Abstract
Thyme has medicinal and aromatic value because of its potent antimicrobial and antioxidant properties. However, the absence of a fully sequenced thyme genome limits functional genomic studies of Chinese native thymes. Thymus quinquecostatus Čelak., which contains large amounts of bioactive monoterpenes such as thymol and carvacrol, is an important wild medicinal and aromatic plant in China. Monoterpenoids are abundant in glandular secretory trichomes. Here, high-fidelity and chromatin conformation capture technologies were used to assemble and annotate the T. quinquecostatus genome at the chromosome level. The 13 chromosomes of T. quinquecostatus had a total length of 528.66 Mb, a contig N50 of 8.06 Mb, and a BUSCO score of 97.34%. We found that T. quinquecostatus had experienced two whole-genome duplications, with the most recent event occurring ∼4.34 million years ago. Deep analyses of the genome, in conjunction with comparative genomic, phylogenetic, transcriptomic, and metabonomic studies, uncovered many regulatory factors and genes related to monoterpenoids and glandular secretory trichome development. Genes encoding terpene synthase (TPS), cytochrome P450 monooxygenases (CYPs), short-chain dehydrogenase/reductase (SDR), R2R3-MYB, and homeodomain-leucine zipper (HD-ZIP) IV were among those present in the T. quinquecostatus genome. Notably, Tq02G002290.1 (TqTPS1) was shown to encode the terpene synthase responsible for catalyzing production of the main monoterpene product γ-terpinene from geranyl diphosphate (GPP). Our study provides significant insight into the mechanisms of glandular secretory trichome formation and monoterpenoid biosynthesis in thyme. This work will facilitate the development of molecular breeding tools to enhance the production of bioactive secondary metabolites in Lamiaceae.
Key words: Thymus quinquecostatus, chromosome-level genome, phylogenetics, glandular secretory trichome, monoterpenoids
Thymus quinquecostatus Čelak. is an important wild medicinal and aromatic plant in China that contains large amounts of bioactive monoterpenes. Assembly and annotation of the T. quinquecostatus genome at the chromosome level will facilitate the development of molecular breeding for thyme. R2R3-MYB-, HD-ZIP IV-, TPS-, CYP-, and SDR-encoding genes are key regulatory factors and genes in the pathways of glandular secretory trichome formation and monoterpenoid biosynthesis.
Introduction
The genus Thymus, which belongs to the Lamiaceae family, is widely distributed worldwide and predominantly found in the Mediterranean basin. Thymus contains more than 300 species of herbaceous perennials or subshrubs with valuable medicinal and aromatic properties (Stahl-Biskup and Venskutonis, 2012). The Chinese native thyme species Thymus quinquecostatus Čelak. is widely used in folk medicine for the treatment of stroke, cold, dyspepsia, toothache, acute gastroenteritis, hypertension, chronic eczema, and other diseases (Kim et al., 2015a). Furthermore, it is used alone or in combination with other herbal medicines in cancer treatment (Hong et al., 2020). In China, T. quinquecostatus is also called di jiao (地椒). In some parts of China, di jiao is used not only in folk medicine but also as a seasoning or as fodder for livestock to improve the taste of their meat. One example in which T. quinquecostatus is used for livestock fodder is with the “di jiao sheep” in the Loess Plateau region. In addition to its medicinal and edible value, T. quinquecostatus has an important ecological function in changing soil microbiological properties during grass litter decomposition (Xiang et al., 2018) because it has relatively short stolons, which can form very strong root networks to prevent soil erosion.
The antibacterial and antioxidant medicinal functions of T. quinquecostatus depend on the bioactive components thymol, carvacrol, citral, geraniol, and nerolidol (Kou et al., 2008; Gavarić et al., 2015; Chan et al., 2016; Tak and Isman, 2016; Benelli et al., 2017; Pavela et al., 2018). The vast majority of these compounds are terpenoids. The evolution of terpenoid biosynthesis can be better understood by investigating terpene synthases (TPSs). Plant terpenoids contain isopentenyl diphosphate (C-5) and isomeric dimethylallyl diphosphate building blocks that are derived from the cytosolic mevalonate (MVA) or plastidial methylerythritol phosphate (MEP) pathways (Tholl, 2006; Dudareva and Pichersky, 2008; Zhou and Pichersky, 2020). TPS catalyzes the formation of the basic monoterpene (C10) and sesquiterpene (C15) skeletons from geranyl diphosphate (GPP) and farnesyl diphosphate (FPP), respectively. In previous studies of thyme, thymol and carvacrol biosynthesis was proposed to cyclize GPP to γ-terpinene, followed by a series of oxidations via p-cymene (Lima et al., 2013). The expression of γ-terpinene synthase genes was found to be related to thymol and carvacrol content. Examples of γ-terpinene synthase genes are OvTPS2 in Origanum vulgare (oregano) (Crocoll et al., 2010), TvTPS2 in Thymus vulgaris (Behnaz et al., 2020), and TcTPS2 in Thymus caespititius (Lima et al., 2013). Furthermore, a new study has shown that the aromatic backbones of thymol and carvacrol are formed by the CYP71D subfamily and short-chain dehydrogenase/reductase (SDR) in combination with dehydrogenases via unstable intermediates (Krause et al., 2021). Although TPSs that form γ-terpinene have been identified and characterized in various European thyme species, there are no reports on phenolic monoterpene precursors and terpene biosynthetic pathways in Chinese native thyme.
Plant secondary metabolites, such as terpenoids, are synthesized and stored in trichomes, which are hair-like extensions of leaf, flower, and stem epidermal cells. Depending on the plant species, trichomes consist of a single cell or multiple cells and are classified as either glandular or nonglandular trichomes (Werker, 2000). Mint, basil, lavender, oregano, and thyme (Lamiaceae) are cultivated for the terpenoids produced in their glandular secretory trichomes (Maleci and Giuliani, 2006). Glandular secretory trichome density was shown to be positively correlated with the production of terpenes (Yan et al., 2017). Consequently, increasing the density of glandular secretory trichomes could be an effective approach for improving the production of these natural metabolites (Tissier, 2012). Knowledge about genes involved in glandular secretory trichome formation comes mostly from work on Artemisia annua, Nicotiana benthamiana (tobacco), and Solanum lycopersicum (tomato) (Robert and Tissier, 2020). Through these studies, transcription factors (R2R3-MYB and HD-ZIP Ⅳ), cell cycle regulators (CycB2), and receptors involved in phytohormone-induced signaling cascades were implicated in glandular secretory trichome development. However, nothing is known about the molecular mechanism underlying glandular secretory trichome formation in Lamiaceae species. Uncovering genetic networks that control glandular secretory trichome formation in Lamiaceae will not only enable in-depth studies of plant secondary metabolism but also guide breeding strategies to improve terpene production (Huchelmann et al., 2017; Chalvin et al., 2020).
Here we report a reference genome sequence of T. quinquecostatus, which was obtained using a combination of high-fidelity (HiFi) sequencing and chromatin conformation capture (Hi-C) technologies. Genome-scale analyses of the sequencing data along with comparative genomic, phylogenetic, transcriptomic, and metabolomic studies revealed mechanisms that underlie glandular secretory trichome formation and monoterpenoid biosynthesis in T. quinquecostatus. The T. quinquecostatus genome presented here provides a resource that will also facilitate research on molecular breeding and functional gene identification related to important characteristics in thyme.
Results and discussion
Sequencing, assembly, and annotation of the T. quinquecostatus genome
Plants in the Lamiaceae family are distributed worldwide. Some members of this family, such as thyme, lavender, mint, basil, rosemary, marjoram, perilla, sage, and skullcap, are medicinal plants that contain a diverse set of specialized metabolites (Wu, 1977). Terpenoids, phenolic acids, and flavonoids are the most common plant secondary metabolites in Lamiaceae (Wu et al., 2016; Zhao et al., 2016). A high-quality genome sequence would provide a key resource for advancing studies on the molecular basis of specialized metabolite diversity in different members of Lamiaceae and would promote a better understanding of the evolution of pathways involved in the biosynthesis of plant secondary compounds (Afendi et al., 2012).
We sequenced the genome of T. quinquecostatus using Illumina HiSeq and PacBio technologies (Supplemental Tables 1–3). We obtained 21.74 Gb of PacBio circular consensus sequencing (CCS) reads (Cheng et al., 2021), amounting to 39.80× coverage of the ∼546.18 Mb genome, whose size was estimated by k-mer distribution analysis (Supplemental Figure 1; Table 1). We measured the genome size to be 536.25 Mb using flow cytometry, which was close to the value given by the k-mer method (Supplemental Figure 1). Finally, a chromosome-level genome was obtained, which contained 13 pseudochromosomes with a total length of 528.66 Mb (Figure 2; Supplemental Figure 2; Table 1), a contig N50 of 8.06 Mb, and a scaffold N50 of 36.44 Mb (Table 1).
Table 1.
Global statistics of T. quinquecostatus genome assembly and annotation
| Parameter | Size or number |
|---|---|
| Estimate of genome size (flow cytometry), Mb | 536.25 |
| Estimate of genome size (survey), Mb | 546.18 |
| Assembled genome size, Mb | 528.66 |
| Total length of contigs, Mb | 528.66 |
| Total number of contigs | 628 |
| N50 of contigs, bp | 8,059,748 |
| Largest contig, bp | 17,334,620 |
| Total length of scaffolds, Mb | 528.68 |
| Total number of scaffolds | 478 |
| N50 of scaffolds, bp | 36,440,660 |
| Largest scaffold, bp | 49,018,360 |
| GC content, % | 40.40 |
| Complete CEGMA, % | 97.82 |
| Complete BUSCOs, % | 97.34 |
| Total length of repeat, Mb | 373.28 |
| Repeat density, % | 70.61 |
| Long terminal repeat (LTR) density, % | 61.47 |
| Microsatellite repeat density, % | 9.14 |
| Number of protein-coding genes | 29,676 |
| Number of annotated genes | 28,862 |
| Number of rRNA | 2557 |
| Number of tRNA | 1138 |
| Number of miRNAs | 58 |
| Number of snRNAs | 86 |
| Number of snoRNAs | 68 |
| Number of pseudogenes | 217 |
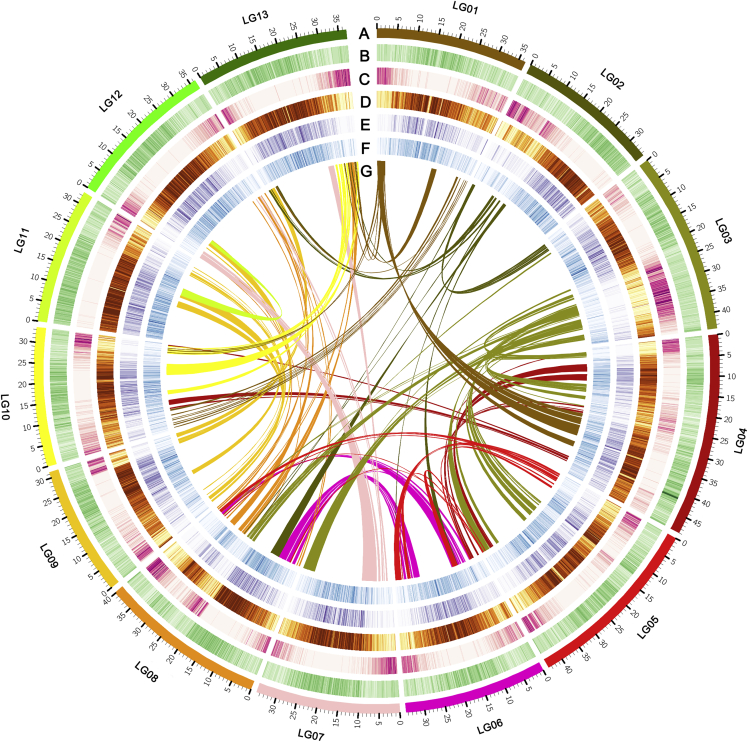
Figure 2.
Circos plot showing T. quinquecostatus genomic features.
(A) Karyotyping results.
(B) GC content.
(C) Protein-coding gene density.
(D) DNA transposable element density.
(E) LTR/Copia transposable element density.
(F) LTR/Gypsy transposable element density.
(G) Schematic presentation of major interchromosomal relationships in the T. quinquecostatus genome. The chromosome size is shown in Mb.
The accuracy and completeness of the assembled T. quinquecostatus genome were validated using short-read sequence alignment, Benchmarking Universal Single-Copy Orthologs (BUSCO), and Core Eukaryotic Genes Mapping Approach (CEGMA). Illumina short reads were mapped to the assembly and showed a mapping rate of 95.26% for the short reads and a mapping coverage of 89.47% for the assembled genome. Furthermore, 91.41% (exon), 6.57% (intergenic), and 2.03% (intron) of the sequences from these transcripts were successfully aligned. BUSCO analysis showed that 97.34% of the conserved genes belonged to complete and single-copy, complete and duplicated, fragmented, and missing categories, which accounted for 1499 (92.87%), 72 (4.46%), 16 (0.99%), and 27 (1.67%) of the total genes, respectively. CEGMA showed that 97.82% of the assembled T. quinquecostatus genome was reliably annotated (Table 1).
A Hi-C heatmap showed that interactions within chromosomes were more frequent than those between chromosomes, which suggested that the Hi-C assembly (Servant et al., 2015) was of high quality (Supplemental Figure 2). A total of 478 scaffolds were constructed, and the longest scaffold was 49.02 Mb (Supplemental Tables 4–6). A total of 506.90 Mb of sequences were anchored onto 13 pseudochromosomes, accounting for 95.88% of the initial assembly (Supplemental Table 6). In addition, Hi-C data mapped against the Hi-C scaffold assembly showed 488.75 Mb of sequences for determining the order and direction in 13 pseudochromosomes and a 96.42% valid rate of assembled sequences (Supplemental Table 6). Taken together, these statistics verified that our genome assembly is precise, complete, and of high quality at the chromosome scale.
The T. quinquecostatus genome was highly repetitive, with a total of 373.28 Mb of repetitive sequences annotated, accounting for 70.61% of the genome (Table 1). Long terminal repeat (LTR) retrotransposons were the dominant repeat type, taking up 324.98 Mb (61.47%) of the genome (Figure 2; Supplemental Table 7). LTRs consisted of two major types, class I (retroelement) and class II (DNA transposon), which represented 54.41% and 7.06% of the assembled genome, respectively. In addition, 239400 tandem repeats were identified, accounting for 48.30 Mb (9.14%) of the genome (Supplemental Table 8).
A total of 29676 protein-coding genes were predicted, with 28862 (97.26%) annotated by incorporating transcriptome, homology, and ab initio prediction (Supplemental Tables 9–11; Supplemental Figure 3) (Haas et al., 2008). Statistical gene information for T. quinquecostatus and the other species, including Arabidopsis thaliana, Ocimum tenuiflorum, Scutellaria baicalensis, Salvia miltiorrhiza, Salvia splendens, and Tectona grandis, showed an average T. quinquecostatus gene length of 3155.30 bp, a coding sequence length of 1315.72 bp, and an exon number of 5.25 (Supplemental Figure 3; Supplemental Table 10). Functional annotation of these genes against the published databases Pfam, Swiss-Prot, TrEMBL, EggNOG (Supplemental Figure 4A), and nr (Supplemental Table 11) resulted in 85.38%, 79.14%, 96.66%, 83.74%, and 96.71% functionally assigned genes, respectively. We further annotated these genes using the EuKaryotic Orthologous Groups (KOG), Gene Ontology (GO) (Supplemental Figure 4B), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Supplemental Table 11). We also identified 58 micro-RNAs (miRNAs), 86 small nuclear RNAs (snRNAs), 68 small nucleolar RNAs (snoRNAs), 1138 tRNAs, 2557 rRNAs, and 217 pseudogenes in the T. quinquecostatus genome (Table 1).
Evolutionary and comparative genomic analyses of T. quinquecostatus
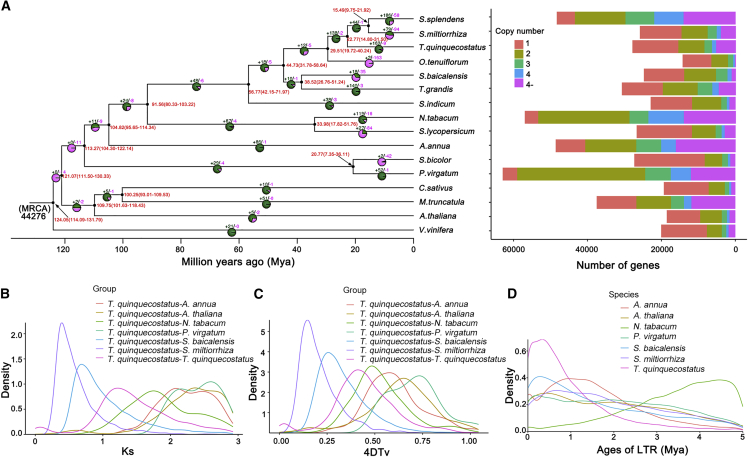
To investigate the evolutionary history of T. quinquecostatus, we analyzed the genomes of 15 selected angiosperm species: A. thaliana, A. annua, Cucumis sativus, Medicago truncatula, Nicotiana tabacum, O. tenuiflorum, Panicum virgatum, S. miltiorrhiza, S. splendens, S. baicalensis, Sesamum indicum, S. lycopersicum, Sorghum bicolor, T. grandis, and Vitis vinifera (Supplemental Table 12). Maximum likelihood-based phylogenetic analyses were performed with V. vinifera as the outgroup. The most recent common ancestor (MRCA) of the 16 species contained 44276 gene families and 951 high-quality single-copy orthologous genes (Figure 3A). The results indicated that T. quinquecostatus is closely related to O. tenuiflorum, S. splendens, and S. miltiorrhiza (Figure 3A). Molecular dating using V. vinifera for fossil calibration indicated that T. quinquecostatus-O. tenuiflorum emerged ∼29.51 (19.72–40.24) million years ago (Mya) and that T. quinquecostatus-two Salvia species (S. splendens and S. miltiorrhiza) emerged ∼22.77 (14.80–31.50) Mya.
Figure 3.
Genome comparison and evolutionary analysis.
(A) Phylogenetic analysis, divergence time estimates, and the number of gene copies and their distribution among 16 plant species. The tree was constructed on the basis of 951 single-copy truly orthologous genes. Divergence times (Mya) are indicated by the red numbers beside the branch nodes. The numbers of gene-family expansion and contraction events are indicated by green and pink numbers, respectively, on each species branch. MRCA, most recent common ancestor.
(B) Distribution of the synonymous substitution rate (Ks) between T. quinquecostatus and A. annua, A. thaliana, N. tabacum, P. virgatum, S. baicalensis, S. miltiorrhiza, and T. quinquecostatus.
(C) Genome duplication in T. quinquecostatus and genomes of related species as revealed by 4DTv analyses.
(D) Distribution of insertion ages of LTR-retrotransposons in the genomes of T. quinquecostatus and genomes of related species. LTR, long terminal repeat; Mya, million years ago.
A total of 65100 gene families were identified in the 16 selected angiosperm species, and 1873 common gene families with 479 unique gene families were uncovered in the T. quinquecostatus genome (Supplemental Figure 5A; Supplemental Table 13). We compared the gene families among five Lamiaceae species. As shown in Supplemental Figure 5B, 7017 gene families were shared among O. tenuiflorum, T. quinquecostatus, S. miltiorrhiza, T. grandis, and S. splendens, and 1127 gene families were specific to T. quinquecostatus. There were 2442 expanded gene families and 8 contracted gene families (Supplemental Figures 7 and 8), suggesting that more T. quinquecostatus gene families experienced expansion than contraction during adaptive evolution. KEGG enrichment analysis showed that most of the rapidly expanded gene families clustered in the flavonoid, photosynthetic, anthocyanin, phenylpropanoid, flavone, flavonol biosynthetic, and phenylalanine metabolism pathways (Supplemental Figure 7E), whereas contracted gene families clustered in the sesquiterpenoid and triterpenoid, monoterpenoid, cutin, suberin, and wax biosynthetic pathways (Supplemental Figure 8E). These metabolic processes may be related to T. quinquecostatus flower color, as well as terpenoid and lignin biosynthesis. Functional annotation of expanded and contracted gene families accounted for various traits of T. quinquecostatus, including flower color and high terpene levels. The expansion and contraction of gene families may have played an essential role in the evolution of T. quinquecostatus, contributing to phenotypic diversification, environmental adaptation, and even speciation.
There are many common events in eudicots, including ancient polyploidizations, that act as an important evolutionary force driving speciation (Wood et al., 2009). To evaluate the evolutionary relationships among T. quinquecostatus and other plant species, we measured the synonymous substitution rates (Ks) of orthologous gene pairs. The distribution of these rates suggested that T. quinquecostatus experienced two whole-genome duplication (WGD) events in its evolutionary history (Zwaenepoel and Van de Peer, 2019). Two WGD events were investigated in the T. quinquecostatus genome on the basis of the distribution of Ks values of ∼0.07 and ∼1.22 between orthologs, corresponding to divergence at ∼4.34 and ∼70.97 Mya, respectively. The analysis revealed peaks of ∼0.39 for T. quinquecostatus-S. miltiorrhiza and ∼0.68 for T. quinquecostatus-S. baicalensis, corresponding to divergence at ∼22.77 and ∼44.73 Mya, respectively (Figure 3B). Analysis of the distribution of sequence divergence (4DTv) values for syntenic duplicate genes revealed two significant peaks for the T. quinquecostatus genome (4DTv ∼0.02 and ∼0.41; Figure 3C), which further confirmed that T. quinquecostatus had experienced two WGD events. A divergence peak value (4DTv ∼0.15 and ∼0.26) was observed for T. quinquecostatus-S. miltiorrhiza and T. quinquecostatus-S. baicalensis in the map (Figure 3C), which suggested that the divergence of T. quinquecostatus-S. miltiorrhiza occurred later than that of T. quinquecostatus-S. baicalensis. During the evolution of plant genomes, the frequency of WGD and polyploidization was high, resulting in a large proportion of duplicated genes and repetitive sequences (Lockton and Gaut, 2005). We found that T. quinquecostatus, like many other flowering plants, experienced two rounds of WGD, with the recent event occurring ∼4.34 Mya, followed by extensive genomic rearrangements that resulted in 13 chromosomes in thyme after its divergence from the common paleopolyploid ancestor (Wei et al., 2018; Xia et al., 2020).
Accumulation of LTR-retrotransposons (LTR-RTs) is an important contributor to genome expansion and diversity (Kidwell and Lisch, 1997). A comparison of the insertion ages for LTR-RTs showed similar insertion profiles among the genomes. We found that most LTR-RT insertion events in the T. quinquecostatus genome occurred recently or less than 1 Mya. Nevertheless, the T.quinquecostatus genome contains fewer ancient insertions (>1 Mya) and more recent LTR-RT insertions(<1 Mya) than that of N. tabacum (Figure 3D). We also observed that the genomes of P. virgatum, S. baicalensis, and T. quinquecostatus carried younger LTR-RTs; the highest proportion of LTR-retrotransposons had insertion times of ∼0.23, 0.32, and 0.33 Mya, respectively. This may have resulted from rapid changes in the environment, such as the effects of pathogens and interference from human activities, that have occurred in recent years. Overall, these findings provide new insights into the evolution of T. quinquecostatus.
To analyze colinear relationships within the T. quinquecostatus, S. baicalensis, and V. vinifera genomes, we identified homologous proteins using BLASTP and syntenic blocks using MCScanX. A total of 935 and 520 syntenic blocks were identified on the basis of the orthologous gene orders between T. quinquecostatus-S. baicalensis and T. quinquecostatus-V. vinifera, corresponding to 19740 and 9577 gene pairs in T. quinquecostatus-S. baicalensis and T. quinquecostatus-V. vinifera, respectively (Supplemental Figures 9–11). A total of 282 syntenic blocks and 3263 gene pairs were identified in intragenomic comparisons of T. quinquecostatus-T. quinquecostatus (Supplemental Figure 11). The frequency of large-scale fragment rearrangements was determined in the three genomes, including inversions and translocations. The dot and bar graphs showed that T. quinquecostatus-S. baicalensis had higher collinearity than T. quinquecostatus-V. vinifera, consistent with their close phylogenetic relationship as members of the Lamiaceae clade (Supplemental Figures 9 and 10).
To date, there are only a few reports on the genomes of S. miltiorrhiza, S. splendens, S. baicalensis, and Lavandula angustifolia from Lamiaceae (Xu et al., 2016a; Dong et al., 2018; Zhao et al., 2019; Li et al., 2021). We now provide a high-quality reference genome sequence for T. quinquecostatus, which is the first genome for the genus Thymus. This genomic information provides a foundation for comparative genomic analysis between members of Lamiaceae.
Transcriptome sequencing and phylogenetic analysis reveal the genetic mechanism of glandular secretory trichome formation
To identify genes that play important roles in glandular secretory trichome formation and monoterpenoid biosynthesis, we performed transcriptomic analyses of T. quinquecostatus and T. vulgaris ‘Elsbeth.’ A total of 33 complementary DNA (cDNA) libraries were processed for transcriptome sequencing, generating 238.49 Gb of clean data (Supplemental Figure 12). After data filtering, the Q30 values were greater than 93.68%. For individual samples, clean reads varied from 5.98 to 9.31 Gb (Supplemental Table 14). The comparison rate of all reads was ≥44.13%, and the unique mapping rate was ≥43.10% (Supplemental Table 15).
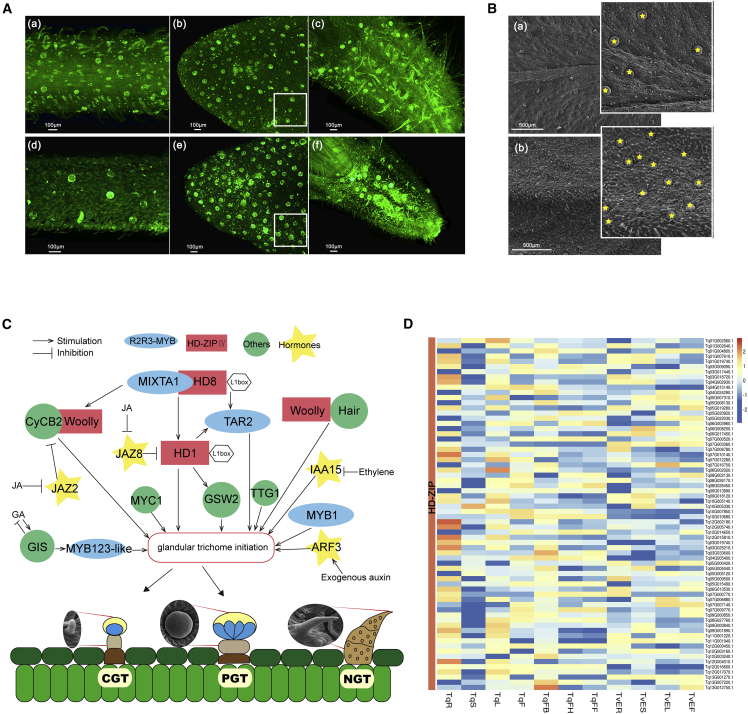
Glandular secretory trichomes can synthesize, store, or secrete monoterpenoids (Maleci and Giuliani, 2006; Yan et al., 2017). The type, size, and density of glandular secretory trichomes are shown in Figures 4A–4C. Glandular secretory trichomes were located mainly in leaves and flowers, and there were a few glandular secretory trichomes on the stems. The quantities of glandular secretory trichomes on leaves in T. vulgaris ‘Elsbeth’ (13 per mm2) were significantly higher than those in T. quinquecostatus (5 per mm2) (Figures 4A and 4B). Thyme contains two types of glandular secretory trichomes: capitate glandular trichomes and peltate glandular trichomes (Figure 4C). By searching and collecting information on regulatory genes related to glandular secretory trichome formation in A. annua, N. benthamiana, and S. lycopersicum, we summarized the genetic network of glandular secretory trichome formation in thyme (Figure 4C). The formation of glandular secretory trichomes has four stages: determination, initiation, morphogenesis, and maturation. Given their common organizational scheme, it has been suggested that glandular secretory trichomes share similar developmental events among different plant species (Chalvin et al., 2020). The initiation of most glandular secretory trichomes is regulated by MYB and HD-ZIP transcription factors (TFs). In A. annua, AaMYB1 positively regulates the development of glandular secretory trichomes (Matías-Hernández et al., 2017). AaMIXTA1 interacts with AaHD8 to form a regulatory complex that directly promotes AaHD1 expression and positively regulates the initiation of glandular secretory trichomes (Yan et al., 2017, 2018; Shi et al., 2018). AaTRICHOME AND ARTEMISININ REGULATOR (AaTAR2) can also positively modulate glandular secretory trichomes, and AaHD1 and AaHD8 enhance the expression of AaTAR2 by directly binding to its promoter (Zhou et al., 2020). In tomato, formation of type Ⅰ capitate glandular trichomes involves the Woolly gene, which encodes an HD-ZIP TF, together with SlCyCB2. In addition, the Hair gene, which encodes a C2H2 zinc finger protein, can interact with Woolly (Chang et al., 2018). In tobacco, NbMYB123-like (homolog of AtMYB123), which encodes a putative R2R3-MYB domain TF, also participates in the development of glandular secretory trichomes (Liu et al., 2018). In addition to TFs, glandular secretory trichome initiation is known to be regulated by plant hormones (Maes and Goossens, 2010) such as jasmonates (JA) (Yan et al., 2017), indole-3-acetic acid (IAA) (Zhang et al., 2015), and gibberellic acid (GA) (Chen et al., 2020).
Figure 4.
Schematic overview of glandular secretory trichome formation in thyme.
(A) The density of glandular secretory trichomes is shown in T. quinquecostatus and T. vulgaris ‘Elsbeth’ by fluorescence microscopy. (a) TqStem. (b) TqLeaf. (c) TqFlower. (d) TvEStem. (e) TvELeaf. (f) TvEFlower. Tq, T. quinquecostatus; TvE, T. vulgaris ‘Elsbeth’; GST, glandular secretory trichome. White frames in (b) and (e) represent 0.4 mm × 0.4 mm.
(B) The density of glandular secretory trichomes is shown in TqLeaf and TvELeaf by scanning electron microscopy. (a) Tq, T. quinquecostatus. (b) TvE, T. vulgaris ‘Elsbeth’. The yellow stars indicate glandular secretory trichomes.
(C) Glandular secretory trichome type and developmental pathway in thyme. CGT, capitate glandular trichome; PGT, peltate glandular trichome; NGT, nonglandular trichomes.
(D) Expression heatmap of DEGs in the HD-ZIP-encoding gene family. HD-ZIP, homeodomain-leucine zipper; DEGs, differentially expressed genes.
We summarized the genetic network that modulates glandular secretory trichome formation in thyme based on identified TFs (Figure 4C). We found that two TFs, including R2R3-MYB (encoded by MYB1, MIXTA1, TAR2, and MYB123-like) and HD-ZIP IV (encoded by HD8, HD1, and Woolly), are involved in thyme glandular secretory trichome development. Sequence similarity search results showed that 175 MYB-encoding and 87 HD-ZIP-encoding genes were identified in the T. quinquecostatus genome. In addition to hormone-related genes (IAA15, JAZ2, JAZ8, and ARF3), genes such as Glandular trichome-Specific WRKY 2 (GSW2), MYC1, B-type cyclin 2 (CyCB2), Glabrous Inflorescence Stems (GIS), Transparent Testa Glabra 1 (TTG1), and Hair were involved in glandular secretory trichome formation. A heatmap showed that 142 MYB-encoding (Supplemental Figure 13A) and 68 HD-ZIP-encoding DEGs (Figure 4D) were involved in glandular secretory trichome formation (Figure 4C). We also identified 62 hormone-related genes (29 IAA15s, 4 JAZ2s, 5 JAZ8s, and 24 ARF3s), 63 GSW2s, 62 MYC1s, 26 CyCB2s, 15 GISs, 11 TTG1s, and three Hairs that were differentially expressed in thyme (Supplemental Figure 13).
Phylogenetic tree analysis (Kumar et al., 2018) of MYB- and HD-ZIP-encoding DEGs in T. quinquecostatus and other species showed that Tq12G014160.1, Tq03G019480.1, Tq08G011440.1, Tq07G006560.1, and Tq02G008300.1 might be R2R3-MYBs (Supplemental Figure 14B), whereas Tq05G008130.1, Tq04G024290.1, Tq07G000520.1, Tq07G008780.1, Tq01G004800.1, Tq10G010680.1, Tq03G017440.1, Tq05G022030.1, Tq06G008250.1, and Tq10G005330.1 might be HD-ZIP IVs (Supplemental Figure 14C). Because glandular secretory trichomes are distributed mainly in leaves and flowers, the expression of Tq01G004800.1, Tq03G017440.1, Tq04G024290.1, Tq05G008130.1, Tq05G022030.1, Tq06G008250.1, Tq07G000520.1, Tq07G008780.1, and Tq10G005330.1 was particularly high in these organs. These genes encode members of the HD-ZIP Ⅳ family that are likely to be related to glandular secretory trichome development in thyme.
The genetic mechanism underlying monoterpenoid biosynthesis, identification of terpenoid-related gene expression patterns, and γ-terpinene synthase
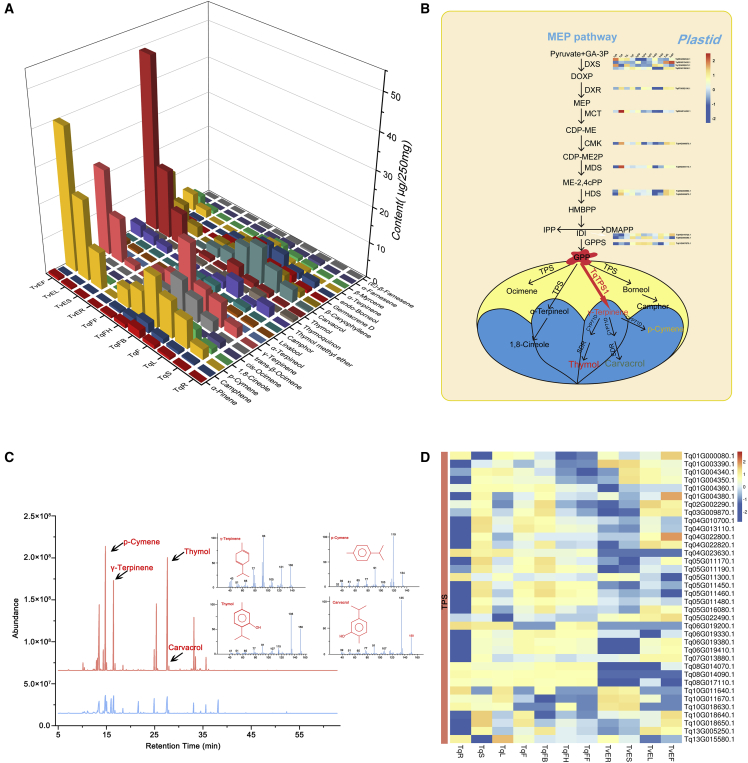
We measured volatiles from roots, stems, leaves, and flowers at different stages (bud, half-open, and full-open) in T. quinquecostatus (Figure 1B–1G) and T. vulgaris ‘Elsbeth’ using headspace solid-phase microextraction coupled to gas chromatography-mass spectrometry (HS-SPME-GC-MS). The composition and content of volatile organic compounds (VOCs) differed between the two species. The absolute contents of thymol, carvacrol, p-cymene, γ-terpinene, α-terpinene, 1,8-cineole, trans-β-ocimene, and endo-borneol were relatively high. VOCs in flowers were higher than those in leaves. Furthermore, VOCs in the bud stage were higher than those in the other flower developmental stages and decreased gradually as the flowers opened (Figure 5A). The mass spectra of major VOCs are shown in Figure 5C, and the chemical standards are shown in Supplemental Figure 15. Results of VOC analysis in T. quinquecostatus and T. vulgaris ‘Elsbeth’ were similar to those in other plant species. For example, VOCs in Cistanche deserticola flower buds gradually decreased or disappeared with flowering (Qiao et al., 2021). Li et al., 2019 found that 14 VOCs accumulated in lavender flower buds and decreased as the flowers matured. In the red apple ‘Pelingo,’ linalool was the most abundant flower volatile, accounting for 43% of volatiles in the flower buds and 27.7% in the mature flowers (Fraternale et al., 2014). Among essential oil constituents, the phenolic monoterpenes thymol and its isomer carvacrol, along with their biogenetic precursors p-cymene and γ-terpinene, are the most frequent chemophenetic and bioactive markers in several Thymus species (Morshedloo et al., 2017; Emami Bistgani et al., 2018).
Figure 1.
Overview of the morphological characteristics of T. quinquecostatus.
(A) Plant at the flowering stage.
(B) Root.
(C) Stem.
(D) Leaf.
(E) Flower at the bud stage.
(F) Flower at the half-open stage.
(G) Flower at the full-open stage.
Figure 5.
Analysis of compounds in different tissues and schematic overview of the monoterpene biosynthetic pathway.
(A) Major volatile compounds in different tissues per 250 mg in T. vulgaris ‘Elsbeth’ and T. quinquecostatus.
(B) Monoterpenoid biosynthesis in the MEP pathway.
(C) GC-MS peaks of flowers in T. vulgaris ‘Elsbeth’ (red) and T. quinquecostatus (blue) and the mass spectra of p-cymene, γ-terpinene, thymol, and carvacrol.
(D) Heatmap of DEGs in the TPS-encoding gene family.
The monoterpenoid biosynthetic pathway of T. quinquecostatus is shown in Figure 5B. A diverse array of differentially expressed genes (DEGs) in the MEP pathway, including members of eight gene families and 13 DEGs encoding four 1-deoxy-D-xylulose 5-phosphate synthases (DXSs), one 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR), one 2-C-methyl-derythritol-4-phosphate cytidylyltransferase (MCT), one 4-(cytidine-5-diphospho)-2-C-methyl-D-erythritol kinase (CMK), one 2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase (MDS), two (E)-4-hydroxy-3-methyl-but-2-enyl-pyrophosphate reductases (HDSs), two isopentenyl diphosphate isomerases (IDIs), and one geranyl diphosphate synthase (GPPS), were identified in the T. quinquecostatus genome (Figure 5B). Our results showed that the copy numbers of these genes were expanded in thyme, especially for DXS, which encodes the crucial rate-limiting enzyme of the MEP pathway.
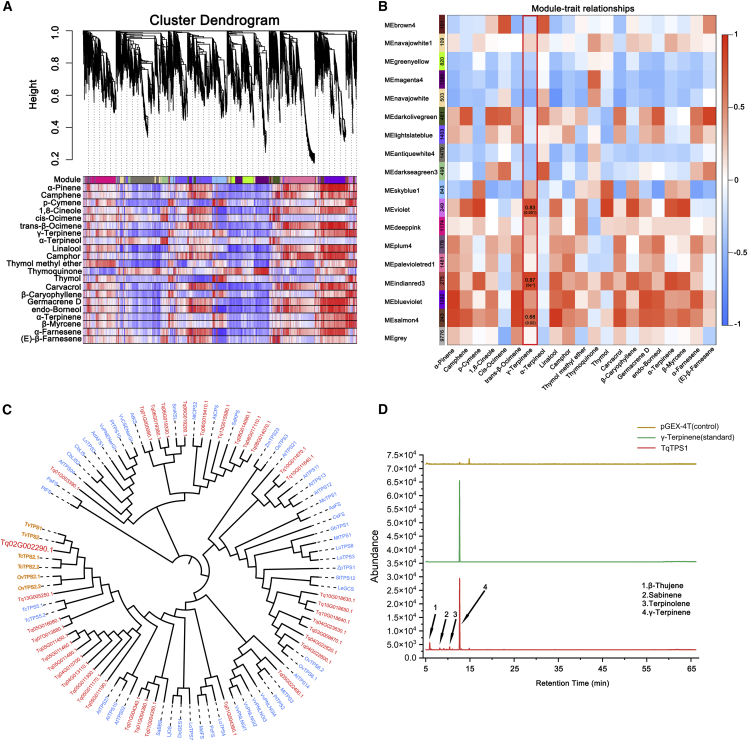
TPSs catalyze the formation of the basic skeleton of monoterpenes (C10) from GPP. We identified 36 DEGs encoding TPSs, which catalyze the synthesis of γ-terpinene, α-terpineol, borneol, and ocimene (Figure 5B). Phylogenetic tree analysis of TPS-encoding DEGs in T. quinquecostatus and other species showed that Tq02G002290.1 and Tq13G005250.1 may be γ-terpinene synthase-encoding genes (Figure 6C). TPS-, CYP-, and SDR-encoding genes are crucial for monoterpenoid biosynthesis and are often found in physical clusters in the genome (Supplemental Table 16). Sequence similarity results showed that 69, 420, and 163 TPS-, CYP-, and SDR-encoding genes were present in T. quinquecostatus. Furthermore, we identified 189 differentially expressed CYP-encoding genes and 47 SDR-encoding genes, which catalyze the formation of carvacrol and thymol from p-cymene (Supplemental Figure 16). Phylogenetic tree analysis of CYP- and SDR-encoding DEGs in T. quinquecostatus and CYP71Ds and SDRs of other species, showed that Tq06G018730.1, Tq06G018750.1, Tq06G018720.1, Tq03G008200.1, Tq13G008670.1, Tq04G023650.1, and Tq05G002240.1 may be CYP71Ds (Supplemental Figure 14D). On the other hand, Tq07G003330.1, Tq03G029330.1, Tq04G008130.1, Tq13G017990.1, Tq13G002980.1, and Tq09G015420.1 may be SDRs (Supplemental Figure 14A). These results show that TPS-, CYP71D-, and SDR-encoding genes are key genes related to thymol and carvacrol biosynthesis and lay the foundation for future research on gene function in thyme.
Figure 6.
Modular analysis of terpenoid-associated genes on the basis of WGCNA and γ-terpinene synthase-encoding gene expression analysis.
(A) The heatmap shows the relatedness of terpenoids with all coexpression modules identified in WGCNA.
(B) Modular organization analysis of terpenoid-related gene expression in thyme. Module–terpenoid correlations and corresponding p values (in parenthesis). The left panel shows 17 modules. The right panel is a color scale for module trait correlation from −1 to 1.
(C) Phylogenetic tree of the TPS-encoding genes (DEGs) in thyme and other species. TvTPS1, TvTPS2, TcTPS2.1, TcTPS2.2, OvTPS2.1, and OvTPS2.2 (orange color) are γ-terpinene synthase-encoding genes in thyme and oregano (Lamiaceae).
(D) Heterologous expression of TqTPS1 in Escherichia coli via in vitro enzyme assay. WGCNA, weighted gene coexpression network analysis.
We used weighted gene coexpression network analysis (WGCNA), which is a systems biology method, to analyze gene expression in thymes. Through WGCNA, we elucidated 21965 DEGs (Supplemental Figure 17A) and analyzed the relationships among modules (Supplemental Figure 17B). Using a set of parameters that produced refined clusters, we detected 17 modules containing 101–1481 DEGs each (fold change [FC] > 2 or < 0.5, false discovery rate [FDR] < 0.01) (Supplemental Figure 17B). Hub gene analyses were useful in revealing important regulatory factors and genes for monoterpenoid biosynthesis and glandular secretory trichome formation. Transcriptome data analyses and WGCNA revealed gene hubs, which were coexpressed with terpenoid- and trichome-related gene modules (Figure 6B). Among the modules were TPS-, CYP71D-, and SDR-encoding genes related to monoterpenoid biosynthesis and R2R3-MYB- and HD-ZIP IV-encoding genes associated with glandular secretory trichome formation. qRT-PCR results also showed that Tq06G018730.1 (CYP71D), Tq13G017990.1 (SDR), Tq05G008130.1 (HD-ZIP IV), and Tq12G014160.1 (R2R3-MYB) played important roles in monoterpenoid biosynthesis and glandular secretory trichome formation, consistent with the heatmap analysis of these DEGs in thymes (Supplemental Figure 18).
To identify the relationship between modules and terpenoids, gene coexpression networks were constructed using 21965 DEGs (Figure 6A). Highly interconnected genes were clustered in the same module, and 17 modules were obtained. The module–terpenoid relationship revealed that each terpenoid was significantly relevant to at least one module (Figure 6A). The investigation of relationships between 17 module eigengenes and terpenoid content revealed that the correlation coefficients ranged from −0.55 to 0.87 for γ-terpinene (Figure 6B). The eigengenes of the MEindianred3, MEviolet, and MEsalmon4 modules showed significant positive correlations with γ-terpinene (R > 0.5, p < 0.05) (Figure 6B). Nine compounds showed significant correlations with MEsalmon4 and seven compounds with MEindianred3. The compounds were negatively correlated with MEgreenyellow and MEmagenta4 and positively correlated with MEblueviolet, and vice versa. The MEindianred3 module consisted of three TPS-encoding genes: Tq02G002290.1, Tq03G009870.1, and Tq13G005250.1.
To investigate the evolutionary relationship between the Tq02G002290.1 gene identified in this study and other TPS-encoding genes, a phylogenetic tree was generated by the neighbor-joining method using amino acid sequences of TPS DEGs of T. quinquecostatus and other plant species. Tq02G002290.1 clustered together with six other previously reported γ-terpinene synthase-encoding genes in oregano and thyme, such as OvTPS2.1, OvTPS2.2, TcTPS2.1, TcTPS2.2, TvTPS1, and TvTPS2 (Crocoll et al., 2010; Lima et al., 2013; Rudolph et al., 2016) (Figure 6C). Multiple alignment revealed that the amino acid sequence of the protein encoded by Tq02G002290.1 was highly similar to those of other functionally characterized TPS proteins, and it contained conserved domains such as DDXXD and NSE/DTE. TPS enzymes catalyze steps in terpene biosynthesis by binding to Mg2+ or Mn2+ cofactors, and RRX8W is involved in cyclization reactions during catalysis (Supplemental Figure 19C). To confirm the activity of the Tq02G002290.1-encoded protein and elucidate its role in monoterpene biosynthesis in T. quinquecostatus, a GPP substrate was used in cell-free systems. Recombinant protein for biochemical analysis was prepared by cloning Tq02G002290.1 and expressing it in the Escherichia coli BL21 strain, with the E. coli expression system containing empty vectors as a control (Supplemental Figure 19A). The Tq02G002290.1 protein product was successfully induced in the supernatant and purified as a homogeneous soluble protein (Supplemental Figure 19B). The product peaks were identified by comparing mass spectra with the National Institute of Standards and Technology (NIST) library or in relation to standards. In general, the protein encoded by Tq02G002290.1 was found to be a versatile enzyme, yielding the main product γ-terpinene along with the by-products β-thujene, sabinene, and terpinolene. As such, Tq02G002290.1 was named TqTPS1 (Figure 6D). TPSs act as metabolic gatekeepers in the biosynthesis of diverse plant terpenoids (Mcgarvey and Croteau, 1995). In Lathyrus odoratus, five LoTPS were found to be C10/C15 multisubstrate enzymes that catalytically generated multiple volatilized terpenes (Bao et al., 2020). As mentioned above, TqTPS1 is capable of catalyzing the formation of multiple products. However, it remains to be determined whether TqTPS1 is a multisubstrate enzyme.
Plants in the genus Thymus are widely used as spices, herbal teas, and insecticides because of their flavor and fragrance. Here, we sequenced the genome of the representative Chinese native thyme T. quinquecostatus. The T. quinquecostatus genome has 13 pseudochromosomes with a total length of 528.66 Mb. Comparative genomic analyses showed that T. quinquecostatus had a close relationship with O. tenuiflorum, S. splendens, and S. miltiorrhiza. Ks and 4DTv analyses indicated that the T. quinquecostatus genome experienced two WGD events. In-depth analyses of the T. quinquecostatus genome revealed the biosynthetic pathway of monoterpene bioactive components and gene networks controlling glandular secretory trichome development, most notably TqTPS1. Our results provide new targets for the synthesis of downstream monoterpene biosynthetic products in thyme, such as thymol and carvacrol. The datasets and analyses presented in this study provide an important resource that will facilitate molecular breeding and functional gene identification in thyme.
Methods
Plant materials
A survey of native thyme species grown in China was carried out using the Flora of China (Li and Ian, 1994). According to ethnobotanical information, all T. quinquecostatus (NCBI TaxID 228974) accessions were collected directly from Jinhekou village in the Hebei province of China in 2018. Herbarium specimens were identified and categorized by the Institute of Botany, Chinese Academy of Sciences (IB-CAS). T. quinquecostatus and T. vulgaris ‘Elsbeth’ were grown at the IB-CAS experimental farm, Beijing, China. T. quinquecostatus was selected for de novo sequencing because it is widely used as a medicinal plant in China. Fresh young leaves of T. quinquecostatus were collected from plants, immediately frozen in liquid nitrogen, and stored at −80°C before DNA extraction.
Genome sequencing
Genomic DNA was extracted from young leaves of T. quinquecostatus using a DNA Secure Plant Kit (Tiangen, China). An Illumina genomic library was constructed according to Illumina’s standard protocol, and paired-end reads (2 × 150 bp) sequenced on an Illumina NovaSeq 6000 platform were used for genome survey and assessment. The genomes were sequenced using the PacBio Sequel II platform (Pacific Biosciences) to produce CCS reads (HiFi) for contig assembly. The Hi-C library (vanBerkum et al., 2010) was constructed using the HindIII restriction enzyme according to the instructions of the BioMarker Technologies Company (Xie et al., 2015) and sequenced on an Illumina NovaSeq 6000 platform for chromosome construction. Four types of live T. quinquecostatus tissues (root, stem, leaf, and flower) were collected for transcriptome sequencing for genome annotation and assessment. RNA-seq libraries were sequenced on an Illumina NovaSeq 6000 platform in paired-end mode. All sequencing services were provided by Biomarker Technologies Co., Ltd. (Beijing, China).
Genome survey and assembly
The genome size, heterozygosity, and repeat content were estimated on the basis of k-mer distribution using 19-mers extracted from the Illumina short reads. The estimated genome size was further validated using flow cytometry. Raw PacBio subreads were filtered and corrected using the PacBio circular consensus sequencing (pbccs) pipeline with default parameters (https://github.com/PacificBiosciences/ccs). The resulting CCS reads were subjected to hifiasm (version 0.12) (Cheng et al., 2021) for de novo assembly. The primary contigs were corrected using Pilon software (version 1.18) (Utturkar et al., 2017). BWA (version 0.7.10-r789) (Li, 2013) and SAMtools (version 1.9) (Li et al., 2009) were used for read alignment and SAM/BAM format conversion. CEGMA (version 2.5) (Parra et al., 2007) and BUSCO (version 4) (Simão et al., 2015) were used to assess the completeness of the genome and gene annotation.
Chromosome assembly using Hi-C
The raw data were filtered using a Perl script as implemented in LACHESIS software (Burton et al., 2013). BWA software (version 0.7.10-r789) (Li, 2013) was used to map the Hi-C reads to the draft assembly, and uniquely mapped reads were selected for further analysis. Our newly developed ALLHiC (Zhang et al., 2019) pipeline was used to link the contigs to 13 pseudochromosomes. HiC-Pro software (version 2.10.0) (Servant et al., 2015) was used to calculate the Hi-C mapping rate and evaluate the quality of the Hi-C scaffolding.
Protein-coding gene prediction
The following three approaches, as incorporated in the Evidence Modeler (EVM) pipeline (version 1.1.1) (Haas et al., 2008), were used to predict high-quality protein-coding genes: ab initio gene predictions, transcript evidence, and homology-based predictions. Augustus (version 2.4) (Stanke et al., 2008) and SNAP (version 2006-07-28) (Korf, 2004) were used for ab initio gene predictions. For homology-based prediction models, six proteomes (A. thaliana, O. tenuiflorum, S. baicalensis, S. miltiorrhiza, S. splendens, and T. grandis) were downloaded from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html). The protein sequences of the T. quinquecostatus genome were aligned to those of the six species using GeMoMa software (version 1.7) (Keilwagen et al., 2016). For transcript evidence, RNA-seq data from different tissues (root, stem, leaf, and flower) were assembled against reference transcripts using HISAT (version 2.0.4) (Kim et al., 2015b), StringTie (version 1.2.3) (Pertea et al., 2015), and GeneMarkS-T (version 5.1) (Tang et al., 2015a). The no-reference transcripts were assembled using Trinity (version 2.11) (Grabherr et al., 2011), and PASA (version 2.0.2) (Haas et al., 2003) was used for gene prediction. Finally, tiers with protein-coding evidence were incorporated in the EVM pipeline (version 1.1.1) (Haas et al., 2008) to predict high-quality protein-coding genes.
Functional annotation
Functional annotations of protein-coding genes from T. quinquecostatus were obtained by performing BLASTP against the public databases EggNOG (Huerta-Cepas et al., 2019), GO (Ashburner et al., 2000), KOG (Koonin et al., 2004), TrEMBL (Boeckmann et al., 2003), nr (Marchler-Bauer et al., 2011), Swiss-Prot (Bairoch and Apweiler, 2000), and KEGG (Kanehisa et al., 2014) with an E-value cut-off of 1.0 × 10−5. Protein domains were identified using InterProScan (version 4.8) (Jones et al., 2014) and HMMER (version 3.3) (Klingenberg et al., 2013) to query against the InterPro and Pfam databases. GO terms were assigned on the basis of InterPro or Pfam entries. In addition, protein-coding genes were searched against the KEGG database to identify possible pathways in which they might be involved. GO enrichment and KEGG pathway analysis were performed using the online platform OmicShare (https://www.omicshare.com/).
Identification of repetitive elements
For de novo prediction, the repeat sequence library of the genome was first customized using the RepeatModeler2 pipeline (version 2.0.1) (Flynn et al., 2020), which used RECON (version 1.0.8) (Bao and Eddy, 2002) and RepeatScout (version 1.0.6) (Price et al., 2005) to obtain the consensus repeat library. RepeatClassifier was then used to identify and cluster repetitive elements with the Repbase (version 19.06) (Jurka et al., 2005), REXdb (version 3.0) (Neumann et al., 2019), and Dfam (version 3.2) (Wheeler et al., 2013) databases. LTR_retriever (version 2.8) (Ou and Jiang, 2017), LTRharvest (version 1.5.9) (Ellinghaus et al., 2008), and LTR_FINDER (version 1.1) (Xu and Wang, 2007) software packages were then used to classify the unknown-type transposon sequences (TEs). Finally, RepeatMasker (version 4.1.0) (Tarailo-Graovac and Chen, 2009) was used to predict the TEs in the genome based on the constructed repeat sequence database. The microsatellite identification tool MISA (version 2.1) (Beier et al., 2017) and the Tandem Repeat Finder version 409 (TRF) package (Benson, 1999) were used to identify tandem repeat sequences in the genome.
Identification of noncoding RNA genes and pseudogenes
tRNAscan-SE software (version 1.3.1) (Chan and Lowe, 2019) was used with eukaryotic parameters to identify tRNA genes. Barrnap (version 0.9) (Singleton et al., 2021) was used to identify rRNA genes mainly on the basis of the Rfam database (version 13.0) (Kalvari et al., 2017). The miRBase database (Griffiths-Jones et al., 2006) was used to identify miRNA genes. Infernal (version 1.1) (Nawrocki and Eddy, 2013) was used with default parameters to annotate snRNA and snoRNA genes on the basis of the Rfam database (version 13.0) (Kalvari et al., 2017). genBlastA (version 1.0.4) (She et al., 2009) and GeneWise (version 2.4.1) (Birney et al., 2004) were used to predict pseudogenes.
Reconstruction of the phylogenetic tree
OrthoFinder (version 2.4.0) (Emms and Kelly, 2019) was used to identify homologous genes in T. quinquecostatus and 15 angiosperm species. MAFFT (version 7.407) (Katoh and Standley, 2013) was used with default parameters to align each orthologous gene sequence. The PANTHER version 15 database (Mi et al., 2019) was used to annotate gene families. Individual gene alignments were processed using in-house Python scripts to extract conserved regions. Conserved regions of individual genes were concatenated into a supermatrix dataset. GO and KEGG enrichment analysis was performed using unique gene families of T. quinquecostatus via clusterProfiler (version 3.14.0) (Yu et al., 2012). ModelFinder (Kalyaanamoorthy et al., 2017), as implemented in IQ-TREE (version 1.6.11) (Nguyen et al., 2015), was used to estimate the best substitution models. Finally, RAxML was used with the best-fit substitution model and 1000 bootstrap replicates to infer the maximum-likelihood tree. Divergence time estimates were calculated using the MCMCTree program in Phylogenetic Analysis by Maximum Likelihood software (PAML version 4.9i) (Yang, 1997), with two secondary calibration points obtained from the TimeTree database (Kumar et al., 2017) (http://www.timetree.org/). The graphical phylogenetic tree was displayed using MCMCTreeR (version 1.1) (Puttick, 2019).
Expansion and contraction of gene families
All the deduced proteins were filtered using in-house Python scripts to remove alternative splicing and redundant genes, and only the longest transcripts were retained. BLASTP software was used to identify gene-family clusters via OrthoMCL (Enright et al., 2002) on the basis of sequence similarity information from the BLAST output. Based on the dated phylogeny, expansions and contractions of orthologous gene families were determined using CAFE software (version 4.2) (Han et al., 2013). Gene families were considered significantly expanded or contracted when they presented p values smaller than 0.05. Genes in significantly expanded families were then used for GO and KEGG enrichment analysis.
Synteny analysis and whole-genome duplication
CIRCOS software (version 0.69-6) (Krzywinski et al., 2009) was used to visualize gene density, G-C content, repeats, and gene synteny on individual pseudochromosomes. BLASTP was used to examine WGD in the T. quinquecostatus genome and identify orthologous and paralogous genes. MCScanX software (Wang et al., 2012) was used to identify collinear blocks. Syntenic blocks were visualized using MCScan, and chromosome lengths were not scaled. Synonymous substitution rates (Ks) of the collinear orthologous gene pairs were calculated using WGD software (version 1.1.1) (Zwaenepoel and Van de Peer, 2019). The synonymous substitutions per site per year as 8.61 × 10−9 (μ) were estimated using divergence time. The mean Ks values of syntenic blocks between T. quinquecostatus and S. miltiorrhiza, and two WGD events in the T. quinquecostatus genome, were investigated using the following equation: time = Ks/2μ. The 4DTv value of each gene pair was calculated and then corrected using the script in the following link: https://github.com/JinfengChen/Scripts.
LTR sequences (set parameter S ≥ 6) in the T. quinquecostatus genome were identified using LTR_FINDER (version 1.07) (Xu and Wang, 2007). Distance K was calculated using the Kimura model in EMBOSS (version 6.6.0) (Rice et al., 2000). The integration times (Mya) of intact LTRs were estimated using the following equation: T = K/2r, where K is the number of nucleotide substitutions per site between each LTR pair, and r is the nucleotide substitution rate, which was set to 7 × 10−9 substitutions per site per year (Ossowski et al., 2010).
DIAMOND (version 0.9.29.130) (Buchfink et al., 2015) was used to compare the gene sequences of the two species to determine similar gene pairs (E < 1e−5, C score > 0.5, where JCVI software was used to filter the C score value). Next, MCScanX (Wang et al., 2012) was used to determine whether similar gene pairs were adjacent on the chromosomes and ultimately to obtain all the genes in the syntenic block. Collinear patterns of each species were drawn using JCVI (version 0.9.13) (Tang et al., 2015b). Chromosome-scale syntenic block scattered point graphs and bar graphs were constructed using VGSC (Xu et al., 2016b).
Phylogenetic analysis of TPS-, CYP-, SDR-, MYB-, and HD-ZIP-encoding genes
TPS-, CYP-, SDR-, MYB-, and HD-ZIP-encoding genes were identified from the scientific literature (Supplemental Tables 17–21). The amino acid sequences of the proteins encoded by these genes were obtained from the National Center for Biotechnology Information (NCBI) website (https://www.ncbi.nlm.nih.gov/). TPS-, CYP-, SDR-, MYB-, and HD-ZIP-encoding genes were identified using the MAKER-P pipeline (Campbell et al., 2014) and Fgenesh (Salamov and Solovyev, 2000). T. quinquecostatus TPS-, CYP-, SDR-, MYB-, and HD-ZIP-encoding genes were predicted using hmmscan (Finn et al., 2010) and NLR-parser (Steuernagel et al., 2015). Phylogenetic trees of DEGs and genes of other species obtained from NCBI were constructed using MEGA X (Kumar et al., 2018).
Transcriptome sequencing, identification of specifically expressed genes, and WGCNA analysis
Raw RNA-seq data from four tissues (root, stem, leaf, and flower) of T. quinquecostatus and T. vulgaris ‘Elsbeth’ and three flower developmental stages (bud, half-bloom, and full-bloom) of T. quinquecostatus were filtered using Trimmomatic software (Bolger et al., 2014). The resulting clean reads were mapped to coding sequences predicted from the genome using Bowtie (version 2.0) (Langmead and Salzberg, 2012). Fragments per kilobase of exon per million fragments mapped (FPKM) were calculated using RSEM software (version 1.3.2) (https://github.com/deweylab/RSEM) implemented in Trinity (Grabherr et al., 2011). Genes with log|FC| > 2 and p < 0.05 were identified as DEGs. Coexpression and module analyses were performed using the R package WGCNA (version 1.51) for all DEGs (Langfelder and Horvath, 2008).
Observation and density of glandular secretory trichomes
The morphology and distribution of glandular secretory trichomes were evaluated using a stereomicroscope (Leica DVM6), fluorescence microscope (Leica DM6 B), and scanning electron microscope (S-4800; Hitachi, Tokyo, Japan). ImageJ software (https://imagej.nih.gov/ij/) was used to count glandular secretory trichomes and measure leaf area. Glandular secretory trichome density was calculated from three plants.
Analysis of volatile organic compounds by headspace solid-phase microextraction (HS-SPME)
VOCs of 33 RNA-seq samples were detected via headspace solid-phase microextraction (HS-SPME). Fresh leaf powder (0.25 g) was weighed and immediately placed into a 20-mL headspace vial (Agilent, Palo Alto, CA) containing 20 μL of internal standard solution (1 mg/mL, 3-octanol, Cas#589-98-o; Aladdin, Shanghai, China). The vials were sealed using crimp-top caps with TFE-silicone headspace septa (Agilent). Each vial was immediately incubated at 40°C for 30 min, and then a 100-μm polydimethylsiloxane-coated fiber (Supelco, Inc., Bellefonte, PA) was exposed to the headspace to absorb the volatiles for 30 min. All volatile components on the coated fiber were analyzed by gas chromatography (GC). A Model 7890A GC instrument and a 7000B mass spectrometer (Agilent) were used to perform GC-mass spectrometry (GC-MS) analysis (Pontes et al., 2009). The determination of the percentage content of each compound was based on normalization of the GC peak areas. Identification of the compounds was based on comparison of retention indices (RIs) relative to a homologous series of n-alkanes (C7–C40), mass spectra (MS) from the NIST library (version 14.0), and data from the scientific literature. Retention indices were based on the formula RI = 100Z + 100[RT(x) − RT(z)]/[RT(z + 1) − RT(z)], RT(x), RT(z) and RT(z + 1) for the composition, and the number of carbons Z and Z + 1 for the retention time of the normal alkane.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from three tissue replicates using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China). Primers were designed using Primer Premier 5.0, and the sequences are listed in Supplemental Table 22. qRT-PCR was performed using the ABI 7300 Real-time PCR System (Framingham, MA) with SYBR Green PCR Master Mix (TaKaRa), following procedures described previously (Wang et al., 2015). All PCR assays were performed in triplicate. The reference gene was 18S rRNA. Relative expression levels were quantified with the 2−ΔΔCt method (Wang et al., 2014).
Heterologous expression of TqTPS1 in Escherichia coli and in vitro enzyme assay
To express TqTPS1 recombinant proteins in E. coli, the full-length sequence of TqTPS1 was amplified with precise primers and then subcloned into the pGEX 4T-1 vector system. Next, an empty vector (as a control) and vectors ligated with TqTPS1 were transformed into E. coli strain BL21 (DE3). Expression of recombinant TqTPS1 was induced using 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 18°C for 12 h in an incubator with constant shaking. Subsequently, the cells were harvested via centrifugation, resuspended in phosphate-buffered saline, and broken down via sonication. Crude proteins were then applied to glutathione beads (Tiandirenhe Biotech, Changzhou, China). The purified protein was incubated with GPP, and the reaction product was detected using GC-MS.
Statistical analysis
All samples were prepared and analyzed in triplicate, and data were expressed as the mean ± SD. Statistical analyses were performed by analysis of variance (ANOVA), and Duncan’s test was used to determine the significance of differences between groups. Differences at p < 0.05 were considered significant. SPSS 18 (SPSS Inc., Chicago, IL) was used for the analysis.
Availability of data and materials
The raw sequence and genome assembly data were deposited in NCBI under project accession number PRJNA690675. The genome assembly and annotation data were also deposited in the China National Genomics Data Center under project accession number GWHBJVA00000000.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA23080603).
Author contributions
M.S. and Y.Z. performed the experiments, analyzed the data, and wrote the manuscript. L.Z. and N.L. helped with thyme VOC analysis. H.B. and G.S. helped with sample collection and analyzed the data. J.Z. helped with sample collection and research design. L.S. was involved in designing the research and revising the manuscript. All authors read and approved the manuscript.
Acknowledgments
We thank Professor Cathie Martin from the John Innes Center of Norwich Research Park in the UK and Dr. Qing Zhao from the Shanghai Chenshan Botanical Garden of the Chinese Academy of Sciences in China for sharing the Scutellaria baicalensis genome-wide annotation files. No conflict of interest declared.
Published: July 16, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Jinzheng Zhang, Email: caohua@ibcas.ac.cn.
Lei Shi, Email: shilei_67@126.com.
Supplemental information
References
- Afendi F.M., Okada T., Yamazaki M., Hirai-Morita A., Nakamura Y., Nakamura K., Ikeda S., Takahashi H., Altaf-Ul-Amin M., Darusman L.K., et al. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao T., Shadrack K., Yang S., Xue X., Li S., Wang N., Wang Q., Wang L., Gao X., Cronk Q. Functional characterization of terpene synthases accounting for the volatilized-terpene heterogeneity in Lathyrus odoratus cultivar flowers. Plant Cell Physiol. 2020;61:1733–1749. doi: 10.1093/pcp/pcaa100. [DOI] [PubMed] [Google Scholar]
- Bao Z., Eddy S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnaz T., Mehdi R., Ahmad A., Helena T. Sequencing and variation of terpene synthase gene (TPS2) as the major gene in biosynthesis fo thymol in different Thymus Species. Phytochemistry. 2020;169:112126. doi: 10.1016/j.phytochem.2019.112126. [DOI] [PubMed] [Google Scholar]
- Beier S., Thiel T., Münch T., Scholz U., Mascher M. Misa-web: a web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benelli G., Pavela R., Canale A., Cianfaglione K., Ciaschetti G., Conti F., Nicoletti M., Senthil-Nathan S., Mehlhorn H., Maggi F. Acute larvicidal toxicity of five essential oils (Pinus nigra, Hyssopus officinalis, Satureja montana, Aloysia citrodora and Pelargonium graveolens) against the filariasis vector Culex quinquefasciatus: synergistic and antagonistic effects. Parasitol. Int. 2017;66:166–171. doi: 10.1016/j.parint.2017.01.012. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.S., Law M., Holt C., Stein J.C., Moghe G.D., Hufnagel D.E., Lei J., Achawanantakun R., Jiao D., Lawrence C.J., et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 2014;164:513–524. doi: 10.1104/pp.113.230144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalvin C., Drevensek S., Dron M., Bendahmane A., Boualem A. Genetic control of glandular trichome development. Trends Plant Sci. 2020;25:477–487. doi: 10.1016/j.tplants.2019.12.025. [DOI] [PubMed] [Google Scholar]
- Chan P.P., Lowe T.M. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.K., Tan L.T.H., Chan K.G., Lee L.H., Goh B.H. Nerolidol: a sesquiterpene alcohol with multi-faceted pharmacological and biological activities. Molecules. 2016;21:529. doi: 10.3390/molecules21050529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Yu T., Yang Q., Li C., Xiong C., Gao S., Xie Q., Zheng F., Li H., Tian Z., et al. Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 2018;96:90–102. doi: 10.1111/tpj.14018. [DOI] [PubMed] [Google Scholar]
- Chen Y., Su D., Li J., Ying S., Deng H., He X., Zhu Y., Li Y., Chen Y., Pirrello J., et al. Overexpression of bHLH95, a basic helix-loop-helix transcription factor family member, impacts trichome formation via regulating gibberellin biosynthesis in tomato. J. Exp. Bot. 2020;71:3450–3462. doi: 10.1093/jxb/eraa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Concepcion G.T., Feng X., Zhang H., Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocoll C., Asbach J., Novak J., Gershenzon J., Degenhardt J. Terpene synthases of oregano (Origanum vulgare L.) and their roles in the pathway and regulation of terpene biosynthesis. Plant Mol. Biol. 2010;73:587–603. doi: 10.1007/s11103-010-9636-1. [DOI] [PubMed] [Google Scholar]
- Dong A.X., Xin H.B., Li Z.J., Liu H., Sun Y.Q., Nie S., Zhao Z.N., Cui R.F., Zhang R.G., Yun Q.Z., et al. High-quality assembly of the reference genome for scarlet sage, Salvia splendens, an economically important ornamental plant. GigaScience. 2018;7 doi: 10.1093/gigascience/giy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N., Pichersky E. Metabolic engineering of plant volatiles. Curr. Opin. Biotechnol. 2008;19:181–189. doi: 10.1016/j.copbio.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S., Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinf. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami Bistgani Z., Ataollah Siadat S., Bakhshandeh A., Ghasemi Pirbalouti A., Hashemi M., Maggi F., Reza Morshedloo M. Application of combined fertilizers improves biomass, essential oil yield, aroma profile, and antioxidant properties of Thymus daenensis Celak. Ind. Crops Prod. 2018;121:434–440. [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E., Gavin O.L., Gunasekaran P., Ceric G., Forslund K., et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.M., Hubley R., Goubert C., Rosen J., Clark A.G., Feschotte C., Smit A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraternale D., Flamini G., Ricci D., Giomaro G. Flowers volatile profile of a rare red apple tree from Marche region (Italy) J. Oleo Sci. 2014;63:1195–1201. doi: 10.5650/jos.ess14088. [DOI] [PubMed] [Google Scholar]
- Gavarić N., Kladar N., Mišan A., Nikolić A., Samojlik I., Mimica-Dukić N., Božin B. Postdistillation waste material of thyme (Thymus vulgaris L., Lamiaceae) as a potential source of biologically active compounds. Ind. Crops Prod. 2015;74:457–464. [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.V., Thomas G.W.C., Lugo-Martinez J., Hahn M.W. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- Hong J.Y., Kim H., Jeon W.J., Baek S., Ha I.H. Antioxidative effects of Thymus quinquecostatus Celak through mitochondrial biogenesis improvement in RAW 264.7 macrophages. Antioxidants. 2020;9:548. doi: 10.3390/antiox9060548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchelmann A., Boutry M., Hachez C. Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol. 2017;175:6–22. doi: 10.1104/pp.17.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kalvari I., Argasinska J., Quinones-Olvera N., Nawrocki E.P., Rivas E., Eddy S.R., Bateman A., Finn R.D., Petrov A.I. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2017;46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J., Wenk M., Erickson J.L., Schattat M.H., Grau J., Hartung F. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016;44:e89. doi: 10.1093/nar/gkw092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M.G., Lisch D. Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA. 1997;94:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Hwang J.W., Sung S.H., Park S.J., Kim Y.T., Kim E.K., Moon S.H., Jeon B.T., Park P.J. Protective effect of carvacrol from Thymus quinquecostatus Celak against tert-butyl hydroperoxide-induced oxidative damage in Chang cells. Food Sci. Biotechnol. 2015;24:735–741. [Google Scholar]
- Klingenberg H., Aßhauer K.P., Lingner T., Meinicke P. Protein signature-based estimation of metagenomic abundances including all domains of life and viruses. Bioinformatics. 2013;29:973–980. doi: 10.1093/bioinformatics/btt077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Fedorova N.D., Jackson J.D., Jacobs A.R., Krylov D.M., Makarova K.S., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Rao B.S., et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinf. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou O., Walia S., Dhaliwal G.S. Essential oils as green pesticides: potential and constrains. Biopestic. Int. 2008;4:63–84. [Google Scholar]
- Krause S.T., Liao P., Crocoll C., Boachon B., Förster C., Leidecker F., Wiese N., Zhao D., Wood J.C., Buell C.R., et al. The biosynthesis of thymol, carvacrol, and thymohydroquinone in Lamiaceae proceeds via cytochrome P450s and a short-chain dehydrogenase. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2110092118. e2110092118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome project data processing Supgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Li J., Dong Y., Hao H., Ling Z., Bai H., Wang H., Cui H., Shi L. Time-series transcriptome provides insights into the gene regulation network involved in the volatile terpenoid metabolism during the flower development of lavender. BMC Plant Biol. 2019;19:313. doi: 10.1186/s12870-019-1908-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang Y., Dong Y., Zhang W., Wang D., Bai H., Li K., Li H., Shi L. The chromosome-based lavender genome provides new insights into Lamiaceae evolution and terpenoid biosynthesis. Hortic. Res. 2021;8:53. doi: 10.1038/s41438-021-00490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.W., Ian C.H. Vol. 17. Science Press (Beijing) and Missouri Botanical Garden Press; 1994. pp. 186–188. (Flora of China). [Google Scholar]
- Lima A.S., Schimmel J., Lukas B., Novak J., Barroso J.G., Figueiredo A.C., Pedro L.G., Degenhardt J., Trindade H. Genomic characterization, molecular cloning and expression analysis of two terpene synthases from Thymus caespititius (Lamiaceae) Planta. 2013;238:191–204. doi: 10.1007/s00425-013-1884-2. [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu D., Khan A.R., Liu B., Wu M., Huang L., Wu J., Song G., Ni H., Ying H., et al. NbGIS regulates glandular trichome initiation through GA signaling in tobacco. Plant Mol. Biol. 2018;98:153–167. doi: 10.1007/s11103-018-0772-3. [DOI] [PubMed] [Google Scholar]
- Lockton S., Gaut B.S. Plant conserved non-coding sequences and paralogue evolution. Trends Genet. 2005;21:60–65. doi: 10.1016/j.tig.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Maes L., Goossens A. Hormone-mediated promotion of trichome initiation in plants is conserved but utilizes species- and trichome-specific regulatory mechanisms. Plant Signal. Behav. 2010;5:205–207. doi: 10.4161/psb.5.2.11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleci B.L., Giuliani C. In: Proceedings of the First International Symposium on the Labiatae: Advances in Production, Biotechnology and Utilisation. Cervelli C., Ruffoni B., Dalla G.C., editors. International Society for Horticultural Science (ISHS); Leuven, Belgium: 2006. The glandular trichomes of the Labiatae. A review; pp. 85–90. Acta Horticulturae 723. [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matías-Hernández L., Jiang W., Yang K., Tang K., Brodelius P.E., Pelaz S. AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 2017;90:520–534. doi: 10.1111/tpj.13509. [DOI] [PubMed] [Google Scholar]
- Mcgarvey D.J., Croteau R. Terpenoid metabolism. Plant Cell. 1995;7:1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshedloo M.R., Craker L.E., Salami A., Nazeri V., Sang H., Maggi F. Effect of prolonged water stress on essential oil content, compositions and gene expression patterns of mono-and sesquiterpene synthesis in two oregano (Origanum vulgare L.) subspecies. Plant Physiol. Biochem. 2017;111:119–128. doi: 10.1016/j.plaphy.2016.11.023. [DOI] [PubMed] [Google Scholar]
- Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P., Novák P., Hoštáková N., Macas J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA. 2019;10:1. doi: 10.1186/s13100-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S., Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal-repeat retrotransposons. Plant Physiol. 2017;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Pavela R., Maggi F., Cianfaglione K., Bruno M., Benelli G. Larvicidal activity of essential oils of five Apiaceae taxa and some of their main constituents against Culex quinquefasciatus. Chem. Biodivers. 2018;15:e1700382. doi: 10.1002/cbdv.201700382. [DOI] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes M., Marques J.C., Câmara J. Headspace solid-phase microextraction-gas chromatography-quadrupole mass spectrometric methodology for the establishment of the volatile composition of Passiflora fruit species. Microchem. J. 2009;93:1–11. [Google Scholar]
- Price A.L., Jones N.C., Pevzner P.A. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Puttick M.N. MCMCtreeR: functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics. 2019;35:5321–5322. doi: 10.1093/bioinformatics/btz554. [DOI] [PubMed] [Google Scholar]
- Qiao H.L., Lu P.F., Xu R., Chen J., Wang X., Ma W.S., Liu T.N. Analysis of volatile compounds of inflorescence by GC-MS from Cistanche deserticola. J. Chin. Med. Mater. 2012;35:573–577. [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Robert S., Tissier A. Glandular trichomes: mircro-organs with model status? New Phytol. 2020;225:2251–2266. doi: 10.1111/nph.16283. [DOI] [PubMed] [Google Scholar]
- Rudolph K., Parthier C., Egerer-Sieber C., Geiger D., Muller Y.A., Kreis W., Müller-Uri F. Expression, crystallization and structure elucidation of γ-terpinene synthase from Thymus vulgaris. Acta Crystallogr. F Struct. Biol. Commun. 2016;72:16–23. doi: 10.1107/S2053230X15023043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov A.A., Solovyev V.V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She R., Chu J.S.C., Wang K., Pei J., Chen N. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 2009;19:143–149. doi: 10.1101/gr.082081.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P., Fu X., Shen Q., Liu M., Pan Q., Tang Y., Jiang W., Lv Z., Yan T., Ma Y., et al. The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol. 2018;217:261–276. doi: 10.1111/nph.14789. [DOI] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Singleton C.M., Petriglieri F., Kristensen J.M., Kirkegaard R.H., Michaelsen T.Y., Andersen M.H., Kondrotaite Z., Karst S.M., Dueholm M.S., Nielsen P.H., et al. Connecting structure to function with the recovery of over 1000 high-quality metagenome-assembled genomes from activated sludge using long-read sequencing. Nat. Commun. 2021;12:2009. doi: 10.1038/s41467-021-22203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl-Biskup E., Venskutonis R.P. Second edition. Vol. 1. Woodhead Publishing Series in Food Science, Technology and Nutrition; 2012. pp. 499–525. (Handbook of herbs and spices). [Google Scholar]
- Stanke M., Diekhans M., Baertsch R., Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Steuernagel B., Jupe F., Witek K., Jones J.D.G., Wulff B.B.H. NLR-parser: rapid annotation of plant NLR complements. Bioinformatics. 2015;31:1665–1667. doi: 10.1093/bioinformatics/btv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak J.H., Isman M.B. Metabolismof citral, the major constituent of lemongrass oil, in the cabbage looper, Trichoplusia ni, and effects of enzyme inhibitors on toxicity and metabolism. Pestic. Biochem. Physiol. 2016;133:20–25. doi: 10.1016/j.pestbp.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Tang H.B., Krishnakumar V., Li J.P., Kim M., Zhang X.T. jcvi: JCVI utility libraries. Zenodo. 2015 doi: 10.5281/zenodo.31631. Preprint at. [DOI] [Google Scholar]
- Tang S., Lomsadze A., Borodovsky M. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2015;43:e78. doi: 10.1093/nar/gkv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinf. 2009;4:4–10. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- Tholl D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006;9:297–304. doi: 10.1016/j.pbi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Tissier A. Glandular trichomes: what comes after expressed sequence tags? Plant J. 2012;70:51–68. doi: 10.1111/j.1365-313X.2012.04913.x. [DOI] [PubMed] [Google Scholar]
- Utturkar S.M., Klingeman D.M., Hurt R.A., Brown S.D. A case study into microbial genome assembly gap sequences and finishing strategies. Front. Microbiol. 2017;8:1272. doi: 10.3389/fmicb.2017.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanBerkum N.L., Lieberman-Aiden E., Williams L., Imakaev M., Gnirke A., Mirny L.A., Dekker J., Lander E.S. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp. 2010;39:1869. doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y., Park S., Kim S., Lee D., Kim G., Kim Y., Park K.H., Lee H. Use of hTERT and HPV E6/E7 mRNA RT-qPCR TaqMan assays in combination for diagnosing high-grade cervical lesions and malignant tumors. Am. J. Clin. Pathol. 2015;143:344–351. doi: 10.1309/AJCPF2XGZ2XIQYQX. [DOI] [PubMed] [Google Scholar]
- Wang X., Xu H.R., Li T., Qu L., Zhao Z.D., Zhang Z.Y. Expression analysis of KAP9.2 and Hoxc13 genes during different cashmere growth stages by qRT-PCR method. Mol. Biol. Rep. 2014;41:5665–5668. doi: 10.1007/s11033-014-3435-8. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Yang H., Wang S., Zhao J., Liu C., Gao L., Xia E., Lu Y., Tai Y., She G., et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA. 2018;115:E4151–E4158. doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werker E. Trichome diversity and development. Adv. Bot. Res. 2000;31:1–35. [Google Scholar]
- Wheeler T.J., Clements J., Eddy S.R., Hubley R., Jones T.A., Jurka J., Smit A.F.A., Finn R.D. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 2013;41:D70–D82. doi: 10.1093/nar/gks1265. –D82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T.E., Takebayashi N., Barker M.S., Mayrose I., Greenspoon P.B., Rieseberg L.H. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.B., Ni Z.Y., Shi Q.W., Dong M., Kiyota H., Gu Y.C., Cong B. Constituents from Salvia species and their biological activities. Chem. Rev. 2012;112:5967–6026. doi: 10.1021/cr200058f. [DOI] [PubMed] [Google Scholar]
- Wu Z.Y. Science Press; Beijing: 1977. Flora of China; pp. 194–198. [Google Scholar]
- Xia E., Tong W., Hou Y., An Y., Chen L., Wu Q., Liu Y., Yu J., Li F., Li R., et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into genome evolution and adaptation of tea plants. Mol. Plant. 2020;13:1013–1026. doi: 10.1016/j.molp.2020.04.010. [DOI] [PubMed] [Google Scholar]
- Xiang Y., An S., Cheng M., Liu L., Xie Y. Changes of soil microbiological properties during grass litter decomposition in Loess Hilly Region, China. Int. J. Environ. Res. Public Health. 2018;15:1797. doi: 10.3390/ijerph15091797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Zheng J.F., Liu S., Peng C., Zhou Y.M., Yang Q.Y., Zhang H.Y. De novo plant genome assembly based on chromatin interactions: a case study of Arabidopsis thaliana. Mol. Plant. 2015;8:489–492. doi: 10.1016/j.molp.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Xu H., Song J., Luo H., Zhang Y., Li Q., Zhu Y., Xu J., Li Y., Song C., Wang B., et al. Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant. 2016;9:949–952. doi: 10.1016/j.molp.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Bi C., Wu G., Wei S., Dai X., Yin T., Ye N. VGSC: a web-based vector graph toolkit of genome synteny and collinearity. BioMed Res. Int. 2016;2019:2150291. doi: 10.1155/2019/2150291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T., Chen M., Shen Q., Li L., Fu X., Pan Q., Tang Y., Shi P., Lv Z., Jiang W., et al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 2017;213:1145–1155. doi: 10.1111/nph.14205. [DOI] [PubMed] [Google Scholar]
- Yan T., Li L., Xie L., Chen M., Shen Q., Pan Q., Fu X., Shi P., Tang Y., Huang H., et al. A novel HD-ZIP-/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol. 2018;218:567–578. doi: 10.1111/nph.15005. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yan F., Tang Y., Yuan Y., Deng W., Li Z. Auxin response gene SlARF3 plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes. Plant Cell Physiol. 2015;56:2110–2124. doi: 10.1093/pcp/pcv136. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang S., Zhao Q., Ming R., Tang H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants. 2019;5:833–845. doi: 10.1038/s41477-019-0487-8. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Chen X.Y., Martin C. Scutellaria baicalensis, the golden herb from the garden of Chinese medicinal plants. Sci. Bull. 2016;61:1391–1398. doi: 10.1007/s11434-016-1136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Yang J., Cui M.Y., Liu J., Fang Y., Yan M., Qiu W., Shang H., Xu Z., Yidiresi R., et al. The reference genome sequence of Scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis. Mol. Plant. 2019;12:935–950. doi: 10.1016/j.molp.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Zhou F., Pichersky E. More is better: the diversity of terpene metabolism in plants. Curr. Opin. Plant Biol. 2020;55:1–10. doi: 10.1016/j.pbi.2020.01.005. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Tan H., Li Q., Li Q., Wang Y., Bu Q., Li Y., Wu Y., Chen W., Zhang L. TRICHOME AND ARTEMISININ REGULATOR 2 positively regulates trichome development and artemisinin biosynthesis in Artemisia annua. New Phytol. 2020;228:932–945. doi: 10.1111/nph.16777. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A., Van de Peer Y. wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 2019;35:2153–2155. doi: 10.1093/bioinformatics/bty915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence and genome assembly data were deposited in NCBI under project accession number PRJNA690675. The genome assembly and annotation data were also deposited in the China National Genomics Data Center under project accession number GWHBJVA00000000.