Abstract
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes diverse clinical manifestations and tissue injuries in multiple organs. However, cellular and molecular understanding of SARS-CoV-2 infection-associated pathology and immune defense features in different organs remains incomplete. Here, we profiled approximately 77 000 single-nucleus transcriptomes of the lung, liver, kidney, and cerebral cortex in rhesus macaques ( Macaca mulatta) infected with SARS-CoV-2 and healthy controls. Integrated analysis of the multi-organ dataset suggested that the liver harbored the strongest global transcriptional alterations. We observed prominent impairment in lung epithelial cells, especially in AT2 and ciliated cells, and evident signs of fibrosis in fibroblasts. These lung injury characteristics are similar to those reported in patients with coronavirus disease 2019 (COVID-19). Furthermore, we found suppressed MHC class I/II molecular activity in the lung, inflammatory response in the liver, and activation of the kynurenine pathway, which induced the development of an immunosuppressive microenvironment. Analysis of the kidney dataset highlighted tropism of tubule cells to SARS-CoV-2, and we found membranous nephropathy (an autoimmune disease) caused by podocyte dysregulation. In addition, we identified the pathological states of astrocytes and oligodendrocytes in the cerebral cortex, providing molecular insights into COVID-19-related neurological implications. Overall, our multi-organ single-nucleus transcriptomic survey of SARS-CoV-2-infected rhesus macaques broadens our understanding of disease features and antiviral immune defects caused by SARS-CoV-2 infection, which may facilitate the development of therapeutic interventions for COVID-19.
Keywords: SARS-CoV-2, Rhesus macaque, Animal model, Single-nucleus RNA sequencing, Antiviral immune defects, Multiple organs
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), continues to threaten global health, with more than 620 million confirmed cases so far (as of 1 November 2022; https://www.who.int/emergencies/diseases/novel-coronavirus-2019). Patients may present with fever, dry cough, and dyspnea, with some developing acute respiratory distress syndrome (ARDS), potentially leading to death in critical cases ( Wang et al., 2020a; Wiersinga et al., 2020). Histopathological examination of autopsy samples shows diffuse alveolar damage (DAD) and exudative inflammation ( Liu et al., 2020a). Liver and kidney function abnormalities have also been observed in COVID-19 patients, as indicated by elevated alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels, proteinuria, and hematuria ( Cheng et al., 2020; Fu et al., 2021). In addition, SARS-CoV-2 infection can affect the central nervous system (CNS), resulting in neurological symptoms in some patients ( Mao et al., 2020). Cognitive deficits ( Liu et al., 2021) and accelerated aging ( Cao et al., 2022), including aging signatures in the brain ( Mavrikaki et al., 2021), have also been reported in people with COVID-19.
Recently, postmortem organ tissue specimens have been used to analyze COVID-19 pathology at single-cell resolution ( Delorey et al., 2021; Melms et al., 2021; Ren et al., 2021; Wang et al., 2021; Yang et al., 2021). However, human subjects show considerable variation in exposure level, exposure route, sampling time relative to exposure, and background health conditions. Postmortem interval may also have an adverse effect on tissue quality. Furthermore, obtaining age-matched healthy tissue controls is a challenge. The limited accessibility of autopsy specimens from patients who died of COVID-19 and matched healthy controls has restricted in-depth pathological study in human subjects.
These limitations can be counteracted by using animal models, although severe disease observed in humans cannot be completely recapitulated. Animals, including monkeys, tree shrews, and ferrets, are permissive to SARS-CoV-2 infection ( Deng et al., 2020; Fan et al., 2022; Kim et al., 2020; Muñoz-Fontela et al., 2020; Munster et al., 2020; Shan et al., 2020; Song et al., 2021; Song et al., 2020; Xu et al., 2020), with various animal models developed to test vaccines ( Gao et al., 2020; Wang et al., 2020b) and understand disease progression and immune responses ( Lee et al., 2021; Nelson et al., 2022). Rhesus macaques are a commonly used non-human primate model for studies on human respiratory viral diseases ( Skinner et al., 2014; Yao et al., 2014). Their inoculation with SARS-CoV-2 can result in mild-to-moderate respiratory disease ( Deng et al., 2020; Munster et al., 2020; Shan et al., 2020; Song et al., 2020), suggesting that rhesus macaques may be a suitable non-human primate model to mimic COVID-19-associated pathology and expand knowledge about this disease.
Current understanding of SARS-CoV-2 infection-induced cellular and molecular pathology remains incomplete ( Merad et al., 2022). To identify cell types critical for the tropism observed in affected organs and to analyze infection-induced dysfunction in a cell-type-specific manner, we profiled single-nucleus transcriptomic atlases of visceral organs (i.e., lung, liver, and kidney) and the CNS (i.e., cerebral cortex) from SARS-CoV-2-infected rhesus macaques. Compared with the healthy macaques, infection-related injuries in the macaque lungs were similar to those reported in human patients, indicating that macaques are reliable models with which to recapitulate SARS-CoV-2-caused pathology. Furthermore, we found several previously unidentified infection-associated disorders and antiviral immune defects, providing insights into potential therapeutic interventions for COVID-19.
MATERIALS AND METHODS
Animals and ethics statement
Chinese rhesus macaques ( Macaca mulatta) were sourced from the Laboratory Animal Center, Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS). All animal experiment procedures were performed at the Kunming National High-Level Biosafety Research Center for Non-Human Primates, KIZ, CAS, and in accordance with the guidelines approved by the Ethics Committee of the Kunming Institute of Zoology (approval number: IACUC20005).
SARS-CoV-2 challenge in rhesus macaques
The SARS-CoV-2 strain 107 (accession number: NMDCN0000HUI) was provided by the Guangdong Provincial Center for Disease Control and Prevention (Guangzhou, China), and has been described in our previous studies ( Song et al., 2021; Song et al., 2020). The virus was propagated in African green monkey kidney epithelial cells (Vero-E6) (ATCC, No. 1586) and titrated. The macaques were intranasally (40%) and endotracheally (60%) challenged with 10 7 TCID 50 (50% tissue culture infectious dose) of SARS-CoV-2 and euthanized on the 7th day post-infection (dpi). All seven lung lobes, liver, kidney, and cerebral cortex (prefrontal) were collected and stored in ultra-low temperature refrigerators or immediately fixed in 4% (wt/vol) formaldehyde solution.
Quantitative real-time polymerase chain reaction (qRT-PCR) for viral load detection
Lung tissue samples were homogenized, and RNA was extracted using TRIzol Reagent (Thermo, USA). For detection of SARS-CoV-2 RNA, a THUNDERBIRD Probe One-Step qRT-PCR Kit (TOYOBO, Japan) was used in accordance with the manufacturer’s protocols. Primers and probes included: forward primer 5'-GGGGAACTTCTCCTGCTAGAAT-3', reverse primer 5'-CAGACATTTTGCTCTCAAGCTG-3', and probe FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3'. In each run, serial dilutions of the SARS-CoV-2 RNA reference standard (National Institute of Metrology, China) were run in parallel to calculate viral copy numbers in each sample.
Tissue processing and nuclei isolation for lung, kidney, and liver tissues
We dissolved one tablet of cOmplete™ EDTA-free Protease Inhibitor Cocktail (Roche, #11873580001, Switzerland) into 1 mL of Nuclei EZ lysis buffer from a Nuclei EZ Prep Isolation Kit (Sigma, Cat #NUC-101, USA), named cOmplete stock (10X). We then prepared lysis buffer containing 1.8 mL of EZ lysis buffer supplemented with 200 μL of cOmplete stock and 50 μL of Ambion™ RNase Inhibitor (Invitrogen, AM2684, USA). We added 2 mL of lysis buffer to C tubes (Miltenyi Biotec, #130-093-237, Germany), which were maintained on ice.
Frozen visceral organ tissue specimens (lower lung, liver, and kidney) were taken out from the ultra-low temperature refrigerator, placed in a clean dish on ice, and thawed for 2 min. We injected 1 mL of lysis buffer into the tissue to “inflate” it with a syringe and moved the needle during injection to distribute the solution evenly. The tissues were then cut into small pieces with scissors. The minced tissue pieces and solution were transferred into a C tube containing 2 mL of the prepared lysis buffer. The C tube was closed and inverted to ensure that all pieces were at the base. The C tube was placed on a gentleMACS™ Dissociator (Miltenyi Biotec, #130-093-235, Germany). The m_lung_01 program was run completely, followed by the m_lung_02 program for no more than 20 s, then stopped. The C tube was immediately placed on ice. The homogenate was foamy and contained some small pieces of tissue that had not been dissociated. The foam and supernatant were discarded using wide bore tips after centrifuging the C Tube in a swinging bucket rotor centrifuge at 500 × g for 5 min at 4 °C. The pellet was resuspended in 2–4 mL of resuspension buffer (consisting of 1×phosphate-buffered saline (PBS), 0.1% bovine serum albumin (BSA), and 0.5 U/μL Ambion™ RNase Inhibitor (Invitrogen, AM2684, USA)). The suspension was filtered through a 70 μm cell strainer into a new 15 mL centrifuge tube to remove the remaining tissue pieces. Nuclei were pelleted by centrifugation at 300 × g for 3 min at 4 °C to avoid blebbing and clumping of nuclei. The supernatant was aspirated thoroughly, and the isolated nuclei were resuspended in 1–2 mL of resuspension buffer. The nuclear suspension was then filtered through a 35 μm cell strainer for further cleaning. The nuclei were counted with a hemocytometer, and nuclear concentration was adjusted to 700–1 200/μL.
Tissue processing and nuclei isolation for cerebral cortex tissues
Frozen prefrontal cortex tissue blocks were taken out from the ultra-low temperature refrigerator and minced into small pieces in a clean dish on ice. The tissue pieces were transferred to pre-chilled glass dounce tissue grinders (Sigma, Cat #D8938, USA) for homogenization in 2 mL of cold Nuclei EZ lysis buffer (Sigma, Cat #NUC-101, USA) on ice, 20 times with pestle A followed by 20 times with pestle B. The homogenate was transferred into a 15 mL centrifuge tube with an additional 2 mL of cold Nuclei EZ lysis buffer, then incubated on ice for 5 min before centrifugation (500 × g for 5 min at 4 °C). The supernatant was discarded, and the sediment was resuspended in 4 mL of cold Nuclei EZ lysis buffer, then incubated on ice. After centrifugation (500 × g for 5 min at 4 °C) and removal of the supernatant, the primitive nuclear products were cleaned via density gradient centrifugation to remove debris. The sediment was resuspended in 3 100 μL of cold PBS and filtered through a 70 μm cell strainer into a new tube. Cold debris removal solution (900 μL) (Miltenyi Biotec, #130-109-398, Germany) was added to the suspension and mixed well. The mixture was overlaid with 4 mL of cold PBS gently, followed by centrifugation at 3 000 × g for 10 min at 4 °C with full acceleration and full brake. The debris aggregated together to form a cloud-like layer at the interphase during centrifugation. The two top phases were discarded, followed by the addition of cold PBS and gentle inversion of the tube three times. The suspension was pelleted by centrifugation at 1 000 × g for 5 min at 4 °C. The supernatant was aspirated completely. The nuclear sediment was twice washed with 2 mL of nuclei suspension buffer (NSB, consisting of 1× PBS, 0.1% BSA, and 0.5 U/μL Ambion™ RNase Inhibitor (Invitrogen, AM2684, USA)) for further purification. Finally, the isolated nuclei were resuspended in an appropriate volume of NSB. The nuclei were counted with a hemocytometer, with the concentration adjusted to 700–1 200/μL.
Single-nucleus RNA-seq
The nuclear suspensions were loaded onto Chromium Single-Cell Chip B (10X Genomics, USA) to generate single-cell Gel Beads-in-Emulsion (GEMs) using the Chromium Single-Cell Controller to capture 5 000–8 000 single cells per reaction. Reverse transcription (first-strand cDNA generation), full-length cDNA amplification, and 3’ library construction were conducted using a Chromium Single-Cell 3’ Reagent Kit v3 (10X Genomics, USA) following the manufacturer’s user guide. The libraries were sequenced on the Illumina NovaSeq sequencing platform with a paired-end read length of 150 bp.
Data processing and quality control
Raw sequencing data were processed using Cell Ranger (v3.1.0, 10X Genomics) with default mapping parameters, except for the use of the pre-mRNA version of the M. mulatta reference genome from Ensembl, which was customized under 10X Genomics official instructions to ensure the capture of intronic reads originating from pre-mRNA transcripts abundant in the nuclear fraction. The feature-barcode matrices generated by Cell Ranger were loaded into the subsequent analysis pipeline.
Cells that passed the following filtration were retained for downstream analysis: number of detected genes per nucleus at least 500, except for some cerebral cortex samples in which the number of detected genes had to be at least 1 500 based on iterative analysis results; unique molecular identifier (UMI) counts per nucleus less than 50 000; percentage of UMIs from mitochondrial genes below 15%; doublet score calculated by Scrublet package ( Wolock et al., 2019) below 0.25.
Dimension reduction and clustering
The Scanpy (v1.4.5.post3) package ( Wolf et al., 2018) was used to perform preprocessing of gene expression data. The filtered gene-cell matrix was normalized to 10 4 molecules/cell for sequencing depth with the “normalize_total” function and log transformed with the “log1p” function. Data variation caused by the number of UMI counts and percentage of UMIs from mitochondrial genes was regressed out using the “regress_out” function. Variable genes were obtained with mean expression levels ranging from 0.0125 to 3 and dispersion greater than 0.5. Uniform manifold approximation and projection (UMAP) was performed with the first 30 principal components obtained from principal component analysis (PCA) (batch effect corrected by Harmony ( Korsunsky et al., 2019)) for visualization of single cells. Cell clustering was performed using the Leiden algorithm.
Analysis of differentially expressed genes (DEGs)
Differential gene expression analysis of each cell type between infected and healthy samples was performed using the “FindMarkers” function in the Seurat package ( Butler et al., 2018) in R. The “MAST” method was applied. We only tested those genes detected in at least 10% of either healthy or infected samples for each cell type. We used “adjusted P-value (Bonferroni corrected) <0.05” as a threshold to filter genes. Log 2FC was calculated and used to rank the DEGs.
To identify marker genes for each cell group and facilitate the construction of dot plots of marker genes for each cell type, the “FindAllMarkers” function in the Seurat package was adopted.
Gene Ontology (GO) term and pathway enrichment analysis of DEGs
Enrichment analysis of GO terms (Biological Process) and pathways for infection-associated DEGs (|log 2FC|>0.25, adjusted P<0.05) was implemented using Enrichr ( Chen et al., 2013; Kuleshov et al., 2016) (https://maayanlab.cloud/Enrichr/). GO terms and pathways were considered significantly enriched at P<0.05.
Significance tests for selected genes
We performed the one-sided Wilcoxon Rank-Sum test to compare differences in gene expression between infected and healthy samples based on log-normalized expression data. Results were considered statistically significant at adjusted P<0.05.
Subclustering of astrocytes
Astrocyte subclustering was performed using scaled data with 4 000 highly variable genes with the Seurat package ( Butler et al., 2018) in R. The “RunPCA” function was used for PCA to reduce dimensionality. Nearest neighbors were computed using the “FindNeighbors” function. Groups were clustered based on the Louvain algorithm using the “FindClusters” function. The “RunUMAP” function was used for visualization.
Trajectory analysis
We performed pseudotime trajectory analysis using the R package Monocle3 ( Cao et al., 2019) to study cell state changes in astrocytes in the cerebral cortex. We first imported the data derived from Seurat into Monocle3, including an input read count matrix with the top variable features selected by the “FindVariableFeatures” function and a UMAP layout. Trajectory was then fitted using the “learn_graph” function and cells were ordered using the “order_cells” function in Monocle3.
Calculating module score
We applied the “AddModuleScore” function in the Seurat package to investigate differences between infected and healthy samples in terms of astrocyte reactivity and microglial activation in the cerebral cortex and necrotic cell death in the lung and kidney. One-sided Wilcoxon rank-sum test was used to check whether infected samples scored higher than healthy controls. Tests with adjusted P<0.05 were considered significant.
Analysis of regulons
We performed SCENIC analysis ( Aibar et al., 2017) to study gene regulons in astrocytes using the “pySCENIC” package in Python. We used count data as input and applied the standard SCENIC workflow (grnboost2 – cistarget – AUCell). After obtaining the regulon activity matrix, we used “MAST” to identify differential regulons between infected and healthy samples of astrocytes. Regulons with P<0.05 were filtered out.
Cell-cell communication analysis
CellChat (v1.1.3) ( Jin et al., 2021) was used to explore changes in cell-cell communication among cell types in lung tissue, with the aim to determine: (1) whether cell-cell communication is enhanced in the infected group compared with the healthy group; (2) which cell-type interactions are significantly altered by infected status; and (3) which signaling pathways mediate changes in communication. First, DEGs were identified for all cell groups between the infected and healthy groups; second, ensemble average expression was calculated for each cell group; third, intercellular communication probability was calculated based on the projection of ligand-receptor expression profiling onto protein-protein networks; last, significant differences in intercellular communication between the infected and healthy groups were identified by a permutation test. The probability of communication at the signaling pathway level was inferred by calculating the probability of communication for all ligand-receptor interactions associated with each signaling pathway. Several signaling pathways were highlighted by ranking signaling from the infected and healthy groups. Cell group and ligand-receptor contributions of these pathways were further computed and visualized. Calculations were performed using the default parameters following the CellChat workflow.
CellPhoneDB software ( Efremova et al., 2020) was used to determine cell-cell communication patterns between cell types in the infected samples of the cerebral cortex. This method mainly evaluates the number of ligand-receptor interactions of every two cell types. Interaction pairs were regarded as significant at P<0.05.
Immunofluorescence staining
Formalin fixed paraffin-embedded (FFPE) tissue sections (5 μm) were dewaxed in xylene, then rehydrated in 100%, 95%, 85%, 75%, and 50% gradient ethanol and distilled water. Heat antigen retrieval was performed by steaming at 98 °C in retrieval solution (10 mmol/L sodium citrate, pH=6.0) for 20 min. Sections were allowed to cool at room temperature. Following antigen retrieval, sections were permeabilized in PBS containing 0.1% Triton X-100, then blocked using 5%–10% donkey serum in PBS for 1 h. The cerebral cortex sections were incubated with a primary antibody (rabbit anti-GFAP, Abcam (UK), ab7260, 1:500) overnight at 4 °C. A secondary antibody (donkey anti-rabbit, Alexa Fluor™ 488, Invitrogen (USA)) diluted in blocking buffer (1:500) was applied for 2 h at room temperature. For staining of macaque immunoglobulin G (IgG), rabbit anti-monkey IgG H&L (Alexa Fluor® 488) antibody (Bioss (China), bs-0335R-AF488, 1:500) was incubated with blocked kidney sections for 2 h at room temperature. Cell nuclei from all sections were counterstained with 4’,6-diamidino-2-phenylindole (DAPI, Invitrogen (USA), D1306). Immunostaining images were acquired using an Olympus laser confocal microscope (FV3000) and analyzed with Olympus confocal software and Adobe Photoshop CC 2018.
RESULTS
Integrative analysis of single-nucleus transcriptomes of multiple macaque organs
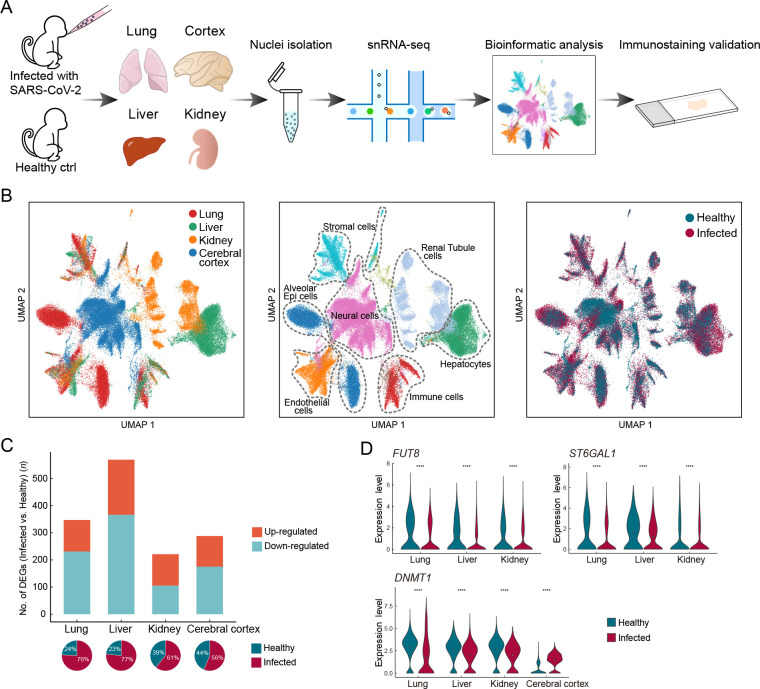
We performed single-nucleus RNA-sequencing (snRNA-seq) of lung, liver, kidney, and cerebral prefrontal cortex tissues obtained from two young rhesus macaques (3 years old, one male, one female) infected with SARS-CoV-2 strain 107 (as described in our previous studies ( Song et al., 2021; Song et al., 2020)) and sacrificed at 7 dpi ( Figure 1A). Viral loads in the lung tissue were detected by qRT-PCR, which confirmed successful infection (Supplementary Figure S1 and Table S1). We performed snRNA-seq using samples of the same organs taken from healthy young macaques as controls (two healthy macaques for cerebral cortex and one macaque for the other three organs) ( Figure 1A). After quality control and batch effect correction, we obtained a total of 77 029 nuclei from all four organs (lung, n=22 141 nuclei; liver, n=17 732 nuclei; kidney, n=13 550 nuclei; cerebral cortex, n=23 606 nuclei) (Supplementary Figure S2 and Table S2). Cell types were annotated with classical marker genes in each organ (Supplementary Figure S3 and Table S3). Integrated data across all four organs were visualized using UMAP ( Figure 1B). Alveolar epithelial cells in the lung, hepatocytes in the liver, tubular cells in the kidney, and neural cells in the cerebral cortex were the exclusive parenchymal cell types identified in the respective organs. Certain cell types were largely mixed and similar in the four organs, e.g., endothelial cells, stromal cells, and immune cells. The liver exhibited the greatest transcriptional disturbance among organs, as indicated by the number of DEGs identified at the all-cell level ( Figure 1C; Supplementary Table S4), which may be the result of the diverse metabolic functions of the liver.
Figure 1.
Overview of single-nucleus transcriptomes from four organs in rhesus macaques
A: Study flowchart. B: UMAP of 77 029 nuclei captured in four organs from infected and healthy macaques, colored by organ (left), broad cell type (middle), and infection status (right). C: Bar plot showing number of infection-associated DEGs (|logFC|>0.7, adjusted P<0.05) at the all-cell level in each organ. Pie charts below show the percentage of nuclei from infected and healthy samples for each organ. D: Violin plots showing dysregulated expression of FUT8, ST6GAL1, and DNMT1 in multiple organs.
Five DEGs were shared among the DEGs in the lung, liver, and kidney. The expression levels of two glycosyltransferase-coding genes, fucosyltransferase FUT8, which is the sole enzyme catalyzing core fucosylation and is required for T cell activation ( Liang et al., 2018), and sialyltransferase ST6GAL1, which enhances B cell-mediated humoral immunity ( Irons et al., 2020), were down-regulated in the three aforementioned visceral organs at the all-cell level ( Figure 1D), suggesting that adaptive immune responses are likely suppressed during SARS-CoV-2 infection. In addition, DNMT1, which encodes DNA methyltransferase 1, was shared by all DEG datasets from the four organs ( Figure 1D). Transcriptional regulation is largely ruled by epigenetic mechanisms, such as DNA methylation ( Zhu et al., 2016), hence the universal dysregulation of DNMT1 in various organs may underlie the infection phenotype of broadly altered transcriptional programs. Therefore, changes in the epigenetic landscape in host cells following infection may indicate a molecular mechanism used by SARS-CoV-2 to perturb host defense programs and physiological functions.
SARS-CoV-2-infected macaque lungs showed epithelial injury and fibrotic signatures similar to those in human COVID-19 patients
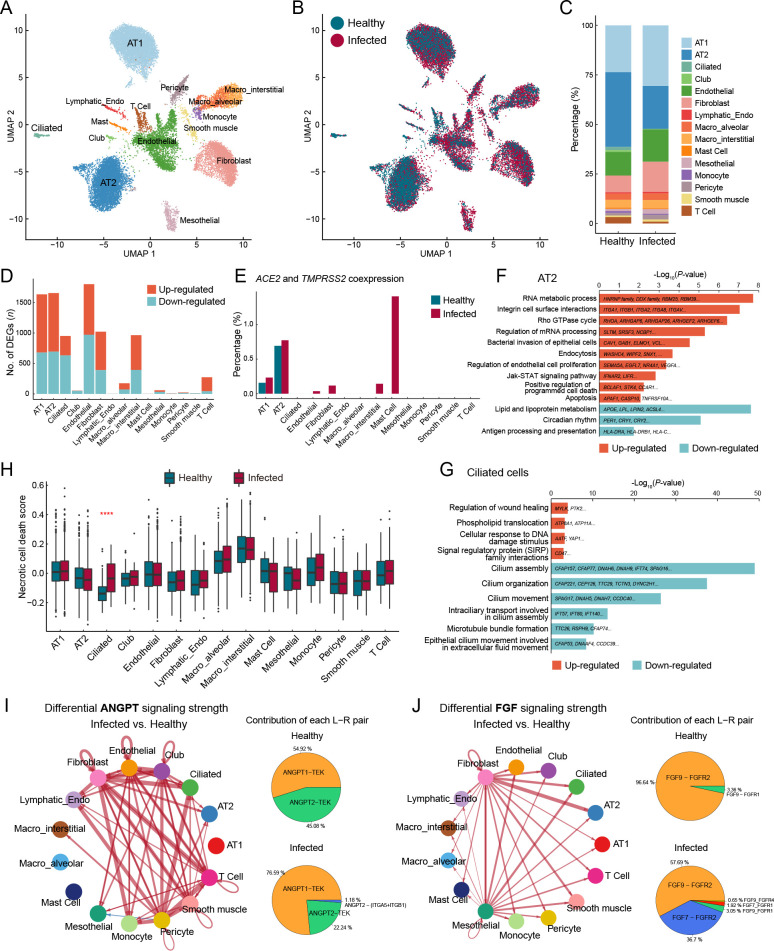
In the lung dataset, alveolar type I (AT1) and type II epithelial cells (AT2) were the most abundant cell types, with other epithelial cells, such as ciliated and club cells, also recovered ( Figure 2A–C). The cell types most affected by SARS-CoV-2 infection included AT2, AT1, ciliated cells, endothelial cells, fibroblasts, and interstitial macrophages ( Figure 2D; Supplementary Table S5). Previous studies have demonstrated that SARS-CoV-2 uses angiotensin converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) as the cell-surface receptor and cellular protease for spike protein binding and priming, respectively ( Hoffmann et al., 2020; Scudellari, 2021; Zhou et al., 2020). Therefore, we measured the coexpression of these two well-known SARS-CoV-2 entry factors (percentage of positive cells with read counts >0 for both genes) and found enrichment in the AT2 cells ( Figure 2E), consistent with previous findings in human and cynomolgus monkeys ( Ma et al., 2021; Muus et al., 2021; Sungnak et al., 2020). The proportion of AT2 cells was markedly reduced in infected samples compared to the healthy control ( Figure 2C), in line with previous single-cell transcriptomic research using autopsy tissue samples from COVID-19 patients ( Delorey et al., 2021; Melms et al., 2021), likely reflecting direct infection-induced apoptosis or cell death as AT2 cells possess entry factor expression. Accordingly, GO (Biological Process) and pathway enrichment analysis of DEGs in the AT2 cells revealed up-regulation of “positive regulation of programmed cell death” and “apoptosis” ( Figure 2F). For instance, APAF1, BCLAF1, TNFRSF10A, TNFRSF21, and caspase-10 ( CASP10), which participate in apoptotic induction, were up-regulated in AT2 cells (Supplementary Figure S4A). In addition, RNA metabolic processes were significantly enriched in up-regulated genes in AT2 cells ( Figure 2F). For example, heterogeneous nuclear ribonucleoproteins (HNRNPs), representing a large family of factors responsible for RNA processing, including RNA splicing, maturation, decay, and translation, were up-regulated in AT2 cells (Supplementary Figure S4A). “Bacterial invasion of epithelial cells” and “endocytosis” also implied direct infection of AT2 cells. Together, these results suggest that AT2 is the potential lung cell type targeted by SARS-CoV-2. The maintenance of alveolar epithelium homeostasis and its regeneration after injury are fueled by surfactant-producing AT2 cells, which can self-renew and differentiate into AT1 cells. Regeneration-related genes critical for lung injury repair were down-regulated in AT2 cells after SARS-CoV-2 infection (Supplementary Figure S4B), suggesting impairment in the differentiation ability of AT2 cells, consistent with that found in COVID-19 patient lungs ( Melms et al., 2021; Wang et al., 2021).
Figure 2.
Epithelial cell impairment and signaling pathway changes in macaque lungs after SARS-CoV-2 infection
A: Clusters and respective cell type assignments in UMAP. AT1, alveolar type 1 cell; AT2, alveolar type 2 cell; Lymphatic_Endo, lymphatic endothelial cell; Macro_interstitial, interstitial macrophage; Macro_alveolar, alveolar macrophage. B: Sample origin status of cells with the same embedding as in (A). In total, 22 141 single nuclei from lung tissues of infected (16 781 nuclei) and healthy (5 360 nuclei) animals were considered. C: Bar plot presenting proportion of each cell type in infected and healthy lungs. D: Bar plot showing numbers of DEGs (infected vs. healthy, |logFC|>0.25, adjusted P<0.05) in different cell types of the lung. E: Bar plot showing percentage of cells coexpressing ACE2 and TMPRSS2 in each cell type. F, G: GO and pathway enrichment analysis of infection-associated DEGs in AT2 cells (F) and ciliated cells (G). Representative genes are shown on the bar. H: Necrotic cell death score computed for different cell types, split by status. Genes in GOBP_NECROTIC_CELL_DEATH (GO: 0070265) were used to calculate the score. Statistical significance was determined by one-sided Wilcoxon rank-sum test, ****: P<0.0001. I, J: Differential cell communication in ANGPT signaling (I) and FGF signaling pathways (J) between infected and healthy samples found by CellChat. Circle plots (left) showing differential signaling strengths of infected samples compared to the healthy control. Red arrows represent enhanced communications, and arrow thickness represents differential amplitude. Pie plots (right) showing contribution of each ligand-receptor pair.
The dysregulated biological processes of AT1 cells showed similarities to those of AT2 cells. Notably, cytokine-associated pathways were elevated in AT1 cells (Supplementary Figure S4C), suggesting activation of the inflammatory response in the lung after infection. LMOD1, a marker gene of idiopathic pulmonary fibrosis ( Selman et al., 2006), was one of the most up-regulated genes in the AT1 cells (log 2FC=2.03). In addition, collagen-related genes were highly expressed in fibroblasts from infected samples (Supplementary Figure S4D), suggesting a contribution to fibrosis. Orphan nuclear receptors NR4A1 and NR4A2, which inhibit fibrosis ( Chen et al., 2015; Palumbo-Zerr et al., 2015), were significantly down-regulated (Supplementary Figure S4E). Moreover, fibrogenic factor TIMP2 and protective antifibrotic factor TIMP3 ( Kassiri et al., 2009; Nie et al., 2004) were both dysregulated in the profibrotic direction (Supplementary Figure S4E). These observations suggest the occurrence of fibrosis in the lungs of SARS-CoV-2-infected macaques.
Ciliated cells exhibited impaired physiological functions associated with cilia beating and mucus removal in the SARS-CoV-2-infected lungs. Numerous genes closely associated with cilium formation and movement were down-regulated ( Figure 2G), consistent with previous study of bronchoalveolar lavage fluid (BALF) in COVID-19 patients ( He et al., 2020). Consistent with the reduction in ciliated cell proportion ( Figure 2C), the necrotic cell death score was significantly higher in ciliated cells from infected samples than from the healthy control, while other cell types were minimally changed by infection ( Figure 2H). CD47, a cell-survival signaling molecule that interacts with SIRPα ( Lehrman et al., 2018; Okazawa et al., 2005), showed significantly higher expression in ciliated cells from infected samples than from the healthy control ( Figure 2G), possibly due to the preferential survival of cells with elevated CD47 expression. These observations suggest that ciliated cells are acutely affected by SARS-CoV-2 infection, leading to necrosis.
Antigen presentation deficiency and signaling pathway alterations in macaque lungs
Although macrophages are professional antigen-presenting cells (APCs), the expression levels of major histocompatibility complex (MHC) component genes HLA-DRA, HLA-DRB1, HLA-DPA1, HLA-C, HLA-B, and chaperone molecule CD74 were surprisingly down-regulated in the lung macrophages after SARS-CoV-2 infection (Supplementary Figure S4F–H). Moreover, we observed reduced expression of MHC class I/II genes in non-classical APCs, including alveolar epithelial cells and endothelial cells (Supplementary Figure S4H). Therefore, we propose that suppression of MHC class I/II-related components responsible for antigen presentation and adaptive immunity may be a potential immune evasion strategy for SARS-CoV-2 to target the MHC system.
To identify changes in global cellular communication in the lung after infection, we used the intercellular communication inference tool “CellChat” ( Jin et al., 2021). We found that angiopoietin (ANGPT) and fibroblast growth factor (FGF) signal intensities were significantly increased in infected samples (Supplementary Figure S5A). The ANGPT signaling pathway was activated in sender cell types other than fibroblasts and pericytes, and in receiver cell types other than endothelial cells ( Figure 2I; Supplementary Figure S5B). The ANGPT1-TEK pair became more dominant in infected samples compared to the control ( Figure 2I). This pair maintains the endothelial barrier and blood vessel integrity and plays a role in anti-inflammation ( David et al., 2013; Huang et al., 2010). Therefore, an increase in their signaling may facilitate tissue injury repair after infection. In addition, the FGF signaling pathway showed increased fibroblast and mesothelial cell activity in the lung ( Figure 2J; Supplementary Figure S5C). Among the most frequent ligand-receptor pairs, the contribution of FGF7-FGFR2 to FGF signaling was substantially increased after SARS-CoV-2 infection ( Figure 2J). FGF7 is thought to regulate pulmonary epithelial proliferation and facilitate lung repair ( Xie et al., 2020; Yano et al., 2000). These results suggest that the spontaneous lung injury repair program was initiated in macaques by 7 dpi or earlier.
Inflammatory response and immune tolerance in macaque liver
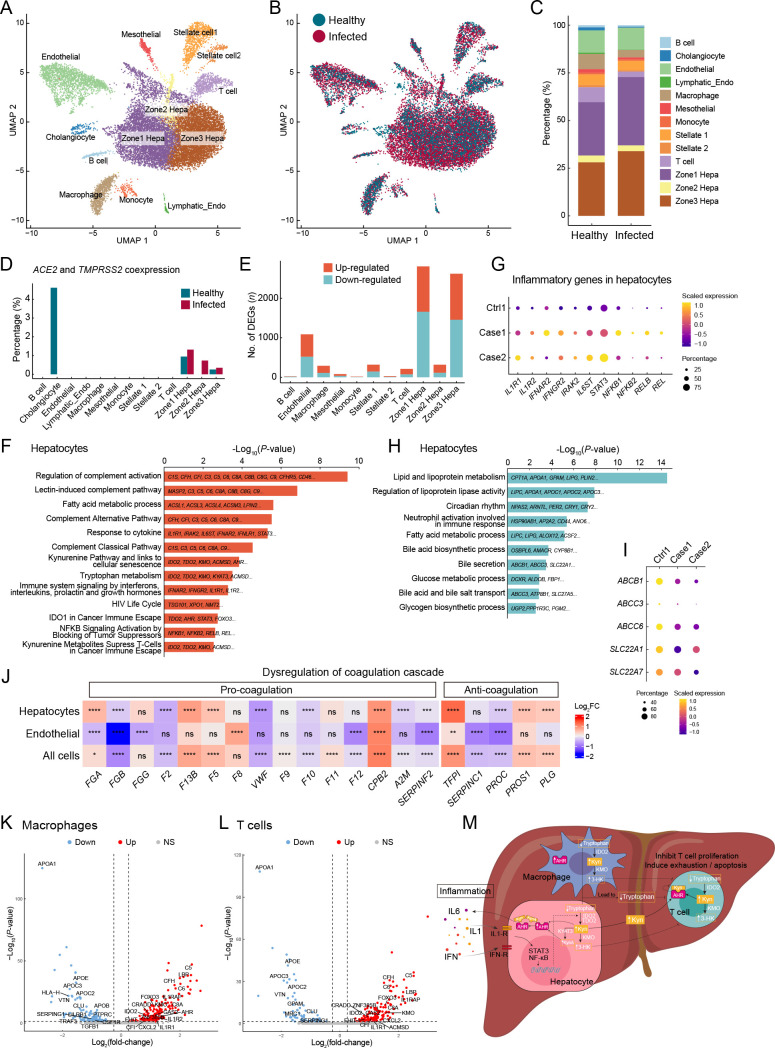
Hepatocytes, which were the predominant cells in the liver ( Figure 3A–C), were enriched in ACE2 and TMPRSS2 coexpression, especially zone 1 hepatocytes ( Figure 3D), and contained the most DEGs between infected and healthy livers ( Figure 3E; Supplementary Table S6). Although the proportion of coexpression was notably higher in the healthy control cholangiocytes, the proportion in infected samples was zero ( Figure 3D), which may be due to the extremely low number of cholangiocytes captured in infected samples ( Figure 3C). Therefore, we inferred that hepatocytes may be the potential target cell type in the liver and preferentially affected during SARS-CoV-2 infection.
Figure 3.
Hepatocyte dysregulations and immune evasion pathway in the liver of SARS-CoV-2-infected macaques
A: Clusters and respective cell type assignments in UMAP. Zone 1 Hepa, hepatocytes in zone 1; Zone 2 Hepa, hepatocytes in zone 2; Zone 3 Hepa, hepatocytes in zone 3; Lymphatic_Endo, lymphatic endothelial cell. B: Sample origin status of cells with the same embedding as in A. In total, 17 732 single nuclei from liver tissues of infected (13 580 nuclei) and healthy (4 152 nuclei) animals were considered. C: Bar plot presenting proportion of cell types in infected and healthy livers. D: Bar plot showing percentage of cells coexpressing ACE2 and TMPRSS2 in each cell type. E: Bar plot showing numbers of DEGs (infected vs. healthy, |logFC|>0.25, adjusted P<0.05) in different cell types of the liver. F: GO and pathway enrichment analysis of up-regulated DEGs in hepatocytes. Representative genes are indicated on the bar. G: Dot plot showing activation of inflammation-related genes in hepatocytes. H: GO and pathway enrichment analysis of down-regulated DEGs in hepatocytes. Representative genes are indicated on the bar. I: Dot plot showing reduced expression of membrane transporters involved in bile secretion in hepatocytes. J: Heatmap showing dysregulation of coagulation cascade-related genes in hepatocytes, endothelial cells, and all cells in the liver. Statistical significance was determined by one-sided Wilcoxon rank-sum test, indicated on each block, ns means not significant; *: P<0.05; **: P<0.01; ***: P<0.001; ****: P<0.0001. K, L: Volcano plots showing transcriptional changes in macrophages (K) and T cells (L). Significance (−Log 10 P) is on the y-axis and magnitude (Log 2(fold-change)) is on the x-axis. M: Model summarizing potential mechanisms contributing to immunosuppressive microenvironment and immune tolerance in the liver after SARS-CoV-2 infection, focusing on the kynurenine pathway and inflammation.
Figure 3.
DEG enrichment analysis in hepatocytes indicated that the complement system was activated in infected samples, as evidenced by the activation of various pathways, including the classical, alternative, and lectin-induced complement pathways ( Figure 3F). The substantial increase in complement component proteins C5, C6, C8A, and C9 (log 2FC=1.94, 1.44, 1.26, and 0.97, respectively) may result in enhanced formation of membrane attack complex (MAC), which is incorporated into the cell membrane to cause cell lysis. “Response to cytokine” was also enriched in the up-regulated genes of hepatocytes ( Figure 3F). Cytokine receptors IL1R1, IL1R2, IFNAR2, and IFNGR2 were elevated, as were IRAK2 and IL6ST, which are regulators and signal transducers that participate in cytokine-induced activation of the JAK-STAT3 and NF-κB signaling pathways. Accordingly, the expression levels of STAT3, NFKB1, NFKB2, RELB, and REL were also up-regulated ( Figure 3G). Therefore, after SARS-CoV-2 infection, innate immune defense-complement system and inflammatory response were activated in the macaque livers, especially in the hepatocytes.
The liver has multiple metabolic functions in the body. Various metabolic processes, including those involving lipids, lipoproteins, fatty acids, glucose, and glycogen, were dysregulated in the liver after SARS-CoV-2 infection ( Figure 3H; Supplementary Figure S6A). Bile is a critical aqueous liver secretion produced by hepatocytes that facilitates dietary fat absorption in the intestinal lumen. Here, bile biosynthesis and secretion-related processes were significantly enriched in down-regulated genes in hepatocytes ( Figure 3H). For example, the expression levels of genes encoding membrane transporters involved in bile secretion, such as ABCB1, ABCC3, ABCC6, SLC22A1, and SLC22A7 ( Boyer, 2013), were decreased in the hepatocytes ( Figure 3I), indicating that bile generation and secretion may be impaired in the liver after SARS-CoV-2 infection.
The liver also plays a central role in the blood coagulation cascade as it produces most coagulation factors. In this study, we found extensive dysregulation of coagulation factors, inhibitors, and fibrinolysis-associated proteins in the liver after infection ( Figure 3J), especially in hepatocytes and endothelial cells, which are the primary sources of these coagulation factors. The F2 gene encoding prothrombin, the active form (thrombin) of which plays a central role in hemostasis ( Crawley et al., 2007) by converting fibrinogen into fibrin to form blood clots, was down-regulated. The PLG gene encoding plasminogen ( Schuster et al., 2007), the active form (plasmin) of which plays a key role in fibrinolysis by dissolving fibrin in blood clots, was up-regulated ( Figure 3J). These observations suggest that SARS-CoV-2 infection may lead to a higher risk of bleeding.
The liver is the largest solid organ in the body and contains many resident immune cells. However, we found no activation signatures of macrophages or T cells, other than up-regulation of complement components ( Figure 3K, L). Surprisingly, tryptophan metabolism along the kynurenine (Kyn) pathway was activated in the liver following SARS-CoV-2 infection. The Kyn pathway is a key mechanism for creating an immunosuppressive microenvironment, which facilitates immune surveillance escape by many types of cancer cells ( Adams et al., 2012; Liu et al., 2020b; Nevler et al., 2019; Rad Pour et al., 2019). Activation of the Kyn pathway has also been reported in viral infection ( Jenabian et al., 2015; Ma et al., 2009; Pizzini et al., 2019).
Indoleamine 2,3-dioxygenase 2 (IDO2) and tryptophan 2,3-dioxygenase (TDO2), which catabolize the substrate tryptophan to form Kyn, are first and rate-limiting enzymes of the Kyn pathway. Here, the expression levels of IDO2 and TDO2 were elevated in hepatocytes ( Figure 3F), and IDO2 was also elevated in macrophages and T cells after SARS-CoV-2 infection ( Figure 3K, L). Moreover, Kyn 3-monooxygenase (KMO), which catalyzes the hydroxylation of Kyn to form 3-hydroxykynurenine (3-HK), and Kyn aminotransferase 3 (KYAT3), which catalyzes the transamination of Kyn to form kynurenic acid (KynA), were up-regulated at the transcriptional level in hepatocytes ( Figure 3F). KMO gene expression was also elevated in immune cells ( Figure 3K, L). Tryptophan is an amino acid required for the survival and proliferation of T cells, such as T helper (Th) cells and cytotoxic T lymphocytes (CTLs) ( Fallarino et al., 2002; Lee et al., 2002). Thus, our results suggest that immune cell populations are suppressed in the tryptophan-deprived microenvironment established by the overexpression of IDO2 and TDO2 after SARS-CoV-2 infection.
Importantly, the aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor, which can be activated by binding to endogenous tryptophan derivatives including Kyn and KynA ( Dinatale et al., 2010; Opitz et al., 2011). The Kyn-AHR signaling pathway suppresses the differentiation and activity of immune cells, resulting in an impaired antitumor immune response ( Liu et al., 2018; Mezrich et al., 2010; Opitz et al., 2011). Along with Kyn accumulation resulting from increased dioxygenases, AHR gene expression was up-regulated in hepatocytes ( Figure 3F), macrophages ( Figure 3K), and T cells (although not significantly in T cells). Consistent with the immunosuppressive effect described above, the proportions of macrophages and T cells were reduced in the infected liver samples ( Figure 3C), and genes associated with apoptosis, including CRADD, GAS2, FHIT, and ZNF385B, were up-regulated in these immune cells ( Figure 3K, L).
The Kyn pathway is induced by proinflammatory cytokines ( Brooks et al., 2016; Dostal et al., 2018; Santhanam et al., 2016). As described previously, cytokine receptor genes, signal transducer genes, NF-κB subunit genes, and cytokine and chemokine genes, such as IL1A, IL6, CXCL2, and CXCL10, were up-regulated in the liver ( Figure 3G, K, L; Supplementary Figure S6B). Therefore, given the integrated activation of the Kyn pathway and its immunoregulatory effects ( Wu et al., 2018), as well as the inflammatory signatures observed in the liver, we propose that inflammation triggered by SARS-CoV-2 infection induced Kyn pathway activation, leading to the establishment of an immunosuppressive microenvironment and immune tolerance in the macaque liver. These mechanisms are summarized in Figure 3M, providing new insights into liver manifestations and novel immunomodulatory therapies for SARS-CoV-2 infection.
Renal tubule tropism and NELL1-associated membranous nephropathy in the kidney
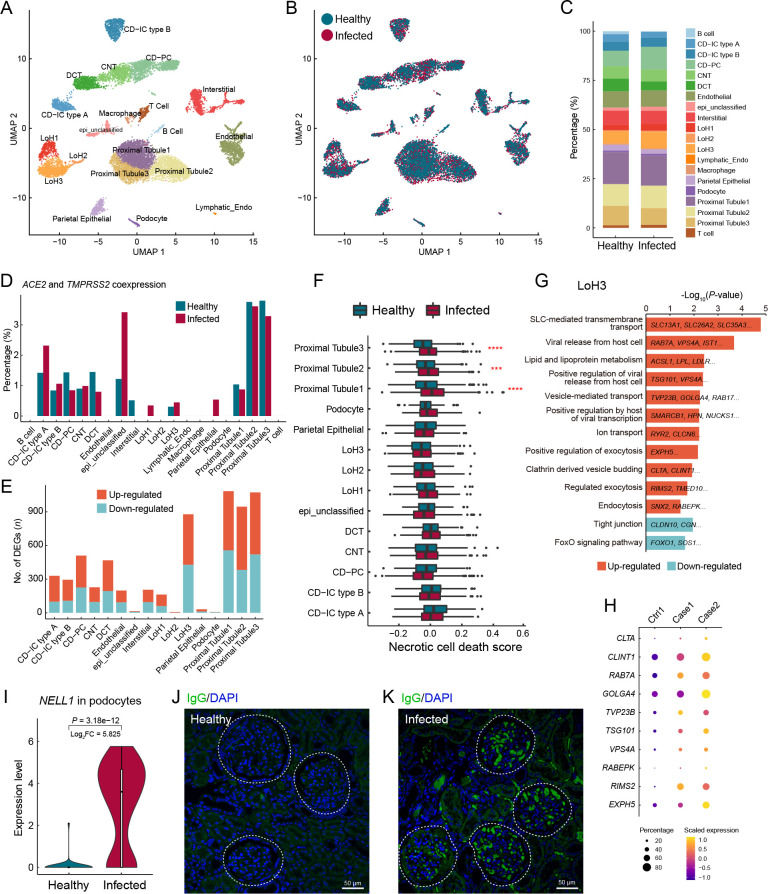
The kidney is a selective filtering organ composed of capillary vessels and renal tubules divided into segments, including proximal tubules, loops of Henle (LoH), distal convoluted tubules, connecting tubules, and collecting ducts ( Figure 4A). Here, the proportion of each parenchymal cell type showed minimal change after infection ( Figure 4B, C). Coexpression of ACE2 and TMPRSS2 was highlighted in the proximal tubule cells ( Figure 4D), indicating that proximal tubules may be more vulnerable to SARS-COV-2 infection, consistent with findings reported in human kidney cells ( Pan et al., 2020). Accordingly, proximal tubule cells were the most severely affected cell type, with more DEGs than any other cell type ( Figure 4E; Supplementary Table S7). Based on the necrotic cell death scores for all parenchymal cell types, the necrosis score was significantly higher for proximal tubule cells in the infection group than in the healthy control ( Figure 4F), suggesting the formation of necrotic lesions in the proximal tubules of infected macaques, in accordance with the renal histopathological features of proximal tubule acute necrotic injury and intraluminal necrotic epithelium debris found in autopsy samples of COVID-19 patients ( Su et al., 2020; Werion et al., 2020).
Figure 4.
Tubule cell tropism and glomerular involvement in the kidney of SARS-CoV-2-infected macaques
A: Clusters and respective cell type assignments in UMAP. LoH, loop of Henle; DCT, distal convoluted tubule; CNT, connecting tubule; CD-PC, collecting duct-principal cell; CD-IC type A, collecting duct-intercalated cell (type A); CD-IC type B, collecting duct-intercalated cell (type B); epi_unclassified, unclassified epithelial cell; Lymphatic_Endo, lymphatic endothelial cell. B: Sample origin status of cells with the same embedding as in A. In total, 13 550 single nuclei from kidney tissues of infected (8 243 nuclei) and healthy (5 307 nuclei) animals were considered. C: Bar plot presenting proportion of cell types in infected and healthy kidneys. D: Bar plot showing percentage of cells coexpressing ACE2 and TMPRSS2 in each cell type. E: Bar plot showing numbers of DEGs (infected vs. healthy, |logFC|>0.25, adjusted P<0.05) in different cell types of the kidney. F: Necrotic cell death score computed for different cell types, split by status. Genes in GOBP_NECROTIC_CELL_DEATH (GO: 0070265) were used to calculate the score. Statistical significance was determined by one-sided Wilcoxon rank-sum test as indicated. ***: P<0.001; ****: P<0.0001. G: GO and pathway enrichment analysis of infection-associated DEGs in LoH3 cluster. Representative genes are shown on the bar. H: Dot plot showing up-regulation of viral life cycle-related genes in LoH3 cells. I: Violin plot showing differential expression of NELL1 gene in podocytes from infected and healthy kidney samples. One-sided Wilcoxon rank-sum test. J, K: Immunofluorescence staining of macaque autoantibodies IgG (green), counterstained with DAPI (blue), in kidney sections from healthy (J) and infected (K) samples. White circles indicate renal corpuscles. Scale bars: 50 μm. Immunostaining performed for the two infected animals showed the same results.
Our results also indicated that LoH cells may be affected, as up-regulated genes in the “LoH3” cluster were enriched in pathways related to the viral life cycle, including “viral release”, “exocytosis”, “vesicle-mediated transport”, “viral transcription”, and “endocytosis” ( Figure 4G). Specifically, CLTA and CLINT1 are involved in the formation and trafficking of clathrin-coated vesicles and are reportedly associated with SARS-CoV-2 entry via endocytosis ( Bayati et al., 2021). RAB7A is an important regulator of vesicular transport and endolysosomal trafficking and is required for vesicular trafficking and cell surface expression of ACE2 ( Daniloski et al., 2021). GOLGA4 plays a key role in vesicular transport at the Golgi apparatus level and is also involved in endosome-to-Golgi trafficking. VPS4A functions in membrane fission events, such as cytokinesis and enveloped virus budding ( Chen & Lamb, 2008), and RIMS2 and EXPH5 are effector proteins involved in exocytosis. Elevated expression of these viral-life-cycle molecules ( Figure 4H) suggests that SARS-CoV-2 likely infected LoH3 cluster cells, where virions underwent internalization, transport, and release. Thus, in addition to the vulnerable proximal tubule in the renal cortex, the LoH in the renal medulla may also be a target of SARS-CoV-2, which deserves further study.
However, the LoH3 cluster did not contain a high percentage of ACE2- and TMPRSS2-coexpressing cells ( Figure 4D). Interestingly, we assessed the expression of several potential coronavirus entry factors ( Singh et al., 2020) in LoH3 cells (Supplementary Figure S7A) and found that the NRP1 gene was significantly increased in these cells of the infected macaques. The protein encoded by NRP1 is a cofactor that can facilitate and potentiate SARS-CoV-2 infectivity ( Cantuti-Castelvetri et al., 2020). In addition, CTSL, a substitutive protease that primes the spike protein of SARS-CoV and possibly SARS-CoV-2 ( Simmons et al., 2013), was markedly up-regulated in LoH3 cells of infected samples. NRP1 and CTSL were also up-regulated in the proximal tubules (Supplementary Figure S7B). Thus, these results suggest that the increase in NRP1 and CTSL expression following infection may be a positive-feedback molecular mechanism for SARS-CoV-2 entry into the kidney.
Podocytes in the renal corpuscle form the final filtration barrier of the glomerulus and play a critical role in preventing the loss of protein in urine. Podocyte dysfunction can easily induce proteinuria, a common manifestation in COVID-19 patients ( Pei et al., 2020). Therefore, we speculated that podocyte dysregulation may also occur in infected macaques. Notably, NELL1 (encoding neural epidermal growth factor-like 1) expression was specifically and highly up-regulated in podocytes of the infection group (log 2FC=5.8) ( Figure 4I). NELL1 is a novel antigen marker for membranous nephropathy (MN), in addition to the well-studied and common antigen marker PLA2R ( Caza et al., 2021; Sethi et al., 2020). In NELL1-associated MN, NELL1 is shed from podocytes and binds to autoantibodies against NELL1 to form antigen-antibody complexes localized in the glomerular basement membrane. Here, immunostaining of macaque IgG in kidney sections showed segmental positive IgG staining in glomeruli of infected samples ( Figure 4K), while IgG staining was negative in the healthy samples ( Figure 4J). This finding suggests that SARS-CoV-2 infection may lead to the development of NELL1-associated MN in macaques. This discovery provides new insights into SARS-CoV-2-associated kidney disease, warranting further attention in the clinical treatment of COVID-19 patients.
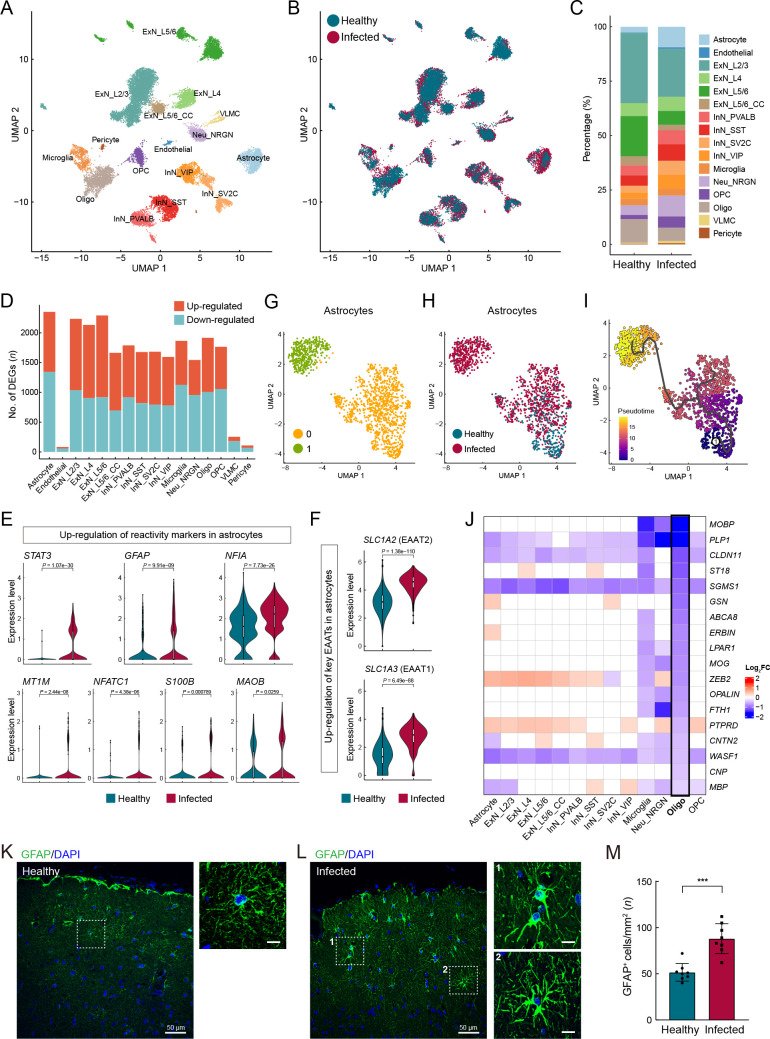
Astrocyte reactivity and myelination defects in macaque cerebral cortex
Various neuronal and glial cell types were captured in our cerebral cortex atlas ( Figure 5A–C), with astrocytes containing the most DEGs ( Figure 5D; Supplementary Table S8). We found that astrocyte reactivity marker genes, including STAT3, GFAP, NFIA, MT1M, NFATC1, S100B, and MAOB ( Escartin et al., 2021; Laug et al., 2019), were significantly up-regulated in astrocytes ( Figure 5E), indicating that astrocytes were in a reactive state in the infected macaques. Reactive astrogliosis may explain the marked increase in the proportion of astrocytes in the infected samples ( Figure 5C). Under physiological conditions, in addition to participating in blood-brain barrier formation with endothelial cells and pericytes, astrocytes regulate neuronal activities via tripartite synapses. Specifically, astrocytes subtly regulate the levels of the excitatory neurotransmitter glutamate in the synaptic cleft via glutamate transporters EAAT1 ( SLC1A3) and EAAT2 ( SLC1A2) located in the astrocyte end-feet. The expression levels of SLC1A3 and SLC1A2 were substantially up-regulated in the astrocytes ( Figure 5F), which may lead to lower glutamate levels and reduced neuronal excitability.
Figure 5.
Pathological states of astrocytes and oligodendrocytes in cerebral cortex of SARS-CoV-2-infected macaques
A: Clusters and respective cell type assignments in UMAP. ExN_L2/3, layer 2/3 excitatory neuron; ExN_L4, layer 4 excitatory neuron; ExN_L5/6, layer 5/6 corticofugal projection neuron; ExN_L5/6_CC, layer 5/6 cortico-cortical projection neuron; Neu_NRGN, NRGN highly expressing neuron; InN_PVALB, parvalbumin interneuron; InN_SST, somatostatin interneuron; InN_VIP, VIP interneuron; InN_SV2C, SV2C interneuron; Oligo, oligodendrocyte; OPC, oligodendrocyte precursor cell; VLMC, vascular and leptomeningeal cell. B: Sample origin status of cells with the same embedding as in A. In total, 23 606 single nuclei from cerebral cortex tissues of infected (13 261 nuclei) and healthy (10 345 nuclei) animals were considered. C: Bar plot presenting proportion of cell types in infected and healthy cerebral cortex samples. D: Bar plot showing numbers of DEGs (infected vs. healthy, |logFC|>0.25, adjusted P<0.05) in different cell types of cerebral cortex. E: Violin plots showing up-regulation of certain classical astrocyte reactivity markers in astrocytes from infected samples compared with healthy samples. F: Violin plots showing increased expression of glutamate transporters EAAT1 ( SLC1A3) and EAAT2 ( SLC1A2) in astrocytes from infected samples compared to healthy samples. G, H: UMAP plots showing astrocyte subclustering, colored by subclusters (G) and status (H). I: Pseudotime trajectory is indicated in graded purple (low) to yellow (high), plotting emergence of astrocyte subpopulation associated with infection. Circled 1 indicates original source population. J: Heatmap showing down-regulation of myelin-associated genes in oligodendrocytes, log 2FC is color coded. K, L: Immunofluorescence staining for GFAP (green) in prefrontal cortex sections from healthy (K) and infected (L) macaques, counterstained with DAPI (blue). Right images (scale bars: 10 μm) show magnification of white boxes on left (scale bars: 50 μm). M: Quantification of GFAP + cell numbers in prefrontal cortex sections of healthy and infected macaques, related to K and L. Data are presented as mean±standard deviation ( SD); ***: P=0.0003 as determined by Mann-Whitney test. Samples of two infected animals and two healthy animals were used for immunostaining, and four fields (mm 2) from different slices of each animal were included.
Figure 5.
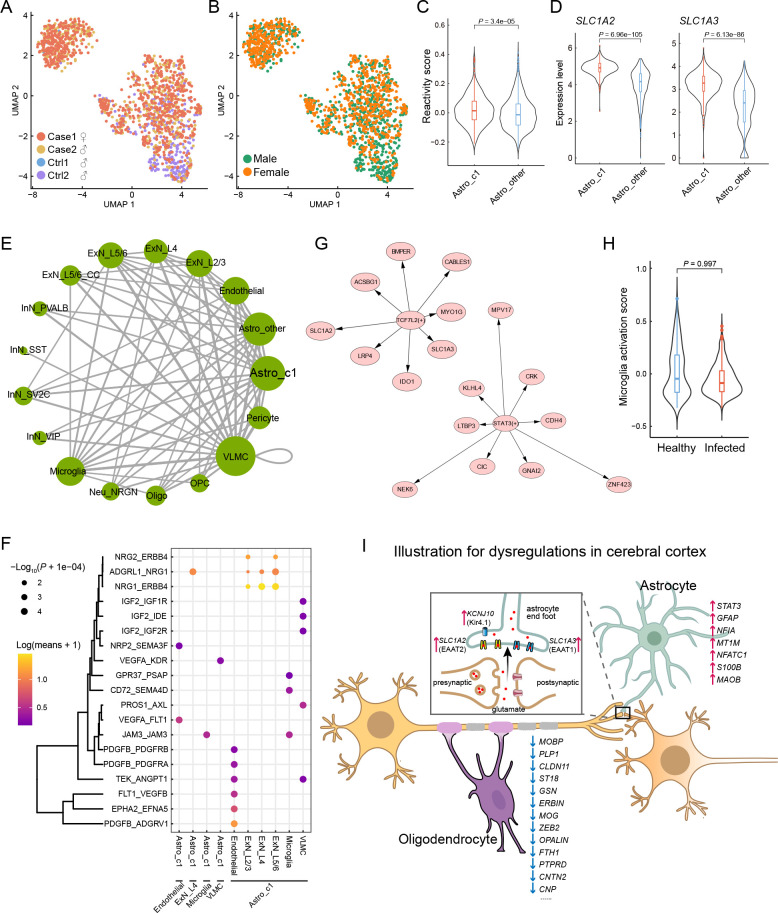
We further investigated astrocytes using unsupervised clustering and identified two major subclusters ( Figure 5G). The two subclusters exhibited relatively even distributions in terms of sample and gender, as shown in UMAP ( Figure 6A, B), indicating minimal technical and/or batch artifacts. Subcluster 1 was an infected sample-specific subpopulation, predominantly composed of cells from infected samples ( Figure 5H). The astrocyte reactivity score, calculated based on the expression levels of the seven reactive marker genes mentioned earlier, was significantly higher in infected sample-specific subcluster 1 than in other astrocytes ( Figure 6C). The expression levels of SLC1A2 and SLC1A3 were also significantly higher in subcluster 1 than in other astrocytes ( Figure 6D). Trajectory analysis revealed that infection-associated subcluster 1 emerged from the parental homeostatic population ( Figure 5I), further suggesting that these astrocytes emerged in response to the pathological environment.
Figure 6.
Evaluation of astrocytes in infected samples and summary of cerebral cortex perturbation
A, B: Subclustering of astrocytes in UMAP, colored by individual (A) and gender (B). C: Comparison of reactivity score between astrocyte subcluster 1 and others, one-sided Wilcoxon rank-sum test. D: Violin plots showing differential expression of SLC1A2 and SLC1A3 between astrocyte subcluster 1 and others, one-sided Wilcoxon rank-sum test. E: Cell-cell interaction network in infected samples computed from CellphoneDB. Each node represents one cell type, each edge between two nodes indicates an interaction between two cell types. Thicker edges mean stronger interactions. Bigger nodes mean stronger total interactions with other cell types. F: Top significant ligand-receptor pairs mediating interactions between astrocyte subcluster 1 and other cell types. G: Regulons highly manifested in astrocytes of infected samples. Length of arrows represents respective relevance for the prediction of target gene expression, shorter arrows mean stronger relevance. H: Comparison of activation scores of microglia between infected and healthy samples, one-sided Wilcoxon rank-sum test. I: Illustration representing SARS-CoV-2 infection-associated perturbations in macaque cerebral cortex, focusing on astrocyte reactivity, glutamate cycling disorder, myelination defects, reduced neuronal excitability, and impaired axonal conduction.
Hypertrophy of cellular processes is a typical phenotype of reactive astrocytes ( Pekny & Pekna, 2014). Here, GFAP immunostaining of prefrontal cortex sections to label astrocyte morphology showed evident signs of characteristic hypertrophy in infected macaques ( Figure 5K, L). The number of GFAP-positive astrocytes in infected animals was significantly higher than that in the healthy macaques ( Figure 5M), further validating reactive astrogliosis. These findings suggest the occurrence of astrocyte reactivity and dysregulation of excitatory synaptic transmission in SARS-CoV-2-infected macaques, with these perturbations in astrocytes inducing an infected sample-specific subpopulation at the molecular level.
We next used CellPhoneDB ( Efremova et al., 2020) to infer the potential cell-cell communication network mediated by ligand-receptor interaction (LRI) pairs in infected cerebral cortex samples ( Figure 6E). Here, we focused on communication between astrocyte subcluster 1 and other cell types to investigate the possible underlying dysregulation. Among the identified significant LRI pairs, we found that the NRG1-ERBB4 interaction pair between astrocyte subcluster 1 and excitatory neurons was quite relevant ( Figure 6F). As NRG1-ERBB4 signaling is implicated in psychiatric disorders, including epilepsy ( Zhu et al., 2017) and schizophrenia ( Banerjee et al., 2010), the increase in NRG1-ERBB4 signaling triggered by the infected sample-enriched astrocyte subcluster may cause psychiatric symptoms in SARS-CoV-2-infected macaques, although this was not tested here. In addition, we identified two highly manifested regulons in astrocytes from the infected samples ( Figure 6G). SLC1A2 and SLC1A3 were predicted to be regulated by TCF7L2 ( Figure 6G) , one of the top DEGs identified in the astrocytes (log 2FC=2.2). TCF7L2 can also activate IDO1 expression ( Figure 6G), which may lead to more activated tryptophan metabolism in the brain, as revealed in the liver, and may explain the lack of activation signatures in microglia ( Figure 6H).
Oligodendrocytes in the CNS are responsible for producing the myelin sheath that insulates axons. Common markers for oligodendrocytes include MOBP, MOG, MBP, and PLP1, which encode structural components of the myelin sheath and play important roles in myelin sheath completion and maintenance. The CLDN11 gene encodes claudin 11, which is specific to oligodendrocytes and facilitates the formation of tight junctions in the myelin paranodal loops ( Morita et al., 1999). The actin disassembly protein gelsolin encoded by GSN promotes actin disassembly and is required for myelin wrapping, with high enrichment in actively myelinating oligodendrocytes ( Zuchero et al., 2015). The Erbb2 interacting protein ( ERBIN) is necessary for remyelination of regenerated axons after injury ( Liang et al., 2012). PTPRD is expressed in oligodendrocytes at postnatal stages and contributes to the initiation of axonal myelination ( Zhu et al., 2015). The cell adhesion molecule encoded by the CNTN2 gene can affect the expression levels of myelin and myelin-regulating genes and is also responsible for oligodendrocyte branching ( Zoupi et al., 2018). The CNP gene encodes a phosphodiesterase that interacts with tubulin and promotes microtubule assembly and is thus necessary for oligodendrocyte process outgrowth and arborization ( Lee et al., 2005). Intriguingly, we found that many genes closely associated with myelin, including those mentioned above, were specifically down-regulated in oligodendrocytes ( Figure 5J), predicting likely myelination defects in the cerebral cortex of infected macaques. As myelin sheaths around neuronal axons enable rapid impulse propagation of electrical signals in the CNS, the hypomyelination or demyelination in the cerebrum of infected animals may impair axonal conduction. Overall, SARS-CoV-2 infection-associated perturbations in the macaque cerebral cortex, including pathological astrocytes and oligodendrocytes, and subsequent neuronal impairment of synaptic excitability transmission and axonal conduction were evident, and may contribute to cognitive impairment and psychiatric symptoms after infection ( Figure 6I).
Endothelial cell dysfunction and circadian rhythm disruption after infection
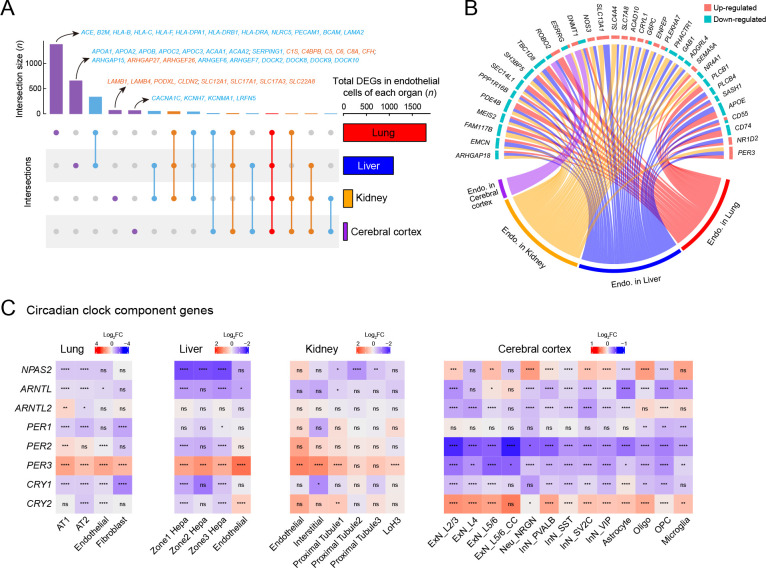
Endothelial cells are highly involved in COVID-19 pathologies ( Varga et al., 2020). Thus, we performed integrative analysis of endothelial cells from the four tested organs and found different cellular perturbations based on organ-specific physiological functions ( Figure 7A). Expression of ACE, responsible for converting angiotensin I into angiotensin II and leading to increased vasoconstriction and inflammation ( Givertz, 2001), was specifically decreased in the lung endothelial cells ( Figure 7A), suggesting a vasoprotective effect, consistent with the observed tendency toward lung injury repair after several days of exposure to SARS-CoV-2. MHC components, including human leukocyte antigen (HLA) class I/II genes, were down-regulated in the lung endothelium ( Figure 7A), further indicating that inhibition of the MHC pathway may be an immune evasion strategy used by SARS-CoV-2.
Figure 7.
Endothelial cell dysfunction and circadian rhythm disruption in multiple organs of SARS-CoV-2 infected rhesus macaques
A: UpSet plot showing a matrix layout of DEGs (infected vs. healthy, |logFC|>0.25, adjusted P<0.05) of endothelial cells shared across organs and specific to one organ. Each matrix column represents either DEGs specific to one organ (single circle with no vertical lines) or DEGs shared between organs (circles with vertical lines). Top, bar graph displaying number of DEGs in each combination. Representative genes are indicated for the four bars, which are specific to one given organ; red, up-regulated; blue, down-regulated. Right, bar graph displaying total number of DEGs in endothelial cells from a given organ. B: Circos plot showing common DEGs in endothelial cells of multiple organs. C: Heatmaps showing disrupted expression of core component genes of circadian clock in four organs. Statistical significance was determined by one-sided Wilcoxon rank-sum test for each block (ns is not significant; *: P<0.05; **: P<0.01; ***: P<0.001; ****: P<0.0001). Cell types that showed no significant changes in these genes were omitted.
Endothelial cells in the liver showed unique dysregulation of Rho GTPase regulators, including inhibitor GAPs and activator GEFs (including DOCKs) ( Figure 7A), which play key roles in regulating vascular barrier function and integrity via the actin cytoskeleton ( Beckers et al., 2010), potentially leading to impaired endothelial barrier function in the liver after infection. PODXL plays a role in orchestrating interactions with extracellular matrix components and basement membranes to regulate vascular integrity and permeability ( Debruin et al., 2014). The levels of PODXL and tight junction protein CLDN2, as well as basement membrane laminin components LAMB1 and LAMB4, were specifically increased in the kidney endothelial cells ( Figure 7A), possibly contributing to glomerular permeability disorder in the kidney, in addition to MN. Endothelial cells in the cerebral cortex exhibited several specific changes in potassium and calcium channels associated with nervous system functions ( Figure 7A).
Endothelial cells in the different organs showed several shared transcriptional changes ( Figure 7B). RIG-I is a key sensor for recognizing nucleic acids derived from RNA viruses, and SEC14L1 encodes a negative regulator of the RIG-I antiviral signal ( Li et al., 2013), and its down-regulation may help attenuate viral replication. Endomucin ( EMCN) prevents leukocyte-endothelial cell adhesion, and thus reduced endomucin is critical to facilitate the adhesion of leukocytes to inflamed tissues ( Zahr et al., 2016). ARHGAP18, an essential inhibitor of inflammation development in the endothelium ( Coleman et al., 2020), and PPP1R16B (also known as TIMAP), a positive regulator of endothelial barrier function ( Csortos et al., 2008), were both down-regulated in the endothelial cells of infected macaques. These changes suggest that endothelial cells exhibited elevated innate antiviral immunity, enhanced leukocyte adherence and extravasation, and impaired barrier function to some extent. DNMT1 gene expression was also dysregulated in the endothelial cells of all four organs ( Figure 7B), consistent with the findings presented in Figure 1D.
Recent studies have revealed an increasing prevalence of anxiety and circadian rhythm disorders during the COVID-19 pandemic ( Boiko et al., 2022; Deng et al., 2021). A follow-up study reported that COVID-19-impacted people still experience fatigue, sleep difficulties, and anxiety or depression six months after acute infection ( Huang et al., 2021). Whether the virus disrupts circadian rhythm and affects sleep behavior needs to be explored. Here, we observed that the expression of core components of the circadian clock was extensively dysregulated in the four organs of the SARS-CoV-2-infected macaques ( Figure 7C). The visceral organs showed a similar pattern of circadian clock disruption: the PER3 gene was up-regulated and other components were down-regulated or changed negligibly. In contrast, PER3 and PER2 were down-regulated in all neural cell types in the cerebral cortex, and the CLOCK paralog NPAS2, which plays a central role in the regulation of circadian rhythm, was disturbed in multiple neural cell types in a discordant manner. Thus, the widespread dysregulation of circadian clock components in our dataset provides molecular evidence linking SARS-CoV-2 infection with disruption of the circadian rhythm.
DISCUSSION
Previous studies have investigated the molecular basis of the pathological hallmarks of COVID-19 using postmortem specimens obtained from critically ill patients ( Delorey et al., 2021; Melms et al., 2021; Wang et al., 2021). However, differences in age, underlying disease, and length of hospital stay and treatment of human subjects have contributed to the heterogeneity of COVID-19 samples and complicated the delineation of pathological mechanisms. Furthermore, responses of non-respiratory organs to SARS-CoV-2 infection are poorly understood at the single-cell level. In the present study, we explored single-cell transcriptomic changes in the lung, liver, kidney, and cerebrum in a rhesus macaque model at 7 dpi. Although we detected no viral reads via snRNA-seq, we observed notable cellular perturbations in all four organs, demonstrating multi-organ involvement in SARS-CoV-2 infection, further supporting the broad threat of the virus to the host body.
In terms of susceptibility, we identified AT2 cells, hepatocytes, and proximal tubule cells as potential target cells of SARS-CoV-2 according to the coexpression percentage of ACE2 and TMPRSS2. A LoH cell subtype (cluster LoH3) was also identified as a probable target cell, as indicated by the up-regulated expression of genes associated with the viral life cycle. In addition, ciliated cells and proximal tubule cells were severely impacted according to necrosis score analysis. Aberrant RNA metabolic processes in AT2 cells, such as up-regulation of HNRNPs, also suggested that these cells may be a target in the lung. The influenza A virus utilizes hnRNP K to alter host splicing and promote infection ( Thompson et al., 2018), and hnRNP A2/B1 interacts with NSP1 (non-structural protein 1) in SARS-CoV-2 to inhibit host interferon β (IFN-β) expression and facilitate β-coronavirus infection ( Zhou et al., 2021). Thus, the up-regulated RNA metabolic processes observed in AT2 cells suggest that AT2 cells are infected, and mRNA processing-associated proteins are leveraged by SARS-CoV-2 to facilitate its transcription and replication.
In the cerebral cortex, we identified a marked increase in astrocyte reactivity and glutamate transporters. Furthermore, substantial down-regulation of myelin structural molecules and regulators pointed to myelination defects. Thus, neuronal perturbations may follow glial dysfunction, together causing neurological involvement. However, the expression level of acknowledged viral entry factors was extremely low in the cerebral cortex (data not shown), and thus the target cell type by which the virus enters the CNS remains unclear.
According to our multi-organ study, SARS-CoV-2 infection results in immune disorder. From a general view, reduced expression of FUT8 and ST6GAL1 was observed in the lung, liver, and kidney, as shown in Figure 1D, indicating that SARS-CoV-2 can restrain the host adaptive immune response to facilitate its own survival. From a multi-organ perspective, we found various adverse immune responses in the different organs.
In the lungs, we found decreased expression of MHC class I/II genes (mainly HLA-C, HLA-DRB1, and CD74) in macrophages, epithelial cells, and endothelial cells after macaque infection, concordant with other studies of peripheral blood mononuclear cells (PBMCs) ( Wilk et al., 2020), pre-enriched dendritic cell subsets from peripheral blood ( Saichi et al., 2021), and nasopharyngeal swabs ( Yoo et al., 2021) from SARS-CoV-2-infected patients. The suppression of MHC components in professional and non-classical APCs in the macaque lungs further confirmed that SARS-CoV-2 targets MHC-related pathways to suppress antigen presentation and facilitate immune evasion. In addition, the sole receptor for IFN-γ ( IFNGR1) was down-regulated in macrophages, with IFN-γ known to be a primary regulator of MHC class I/II pathway activation ( Goñalons et al., 1998; Zhou, 2009). Thus, we reasoned that the virus inhibits IFN-γ signaling, leading to insufficient MHC levels. Indeed, previous studies have demonstrated that SARS-CoV-2 can inhibit IFN signaling pathways (including types I, III, and II) in human patients ( Blanco-Melo et al., 2020; Galani et al., 2021; Hadjadj et al., 2020; Yoo et al., 2021; Zhang et al., 2020), especially in severe cases, and an impaired IFN response is a driving feature of disease progression.
In addition to inhibiting antigen presentation in the lung, we also found that the Kyn pathway was nearly globally activated in the liver, as indicated by the up-regulation of IDO2, TDO2, KMO, and AHR in hepatocytes and immune cells, including macrophages and T cells. In the Kyn pathway, key enzymes play significant roles in facilitating immune evasion in a variety of cancers, including hepatocarcinoma ( Jin et al., 2015; Li et al., 2020). In addition to the potent immunosuppressive effects of IDO2/TDO2/KMO up-regulation-induced local tryptophan depletion and biologically active tryptophan catabolites, AHR activation by binding to the endogenous ligand Kyn suppresses T cell proliferation and function. Tryptophan catabolism has also been implicated in human immunodeficiency virus (HIV) ( Jenabian et al., 2015), hepatitis B virus (HBV) ( Ma et al., 2009), and influenza virus infections ( Pizzini et al., 2019). Therefore, considering that the Kyn pathway was activated in the SARS-CoV-2-infected macaque livers, and given the immunomodulatory role of the Kyn pathway ( Wu et al., 2018), we propose that SARS-CoV-2 infection can lead to immunosuppression or tolerance in the liver by manipulating the Kyn pathway.
Recent metabolomic analysis of blood plasma and serum based on liquid chromatography-mass spectrometry revealed reduced tryptophan levels and increased Kyn levels in the peripheral blood of COVID-19 patients ( Lawler et al., 2021; Lionetto et al., 2021; Thomas et al., 2020). Our findings showing up-regulation of IDO2 and TDO2 in the macaque livers further corroborated the activation of tryptophan metabolism along the Kyn pathway during SARS-CoV-2 infection. The immune evasion strategies of SARS-CoV-2 lead to a higher risk of prolonged infection and long-term symptoms. Therefore, it may be necessary for COVID-19-infected people and survivors to monitor their nutritional status, particularly levels of tryptophan and its metabolites.
Furthermore, immune disturbance caused by SARS-CoV-2 infection was also reflected in the discovery of MN in the kidney. MN is an autoimmune disease in which immune complexes inappropriately attack the filter membranes. PLA2R is the most common target antigen in primary MN ( Ronco et al., 2021), while secondary MN can be secondary to infection, drugs, and malignancy. In our study, NELL1-associated MN was found in the kidney, and the activated complement system in the liver exacerbated MN by MAC ( Cravedi, 2021). MN has rarely been reported in human COVID-19 infection ( Guo et al., 2022) or vaccination ( Aydin et al., 2021), with only a small fraction of these cases being PLA2R-positive. Hence, the observation of MN in our study provides new insights into SARS-CoV-2 infection-associated renal manifestations, although whether the MN in macaques is primary or secondary remains to be elucidated.
The current study has several limitations. First, we only included samples at 7 dpi for analysis. Longitudinal studies using samples collected at different times of infection and compared to infected groups after vaccination should help elucidate cell-type responses and gene expression patterns under such conditions. Second, although not performed in the current study, we will explore functional characterizations of the abovementioned genes to confirm their putative roles and test their potential as valid targets for treatment. We believe that the transcriptomic profiling landscape of single cells in different organs in this report should provide a solid basis for future studies.
Overall, this study provides evidence of multi-organ pathological changes in rhesus macaques following SARS-CoV-2 infection, as well as novel findings not previously reported in human subjects. We identified potential target cell types and genes with dysregulated expression and enriched pathways. Our study supports rhesus macaques as useful models for studying SARS-CoV-2 pathology and highlights potential therapeutic targets for COVID-19. Future studies with larger sample sizes are expected to provide a more comprehensive understanding of the pathogenesis of SARS-CoV-2 and host defense, particularly local adaptive immune responses in multiple organs.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
ACKNOWLEDGEMENTS
We thank Dr. Changwen Ke (Guangdong Provincial Center for Disease Control and Prevention) for providing the SARS-CoV-2 strain.
Funding Statement
This work was supported by the National Basic Research Program of China (2020YFA0804000, 2020YFC0842000, 2020YFA0112200, 2021YFC2301703); Strategic Priority Research Program of the Chinese Academy of Sciences (XDB32010100); Special Associate Research Program of the Chinese Academy of Sciences (E1290601); National Natural Science Foundation of China (32122037, 81891001, 32192411, 32100512, U1902215); Collaborative Research Fund of the Chinese Institute for Brain Research, Beijing (2020-NKX-PT-03); CAS Project for Young Scientists in Basic Research (YSBR-013); Young Elite Scientist Sponsorship Program by the China Association for Science and Technology (2020QNRC001); and National Resource Center for Non-Human Primates
Contributor Information
Yong-Gang Yao, Email: yaoyg@mail.kiz.ac.cn.
Yong-Tang Zheng, Email: zhengyt@mail.kiz.ac.cn.
Xiaoqun Wang, Email: xiaoqunwang@bnu.edu.cn.
References
- Adams S, Braidy N, Bessesde A, Brew BJ, Grant R, Teo C, et al The kynurenine pathway in brain tumor pathogenesis. Cancer Research. 2012;72(22):5649–5657. doi: 10.1158/0008-5472.CAN-12-0549. [DOI] [PubMed] [Google Scholar]
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al SCENIC: single-cell regulatory network inference and clustering. Nature Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin MF, Yıldız A, Oruç A, Sezen M, Dilek K, Güllülü M, et al Relapse of primary membranous nephropathy after inactivated SARS-CoV-2 virus vaccination. Kidney International. 2021;100(2):464–465. doi: 10.1016/j.kint.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Macdonald ML, Borgmann-Winter KE, Hahn CG. 2010. Neuregulin 1-erbB4 pathway in schizophrenia: from genes to an interactome. Brain Research Bulletin, 83(3–4): 132–139.
- Bayati A, Kumar R, Francis V, Mcpherson PS SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. Journal of Biological Chemistry. 2021;296:100306. doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers CML, Van Hinsbergh VWM, Van Nieuw Amerongen GP Driving Rho GTPase activity in endothelial cells regulates barrier integrity. Thrombosis and Haemostasis. 2010;103(1):40–55. doi: 10.1160/TH09-06-0403. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko DI, Skrypnikov AM, Shkodina AD, Hasan MM, Ashraf GM, Rahman MH Circadian rhythm disorder and anxiety as mental health complications in post-COVID-19. Environmental Science and Pollution Research. 2022;29(19):28062–28069. doi: 10.1007/s11356-021-18384-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Boyer JL Bile formation and secretion. Comprehensive Physiology. 2013;3(3):1035–1078. doi: 10.1002/cphy.c120027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AK, Lawson MA, Smith RA, Janda TM, Kelley KW, Mccusker RH Interactions between inflammatory mediators and corticosteroids regulate transcription of genes within the Kynurenine pathway in the mouse hippocampus. Journal of Neuroinflammation. 2016;13(1):98. doi: 10.1186/s12974-016-0563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao JY, Spielmann M, Qiu XJ, Huang XF, Ibrahim DM, Hill AJ, et al The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Li WJ, Wang T, Ran DZ, Davalos V, Planas-Serra L, et al Accelerated biological aging in COVID-19 patients. Nature Communications. 2022;13(1):2135. doi: 10.1038/s41467-022-29801-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza TN, Hassen SI, Dvanajscak Z, Kuperman M, Edmondson R, Herzog C, et al NELL1 is a target antigen in malignancy-associated membranous nephropathy. Kidney International. 2021;99(4):967–976. doi: 10.1016/j.kint.2020.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372(2):221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan QN, Wang ZC, Meirelles GV, et al Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PG, Li J, Huo Y, Lu J, Wan LL, Li B, et al Orphan nuclear receptor NR4A2 inhibits hepatic stellate cell proliferation through MAPK pathway in liver fibrosis. PeerJ. 2015;3:e1518. doi: 10.7717/peerj.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YC, Luo R, Wang K, Zhang M, Wang ZX, Dong L, et al Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney International. 2020;97(5):829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman PR, Lay AJ, Ting KK, Zhao Y, Li J, Jarrah S, et al YAP and the RhoC regulator ARHGAP18, are required to mediate flow-dependent endothelial cell alignment. Cell Communication and Signaling. 2020;18(1):18. doi: 10.1186/s12964-020-0511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravedi P Complement in membranous nephropathy: what we thought we knew and what we really know. Kidney International. 2021;100(3):499–501. doi: 10.1016/j.kint.2021.03.010. [DOI] [PubMed] [Google Scholar]
- Crawley JTB, Zanardelli S, Chion CKNK, Lane DA. 2007. The central role of thrombin in hemostasis. Journal of Thrombosis and Haemostasis, 5 Suppl 1: 95–101.
- Csortos C, Czikora I, Bogatcheva NV, Adyshev DM, Poirier C, Olah G, et al TIMAP is a positive regulator of pulmonary endothelial barrier function. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2008;295(3):L440–L450. doi: 10.1152/ajplung.00325.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z, Jordan TX, Wessels HH, Hoagland DA, Kasela S, Legut M, et al Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184(1):92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S, Kümpers P, Van Slyke P, Parikh SM Mending leaky blood vessels: the angiopoietin-Tie2 pathway in sepsis. Journal of Pharmacology and Experimental Therapeutics. 2013;345(1):2–6. doi: 10.1124/jpet.112.201061. [DOI] [PubMed] [Google Scholar]
- Debruin EJ, Hughes MR, Sina C, Lu A, Cait J, Jian ZQ, et al Podocalyxin regulates murine lung vascular permeability by altering endothelial cell adhesion. PLoS One. 2014;9(10):e108881. doi: 10.1371/journal.pone.0108881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorey TM, Ziegler CGK, Heimberg G, Normand R, Yang YM, Segerstolpe Å, et al COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595(7865):107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JW, Zhou FW, Hou WT, Silver Z, Wong CY, Chang O, et al The prevalence of depression, anxiety, and sleep disturbances in COVID-19 patients: a meta-analysis. Annals of the New York Academy of Sciences. 2021;1486(1):90–111. doi: 10.1111/nyas.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Bao LL, Gao H, Xiang ZG, Qu YJ, Song ZQ, et al Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in rhesus macaques. Nature Communications. 2020;11(1):4400. doi: 10.1038/s41467-020-18149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, et al Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicological Sciences. 2010;115(1):89–97. doi: 10.1093/toxsci/kfq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostal CR, Gamsby NS, Lawson MA, Mccusker RH Glia- and tissue-specific changes in the Kynurenine pathway after treatment of mice with lipopolysaccharide and dexamethasone . Brain, Behavior, and Immunity. 2018;69:321–335. doi: 10.1016/j.bbi.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nature Protocols. 2020;15(4):1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- Escartin C, Galea E, Lakatos A, O'Callaghan JP, Petzold GC, Serrano-Pozo A, et al Reactive astrocyte nomenclature, definitions, and future directions. Nature Neuroscience. 2021;24(3):312–325. doi: 10.1038/s41593-020-00783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallarino F, Grohmann U, Vacca C, Bianchi R, Orabona C, Spreca A, et al T cell apoptosis by tryptophan catabolism. Cell Death & Differentiation. 2002;9(10):1069–1077. doi: 10.1038/sj.cdd.4401073. [DOI] [PubMed] [Google Scholar]
- Fan CF, Wu Y, Rui X, Yang YS, Ling C, Liu SS, et al Animal models for COVID-19: advances, gaps and perspectives. Signal Transduction and Targeted Therapy. 2022;7(1):220. doi: 10.1038/s41392-022-01087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhu R, Bai T, Han P, He Q, Jing MJ, et al Clinical features of patients infected with coronavirus disease 2019 with elevated liver biochemistries: a multicenter, retrospective study. Hepatology. 2021;73(4):1509–1520. doi: 10.1002/hep.31446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani IE, Rovina N, Lampropoulou V, Triantafyllia V, Manioudaki M, Pavlos E, et al Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nature Immunology. 2021;22(1):32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- Gao Q, Bao LL, Mao HY, Wang L, Xu KW, Yang MN, et al Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givertz MM Manipulation of the renin-angiotensin system. Circulation. 2001;104(5):e14–e18. doi: 10.1161/hc3001.094733. [DOI] [PubMed] [Google Scholar]
- Goñalons E, Barrachina M, García-Sanz JA, Celada A Translational control of MHC class II I-A molecules by IFN-γ. Journal of Immunology. 1998;161(4):1837–1843. [PubMed] [Google Scholar]
- Guo WW, Tan PH, Baikunje S Membranous nephropathy in a patient with COVID-19 infection. Journal of Nephrology. 2022;35(1):351–355. doi: 10.1007/s40620-021-01165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JP, Cai SJ, Feng HJ, Cai BM, Lin LH, Mai Y, et al Single-cell analysis reveals bronchoalveolar epithelial dysfunction in COVID-19 patients. Protein & Cell. 2020;11(9):680–687. doi: 10.1007/s13238-020-00752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CL, Huang LX, Wang YM, Li X, Ren LL, Gu XY, et al 6-Month consequences of COVID-19 in patients discharged from hospital: a cohort study. The Lancet. 2021;397(10270):220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HH, Bhat A, Woodnutt G, Lappe R Targeting the ANGPT-TIE2 pathway in malignancy. Nature Reviews Cancer. 2010;10(8):575–585. doi: 10.1038/nrc2894. [DOI] [PubMed] [Google Scholar]
- Irons EE, Punch PR, Lau JTY Blood-borne ST6GAL1 regulates immunoglobulin production in B cells. Frontiers in Immunology. 2020;11:617. doi: 10.3389/fimmu.2020.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenabian MA, El-Far M, Vyboh K, Kema I, Costiniuk CT, Thomas R, et al Immunosuppressive tryptophan catabolism and gut mucosal dysfunction following early HIV infection. Journal of Infectious Diseases. 2015;212(3):355–366. doi: 10.1093/infdis/jiv037. [DOI] [PubMed] [Google Scholar]
- Jin HJ, Zhang YR, You HY, Tao XM, Wang C, Jin GZ, et al Prognostic significance of kynurenine 3-monooxygenase and effects on proliferation, migration and invasion of human hepatocellular carcinoma. Scientific Reports. 2015;5:10466. doi: 10.1038/srep10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SQ, Guerrero-Juarez CF, Zhang LH, Chang I, Ramos R, Kuan CH, et al Inference and analysis of cell-cell communication using CellChat. Nature Communications. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiri Z, Oudit GY, Kandalam V, Awad A, Wang XH, Ziou X, et al Loss of TIMP3 enhances interstitial nephritis and fibrosis. Journal of the American Society of Nephrology. 2009;20(6):1223–1235. doi: 10.1681/ASN.2008050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, et al Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host & Microbe. 2020;27(5):704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan QN, Wang ZC, et al Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laug D, Huang TW, Huerta NAB, Huang AYS, Sardar D, Ortiz-Guzman J, et al Nuclear factor I-A regulates diverse reactive astrocyte responses after CNS injury. The Journal of Clinical Investigation. 2019;129(10):4408–4418. doi: 10.1172/JCI127492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler NG, Gray N, Kimhofer T, Boughton B, Gay M, Yang RC, et al Systemic perturbations in amine and kynurenine metabolism associated with acute SARS-CoV-2 infection and inflammatory cytokine responses. Journal of Proteome Research. 2021;20(5):2796–2811. doi: 10.1021/acs.jproteome.1c00052. [DOI] [PubMed] [Google Scholar]
- Lee GK, Park HJ, Macleod M, Chandler P, Munn DH, Mellor AL Tryptophan deprivation sensitizes activated T cells to apoptosis prior to cell division. Immunology. 2002;107(4):452–460. doi: 10.1046/j.1365-2567.2002.01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Gravel M, Zhang RL, Thibault P, Braun PE Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. Journal of Cell Biology. 2005;170(4):661–673. doi: 10.1083/jcb.200411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Koh JY, Yi K, Kim YI, Park SJ, Kim EH, et al Single-cell transcriptome of bronchoalveolar lavage fluid reveals sequential change of macrophages during SARS-CoV-2 infection in ferrets. Nature Communications. 2021;12(1):4567. doi: 10.1038/s41467-021-24807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman EK, Wilton DK, Litvina EY, Welsh CA, Chang ST, Frouin A, et al CD47 protects synapses from excess microglia-mediated pruning during development. Neuron. 2018;100(1):120–134.e6. doi: 10.1016/j.neuron.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MT, Di W, Xu H, Yang YK, Chen HW, Zhang FX, et al Negative regulation of RIG-I-mediated innate antiviral signaling by SEC14L1. Journal of Virology. 2013;87(18):10037–10046. doi: 10.1128/JVI.01073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SB, Li L, Wu JY, Song FB, Qin ZW, Hou L, et al TDO promotes hepatocellular carcinoma progression. OncoTargets and Therapy. 2020;13:5845–5855. doi: 10.2147/OTT.S252929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Tao YM, Shen CY, Tan Z, Xiong WC, Mei L Erbin is required for myelination in regenerated axons after injury. Journal of Neuroscience. 2012;32(43):15169–15180. doi: 10.1523/JNEUROSCI.2466-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Mao SS, Sun SJ, Li M, Li Z, Yu R, et al Core fucosylation of the T cell receptor is required for T cell activation. Frontiers in Immunology. 2018;9:78. doi: 10.3389/fimmu.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetto L, Ulivieri M, Capi M, De Bernardini D, Fazio F, Petrucca A, et al Increased kynurenine-to-tryptophan ratio in the serum of patients infected with SARS-CoV2: an observational cohort study. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2021;1867(3):166042. doi: 10.1016/j.bbadis.2020.166042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Shi Y, Cai J, Duan YQ, Wang RS, Zhang HY, et al Pathological changes in the lungs and lymphatic organs of 12 COVID-19 autopsy cases. National Science Review. 2020a;7(12):1868–1878. doi: 10.1093/nsr/nwaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhai J, Kong XY, Wang XY, Wang ZZ, Fang Y, et al Comprehensive analysis of the expression and prognosis for TDO2 in breast cancer. Molecular Therapy Oncolytics. 2020b;17:153–168. doi: 10.1016/j.omto.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Wang YR, Wang QH, Chen Y, Chen X, Li Y, et al Post-infection cognitive impairments in a cohort of elderly patients with COVID-19. Molecular Neurodegeneration. 2021;16(1):48. doi: 10.1186/s13024-021-00469-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Liang XY, Dong WQ, Fang Y, Lv JD, Zhang TZ, et al Tumor-repopulating cells induce PD-1 expression in CD8 + T cells by transferring kynurenine and AhR activation . Cancer Cell. 2018;33(3):480–494.e7. doi: 10.1016/j.ccell.2018.02.005. [DOI] [PubMed] [Google Scholar]
- Ma L, Xu B, Wang WJ, Deng WP, Ding M. 2009. Analysis of tryptophan catabolism in HBV patients by HPLC with programmed wavelength ultraviolet detection. Clinica Chimica Acta, 405(1–2): 94–96.
- Ma S, Sun SH, Li JM, Fan YL, Qu J, Sun L, et al Single-cell transcriptomic atlas of primate cardiopulmonary aging. Cell Research. 2021;31(4):415–432. doi: 10.1038/s41422-020-00412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Jin HJ, Wang MD, Hu Y, Chen SC, He QW, et al Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurology. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrikaki M, Lee JD, Solomon IH, Slack FJ. 2021. Severe COVID-19 induces molecular signatures of aging in the human brain. medRxiv,doi: 10.1101/2021.11.24.21266779.
- Melms JC, Biermann J, Huang HC, Wang YP, Nair A, Tagore S, et al A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;595(7865):114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Blish CA, Sallusto F, Iwasaki A The immunology and immunopathology of COVID-19. Science. 2022;375(6585):1122–1127. doi: 10.1126/science.abm8108. [DOI] [PubMed] [Google Scholar]
- Mezrich JD, Fechner JH, Zhang XJ, Johnson BP, Burlingham WJ, Bradfield CA An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. The Journal of Immunology. 2010;185(6):3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. The Journal of Cell Biology. 1999;145(3):579–588. doi: 10.1083/jcb.145.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, et al Animal models for COVID-19. Nature. 2020;586(7830):509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Feldmann F, Williamson BN, Van Doremalen N, Pérez-Pérez L, Schulz J, et al Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585(7824):268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muus C, Luecken MD, Eraslan G, Sikkema L, Waghray A, Heimberg G, et al Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nature Medicine. 2021;27(3):546–559. doi: 10.1038/s41591-020-01227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Namasivayam S, Foreman TW, Kauffman KD, Sakai S, Dorosky DE, et al Mild SARS-CoV-2 infection in rhesus macaques is associated with viral control prior to antigen-specific T cell responses in tissues. Science Immunology. 2022;7:eabo0535. doi: 10.1126/sciimmunol.abo0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevler A, Muller AJ, Sutanto-Ward E, Duhadaway JB, Nagatomo K, Londin E, et al Host IDO2 gene status influences tumor progression and radiotherapy response in KRAS-driven sporadic pancreatic cancers . Clinical Cancer Research. 2019;25(2):724–734. doi: 10.1158/1078-0432.CCR-18-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie QH, Duan GR, Luo XD, Xie YM, Luo H, Zhou YX, et al Expression of TIMP-1 and TIMP-2 in rats with hepatic fibrosis. World Journal of Gastroenterology. 2004;10(1):86–90. doi: 10.3748/wjg.v10.i1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa H, Motegi SI, Ohyama N, Ohnishi H, Tomizawa T, Kaneko Y, et al Negative regulation of phagocytosis in macrophages by the CD47-SHPS-1 system. The Journal of Immunology. 2005;174(4):2004–2011. doi: 10.4049/jimmunol.174.4.2004. [DOI] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, et al An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478(7368):197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- Palumbo-Zerr K, Zerr P, Distler A, Fliehr J, Mancuso R, Huang JG, et al Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nature Medicine. 2015;21(2):150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- Pan XW, Xu D, Zhang H, Zhou W, Wang LH, Cui XG Identification of a potential mechanism of acute kidney injury during the COVID-19 outbreak: a study based on single-cell transcriptome analysis. Intensive Care Medicine. 2020;46(6):1114–1116. doi: 10.1007/s00134-020-06026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei GC, Zhang ZG, Peng J, Liu L, Zhang CX, Yu C, et al Renal involvement and early prognosis in patients with COVID-19 pneumonia. Journal of the American Society of Nephrology. 2020;31(6):1157–1165. doi: 10.1681/ASN.2020030276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Pekna M Astrocyte reactivity and reactive astrogliosis: costs and benefits. Physiological Reviews. 2014;94(4):1077–1098. doi: 10.1152/physrev.00041.2013. [DOI] [PubMed] [Google Scholar]
- Pizzini A, Kurz K, Santifaller J, Tschurtschenthaler C, Theurl I, Fuchs D, et al Assessment of neopterin and indoleamine 2, 3-dioxygenase activity in patients with seasonal influenza: a pilot study. Influenza and Other Respiratory Viruses. 2019;13(6):603–609. doi: 10.1111/irv.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad Pour S, Morikawa H, Kiani NA, Yang MY, Azimi A, Shafi G, et al Exhaustion of CD4+ T-cells mediated by the Kynurenine pathway in melanoma. Scientific Reports. 2019;9(1):12150. doi: 10.1038/s41598-019-48635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XW, Wen W, Fan XY, Hou WH, Su B, Cai PF, et al COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184(7):1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronco P, Beck L, Debiec H, Fervenza FC, Hou FF, Jha V, et al Membranous nephropathy. Nature Reviews Disease Primers. 2021;7(1):69. doi: 10.1038/s41572-021-00303-z. [DOI] [PubMed] [Google Scholar]
- Saichi M, Ladjemi MZ, Korniotis S, Rousseau C, Ait Hamou Z, Massenet-Regad L, et al Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nature Cell Biology. 2021;23(5):538–551. doi: 10.1038/s41556-021-00681-2. [DOI] [PubMed] [Google Scholar]
- Santhanam S, Alvarado DM, Ciorba MA Therapeutic targeting of inflammation and tryptophan metabolism in colon and gastrointestinal cancer. Translational Research. 2016;167(1):67–79. doi: 10.1016/j.trsl.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster V, Hügle B, Tefs K Plasminogen deficiency. Journal of Thrombosis and Haemostasis. 2007;5(12):2315–2322. doi: 10.1111/j.1538-7836.2007.02776.x. [DOI] [PubMed] [Google Scholar]
- Scudellari M How the coronavirus infects cells - and why delta is so dangerous. Nature. 2021;595(7869):640–644. doi: 10.1038/d41586-021-02039-y. [DOI] [PubMed] [Google Scholar]
- Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, et al Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. American Journal of Respiratory and Critical Care Medicine. 2006;173(2):188–198. doi: 10.1164/rccm.200504-644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S, Debiec H, Madden B, Charlesworth MC, Morelle J, Gross L, et al Neural epidermal growth factor-like 1 protein (NELL-1) associated membranous nephropathy. Kidney International. 2020;97(1):163–174. doi: 10.1016/j.kint.2019.09.014. [DOI] [PubMed] [Google Scholar]
- Shan C, Yao YF, Yang XL, Zhou YW, Gao G, Peng Y, et al. 2020. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Research, 30(8): 670–677.
- Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Research. 2013;100(3):605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Bansal V, Feschotte C A single-cell RNA expression map of human coronavirus entry factors. Cell Reports. 2020;32(12):108175. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JA, Zurawski SM, Sugimoto C, Vinet-Oliphant H, Vinod P, Xue YM, et al Immunologic characterization of a rhesus macaque H1N1 challenge model for candidate influenza virus vaccine assessment. Clinical and Vaccine Immunology. 2014;21(12):1668–1680. doi: 10.1128/CVI.00547-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song TZ, Zheng HY, Han JB, Feng XL, Liu FL, Yang X, et al Northern pig-tailed macaques ( Macaca leonina) infected with SARS-CoV-2 show rapid viral clearance and persistent immune response . Zoological Research. 2021;42(3):350–353. doi: 10.24272/j.issn.2095-8137.2020.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song TZ, Zheng HY, Han JB, Jin L, Yang X, Liu FL, et al Delayed severe cytokine storm and immune cell infiltration in SARS-CoV-2-infected aged Chinese rhesus macaques. Zoological Research. 2020;41(5):503–516. doi: 10.24272/j.issn.2095-8137.2020.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Yang M, Wan C, Yi LX, Tang F, Zhu HY, et al Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney International. 2020;98(1):219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, et al SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature Medicine. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Stefanoni D, Reisz JA, Nemkov T, Bertolone L, Francis RO, et al COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5(14):e140327. doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MG, Muñoz-Moreno R, Bhat P, Roytenberg R, Lindberg J, Gazzara MR, et al Co-regulatory activity of hnRNP K and NS1-BP in influenza and human mRNA splicing. Nature Communications. 2018;9(1):2407. doi: 10.1038/s41467-018-04779-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, et al Endothelial cell infection and endotheliitis in COVID-19. The Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DW, Hu B, Hu C, Zhu FF, Liu X, Zhang J, et al Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020a;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang YT, Huang BY, Deng W, Quan YR, Wang WL, et al Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020b;182(3):713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yao XH, Ma S, Ping YF, Fan YL, Sun SH, et al A single-cell transcriptomic landscape of the lungs of patients with COVID-19. Nature Cell Biology. 2021;23(12):1314–1328. doi: 10.1038/s41556-021-00796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werion A, Belkhir L, Perrot M, Schmit G, Aydin S, Chen ZY, et al SARS-CoV-2 causes a specific dysfunction of the kidney proximal tubule. Kidney International. 2020;98(5):1296–1307. doi: 10.1016/j.kint.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, Mckechnie JL, et al A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nature Medicine. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biology. 2018;19(1):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock SL, Lopez R, Klein AM Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Systems. 2019;8(4):281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Gong JP, Liu Y Indoleamine 2, 3-dioxygenase regulation of immune response (review) Molecular Medicine Reports. 2018;17(4):4867–4873. doi: 10.3892/mmr.2018.8537. [DOI] [PubMed] [Google Scholar]
- Xie YL, Su N, Yang J, Tan QY, Huang S, Jin M, et al FGF/FGFR signaling in health and disease. Signal Transduction and Targeted Therapy. 2020;5(1):181. doi: 10.1038/s41392-020-00222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yu DD, Ma YH, Yao YL, Luo RH, Feng XL, et al COVID-19-like symptoms observed in Chinese tree shrews infected with SARS-CoV-2. Zoological Research. 2020;41(5):517–526. doi: 10.24272/j.issn.2095-8137.2020.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AC, Kern F, Losada PM, Agam MR, Maat CA, Schmartz GP, et al Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature. 2021;595(7868):565–571. doi: 10.1038/s41586-021-03710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Mason RJ, Pan TL, Deterding RR, Nielsen LD, Shannon JM KGF regulates pulmonary epithelial proliferation and surfactant protein gene expression in adult rat lung. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2000;279(6):L1146–L1158. doi: 10.1152/ajplung.2000.279.6.L1146. [DOI] [PubMed] [Google Scholar]
- Yao YF, Bao LL, Deng W, Xu LL, Li FD, Lv Q, et al An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. The Journal of Infectious Diseases. 2014;209(2):236–242. doi: 10.1093/infdis/jit590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JS, Sasaki M, Cho SX, Kasuga Y, Zhu BH, Ouda R, et al SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nature Communications. 2021;12(1):6602. doi: 10.1038/s41467-021-26910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr A, Alcaide P, Yang JL, Jones A, Gregory M, Dela Paz NG, et al Endomucin prevents leukocyte-endothelial cell adhesion and has a critical role under resting and inflammatory conditions. Nature Communications. 2016;7:10363. doi: 10.1038/ncomms10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bastard P, Liu ZY, Le Pen J, Moncada-Velez M, Chen J, et al Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F. 2009. Molecular mechanisms of IFN-γ to up-regulate MHC class I antigen processing and presentation. International Reviews of Immunology, 28(3–4): 239–260.
- Zhou FH, Wan QY, Chen S, Chen Y, Wang PH, Yao X, et al Attenuating innate immunity and facilitating β-coronavirus infection by NSP1 of SARS-CoV-2 through specific redistributing hnRNP A2/B1 cellular localization. Signal Transduction and Targeted Therapy. 2021;6(1):371. doi: 10.1038/s41392-021-00786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wang GH, Qian J Transcription factors as readers and effectors of DNA methylation. Nature Reviews Genetics. 2016;17(9):551–565. doi: 10.1038/nrg.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JM, Li KX, Cao SX, Chen XJ, Shen CJ, Zhang Y, et al Increased NRG1-ErbB4 signaling in human symptomatic epilepsy. Scientific Reports. 2017;7(1):141. doi: 10.1038/s41598-017-00207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Tan Z, Zhao SF, Huang H, Zhao XF, Hu XM, et al Developmental expression and function analysis of protein tyrosine phosphatase receptor type D in oligodendrocyte myelination. Neuroscience. 2015;308:106–114. doi: 10.1016/j.neuroscience.2015.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoupi L, Savvaki M, Kalemaki K, Kalafatakis I, Sidiropoulou K, Karagogeos D The function of contactin-2/TAG-1 in oligodendrocytes in health and demyelinating pathology. Glia. 2018;66(3):576–591. doi: 10.1002/glia.23266. [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Fu MM, Sloan SA, Ibrahim A, Olson A, Zaremba A, et al CNS myelin wrapping is driven by actin disassembly. Developmental Cell. 2015;34(2):152–167. doi: 10.1016/j.devcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.