Abstract
The role of the Klebsiella pneumoniae capsular polysaccharide (K antigen) during colonization of the mouse large intestine was assessed with wild-type K. pneumoniae LM21 and its isogenic capsule-defective mutant. When bacterial strains were fed alone to mice, the capsulated bacteria persisted in the intestinal tract at levels of 108 CFU/g of feces while the capsule-defective strain colonized at low levels, 104 CFU/g of feces. In mixed-infection experiments, the mutant was rapidly outcompeted by the wild type. In situ hybridization on colonic sections revealed that bacterial cells of both strains were evenly distributed in the mucus layer at day 1 after infection, while at day 20 the wild type remained dispersed and the capsule-defective strain was seen in clusters in the mucus layer. These results suggest that capsular polysaccharide plays an important role in the gut colonization ability of K. pneumoniae.
Colonization of the intestinal tract is the first task that an enteric pathogen has to perform before infecting the host system (9). Pathogens face a complex ecosystem composed of 1012 viable bacteria per gram of gut contents and consisting of up to 400 to 500 different bacterial species. Epidemiological studies have revealed that the initial step in infection by Klebsiella pneumoniae strains involved in nosocomial infections is colonization of the gastrointestinal tract (4, 19). It is generally assumed that the ability of microorganisms to initiate colonization depends on the ability to adhere to mucosal surfaces in the gastrointestinal tract. Bacterial surface structures are likely to be involved in these interactions, and a number of proteinaceous adhesins from K. pneumoniae strains have been characterized (3, 5, 7, 12). In spite of the fact that the majority of K. pneumoniae clinical isolates produce a very characteristic, heavy polysaccharide capsule (K antigen) covering the entire bacterial surface, very few studies have been performed to assess the role of this component during colonization of mucosal surfaces. The importance of Escherichia coli K5 capsule expression was demonstrated in gnotobiotic rats (11), whereas the K. pneumoniae capsule was found to play no role in colonization of the gut of germfree chickens (2).
In the present study, the role of the K. pneumoniae capsule in colonization of the mouse intestinal tract was investigated by comparing a wild-type strain and its isogenic mutant defective in synthesis of the capsule. The present work concerns (i) the importance of the capsule in colonization of the intestinal tract of mice and (ii) the spatial distribution of K. pneumoniae strains in the gut as determined by in situ hybridization.
Clinical isolate K. pneumoniae LM21, of serotype K35 (typed at the World Health Organization International Escherichia and Klebsiella Centre, Copenhagen, Denmark), and its derivative mutant LM21(cps), defective in synthesis of the capsule, have been described previously (8). A spontaneous streptomycin-resistant (Strr) mutant and a streptomycin–nalidixic acid-resistant (Strr Nalr) double mutant of wild-type K. pneumoniae LM21 and a spontaneous streptomycin-resistant mutant of K. pneumoniae LM21(cps) were used throughout this study. The antibiotic-resistant isolates were found to be identical to their parents in respect to growth physiology, biochemical reactions, serotype, and colonization ability of the mouse gut. Moreover, in vitro growth rate studies were performed in minimal medium M9 (18) supplemented either with 0.4% acetate or with 0.4% glucose and 1% Casamino Acids; in neither of the media did the generation times differ significantly.
Gut colonization ability was assayed by the streptomycin-treated mouse colonization model, which was previously used to study the colonization abilities and growth physiologies of E. coli and Salmonella typhimurium in the intestinal environment (13, 15–17). Capsule formation on the wild-type strain after colonization was confirmed visually and by specific K antiserum.
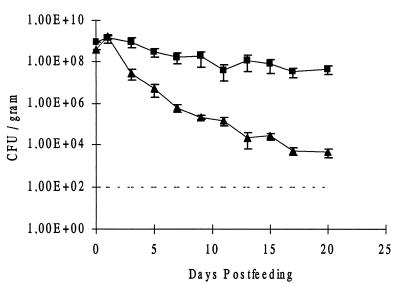
When mice were fed wild-type K. pneumoniae LM21 Strr Nalr at 1 × 109 CFU, the strain persisted in the gut at levels of 5 × 108 CFU per g of feces (Fig. 1). Colonization experiments with the same number of K. pneumoniae LM21 Strr organisms fed to another group of mice gave the same results, confirming that resistance to nalidixic acid had no effect on the ability of the wild-type strain to persist in the intestinal gut of mice (data not shown).
FIG. 1.
Colonization by wild-type K. pneumoniae LM21 (■) and its capsule-defective mutant, K. pneumoniae LM21(cps) (▴), of the intestines of streptomycin-treated mice. The strains were fed separately to each of three streptomycin-treated mice. Symbols for day 0 represent the sizes of the inocula. At the indicated times, fecal samples were plated as described in the text. The detection limit is marked by a broken line. Bars represent standard errors of the means.
When the K. pneumoniae LM21(cps) Strr isogenic mutant was fed to another group of mice at 6 × 108 CFU, the capsule-defective mutant strain was able to persist only at low levels, 1 × 104 CFU per gram of feces, at day 20 (Fig. 1). From these experiments, it was clear that the encapsulated strain had an advantage in persisting in high numbers in the intestinal tract.
On day 20 after feeding with bacterial strains, mice were sacrificed and a 1-cm-long piece of the ileum and jejunum was removed; the contents were weighed, diluted in cold 0.9% (wt/vol) sodium chloride, and plated. The ceca were nicked, and cecal contents were removed, weighed, diluted, and plated. The ceca were gently washed in saline solution, and the mucus layer was scraped off with a spatula, diluted, and plated. Platings were performed on antibiotic-containing L-broth agar plates.
For the wild-type strain K. pneumoniae LM21 Strr Nalr, 5 × 107 CFU per gram of cecal contents, similar to the concentration in feces, and 5.3 × 106 CFU per gram of cecal mucus were recovered. In the jejunum and ileum, 1 × 104 and 1.35 × 106 CFU per gram of contents, respectively, were detected. Similar results were observed in experiments performed with the K. pneumoniae LM21 Strr strain (data not shown). The K. pneumoniae LM21(cps) Strr capsule-defective mutant was detected at levels of 1 × 104 CFU per gram of cecal contents and 1.2 × 103 CFU per gram of cecal mucus. The numbers of bacteria present in the ileum and jejunum were less than 102 per gram of contents.
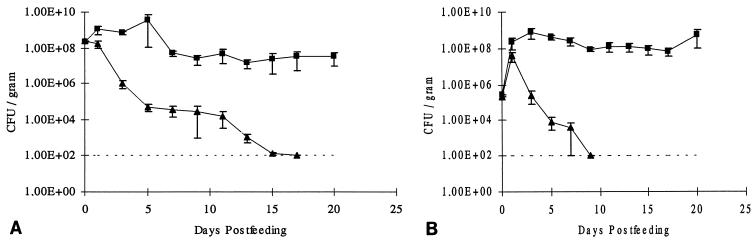
The role of the capsule in intestinal colonization was confirmed by competition experiments; i.e., the wild-type strain and the defective mutant were given simultaneously to the mice. First, co-colonization experiments were performed by feeding the mice simultaneously K. pneumoniae LM21 Strr Nalr and its isogenic mutant K. pneumoniae LM21(cps) Strr, at 3 × 108 CFU of each strain. The numbers of bacteria in feces were determined for 20 days thereafter (Fig. 2A). The CFU per gram of feces decreased rapidly for the capsule-defective mutant at day 15, being below the detection limit. The wild-type strain was maintained in the gut during the entire period at a level of 5 × 107 CFU per gram of feces.
FIG. 2.
Co-colonization by wild-type K. pneumoniae LM21 (■) and its capsule-defective mutant, K. pneumoniae LM21(cps) (▴), of the intestines of streptomycin-treated mice. (A) The strains were fed simultaneously at 108 CFU to streptomycin-treated mice. (B) The strains were fed simultaneously at 105 CFU to streptomycin-treated mice. Symbols for day 0 represent the sizes of the inocula. At the indicated times, fecal samples were plated as described in the text. Bars represent standard errors of the means.
Another set of mice was simultaneously fed the two strains at 2 × 105 CFU each (Fig. 2B). At day 1, the two strains grew equally well, reaching levels of approximately 108 CFU per gram of feces. Then, the numbers of the capsule-defective mutant decreased rapidly and were not detectable at day 9. The wild-type strain persisted in the gut at a level of 108 CFU per gram during the 3-week experimental period.
Independently of the inoculum size, the capsule-defective mutant was outcompeted by the wild-type strain K. pneumoniae LM21. Competition for nutrients and a difference in growth rate could explain the elimination of the capsule-defective mutant LM21(cps) by the wild-type strain K. pneumoniae LM21. However, when mice were fed the strains at low numbers, both strains reached levels of 108 CFU per gram of feces within 1 day, suggesting that they grow equally well in the gut (Fig. 2B). Thereafter, the two strains might compete in order to occupy the same spatial niches in the intestine, resulting in outcompetition of the mutant. Another hypothesis could be that the capsule-defective mutant is more susceptible to host defense factors such as phagocytic cells or more susceptible to bacteriocins produced by the resident flora. Furthermore, the presence of the capsule could enhance penetration through mucus and/or adhesion to the mucus layer, giving an advantage to the wild type during establishment of the strains in the gut. Colonization of the mouse intestine by a smooth strain of S. typhimurium and its lipopolysaccharide-defective mutant (13, 15) showed that the wild type had an advantage versus its mutant, emphasizing the importance of surface structures in vivo.
The spatial distribution of the wild-type strain K. pneumoniae LM21 and its mutant during colonization of the gut was assayed by in situ hybridization at day 1 and at day 20 after feeding. Probe EC1531, designed to hybridize to the 23S rRNA from E. coli, was previously described (16). By evaluating the probe in an updated database with the ARB software package (Lehrstuhl für Mikrobiologie, Technical University, Munich, Germany), developed for rapid handling and exploitation of rRNA sequence data, it was found that the probe hybridizes to a number of Enterobacteriaceae, among them K. pneumoniae and S. typhimurium strains (6). This probe was labeled at the 5′ end with (red) CY3 fluorescent dye (cyanine dye CY3.29-OSu; Biological Detection Systems, Pittsburgh, Pa.). Probe EUB338 (1), specific for the eubacterial domain, was also used and was labeled with (green) fluorescein (Peninsula Laboratories, Inc., Belmont, Calif.) at the 5′ end.
Hybridizations were initially performed on pure cultures. Wild-type K. pneumoniae LM21, the LM21 Strr Nalr mutant, the capsule-defective mutant LM21(cps), and LM21(cps) Strr were hybridized to probe EC1531, and no differences were observed. Bacterial cells from all strains had similar intensities and were evenly dispersed with no aggregation of the cells. Hybridizations on bacterial smears from parts of the mouse gut, fecal samples, and colonic histological sections were performed as previously described (13, 16, 17). In all sections, the tissue appeared to be autofluorescent at red, green, and blue wavelengths, whereas fluorescence from the probed bacteria could be seen only through the filter corresponding to fluorochrome, as previously observed (16). Hybridization of both colonic sections and fecal samples from streptomycin-treated uninfected mice did not reveal any hybridization to the remaining gram-positive flora by the fluorescently labeled probe (EC1531) that detects Enterobacteriaceae.
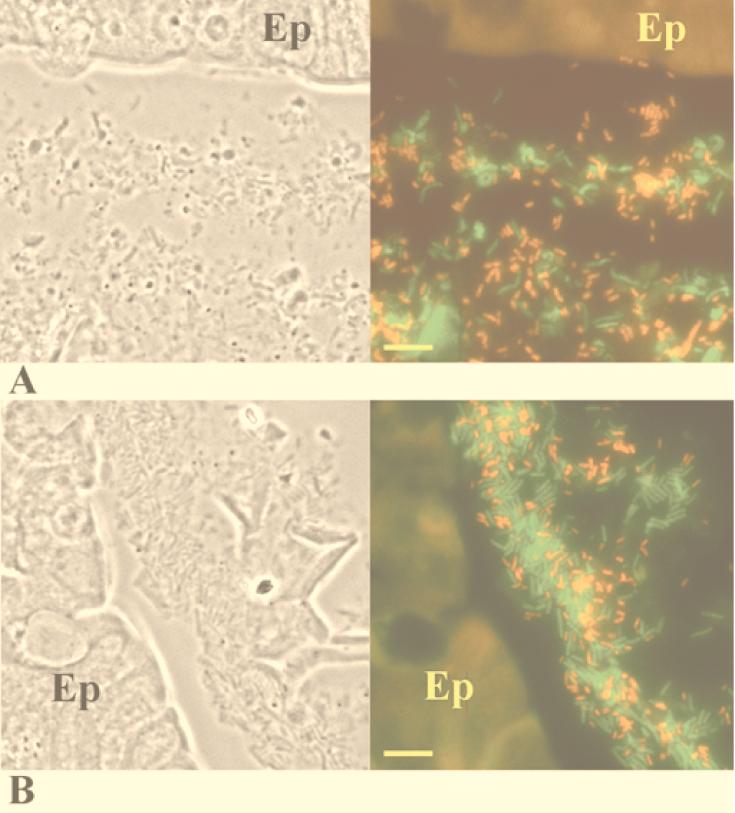
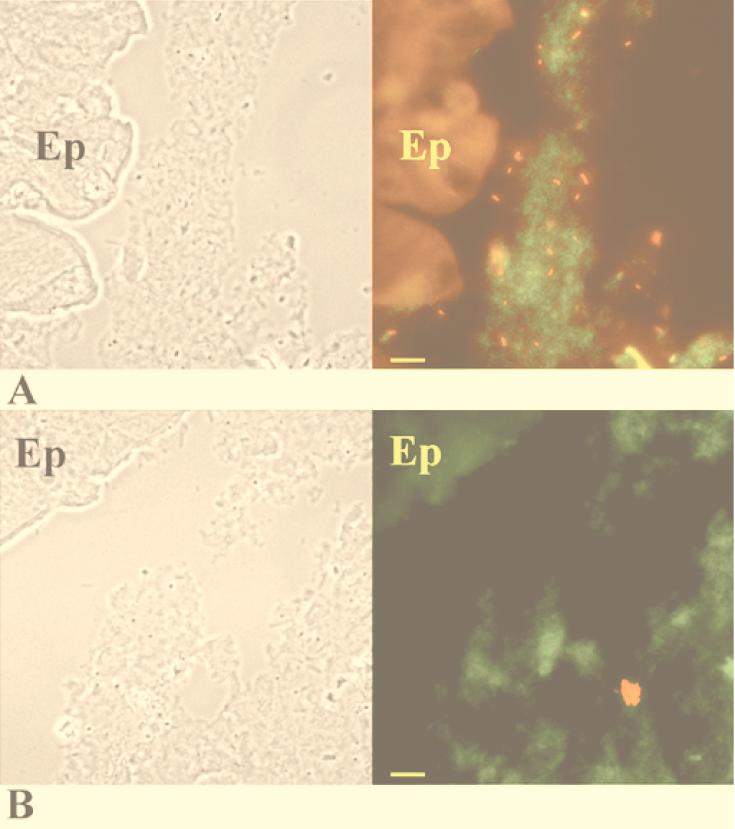
At day 1 after infection with the K. pneumoniae LM21 Strr wild-type strain, microscopic observations of the in situ hybridization performed with both probes simultaneously revealed that the wild-type cells were uniformly distributed in the mucus layer (Fig. 3A). Only a few bacteria were found in contact with epithelial cells. Similarly, in mice colonized by the capsule-defective mutant strain, the bacterial cells were seen evenly distributed in the mucus layer (Fig. 3B). At day 20 from the onset of colonization by wild-type LM21, the bacterial cells remained evenly distributed as single cells in the mucus layer (Fig. 4A). The isogenic mutant strain was also localized in the mucus layer (Fig. 4B), but cells were present in small clusters; very few bacterial cells were detected as single cells. The numbers of bacteria inside a single cluster varied from 3 to 20 single cells.
FIG. 3.
Colonic sections of mice fed wild-type K. pneumoniae LM21 (A) and the K. pneumoniae LM21(cps) capsule-defective mutant (B) on day 1 after onset of infection. (Left) Phase-contrast picture of the corresponding area of in situ hybridization. (Right) In situ hybridization with fluorescence-labeled oligonucleotide probes. K. pneumoniae bacteria appear red, while other eubacteria appear green. The colonic epithelium (Ep) is fluorescent because of the autofluorescence of the eucaryotic tissue. Bars, 10 μm.
FIG. 4.
Colonic sections of mice fed wild-type K. pneumoniae LM21 (A) and the K. pneumoniae LM21(cps) capsule-defective mutant (B) on day 20 after infection. (Left) Phase-contrast picture of the corresponding area of in situ hybridization. (Right) In situ hybridization with fluorescence-labeled oligonucleotide probes. K. pneumoniae bacteria appear red, while other eubacteria appear green. The colonic epithelium (Ep) is fluorescent because of the autofluorescence of the eucaryotic tissue. Bars, 10 μm.
The numbers of single and clustered bacterial cells were determined for the wild-type strain and the mutant strain (assuming that bacterial cells that were not single represented a cluster). With the K. pneumoniae LM21 wild-type strain, the ratio of clusters to single cells was 1:17, whereas with the K. pneumoniae LM21(cps) capsule-defective mutant, the ratio was 1:0.69. At least five images from six different colonic sections of each colonization experiment were counted. This clustering was observed only in vivo in intestinal sections from colonized mice and never during in vitro growth. In situ hybridization on smears from fecal samples obtained at day 20 after the onset of colonization revealed no clustering, either for the wild-type strain or for the capsule-defective mutant (data not shown). Therefore, the intestinal environment seems to cause formation of bacterial clusters of the capsule-defective K. pneumoniae LM21(cps) mutant strain. It is not clear whether this clustering is a result of growth, i.e., microcolonies, or is a type of autoflocculation in the mucus layer perhaps due to hydrophobicity.
The in vivo formation of clusters might explain the lesser colonization ability of the mutant. It can be assumed that during the natural mucus turnover, a higher number of bacterial cells of the mutant strain than of the wild type are exfoliated and swept away. The mechanisms involved in clustering in vivo are very intriguing and will be the subject of further in vivo studies.
Acknowledgments
We thank Anni Ravn and Ulla Andreasen (Statens Veterinære Serumlaboratorium, Copenhagen, Denmark) for excellent technical assistance in preparation of histological sections.
This study was partly supported by the Danish Medical Research Council (K.A.K.). S.F.-B. received a short-term fellowship from the European Molecular Biology Organization (EMBO) and a visiting grant from The Danish Rectors' Conference (Rektorkollegiet).
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camprubi S, Merino S, Guillot J F, Tomas J M. The role of the O-antigen lipopolysaccharide on the colonization in vivo of the germfree chicken gut by Klebsiella pneumoniae. Microb Pathog. 1993;14:433–440. doi: 10.1006/mpat.1993.1042. [DOI] [PubMed] [Google Scholar]
- 3.Darfeuille-Michaud A, Jallat C, Aubel D, Sirot D, Rich C, Sirot J, Joly B. R-plasmid-encoded adhesive factor in Klebsiella pneumoniae strains responsible for human nosocomial infections. Infect Immun. 1992;60:44–45. doi: 10.1128/iai.60.1.44-55.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dechamps C, Sauvant M P, Chanal C, Sirot D, Gazui N, Malhuret R, Baguet J C, Sirot J. Prospective survey of colonization and infection caused by expanded-spectrum β-lactamase-producing members of the family Enterobacteriaceae in an intensive care unit. J Clin Microbiol. 1989;27:2887–2890. doi: 10.1128/jcm.27.12.2887-2890.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Martino P, Livrelli V, Sirot D, Joly B, Darfeuille-Michaud A. A new fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect Immun. 1996;64:2266–2273. doi: 10.1128/iai.64.6.2266-2273.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erbel, L. (Lehrstuhl für Mikrobiologie, Technical University, Munich, Germany). Personal communication.
- 7.Fader R C, Gondesen K, Tolley B, Ritchie D G, Moller P. Evidence that in vitro adherence of Klebsiella pneumoniae to ciliated hamster tracheal cells is mediated by type 1 fimbriae. Infect Immun. 1988;56:3011–3013. doi: 10.1128/iai.56.11.3011-3013.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favre-Bonté S, Joly B, Forestier C. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect Immun. 1999;67:554–561. doi: 10.1128/iai.67.2.554-561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freter R. Mechanisms of bacterial colonization of the mucosal surfaces in the gut. In: Roth J A, editor. Virulence mechanisms of bacterial pathogens. Washington, D.C: ASM Press; 1988. pp. 45–60. [Google Scholar]
- 10.Freter R, Brickner H, Fekete J, Vickerman M M, Carey K E. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun. 1983;39:683–703. doi: 10.1128/iai.39.2.686-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herias M V, Midtvedt T, Hanson L Å, Wold A E. Escherichia coli K5 capsule expression enhances colonization of the large intestine in the gnotobiotic rat. Infect Immun. 1997;65:531–536. doi: 10.1128/iai.65.2.531-536.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornick D B, Allen B L, Horn M A, Clegg S. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect Immun. 1992;60:1577–1588. doi: 10.1128/iai.60.4.1577-1588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licht T R, Krogfelt K A, Cohen P S, Poulsen L K, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myhal M L, Laux D C, Cohen P S. Relative colonizing abilities of human fecal and K12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur J Clin Microbiol. 1982;1:186–192. doi: 10.1007/BF02019621. [DOI] [PubMed] [Google Scholar]
- 15.Nevola J J, Stocker B A D, Laux D C, Cohen P S. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect Immun. 1985;50:152–159. doi: 10.1128/iai.50.1.152-159.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulsen L K, Lan F, Kristensen C S, Hobolth P, Molin S, Krogfelt K A. Spatial distribution of Escherichia coli in the mouse large intestine from rRNA in situ hybridization. Infect Immun. 1994;62:5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poulsen L K, Licht T R, Rang C, Krogfelt K A, Molin S. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol. 1995;177:5840–5845. doi: 10.1128/jb.177.20.5840-5845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Selden R, Lee S, Wenlanlou W, Bennett J V, Eickhoff T C. Nosocomial Klebsiella infections: intestinal colonization as a reservoir. Ann Intern Med. 1971;74:657–664. doi: 10.7326/0003-4819-74-5-657. [DOI] [PubMed] [Google Scholar]