Abstract
Genome-wide association studies (GWAS) have identified hundreds of loci associated with Crohn’s disease (CD). However, as with all complex diseases, robust identification of the genes dysregulated by non-coding variants typically driving GWAS discoveries has been challenging. Here, to complement GWAS and better define actionable biological targets, we analyzed sequence data from more than 30,000 CD patients and 80,000 population controls. We directly implicate ten genes in general onset CD for the first time via association to coding variation, four of which lie within established CD GWAS loci. In nine instances a single coding variant is significantly associated and in the tenth, ATG4C, we see additionally a significantly increased burden of very rare coding variants in Crohn’s disease cases. In addition to reiterating the central role of innate and adaptive immune cells as well as autophagy in CD pathogenesis, these newly associated genes highlight the emerging role of mesenchymal cells in the development and maintenance of intestinal inflammation.
Introduction
GWAS in Crohn’s Disease (CD), and inflammatory bowel disease (IBD) more generally, have successfully identified more than 200 loci contributing to risk of disease1–4. While most GWAS hits do not immediately implicate an obvious functional variant or gene, a subset have been directly mapped to coding variants (e.g., NOD2, IL23R, ATG16L1, SLC39A8, FUT2, TYK2, IFIH1, SLAMF8, PLCG2)5, providing more direct clues to pathogenesis. Further, targeted and genome-wide sequencing approaches have revealed additional, lower-frequency, disease-associated coding variants (e.g., CARD9, RNF186, ADCY7, INAVA/C1orf106, SLC39A8, NOD2)6–9 originally undetected by GWAS. Such coding variants, common and rare, have led to functional follow-up experiments demonstrating causal mechanisms for at least ten genes and have provided the most direct biological insights to emerge from genetic studies of IBD10–13.
Results
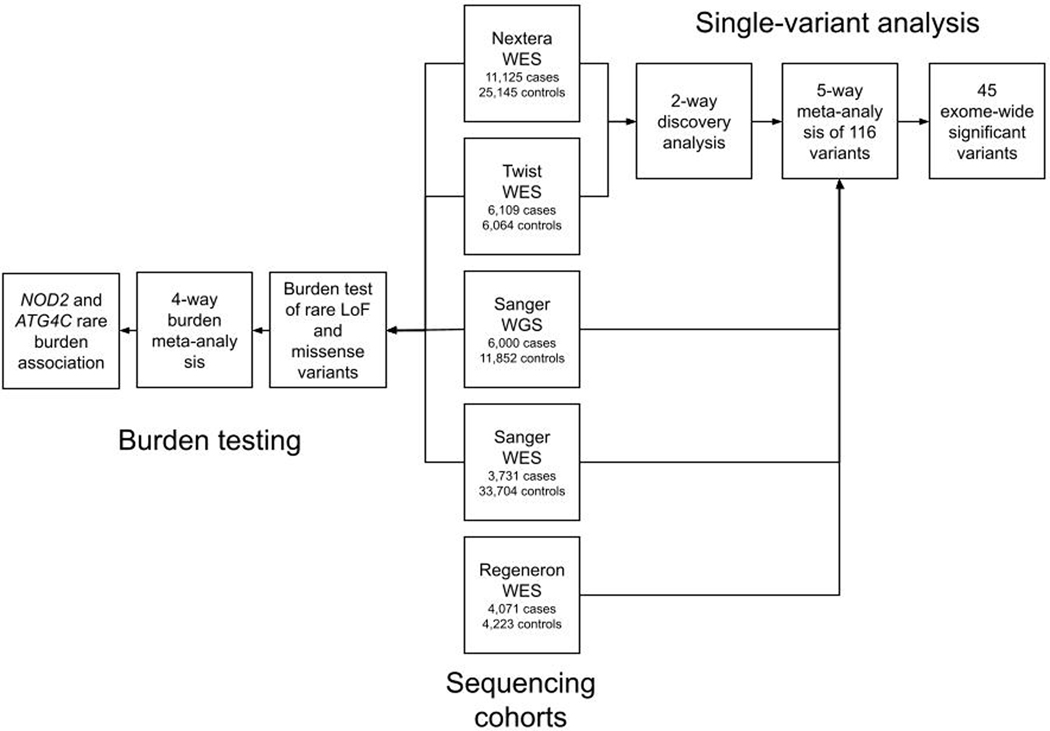
To further advance the interpretation of GWAS loci — and to define novel CD associated genes using variation rarer than that routinely detected by GWAS — we pursued large-scale exome sequencing using CD case and control collections from more than 35 centers in the International IBD Genetics Consortium. The primary analysis consisted of exome sequencing of 18,816 CD cases across 35 IBD studies and 13,412 non-IBD control samples from the same studies. These samples were all sequenced at the Broad Institute and were supplemented with 22,536 population controls from approved non-IBD studies sequenced contemporaneously at the Broad Institute and accessed from dbGAP (Supplementary Table 1 and Extended Data Figure 1). Two different exome capture platforms were employed during the course of the study (referred to hereafter as Nextera [Illumina] and Twist [Twist Biosciences]). Details of capture and sequencing of these cohorts (and those subsequently used in follow-up) are provided in Online Methods and Supplementary Information.
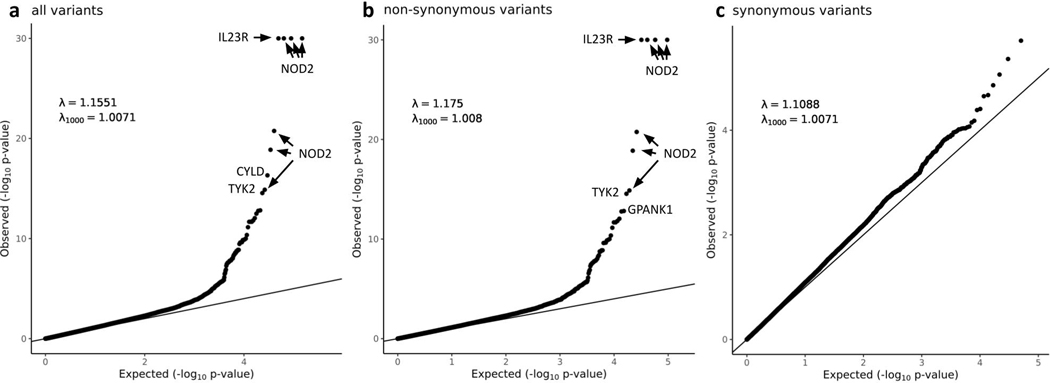
Calling and quality control (QC) of data from the two exome capture platforms were conducted in parallel (Table 1; Extended Data Figure 2 and Online Methods). Sensitivity to detect low-frequency coding variants was evaluated in each callset post-QC by comparison to passing sites in gnomAD v2.1 that had 0.0001 < non-Finnish European (NFE) minor allele frequency (MAF) < 0.1 (Online Methods). We observed that 84% of all exonic single nucleotide polymorphisms (SNPs) in this frequency range were detected in both CD datasets with sufficiently high quality to enter meta-analysis. Analysis of each dataset was conducted in SAIGE using a logistic mixed-model14, and a standard inverse-variance weighted (IVW) meta-analysis conducted across 164,149 non-synonymous variants with minor allele frequency (gnomAD NFE) between 0.0001 – 0.1. A study-wide significance threshold of 3 × 10−7 was applied (Supplementary Table 2). As a control for the entire study, we also analysed 96,326 synonymous variants. Forty-three sites (Supplementary Table 3) failed a heterogeneity-of-effect test between the Nextera and Twist discovery cohorts (IVW pHET < 0.0001) and were eliminated from further analysis. We did not observe an inflation in the exome-wide distribution of test statistics (Extended Data Figure 3).
Table 1.
Sample characteristics. Numbers listed are post-QC and are of samples entered into analysis.
| Sequencing Center | Data Type | Exome Capture | Study Stage | # CD Cases | # Controls |
|---|---|---|---|---|---|
| Broad | WES | Nextera | Discovery | 11,125 | 25,145 |
| Broad | WES | Twist | Discovery | 6,109 | 6,064 |
| Sanger | WGS | n/a | Follow-up | 6,000 | 11,852 |
| Sanger | WES | Agilent | Follow-up | 3,731 | 33,704* |
| Regeneron | WES | Agilent | Follow-up | 4,071 | 4,223 |
Whole-exome sequencing data derived from the UK Biobank cohort, sequenced by Regeneron using the IDT xGen Exome Research Panel v1.0 including supplemental probes, were used as population controls for the Sanger WES cases (Online Methods).
Exonic variants significantly associated with IBD
The most significantly-associated variants (p < 10−10) in the Discovery stage were previously-known CD variants (or variants in linkage disequilibrium [LD] with them), indicating the QC and analysis pipeline removed highly associated false positives. Twenty-eight variants achieved study-wide significance, including known variants within CD genes established in prior GWAS and sequencing efforts: NOD2, IL23R, LRRK2, TYK2, SLC39A8, IRGM and CARD9. Excluding synonymous variants in LD with these known associated non-synonymous variants, synonymous variants showed little deviation from the null and none reached study-wide significance (Extended Data Figure 3). Encouraged by this, we then nominated a list of 116 variants (including known variants) with p < 0.0002 for further evaluation in three follow-up cohorts.
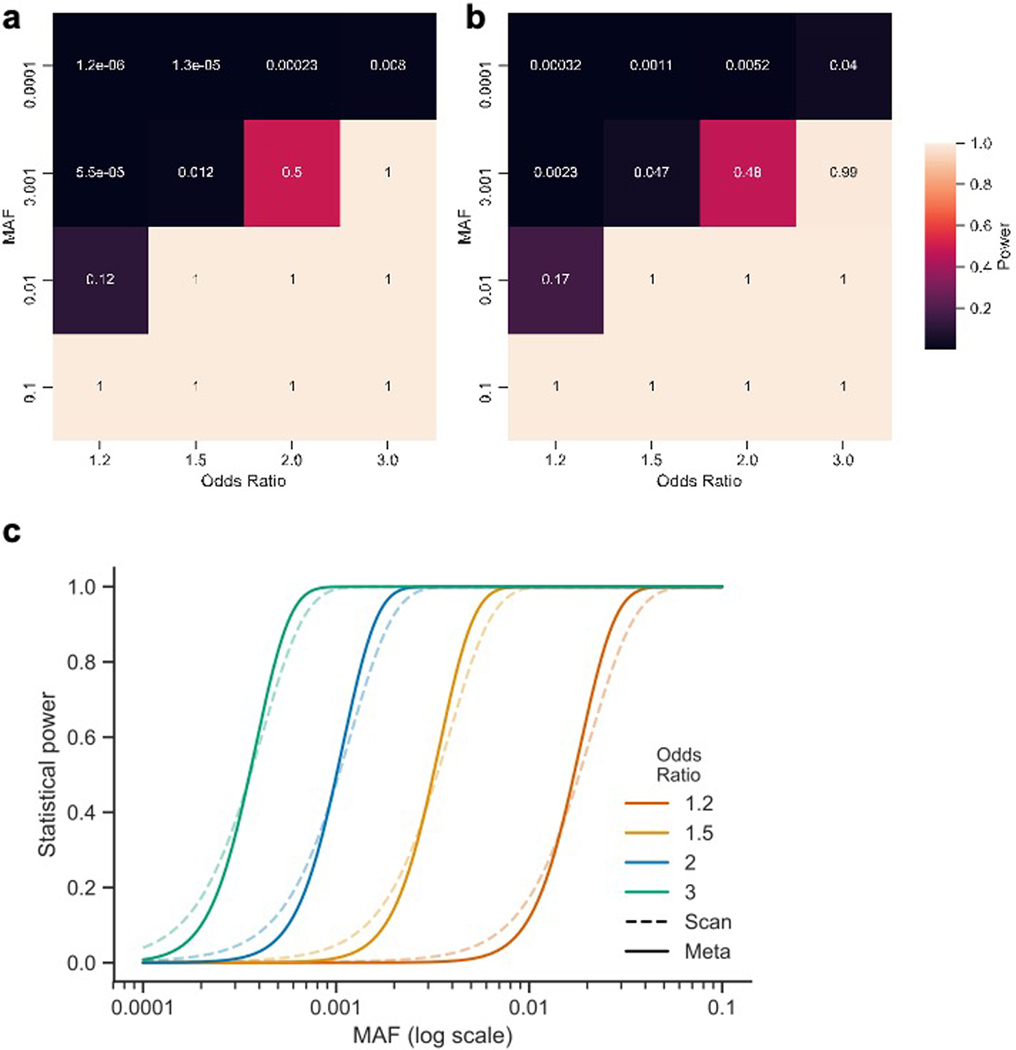
Additional exome and genome sequencing was undertaken at the Sanger Institute on an independent cohort of 9,731 CD cases ascertained by the UK IBD Genetics Consortium and IBD BioResource. Genome sequencing with a target depth of 15x was performed on 6,000 CD patients. Whole-genome sequences from 11,852 individuals from the INTERVAL blood donor cohort were used as population controls. Another 3,731 CD patients were exome sequenced using the Agilent SureSelect Human All Exon V5 capture. 33,704 individuals without IBD or other related diseases from the UK Biobank were used as controls for the Sanger WES cases. These UK Biobank samples were sequenced by Regeneron using the IDT xGen Exome Research Panel v1.0 (including supplemental probes), and thus QC and subsequent analyses were restricted to the intersection of the Agilent and the IDT capture regions. Exome and genome datasets were processed in parallel with similar QC parameters (Online Methods). Association analyses were performed using a logistic mixed-effects model implemented in the REGENIE v1.0.6.7 and v2.0.2 software, correcting for the case-control imbalance using the Firth correction. 28 of 116 variants were associated (p < 4.3 × 10−4 [0.05/116]) with CD in the meta-analysis of the two Sanger cohorts and 94 replicated the direction of effect seen in the discovery cohort (p = 3 × 10−12, binomial test). Summary statistics from a German dataset of 4,071 CD cases and 4,223 controls exome-sequenced at Regeneron (Online Methods) were also ascertained and an inverse-variance weighted meta-analysis carried out across all five cohorts (Table 1). 45 of the 116 variants exceeded the study-wide significance threshold, p < 3 × 10−7 (Supplementary Table 4 and Supplementary Data 1). Of note, all 45 study-wide significant sites in the discovery stage exome-wide scan showed stronger evidence of association in the meta-analysis, and none showed significant evidence of heterogeneity of effect across studies. The ‘scan’ exome-wide and the combined meta-analyses have similar power to detect the same true associations at their respective significance thresholds (Extended Data Figure 4).
Among the 164,149 low frequency non-synonymous variants tested for association in this study, 14 were mapped to a credible set with posterior inclusion probability (PIP) > 5% from a previous IBD fine-mapping study5. 8 of the 14 variants reached exome-wide significance (in NOD2, IL23R, TYK2, PTPN22, and CARD9). The remaining 6 variants (in GPR65, MST1, NOD2, and SMAD3) have genetic effects consistent with those previously reported, with P-values ranging from 0.025 to 8 × 10−5 in the WES discovery cohort. Together, these results demonstrate the accuracy of our exome sequencing study in the lowest frequency range covered by previous GWAS approaches.
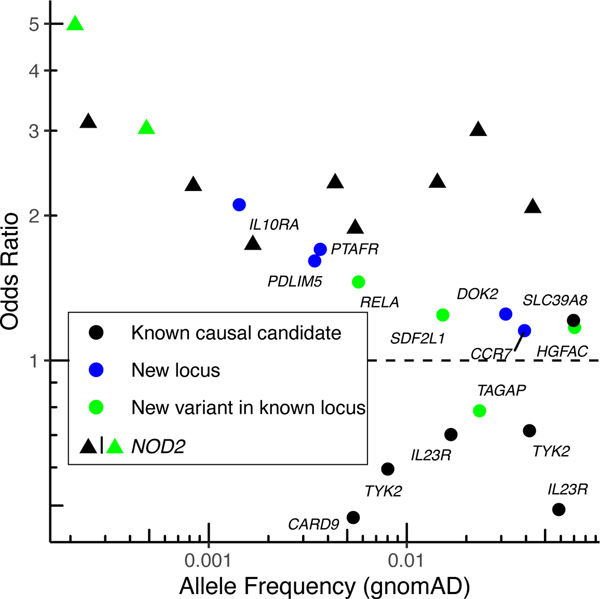
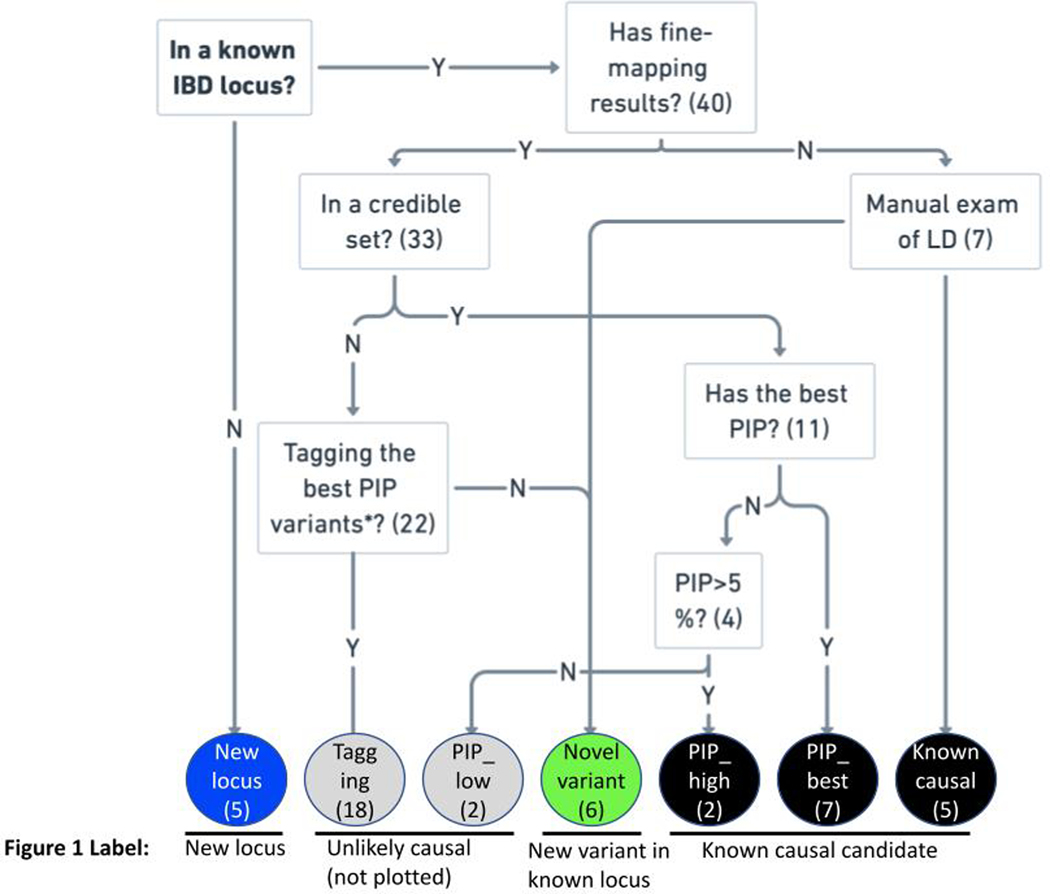
To identify new loci not yet implicated in CD and independent exonic association signals at known loci, we accounted for the LD between the 45 exome-wide significant variants and previously-reported IBD GWAS hits, as well as prior rare variant discoveries (Online Methods). We identified five coding variants in genes not previously implicated in IBD, even by their proximity to previous GWAS signals. We also discovered six independently associated novel exonic variants in genes previously known to harbor coding mutations underpinning CD or IBD risk, two of which are in NOD2 (“New locus” and “New variant in known locus” in Figure 1, Supplementary Table 4 and Extended Data Figure 5). 14 significant variants recaptured known IBD causal candidates from fine-mapping, including variants in CARD9, IL23R and NOD2, and the remaining 20 variants either tag the known causal variants through LD or have very small PIP from fine-mapping and thus are highly unlikely to be CD causal variants (“Known causal candidate” and “Unlikely causal” in Figure 1, Supplementary Table 4 and Extended Data Figure 5). A harmonized summary with findings from this study and the fine-mapping study5 for genes implicated by the 45 exome-wide significant variants is available in Supplementary Table 5. Of note, evidence from earlier GWAS and this study cannot be considered independent and trivially combined since there is considerable sample overlap - while 46% of scan samples come from clinical sites or national cohorts not previously involved in the largest GWAS4, the remainder are from sites which also contributed earlier recruited samples (generally 10 or more years ago) to previously reported GWAS studies and have been exome sequenced in this study. Similarly, at least 10% of the Sanger WGS samples used for meta-analysis were previously included in past genotyping studies.
Figure 1. Odds ratio and minor allele frequency for exome-wide significant findings that are not tagging stronger, established non-coding association signals.
Known causal candidate: in a credible set from a fine-mapping study5 with posterior inclusion probability > 5% or reported in previous studies6,8 (Online Methods). New locus: in a locus not yet implicated by GWAS. New variant in known locus: in a known GWAS locus, but represents an association independent from previously-reported IBD putative causal variants (Online Methods).
Some of the newly implicated CD genes (Table 2) contribute to biological pathways previously implicated via GWAS, such as autophagy (ATG4C), or Mendelian forms of IBD, such as the IL-10 signaling pathway15, or by extensive functional studies of inflammatory response in IBD, such as the NF-kB family of transcriptional regulators16. In contrast, many of the newly associated genes appear to be linked to the roles of mesenchymal cells in intestinal homeostasis, a pathway not previously implicated by genetic studies. The mesenchyme is composed of non-hematopoietic, non-endothelial and non-epithelial cells such as fibroblasts, myofibroblasts (stromal cells) and pericytes17. In the intestine, mucosal mesenchymal cells (MCs) act as a second barrier through their interactions with both immune and epithelial cells18. Under physiological conditions, mesenchymal cells (MCs) regulate immune cell maturation, migration and recruitment of immune cells19 as well as maintenance of the stem cell niche in the intestinal crypt and mucosal repair through epithelial to mesenchymal transition (EMT)17,18,20. MCs are highly activated by pro-inflammatory signals during chronic inflammation resulting in sub-epithelial myofibroblasts proliferation and extracellular matrix (ECM) production, and, not surprisingly, are involved in the development of fibrotic disorders21.
Table 2. Novel variants achieving study-wide significance that implicate genes directly in general onset CD.
Four of these variants (in TAGAP1,46, SDF2L11, RELA1 and HGFAC2) are in regions highlighted in prior GWAS but represent independent associations directly implicating these genes (Online Methods). Pos: genomic position in hg38; Scan p: p-value from the exome-wide discovery, including subjects exome sequenced at the Broad Institute; Meta p: p-value from the full meta-analysis of the five cohorts shown in Table 1; AA Conseq.: consequence on the amino acid; A0: reference allele; A1: tested/effect allele; spl reg: splice-site region
| Chr | Pos | A0 | A1 | MAF | Scan p | Meta p | OR | Gene | AA Conseq. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 28,150,681 | T | C | 0.0037 | 1.68E-07 | 2.96E-12 | 1.702 | PTAFR | N114S |
| 4 | 3,447,925 | G | A | 0.0704 | 1.37E-10 | 6.92E-15 | 1.170 | HGFAC | R516H |
| 4 | 94,573,345 | C | T | 0.0034 | 1.79E-05 | 2.55E-07 | 1.610 | PDLIM5 | spl reg |
| 6 | 159,041,392 | C | T | 0.0233 | 7.60E-06 | 4.04E-10 | 0.786 | TAGAP | E147K |
| 8 | 21,909,729 | G | A | 0.0316 | 1.34E-04 | 2.09E-13 | 1.248 | DOK2 | P274L |
| 11 | 65,658,293 | C | T | 0.0057 | 6.23E-05 | 2.31E-07 | 1.457 | RELA | D288N |
| 11 | 117,998,788 | C | T | 0.0014 | 1.13E-05 | 6.29E-09 | 2.107 | IL10RA | P295L |
| 17 | 40,558,934 | T | C | 0.0393 | 6.16E-06 | 4.70E-08 | 1.153 | CCR7 | M7V/M1? |
| 22 | 21,643,991 | G | A | 0.0152 | 2.44E-05 | 2.21E-07 | 1.242 | SDF2L1 | R161H |
Among the newly discovered IBD genes, PDLIM5 is highly expressed in sub-epithelial myofibroblasts31. PDLIM5 is a cytoskeleton-associated protein well known as a regulator of EMT through TGF-β1/SMAD3 signaling28. Knocking out its expression also leads to an alteration in ECM assembly, specifically by decreasing collagen IV network density28,32–34 (Figure 2). Interestingly, SDF2L1, also among the newly identified CD genes, has previously been shown to be elevated in plasma cells within the lamina propria of patients with CD failing to achieve durable remission on anti-TNF therapy35. SDF2L1, which is induced in response to endoplasmic reticulum (ER) stress, is also expressed in Paneth and goblet cells36. Impairment of ER stress is closely linked to intestinal inflammation37. Therefore, a mutation in this gene could putatively impair epithelial homeostasis in many ways such as preventing goblet cell differentiation, migration and proper production of mucus38 or perturbing the production of antibodies by plasma cells39.
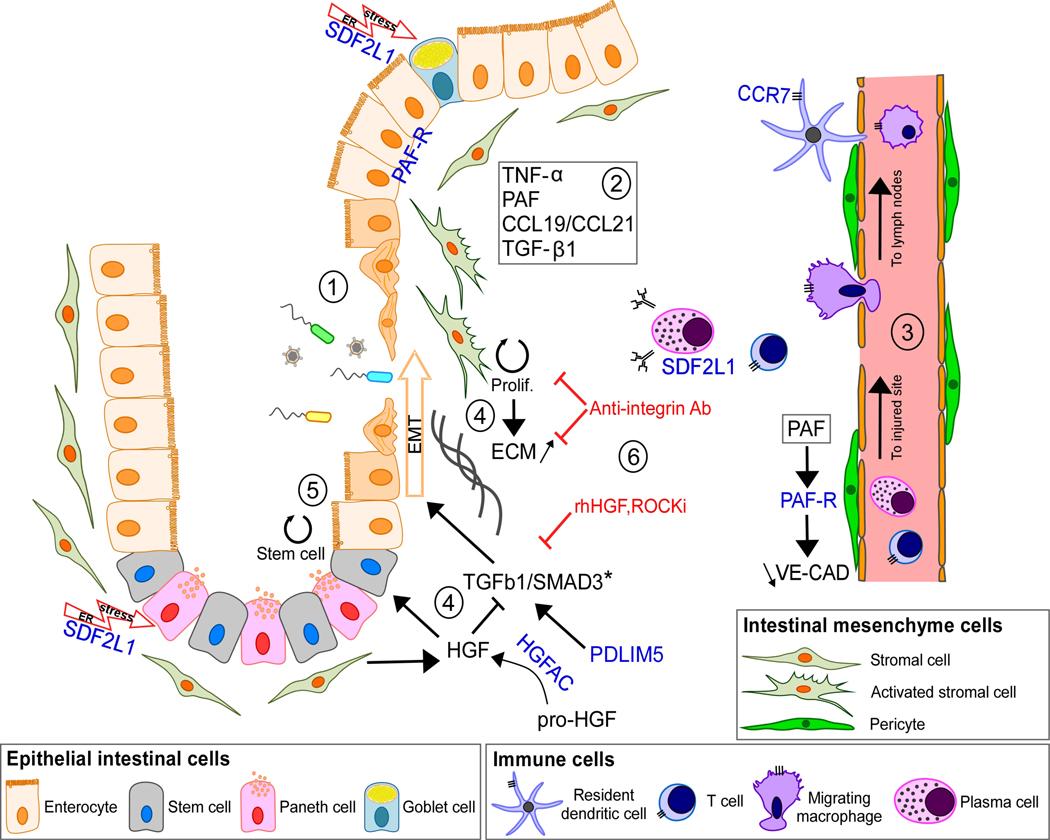
Figure 2. Schematic representation of inflamed mucosa showing the mesenchymal related genes with newly identified mutations.
(1) Following mucosal injury, mesenchymal cells (MCs) are highly activated by pro-inflammatory signals such as TNF-α, CCL19/CCL21, PAF and TGF-β1. (2) Among these, TNF-α increases PAF-R expression in intestinal epithelial cells during wound repair22. However, prolonged exposure to PAF dissolves cell junctions and increases epithelial permeability23. In endothelial cells, the PAF-R/PAF axis has a similar effect on Vascular Endothelial-Cadherin (VE-CAD) assembly24. A leaky endothelium can result in an increase in immune cell infiltration and aggravate inflammation at injured sites25. (3) Secretion of CCL19/CCL21 by activated stromal fibroblasts in response to epithelial damage or infection attracts dendritic cells (DC) and other immune cells which then migrate to mesenteric lymph nodes, where the immune response is coordinated. CCR7+ DC, macrophages and T cells also exacerbate inflammation through pro-inflammatory mediators such as CCL19/CCL21. Plasma cells express SDF2L1 in response to inflammation and ER stress due to massive antibody production26. (4) Mucosal repair mediated by TGF-β1/SMAD3 signaling has a key role in epithelial homeostasis after tissue injury27. Importantly, a causal variant in SMAD3* further supports the importance of this pathway in disease susceptibility5. PDLIM5 is a known regulator of SMAD3 stability during EMT28. Uncontrolled EMT increases fibroblast proliferation and excessive ECM production leading to fibrosis29. Active HGF released by HGFAC antagonizes TGF-β1 resulting in a decrease of EMT. (5) HGF secreted by fibroblasts play a role in maintaining the stem cell niche in intestinal crypts30. SDF2L1 expressed by Paneth cells in response to ER stress may participate in this process. (6) The current genetic findings provide support to the scientific rationale for targeting EMT and fibrosis for the treatment of CD, such as with anti-integrin antibodies (anti-αvβ6), recombinant human HGF (rhHGF) and Rho Kinase Inhibitor (ROCKi); shown in red31.
HGFAC, PAF-R and CCR7 can be linked to IBD-relevant MC functions via their known ligands. Specifically, HGFAC is a serine protease that cleaves Hepatocyte Growth Factor (HGF) to its active form. It has previously been shown that HGF is a paracrine factor secreted by stromal cells (fibroblasts and myofibroblasts) that regulates epithelial homeostasis, in particular the balance between epithelial proliferation, differentiation and apoptosis, and has been shown to be elevated in the serum of CD patients20,40. In human kidney epithelial cells, HGF has been shown to have antifibrotic properties by upregulating SMAD co-repressor SnoN resulting in inhibition of EMT and likely plays a similar role in the intestinal environment41. Platelet activating factor receptor (PAF-R) is expressed in epithelial and in endothelial cells as well as in pericytes, a population of MCs surrounding blood vessels that regulates angiogenesis24,42. PAF-R is a G-protein-coupled receptor for PAF, a pro-inflammatory lipid that is elevated in the mucosa of CD patients, potentially reflecting disease activity43. PAF-R/PAF axis is known to regulate endothelial and epithelial permeability that is associated with inflammatory diseases23,44. Finally, CCR7+ immune cells have a macrophage-like or a dendritic (DC) morphology. It is known that mesenchymal stromal cells induce CCR7+ DC migration to mesenteric lymph nodes within inflamed mucosa of IBD patients45. It is known that CCR7 ligands, CCL19/CCL21, are highly expressed in a recently identified population of proinflammatory stromal cells that appear to prevent the resolution phase that is normally found as part of the wound healing process31. Our identification of CD-associated rare coding variants in these genes suggests that perturbation of these finely-balanced cellular processes that are key to intestinal homeostasis causally contributes to CD susceptibility.
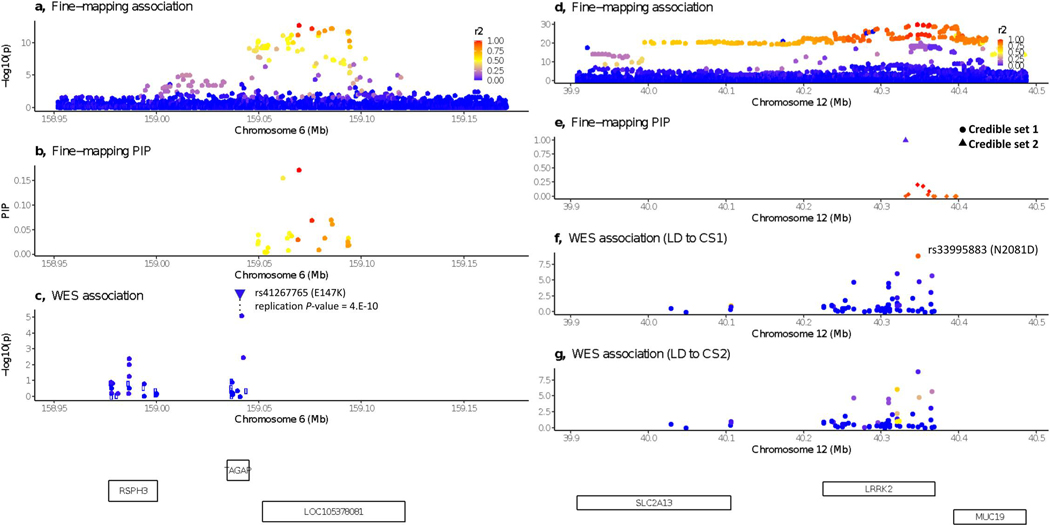
The identified coding variants in RELA, TAGAP, and SDF2L1 are close to, but not in LD (r2 < 0.05) with common non-coding variants significantly associated with IBD risk via GWAS (Online Methods and TAGAP as an example in Extended Data Figure 6a-c). These very likely pinpoint the genes dysregulated by the associated common variant and provide a focus for uncovering the function of those variants, perhaps leading to allelic series of perturbations further informing on the mechanism of their contribution to CD pathogenesis.The associated missense variant in HGFAC is in partial LD (r2 = 0.35 in 1000 Genomes non-Finnish European populations) with a common non-coding variant (rs3752440) previously reported as associated with CD2.Unfortunately, the missense variant was not included in this previous study — precluding formal assessment of whether this explains the previously observed association signal or represents an independent variant directly implicating HGFAC — though we note the missense variant here has a higher odds-ratio and greater significance than the variant in the previous report. The two novel NOD2 associations are not in LD with previously reported putative causal variants; One modestly reduces basal activity and has at least 2-fold reduction in peptidoglycan induced NF-κB response47, while the other is a splice donor variant (Supplementary Table 6). None of the variants described in Table 2 have reached genome-wide significance in previously published GWAS (variant in HGFAC has almost reached significance in ref 4– p=5e-8 for IBD). The nine new CD-associated variants all had an info score of 1 in the UKBB GWAS imputation48, except PTAFR and PDLIM5, which had info scores of 0.72 and 0.9, respectively. Novel variants described in Table 2, together explain around 0.12% of the variance on the liability scale (0.3% on the observed scale). In comparison, the 25 independent coding variants that were included in the meta-analysis together explained 2.1% of variance (5.1% on the observed scale). We performed two gene-based rare-variant (MAF < 0.001) burden tests in the full-exome Nextera and Twist data sets using SAIGE-GENE49, one restricted to loss-of-function variants and another using all non-synonymous variants (Supplementary Table 7). The burden test meta-analysis was performed on 15,823 genes for the non-synonymous variant analysis and 3,953 for the pLoF variant only analysis. Correcting for 20,000 genes, associations with p < 2.5 × 10−6 were considered statistically significant. NOD2 unsurprisingly stood out far above the expected distribution (LoFp = 7.7 × 10−7; NonSynp < 10−16). Only one other gene in either analysis exceeded the threshold expected once in the study by chance (ATG4C, NonSynp = 3.3 × 10−6). This potentially novel signal in ATG4C was driven by three distinct missense variants with individual p < 0.01 (N75S, R80H and C367Y) (Supplementary Table 8) along with two others with p < 0.05 (K371R, R381X). The ATG4C gene burden signal was examined in the Sanger data sets and replicated, with the meta-analysis reaching exome-wide significance (p = 1.5 × 10−7) driven by several of the same variants. Further examination of results from the single variant tests in ATG4C identified a frameshift variant with frequency of 0.002 (1:62834058-TTG-T) — too high to be included in our burden test — that just missed our threshold for testing in the follow-up cohorts (p = 0.0003, Beta = 0.55 in the Broad meta-analysis). This variant also showed evidence of association in the meta-analysis of the Sanger cohorts (p = 1.3 × 10−5), and also exceeded our study-wide significance threshold in the five-way meta-analysis of all cohorts (p = 1.55 × 10−8). Of further note, an additional ATG4C frameshift variant specifically enriched in Finland (1:62819215:C:CT) is associated with IBD (p = 6.91 × 10−8, Beta = 1.20) in the publicly released FinnGen resource (r5.finngen.fi). All variants in burden and individual tests increase risk, and the presence of four truncating variants in these analyses suggests that loss-of-function variants in ATG4C strongly increase CD risk.
Discussion
Here, we demonstrate that large-scale exome sequencing can complement GWAS by pinpointing specific genes both indirectly implicated by GWAS as well as those not yet observed in GWAS. With high sensitivity to directly test individual variants down to 0.01% minor allele frequency, as well as assess burden of ultra-rare mutations, we begin to fill in the low-frequency and rare-variant component of the genetic architecture of Crohn’s disease. This component was not observable by earlier generations of CD GWAS meta-analyses, which have had more limited coverage of low-frequency and rare variation.
Past findings in IBD5, and most other complex diseases, suggest that while coding variants are vastly outnumbered by non-coding variation, they are highly enriched for associations to common and rare disease. Furthermore, associated coding variants tend to have stronger effects than their non-coding counterparts, often keeping them lower in frequency via natural selection. While this alone validates the use of exome sequencing for efficiency’s sake, the primary advantage of targeting coding regions for discovery is that coding variants uniquely pinpoint genes, and often pathogenetic mechanisms, in a fashion that is at present far more challenging to achieve routinely for non-coding associations. In the case of several of the new findings (e.g., RELA, TAGAP), the coding variation here provides concrete evidence of genes previously indirectly implicated by independent non-coding GWAS associations. These identify the likely gene underlying these associations and build allelic series of natural perturbations at these genes. Moreover, IL10RA and RELA are known to harbor mutations causing rare, Mendelian, inflammatory GI disorders, and this study extends the phenotypic spectrum resulting from perturbing genetic variation to more complex forms of CD. From a functional perspective, the novel genes identified in the current study reiterate the central role of innate and adaptive immune cells as well as autophagy in CD pathogenesis. Moreover, the involvement of PDLIM5, SDF2L1 HGFAC, PAF-R and CCR7 pathways, in addition to the previously reported causal variant in SMAD35, highlights the emerging role of mesenchymal cells in the development and maintenance of intestinal inflammation (Fig. 2)18. Moreover, while previous studies have demonstrated the disruption of mesenchymal cell biology in IBD, the current findings of coding variants in these genes demonstrates that these cells and functions causally contribute to disease susceptibility. Furthermore, the association of these pathways to CD pathogenesis provides additional rationale for development of therapeutic modalities that can re-establish the balance to the mesenchymal niche, as it is believed that genetic evidence for a drug target has a measurable impact drug development29,50.
We expect that, in the next year, expanded sequencing efforts underway in ulcerative colitis will come to completion, enabling a more comprehensive survey of low-frequency and rare variation in ulcerative colitis, and IBD in general. Integrated with a much larger GWAS spearheaded in parallel by the IIBDGC, we expect a substantial number of conclusively linked genes and informative allelic series to emerge.
Online Methods
Ethics declarations
All relevant ethical guidelines have been followed, and any necessary IRB and/or ethics committee approvals have been obtained. The details of the IRB/oversight body that provided approval or exemption for the research described are given below:
Study Protocol 2013P002634, The Broad Institute Study of Inflammatory Bowel Disease Genetics, undergoes annual continuing review by the Mass General Brigham Human Research Committee (MGBHRC) Institutional Review Board (IRB) of Mass General Brigham. Ethical approval was given on January 27, 2021, for this study. Mass General Brigham IRB, Mass General Brigham, 399 Revolution Drive, Suite 710, Somerville, MA 02145.
All informed consent from participants has been obtained and the appropriate institutional forms have been archived.
DNA samples sequenced at the Sanger Institute were ascertained under the following ethical approvals: 12/EE/0482, 12/YH/0172, 16/YH/0247, 09/H1204/30, 17/EE/0265, 16/WM/0152, 09/H0504/125, 15/EE/0286, 11/YH/0020, 09/H0717/4, REC 22/02, 03/5/012, 03/5/012, 2000/4/192, 05/Q1407/274, 05/Q0502/127, 08/H0802/147, LREC/2002/6/18, GREC/03/0273 and YREC/P12/03.
Broad Institute sequencing pipeline
Sample processing.
Exome sequencing was performed at the Broad Institute. The sequencing process included sample prep (Illumina Nextera, IIlumina TruSeq, and Kapa Hyperprep), hybrid capture (Illumina Rapid Capture Enrichment (Nextera) - 37Mb target, and Twist Custom Capture - 37Mb target), and sequencing (Illumina HiSeq2000, Illumina HiSeq2500, Illumina HiSeq4000, Illumina HiSeqX, Illumina NovaSeq6000 – 76bp and 150bp paired reads). Sequencing was performed at a median depth of 85% targeted bases at > 20X. Sequencing reads were mapped by BWA-MEM to the hg38 reference using a “functional equivalence” pipeline. The mapped reads were then marked for duplicates, and base quality scores were recalibrated. They were then converted to CRAMs using Picard 2.16.0-SNAPSHOT and GATK 4.0.11.0. The CRAMs were then further compressed using ref-blocking to generate gVCFs. These CRAMs and gVCFs were then used as inputs for joint calling. To perform joint calling, the single-sample gVCFs were hierarchically merged (separately for samples using Nextera and Twist exome capture).
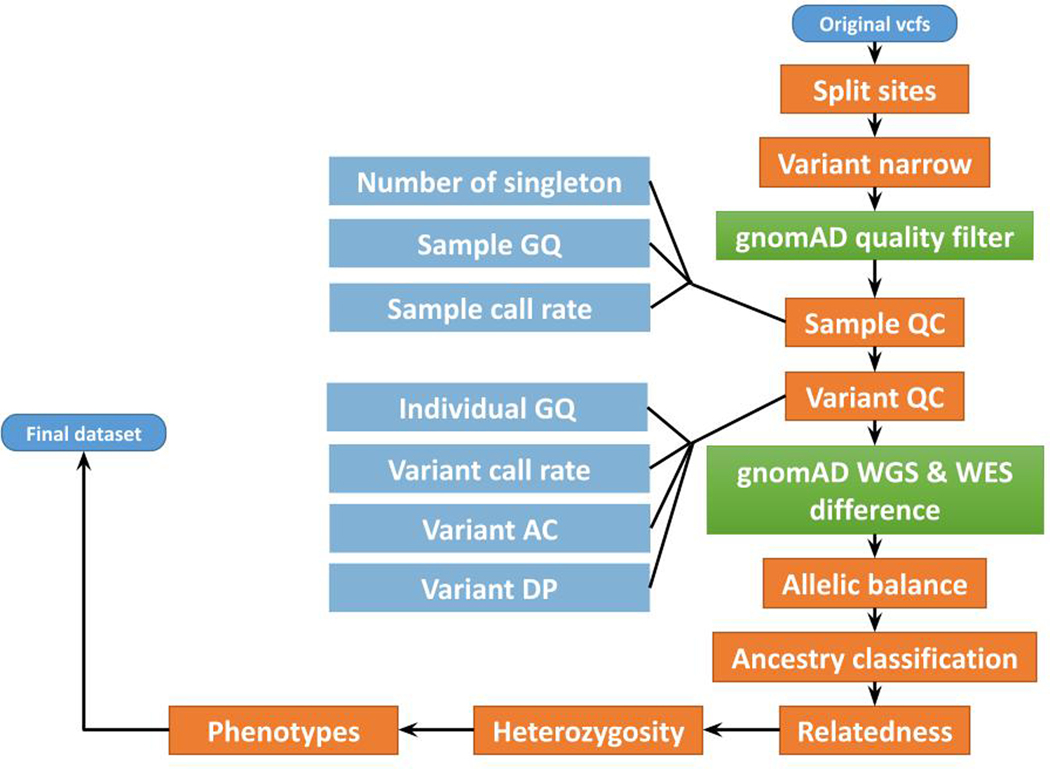
Quality control.
Quality control (QC) analyses were conducted in Hail v0.2.47 (Extended Data Figure 2). We first split multiallelic sites and coded genotypes with genotype quality (GQ) < 20 as missing. Variants not annotated as frameshift, inframe deletion, inframe insertion, stop lost, stop gained, start lost, splice acceptor, splice donor, splice region, missense, or synonymous were removed from the following analysis. We also removed variants that have known quality issues (have a non-empty QUAL column) in the gnomAD dataset. Sample QC: poor-quality samples that met the following criteria were identified and removed: 1) samples with an extremely large number of singletons (≥ 500); 2) samples with mean GQ < 40; and 3) samples with missingness rates > 10%. Variant QC: low-quality variants that met the following criteria were identified and removed: 1) variants with missingness rate > 5%; 2) variants with mean read depth (DP) < 10; 3) variants that failed the Hardy-Weinberg Equilibrium (HWE) test for controls with p < 1 × 10−4; and 4) variants with > 10% samples that were heterozygous and with an allelic balance ratio < 0.3 or > 0.7. Variants with different genotypes in WES and WGS in gnomAD were also removed. For Twist exome capture samples, we additionally removed 1) samples that had a significantly high or low inbreeding coefficient (> 0.2 or < −0.2); 2) samples that had a high heterozygosity away from mean (± 5 standard deviations); and 3) related samples, which were removed sequentially by removing the individual with the largest number of related samples (in PLINK, the individual with PI_HAT > 0.2 when using the “--genome” option) until no related samples remained. For Nextera capture samples, we additionally removed variants showing a significant heterogeneous effect across Ashkenazi Jewish (AJ), Lithuanian (LIT), Finnish (FIN), and non-Finnish European (NFE) samples (see “Population Assignment” below).
Population assignment.
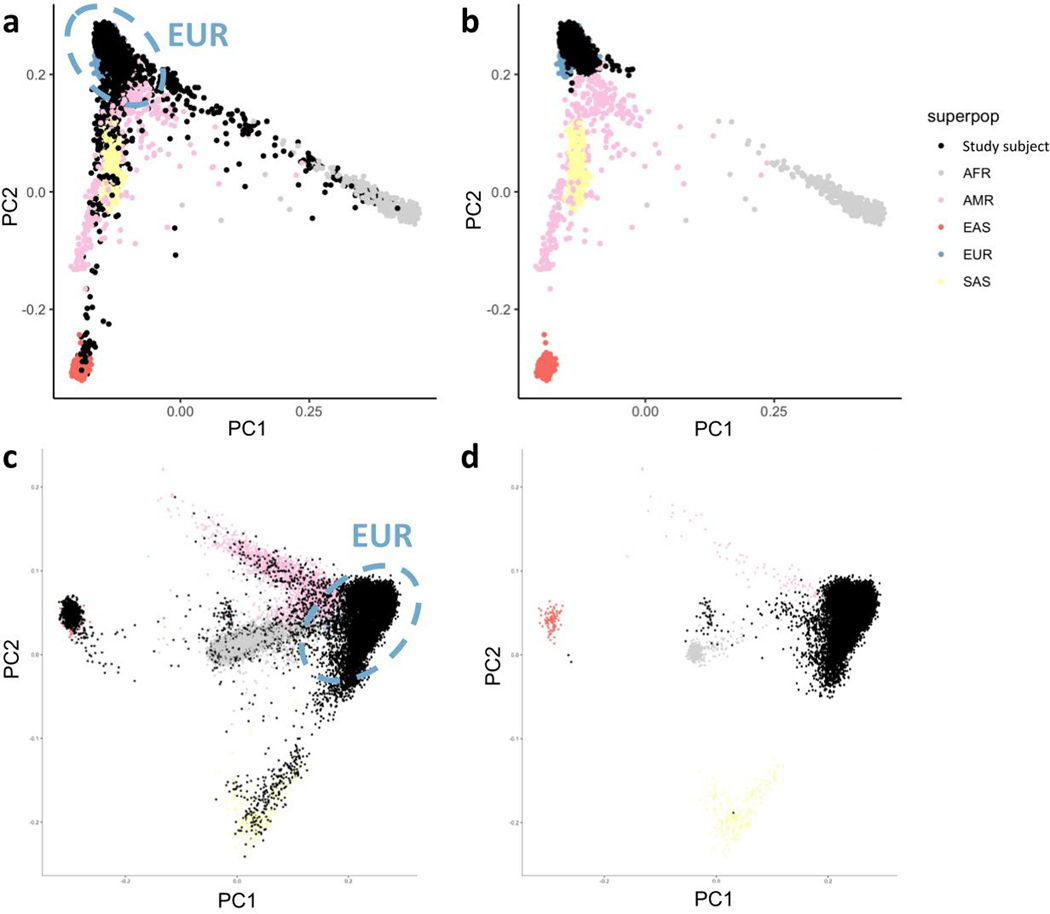
We projected all samples onto principal component (PC) axes generated from the 1000 Genomes Project Phase 3 common variants, and classified their ancestry using a random forest method to the European (CEU, TSI, FIN, GBR, IBS), African (YRI, LWK, GWD, MSL, ESN, ASW, ACB), East Asian (CHB, JPT, CHS, CDX, KHV), South Asian (GIH, PJL, BEB, STU, ITU) and American (MXL, PUR, CLM, PEL) samples. We kept samples that were classified as European with prediction probability greater than 80% (Extended Data Figure 7). For Nextera samples, we used a second random forest classifier to assign EUR samples to AJ, LIT, FIN, or NFE, and a third random forest classifier to clean the AJ/NFE split.
Meta-analysis.
We used METAL87 with the inverse variance weighted (IVW) fixed-effect model to meta-analyse the SAIGE association statistics from Nextera and Twist samples (Table 1). The heterogeneity test was performed using Cochran’s Q with one degree of freedom.
Sanger Institute sequencing pipeline
Sample processing.
Genome sequencing was performed at the Sanger Institute using the Illumina HiSeq X platform with a combination of PCR (n=4751, controls only) and PCR-free library preparation protocols. Sequencing was performed at a median depth of 18.6X. Exome sequencing of cases was performed at the Sanger Institute using the Illumina NovaSeq 6000 and the Agilent SureSelect Human All Exon V5 capture set. Controls from the UK Biobank were sequenced separately as a part of the UKBB WES50K release using Illumina NovaSeq and the IDT xGen Exome Research Panel v1.0 capture set (including supplemental probes). 33,704 UKBB participants were selected for use as controls, excluding participants with recorded or self-reported CD, UC, unspecified noninfective gastroenteritis or colitis, any other immune-mediated disorders, or a history of being prescribed any drugs used to treat IBD. Exome and genome datasets were analysed separately but followed a similar analysis protocol.
Reads were mapped to hg38 reference using BWA-MEM versions 0.7.12 (WGS) and 0.7.17 (WES). Variant calls were performed using a GATK Best Practices-like pipeline (versions 4.0.10.1 (WGS) and 4.1.8 (WES); per-sample intermediate variant calling was followed by joint genotyping across the individual genome and exome cohorts. For the exome cohort, variant calling was limited to Agilent extended target regions. Per-region VCF shards were imported into the Hail software and combined. Multi-allelic sites were split. For the exome cohort, we subsetted the calls to the intersection of Agilent and IDT exome captures, further excluding regions recommended for exclusion by the UKBB due to an error in read mapping that results in no variant calls made.
Population assignment.
We selected a set of ∼14,000 well-genotyped common variants to identify the genetic ancestry of individual participants through the projection of 1000 Genomes Project cohort-derived principal components. For genomes, due to primarily European genetic ancestry of the controls, we excluded samples outside of four median absolute deviations from the median point of the European ancestry cluster of 1000G. For exomes, we implemented a Random Forest technique that classified samples based on principal components into broad genetic ancestry groups (EUR, AFR, SAS, EAS, admixed), with self-reported ancestry as training labels. For these analyses, we only retained the EUR samples, as the number of cases for other groups was too small for robust association analysis.
Quality control.
A combination of hard-cutoff filters and per-ancestry/per-batch outlier filters were used to identify low-quality samples. We applied hard-filters for sample depth (> 12x genomes, > 15x exomes), call rate (> 0.95), chimerism < 0.5 (WGS) and FREEMIX < 0.02 (WGS). We excluded genotype calls with an allelic imbalance (for hets, (ab < 0.20) | (ab > 0.80)), low depth (< 2x), and low GQ (< 20). We then performed per-ancestry and per-sequencing protocol (AGILENT vs IDT for WES, PCR vs PCR-free for WES) filtering of samples falling outside 4 MAD from the median per-batch heterozygosity rate, Ti/Tv rate, number of called SNPs and INTELs, and insertion and deletion counts/ratio.
An ancestry-aware relatedness calculation (pc-relate method in Hail88) was used to identify related samples. As our association approach (logistic mixed-models) can control for residual relatedness, we only excluded duplicates or MZ twins from within the cohorts and excluded second and third degree relatives when the kinship was across the cohorts (e.g., parent in WGS, child in WES; kinship metric > 0.1 calculated via PC-Relate method using 10 principal components). In addition, we removed samples that were also present in the Broad Institute’s cohorts.
Association testing.
Association analysis was performed using a logistic mixed-model implemented in the REGENIE software v1.0.6.7 (singe-variant) and v2.0.2 (burden). A set of high-confidence variants (> 1% MAF, 99% call rate, and in Hardy-Weinberg Equilibrium) was used for t-fitting. To control for case-control imbalance, Firth correction was applied to p-values < 0.05. To control for residual ancestry and sequencing heterogeneity, we calculated 10 principal components on a set of well-genotyped common SNPs, excluding regions with known long-range LD. These were used as covariates for association analyses. Only variants with call-rate above 90% after filtering poor calls were included in the association analysis. For WES, we verified that the > 90% call-rate condition holds true in both AGILENT and IDT samples. Association analysis was performed on QC-passing calls.
Kiel/Regeneron sequencing pipeline
Sample Preparation and Sequencing.
The DNA samples were normalized and 100ng of genomic DNA was prepared for exome capture with custom reagents from New England Biolabs, Roche/Kapa, and IDT using a fully-automated approach developed at the Regeneron Genetics Center. Unique, asymmetric 10 base pair barcodes were added to each side of the DNA fragment during library preparation to facilitate multiplexed exome capture and sequencing. Equal amounts of sample were pooled prior to exome capture with a slightly modified version of IDT’s xGen v1 probes; supplemental probes were added to capture regions of the genome well-covered by a previous capture reagent (NimbleGen VCRome) but poorly covered by the standard xGen probes, the same as the probe libary used in UK Biobank exome sequencing. These supplemental probes were included in QC but excluded in the final analysis as we only looked up variants that were in the standard exome captures and reached the nominal significance for replication (Extended Data Figure 1). Captured fragments were bound to streptavidin-conjugated beads and non-specific DNA fragments were removed by a series of stringent washes according to the manufacturer’s recommended protocol (IDT). The captured DNA was PCR amplified and quantified by qRT-PCR (Kapa Biosystems). The multiplexed samples were pooled and then sequenced using 75 bp paired-end reads with two index reads on the Illumina NovaSeq 6000 platform using S2 flow cells.
Variant calling and quality control.
Sample read mapping and variant calling, aggregation and quality control were performed via the SPB protocol described in Van Hout et al.89. Briefly, for each sample, NovaSeq WES reads are mapped with BWA MEM 0.7.17-r1188 to the hg38 reference genome. Small variants are identified with WeCall v1.1.2 and reported as per-sample gVCFs. These gVCFs are aggregated with GLnexus into a joint-genotyped, multi-sample VCF (pVCF). SNV genotypes with read depth (DP) less than seven and indel genotypes with read depth less than ten are changed to no-call genotypes. After the application of the DP genotype filter, a variant-level allele balance filter is applied, retaining only variants that meet either of the following criteria: (i) at least one homozygous variant carrier or (ii) at least one heterozygous variant carrier with an allele balance (AB) greater than the cutoff.
Analysis.
We combined the gvcf files with bcftools 1.11 using the “merge” command, then imported the joint vcf into Hail. We then split the multiallelic variants and removed variants with “<NON_REF>” alternative alleles. We applied the QC steps and assigned populations as in the Broad Institute sequencing pipeline.
Statistics & Reproducibility
Previous studies show that a large sample size is needed for IBD genetic studies. We have thus included all samples available to us. We excluded samples of non-European ancestries due to their very limited sample size when properly matched between cases and controls (Extended Data Figure 7). We also excluded data of poor quality from the analysis (Extended Data Figure 2). These exclusions were necessary to ensure the quality of this study. All criteria were pre-established. We used the logistic mixed-model for the association analysis, followed by the fixed-effect meta-analysis to combine multiple cohorts. We have multiple cohorts in the study that serve the purpose of replication. Two large cohorts done at Broad Institute of different exome capture platform were used to discover candidate variants. Two independent cohorts at Sanger and one Kiel/Regeneron cohort were used to replicate the findings (Extended Data Figure 1). All reported findings have been replicated. No randomization was conducted. No blinding was carried out. Code and pipelines to reproduce our analysis are available on Zenodo90.
Cross-cohort meta-analysis
We used the Cochran–Mantel–Haenszel (CMH) test to combine association summary statistics between the Broad Institute, Sanger Institute and Kiel/Regeneron cohorts.
Relation to known IBD causal variants
We assigned the 45 study-wide significant variants to one of the four categories based on their relation with known IBD associations and/or fine-mapping results (Extended Data Figure 5 and Supplementary Table 4: 1) Known causal candidate: variants in a fine-mapping credible set5 with PIP > 5%, or reported in the earlier sequencing studies after manual review6,8. 2) New locus: variants implicating a genetic locus in general onset CD that have not been previously reported. 3) Unlikely causal : variants with PIP < 5%, or variants tagging the best PIP variants using conditional analysis (Conditional analysis, LRRK2 shown as an example in Extended Data Figure 6d-g). 4) New variant in known locus: variants in known GWAS loci with MAF < 0.5% (and thus, no LD to evaluate tagging), remain study-wide significant after conditional analysis using the LD from gnomAD (TAGAP shown as an example in Extended Data Figure 6a-c), or after manual review (Exceptions and notes).
Variance explained
Using the Sanger WGS data (6,000 cases, 11,852 controls) we fitted a series of univariate logistic regressions (is_case ∼ variant_genotype) models and estimated the pseudo-r2. Pseudo-r2 estimates were summed up to estimate the observed-scale variance explained by a group of variants. To convert the estimate into an estimate of heritability on the liability scale, we assumed the prevalence of Crohn’s disease is 276 in 100,000 (UK estimate from ref 91).
Conditional analysis
For study-wide significant variants not in a previously reported credible set5, we performed a conditional analysis to test whether they are independent from or tagging the known causal variants5. We first classified variants as “tagging” if they had r2 > 0.8 with any variants in the reported credible sets5. For other variants, we performed a conditional analysis using 1) the p-value estimates from previous fine-mapping studies for credible set variants and 2) the LD calculated from gnomAD. We were unable to directly fit a multivariate model or use the LD from study subjects, because exome sequencing does not cover the non-coding putative causal variants, and the ImmunoChip does not have good quality for rare coding variants. The conditional z statistic, z'Seq, for a variant with marginal statistic of zSeq from our study, was calculated as follows:
in which zFMi is the z statistic of the variant with the best PIP in the credible set i from the fine-mapping study, ri is the LD between the two variants, and NSeq and NFM are the effective sample sizes for our study and the fine-mapping study respectively. We used the absolute value in this equation because of the challenges to align the alleles across sequencing, the fine-mapping study, and the gnomAD reference panel. Taking the absolute value is a conservative approximation (less likely to declare a variant as novel association) because it assumes that the putative causal variants from fine-mapping have the same direction of effect as the variant being tested when they are in LD. This is very likely to be correct. The effective sample size was calculated as 4/(1/Ncase + 1/Ncontrol, in which Ncase and Ncontrol are the sample sizes for cases and controls respectively. For each variant, we summed the effective sample sizes across all cohorts in which the variant is observed (thus, NSeq can differ from variant to variant). We calculated the conditional p-value of z'seqi under the standard Gaussian distribution. A variant was classified as “tagging” if the conditional p-value failed to reach study-wide significance at 3 × 10−7.
Exceptions and notes
HGFAC: despite this locus having been reported in an earlier GWAS2, the coding variant we identify was not tested for association due to incomplete coverage of this region, and is thus reported in this study as directly implicating this gene (r2 = 0.35 with the previously reported GWAS SNP, rs2073505). We thus assign this variant as “New variant in known locus”. RELA: similarly to HGFAC, this locus has been reported in an earlier GWAS2, but the coding variant we identify was not tested for association due to incomplete coverage of this region, and thus is reported in this study as directly implicating this gene (r2 = 0.002 with the previously reported GWAS SNP, rs568617). We thus assign this variant as “New variant in known locus”. SLC39A8: the SLC39A8 A391T variant was not reported in the fine-mapping paper, as its genetic region was not included in the ImmunoChip design. Because this variant has been published in several papers as an IBD variant with genetic and functional evidence92–94, we assign this variant as “Known causal candidate”. TYK2: the TYK2 A928V was not reported in the fine-mapping paper5, likely due to a lack of power. Because this variant has been known to be a causal variant for several autoimmune disorders95 and in another IBD study96, we assign this variant as “Known causal candidate”. SDF2L1: this variant has marginal p-value = 2 × 10−7 and conditional p = 3.4 × 10-4. The r2 between this variant and the non-coding variant with the best PIP from fine-mapping is 0.045. We manually assigned this variant to “New variants in known locus”, as this is a missense variant. NOD2: a) Previous studies5–7 have shown evidence that the NOD2 S431L variant tags the NOD2 V793M variant, with the latter more likely to be the CD causal variant. In this study, however, S431L reached study-wide significance, but V793M failed to meet the significance cutoff. We therefore retained S431L in Figure 1 for the purpose of keeping this association signal. b) Due to the complexity of the NOD2 locus, we conducted a haplotype analysis using the Twist subjects and additionally classified signed variants that share the same haplotype with known IBD variants as “tagging”. We found that for the NOD2 S47L variant, 18 out of 19 copies of the T allele are on the same haplotype as the fs1007insC variant. We therefore classify S47L as “tagging”. c) The NOD2 A755V variant is in LD with rs184788345, the best PIP variant from fine-mapping ( r2 = 0.85). The marginal p-value for A755V is one order of magnitude less significant than rs184788345. Considering A755V is a missense variant while none of the variants in the credible set defined by rs184788345 are coding, we assign A755V as a likely “Known causal candidate”.
Data availability
We describe all datasets in the manuscript or its Supplementary Information. Genome Reference Consortium Human Build 38 can be accessed at https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.40/. Sequence data used in this study has been made publicly available in dbGaP Study Accession: phs001642.v1.p1 - Center for Common Disease Genomics [CCDG] - Autoimmune: Inflammatory Bowel Disease (IBD) Exomes and Genomes (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001642.v1.p1). The summary statistics of Nextera and Twist meta-analysis have been deposited on GitHub (https://github.com/iibdgc/Crohn-s-Disease-WES-meta) (10.5281/zenodo.6564928).
This research has been conducted using the UK Biobank Resource and controls made publicly available by dbGaP (phs001000.v1.p1, phs000806.v1.p1 - Myocardial Infarction Genetics Consortium (MIGen), phs000401.v1.p1 - NHLBI GO-ESP Project, phs000298.v4.p3 - Autism Sequencing Consortium (ASC), phs000572.v8.p4 - Alzheimer’s Disease Sequencing Project (ADSP), phs001489.v1.p1 - Epi25 Consortium, phs001095.v1.p1 - T2D-GENES) as well as additional controls from the 1000 Genomes Project, the Epi25 Collaborative, UK-Ireland Collaborators (A. McQuillin, D. Blackwood, A. McIntosh), and collaborators A. Pulver, H. Ostrer, D. Chung, M. Hiltunen, and A. Palotie (H2000 and SUPER cohorts) (Supplementary Table 1).
Code availability
The software and code used are described throughout the Supplementary Methods and can be found at https://github.com/iibdgc/Crohn-s-Disease-WES-meta (10.5281/zenodo.6564928).
Extended Data
Extended Data Fig. 1. Overview of the study design.
We utilized a logistic mixed-model for the association analysis, followed by the fixed-effect meta-analysis to combine multiple cohorts. Multiple cohorts serve the purpose of replication. Two large cohorts at Broad Institute of different exome capture platforms were used to discover candidate variants (Nextera WES and Twist WES ). Two independent cohorts at Sanger (Sanger WGS and Sanger WES) and one Kiel/Regeneron cohort (Regeneron WES) were used to replicate the findings.
Extended Data Fig. 2. Quality control procedures applied in the Broad sequencing pipeline.
We show as an example the quality control steps performed on variants and subjects from the Broad sequencing platform. Quality controls performed on data from other platforms follow a similar plan and are described in Methods. Quality control steps using external information from gnomAD were colored green. Thresholds and details can be found in Methods.
Extended Data Fig. 3. QQ plots of the heterogeneity-of-effect test between the Nextera and Twist discovery cohorts.
Only QC passed variants with minor allele frequency in NFE between 0.0001 and 0.10 were included. a, all variants. b, non-synonymous variants. c, synonymous variants. In a and b, the y axis is capped at -log10 p = 30 while the top four variants (three in NOD2 and one in IL23R) have -log10 p > 100. In c, to remove the synonymous variants that tag causal non-synonymous variants and artifacts through LD, we removed loci hosting large-effect coding variants (IL23R, NOD2, LRRK2, TYK2, ATG16L1, SLC39A8, PTGER4, IRGM, CARD9), implicated by variants removed in the heterogeneous test (AHNAK2, LILRA), and with long range LD (MHC).
Extended Data Fig. 4. Power to detect single variant associations.
We performed a series of power calculations using the methodology described by Johnson and Abecasis (2017). Our initial ‘exome-wide scan’ (two cohorts) had fewer samples and a more lenient significance threshold than subsequent meta-analysis (five cohorts). However, both analyses had similar power to detect true associations at their respective significance levels. Our single-variant association analyses did not have the power to uncover association to variants with a MAF = 0.0001 and below (unless the variant has a very strong effect, e.g. 0.76 power at OR = 8). Similarly, the exome-wide scan had limited power to detect association to variants with a MAF = 0.001 and OR < 2, but was well-powered above these thresholds. a, Power of the exome-wide scan analysis b, Power of the meta-analysis. c, Power to detect single-variant associations at different minor allele frequencies at α = 0.0002 (‘scan’; dashed lines) and 3 x 10–7 (‘meta’; solid lines) and assuming Crohn’s disease population prevalence of 276 in 100,000, and an additive effect model.
Extended Data Fig. 5. Relation to known IBD associations.
Numbers in brackets are the number of variants assigned to the categories out of the 45 exome-wide significant variants.
Extended Data Fig. 6. WES variants from this study implicating known IBD loci.
a-c: a novel CD variant implicating TAGAP. d-g: CD variants tagging fine-mapped IBD associations in LRRK2. a and d, P-value for variants from the fine-mapping study5. b and e, PIP from fine-mapping. c, f and g, P-value for variants from this study. Open circle indicating LD information is missing. LD calculated between the plotted variant and the best variant in b for panel c, and variants with best PIP in credible sets 1 and 2 (panel e) respectively for panels f and g.
Extended Data Fig. 7. Nextera and Twist callset population assignment.
Principal components for a, c, before removing non-European samples for Twist and Nextera respectively. b, d, after removing non-European samples for Twist and Nextera respectively. Principal components generated from the 1000 Genome Project Phase III data and different colors stand for different continental / superpopulations. Study subjects (black dots) were projected onto principal components.
Supplementary Material
Box 1. Genes newly implicated in Crohn’s disease risk.
DOK2
(Docking Protein 2, Downstream of Tyrosine Kinase 2) encodes a cytoplasmic signaling protein highly expressed in macrophages and T-cells in the terminal ileum. Loss of Dok-2 in mice causes severe DSS-induced colitis with reduced IL-17A and IL-22 expression51, and DOK2 is known to be differentially methylated in colonic tissue of IBD patients52. DOK2 regulates both TLR2-induced inflammatory signaling and NK cell development, and DOK2 loss-of-function is associated with increased IFN-ɣ production53,54. The P274L variant has previously been implicated in atopic eczema where the rare allele was significantly protective for atopic eczema likely by disturbing the RasGAP activation of DOK2, and transcriptomic analyses also suggest that DOK2 is a central hub interacting with CD200R1, IL6R, and STAT355.
TAGAP
(T-Cell Activation RhoGTPase Activating Protein) has a pivotal role in TH17 development and modulates the risk of autoimmunity through influencing thymocyte migration in thymic selection56,57. TAGAP expression is upregulated in rectal tissue in IBD patients, and TAGAP is required for Dectin-induced anti-fungal signaling and proinflammatory cytokine production in myeloid cells58,59.
PTAFR
(Platelet Activating Factor Receptor), a hypoxia response gene, has an affinity for bacterial phosphorylcholine (ChoP) moieties60 and influences development of cigarette-induced Chronic Obstructive Pulmonary Disease (COPD) by inducing neutrophil autophagic death in mice61. PTAFR regulates colitis-induced pulmonary inflammation through the NLRP3 inflammasome62.
PDLIM5
(PDZ And LIM Domain 5) encodes a kidney anion exchanger and scaffolding protein. Genetic variation in this gene is associated with prostate cancer, schizophrenia, diverticular disease, diverticulosis, colorectal cancer, testicular cancer, and self-reported angina63. PDLIM5 has been reported to be a STAT3 interaction partner involved in actin binding64, with STAT3 previously being identified as an IBD gene65. PDLIM5 is highly expressed in myofibroblast cells, which are important mesenchymal cells of the intestinal lamina propria31.
SDF2L1
(Stromal Cell Derived Factor 2 Like 1) has been recently identified to be expressed in the ER stress response in primary intestinal epithelial cells66. SDF2L1 is an ER resident protein that functions within a large protein complex regulating the BiP and Erdj3 chaperone cycle to promote protein folding and secretion67–69. Structurally, Sdf2l1 contains an N-terminal signal peptide for entry into the ER lumen and a C-terminal ER retention signal flanking 3 MIR domains that promote complex assembly. The CD risk variant R161H is located in the third MIR domain. In murine models, deletion of SDF2L1 in the liver resulted in prolonged ER stress and insulin resistance70. In the intestine, single cell transcriptional profiling revealed that SDF2L1 is predominantly expressed in highly secretory cell lineages, including mucin-secreting goblet cells and immunoglobulin-secreting plasma cells71. Moreover, SDF2L1 expression is dynamically regulated and specifically induced during the acute phase of the unfolded protein response (UPR)66. Together, these observations suggest a critical role for SDF2L1 in maintaining ER homeostasis and secretory capacity, which may promote barrier function at the level of mucus integrity and/or neutralization of immunoglobulins and antimicrobial peptides that collectively limit interactions between luminal microbes and the host immune system.
CCR7
(chemokine receptor 7) encodes a chemokine receptor. CCR7 and its ligands CCL19/CCL21 promote homing of T-cells and dendritic cells to T-cell areas of lymphoid tissues where T-cell priming occurs. CCR7 also contributes to adaptive immune functions including thymocyte development, secondary lymphoid organogenesis, high affinity antibody responses, regulatory and memory T-cell function, and lymphocyte egress from tissues. CCR7 expression is upregulated in an inflamed gut in CD72, and CCR7 regulates the intestinal TH1/TH17/Treg balance during Crohn’s-like murine ileitis73. Genetic variation in CCR7 is associated with atopy, asthma, COPD, and IBD in African-Americans63. CCL19 and CCL21 are highly expressed in a population of stromal cells (designated as S4) that are expanded in IBD inflamed tissues and that continually produce proinflammatory factors, preventing the resolution phase of the wound-healing response31.
IL10RA
(Interleukin 10 receptor A) is a potent regulator of innate and adaptive immune responses, and IL10RA genetic variants are associated with Very-Early Onset IBD (VEOIBD) cases; a subset of VEOIBD refractory patients respond to hematopoietic stem cell transplantation74. IL10RA (also known as Il10r1) knockout mice are susceptible to chemical-induced colitis75.
RELA
(Nuclear Factor NF-Kappa-B P65 Subunit). NFkB is a ubiquitous transcription factor, and its most abundant form is NFKB1 complexed together with RELA. RELA regulates the Th17 pathway in autoimmune disease models76, and the FOXO3-NF-κB RelA protein complexes reduce proinflammatory cell signaling and function77. RELA haploinsufficiency causes autosomal dominant chronic mucotaneous ulceration78, and RELA is a master transcriptional regulator of epithelial-mesenchymal transition in epithelial cells79. Genetic variation in RELA has been associated with SLE, type 2 diabetes, psoriasis, obesity, asthma, and atopic dermatitis63.
ATG4C –
(Autophagy-Related 4C Cysteine Peptidase) defective autophagy is established as a mechanism contributing to CD risk. This gene encodes one of four Atg4 isoforms (Atg4A, B, C, and D) that prime pro-LC3 and GABARAP (orthologues of yeast Atg8), essential proteins required for autophagosome biogenesis80,81. These Atg4 proteins, including Atg4C, are involved with proteolytic cleavage of Atg8’s C-terminus, thus exposing a specific Atg8 glycine residue necessary for phospholipid covalent binding to Atg8. Atg8 lipidation is necessary for autophagosome formation82.
HGFAC –
Hepatocyte Growth Factor Activator is a serine endopeptidase that converts Hepatocyte Growth Factor (HGF) to its active form in response to thrombin and kallikrein endopeptidases. HGF contributes to neutrophil recruitment. HGF expression is increased in active UC with animal models, suggesting that HGF-MET signaling exacerbates intestinal inflammation83,84. Furthermore, HGF promotes colonic epithelial regeneration and mucosal repair85,86. HGFAC variation is also associated with tuberculosis susceptibility87.
Acknowledgements
We thank all the principal investigators, local staff from individual cohorts, and all of the patients who kindly donated samples used in the study for making possible this global collaboration and resource to advance IBD genetics research. This research was funded in whole, or in part, by the US National Institutes of Health Grants U54HG003067 and 5UM1HG008895, the Wellcome Trust Grants 206194 and 108413/A/15/D, and The Leona M. & Harry B. Helmsley Charitable Trust 2015PG-IBD001. We thank the Broad Institute Genomics Platform for genomic data generation efforts and the Stanley Center for Psychiatric Research at the Broad Institute for supporting control sample aggregation. M.A.R. is in part supported by the NHGRI of the NIH under award R01HG010140 (M.A.R.) and an NIH Center for Multi- and Trans-ethnic Mapping of Mendelian and Complex Diseases grant (5U01HG009080). H.H. acknowledges support from NIDDK K01DK114379, P30DK043351 and the Stanley Center for Psychiatric Research. H.S.W. receives philanthropic support from Martin Schlaff, James Brooks, and the B. Hasso Family Foundation. H.H.U. and A.S. are supported by the NIHR Oxford Biomedical Research Centre and by The Leona M. and Harry B. Helmsley Charitable Trust. A.P is in part supported by the Academy of Finland Centre of Excellence in Complex Disease Genetics Grants 312074 and 336824.
Individual studies contributing to this meta-analysis acknowledge support from NIH grants DK062431, DK062432, DK087694, K23DK117054, R01DK111843, P01DK094779, R01HG010140, 5U01HG009080, and DK062420, NIDDK Grants P01DK046763, U01DK062413, and R01DK104844.
Additional acknowledgments pertaining to participating programs and consortia are included in Supplementary Tables. Acknowledgments of participating consortia and programs.
Footnotes
Competing Interests Statement
A.B., M.F, J.E.H., D.S are current or former employees and/or stockholders of Regeneron Genetics Center or Regeneron Pharmaceuticals. M.A. is consulting for or part of the advisory board for AbbVie Inc, Bellatrix Pharmaceuticals, Bristol Myers Squibb, Eli Lilly Pharmaceuticals, Gilead, Janssen Ortho, LLC, and Prometheus Biosciences; teaching, lecturing, or speaking at Alimentiv, Arena Pharmaceuticals, Janssen, Prime CME, Takeda Pharmaceuticals. A.B is an employee of Regeneron and owns stock in Regeneron. O.M.D. has served in the IBD fellowship funding committee for Pfizer and has a funded research project by Pfizer. H.K. receives grant funding from Takeda and Pfizer and has received consulting fees from Takeda. A.P. is a member of Astra Zenecas Genomics Advisory Board. M.A.R. is on the SAB of 54gene and has advised BioMarin, Third Rock Ventures, MazeTx, and Related Sciences. G.A.H. is an employee of Takeda, former employee of AbbVie and owns stock in Takeda and AbbVie. C.A.L. reports grants from Genentech, grants and personal fees from Janssen, grants and personal fees from Takeda, grants from AbbVie, personal fees from Ferring, grants from Eli Lilly, grants from Pfizer, grants from Roche, grants from UCB Biopharma, grants from Sanofi Aventis, grants from Biogen IDEC, grants from Orion OYJ, personal fees from Dr Falk Pharma, grants from AstraZeneca, outside the submitted work. H.H.U reports research collaboration or consultancy with Janssen, Eli Lilly, UCB Pharma, Celgene, MiroBio, OMass, and Mestag. D.P.B.M. has consulted for Takeda, Boehringer Ingelheim, Palatin Technologies, Bridge Biotherapeutics, Pfizer, and Gilead. M.P. received an unrestricted research grant from Pfizer UK and speaker fees from Janssen. P.I. received lecture fees from AbbVie, BMS, Celgene, Celltrion, Falk Pharma, Ferring, Galapagos, Gilead, MSD, Janssen, Pfizer, Takeda, Tillotts, Sapphire Medical, Sandoz, Shire and Warner Chilcott; financial support for research from Celltrion, MSD, Pfizer and Takeda; advisory fees from AbbVie, Arena, Boehringer-Ingelheim, BMS, Celgene, Celltrion, Genentech, Gilead, Hospira, Janssen, Lilly, MSD, Pfizer, Pharmacosmos, Prometheus, Roche, Sandoz, Samsung Bioepis, Takeda, Topivert, VH2, Vifor Pharma and Warner Chilcott. Cedars-Sinai and D.P.B.M. have financial interests in Prometheus Biosciences, a company which has access to the data and specimens in Cedars-Sinais MIRIAD Biobank (including the Cedars-Sinai data and specimens used in this study) and seeks to develop commercial products. H.H. has received consultancy fees from Ono Pharmaceutical and honorarium from Xian Janssen Pharmaceutical. C.A.A. has received consultancy fees from Genomics plc and BridgeBio Inc. and lecture fees from GSK. M.J.D. is a founder of Maze Therapeutics. The remaining authors declare no competing interests.
References
- 1.Jostins L et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu JZ et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo Y et al. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat. Genet. 49, 186–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lange KM et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivas MA et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 43, 1066–1073 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivas MA et al. A protein-truncating R179X variant in RNF186 confers protection against ulcerative colitis. Nature Communications vol. 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivas MA et al. Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLoS Genet. 14, e1007329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaudoin M et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 9, e1003723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Z et al. Ubiquitin Ligase TRIM62 Regulates CARD9-Mediated Anti-fungal Immunity and Intestinal Inflammation. Immunity 43, 715–726 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leshchiner ES et al. Small-molecule inhibitors directly target CARD9 and mimic its protective variant in inflammatory bowel disease. Proc. Natl. Acad. Sci. U. S. A. 114, 11392–11397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivanesan D et al. IL23R (Interleukin 23 Receptor) Variants Protective against Inflammatory Bowel Diseases (IBD) Display Loss of Function due to Impaired Protein Stability and Intracellular Trafficking. J. Biol. Chem. 291, 8673–8685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohanan V et al. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science 359, 1161–1166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glocker E-O et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Zhang L, Joo D & Sun S-C NF-κB signaling in inflammation. Signal Transduct Target Ther 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koliaraki V, Prados A, Armaka M & Kollias G The mesenchymal context in inflammation, immunity and cancer. Nat. Immunol. 21, 974–982 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Kurashima Y et al. Mucosal Mesenchymal Cells: Secondary Barrier and Peripheral Educator for the Gut Immune System. Front. Immunol. 8, 1787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson CA, Nibbs RJ, McCoy KD & Mowat AM Immunological roles of intestinal mesenchymal cells. Immunology 160, 313–324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koliaraki V, Pallangyo CK, Greten FR & Kollias G Mesenchymal Cells in Colon Cancer. Gastroenterology 152, 964–979 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Li C & Kuemmerle JF The fate of myofibroblasts during the development of fibrosis in Crohn’s disease. J. Dig. Dis. 21, 326–331 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Birkl D et al. TNFα promotes mucosal wound repair through enhanced platelet activating factor receptor signaling in the epithelium. Mucosal Immunol. 12, 909–918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakravarty V et al. Prolonged Exposure to Platelet Activating Factor Transforms Breast Epithelial Cells. Front. Genet. 12, 634938 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudry-Clergeon H, Stengel D, Ninio E & Vilgrain I Platelet-activating factor increases VE-cadherin tyrosine phosphorylation in mouse endothelial cells and its association with the PtdIns3’-kinase. FASEB J. 19, 512–520 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cromer WE, Mathis JM, Granger DN, Chaitanya GV & Alexander JS Role of the endothelium in inflammatory bowel diseases. World J. Gastroenterol. 17, 578–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gommerman JL, Rojas OL & Fritz JH Re-thinking the functions of IgA(+) plasma cells. Gut Microbes 5, 652–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone RC et al. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res. 365, 495–506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y et al. PDLIM5 inhibits STUB1-mediated degradation of SMAD3 and promotes the migration and invasion of lung cancer cells. J. Biol. Chem. 295, 13798–13811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pariente B et al. Treatments for Crohn’s Disease-Associated Bowel Damage: A Systematic Review. Clin. Gastroenterol. Hepatol. 17, 847–856 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Fukushima T, Uchiyama S, Tanaka H & Kataoka H Hepatocyte Growth Factor Activator: A Proteinase Linking Tissue Injury with Repair. Int. J. Mol. Sci. 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinchen J et al. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 175, 372–386.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maier JI et al. EPB41L5 controls podocyte extracellular matrix assembly by adhesome-dependent force transmission. Cell Rep. 34, 108883 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Yuda A, Lee WS, Petrovic P & McCulloch CA Novel proteins that regulate cell extension formation in fibroblasts. Exp. Cell Res. 365, 85–96 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Pompili S, Latella G, Gaudio E, Sferra R & Vetuschi A The Charming World of the Extracellular Matrix: A Dynamic and Protective Network of the Intestinal Wall. Front. Med. 8, 610189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin JC et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 178, 1493–1508.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treveil A et al. Regulatory network analysis of Paneth cell and goblet cell enriched gut organoids using transcriptomics approaches. Mol Omics 16, 39–58 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Kaser A & Blumberg RS Endoplasmic reticulum stress in the intestinal epithelium and inflammatory bowel disease. Semin. Immunol. 21, 156–163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M & Wu C The relationship between intestinal goblet cells and the immune response. Biosci. Rep. 40, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X et al. Function and dysfunction of plasma cells in intestine. Cell Biosci. 9, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boucher G et al. Serum Analyte Profiles Associated With Crohn’s Disease and Disease Location. Inflamm. Bowel Dis. (2021) doi: 10.1093/ibd/izab123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Dai C & Liu Y A novel mechanism by which hepatocyte growth factor blocks tubular epithelial to mesenchymal transition. J. Am. Soc. Nephrol. 16, 68–78 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Meran L, Baulies A & Li VSW Intestinal Stem Cell Niche: The Extracellular Matrix and Cellular Components. Stem Cells Int. 2017, 7970385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobhani I et al. Raised concentrations of platelet activating factor in colonic mucosa of Crohn’s disease patients. Gut 33, 1220–1225 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knezevic II et al. Tiam1 and Rac1 are required for platelet-activating factor-induced endothelial junctional disassembly and increase in vascular permeability. J. Biol. Chem. 284, 5381–5394 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang MH et al. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J. Immunol. 176, 803–810 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Festen EAM et al. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet. 7, e1001283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chamaillard M et al. Gene–environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 100, 3455–3460 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou W et al. Scalable generalized linear mixed model for region-based association tests in large biobanks and cohorts. doi: 10.1101/583278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Waseda M, Arimura S, Shimura E, Nakae S & Yamanashi Y Loss of Dok-1 and Dok-2 in mice causes severe experimental colitis accompanied by reduced expression of IL-17A and IL-22. Biochem. Biophys. Res. Commun. 478, 135–142 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Cooke J et al. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm. Bowel Dis. 18, 2128–2137 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Rhodes J Erythrocyte rosettes provide an analogue for Schiff base formation in specific T cell activation. J. Immunol. 145, 463–469 (1990). [PubMed] [Google Scholar]
- 54.Celis-Gutierrez J et al. Dok1 and Dok2 proteins regulate natural killer cell development and function. EMBO J. 33, 1928–1940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mucha S et al. Protein-coding variants contribute to the risk of atopic dermatitis and skin-specific gene expression. J. Allergy Clin. Immunol. 145, 1208–1218 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Tamehiro N et al. T-cell activation RhoGTPase-activating protein plays an important role in TH17-cell differentiation. Immunol. Cell Biol. 95, 729–735 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Duke-Cohan JS et al. Regulation of thymocyte trafficking by Tagap, a GAP domain protein linked to human autoimmunity. Sci. Signal. 11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medrano LM et al. Expression patterns common and unique to ulcerative colitis and celiac disease. Ann. Hum. Genet. 83, 86–94 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Chen J et al. TAGAP instructs Th17 differentiation by bridging Dectin activation to EPHB2 signaling in innate antifungal response. Nat. Commun. 11, 1913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark SE & Weiser JN Microbial modulation of host immunity with the small molecule phosphorylcholine. Infect. Immun. 81, 392–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lv X-X et al. Cigarette smoke promotes COPD by activating platelet-activating factor receptor and inducing neutrophil autophagic death in mice. Oncotarget 8, 74720–74735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu G et al. Platelet activating factor receptor regulates colitis-induced pulmonary inflammation through the NLRP3 inflammasome. Mucosal Immunol. 12, 862–873 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Ochoa D et al. Open Targets Platform: supporting systematic drug–target identification and prioritisation. Nucleic Acids Res. 49, D1302–D1310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blumert C et al. Analysis of the STAT3 interactome using in-situ biotinylation and SILAC. J. Proteomics 94, 370–386 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Barrett JC et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40, 955–962 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.You K et al. QRICH1 dictates the outcome of ER stress through transcriptional control of proteostasis. Science 371, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujimori T et al. Endoplasmic reticulum proteins SDF2 and SDF2L1 act as components of the BiP chaperone cycle to prevent protein aggregation. Genes Cells 22, 684–698 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Meunier L, Usherwood Y-K, Chung KT & Hendershot LM A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13, 4456–4469 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanafusa K, Wada I & Hosokawa N SDF2-like protein 1 (SDF2L1) regulates the endoplasmic reticulum localization and chaperone activity of ERdj3 protein. J. Biol. Chem. 294, 19335–19348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sasako T et al. Hepatic Sdf2l1 controls feeding-induced ER stress and regulates metabolism. Nat. Commun. 10, 947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smillie CS et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Autschbach F, Funke B, Katzenmeier M & Gassler N Expression of chemokine receptors in normal and inflamed human intestine, tonsil, and liver--an immunohistochemical analysis with new monoclonal antibodies from the 8th international workshop and conference on human leucocyte differentiation antigens. Cell. Immunol. 236, 110–114 (2005). [DOI] [PubMed] [Google Scholar]
- 73.McNamee EN et al. Chemokine receptor CCR7 regulates the intestinal TH1/TH17/Treg balance during Crohn’s-like murine ileitis. J. Leukoc. Biol. 97, 1011–1022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murugan D et al. Very early onset inflammatory bowel disease associated with aberrant trafficking of IL-10R1 and cure by T cell replete haploidentical bone marrow transplantation. J. Clin. Immunol. 34, 331–339 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Pils MC et al. Monocytes/macrophages and/or neutrophils are the target of IL-10 in the LPS endotoxemia model. Eur. J. Immunol. 40, 443–448 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Qu X et al. TLR4-RelA-miR-30a signal pathway regulates Th17 differentiation during experimental autoimmune encephalomyelitis development. J. Neuroinflammation 16, 183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson MG et al. FOXO3-NF-κB RelA Protein Complexes Reduce Proinflammatory Cell Signaling and Function. J. Immunol. 195, 5637–5647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Badran YR et al. Human RELA haploinsufficiency results in autosomal-dominant chronic mucocutaneous ulceration. J. Exp. Med. 214, 1937–1947 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tian B et al. The NFκB subunit RELA is a master transcriptional regulator of the committed epithelial-mesenchymal transition in airway epithelial cells. J. Biol. Chem. 293, 16528–16545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rioux JD et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCarroll SA et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 40, 1107–1112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Agrotis A, Pengo N, Burden JJ & Ketteler R Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells. Autophagy 15, 976–997 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finisguerra V et al. MET is required for the recruitment of anti-tumoural neutrophils. Nature 522, 349–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stakenborg M et al. Neutrophilic HGF-MET signaling exacerbates intestinal inflammation. J. Crohn’s Colitis (2020) doi: 10.1093/ecco-jcc/jjaa121. [DOI] [PubMed] [Google Scholar]
- 85.Kanayama M et al. Hepatocyte growth factor promotes colonic epithelial regeneration via Akt signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G230–9 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Tahara Y et al. Hepatocyte growth factor facilitates colonic mucosal repair in experimental ulcerative colitis in rats. J. Pharmacol. Exp. Ther. 307, 146–151 (2003). [DOI] [PubMed] [Google Scholar]
- 87.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conomos MP, Reiner AP, Weir BS & Thornton TA Model-free Estimation of Recent Genetic Relatedness. Am. J. Hum. Genet. 98, 127–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Hout CV et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature 586, 749–756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Venkataraman Guhan R., Yuan Kai, Huang Hailiang. Crohn’s Disease WES meta-analysis. [Computer software]. Zenodo. 10.5281/zenodo.6564928 [DOI] [Google Scholar]
- 91.Pasvol TJ et al. Incidence and prevalence of inflammatory bowel disease in UK primary care: a population-based cohort study. BMJ Open 10, e036584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakata T et al. A missense variant in SLC39A8 confers risk for Crohn’s disease by disrupting manganese homeostasis and intestinal barrier integrity. Proc. Natl. Acad. Sci. U. S. A. 117, 28930–28938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li D et al. A Pleiotropic Missense Variant in SLC39A8 Is Associated With Crohn’s Disease and Human Gut Microbiome Composition. Gastroenterology vol. 151 724–732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sunuwar L et al. Pleiotropic ZIP8 A391T implicates abnormal manganese homeostasis in complex human disease. JCI Insight 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ellinghaus D et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48, 510–518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Diogo D et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS One 10, e0122271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We describe all datasets in the manuscript or its Supplementary Information. Genome Reference Consortium Human Build 38 can be accessed at https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.40/. Sequence data used in this study has been made publicly available in dbGaP Study Accession: phs001642.v1.p1 - Center for Common Disease Genomics [CCDG] - Autoimmune: Inflammatory Bowel Disease (IBD) Exomes and Genomes (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001642.v1.p1). The summary statistics of Nextera and Twist meta-analysis have been deposited on GitHub (https://github.com/iibdgc/Crohn-s-Disease-WES-meta) (10.5281/zenodo.6564928).
This research has been conducted using the UK Biobank Resource and controls made publicly available by dbGaP (phs001000.v1.p1, phs000806.v1.p1 - Myocardial Infarction Genetics Consortium (MIGen), phs000401.v1.p1 - NHLBI GO-ESP Project, phs000298.v4.p3 - Autism Sequencing Consortium (ASC), phs000572.v8.p4 - Alzheimer’s Disease Sequencing Project (ADSP), phs001489.v1.p1 - Epi25 Consortium, phs001095.v1.p1 - T2D-GENES) as well as additional controls from the 1000 Genomes Project, the Epi25 Collaborative, UK-Ireland Collaborators (A. McQuillin, D. Blackwood, A. McIntosh), and collaborators A. Pulver, H. Ostrer, D. Chung, M. Hiltunen, and A. Palotie (H2000 and SUPER cohorts) (Supplementary Table 1).