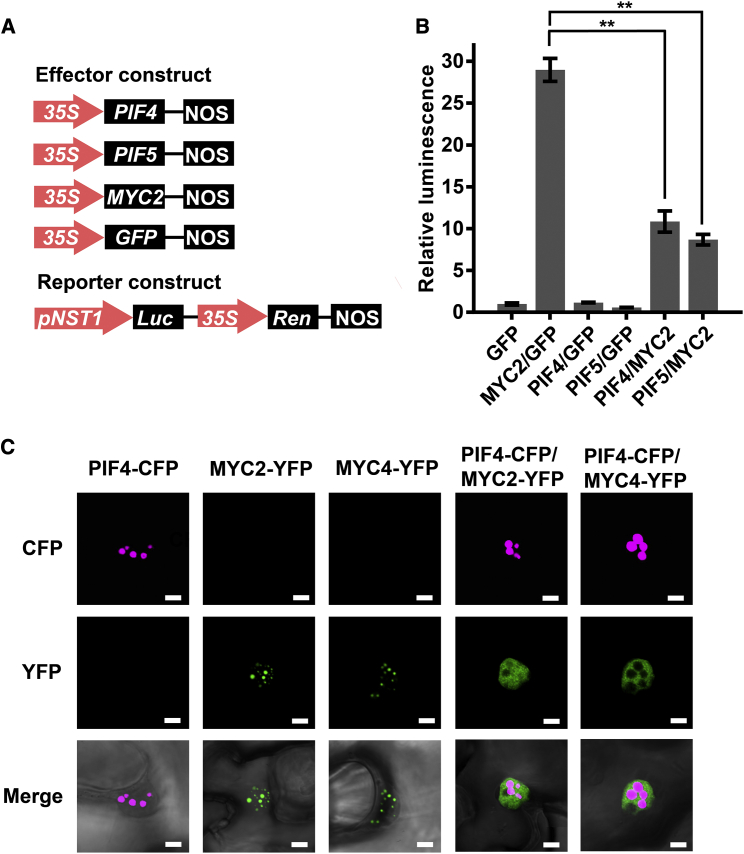
MYC2 and MYC4 are known to redundantly regulate SCW thickening through their binding to the NST1 promoter (Zhang et al., 2018a). We therefore analyzed how PIFs affect MYC2 transcriptional activity on the NST1 promoter. PIF4, PIF5, and MYC2 were expressed using a dual-luciferase (LUC) reporter assay in Arabidopsis protoplasts. MYC2, but not PIF4 or PIF5, was able to activate the NST1 promoter (Figures 5A and 5B). When MYC2 was co-expressed with either PIF4 or PIF5, MYC2 activity on the NST1 promoter was reduced (Figure 5B), suggesting that MYC2 transcriptional activity is repressed by either PIF4 or PIF5.
Figure 5.
PIF4 represses MYC2 transcriptional activity.
(A) Schematic representation of the NST1 promoter-driven dual-LUC reporter gene and three effector gene constructs. 35S promoter, NST1 promoter (−1 to −3711 bp from ATG), Renilla luciferase (REN), and firefly luciferase (LUC) are indicated in reporter constructs. In effector constructs, PIF4, PIF5, and MYC2 are driven by the 35S promoter.
(B) PIF4/PIF5 inhibit MYC2 activation of the NST1 promoter. Arabidopsis protoplasts were transfected with the reporter constructs in combination with different effector constructs. After transfection, the protoplasts were kept in the dark for 16 h. Relative luminescence was normalized to that of protoplasts transformed with the reporter and empty effector (GFP). Tukey’s honestly significant difference (HSD) test (∗∗P < 0.01) was used for statistical analysis, n = 3, mean ± SD.
(C) Subcellular localization of PIF4 and MYC2/MYC4. Constructs of PIF4-CFP, MYC2-YFP, and MYC4-YFP were transferred to tobacco leaves, separately or together, by agroinfiltration. The tobacco leaves were then kept in the dark for 12 h before fluorescence observation. Scale bar, 5 μm.
Although previous studies have reported interactions between PIFs and MYC2 (Zhang et al., 2018b; Zhao et al., 2021), we carried out pulldown and yeast two-hybrid assays for verification in our study. PIF4 was able to pull down MYC2 (Supplemental Figure 8A), and its N terminus may mediate its interaction with MYC2 and MYC4 (Supplemental Figures 8B and 8C).
Then, the subcellular localization of MYC2 and MYC4 along with PIF4 was examined. Tobacco leaf cells were used to express MYC2/MYC4-YFP and PIF4-CFP proteins and then exposed to dark conditions. When they were expressed separately, PIF4 and MYC2/MYC4 proteins were localized in the nucleus, visible in the shape of distinct dots. However, when PIF4 was co-expressed with either MYC2 or MYC4, the nuclear localization of PIF4 was unchanged, whereas MYC2 and MYC4 became diffused in the nucleus (Figure 5B). These results suggest that PIF4 affects MYC2/MYC4 nuclear localization and/or stability.
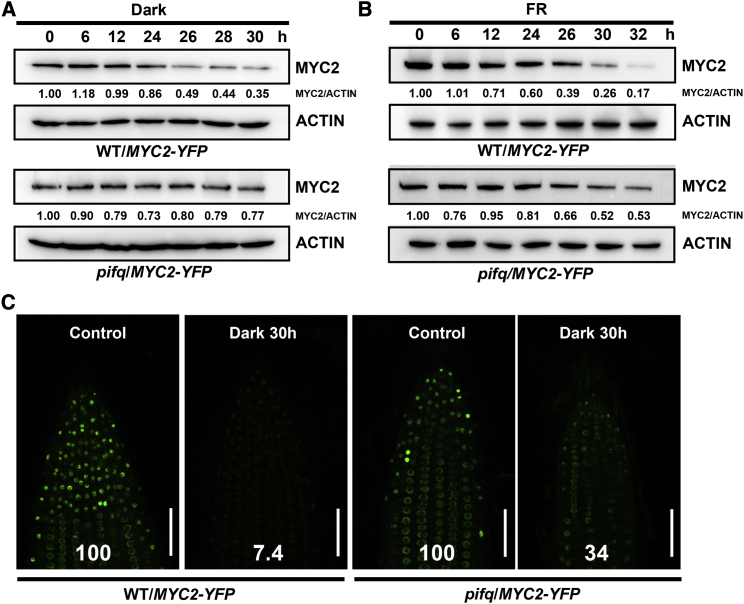
Next, MYC2 protein stability was tested in planta. MYC2-YFP was expressed in WT and pifq mutant plants. Immunoblot analysis showed that MYC2 abundance was reduced in the transgenic plants under both dark and far-red light conditions, and MYC2 was more stable in the pifq mutant than in the WT background (Figures 6A and 6B). The MYC2-YFP fluorescence signal was also observed in root tip cells. After dark treatment, the signal density was substantially decreased in the WT background, but this decrease was mitigated in the pifq mutant background (Figure 6C). These results suggest that PIFs affect MYC2 protein stability in planta.
Figure 6.
PIF4 affects MYC2 stability in the dark and in far-red light. MYC2 stability was examined in the WT and pifq backgrounds.
(A) Transgenic plants (WT/MYC2-YFP and pifq/MYC2-YFP) were grown in WL conditions for 10 days and then exposed to dark conditions. MYC2 and ACTIN were immunoblotted. Normalized MYC2 abundance relative to ACTIN is shown as MYC2/ACTIN.
(B) Transgenic plants (WT/MYC2-YFP and pifq/MYC2-YFP) were grown in WL conditions for 10 days and then exposed to far-red light. MYC2 and ACTIN were immunoblotted. Normalized MYC2 abundance relative to ACTIN is shown as MYC2/ACTIN.
(C) Transgenic plants (WT/MYC2-YFP and pifq/MYC2-YFP) were grown in WL conditions for 4 days and then treated with/without darkness for 30 h. YFP fluorescence was observed in the root tip, and the relative change in signal intensity was quantified. Scale bar, 50 μm.
MYC2/MYC4 act downstream of PHYB and PIFs in a genetic pathway to regulate stem SCW thickening
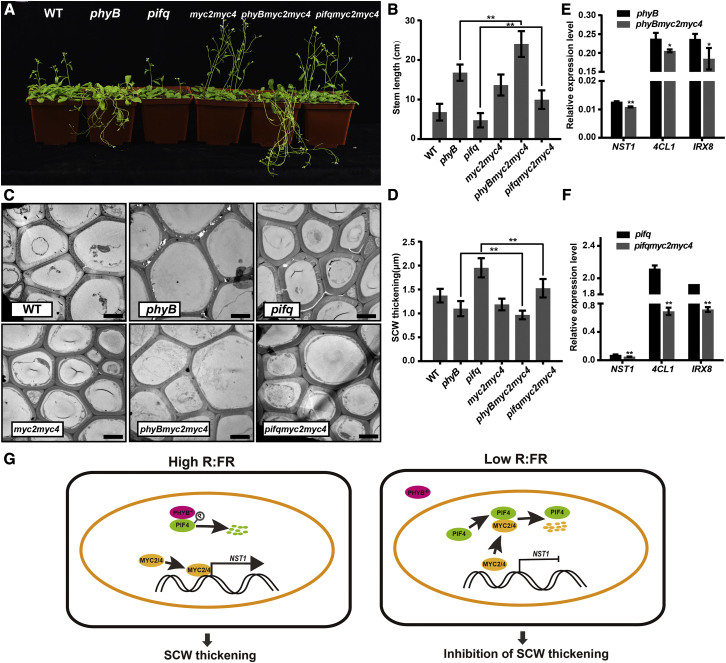
Next, myc2myc4 double mutants were crossed with pifq and phyB mutants to test whether MYC2/MYC4 act in the same genetic pathway with PHYB and PIFs in the regulation of SCW thickening. The pifq phenotype of a shorter inflorescence stem relative to the WT (Figures 2A and 2B) was partially restored in the sextuple pifqmyc2myc4 mutant under WL (Figures 7A and 7B). Consistent with this result, both fiber cell thickness (Figures 7C and 7D) and expression of SCW thickening-related genes such as NST1, 4CL1, and IRX8 were decreased in pifqmyc2myc4 plants compared with pifq (Figures 7E and 7F). Furthermore, rosette leaf growth and hypocotyl elongation, which were inhibited in pifq (Leivar and Quail, 2011), were rescued in pifqmyc2myc4 plants (Supplemental Figures 9A, 9D, and 9E). These results indicate that MYC2/MYC4 are genetically downstream of the PHYB-PIFs signaling module. Nevertheless, the faster stem elongation and thinner SCW phenotypes of phyB mutants were enhanced in phyBmyc2myc4 triple mutants (Figures 7A–7D), and expression of SCW regulatory and biosynthesis-related genes was slightly downregulated in the triple mutants compared with that in phyB (Figure 7E). In addition, phyBmyc2myc4 mutants exhibited greater hypocotyl elongation than phyB seedlings (Supplemental Figures 9B and 9C). Together, these data indicate that the PHYB-PIFs signaling module affects SCW thickening in the inflorescence stem of Arabidopsis. MYC2/MYC4 act downstream of PIFs to modify the transcriptional network that regulates SCW thickening.
Figure 7.
MYC2/MYC4 genetically interact with PIFs.
(A) Mutant plants (phyB, pifq, myc2myc4, phyBmyc2myc4, pifqmyc2myc4) were grown in WL until 4 weeks of age. Scale bar, 5 cm.
(B) Inflorescence stem length in various mutants. Tukey’s HSD test (∗∗P < 0.01) was used for statistical analysis, n > 10, mean ± SD.
(C) Transmission electron micrographs of stem cross sections showing interfascicular fiber cells. Scale bar, 5 μm.
(D) Statistics of SCW thickness in interfascicular fiber cells in (C). Data were collected from three biological replicates, and more than 10 cells were measured per biological replicate. Tukey’s HSD test (∗∗P < 0.01) was used for statistical analysis, mean ± SD.
(E and F) Expression of the key SCW regulatory (NST1) and biosynthesis-related (4CL1 and IRX8) genes in mutant plants. Analysis was performed on three biological replicates. Student’s t-test (∗∗P < 0.01, ∗P < 0.05) was used for statistical analysis, mean ± SD.
(G) WL enhances SCW thickening in fiber cells of the inflorescence stem. Under WL (high R:FR), PHYB is activated to its Pfr form, which enters the nucleus to inhibit PIF activity, and MYC2 is available to bind to the NST1 promoter to activate the NST1-directed SCW thickening process. In the shade (low R:FR), PHYB reverts to its inactive Pr form. PHYB cannot enter the nucleus, and PIF proteins interact with MYC2, displacing its binding to the NST1 promoter. Thus, the NST1-directed SCW thickening process is suppressed.
Discussion
Cell elongation and SCW thickening in the Arabidopsis inflorescence stem are coordinately regulated under shaded light
Light plays a key role in growth and development throughout the entire plant life cycle. Plants grow longer inflorescences with weaker stem strength and thinner SCWs under shaded conditions (Kozuka et al., 2010; Casal, 2012; Wu et al., 2017). In the stem, SCW thickening in xylem cells is regulated by developmental signals as well as environmental conditions such as light (Didi et al., 2015; Zhang et al., 2018a; Huang et al., 2018; Hori et al., 2020); however, the mechanisms by which shaded conditions modulate SCW thickening and mechanical properties are not well defined.
A characteristic feature of shaded light conditions is the decreased R:FR light ratio (Hersch et al., 2014). Using the Arabidopsis inflorescence stem as a model, we examined the effect of R:FR ratio on SCW thickening of interfascicular fibers and vessel cells, the major cell types with SCWs in the inflorescence stem. A low R:FR light ratio induced rapid elongation of the inflorescence stem, resulting in a lodging phenotype, consistent with the reported effect of shaded light on stem growth (Casal, 2012) and indicating that a low R:FR light ratio can be used as an experimental proxy to simulate shaded light conditions. Anatomical analyses revealed that shaded light resulted in plants with thinner SCWs in stem fiber cells and weaker mechanical strength of the inflorescence stem. Expression of SCW regulatory and biosynthesis-related genes was downregulated. Many plants respond to low R:FR light with the shade avoidance syndrome (SAS), displaying a series of well-described morphological changes such as enhanced elongation of the hypocotyl, internode, and petiole (Liu et al., 2021). This study indicates that SCW thickening and cell elongation in the stem are tightly coordinated and regulated by shaded light conditions.
PHYB-PIFs signaling module mediates regulation of SCW thickening in fiber cells of the Arabidopsis inflorescence stem
In Arabidopsis inflorescence stems, SCW formation in the fiber and vessel cells is initiated under the control of the master TF switches NST1/SND1 (Zhong et al., 2006; Mitsuda et al., 2007) and VND6/VND7 (Kubo et al., 2005; Yamaguchi et al., 2010), respectively. NST1/SND1 and VND6/VND7 regulate several downstream genes to control SCW thickening (Taylor-Teeples et al., 2015) through highly cell-type-specific spatio-temporal regulation modulated by distinct regulatory signals (Zhong et al., 2006, 2008; Zhu and Li, 2021). To examine how shade affects SCW thickening, inflorescence stems were exposed to low R:FR light after the plants had grown to bolting. Low R:FR light resulted in thinner SCWs in interfascicular fiber cells of the inflorescence stems, whereas the vessel SCWs were largely unaffected.
Shaded light inactivates PHYB and subsequently diminishes its action on the degradation of PIFs, leading to regulation of downstream genes to modulate plant growth and development (Lorrain et al., 2008; Jia et al., 2020). Whether PHYB and PIFs are directly involved in the regulation of SCW thickening by shaded light is unresolved. Here, we examined the effect of PHYB and PIFs on cell wall formation in the Arabidopsis inflorescence stem. Monitoring of inflorescence stem growth indicated that the phyB and pifq mutants have contrasting phenotypes of SCW thickening and mechanical strength properties in the inflorescence stem. Dissection of cell wall properties in the inflorescence stem also indicated that the phyB and pifq mutations affect SCW thickening primarily in the fiber cells but have little effect on the vessel cells. Interestingly, the blue light signal dramatically affects SCW thickening in fiber cells with little effect on vessel cells (Zhang et al., 2018a). It appears that light-regulated SCW thickening occurs mainly in fibers rather than vessel cells. Nevertheless, the evidence here indicates that the process of SCW thickening is positively regulated by PHYB and negatively regulated by PIFs.
Shaded light inhibits SCW thickening through PIF4 inactivation of MYC2 activity
The CRY1 signaling of blue light induces MYC2 and MYC4 expression and subsequently activates the NST1-directed transcriptional network to enhance SCW thickening in the inflorescence stem (Zhang et al., 2018a). In this study, experimental evidence demonstrates the involvement of the PHYB-PIFs signaling module in regulation of SCW thickening. Naturally, the question of how the two light signals are joined in the SCW thickening regulatory pathway should be addressed. Our results showed that PIFs are able to interact with MYC2 and MYC4, supporting the interaction reported in other studies (Zhang et al., 2018b; Zhao et al., 2021). PIF4, MYC2, and MYC4 proteins were observed as distinct dots in the nucleus when they were expressed individually, similar to observations reported previously (Withers et al., 2012; Luo et al., 2014). However, when PIF4 was co-expressed with MYC2 or MYC4, PIF4 localization in the nucleus showed no change, but the dotted localization of MYC2 and MYC4 became diffused in the nucleus. That is, interaction of PIF4 with MYC2/MYC4 altered the localization status of the latter in the nucleus. On the other hand, MYC2 in plants is destabilized under far-red light and dark conditions in the WT but is more stable in the pifq mutant. It is likely that the PHYB-PIFs signaling module affects MYC2 protein stability. MYC2 and MYC4 are known to bind directly to the NST1 promoter to enhance SCW thickening (Zhang et al., 2018a). Evidence in this study revealed that the interaction of PIF4 with MYC2 resulted in inhibition of MYC2 activity on NST1 transcription. Thus, CRY1 signaling and PHYB-PIFs signaling join at MYC2, probably together with MYC4, as previous and present genetic evidence indicates that MYC2 and MYC4 are redundantly involved in light-regulated SCW thickening of the Arabidopsis inflorescence stem (Zhang et al., 2018a). MYC2 is considered to be a transcriptional regulatory hub that interconnects a variety of biological processes in growth and environmental responses (Yadav et al., 2005; Chen et al., 2012; Kazan and Manners, 2013; Song et al., 2014). Our study reveals that this transcriptional regulatory hub acts to connect light signaling with the transcriptional regulatory networks for SCW thickening. PHYB-PIFs signaling modifies MYC2/MYC4 activity on NST1 transcription, possibly through interaction with PIF4 to affect MYC2/MYC4 stability; however, more details of the mechanism by which PIF4 regulates MYC2/MYC4 activity in SCW thickening remain to be elucidated.
To summarize the findings of this study, we propose a model for the shaded light inhibition of SCW thickening (Figure 7G). Under normal light conditions (high R:FR ratio), PHYB undergoes a light-dependent conformational change and translocation to the nucleus to promote phosphorylation and degradation of PIFs (Chen et al., 2005), thus resulting in MYC2/MYC4 stabilization and NST1 transcriptional activation to enhance SCW thickening. Upon perception of shaded light (low R:FR ratio), PHYB undergoes reversion from Pfr to its inactivated Pr form and returns to its cytoplasmic location (Franklin, 2008), allowing accumulation of PIFs in the nucleus where they interact with MYC2/MYC4. Such interaction inhibits MYC2/MYC4 activity on NST1 transcription, leading to the suppression of SCW thickening (Figure 7G). These findings reveal a mechanism by which shaded light inhibits SCW thickening.
This study shows that regulation of SCW thickening is coordinated with cell elongation in stem growth, and such coordination is accurately regulated by light signaling modules. The finding has important implications for our understanding of plant growth and development and suggests a pathway by which to begin modifying the mechanical SCW properties that are crucial to both forestry (wood and wood product properties) and agricultural industries (crop improvement, for example, through the prevention of lodging).
Materials and methods
Plant materials and growth conditions
The Arabidopsis WT ecotype used in this study is in the Columbia-0 (Col-0) background. The phyB, pifq, PIF4-OE, PHYB-OE, and myc2myc4 plants were generated as described previously (Wang et al., 2010; Jia et al., 2014; Ma et al., 2016; Zhang et al., 2018a). The phyBmyc2myc4 and pifqmyc2myc4 mutant plants were generated by crossing myc2myc4 with either phyB or pifq and were genotyped using primers listed in Supplemental Table 2. To generate WT/35S:MYC2-YFP and pifq/35S:MYC2-YFP plants, the MYC2 coding sequence was cloned into the pHB-X-YFP vector and transferred to either WT or pifq Arabidopsis by the floral dip method with Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998). Plants were grown in a phytotron with a light (fluorescent lamp, 80 μE/s·m2)/dark cycle of 16 h/8 h at 22°C. For light treatment of inflorescence stem growth, plants were grown under WL until bolting and then transferred to various light conditions. A red-light condition was achieved with a light-emitting diode (LED) light incubator (Percival E-30LED; red-light wavelength = 670 nm, 30 μE/s·m2). WL (high R:FR) was provided by a WL LED with an R:FR ratio of 13 (Qiding Technology, 67 μE/s·m2). WL + far-red (low R:FR) treatment was achieved using supplemental far-red LEDs (Qiding Technology, far-red light wavelength = 720–750 nm, 47 μE/s·m2) to a R:FR ratio of 0.066. Far-red light was provided by far-red LEDs (Qiding Technology, 47 μE/s·m2). All light parameters were measured with an ILT1400 Radiometer Photometer and an Ocean Optics HR2000+CG spectrophotometer.
Stem section analysis
Arabidopsis inflorescence stems grew to about 11 cm in length. One centimeter of the inflorescence stem was sampled 0.5 cm from the base and used for analysis.
Paraffin sections were prepared as previously described (Zhang et al., 2018a). The stem sample was cut into a 5-mm segment and fixed in formalin-aceto-alcohol (FAA) solution under vacuum, stored at 4°C overnight, dehydrated in a graded ethanol series, and embedded into paraffin. Samples were sectioned to a 10-μm thickness using a Leica RM2235 rotary microtome. The sections were then stained with toluidine blue and observed under a light microscope (Olympus, BX53).
For transmission electron microscope analysis of cell wall thickness, the sample was cut into 2-mm segments, fixed in 3% (v/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde in PBS (0.1 M, pH 7.4), dehydrated in a graded ethanol series, embedded in Epon812, and sectioned. The samples were stained with 2% (w/v) uranyl acetate and lead citrate and observed under a transmission electron microscope (Hitachi H-7650) as previously described (Zhao et al., 2014). SCW thickness was measured with ImageJ software (https://imagej.nih.gov/ij/download.html).
Fiber cell length measurement
The basal part of the inflorescence stem was cut into a 2-cm length and disaggregated by submerging in a glacial acetic acid/30% hydrogen peroxide (v/v 1:1) solution at 60°C overnight, stained with 1% (w/v) safranine for 10 min, and photographed under a light microscope (Olympus, BX53). Fiber cell length was measured with ImageJ software.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from different tissues of Arabidopsis plants using the E.Z.N.A. Plant RNA Kit (Omega, R6827-02). First-strand cDNA was synthesized using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, AT311-03) for real-time quantitative reverse transcription PCR (qRT-PCR) analysis of transcript abundance. qRT-PCR was performed using SYBR Green (TransStart Tip Green qPCR supermix) with a QuantStudio 3 Real-Time PCR System (Applied Biosystems). Gene expression was normalized using ACT2 as an internal control.
Analysis of stem tensile strength and cell wall components
The basal part of the inflorescence stem was used for tensile strength measurement as previously described (Zhang et al., 2018a). Relative tensile strength was normalized against that of the WT. For analysis of cell wall components, the basal part of the inflorescence stem was collected and analyzed as described by Xi et al. (2017). Collected stems were ground to a fine powder in liquid nitrogen. Alcohol-insoluble residue (AIR) was obtained by successively washing the powder with 70% (v/v) ethanol, chloroform/methanol (1:1 v/v), and acetone (Pettolino et al., 2012). After de-starching, AIR was washed with water and acetone, then dried for determination of lignin and crystalline cellulose content.
Immunoblotting
Proteins extracted from transgenic plants were separated with 10% (w/v) SDS–PAGE gels and blotted onto polyvinylidene fluoride membranes (Bio-Rad). Protein blots were then analyzed using either anti-GFP/Myc (1:2000 dilution; Abmart) or anti-ACTIN (1:2000 dilution; Abmart) monoclonal antibodies, followed by horseradish peroxidase (HRP)-conjugated goat-anti-mouse antibodies (1:5000 dilution, Thermo Fisher). Blots were developed in a Tanon Imaging System (Tanon 5200CE) using ECL Western Blotting Substrate (Tanon, 180-501). Protein level was measured with ImageJ software.
Protein subcellular localization
For protein subcellular localization analyses, the MYC2/MYC4 and PIF4 coding sequences were PCR amplified with the proofreading enzyme PHANTA (Vazyme, P520) from cDNA and cloned into pHB-X-YFP and pHB-X-CFP vectors in frame with the YFP/CFP (Luo et al., 2014), respectively. The constructs were then agroinfiltrated into Nicotiana benthamiana leaves according to the method of Gui et al. (2016). After incubation for 48 h in the dark, abaxial epidermal cells of the leaf were observed under a confocal microscope (Leica TSC SP8 STED 3X).
Confocal microscopy and fluorescence quantification
Seeds of MYC2-GFP overexpression lines were sterilized in 10% (v/v) bleach and imbibed in sterile water for 2 days at 4°C in the dark, then placed on Murashige and Skoog (MS) medium containing 0.8% (w/v) agar. Plates were incubated in long day (LD) conditions for 4 days, followed by dark treatment. Confocal imaging of fluorescence signals in seedling roots was performed on a Leica TCS SP8 equipped with 514-nm (YFP) lasers. The laser power was set to 20% for fluorescence imaging, and fluorescence intensity in the confocal microscopy images was measured with ImageJ software.
Yeast two-hybrid assay
The coding sequences of MYC2 and MYC4 were PCR amplified from cDNA using PHANTA and fused with the GAL4 DNA-binding domain (BD) of the bait vector pGBKT7 (Clontech). The N terminus containing transcription activation domain (TAD) and the C terminus containing bHLH of PIF4 were PCR amplified and fused with the GAL4 activation domain (AD) of the prey vector pGADT7 (Clontech). Bait and prey vectors were co-transformed into the Y2H Gold yeast strain, then grown on SD-Trp-Leu and SD-Trp-Leu-His plates (Clontech).
Protein pulldown assay
Recombinant MYC2-His and GST-PIF4 proteins were expressed in Escherichia coli and purified with Ni-NTA agarose (Qiagen) and Pierce GST agarose (Abclonal) according to the manufacturers’ instructions. Purified GST and GST-PIF4 were first incubated on the GST agarose (Abclonal) for 2 h at 4°C. Purified MYC2-His proteins were added and incubated for another 2 h at 4°C. After washing, the proteins were eluted with 2× SDS loading buffer and boiled at 100°C for 10 min. Proteins were then separated by SDS–PAGE and detected with anti-GST (Cell Signaling Technology, Inc [CST]) or anti-HIS (CST) antibodies.
PIFs and MYC2 transcriptional activity assay
The interaction of MYC2 and PIF4 activity was assayed using a dual-LUC reporter assay system (Promega) through Arabidopsis protoplast transfection. The PIF4/PIF5/MYC2 and GFP coding sequences were cloned into the pA7 vector under the control of the 35S promoter and used as an effector. The NST1 promoter sequence (−1 to −3711 bp from ATG) was cloned into the pGreenII 0800-LUC vector upstream and in frame with the luciferase (LUC) gene and was used as a reporter. The renilla luciferase (REN) gene in the pGreenII0800-LUC vector was used as an internal control. Protoplasts from Arabidopsis mesophyll cells were isolated and transformed as described previously (Zhang et al., 2018a).
Promoter-GUS activity analysis
The PIF4 promoter (−1 to −4312 bp from ATG) was cloned into the pORE-R2 vector (Fang et al., 2021) to drive GUS expression, and the promoter construct was transformed into WT Arabidopsis by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS medium containing 50 μg/ml hygromycin and then genotyped. Positive T2 transgenic plants were used for analysis of GUS staining activity as described in Gui et al. (2016).
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 32130072, 31630014) and the Chinese Academy of Sciences (grant no. XDB27020104).
Author contributions
F.L., Q.Z., H.L., H.Y., and L.L. designed the research. F.L. and Q.Z. performed the experiments. F.L., Q.Z., H.L., H.Y., M.S.D., A.B., and L.L. analyzed the data. F.L., Q.Z., M.S.D., A.B., and L.L. wrote the paper. All authors read and approved the article.
Acknowledgments
We thank Dr. Daoxin Xie (Tsinghua University) for providing the myc2 myc4 mutants. We thank Ni Fan and Dr. Xiaoshu Gao for their help with confocal microscopy. We thank Xiaoyan Gao, Zhiping Zhang, Jiqin Li, and Ling Ge for assistance with transmission electron microscopy. M.S.D. and A.B. would like to acknowledge the support of funds from La Trobe University and the Chinese national and provincial governments to the Sino-Australia Cell Wall Research Centre, Zhejiang Agriculture and Forestry University (ZAFU). No conflict of interest is declared.
Published: August 4, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
Sequence data from this article can be found at https://www.arabidopsis.org with the following accession numbers: PHYB (AT2G18790), PIF1 (AT2G20180), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), MYC2 (AT1G32640), MYC4 (AT4G17880), NST1 (AT2G46770), SND1 (AT1G32770), VND6 (AT5G62380), VND7 (AT1G71930), MYB103 (AT1G63910), CESA4 (AT5G44030), IRX8 (AT5G54690), 4CL1 (AT1G51680), LAC4 (AT2G38080), PER64 (AT5G42180), F5H (AT4G36220), PIL1 (AT2G46970), ATHB2 (AT4G16780), BBX28 (AT4G27310), PSY (AT5G17230), PORC (AT1G03630), GUN5 (AT5G13630), XTH27 (AT2G01850), XTH22 (AT5G57560), XTH30 (AT1G32170), EXPA1 (AT1G69530), and ACT2 (AT3G18780).
Supplemental information
References
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C.S., Adám E., Fejes E., Schäfer E., et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J. Shade avoidance. Arabidopsis Book. 2012;10:e0157. doi: 10.1199/tab.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Tao Y., Lim J., Shaw A., Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 2005;15:637–642. doi: 10.1016/j.cub.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell. 2012;24:2898–2916. doi: 10.1105/tpc.112.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claisse G., Charrier B., Kreis M. The Arabidopsis thaliana GSK3/Shaggy like kinase AtSK3-2 modulates floral cell expansion. Plant Mol. Biol. 2007;64:113–124. doi: 10.1007/s11103-007-9138-y. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Didi V., Jackson P., Hejátko J. Hormonal regulation of secondary cell wall formation. J. Exp. Bot. 2015;66:5015–5027. doi: 10.1093/jxb/erv222. [DOI] [PubMed] [Google Scholar]
- Doblin M.S., Pettolino F., Bacic A. Plant cell walls: the skeleton of the plant world. Funct. Plant Biol. 2010;37:357–381. doi: 10.1071/Fp09279. [DOI] [Google Scholar]
- Fang Q., Zhou F., Zhang Y., Singh S., Huang C.F. STOP1 degradation mediated by the F-box proteins RAH1 and RAE1 balances aluminum resistance and plant growth in Arabidopsis thaliana. Plant J. 2021 doi: 10.1111/tpj.15181. [DOI] [PubMed] [Google Scholar]
- Franklin K.A. Shade avoidance. New Phytol. 2008;179:930–944. doi: 10.1111/j.1469-8137.2008.02507.x. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh H.H., Sloan J., Dorca-Fornell C., Fleming A. Inducible repression of multiple expansin genes leads to growth suppression during leaf development. Plant Physiol. 2012;159:1759–1770. doi: 10.1104/pp.112.200881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J., Zheng S., Liu C., Shen J., Li J., Li L. OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell. 2016;38:201–213. doi: 10.1016/j.devcel.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Hao Z., Avci U., Tan L., Zhu X., Glushka J., Pattathil S., Eberhard S., Sholes T., Rothstein G.E., Lukowitz W., et al. Loss of Arabidopsis GAUT12/IRX8 causes anther indehiscence and leads to reduced G lignin associated with altered matrix polysaccharide deposition. Front. Plant Sci. 2014;5:357. doi: 10.3389/fpls.2014.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M., Lorrain S., de Wit M., Trevisan M., Ljung K., Bergmann S., Fankhauser C. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:6515–6520. doi: 10.1073/pnas.1320355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 2012;24:2635–2648. doi: 10.1105/tpc.112.098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori C., Yu X., Mortimer J.C., Sano R., Matsumoto T., Kikuchi J., Demura T., Ohtani M. Impact of abiotic stress on the regulation of cell wall biosynthesis in Populus trichocarpa. Plant Biotechnol. 2020;37:273–283. doi: 10.5511/plantbiotechnology.20.0326a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- Huang C., Zhang R., Gui J., Zhong Y., Li L. The receptor-like kinase AtVRLK1 regulates secondary cell wall thickening. Plant Physiol. 2018;177:671–683. doi: 10.1104/pp.17.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K.-P., Luo Q., He S.-B., Lu X.-D., Yang H.-Q. Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol. Plant. 2014;7:528–540. doi: 10.1093/mp/sst093. [DOI] [PubMed] [Google Scholar]
- Jia Y., Kong X., Hu K., Cao M., Liu J., Ma C., Guo S., Yuan X., Zhao S., Robert H.S., et al. PIFs coordinate shade avoidance by inhibiting auxin repressor ARF18 and metabolic regulator QQS. New Phytol. 2020;228:609–621. doi: 10.1111/nph.16732. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. MYC2: the master in action. Mol. Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- Kozuka T., Kobayashi J., Horiguchi G., Demura T., Sakakibara H., Tsukaya H., Nagatani A. Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 2010;153:1608–1618. doi: 10.1104/pp.110.156802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall H., Philippe F., Domon J.M., Gillet F., Pelloux J., Rayon C. Cell wall metabolism in response to abiotic stress. Plants. 2015;4:112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Meyer K., Chapple C., Douglas C.J. Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell. 1997;9:1985–1998. doi: 10.1105/tpc.9.11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Cohn M.M., Monte E., Al-Sady B., Erickson E., Quail P.H. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell. 2012;24:1398–1419. doi: 10.1105/tpc.112.095711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Jafari F., Wang H. Integration of light and hormone signaling pathways in the regulation of plant shade avoidance syndrome. aBIOTECH. 2021;2:131–145. doi: 10.1007/s42994-021-00038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Luo Q., Lian H.L., He S.B., Li L., Jia K.P., Yang H.Q. COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell. 2014;26:2441–2456. doi: 10.1105/tpc.113.121657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., Noel J.P., Liu H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA. 2016;113:224–229. doi: 10.1073/pnas.1511437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A., Yokoyama R., Seki M., Ito T., Shinozaki K., Takahashi T., Komeda Y., Nishitani K. AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J. 2005;42:525–534. doi: 10.1111/j.1365-313X.2005.02395.x. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., Ohme-Takagi M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16091–16098. doi: 10.1073/pnas.0407107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettolino F.A., Walsh C., Fincher G.B., Bacic A. Determining the polysaccharide composition of plant cell walls. Nat. Protoc. 2012;7:1590–1607. doi: 10.1038/nprot.2012.081. [DOI] [PubMed] [Google Scholar]
- Pham V.N., Kathare P.K., Huq E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018;176:1025–1038. doi: 10.1104/pp.17.01384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C., Schäfer E. Seed germination of Arabidopsis thaliana phyA/phyB double mutants is under phytochrome control. Plant Physiol. 1997;114:1487–1492. doi: 10.1104/pp.114.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. PHYTOCHROME: a light-activated molecular switch that regulates plant gene expression. Annu. Rev. Genet. 1991;25:389–409. doi: 10.1146/annurev.ge.25.120191.002133. [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Voesenek L.A.C.J., Pierik R. The regulation of cell wall extensibility during shade avoidance: a study using two contrasting ecotypes of Stellaria longipes. Plant Physiol. 2008;148:1557–1569. doi: 10.1104/pp.108.125518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Staal M., Elzenga J.T.M., Yokoyama R., Nishitani K., Voesenek L.A.C.J., Pierik R. Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 2010;154:978–990. doi: 10.1104/pp.110.162057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Khanna R., Carle C.M., Quail P.H. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Sharrock R.A., Tepperman J.M., Quail P.H. The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Huang H., Gao H., Wang J., Wu D., Liu X., Yang S., Zhai Q., Li C., Qi T., et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell. 2014;26:263–279. doi: 10.1105/tpc.113.120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B., Sánchez-Lamas M., Yanovsky M.J., Casal J.J., Cerdán P.D. Arabidopsis thaliana life without phytochromes. Proc. Natl. Acad. Sci. USA. 2010;107:4776–4781. doi: 10.1073/pnas.0910446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M., Lin L., de Lucas M., Turco G., Toal T.W., Gaudinier A., Young N.F., Trabucco G.M., Veling M.T., Lamothe R., et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517:571–575. doi: 10.1038/nature14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Johansson H., Lee K.P., Bou-Torrent J., Stewart K., Steel G., Rodríguez-Concepción M., Halliday K.J. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014;10:e1004416. doi: 10.1371/journal.pgen.1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.F., Lian H.L., Kang C.Y., Yang H.Q. Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol. Plant. 2010;3:246–259. doi: 10.1093/mp/ssp097. [DOI] [PubMed] [Google Scholar]
- Withers J., Yao J., Mecey C., Howe G.A., Melotto M., He S.Y. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc. Natl. Acad. Sci. USA. 2012;109:20148–20153. doi: 10.1073/pnas.1210054109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhang W., Ding Y., Zhang J., Cambula E.D., Weng F., Liu Z., Ding C., Tang S., Chen L., et al. Shading contributes to the reduction of stem mechanical strength by decreasing cell wall synthesis in japonica rice (Oryza sativa L.) Front. Plant Sci. 2017;8:881. doi: 10.3389/fpls.2017.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W., Song D., Sun J., Shen J., Li L. Formation of wood secondary cell wall may involve two type cellulose synthase complexes in Populus. Plant Mol. Biol. 2017;93:419–429. doi: 10.1007/s11103-016-0570-8. [DOI] [PubMed] [Google Scholar]
- Yadav V., Mallappa C., Gangappa S.N., Bhatia S., Chattopadhyay S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue lightmediated photomorphogenic growth. Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Goué N., Igarashi H., Ohtani M., Nakano Y., Mortimer J.C., Nishikubo N., Kubo M., Katayama Y., Kakegawa K., et al. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 2010;153:906–914. doi: 10.1104/pp.110.154013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Xie Z., Zhang R., Xu P., Liu H., Yang H., Doblin M.S., Bacic A., Li L. Blue light regulates secondary cell wall thickening via MYC2/MYC4 activation of the NST1-directed transcriptional network in Arabidopsis. Plant Cell. 2018;30:2512–2528. doi: 10.1105/tpc.18.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ji Y., Xue C., Ma H., Xi Y., Huang P., Wang H., An F., Li B., Wang Y., et al. Integrated regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell. 2018;30:1971–1988. doi: 10.1105/tpc.18.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9:e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang X., Gong Y., Wang D., Xu D., Wang N., Sun Y., Gao L., Liu S.S., Deng X.W., et al. Red-light is an environmental effector for mutualism between begomovirus and its vector whitefly. PLoS Pathog. 2021;17:e1008770. doi: 10.1371/journal.ppat.1008770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Sun J., Xu P., Zhang R., Li L. Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 2014;164:765–776. doi: 10.1104/pp.113.231134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H. Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 2015;56:195–214. doi: 10.1093/pcp/pcu140. [DOI] [PubMed] [Google Scholar]
- Zhong R., Demura T., Ye Z.H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li L. Multi-layered regulation of plant cell wall thickening. Plant Cell Physiol. 2021;62:1867–1873. doi: 10.1093/pcp/pcab152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.