Abstract
Hybrid derivatives of closely related bacteria may be used to dissect strain-specific functions that contribute to virulence within a host. However, mismatches between DNA sequences are a potent barrier to recombination. Recipients with mutS and recD mutations overcome this barrier, allowing construction of genetic hybrids. To determine whether Salmonella hybrids constructed in a mutS recD host can be used to study virulence, we assayed the effect of mutS and recD mutations on the virulence of Salmonella typhimurium 14028s in mice. Mutants defective in either mutS or recD do not affect the time course or the 50% lethal dose (LD50) of the infection. In contrast, the inactivation of both mutS and recD results in a synthetic phenotype which substantially increases the time required to cause a lethal infection without changing the LD50. This phenotype results from an inability of mutS recD double mutants to rapidly adapt to purine-limiting conditions present within macrophages. Although the disease progression is slower, S. typhimurium mutS recD mutants retain the ability to cause lethal infections, and, thus, hybrids constructed in mutS recD hosts may permit the analysis of virulence factors in a surrogate animal model.
Salmonella serovars are closely related at the DNA level, and yet show substantial variations in the hosts they are able to infect and the types of diseases they elicit within each host. Certain strains of Salmonella typhimurium have been extensively studied, providing well-defined genetic and physical maps, a wide collection of mutant strains and selectable genetic markers, efficient mechanisms for gene transfer, and an excellent animal model for virulence (14, 23, 24). In contrast, other Salmonella serovars are less amenable to experimental analysis. For example, Salmonella typhi causes typhoid fever in humans but does not infect other animals. The lack of a simple in vivo assay for virulence has hampered the genetic analysis of S. typhi. Instead, most research has focused on the virulence of S. typhimurium in mice as a model for typhoid fever in humans. Overall S. typhimurium and S. typhi are nearly identical at the nucleotide level and cause similar diseases in the appropriate hosts. Thus, it is not surprising that many of the virulence factors identified in S. typhimurium are also present in S. typhi (1). However, there are clearly some important differences between virulence factors in the two serovars. For example, some virulence factors in S. typhimurium are located on a large plasmid which is absent from S. typhi (11). In addition, the genetic determinants specifying host range must differ between these Salmonella serovars (6).
Although these two serovars are closely related, regions of DNA from the S. typhimurium chromosome cannot be freely replaced with the corresponding chromosomal region from S. typhi due to strong barriers to genetic recombination. The recombination of cognate DNA sequences that are similar but not identical is termed “homeologous” recombination (17, 18). Although S. typhimurium and S. typhi are 98 to 99% identical at the nucleotide level, they undergo homeologous recombination at frequencies which are often undetectable (<10−9). The barriers to homeologous recombination include restriction systems, components of the mismatch repair system (mutSL), and the exonuclease activity of the RecBCD complex (recD). Restriction barriers can be overcome by brief exposure to high temperature, which temporarily inactivates restriction endonucleases (7). However, overcoming the other two barriers requires inactivation of the mutS and recD genes (29). Inactivation of either the mutS or recD gene increases the homeologous recombination frequency about 103-fold. Inactivation of both the mutS and recD genes increases the recombination frequency to levels normally observed during homologous recombination (about 106-fold) and increases the length of DNA which is exchanged.
We reasoned that the inactivation of barriers to recombination would allow the construction of genetic hybrids between S. typhi and S. typhimurium, allowing potential virulence genes from S. typhi to be directly studied in vivo in a surrogate S. typhimurium host (15). However, certain genes involved in DNA repair and recombination are required for growth or survival of Salmonella in animals. For example, the recA and recBC gene products are required for homologous recombination, and mutations in these genes attenuate the virulence of S. typhimurium both in mice and in murine macrophages (3, 4). The recD gene product is not required for homologous recombination, but it is induced during growth of S. typhimurium in murine macrophages (12). To determine whether hybrids constructed in a mutS recD host can be used to study Salmonella pathogenesis in an animal model, we assayed the effect of mutS and recD mutations on virulence of S. typhimurium 14028s in mice.
Effect of mutS and recD mutations on virulence of S. typhimurium in mice.
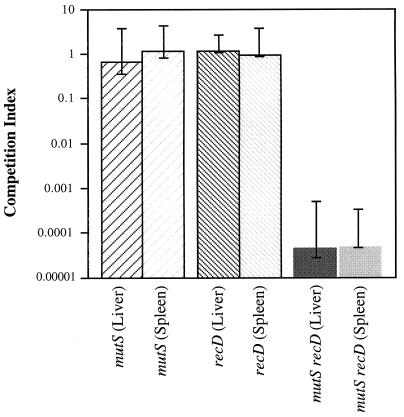
In vivo competition assays were initially used to compare the virulence of mutS, recD, and mutS recD mutant strains to that of wild-type S. typhimurium 14028s (Table 1). Mice which were infected with the mutS or recD derivatives had similar ratios of wild-type to mutant bacteria in both the spleen and liver compared to the ratio of bacteria in the initial inoculum (Fig. 1). In contrast, the ratio of mutS recD to wild-type bacteria recovered from the spleen or liver was diminished 103- to 104-fold relative to the initial infecting ratio (Fig. 1). These results indicate that 4 to 5 days after infection, the interval required for mice to succumb following inoculation with wild-type S. typhimurium 14028s, the mutS recD mutant was unable to establish a lethal infection.
TABLE 1.
Bacterial strains used in this study
| S. typhimurium strain | Genotypea | Source or reference |
|---|---|---|
| LT2 derivatives | ||
| TT16813 | recD542::Tn10dCam | 19 |
| MST3063 | leuA414(Am) hsdL (r− m+) FelS−mutS121::Tn10 | 29 |
| ATCC 14028s derivatives | ||
| MST3083 | Wild type | ATCCb |
| MST4172 | mutS121::Tn10 | This study |
| MST4174 | recD542::Tn10dCam | This study |
| MST4175 | mutS121::Tn10 recD542::Tn10dCam | This study |
| JS110 | recA1 | 16 |
All of the strains used were derivatives of S. typhimurium LT2 or S. typhimurium ATCC 14028s. mutS, recD, or mutS recD derivatives of S. typhimurium 14028s were constructed by P22-mediated transduction of DNA from S. typhimurium LT2 (29). Tn10dCam refers to the transposition-defective derivative of Tn10, Tn10Δ16Δ17 (5). Bacterial strains were routinely grown in rich medium (nutrient broth) composed of 0.8% Difco nutrient broth and 0.5% NaCl, or minimal medium composed of E salts (28) and 0.2% glucose. For solid medium, 1.5% Difco Bacto-agar was added. TBSA top agar contained 1% tryptone, 0.5% NaCl, and 0.7% agar. Tetracycline and chloramphenicol were each added at 20 μg/ml. Bacteria were diluted in 0.85% NaCl unless otherwise indicated.
ATCC, American Type Culture Collection, Manassas, Va.
FIG. 1.
Competition between mutant and wild-type strains in infected mice. Strains were grown overnight in rich medium prior to inoculation. Wild-type and mutant bacteria were administered intraperitoneally at 0.2 ml with 100 to 500 CFU of bacteria to groups of five 6- to 8-week-old female BALB/c mice (Harlan Sprague, Indianapolis, Ind.). The ratios of mutant to wild-type bacteria used ranged from 0.9:1 to 5:1. Mice were sacrificed 4 to 5 days after infection. Bacteria were recovered from the spleen and liver by homogenizing the tissues in 0.85% NaCl and plating serial dilutions onto rich medium. The number of mutant bacteria recovered was determined by replica plating onto rich medium supplemented with the appropriate antibiotic. The competition index is expressed as the (CFU of mutant recovered/CFU of wild type recovered)/(CFU of mutant inoculated/CFU of wild type inoculated). The value reported is the median competition index for five separate experiments. The error bars represent the maximal and minimal competition indices for each mutant versus wild-type bacteria in individual mice.
To determine whether those mutS recD mutants recovered from the spleen or liver had acquired additional mutations which allowed them to survive during the initial infection, we repeated the competition assay by using mutS recD mutants which were reisolated from the spleens of previously infected mice. After overnight growth in rich medium, the infection was repeated, and the number of mutS recD bacteria recovered from the spleen and liver remained 103- to 104-fold lower than the number of wild-type bacteria recovered (data not shown). Thus, the mutS recD mutants recovered from the host have not acquired secondary mutations that suppress the mutS recD phenotype.
To determine whether mutations in mutS and recD affected the number of bacteria required to cause a lethal infection in mice, we compared the 50% lethal doses (LD50s) of wild-type, mutS, recD, and mutS recD mutant derivatives of S. typhimurium 14028s. Serial fivefold dilutions (from 0 to approximately 1,500 CFU) of wild-type or mutant derivatives of S. typhimurium 14028s grown overnight in rich medium were administered intraperitoneally in 0.2-ml inoculums to groups of five 6- to 8-week-old female BALB/c mice. The mice were observed daily through 4 weeks postinoculation. The log LD50 was calculated by the method of Reed and Muench (22). For each of the strains tested, less than 10 CFU produced a lethal infection in 50% of the mice. However, mice infected with the wild-type bacteria succumbed to the infection in 4 to 7 days, while mice infected with mutS recD double mutants took approximately twice as long to succumb.
Taken together, these results indicate that although neither mutS or recD mutations alone have an observable effect on virulence, mutS recD double mutants acquire a distinct, synthetic phenotype that substantially slows the time required to cause lethal infection in BALB/c mice. Although BALB/c mice are somewhat immunocompromised due to a mutation in Nramp1 (Itys) (21, 26), similar phenotypes were also obtained following infection of C3H Nramp1+ (Ityr) mice with mutS recD bacteria (data not shown).
Survival of mutS recD mutants in murine peritoneal macrophages.
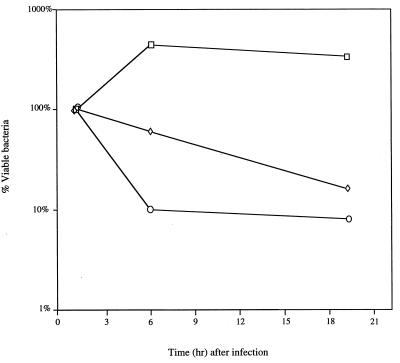
Survival of Salmonella within macrophages is essential for virulence (9) and involves a dynamic equilibrium between bacterial growth and death (2). To determine whether the mutS recD phenotype was due to diminished survival in macrophages, we compared the survival of wild-type and mutant strains of S. typhimurium in murine peritoneal macrophages in vitro (Fig. 2). As expected, the number of wild-type bacteria recovered from infected macrophages increased with time after infection. The number of recA bacteria recovered from infected macrophages decreased about 10-fold by 6 h after infection, reflecting the increased sensitivity of recA mutants to the initial oxidative attack by macrophages. In contrast, the number of mutS recD bacteria recovered from macrophages remained nearly constant for the first 6 h after infection, suggesting that these mutants fail to proliferate in macrophages but are not rapidly killed by the oxidative burst (Fig. 2). Furthermore, although mutants defective for DNA repair and recombination are often sensitive to oxidative DNA damage (3, 4), wild-type and mutS recD strains showed similar sensitivities to H2O2 in vitro (data not shown). Thus, the delay in virulence observed in vivo is probably due to an inability of mutS recD mutants to multiply within the host macrophages.
FIG. 2.
Survival of S. typhimurium derivatives in murine peritoneal macrophages. Macrophages were harvested from the peritoneal cavity of 6- to 8-week-old BALB/c mice 3 days after intraperitoneal injection with 1.5 ml of 10% sterilized proteose peptone (Difco). Macrophages removed from the peritoneal cavity with a 23-gauge needle and 30-ml syringe were resuspended in endotoxin-free RPMI medium containing 5% fetal calf serum and 1 U of heparin per ml. Contaminating erythrocytes were removed by centrifugation, and the number of viable macrophages was quantitated on a hemocytometer following the addition of trypan blue. Approximately 105 peritoneal macrophages were seeded per well in 48-well tissue culture plates. Macrophages were allowed to adhere for 1 h in a 37°C incubator containing 5% CO2. Bacteria were diluted in 0.85% NaCl, centrifuged, and resuspended in 25% mouse serum. After opsonization in mouse serum at 37°C for 15 min, RPMI medium was added to give a final multiplicity of infection of bacteria to macrophages of between 1 and 5. Immediately after the addition of bacteria, tissue culture plates were centrifuged to promote bacterial adherence. Bacterial internalization was allowed to proceed for approximately 30 min, and then each reaction mixture was divided into two aliquots. In one aliquot, extracellular bacteria were killed by the addition of RPMI buffer containing 12.5 μg of gentamicin per ml. Macrophages were lysed with 0.5% deoxycholate, and the number of bacteria in each aliquot was determined by spotting serial dilutions onto rich medium plates. The number of internalized bacteria was initially determined 1.5 h after gentamicin treatment. Survival in macrophages was determined after 6 and 19 h of incubation. The number of surviving bacteria at these times is expressed as a percentage of the number of bacteria internalized. Values represent the mean from experiments performed in triplicate. The standard error was <2.0% of the mean for all values. Strains were wild-type (□), mutS recD (◊), or recA (○) derivatives of S. typhimurium 14028s.
Ability of mutS recD mutants to adapt to nutrient-limiting conditions.
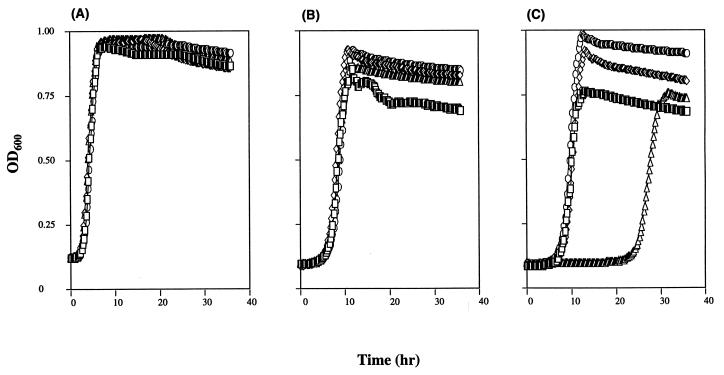
To determine if the differential survival of wild-type bacteria relative to mutS recD mutants was due to differences in bacterial growth, we compared the growth characteristics of wild-type, mutS, recD, and mutS recD strains in vitro (Fig. 3). The wild-type and mutant strains showed similar growth rates when continuously cultured in rich medium (Fig. 3A) or when continuously cultured in minimal medium (Fig. 3B). The wild-type, mutS, and recD strains also exhibited similar lag times and growth rates when grown in rich medium and subcultured into minimal medium (Fig. 3C). However, mutS recD strains exhibited a much longer lag time when grown initially in rich medium and subcultured into minimal medium (Fig. 3C). This inability to adapt was not simply the result of stationary-phase growth, because mutS recD derivatives immediately resumed growth and behaved similarly to the wild-type strain when strains were continuously subcultured in the same type of medium (Fig. 3A and B). Thus, mutS recD mutants appear unable to readily adapt to nutrient-limiting conditions in vitro.
FIG. 3.
Growth of S. typhimurium 14028s derivatives in vitro. The growth rate was determined by measuring the optical density at 600 nm (OD600) as a function of time. Wild-type (□), mutS (◊), recD (○), or mutS recD (▵) strains were either grown in rich medium and subcultured into rich medium (A), grown in minimal medium and subcultured into minimal medium (B), or grown in rich medium and subcultured into minimal medium (C). All assays were performed in 96-well microtiter dishes with a BioTek Instruments ELx808 Automated Microdish Reader. Strains were routinely grown in 200-μl volumes and incubated at 37°C with shaking. Growth was monitored over 36 h by measuring the OD600 at 15-min intervals. The values shown represent the mean optical densities from cultures grown in triplicate. The standard error was <5% of the mean for all time points measured.
Purine supplementation alleviates the mutS recD-dependent growth phenotype.
Growth of Salmonella in macrophages requires the ability to synthesize certain metabolites that are limiting in the host (27). To determine if a common metabolite could restore the ability of mutS recD strains to adapt to nutrient downshift, we analyzed the lag time required to resume growth in minimal medium supplemented with various pools of amino acids, nucleotides, cofactors, or vitamins (13). When subcultured from rich medium into minimal medium supplemented with pools of nutrients that included purines or minimal medium supplemented with only adenine, guanine, or adenine plus guanine, the growth lag for mutS recD strains was eliminated (Table 2). Thus, mutations in mutS and recD affect the ability of S. typhimurium to grow under conditions in which purines are limiting.
TABLE 2.
Effect of nutritional supplements on growth of the mutS recD strain in minimal medium
| Supplement(s)a | Difference in lag time (h)b | % Reduction in lag timec |
|---|---|---|
| None | 27.8 | |
| Guanine | 0.1 | 99 |
| Adenosine | 1.6 | 85 |
| Guanine + adenosine | −1.4 | 119 |
One microliter of each supplement was added per 200 μl of minimal medium in a microtiter dish. The stock concentration of supplements used was described previously (13).
The difference in lag time equals the time required for the mutS recD strain to resume growth after subculturing minus the time required for the wild-type strain to resume growth under the same conditions.
Percent reduction in lag time is the relative decrease in lag time upon subculturing of the mutS recD mutants into minimal medium with the nutritional supplement indicated. One hundred percent reduction in lag time indicates that the supplement completely eliminated the disproportionate growth lag observed upon subculturing of the mutS recD strain from rich to minimal medium.
Conclusions.
The results of the in vivo competition and LD50 assays indicate that the development of a systemic infection is slower for the mutS recD strain than for the isogenic wild-type strain. The phenotype of mutS recD mutants observed in vivo was not simply the result of the disruption of either the mutS or recD genes, because derivatives of S. typhimurium carrying mutations in either one of these genes behaved similarly to the wild-type strain (Fig. 1). Nor was the alteration in virulence simply the result of a silent secondary mutation, because the same phenotype was also observed after these mutations were backcrossed into a wild-type S. typhimurium 14028s background (data not shown). Rather, the alteration in virulence was the direct result of the inactivation of both the mutS and recD gene products. The results indicate that mutS recD double mutants experience a general downshift in purine metabolism. The reasons for the purine requirement in the double mutants are not yet clear. However, these results explain the slower progression of the double mutants observed in vivo, because the environment which S. typhimurium encounters during an infection, probably within the resident macrophages, is nutrient limiting for purines (2, 27).
Many of the genes required for Salmonella pathogenesis encode proteins involved in normal housekeeping functions of the cell. These include genes involved in DNA metabolism and amino acid biosynthesis, as well as genes involved in iron regulation (10, 25) and stationary-phase growth (8, 20). Mutations in these housekeeping genes can attenuate the virulence of Salmonella and allow the immune system to rapidly clear the infecting organisms. In contrast, disruption of mutS and recD does affect the pathogenesis of S. typhimurium, but without attenuating virulence or increasing the clearance of infecting organisms by the host. Such mutations would go unrecognized from the typical genetic selections or screens developed to identify genes involved in pathogenesis, because the manifestation of the phenotype requires the simultaneous acquisition of two mutations, neither of which has any virulence phenotype on its own (i.e., a “synthetic phenotype”). Thus, such synergistic mutations may identify a potentially new class of virulence determinants which have previously gone uncharacterized.
In summary, strains of S. typhimurium which lack both MutS-dependent mismatch repair and RecBCD exonuclease activity are unable to efficiently adapt to purine-limiting conditions in mice. The lag in nutrient adaptation results in the attenuation of the time required to cause systemic disease, but does not change the LD50, indicating that the slower time course of the disease does not promote increased clearance by the host immune system. Thus, the BALB/c mouse model commonly used to study S. typhimurium virulence may be suitable for studying genetic hybrids constructed by recombination with an S. typhimurium mutS recD surrogate host.
Acknowledgments
We thank Don Guiney, Jim Imlay, Steve Libby, and Jim Slauch for experimental advice and resources; Jennifer Neitzer and Anne Thierauf for contributions to some of the experiments; and Rob Edwards and R. Allen Helm for comments on the manuscript.
REFERENCES
- 1.Baumler A J, Tsolis R M, Ficht T A, Adams L G. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–4587. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchmeier N A, Libby S J. Dynamics of growth and death within a Salmonella typhimurium population during infection of macrophages. Can J Microbiol. 1997;43:29–34. doi: 10.1139/m97-005. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier N A, Libby S J, Xu Y, Loewen P C, Switala J, Guiney D G, Fang F C. DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Investig. 1995;95:1047–1053. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier N A, Lipps C J, So M, Heffron F. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 5.Castilho B A, Olfson P, Casadaban M J. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conner C P, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards R, Helm R, Maloy S. Increasing DNA transfer efficiency by temporary inactivation of host restriction. BioTechniques. 1999;26:892–900. doi: 10.2144/99265st02. [DOI] [PubMed] [Google Scholar]
- 8.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternate ς factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within macrophages are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furman M, Fica A, Saxena M, Di Fabio J L, Cabello F C. Salmonella typhi iron uptake mutants are attenuated in mice. Infect Immun. 1994;62:4091–4094. doi: 10.1128/iai.62.9.4091-4094.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guiney D G, Fang F C, Krause M, Libby S, Buchmeier N A, Fierer J. Biology and clinical significance of virulence plasmids in Salmonella serovars. Clin Infect Dis. 1995;21(Suppl. 2):S146–S151. doi: 10.1093/clinids/21.supplement_2.s146. [DOI] [PubMed] [Google Scholar]
- 12.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maloy S R. Experimental techniques in bacterial genetics. Boston, Mass: Jones and Bartlett Publishers; 1990. [Google Scholar]
- 14.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. New York, N.Y: Cold Spring Harbor Laboratory; 1996. [Google Scholar]
- 15.Maloy, S. R., and T. Zahrt. Surrogate genetics: the use of bacterial hybrids as a genetic tool. Methods, in press. [DOI] [PubMed]
- 16.Mann B, Slauch J. Transduction of low-copy number plasmids by bacteriophage P22. Genetics. 1997;146:447–456. doi: 10.1093/genetics/146.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 18.Matic I, Taddei F, Radman M. Genetic barriers among bacteria. Trends Microbiol. 1996;4:69–73. doi: 10.1016/0966-842X(96)81514-9. [DOI] [PubMed] [Google Scholar]
- 19.Miesel L, Roth J R. Salmonella recD mutations increase recombination in a short sequence transduction assay. J Bacteriol. 1994;176:4092–4103. doi: 10.1128/jb.176.13.4092-4103.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickerson C A, Curtiss R., III Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–1823. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien A D. Innate resistance of mice to Salmonella typhi infections. Infect Immun. 1982;38:948–952. doi: 10.1128/iai.38.3.948-952.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed L J, Muench H. A simple method for estimating fifty percent end points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 23.Sanderson K, Hessel A, Liu S, Rudd K E. The genetic map of Salmonella typhimurium, edition VIII. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium. Cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 1903–1999. [Google Scholar]
- 24.Sanderson K, Liu S-L, Hessel A, McClelland M. Genome evolution in the salmonellae. In: de Bruijin F, Lupski J, Weinstock G, editors. Bacterial genomes. New York, N.Y: Chapman and Hall; 1998. pp. 230–239. [Google Scholar]
- 25.Sawatzki G, Hoffmann F A, Kubanek B. Acute iron overload in mice: pathogenesis of Salmonella typhimurium infection. Infect Immun. 1983;39:659–665. doi: 10.1128/iai.39.2.659-665.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skamene E, Schurr E, Gros P. Infection genomics: Nramp1 as a major determinant of natural resistance to intracellular infections. Annu Rev Med. 1998;49:275–287. doi: 10.1146/annurev.med.49.1.275. [DOI] [PubMed] [Google Scholar]
- 27.Stocker B A D. Attenuation of Salmonella by auxotrophy. In: Cabello F, Hormaeche C, Mastroeni P, Bonina L, editors. Biology of Salmonella. New York, N.Y: Plenum Press; 1993. pp. 309–322. [Google Scholar]
- 28.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 29.Zahrt T C, Maloy S. Barriers to recombination between closely related bacteria: MutS and RecBCD inhibit recombination between Salmonella typhimurium and Salmonella typhi. Proc Natl Acad Sci USA. 1997;94:9786–9791. doi: 10.1073/pnas.94.18.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]