Summary
Our recent study demonstrated the generation of induced tissue-specific stem/progenitor (iTS/iTP) cells by the transient overexpression of reprogramming factors combined with tissue-specific selection. Here, we present a protocol to reprogram human hepatocytes to generate human induced tissue-specific liver stem (iTS-L) cells. Human hepatocytes are transfected with Sendai virus vectors (SeV) expressing OCT3/4, SOX2, KLF4, and c-MYC. iTS-L cells continuously express mRNA of hepatocyte-specific markers (HNF1β and HNF4α) and do not form teratomas.
For complete details on the use and execution of this protocol, please refer to Nakashima et al. (2022).1
Subject areas: Cell culture, Health Sciences, Stem Cells, Cell Differentiation
Graphical abstract
Highlights
-
•
A protocol for reprogramming human hepatocytes to iTS-L cells
-
•
Using Sendai virus vectors expressing OCT3/4, SOX2, KLF4, and c-MYC
-
•
Steps for culture of human hepatocytes, viral transfection, and culture of iTS-L cells
-
•
A protocol for the selection of iPS and iTS-L cells
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Our recent study demonstrated the generation of induced tissue-specific stem/progenitor (iTS/iTP) cells by the transient overexpression of reprogramming factors combined with tissue-specific selection. Here, we present a protocol to reprogram human hepatocytes to generate human induced tissue-specific liver stem (iTS-L) cells. Human hepatocytes are transfected with Sendai virus vectors (SeV) expressing OCT3/4, SOX2, KLF4, and C-MYC. iTS-L cells continuously express mRNA of hepatocyte-specific markers (HNF1b and HNF4a) and do not form teratomas.
Before you begin
Prepare the media below. Prewarm the medium intended for cell culture at 37°C at least 30 min prior to beginning each section of this protocol. Refer to the key resources table for a complete list of materials.
-
1.
Human hepatocyte culture medium: Kaly-Cell Thawing Medium (KLC-TM), Kaly-Cell Seeding Medium (KLC-SM), hepatocyte basal medium.
-
2.
Human embryonic stem (ES)/iPS/iTS-L cell culture medium: Primate ES Cell Medium with 5 ng/mL bFGF.
-
3.
Mouse embryonic fibroblast (MEF) culture medium: Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S).
-
4.
All cell types are cultured in an incubator at 37°C, 5% CO2, and 85% humidity.
Institutional permissions
All experimental protocols were in accordance with the guidelines for the care and use of laboratory animals set by Research Laboratory Center, Faculty of Medicine, and the Institute for Animal Experiments, Faculty of Medicine, University of the Ryukyus (Okinawa, Japan).
Mouse embryonic fibroblasts (MEFs) thawing and culturing
Timing: 1 day
-
5.
Add 1 mL of 0.1% gelatin solution to the 60 mm dishes and incubate the dish for 30 min at 37°C.
-
6.
MEFs (inactivated by mitomycin) are obtained from a vender; see key resources table in this protocol. Thaw one frozen vial of murine embryonic fibroblasts (MEFs) (3.0 × 106 cells) in a 37°C water bath.
-
7.
Transfer the content of the vial into a 15 mL tube containing 10 mL of MEF culture medium.
-
8.
Centrifuge the samples at 700 × g for 5 min to pellet the cells.
-
9.
Remove the supernatant.
-
10.
Resuspend the cell pellet in 5 mL of MEF culture medium using a 5 mL pipette to a single cell suspension, by pipetting up and down 5–7 times.
-
11.
Aspirate the gelatin solution from the 60 mm dishes.
-
12.
Transfer the cell suspension into 60 mm dishes (6.0 × 105 cells/dish).
-
13.
Place the MEFs in an incubator at 37°C, 5% CO2, and 85% humidity. Feeders can be used up to 5 days after preparation. The cells are then renewed with fresh MEF culture medium every two days.
Human hepatocyte thawing and culturing
Timing: 1 week
-
14.
Human hepatocytes are obtained from a vender; see the key resources table in this protocol. Thaw one frozen vial of human hepatocytes (3.0 × 106 cells) in a 37°C water bath.
-
15.
Transfer the content of the vial into a 15 mL tube containing 10 mL of KLC-TM medium.
-
16.
Centrifuge the samples at 700 × g for 5 min to pellet the cells.
-
17.
Remove the supernatant.
-
18.
Resuspend the cell pellet in 5 mL of KLC-SM medium using a 5 mL pipette to a single cell suspension pipetting up and down 5–7 times.
-
19.
Transfer the cell suspension into two collagen-coated 100 mm dishes and add 7.5 mL of KLC-SM medium to each dish (final 10 mL/dish).
-
20.
Place the human hepatocytes in an incubator at 37°C, 5% CO2, and 85% humidity. Renew the cells with fresh KLC-SM medium after 6 and 24 h. After 48 h, change the medium to hepatocyte basal medium. Renew with fresh hepatocyte basal medium every two days.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Gelatin | FUJIFILM Wako Pure Chemical Corporation | Cat# 190-15805 |
| Y-27632 | FUJIFILM Wako Pure Chemical Corporation | Cat# 257-00511 |
| KLC-TM medium | KaLy-Cell | Cat# KLC-TM |
| KLC-SM medium | KaLy-Cell | Cat# KLC-SM |
| Hepatocyte basal medium | Lonza | Cat# CC-3199 |
| Primate ES Cell Medium | ReproCELL | Cat# RCHEMD001 |
| Freezing medium for human ES/iPS cells (DAP213) | ReproCELL | Cat# RCHEFM001 |
| Recombinant human bFGF (FGF2) | ReproCELL | Cat# RCHEOT002 |
| D-PBS(-) | Nacalai Tesque | Cat# 11482-15 |
| 0.05% trypsin/EDTA | Thermo Fisher Scientific | Cat# 25300054 |
| DMEM | Wako | Cat# 043-30085 |
| Fetal bovine serum | Thermo Fisher Scientific | Cat# 10270-106 |
| CytoTune-iPS 2.0 | Medical & Biological Laboratories Co., Ltd. | Cat# IDT-DV0304 |
| Penicillin‒streptomycin solution (×100) | FUJIFILM Wako Pure Chemical Corporation | Cat# 16823191 |
| Hanks’ Balanced Salt Solution (HBSS) | Life Technologies | Cat# 14025092 |
| Critical commercial assays | ||
| SuperPREP II Cell Lysis & RT Kit for quantitative PCR | TOYOBO CO., LTD. | Cat# SCQ-401 |
| Luna Universal qPCR Master Mix | New England Biolabs Inc. | Cat# M3003E |
| TaqMan Array 96-Well FAST Plate(Human Stem Cell Pluripotency) | Applied Biosystems | Cat# 4418722 |
| TaqMan™ Fast Advanced Master Mix | Thermo Fisher Scientific | Cat# 4444963 |
| Experimental models: Cell lines | ||
| Cryopreserved Hepatocytes Species:Human, Lot#S1412T, Lot#S1238 and Lot#S1350 | KaLy-Cell | Cat# HHCPC-2 M |
| hiPSC Lines 201B7 | CiRA Foundation | N/A |
| MEF cells | ReproCELL Inc. | Cat# RCHEFC003 |
| Oligonucleotides | ||
| human OCT3/4 forward, GACAGGGGGAGGGGAGGAGCTAGG, human OCT3/4 reverse, CTTCCCTCCAACCAGTTGCCCCAAAC, |
Takahashi et al.2 | N/A |
| human SOX2 forward, GGGAAATGGGAGGGGTGCAAAAGAGG, human SOX2 reverse, TTGCGTGAGTGTGGATGGGATTGGTG, |
Takahashi et al.2 | N/A |
| human KLF4 forward, TGATTGTAGTGCTTTCTGGCTGGGCTCC, human KLF4 reverse, ACGATCGTGGCCCCGGAAAAGGACC, |
Takahashi et al.2 | N/A |
| human c-MYC forward, GCGTCCTGGGAAGGGAGATCCGGAGC, human c-MYC reverse, TTGAGGGGCATCGTCGCGGGAGGCTG, |
Takahashi et al.2 | N/A |
| human NANOG forward, CAGCCCCGATTCTTCCACCAGTCCC, human NANOG reverse, CGGAAGATTCCCAGTCGGGTTCACC, |
Takahashi et al.2 | N/A |
| human GDF3 forward, CTTATGCTACGTAAAGGAGCTGGG, human GDF3 reverse, GTGCCAACCCAGGTCCCGGAAGTT, |
Takahashi et al.2 | N/A |
| human REX1 forward, CAGATCCTAAACAGCTCGCAGAAT, human REX1 reverse, GCGTACGCAAATTAAAGTCCAGA, |
Takahashi et al.2 | N/A |
| human DNMT3b forward, TGCTGCTCACAGGGCCCGATACTTC, human DNMT3b reverse, TCCTTTCGAGCTCAGTGCACCACAAAAC, |
Takahashi et al.2 | N/A |
| human GAPDH forward, ACCACAGTCCATGCCATCAC, human GAPDH reverse, TCCACCACCCTGTTGCTGTA, |
NCBI Reference Sequence | NM 004048 |
| human β-ACTIN forward, CAACCGCGAGAAGATGAC, human β-ACTIN reverse, AGGAAGGCTGGAAGAGTG, |
Kajihara et al.3 | N/A |
| human HNF1β forward, CTGACTACCAGCTAACTCCAGTCTC, human HNF1β reverse, GACTGCAACTTTTTCTTCTGCTATC, |
NCBI Reference Sequence | NM_000458.3 |
| human HNF4α forward, GAACAGGAGCTCTTAACTACAGTGG, human HNF4α reverse, CTGTCAAGAGTCATGAATTCTCCTT, |
NCBI Reference Sequence | NM_000457.4 |
| human β-ACTIN forward, TGGCACCCAGCACAATGAA, human β-ACTIN reverse, CTAAGTCATAGTCCGCCTAGAAGCA, |
NCBI Reference Sequence | NM_001101 |
| Other | ||
| LightCycler 96 Real-Time PCR system | Roche | Cat# 05 815 916 001 |
| InvitrogenTM EVOSTM FL Auto Imaging System | Thermo Fisher Scientific | Cat# AMAFD1000 |
| C.B-17/Icr-scid/scidJcl, male, 8 week-old | CLEA Japan | N/A |
Step-by-step method details
Reprogramming of human hepatocytes
Timing: 3–4 weeks
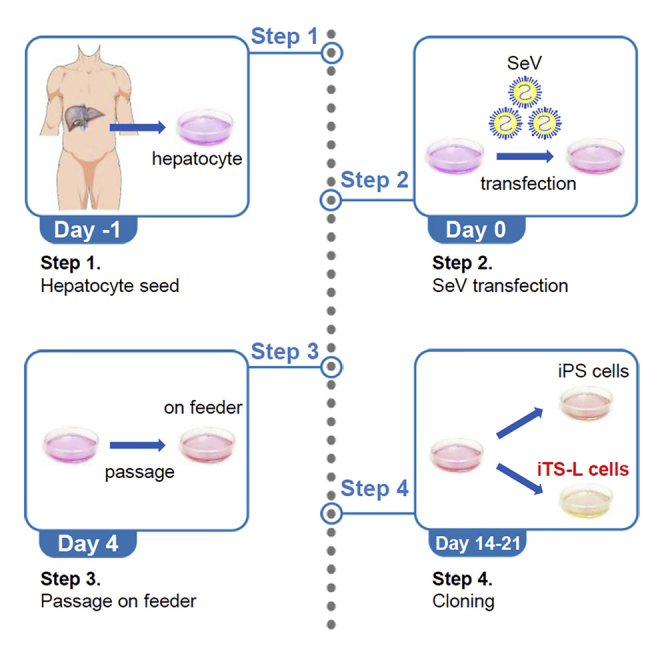
Human hepatocytes are reprogrammed into iPS/iTS-L cells using Sendai virus (SeV) vectors expressing OCT3/4, SOX2, KLF4, and c-MYC from a vender; see key resources table in this protocol.
-
1.
On Day -1, wash human hepatocytes twice with 10 mL of phosphate buffered saline (PBS).
-
2.
The cells are then dissociated with 0.05% trypsin/EDTA for 5 min.
-
3.
Add 5 mL of hepatocyte basal medium and break up cell aggregates by pipetting up and down with a 5 mL pipette.
-
4.
The samples are then centrifuged at 700 × g for 5 min to pellet the cells.
-
5.
Remove the supernatant.
-
6.
Count cells and dilute to 1.0 × 105 cells/mL in hepatocyte basal medium.
-
7.
Plate cells (1.0 × 105 cells/well) in 6-well plates.
-
8.
Human hepatocytes are placed in an incubator at 37°C, 5% CO2, and 85% humidity for 24 h.
-
9.
On Day 0, prepare a 15 mL tube containing 2 mL of hepatocyte basal medium with 10 μL of SeV KOS (5.0 × 105 CIU), 10 μL of SeV Klf4 (5.0 × 105 CIU), and 10 μL of SeV Myc (5.0 × 105 CIU) ((5.0 × 105 CIU)/(1.0 × 105 cells)=5 multiplicity of infection (MOI)) (Figure 1).
-
10.
Aspirate the culture medium.
-
11.
Add 2 mL hepatocyte basal medium with SeV.
-
12.
Place the 6-well plate in an incubator at 37°C, 5% CO2, and 85% humidity for 24 h.
-
13.
On Day 1, aspirate the culture medium and renew with fresh hepatocyte basal medium.
-
14.
Renew with fresh hepatocyte basal medium daily for 3 days.
-
15.
On Day 4, dissociate the hepatocytes and plate the hepatocytes in 60 mm dish containing MEFs using human ES/iPS/iTS-L cell culture medium.
-
16.
The 60 mm dish is then placed in an incubator at 37°C, 5% CO2, and 85% humidity.
-
17.
Change human ES/iPS/iTS-L cell culture medium daily.
-
18.
On Days 14–21, the reprogrammed cells should now transform to a round morphology. Every single clone should be independently expanded for characterization and freezing (Figure 2).
Figure 1.
Schematic Representation of Sendai Virus (SeV) vectors
(A) SeV vector that allows the expression of human KLF4, OCT3/4, and SOX2 proteins.
(B) SeV vector that allows the expression of human KLF4 protein.
(C) SeV vector that allows expression of human c-MYC protein. NP; Nucleocapsid Protein. P; Phosphoprotein. M; Matrix protein. HN; Hemagglutinin-Neuraminidase. L; Large protein.
Figure 2.
Time schedules for the induction of iPS/iTS-L cells transfected with SeV vectors
Open arrowheads indicate the times of cell seeding, passaging, and colony selection. Solid arrowheads indicate the times of transfection.
Teratoma formation assay
Timing: 10–15 weeks
The colonies similar to human ES cells or gut tube endodermal (GTE) cells (Figure 3) are selected for further cultivation and evaluation. Colonies similar to human ES cells should be iPS cells and generate teratomas. The colonies similar to GTE cells should be iTS-L cells and generate no teratomas.
-
19.
Immunodeficient male mice (age: 7 weeks; C. B-17/Icr-scid/scidJcl) are anesthetized with isoflurane inhalation.
-
20.
A total of 1.0 × 106 or more cells in 0.1 mL of cold Hanks' balanced salt solution (HBSS) are subcutaneously injected into the shoulders and buttocks using a 22 G injection needle.
-
21.
The mice are examined daily, and tumors are extracted at 10 or 15 weeks after surgery.
Figure 3.
The morphologies of iPS and iTS-L cells
(A) The morphology of iPS cells.
(B) The morphology of iTS-L cells. The morphologies of iPS cells are similar to those of human ES cells. The morphologies of iTS-L cells are similar to those of gut tube endodermal cells. Scale bars = 200 μm.
Quantitative RT‒PCR
Timing: 1–2 days
The iTS-L cells continuously express HNF1β and HNF4α mRNA but not iPS cells.
-
22.
The iPS/iTS-L cells are cultured in Primate ES Cell Medium to approximately 80% confluence.
-
23.
RNA is prepared using a SuperPREP II Cell Lysis & RT Kit for quantitative PCR according to the manufacturer’s instructions.
-
24.
Real-time PCR analyses are performed using a LightCycler 96 Real-Time PCR system. The PCR protocol is as follows. Luna Universal qPCR Master Mix is used according to the manufacturer’s instructions.
PCR master mix
| Reagent | Final concentration | Amount |
|---|---|---|
| Luna Universal qPCR Master Mix | 1× | 10 μL |
| Forward primer (10 μM) | 0.25 μM | 0.5 μL |
| Forward primer (10 μM) | 0.25 μM | 0.5 μL |
| Template DNA | < 100 ng | variable |
| ddH2O | N/A | variable |
| Total | N/A | 20 μL |
PCR cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Initial Denaturation | 95°C | 10 min | 1 |
| Denaturation | 95°C | 15 s | 40 cycles |
| Annealing | 60°C | 60 s | |
| Denaturation | 95°C | 15 s | 1 [Melt Curve Stage] |
| Annealing | 60°C | 60 s | |
| Denaturation | 95°C | 15 s |
Expected outcomes
iPS/iTS-L cells can be generated and passaged within 3–4 weeks. iPS/iTS-L clones can be expanded for characterization. We recommend the following characterization assays for distinguishing iPS/iTS-L cells: quantitative RT‒PCR for the detection of markers of hepatic stem cells (iPS: NANOG(+), OCT3/4(+), HNF1β(-), and HNF4α(-)/iTS-L: NANOG(±) (less than 1/4 that of iPS), OCT3/4(±) (less than 1/4 that of iPS), HNF1β(+) and HNF4α(+)) and teratoma formation using immunodeficient mice.1 Commercially available iPS cells and original hepatocytes should be used as positive/negative controls for expression.
Limitations
Although the generation efficiency of human iPS cells is low and reprogramming rates vary from 10% to 0.0001%,4,5 the efficiency of iTS cells is relatively higher.6
The following limitations should be mentioned specifically. First, iPS/iTS-L clones should be expanded 3–5 passages before characterization to distinguish iPS/iTS-L cells. In low-passage iPS/iTS-L cells, transgenes derived from SeV may remain. The remaining reprogramming genes may change the characterization of iPS/iTS-L cells. Second, based on our experience, low passage hepatocytes (passages 2–5) should be used for reprogramming. Enzymatic dissociation or passaging and long-term culture have been described to affect the epigenetic state of the cell and to hinder efficient reprogramming.7,8,9 Third, this protocol renders efficient reprogramming when using hepatocytes; other cell types for the generation of other induced tissue-specific stem cells may require further optimization.
Troubleshooting
Problem 1
Human hepatocytes do not proliferate properly (related to step 18 of “before you begin”).
Potential solution
The cells are not diluted to less than 1 × 104 cells/cm2. Low confluency gives rise to poor cell proliferation and early senescence. When human hepatocytes do not proliferate properly after 3–5 days of cell culture, we recommend replating the cells into new dishes at a higher cell density.
Problem 2
Excessive cell death after SeV transfection (related to step 9).
Potential solution
Check the confluency of human hepatocytes at the moment of transfection. The uneven distribution of human hepatocytes may result in cell death after SeV transfection. It should be over 50% for proper survival after transduction.
Problem 3
The generation efficiency of human iPS/iTS-L cells is extremely low (related to step 9).
Potential solution
Increase SeV at 6–10 MOI.
Problem 4
iTS-L cells do not grow well (related to step 18).
Potential solution
Increase the number of cells initially applied to the well and thereby increase cell density.
Problem 5
iTS-L cells spontaneously differentiate (related to step 18).
Potential solution
New bFGF and Primate ES Cell Medium are prepared.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hirofumi Noguchi noguchih@med.u-ryukyu.ac.jp.
Materials availability
All material used are listed in the key resources table, and any further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact.
Acknowledgments
This work was supported in part by JSPS KAKENHI (grant numbers JP22K08759, JP21K19537, JP20H03745, and JP19K09051), the Okinawa Science and Technology Innovation System Construction Project (OSTC), and the Japan Agency for Medical Research and Development (AMED) (grant number JP22bm0104001).
Author contributions
Conceived and Designed Experiments, H.N.; Performed Experiments, H.N., Y.N., C.S.; Analyzed the Data, H.N., Y.N., C.S.; Wrote the Manuscript, H.N.; Funding Acquisition, H.N., M.W., M.M., M.T., I.S.
Declaration of interests
The authors declare no competing interests.
Data and code availability
This protocol does not include the generation of datasets.
References
- 1.Nakashima Y., Miyagi-Shiohira C., Saitoh I., Watanabe M., Matsushita M., Tsukahara M., Noguchi H. Induced hepatic stem cells are suitable for human hepatocyte production. iScience. 2022;25:105052. doi: 10.1016/j.isci.2022.105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Kajihara R., Numakawa T., Odaka H., Yaginuma Y., Fusaki N., Okumiya T., et al. Novel Drug Candidates Improve Ganglioside Accumulation and Neural Dysfunction in GM1 Gangliosidosis Models with Autophagy Activation. Stem Cell Reports. 2020;14:909–923. doi: 10.1016/j.stemcr.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Muñoz E., Arboleda-Estudillo Y., Otu H.H., Cibelli J.B. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345:822–825. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Munoz E., Cibelli J.B. Somatic cell reprogramming informed by the oocyte. Stem Cells Dev. 2018;27:871–887. doi: 10.1089/scd.2018.0066. [DOI] [PubMed] [Google Scholar]
- 6.Noguchi H., Miyagi-Shiohira C., Nakashima Y., Kinjo T., Kobayashi N., Saitoh I., Watanabe M., Shapiro A.M.J., Kin T. Induction of expandable tissue-specific progenitor cells from human pancreatic tissue through transient expression of defined factors. Mol. Ther. Methods Clin. Dev. 2019;13:243–252. doi: 10.1016/j.omtm.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halley-Stott R.P., Gurdon J.B. Epigenetic memory in the context of nuclear reprogramming and cancer. Brief. Funct. Genomics. 2013;12:164–173. doi: 10.1093/bfgp/elt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I.R., et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Streckfuss-Bömeke K., Wolf F., Azizian A., Stauske M., Tiburcy M., Wagner S., Hübscher D., Dressel R., Chen S., Jende J., et al. Comparative study of human-induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes, and skin fibroblasts. Eur. Heart J. 2012;34:2618–2629. doi: 10.1093/eurheartj/ehs203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol does not include the generation of datasets.





