Abstract
Purpose
Beta-lactamase-producing Klebsiella pneumoniae is common in the clinic, but research associated with the co-existence of KPC-2, LAP-2, and CTX-M-65 in K. pneumoniae is still rare. In this study, the phenotypic and genetic characteristics of a multidrug-resistant K. pneumoniae strain SJ25 co-harboring blaKPC-2, blaLAP-2, and blaCTX-M-65 with rare ST1469 were investigated.
Methods and Results
Antimicrobial susceptibility testing revealed that strain SJ25 was resistant to various common antibiotics, except ciprofloxacin, fosfomycin, colistin, and tigecycline. Whole-genome analysis revealed that strain SJ25 carries a variety of antimicrobial resistance genes and virulence determinants. Plasmid analysis confirmed that the blaKPC-2 and blaCTX-M-65 genes were located on an ~136 kb transferrable IncFII/IncR plasmid and that blaLAP-2 was located on an untypeable plasmid.
Conclusion
Our findings emphasized the need for continuous surveillance of β-lactamase-bearing K. pneumoniae in the clinic to control potential dissemination and outbreak.
Keywords: Klebsiella pneumoniae, multidrug-resistant, bla KPC-2 , bla LAP-2 , bla CTX-M-65
Introduction
Increasing antibiotic resistance in Klebsiella pneumoniae is of great concern worldwide. Resistant plasmids with horizontal transfer ability play a pivotal role in the dissemination of resistant strains.1,2 Regarding K. pneumoniae, resistance toward β-lactam antibiotics is a leading clinical problem because this group of antibiotics is the frontline drug for anti-infection therapy.3
K. pneumoniae carbapenemase (KPC)-type carbapenemase, classified as Ambler class A, is one of the most critical carbapenemases commonly found in K. pneumoniae, and contributes to resistance to all β-lactam antibiotics.4 LAP β-lactamase, which belongs to the Ambler class A group, shows a narrow hydrolysis spectrum against β-lactam antibiotics but reports are limited.5,6 CTX-M-type β-lactamases have become the most common extended-spectrum β-lactamases (ESBLs) among Enterobacteriaceae worldwide.7 By replacing the CTX-M-2 group, which was prevalent before 2011, CTX-M-9 group enzymes (especially CTX-M-65) were found to be largely dominant.7,8 To date, several studies have reported the co-location of blaKPC-2 and blaCTX-M-65 in K. pneumoniae.9,10 Notably, most of these studies were performed in China, which gained our attention. Furthermore, the co-existence of blaKPC-2, blaCTX-M-65 and blaLAP-2 in K. pneumoniae is rare.11
In this study, we reported a multidrug-resistant (MDR) strain ST1469 K. pneumoniae SJ25 co-harboring KPC-2, CTX-M-65, and LAP-2 that was isolated from the clinic. Whole-genome sequencing (WGS) revealed the genetic background of the isolate. In our study, we aimed to evaluate the phenotypic characteristics of the blaKPC-2, blaCTX-M-65 and blaLAP-2-carrying K. pneumoniae, and emphasized the importance of further monitoring of β-lactamases-carrying K. pneumoniae in the clinic.
Materials and Methods
During routine screening of the carbapenems-resistant bacteria in a tertiary hospital in Zhejiang Province, China, a carbapenemase-resistant K. pneumoniae was isolated from sputum (designated as strain SJ25). Subsequently, the identity of the species in the isolate and carbapenemase-encoding genes were identified by matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker, Bremen, Germany) and PCR amplification as described previously.12,13
Antimicrobial susceptibility testing (AST) was performed using both the agar dilution method and the broth microdilution method. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) standards (https://clsi.org), except for colistin and tigecycline, which were interpreted following the EUCAST clinical breakpoints (https://www.eucast.org/).14 Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality controls.
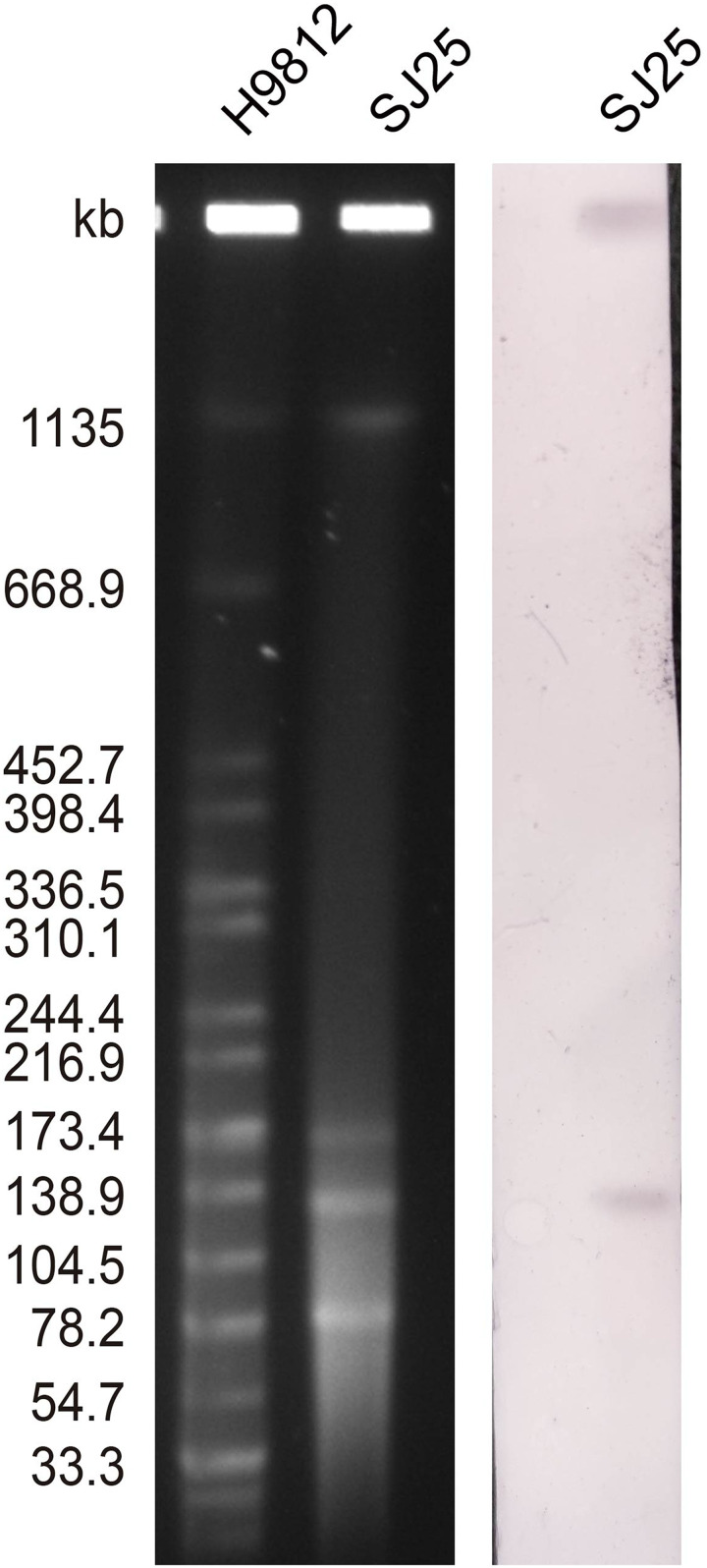
The number and size of the plasmids of K. pneumoniae strain SJ25 were confirmed by the S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) method.12 Southern blotting and hybridization using a DIG-labeled blaKPC-specific probe were performed to estimate the location of the blaKPC gene. To verify the transferability of the blaKPC-2-bearing plasmid, conjugation experiments were conducted using rifampin-resistant P. aeruginosa PAO1Ri as recipients, as described previously.15 The transconjugants were selected on Mueller-Hinton agar plates (OXOID, Hampshire, United Kingdom), containing 200 mg/L rifampicin and 1 mg/L meropenem. The AST results of transconjugant SJ25-P indicated that the plasmid pSJ25_KPC_CTX was successfully transferred into the recipient and impacted bacterial resistance (Table 1).
Table 1.
Antimicrobial Susceptibilities of K. pneumoniae Strain SJ25 and Transconjugant SJ25-P
| Antimicrobials | MIC Values (mg/L) | |
|---|---|---|
| SJ25 | SJ25-P | |
| Amoxicillin/clavulanate | 128 | 128 |
| Piperacillin/tazobactama | >128 | 2 |
| Ceftriaxone | >128 | 16 |
| Cefotaxime | >128 | 16 |
| Cefepime | 16 | 1 |
| Ceftazidime | 128 | 1 |
| Ciprofloxacin | 0.03 | 2 |
| Levofloxacin | 2 | 8 |
| Imipenem | 4 | 4 |
| Meropenem | 2 | 2 |
| Ertapenem | 2 | 2 |
| Trimethoprim/ sulfamethoxazole | >8/152 | 8/152 |
| Amikacin | >128 | 1 |
| Gentamicin | >128 | 1 |
| Aztreonam | 64 | 8 |
| Chloramphenicol | 32 | 128 |
| Fosfomycin | 4 | 64 |
| Colistin | 2 | 1 |
| Tigecycline | 1 | 2 |
Note: aTazobactam at a fixed concentration of 4mg/L.Abbreviation: MIC, minimal inhibitory concentration.
Total DNA was extracted and purified using the OMEGA Bacterial DNA kit (Omega Bio-tek, Norcross, GA, USA). Subsequently, the DNA was sequenced using Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA) and Oxford Nanopore (Oxford Nanopore Technologies, Oxford, UK) platforms.16,17 The assembly of the complete genome was performed using Unicycler v0.4.2. The genome was annotated using Prokka.18 The acquired antibiotic resistance genes (ARGs) and plasmid incompatibility types were determined using ResFinder and PlasmidFinder databases, and the multilocus sequence typing (MLST) was determined by pubMLST (http://pubmlst.org/ecloacae). The detection of virulence factors was performed with VFDB.19 Finally, circular maps of the plasmids pSJ25_KPC_CTX and pSJ25_LAP were plotted using the BLAST Ring Image Generator (BRIG).20
Results and Discussion
Strain K. pneumoniae SJ25 was isolated from a 62-year-old male patient. The patient had received a lung transplant six months earlier, and had been admitted to the hospital for anti-infective treatment for sepsis after surgery. In February 2022, the patient was admitted to the hospital for chest tightness and shortness of breath. The patient was subsequently diagnosed with a pulmonary infection. During hospitalization, amikacin, tigecycline, and ceftazidime/avibactam were given for anti-infection treatment. One month after the patient was admitted, strain SJ25 was isolated from sputum and identified as K. pneumoniae. Three months after admission, the patient’s indicators returned to normal, and he was discharged home.
By referencing the pubMLST database, K. pneumoniae strain SJ25 was classified as ST1469, which is a rarely reported type. Based on the WGS data, strain SJ25 contained a chromosome and five plasmids. Among them, there was a chromosome, 5,416,718 bp in length with a GC content of 57.6%, a plasmid carrying KPC-2 and CTX-M-65 (pSJ25_KPC_CTX) of 136,248 bp in length with a GC content of 53.3%, and a plasmid carrying LAP-2 (pSJ25_LAP) of 86,057 bp in length with a GC content of 53.9%. Furthermore, AST revealed that the strain was resistant to multiple antibiotics, including amoxicillin/clavulanate (minimal inhibitory concentration (MIC) = 128 μg/mL), piperacillin/tazobactam (MIC >128 μg/mL), ceftriaxone (MIC >128 μg/mL), cefotaxime (MIC >128 μg/mL), cefepime (MIC =16 μg/mL), ceftazidime (MIC = 128 μg/mL), levofloxacin (MIC = 2 μg/mL), imipenem (MIC =4 μg/mL), ertapenem (MIC =2 μg/mL), trimethoprim/ sulfamethoxazole (MIC >8/152 μg/mL), amikacin (MIC >128 μg/mL), aztreonam (MIC =64 μg/mL), gentamicin (MIC >128 μg/mL), and chloramphenicol (MIC =32 μg/mL). The strain was only found to be susceptible to ciprofloxacin (MIC =0.03 μg/mL), fosfomycin (MIC =4 μg/mL) and tigecycline (MIC =1 μg/mL), and intermediate to colistin (MIC = 2 μg/mL) and meropenem (MIC =2 μg/mL). Based on the results of ResFinder, strain SJ25 harbored various ARGs, including rmtB, aadA1, blaKPC-2, blaSHV-12, blaCTX-M-65, blaOXA-10, blaLEN2, blaTEM-1B, blaLAP-2, sul2, dfrA14, oqxB, oqxA, qnrS1, tet(A), arr-2 and fosA. These findings were consistent with drug-resistant phenotypes (Table 1). Additionally, screening of virulence factors showed that strain SJ25 harbored several virulence genes encoding multiple functions, such as type VI secretion system ATPase (clpV), type VI secretion protein (icmF), type 3 fimbriae (mrkC), type I fimbriae (fimD), acriflavine resistance protein B (acrB), outer membrane receptor (fepA), and enterobactin synthase subunit F (entF). All these factors contributed to the potential virulence of strain SJ25.
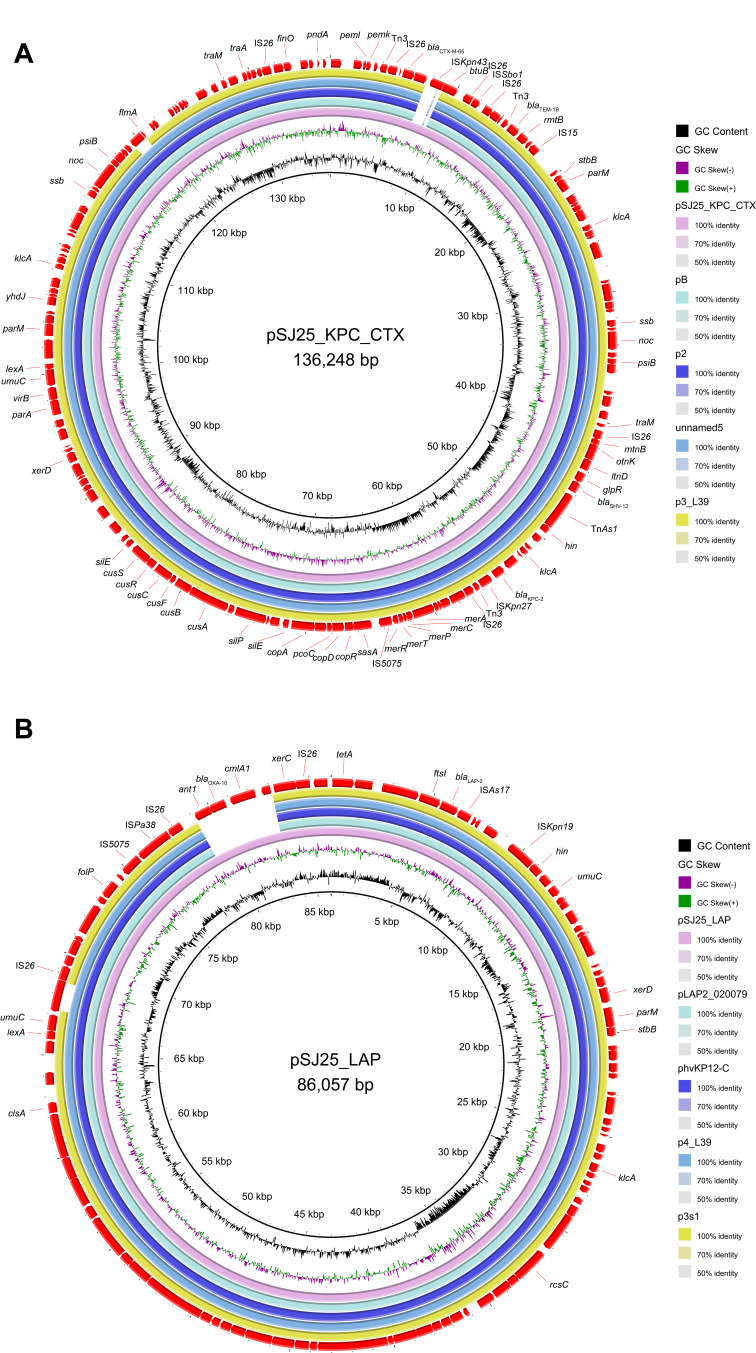
S1-PFGE and Southern blot analysis confirmed the existence of ~136 kb IncFII/IncR pSJ25_KPC_CTX and ~86 kb untypeable plasmid pSJ25_LAP (Figures 1 and 2). The transconjugant SJ25-P carrying the ~136 kb pSJ25_KPC_CTX plasmid showed a MIC of imipenem of 4 mg/L, thereby suggesting that the IncFII/IncR pSJ25_KPC_CTX plasmid is both conjugative and responsible for carbapenem resistance. NCBI BLAST analysis revealed that the complete sequence of pSJ25_KPC_CTX shared the highest degree of genetic similarity (query coverage and identity over 99%) with K. pneumoniae plasmids pB (CP090434.1), p2 (CP090464.1), unnamed5 (CP101775.1) and p3_L39 (CP033956.1). Genetic context characterization demonstrated a conserved structure sequence (IS5075-merR-merT-merP-merC-merA-IS26-Tn3-ISKpn27-blaKPC-2-klcA-hin-TnAs1) in these plasmids. Of note, all five plasmids were identified from clinical isolates in China. In a previous study, it was reported that KPC-2- and CTX-M-65- co-carrying plasmids mainly belonged to incompatibility type IncFII.9 In this study, plasmid pSJ25_KPC_CTX was identified as IncFII/IncR, which indicated the further spread of the co-occurrence of blaKPC-2 and blaCTX-M-65 in different plasmid types. Taken together, these results highlight that an effective prevention strategy should be taken to curb further dissemination of KPC-2- and CTX-M-65-bearing plasmids.
Figure 1.
Plasmid profiles and Southern blot-hybridization of K. pneumoniae strain SJ25. Southern blot-hybridization of S1-nuclease digested DNA using a specific probe (blaKPC). M: XbaI digested total DNA of Salmonella enterica serotype Braenderup H9812 as a size marker.
Figure 2.
Major structural features and comparison of beta-lactamase-encoding plasmids. (A) Genomic map of the blaKPC-2 and blaCTX-M-65 co-producing IncFII/IncR pSJ25_KPC_CTX plasmid with four closely related plasmids (CP090434.1, CP090464.1, CP101775.1, CP033956.1). (B) Circular genome alignment of pSJ25_LAP with four blaLAP-bearing plasmids (CP029382.1, CP103318.1, CP033957.1, and CP034126.1). Open reading frames (ORFs) are indicated by arrows and colored according to their putative functions. Alignment of plasmids was performed and visualized by BLAST ring image generator (BRIG) software.
According to the results of Plasmidfinder analysis, plasmid pSJ25_LAP was untypeable, because no hit was found in the database. In pSJ25_LAP using ResFinder, a total of 9 ARGs were detected, which enabled pSJ25_LAP to exhibit resistance to several antimicrobial agents, including β-lactamase (blaLAP-2 and blaOXA-10), fluoroquinolone (qnrS1), sulphonamide, and trimethoprim (dfrA14 and sul2), chloramphenicol (cmlA1), aminoglycoside (aadA1), tetracycline (tet(A)) and rifampin (ARR-2). By using the BLAST tool in the NCBI database, pSJ25_LAP shared the highest similarity (100% identity with over 93% coverage) with K. pneumoniae plasmids pLAP2_020079 (CP029382.1), phvKP12-C (CP103318.1), p4_L39 (CP033957.1) and p3s1 (CP034126.1). It is worth noting that all these plasmids were identified in the clinic in China, but only few reports are available. pSJ25_LAP was perfectly mapped to these four plasmids, except for a resistance region containing ARGs blaOXA-10 and cmlA1 (Figure 2B). Furthermore, LAP-2 was flanked by multiple mobile elements (IS26, ISAs17, and ISKpn19). These elements could cluster and be combined with resistance genes, thus contributing to multiple resistance transfers of plasmids.21
In this study, an MDR ST1469 K. pneumoniae strain SJ25 co-carrying KPC-2, CTX-M-65 and LAP-2 was isolated from a patient in the clinic in China. The genetic environment and plasmid transfer mechanism of strain SJ25 were elucidated. K. pneumoniae with β-lactam antibiotic drug resistance may cause an infection in patients resulting in a prolonged hospital stay and poor prognosis. Moreover, MDR bacteria make clinical choices extremely limited if the economic cost and availability of medication were taken into consideration.21 Our findings emphasized the need for continuous surveillance of β-lactamase-bearing K. pneumoniae in the clinic to control potential dissemination and outbreak.
Acknowledgments
This work was supported by research grants from the National Natural Science Foundation of China (No. 82072314) and the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022011B).
Nucleotide Sequence Accession Numbers
The whole-genome sequences of the K. pneumoniae were submitted to GenBank under the following BioProject number: PRJNA885055.
Ethics Approval and Consent to Participate
This study was conducted following the Declaration of Helsinki and obtained approval from the clinical research ethics committee of the First Affiliated Hospital, Zhejiang University School of Medicine [number 2020-IIT-660]. The patient provided written informed consent to allow the case details to be published.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):736–755. doi: 10.1111/j.1574-6976.2011.00268.x [DOI] [PubMed] [Google Scholar]
- 2.Carattoli A. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol. 2011;301(8):654–658. doi: 10.1016/j.ijmm.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 3.Zheng B, Tan S, Gao J, et al. An unexpected similarity between antibiotic-resistant NDM-1 and beta-lactamase II from Erythrobacter litoralis. Protein Cell. 2011;2(3):250–258. doi: 10.1007/s13238-011-1027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, Zhao Z, Ge Y, He F. Rapid emergence of a pandrug-resistant Klebsiella pneumoniae st11 isolate in an inpatient in a teaching hospital in China after treatment with multiple broad-spectrum antibiotics. Infect Drug Resist. 2020;13:799–804. doi: 10.2147/IDR.S243334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Leviandier C, Nordmann P. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob Agents Chemother. 2006;50(12). doi: 10.1128/AAC.00597-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kocsis B, Kocsis E, Fontana R, Cornaglia G, Mazzariol A. Identification of blaLAP-2 and qnrS1 genes in the internationally successful Klebsiella pneumoniae ST147 clone. J Med Microbiol. 2013;62(Pt2):269–273. doi: 10.1099/jmm.0.050542-0 [DOI] [PubMed] [Google Scholar]
- 7.Riccobono E, Di Pilato V, Di Maggio T, et al. Characterization of IncI1 sequence type 71 epidemic plasmid lineage responsible for the recent dissemination of CTX-M-65 extended-spectrum β-lactamase in the Bolivian Chaco region. Antimicrob Agents Chemother. 2015;59(9):5340–5347. doi: 10.1128/AAC.00589-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartoloni A, Pallecchi L, Riccobono E, et al. Relentless increase of resistance to fluoroquinolones and expanded-spectrum cephalosporins in Escherichia coli: 20 years of surveillance in resource-limited settings from Latin America. Clin Microbiol Infect. 2013;19(4):356–361. doi: 10.1111/j.1469-0691.2012.03807.x [DOI] [PubMed] [Google Scholar]
- 9.Fu L, Tang L, Wang S, et al. Co-location of the blaKPC-2, blaCTX-M-65, rmtB and virulence relevant factors in an IncFII plasmid from a hypermucoviscous Klebsiella pneumoniae isolate. Microb Pathog. 2018;124:301–304. doi: 10.1016/j.micpath.2018.08.055 [DOI] [PubMed] [Google Scholar]
- 10.Nishida S, Ono Y. Genomic analysis of a pan-resistant Klebsiella pneumoniae sequence type 11 identified in Japan in 2016. Int J Antimicrob Agents. 2020;55(4):105854. doi: 10.1016/j.ijantimicag.2019.11.011 [DOI] [PubMed] [Google Scholar]
- 11.Liu S, Ding Y, Xu Y, Li Z, Zeng Z, Liu J. An outbreak of extensively drug-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a teaching hospital in Southwest China. Front Cell Infect Microbiol. 2022;12:979219. doi: 10.3389/fcimb.2022.979219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng B, Zhang J, Ji J, et al. Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob Agents Chemother. 2015;59(11):7086–7089. doi: 10.1128/AAC.01363-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng B, Xu H, Lv T, et al. Stool samples of acute diarrhea inpatients as a reservoir of ST11 hypervirulent KPC-2-producing Klebsiella pneumoniae. mSystems. 2020;5(3). doi: 10.1128/mSystems.00498-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gou JJ, Liu N, Guo LH, et al. Carbapenem-resistant Enterobacter hormaechei ST1103 with IMP-26 carbapenemase and ESBL gene bla SHV-178. Infect Drug Resist. 2020;13:597–605. doi: 10.2147/IDR.S232514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Xu H, Guo X, et al. Emergence and genetic characterization of plasmid-encoded VIM-2-producing pseudomonas stutzeri with novel integron In1998 isolated from cerebrospinal fluid. Infect Drug Resist. 2021;14:3415–3424. doi: 10.2147/IDR.S320294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng B, Yu X, Xu H, et al. Complete genome sequencing and genomic characterization of two Escherichia coli strains co-producing MCR-1 and NDM-1 from bloodstream infection. Sci Rep. 2017;7(1):17885. doi: 10.1038/s41598-017-18273-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X, Liu W, Zheng B. Complete genome sequencing of Comamonas kerstersii 8943, a causative agent for peritonitis. Sci Data. 2018;5:180222. doi: 10.1038/sdata.2018.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R, Xu H, Guo X, et al. Genomic characterization of two Escherichia fergusonii isolates harboring mcr-1 gene from farm environment. Front Cell Infect Microbiol. 2022;12:774494. doi: 10.3389/fcimb.2022.774494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Zheng D, Zhou S, Chen L, Yang J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022;50(D1):D912–D917. doi: 10.1093/nar/gkab1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Q, Han R, Guo Y, et al. Emergence of ST15 Klebsiella pneumoniae clinical isolates producing plasmids-mediated RmtF and OXA-232 in China. Infect Drug Resist. 2020;13:3125–3129. doi: 10.2147/IDR.S257298 [DOI] [PMC free article] [PubMed] [Google Scholar]