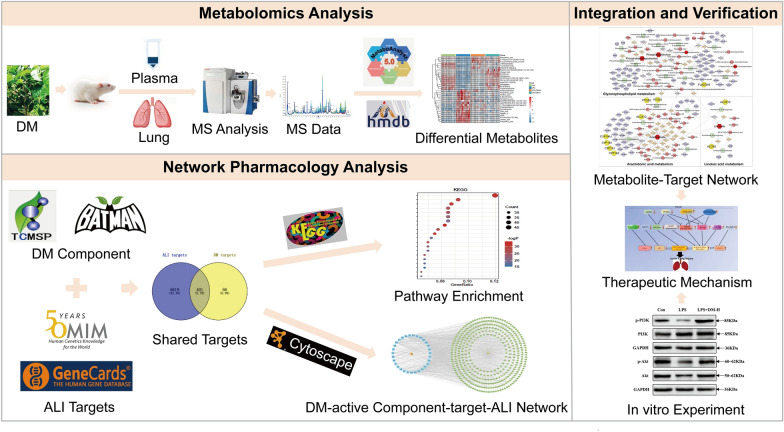
Abstract
Background
Acute lung injury (ALI) is a severe inflammatory disease, underscoring the urgent need for novel treatments. Nauclea officinalis Pierre ex Pitard (Danmu in Chinese, DM) is effective in treating inflammatory respiratory diseases. However, there is still no evidence of its protective effect against ALI.
Methods
Metabolomics was applied to identify the potential biomarkers and pathways in ALI treated with DM. Further, network pharmacology was introduced to predict the key targets of DM against ALI. Then, the potential pathways and key targets were further verified by immunohistochemistry and western blot assays.
Results
DM significantly improved lung histopathological characteristics and inflammatory response in LPS-induced ALI. Metabolomics analysis showed that 16 and 19 differential metabolites were identified in plasma and lung tissue, respectively, and most of these metabolites tended to recover after DM treatment. Network pharmacology analysis revealed that the PI3K/Akt pathway may be the main signaling pathway of DM against ALI. The integrated analysis of metabolomics and network pharmacology identified 10 key genes. These genes are closely related to inflammatory response and cell apoptosis of lipopolysaccharide (LPS)-induced ALI in mice. Furthermore, immunohistochemistry and western blot verified that DM could regulate inflammatory response and cell apoptosis by affecting the PI3K/Akt pathway, and expression changes in Bax and Bcl-2 were also triggered.
Conclusion
This study first integrated metabolomics, network pharmacology and biological verification to investigate the potential mechanism of DM in treating ALI, which is related to the regulation of inflammatory response and cell apoptosis. And the integrated analysis can provide new strategies and ideas for the study of traditional Chinese medicines in the treatment of ALI.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13020-022-00685-6.
Keywords: Acute lung injury, Nauclea officinalis, Metabolomics, Network pharmacology, Mechanisms
Background
Acute lung injury (ALI) is a critical disease with a complex pathology that is mainly characterized by inflammatory dysregulation, disruption of pulmonary endothelial and epithelial barriers, and gas exchange disorders [1, 2]. At present, the common treatments for ALI are reducing pulmonary inflammatory damage and relieving respiratory failure, and conventional treatment drugs include steroids, statins, and glucocorticoids. However, these drugs can only relieve clinical symptoms and are unable to reduce mortality from ALI [3, 4]. The mortality rate of ALI is 40% and above [5]. Therefore, it is urgent to discover effective therapeutic strategies for the treatment of ALI.
Numerous Chinese medicines have shown favorable therapeutic effects on ALI in clinical practice [6, 7]. Nauclea officinalis Pierre ex Pitard (Danmu in Chinese, DM) is a Li-folk Chinese medicine that has been used to treat inflammatory and infectious diseases for over 40 years in Hainan Province [8]. A variety of pharmacological activity studies have been conducted to determine the internal mechanism of DM. The results show that DM has comprehensive biological activities such as anti-inflammatory, antibacterial, antiviral, antioxidant, immune regulation, and promotion of cell proliferation [9]. Our previous study also demonstrated that DM extract could inhibit the release of inflammatory cytokines in RAW264.7 cells and reduce the expression of NO, TNF-α, IL-1β and IL-6 [10]. Further studies have shown that DM may repair the endothelial cell barrier of lung injury by inhibiting the inflammatory response, improving intercellular connections, and promoting angiogenesis [11]. These studies partly explain the excellent effects of DM on inflammatory diseases. However, the protective effects of DM on ALI and its underlying mechanisms remain unclear. Therefore, the study of the mechanisms of DM on ALI may be helpful in providing more options for ALI treatment.
Given the complexity of ALI pathogenesis, it is difficult to reveal the mechanisms of DM on ALI using traditional pharmacological methods. By monitoring the overall dynamic changes in metabolites in response to drug treatment, metabolomics can comprehensively reflect the development of disease and is a powerful tool for the diagnosis and treatment of complex diseases [12, 13]. Therefore, metabolomics is very suitable for revealing the mechanism of Chinese medicine in the treatment of ALI [14, 15]. However, metabolomics only reflect the changes in endogenous small molecule metabolites. The mechanism of metabolite changes remains unclear [16]. Therefore, metabolomics alone has difficulty revealing the complete therapeutic mechanism of DM on ALI.
In recent years, network pharmacology has developed rapidly in the speculation of the mechanisms of Chinese medicine [17]. Network pharmacology is frequently used to predict the targets of Chinese medicine, identify biomarkers of disease, and then speculate on the regulatory mechanisms of Chinese medicine [18, 19]. In previous literature, network pharmacology was employed to explore the therapeutic effect and potential mechanism of ginseng on ALI. The results revealed that ginseng could inhibit the PI3K/Akt and MAPK signaling pathways by downregulating the expression of STAT3, VEGFA, FGF2, PIK3CA, MAPK1 and IL2, which provides a good example of network pharmacology applied to ALI [20]. However, network pharmacology relies on public databases, and database updates seriously affect the reliability of network pharmacology results. Moreover, network pharmacology could predict potential targets and related pathways, but it is still unclear whether the predicted results are accurate. Therefore, the results also need to be verified with other methods [21].
Therefore, in this study, we integrated metabolomics, network pharmacology, and biological experimental methods of verification to reveal the influences of DM on ALI and the underlying mechanisms. First, untargeted metabolomics was applied to determine the effects of DM on ALI, and candidate metabolites and related metabolic pathways were screened. Subsequently, network pharmacology was used to predict the targets of DM active components in ALI and the pathways involved, and the regulatory metabolite-related proteins and responses were further integrated and analyzed to predict the primary mechanism of DM treatment in ALI. Finally, biological experiments were used to verify the accuracy of the predictions. In brief, this comprehensive strategy compensates for the individual defects of network pharmacology and metabolomics. This study helps us better understand the therapeutic principles of DM in the prevention and treatment of ALI.
Materials and methods
Reagents
LPS was obtained from Solarbio (China). Methanol, acetonitrile, and formic acid (chromatography pure) were obtained from Merck (Germany). PI3K mouse monoclonal antibody, p-PI3K rabbit monoclonal antibody, Akt rabbit polyclonal antibody, p-Akt mouse monoclonal antibody, Bcl-2 mouse monoclonal antibody, Bax mouse monoclonal antibody, GAPDH rabbit monoclonal antibody, HRP-conjugated AffiniPure goat anti-mouse IgG (H + L) and goat anti-rabbit IgG (H + L) were obtained from Proteintech Biotechnology (China).
Source and preparation of DM
Nauclea officinalis Pierre ex Pitard stems were collected from Wuzhishan City, Hainan Province, in September 2019 and were identified by Dr. Yong-hui Li, Hainan Medical University. The voucher specimen (No. DM-20190920) was deposited at Hainan Provincial Key Lab of Research and Development on Tropic Herbs. The 5 kg DM stems were ground and extracted with water (1/10, w/v) for 2 h each time. The solutions were combined and concentrated to obtain 159 g of DM extract. The extracts were stored at 4 °C for further study.
Animals and the lipopolysaccharide (LPS) model
Male C57BL/6J mice weighing 22–25 g were purchased from Changsha Tianqin Biotechnology Co., Ltd (Certificate number: SCXK Xiang 2022–0015). The mice were fed a standard diet and provided with water ad libitum. The animal experiment project was conducted in strict accordance with the guidelines for the care and use of laboratory animals and approved by the Ethics Committee of Hainan Medical University (No. HYLL-2022-019).
After acclimatization feeding, 90 mice were randomly divided into 5 groups (n = 18 per group): control group, LPS group, low-dose treatment group (LPS + DM-L, 100 mg/kg), medium-dose treatment group (LPS + DM-M, 200 mg/kg), and high-dose treatment group (LPS + DM-H, 400 mg/kg). The ALI model was induced by intratracheal instillation of 50 μl of LPS (5 mg/kg). The control group received normal saline. The LPS group received LPS stimulation, and the LPS + DM groups received LPS stimulation and DM treatment. After treatment for 6 h, six mice in each group were anesthetized using 3% isoflurane, and bronchoalveolar lavage fluid (BALF) was collected. After treatment for 24 h, twelve mice in each group were anesthetized, and plasma and lung tissues were collected and stored at − 80 °C for further study.
Lung histopathology
Mouse lung tissues were immersed in 4% paraformaldehyde. The lung tissues were then dehydrated, embedded in paraffin, sliced into 4 μm sections, and stained with hematoxylin–eosin (H and E). Finally, the sections were imaged under a light microscope.
Cytokines analysis
The levels of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6) and interleukin-1β (IL-1β) in BALF were detected using ELISA kits from Jiancheng Bioengineering Institute (Nanjing, China), according to the manufacturer's instructions.
Plasma and lung tissue sample preparation
Blood samples were collected in heparin-coated Eppendorf tubes, held for 2 h and then centrifuged at 4 °C and 3000 rpm for 10 min to retain the supernatants as plasma samples. Lung tissue samples were homogenized in a tenfold volume of saline.
One hundred microliters of serum or 1 ml of lung tissue homogenate were diluted with a threefold volume of methanol: acetonitrile (1:1). The mixture was vortexed for 5 min, centrifuged at 4 °C and 12,000 rpm for 15 min to obtain the supernatant, and then dried at 30 °C with N2. Finally, the samples were reconstituted with 120 μl of 50% acetonitrile, vortexed for 5 min and centrifuged (12,000 rpm at 4 °C for 10 min) to obtain the supernatant for injection. A quality control (QC) sample was prepared from 10 μL of each test sample.
UPLC-Q-TOF/MS analysis
The metabolomic analysis of ALI mouse plasma and lung tissue was performed using a UHPLC system (Agilent 1290, USA) coupled with a high-resolution mass spectrometer (AB Sciex Triple TOF 4600) in positive and negative ion modes. The chromatographic columns were Phenomenex Kinetex 1.7 μm XB-C18 (100A, 2.1 mm × 50 mm) and Waters Acquity UPLC BEH C18 (1.7 μm, 2.1 mm × 100 mm).
For chromatography, the mobile phase was composed of acetonitrile (A) and 0.1% formic acid (B). The gradient elution conditions for plasma were as follows: 0–1 min, 5–30% A; 1–3 min, 30–40% A; 3–5 min, 40–60% A; 5–11 min, 60–70% A; 11–13 min, 70–95% A; 13–16 min, 95% A; 16–16.1 min, 95–5% A; 16.1–20 min, 5% A. For lung tissue, the conditions were 0–1 min, 5–15% A; 1–9 min, 15–40% A; 9–10 min, 40–50% A; 10–24 min, 50–75% A; 24–25 min, 75–95% A; 25–27 min, 95% A; 27–27.1 min, 95–5% A; 27.1–32 min, 5% A. The column temperature was set at 40 °C, the flow rate was 0.3 ml/min, and the injection volume was 3 μl.
The MS parameters were as follows: the positive and negative ion modes were detected by an ESI ion source, the mass range was m/z 100–1000 Da, the ion spray voltage was + 5500/− 4500 V, and the source temperature was 550 °C. The declustering potential (DP) was ± 80 V, and the collision energy was ± 35 eV; ion source gas 1 and ion source gas 2 were set at 50 psi, and the curtain gas was set at 30 psi.
Data processing and analysis
The raw MS data were preprocessed with Markerview 1.3.1 (AB Sciex), including peak identification, peak extraction, and peak area normalization [22]. The preprocessed data were then imported into SIMCA-P14.0 (Umetrics, Umea, Sweden) for multivariate statistical analysis and based on two parameters, R2Y and Q2, to evaluate the model truth and predictive ability. Variables were identified as differential metabolites when they satisfied the criteria VIP > 1, P < 0.05, FC > 1.2, or FC < 0.8 [23]. Subsequently, the secondary mass spectra of the potential metabolites were screened with Peakview 1.2.1 software and then identified by HMDB (http://www.hmdb.ca/) and KEGG (http://www.kegg.ca/). Finally, the differential metabolites were imported into MetaboAnalyst 5.0 (http://www.metaboanalyst.ca/) for metabolic pathway enrichment analysis [14].
Network pharmacology analysis
First, all the chemical compounds of DM were obtained from the TCMSP (http://tcmspw.com/tcmsp.php) and literature review [15]. The compounds with OB ≥ 25% were selected as the active compounds of DM for further analysis. Next, the component targets were collected from BATMAN-TCM (http://bionet.ncpsb.org/batman-tcm/) [24], DrugBank (https://www.drugbank.ca/) and Swiss Target Prediction (http://www.swisstargetprediction.ch/) databases [25]. The SMILES of each compound was searched in PubChem (https://pubchem.ncbi.nlm.nih.gov/) and imported into Swiss Target Prediction (http://www.swisstargetprediction.ch/) to acquire the related targets. Moreover, DrugBank (https://www.drugbank.ca/), Gene Cards (https://www.genecards.org/), OMIM (https://omim.org/) and TTD (http://db.idrblab.net/ttd/) [26] were used to search for genes associated with ALI.
Second, the shared targets of DM active ingredients and ALI were selected, which were considered potential therapeutic targets of DM against ALI. The shared targets were imported into the STRING (https://cn.string-db.org/) database to construct a protein–protein interaction (PPI) network, and the hub genes were screened.
Finally, The Cytoscape 3.9.1 was then used to construct the “DM-active component-target-ALI” network. The target genes were then all subjected to the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) to carry out GO and KEGG enrichment for functional enrichment analysis. P-values were derived from the DAVID database and set to below 0.05 [27].
Comprehensive analysis of metabolomics and network pharmacology
The differential metabolites identified in metabolomics were imported into MetScape 3.1.3 to construct a “Metabolite-Reaction-Enzyme-Gene” network [28]. The related genes of metabolites were then combined with the target genes and metabolic pathways screened by network pharmacology to identify key metabolic pathways and core targets.
Immunohistochemistry assay
The lung tissue sections were dewaxed and then soaked sequentially in xylene for 15 min, followed by 5 min each in different concentration of EtOH (95%, 80%, 70%, and 50%). The tissue sections were then subjected to antigen retrieval for 24 min and incubated by adding the corresponding primary and secondary antibodies. Finally, the washed sections were observed under a microscope after adding DAB chromogenic solution.
Western blot assay
Fifty milligrams of lung tissue were homogenized with RIPA lysis buffer (containing phosphatase inhibitor). The protein content of lung tissue was quantified, and then the separation gel and concentrated gel were prepared based on the molecular weight, and a suitable sample was taken for loading. Subsequently, electrophoresis and membrane transfer were performed. The PVDF membrane was removed and blocked with 5% skim milk for 2 h, combined with the primary antibody overnight at 4 °C, and incubated with the secondary antibody for 2 h. The membrane was washed 3 times with TBST for 5 min each time and the immunoreactivity bands were detected by ChemiDoc XRS + chemiluminescence imaging system (Bio-Rad, USA).
Statistical analysis
All data were analyzed by GraphPad Prism and presented as Mean ± SD. The different groups were evaluated by one-way ANOVA tests followed by Tukey’s multiple comparisons. P < 0.05 was accepted as statistically significant.
Results
Effects of DM on histological changes in ALI mice
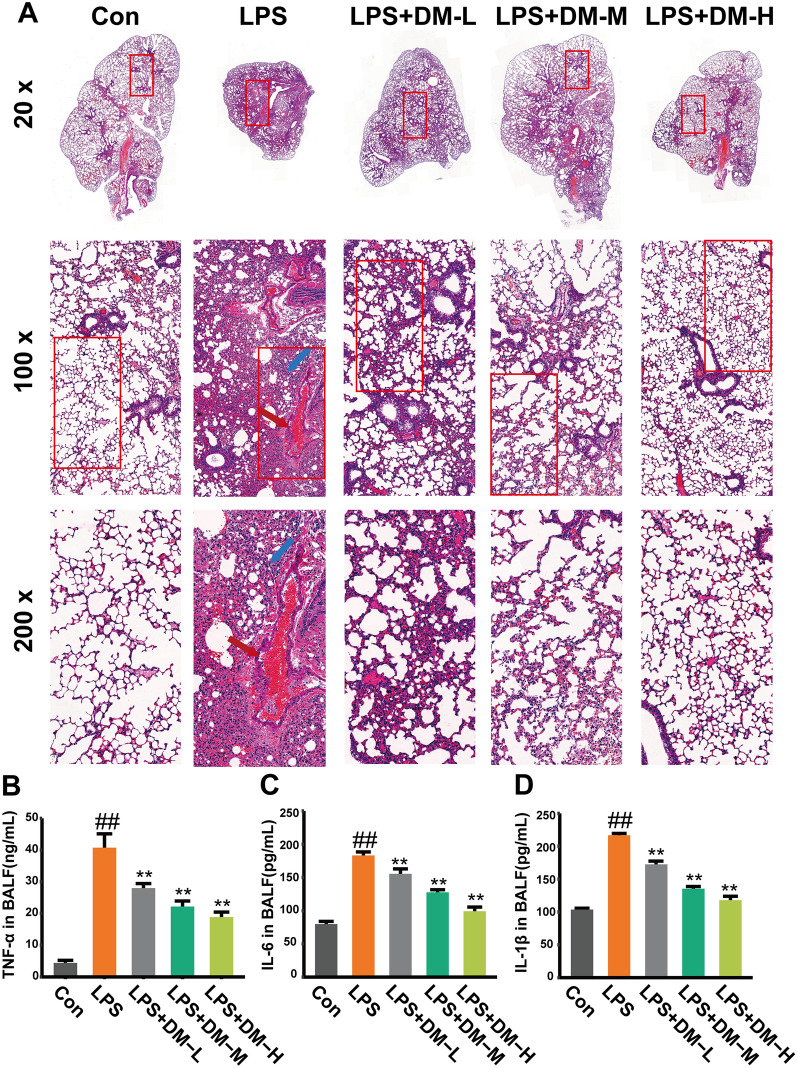
To observe the effect of DM on the structural integrity of lung tissue in ALI mice, hematoxylin–eosin (H&E) staining was applied to evaluate the histological characteristics of mouse lungs. As shown in Fig. 1A, the alveolar walls of mice in the LPS group were extensively thickened, the alveolar sizes varied, the capillaries in the alveolar walls were congested (red arrow), and there was massive infiltration of inflammatory cells (blue arrow). However, lung injury was significantly improved after DM treatment. It was revealed that DM has a therapeutic effect on LPS-induced lung injury.
Fig. 1.
Effect of DM on histopathology and cytokines. A H&E staining of lung tissue in mice. Original magnification, × 20, × 100, × 200.The capillaries in the alveolar walls were congested (red arrow), and massive inflammatory cells infiltrated (blue arrow). B The levels of TNF-ɑ in BALF. C The levels of IL-6 in BALF. D The levels of IL-1β in BALF
DM inhibits inflammatory response in LPS-induced ALI
Pro-inflammatory TNF-α, IL-6, and IL-1β can expeditiously involve in the early phase of inflammatory response and they play a key role in the progression and pathogenesis of ALI. Therefore, their levels were quantified in BALF by ELISA (Fig. 1B–D). The levels of TNF-ɑ, IL-6, and IL-1β in the DM groups were dramatically decreased after drug treatment compared with those in the LPS model group (P < 0.01). The results showed that DM could improve the inflammatory response in LPS-induced ALI.
Plasma and lung tissue metabolic profiling
When testing samples, a QC sample was inserted for every eight test samples in the injection batches to check the stability and repeatability of the instrument and method. Additional file 2: Fig. S3A–D shows the total ion chromatograms (TICs) of plasma and lung tissue in positive and negative ion modes. The overlap between each QC sample indicated that the system was stable and the operating error was small. After PCA of the QC samples, all QC samples in plasma and lung tissue showed obvious clusters (Additional file 2: Fig. S3E, F). As shown in Additional file 2: Fig. S3G, H, 88.92% and 80.14% of metabolites had an RSD% < 30% in plasma and lung tissue, respectively. These results revealed that the detection system was stable and the data were reliable.
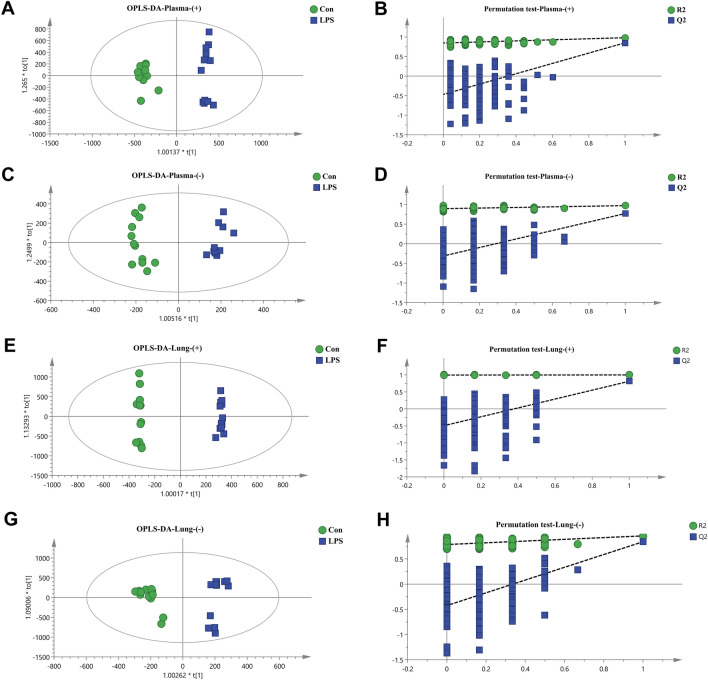
Additional file 2: Fig. S4 shows the TICs of plasma and lung tissue samples in positive and negative ion modes. To investigate the separation among the control, LPS and DM groups, we performed PCA (Fig. 2A–D) and PLS-DA (Additional file 2: Fig. S5A–D). The samples of different groups of plasma and lung tissue of mice were well separated under the two modes, and the R2 and Q2 of the PLS-DA analysis were greater than 0.5 (Additional file 2: Table S1), which further indicated that DM had an excellent regulatory effect on the metabolism of ALI mice.
Fig. 2.
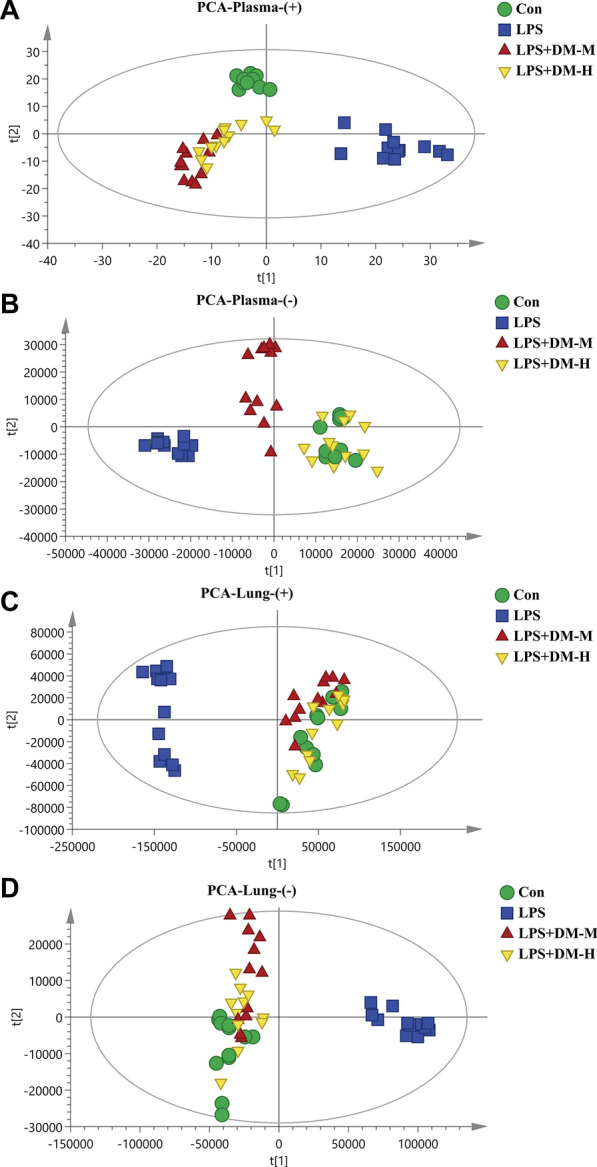
PCA analysis of plasma and lung tissue in mice. A Plasma positive ion mode PCA diagram; B plasma negative ion mode PCA diagram; C lung tissue positive ion mode PCA diagram; D lung tissue negative ion mode PCA diagram
To further investigate the metabolic differences in ALI mice, the plasma and lung tissues from mice in the Con and LPS groups were analyzed by OPLS-DA. As shown in Fig. 3A–H, the samples of the Con and LPS groups were significantly separated in the positive and negative ion modes, indicating that the metabolism of the plasma and lung tissue of ALI mice was abnormal, the R2Y in the model was close to 1, and the Q2 was greater than 0.5, indicating that the model predictions were accurate and that the data were faithful (Additional file 2: Table S1). The permutation test showed that the models were no overfitting and reliable. Furthermore, the red dots in the S-plot away from the center might represent potential differential metabolites (Additional file 2: Fig. S6A–D).
Fig. 3.
OPLS-DA and permutation test analysis of mouse plasma and lung tissue. A Plasma positive ion mode OPLS-DA diagram; B plasma positive ion mode permutation test diagram; C plasma negative ion mode OPLS-DA diagram; D plasma negative ion mode permutation test diagram; E positive lung tissue ion mode OPLS-DA diagram; F lung tissue positive ion mode permutation test diagram; G lung tissue negative ion mode OPLS-DA diagram; H lung tissue negative ion mode permutation test diagram
Identification of potential biomarkers
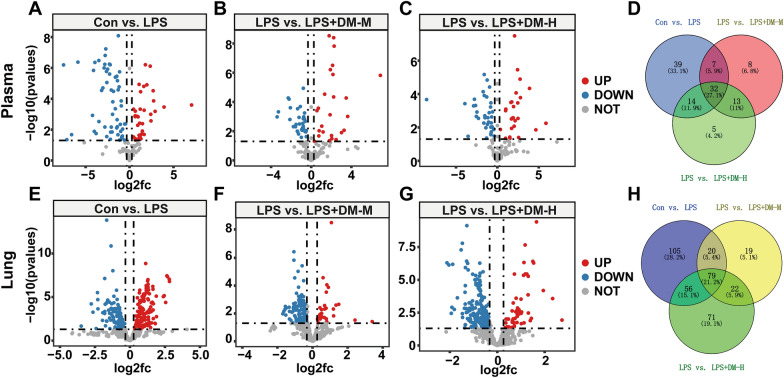
Based on a VIP > 1, p < 0.05, FC > 1.2 or FC < 0.8, in mouse plasma samples, 36 metabolites were upregulated and 59 were downregulated in the LPS group compared with the Con group (Fig. 4A). Compared with the LPS group, there were 25 upregulated and 36 downregulated metabolites in the LPS + DM-M group (Fig. 4B) and 27 upregulated and 39 downregulated metabolites in the LPS + DM-H group (Fig. 4C). The Venn diagram showed that there were 32 differential metabolites in the 4 groups of samples (Fig. 4D). In mouse lung tissue samples, 178 metabolites were upregulated, and 150 were downregulated in the LPS group compared with the control group (Fig. 4E). Compared with the LPS group, there were 37 upregulated and 140 downregulated metabolites in the LPS + DM-M group (Fig. 4F) and 68 upregulated and 217 downregulated metabolites in the LPS + DM-H group (Fig. 4G). The Venn diagram showed 79 differential metabolites in the 4 groups of samples (Fig. 4H).
Fig. 4.
Volcano gram analysis of differential metabolites in plasma, A Con vs. LPS; B LPS vs. LPS + DM-M; C LPS vs. LPS + DM-H; D Venn diagram of differential metabolites in plasma samples. Volcano gram analysis of differential metabolites lung tissue, E Con vs. LPS; F LPS vs. LPS + DM-M; G LPS vs. LPS + DM-H; H Venn diagram of differential metabolites in lung tissue samples
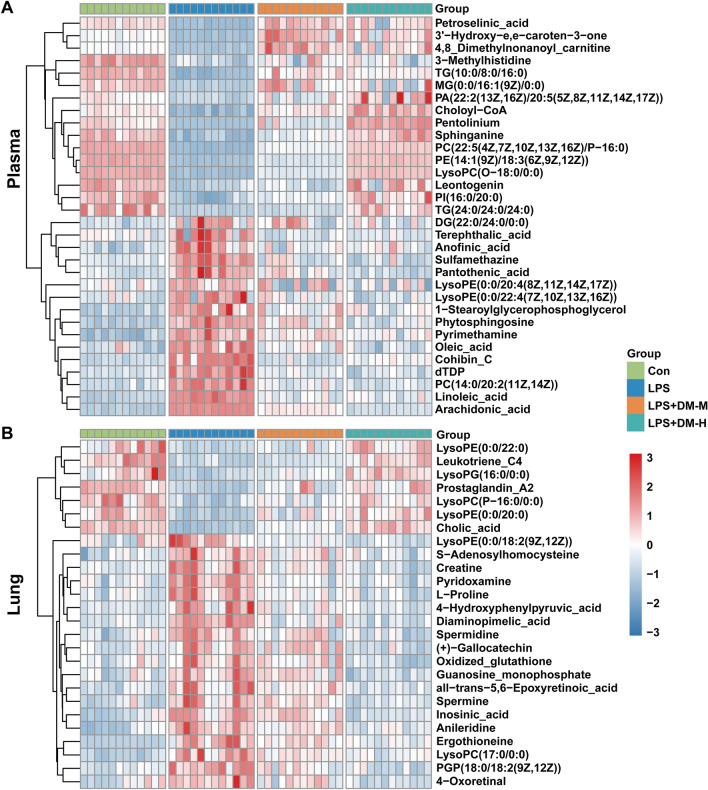
The differential metabolites were identified by HMDB, and 32 metabolites in the plasma (Table 1) and 26 metabolites in the lung tissues (Table 2) were screened. To observe the expression of differential metabolites in the four groups of samples, the differential metabolites were visualized with a heatmap analysis (Fig. 5A, B), in which the metabolites changed significantly between the control and LPS groups, and most of these were reversed in the DM treatment group.
Table 1.
The differential metabolites in plasma of mice
| NO. | Metabolites | Formula | Rt (min) | M/Z | VIP | Con vs. LPS | LPS vs.DM-M | LPS vs.DM-H | Fragmentations (m/z) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | FC | P | FC | P | FC | |||||||
| 1 | PI(20:0/16:0) | C45H87O13P | 17.262 | 866.3584 | 1.21076 | 5.05E-10 | 0.43788 | 2.42E-05 | 1.59392 | 3.64E-07 | 2.15291 | 255.2329; 627.3515 |
| 2 | PE(14:1(9Z)/18:3(6Z,9Z,12Z)) | C37H66NO8P | 2.452 | 701.4933 | 3.70328 | 4.73E-26 | 0.39028 | 6.18E-15 | 1.48864 | 6.24E-23 | 2.34811 | 565.4227; 663.3996 |
| 3 | Sphinganine | C18H39NO2 | 2.386 | 340.2589 | 1.26835 | 1.17E-12 | 0.37635 | 6.71E-08 | 1.84174 | 2.33E-11 | 2.69873 | 60.0443; 284.2973 |
| 4 | PC(22:5(4Z,7Z,10Z,13Z,16Z)/P-16:0) | C46H82NO7P | 2.506 | 814.5797 | 4.90904 | 1.68E-28 | 0.39893 | 1.52E-21 | 1.74587 | 9.63E-27 | 2.24652 | 184.0733; 792.5902 |
| 5 | Choloyl-CoA | C45H74N7O20P3S | 2.517 | 1148.7852 | 1.48302 | 3.66E-13 | 0.59524 | 4.62E-13 | 1.68328 | 8.87E-13 | 1.9727 | 1122.3784;1140.3889 |
| 6 | LysoPC(O-18:0/0:0) | C26H56NO6P | 2.64 | 510.3876 | 1.02389 | 4.10E-25 | 0.40162 | 3.97E-17 | 1.57946 | 9.81E-25 | 2.30119 | 104.0912; 510.3880 |
| 7 | Phytosphingosine | C18H39NO3 | 4.147 | 318.3007 | 2.15063 | 2.22E-10 | 2.62828 | 0.00088 | 0.76484 | 1.16E-08 | 0.52956 | 60.0452; 318.3008 |
| 8 | Pyrimethamine | C12H13C1lN4 | 5.389 | 248.1071 | 2.29407 | 6.02E-10 | 1.83568 | 0.00067 | 0.83932 | 1.74E-05 | 0.76639 | 198.0808; 233.0664 |
| 9 | Sulfamethazine | C12H14N4O2S | 7.174 | 279.0921 | 2.75416 | 1.94E-08 | 1.45655 | 7.71E-07 | 0.71174 | 6.10E-09 | 0.63607 | 124.0271; 149.0230 |
| 10 | Pantothenic acid | C9H17NO5 | 7.348 | 219.1324 | 1.06118 | 2.19E-06 | 1.57678 | 0.00023 | 0.74936 | 6.16E-07 | 0.595687 | 90.0550; 220.1178 |
| 11 | dTDP | C10H16N2O11P2 | 7.272 | 435.0549 | 1.00089 | 2.82E-08 | 2.1878 | 7.15E-08 | 0.52578 | 1.99E-09 | 0.441967 | 127.0586 |
| 12 | Linoleic acid | C18H32O2 | 11.983 | 280.3053 | 1.65539 | 2.77E-16 | 2.74883 | 4.69E-11 | 0.52958 | 4.61E-10 | 0.53843 | 41.0810; 263.1666 |
| 13 | PC(14:0/20:2(11Z,14Z)) | C42H80NO8P | 13.172 | 758.5703 | 2.46195 | 5.09E-10 | 1.9892 | 1.39E-08 | 0.59024 | 2.39E-09 | 0.52511 | 184.0732; 758.5725 |
| 14 | 3'-Hydroxy-e,e-caroten-3-one | C40H54O2 | 2.331 | 566.43 | 2.61367 | 0.000157 | 0.266704 | 0.019972 | 4.068175 | 3.50E-06 | 5.407536 | 287.1794; 349.2458 |
| 15 | TG(10:0/8:0/16:0) | C37H70O6 | 7.152 | 679.5111 | 3.82137 | 6.23E-07 | 0.216653 | 0.533412 | 1.454168 | 0.000318 | 3.093924 | 127.1117; 611.5245 |
| 16 | MG(0:0/16:1(9Z)/0:0) | C19H36O4 | 2.371 | 351.2506 | 1.18789 | 3.31E-07 | 0.088711 | 5.35E-05 | 11.18208 | 0.039996 | 5.920102 | 351.2506 |
| 17 | 4,8 Dimethylnonanoyl carnitine | C18H35NO4 | 2.435 | 352.2458 | 1.15007 | 1.70E-06 | 0.161729 | 0.037749 | 5.573445 | 0.102297 | 3.884997 | 85.0286; 330.2638 |
| 18 | TG(24:0/24:0/24:0) | C75H146O6 | 2.527 | 1143.8027 | 1.9883 | 0.0043 | 0.680223 | 2.95E-09 | 3.449353 | 3.28E-08 | 4.221253 | 775.7538; 1144.1192 |
| 19 | 3-Methylhistidine | C7H11N3O2 | 5.389 | 867.1071 | 2.29407 | 0.001348 | 1.774902 | 0.054547 | 0.701298 | 0.009556 | 0.577705 | 865.5811 |
| 20 | Terephthalic acid | C8H6O4 | 7.19 | 149.0231 | 1.94229 | 0.03816 | 1.556249 | 0.00322 | 0.444208 | 0.004775 | 0.47752 | 121.0295 |
| 21 | Anofinic acid | C12H12O3 | 7.19 | 205.0859 | 1.49016 | 1.16E-05 | 2.118335 | 0.00057 | 0.63091 | 0.001171 | 0.665832 | 187.0754; 205.0859 |
| 22 | 1-Stearoylglycerophosphoglycerol | C24H49O9P | 9.283 | 530.3452 | 2.62522 | 0.012386 | 2.746449 | 0.00105 | 0.597754 | 0.855365 | 1.057998 | 267.2682; 341.3050 |
| 23 | DG(22:0/24:0/0:0) | C49H96O5 | 9.153 | 763.2448 | 1.08291 | 0.003817 | 2.208159 | 0.175033 | 0.71258 | 0.024067 | 0.57913 | 397.3676; 425.3989 |
| 24 | Pentolinium | C15H32N2 | 9.578 | 263.2458 | 1.30865 | 0.00047 | 2.313994 | 0.004618 | 0.577774 | 0.001309 | 0.503244 | 96.0810; 1,140,920 |
| 25 | Cohibin C | C37H68O4 | 15.859 | 577.519 | 1.06788 | 0.000343 | 15.15793 | 0.010844 | 0.391416 | 0.00119 | 0.180381 | 81.0355; 541.4979 |
| 26 | PA(22:2(13Z,16Z)/20:5(5Z,8Z,11Z,14Z,17Z)) | C45H75O8P | 17.227 | 778.7852 | 1.840872 | 2.64E-05 | 0.406488 | 0.004533 | 2.07273 | 0.002669 | 3.610348 | 699.5323; 797.5092 |
| 27 | Leontogenin | C27H42O5 | 17.217 | 464.3371 | 1.464973 | 2.50E-06 | 0.207219 | 0.096437 | 1.677627 | 0.045176 | 2.848881 | 4.0386; 429.2999 |
| 28 | PI(16:0/20:0) | C45H87O13P | 2.242 | 865.3286 | 1.815752 | 4.00E-06 | 0.325564 | 0.011288 | 2.267752 | 0.002873 | 5.618772 | 311.2955; 571.2889 |
| 29 | LysoPE(0:0/20:4(8Z,11Z,14Z,17Z)) | C25H44NO7P | 6.098 | 524.2748 | 4.70183 | 0.000472 | 1.368666 | 0.055892 | 0.7098 | 0.002355 | 0.620224 | 44.0495; 361.2737 |
| 30 | LysoPE(0:0/22:4(7Z,10Z,13Z,16Z)) | C27H48NO7P | 6.346 | 552.3022 | 1.5285 | 0.001548 | 1.62543 | 0.0153 | 0.72558 | 0.00071 | 0.57564 | 389.3050; 487.2819 |
| 31 | Arachidonic acid | C20H32O2 | 10.543 | 327.2306 | 4.0233 | 2.86E-23 | 1.75205 | 7.40E-15 | 0.79066 | 4.25E-22 | 0.64154 | 303.2335 |
| 32 | Oleic acid | C18H34O2 | 10.654 | 283.2625 | 1.01725 | 6.71E-07 | 1.82879 | 7.30E-06 | 0.66678 | 3.83E-08 | 0.53553 | 69.0000; 247.0245 |
Table 2.
The differential metabolites in lung tissue of mice
| NO. | Metabolites | Formula | Rt (min) | M/Z | VIP | Con vs. LPS | LPS vs.DM-M | LPS vs.DM-H | Fragmentations (m/z) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | FC | P | FC | P | FC | |||||||
| 1 | Ergothioneine | C9H15N3O2S | 0.94 | 230.0975 | 6.85519 | 8.89E-06 | 5.223586 | 0.028336 | 0.722204 | 0.000399 | 0.484999 | 69.0580; 127.0362 |
| 2 | ( +)-Gallocatechin | C15H14O7 | 1.07 | 307.0829 | 5.01184 | 3.85E-14 | 1.539765 | 2.77E-09 | 0.776547 | 9.53E-09 | 0.573716 | 139.0390; 307.08113 |
| 3 | Oxidized glutathione | C20H32N6O12S2 | 1.08 | 635.142 | 1.24098 | 0.001474 | 1.349826 | 0.063388 | 0.857859 | 1.44E-05 | 0.603938 | 355.0759; 484.1187 |
| 4 | Inosinic acid | C10H13N4O8P | 0.97 | 349.056 | 3.46688 | 1.50E-06 | 2.506744 | 0.06727 | 0.82939 | 1.75E-06 | 0.426869 | 97.0298; 137.0457 |
| 5 | Guanosine monophosphate | C10H14N5O8P | 0.95 | 364.0649 | 3.05321 | 0.00017 | 1.608101 | 0.084982 | 0.856493 | 4.66E-06 | 0.533426 | 152.0562 |
| 6 | S-Adenosylhomocysteine | C14H20N6O5S | 1.05 | 385.1303 | 2.64985 | 2.61E-05 | 1.629015 | 0.016513 | 0.811106 | 0.00022 | 0.686493 | 112.0183; 170.0849 |
| 7 | all-trans-5,6-Epoxyretinoic acid | C20H28O3 | 13.52 | 317.2117 | 2.30797 | 0.000264 | 1.933585 | 0.045855 | 0.778557 | 0.000488 | 0.550082 | 79.0542; 281.1900 |
| 8 | Creatine | C4H9N3O2 | 0.91 | 132.0775 | 1.79766 | 6.98E-05 | 1.485564 | 0.002392 | 0.766444 | 5.19E-06 | 0.595529 | 44.1000; 55.2000 |
| 9 | LysoPC(17:0/0:0) | C25H52NO7P | 18.32 | 527.3804 | 1.36725 | 7.89E-06 | 3.681324 | 0.018519 | 0.721019 | 0.000698 | 0.545856 | 184.0733; 492.3449 |
| 10 | Pyridoxamine | C8H12N2O2 | 0.78 | 191.0781 | 1.1411 | 1.63E-06 | 1.624016 | 3.72E-06 | 0.662852 | 5.36E-07 | 0.599696 | 134.0606; 151.0871 |
| 11 | Spermidine | C7H19N3 | 0.72 | 146.1654 | 1.13866 | 1.21E-05 | 1.445056 | 0.007625 | 0.818006 | 2.44E-06 | 0.659883 | 72.0820; 84.0840 |
| 12 | L-Proline | C5H9NO2 | 0.91 | 116.0703 | 1.06339 | 3.29E-06 | 1.593842 | 4.40E-05 | 0.70307 | 3.14E-06 | 0.605171 | 70.0651; 41.0386 |
| 13 | Spermine | C10H26N4 | 13.76 | 241.18 | 1.05343 | 4.59E-05 | 2.891314 | 0.077649 | 0.785466 | 8.89E-05 | 0.389801 | 58.0657; 186.1970 |
| 14 | 4-Hydroxyphenylpyruvic acid | C9H8O4 | 3.68 | 181.0491 | 1.19548 | 0.000492 | 2.762776 | 0.011312 | 0.58101 | 0.000701 | 0.392195 | 77.0391; 107.0497 |
| 15 | Diaminopimelic acid | C7H14N2O4 | 0.79 | 213.0852 | 1.8311 | 1.40E-09 | 2.180373 | 9.35E-06 | 0.565259 | 7.44E-10 | 0.411171 | 82.0652; 128.0705 |
| 16 | LysoPC(P-16:0/0:0) | C24H50NO6P | 19 | 480.3413 | 1.87704 | 1.64E-06 | 0.514088 | 0.003854 | 1.339187 | 3.65E-06 | 1.649673 | 180.0188; 462.3343 |
| 17 | Leukotriene C4 | C30H47N3O9S | 12.14 | 626.3128 | 1.31377 | 7.35E-08 | 0.286374 | 0.024872 | 1.351636 | 3.93E-07 | 2.82934 | 189.1638; 319.2268 |
| 18 | LysoPE(0:0/18:2(9Z,12Z)) | C23H44NO7P | 16.79 | 478.2935 | 1.615482 | 0.002276 | 1.730487 | 0.000351 | 0.530287 | 2.76E-05 | 0.393158 | 263.2369; 337.2737 |
| 19 | PGP(18:0/18:2(9Z,12Z)) | C42H80O13P2 | 29.14 | 872.5415 | 1.854269 | 3.27E-06 | 2.225789 | 0.000646 | 0.628777 | 1.16E-06 | 0.394302 | 603.5347; 757.5378 |
| 20 | Anileridine | C22H28N2O2 | 18.7 | 353.2236 | 1.5043 | 4.90E-07 | 2.232817 | 0.004675 | 0.783746 | 1.08E-05 | 0.600209 | 120.0808; 279.1856 |
| 21 | 4-Oxoretinal | C20H26O2 | 13.32 | 299.2014 | 1.25101 | 0.000157 | 1.724575 | 0.008411 | 0.73029 | 3.73E-06 | 0.487338 | 137.0961; 189.1274 |
| 22 | LysoPE(0:0/20:0) | C25H52NO7P | 19.76 | 510.3511 | 2.43664 | 9.46E-09 | 0.428477 | 0.000292 | 1.721601 | 4.79E-07 | 2.180142 | 44.0495; 369.3363 |
| 23 | LysoPE(0:0/22:0) | C27H56NO7P | 23.79 | 538.3835 | 1.752036 | 0.000226 | 0.435181 | 0.009919 | 1.432045 | 3.59E-06 | 2.272196 | 44.0495; 397.3676 |
| 24 | LysoPG(16:0/0:0) | C22H45O9P | 21.28 | 483.2727 | 4.0505 | 0.000257 | 0.438891 | 2.68E-05 | 1.515575 | 2.20E-08 | 2.256267 | 483.2728 |
| 25 | Cholic acid | C24H40O5 | 13.08 | 407.2826 | 2.24678 | 1.49E-14 | 0.315424 | 2.92E-09 | 2.107035 | 3.85E-10 | 3.146423 | 345.2794; 373.2743 |
| 26 | Prostaglandin A2 | C20H30O4 | 14.39 | 333.2069 | 1.57342 | 1.37E-11 | 0.391936 | 0.004173 | 1.707462 | 4.52E-06 | 2.156729 | 67.0542; 299.2006 |
Fig. 5.
The expression of differential metabolites in mouse plasma (A) and lung tissue (B)
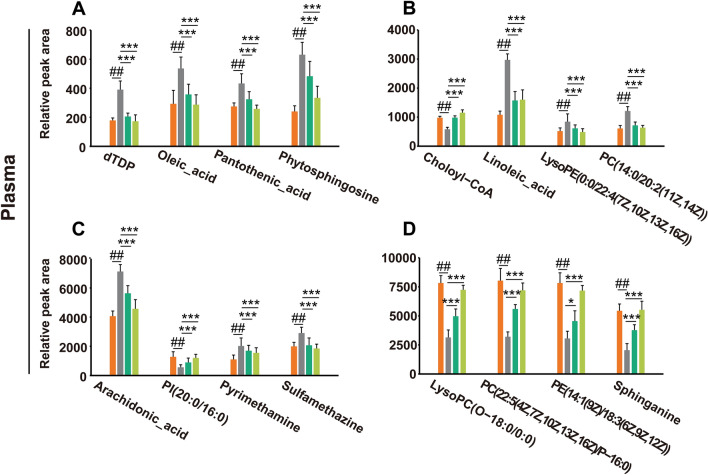
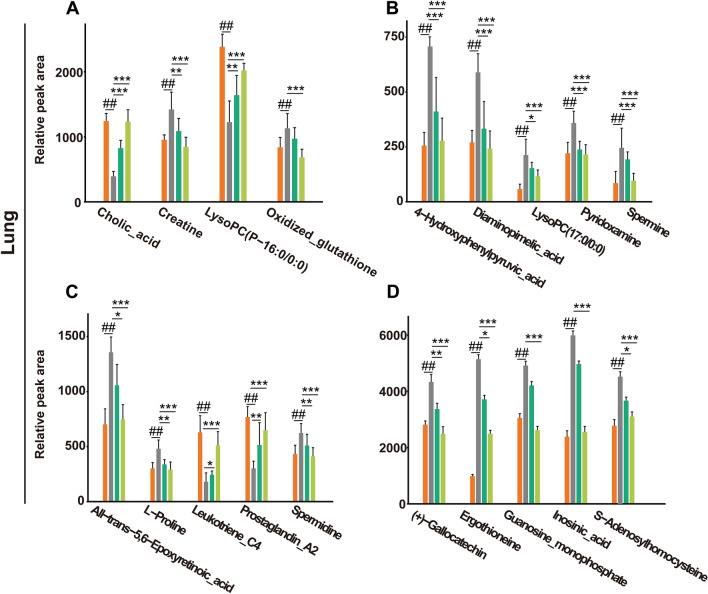
A total of 16 potential metabolites in mouse plasma samples were identified (Fig. 6A–D). Compared with the control group, phytosphingosine, pyrimethamine, sulfamethazine, pantothenic acid, dTDP, linoleic acid, PC(14:0/20:2(11Z,14Z)), LysoPE (0:0/22:4(7Z,10Z,13Z,16Z)), arachidonic acid and oleic acid were significantly upregulated in the LPS group, whereas sphinganine, Choloyl-CoA, PI(20:0/16:0), PE(14:1(9Z)/18:3(6Z,9Z,12Z)), PC(22:5(4Z,7Z,10Z,13Z,16Z)/P-16:0), and LysoPC (O-18:0/0:0) were downregulated. Moreover, as shown in Fig. 7A–D, a total of 19 potential metabolites in the lung tissues were identified in which ergothioneine, ( +)-gallocatechin, oxidized glutathione, inosinic acid, guanosine monophosphate, S-adenosylhomocysteine, all-trans-5,6-epoxyretinoic acid, creatine, lysoPC(17:0/0:0), pyridoxamine, spermidine, L-proline, spermine, 4-hydroxyphenylpyruvic acid, and diaminopimelic acid were upregulated in the LPS group compared with those in the control group, and lysoPC (P-16:0/0:0), leukotriene C4, cholic acid and prostaglandin A2 were downregulated. After treatment with DM, the levels of these potential metabolites returned to normal, which indicated that DM treatment could effectively improve the metabolic disorder of ALI mice.
Fig. 6.
The ionic strength of potential metabolites in mouse plasma (A–D). Data are displayed as Mean ± SD. # P < 0.05, ## P < 0.01 vs. control group, *P < 0.05, **P < 0.01 vs. LPS group
Fig. 7.
The ionic strength of potential metabolites in mouse lung tissues (A–D). Data are displayed as Mean ± SD. #P < 0.05, ##P < 0.01 vs. control group, *P < 0.05, **P < 0.01 vs. LPS group
Metabolic pathway analysis
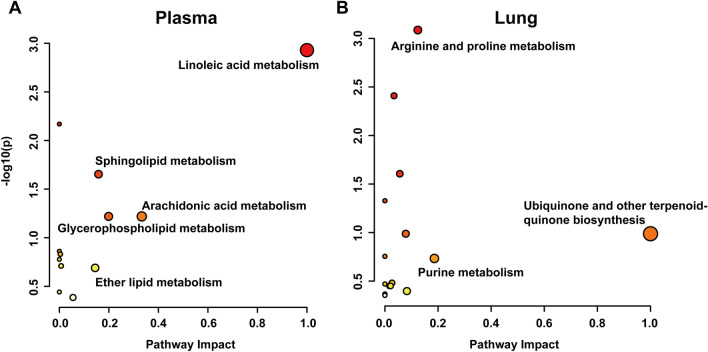
The candidate metabolites were imported into the MetaboAnalyst database for metabolic pathway enrichment analysis, and 12 metabolic pathways were found in plasma and 15 were found in lung tissue, among which glycerophospholipid metabolism, arachidonic acid metabolism, and primary bile acid biosynthesis were shared in mouse plasma and lung tissue (Additional file 2: Table S2). Based on a pathway impact > 0.1, linoleic acid metabolism, arachidonic acid metabolism, glycerophospholipid metabolism, sphingolipid metabolism, and ether lipid metabolism were significantly affected in the plasma (Fig. 8A). Moreover, ubiquinone and other terpenoid quinone biosynthesis, purine metabolism, and arginine and proline metabolism were notably affected in the lung tissue (Fig. 8B).
Fig. 8.
Enrichment analysis of potential metabolic pathways in mouse plasma (A) and lung tissue (B)
Network pharmacology analysis
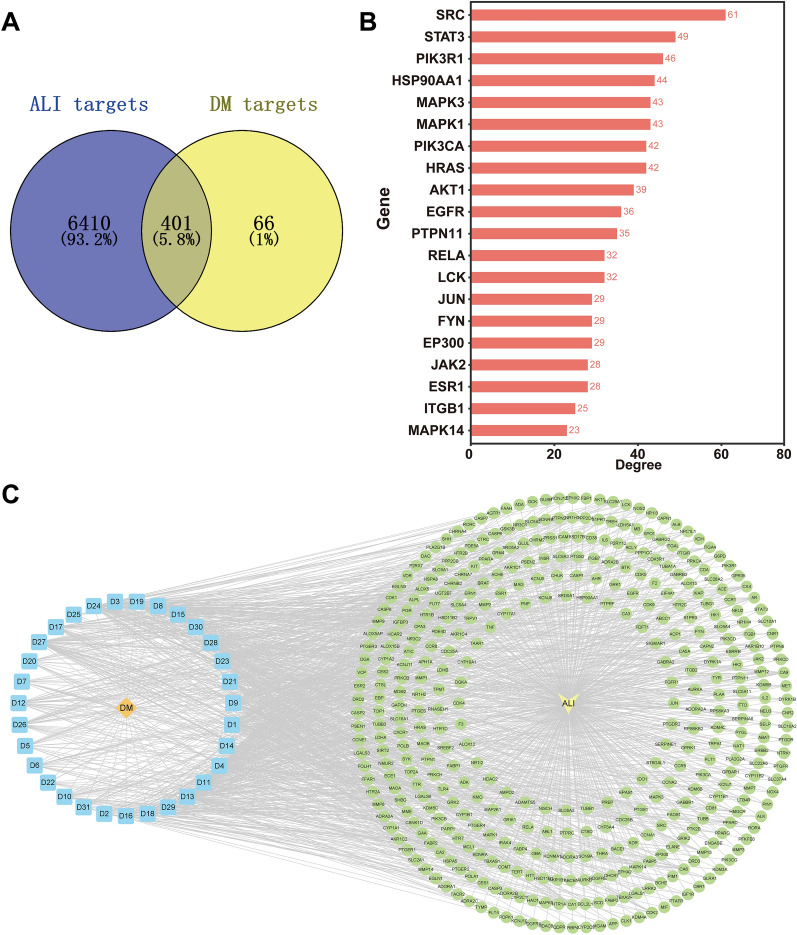
To further explore the mechanism of DM against ALI, we performed a network pharmacology analysis. First, we searched the literature for 166 chemical constituents in DM, mainly alkaloids, phenolic acids, glycosides, terpenoids, flavonoids, and other compounds (Additional file 3: Table S3). Next, 467 targets related to 31 active components of DM were collected from BATMAN-TCM, DrugBank and Swiss Target Prediction. Subsequently, 6811 related targets of ALI were obtained from the DrugBank, DisGeNET, OMIM, and TTD databases. Ultimately, 401 targets related to DM and ALI were identified as potential targets of DM to treat ALI (Fig. 9A).
Fig. 9.
Network pharmacological analysis of DM in the treatment of ALI. A Shared targets of DM and ALI. B Core genes for DM treatment of ALI. C “DM- active components -target-ALI” network, orange: DM, blue: active components (31), yellow: ALI, green: shared target (401)
To obtain the protein–protein interaction relationship between DM active components and ALI targets, the 401 targets were imported into the STRING database for PPI network analysis, and the top 20 core genes were screened (Fig. 9B). The common targets of DM active components and ALI were imported into Cytoscape 3.9.1 software to construct the “DM-active ingredient-target-ALI” network. Figure 9C shows the 31 active components and 401 potential targets of DM in the treatment of ALI, and the importance of active components was evaluated by degree, betweenness, and closeness in the network (Additional file 3: Table S4).
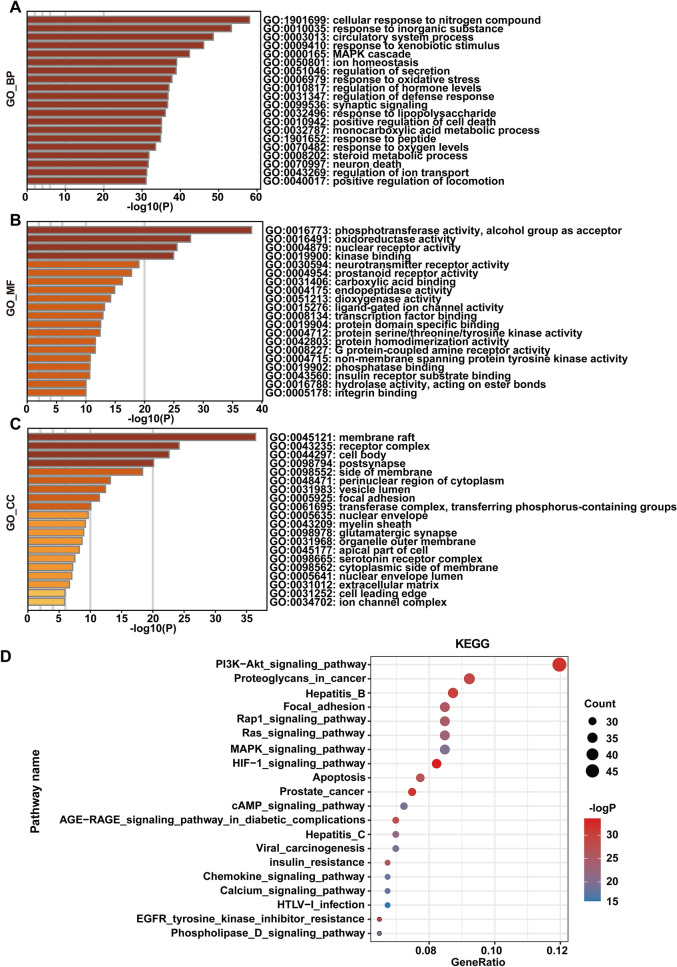
To predict the therapeutic mechanism of DM on ALI, we carried out GO and KEGG enrichment analyses, in which the GO enrichment analysis included biological processes (BP), cellular components (CC) and molecular functions (MF), which explained the biological function of genes at different levels. Biological processes mainly included cellular response to nitrogen compounds, the MAPK cascade, response to oxidative stress, etc. (Fig. 10A). Molecular functions mainly included phosphotransferase activity, the alcohol group as acceptor, oxidoreductase activity, nuclear receptor activity, etc. (Fig. 10B). The cell components mainly included the membrane raft, receptor complex, cell body, etc. (Fig. 10C). According to the KEGG enrichment analysis, 20 pathways were significantly affected, including the PI3K-Akt signaling pathway, MAPK signaling pathway, HIF-1 signaling pathway, and apoptosis, of which PI3K/Akt was identified as a potential pathway for DM to treat ALI (Fig. 10D).
Fig. 10.
Enrichment analysis of GO and KEGG pathways. A biological process B molecular function C cell composition D KEGG enrichment analysis
Comprehensive analysis of metabolomics and network pharmacology
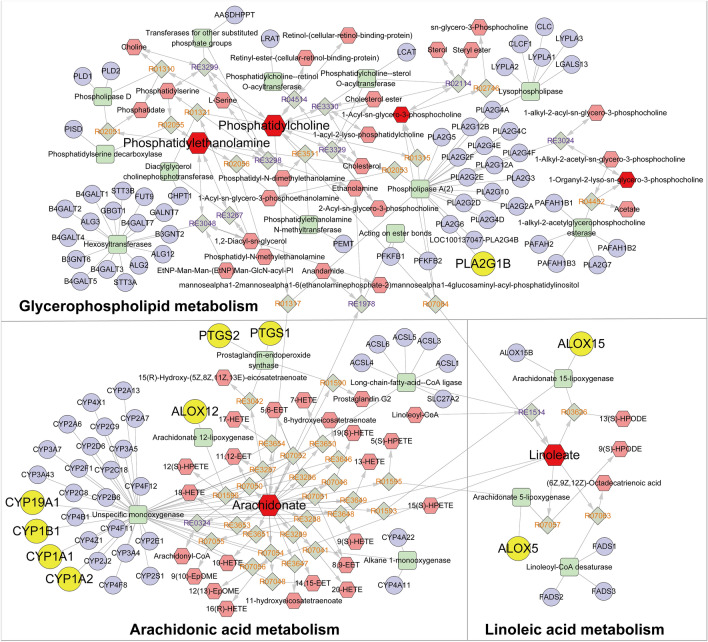
Metabolomics and network pharmacology were integrated to construct an interaction network. First, we introduced candidate metabolites from plasma and lung tissues into MetScape to construct a “Metabolite-Reaction-Enzyme-Gene” network (Fig. 11). Additionally, we matched the genes associated with the candidate metabolites with the potential targets of network pharmacology and found 10 key targets, including PTGS2, PLA2G1B, CYP1A2, ALOX15, PTGS1, ALOX5, ALOX12, CYP1A1, CYP1B1, and CYP19A1 (Additional file 5: Fig. S7). The affected pathways were arachidonic acid metabolism, linoleic acid metabolism, and glycerophospholipid metabolism. These pathways may be involved in inflammation and cell apoptosis.
Fig. 11.
“Potential metabolite-reaction-enzyme-gene” interaction network. Red hexagons represent metabolites, gray diamonds represent reactions, green rectangles represent enzymes, and purple circles represent genes
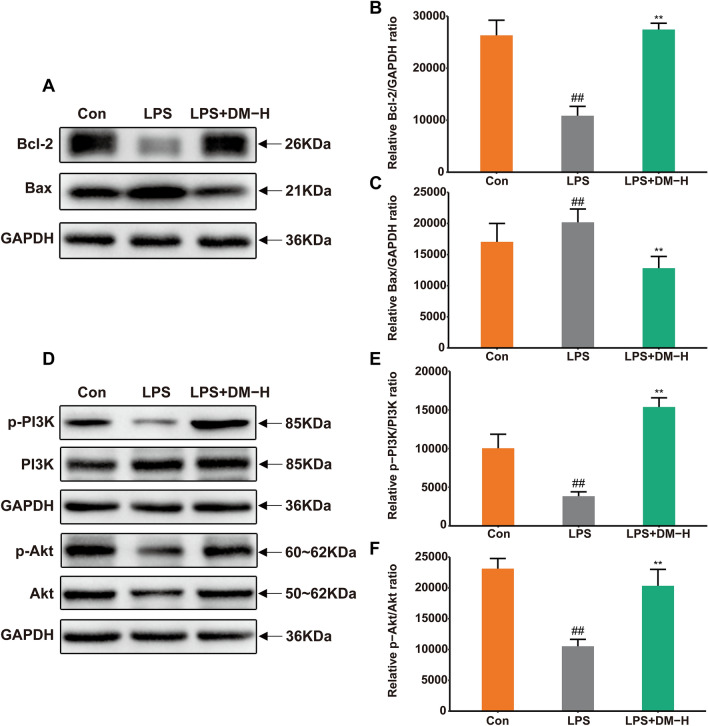
Biological experimental verification
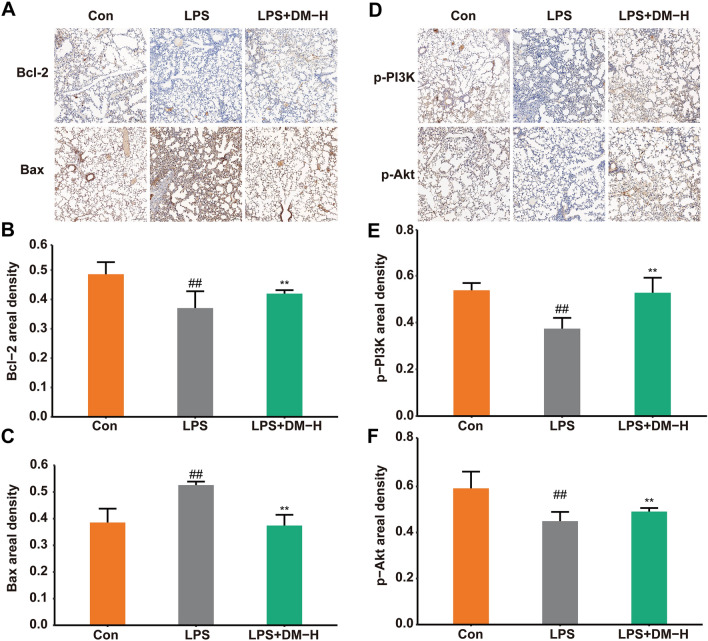
To evaluate the accuracy of the method and results of the integrated analysis of network pharmacology and metabolomics, we used immunohistochemistry and western blot methods to determine the proteins (PI3K/Akt and Bax/Bcl-2) associated with inflammation and cell apoptosis as validation.
Immunohistochemistry was used to investigate the effect of DM on LPS-induced ALI mouse lung tissue apoptosis. As shown in Fig. 12A–C, the expression of Bcl-2 was downregulated, and Bax was upregulated after LPS stimulation. However, after DM treatment, the expression of Bcl-2 was promoted, and Bax was inhibited, indicating that DM may have an antiapoptotic effect on LPS-induced ALI mice. The PI3K/Akt pathway regulates cell proliferation, apoptosis, and the release of anti-inflammatory cytokines. Therefore, we also examined the phosphorylation levels of PI3K and Akt in the lung tissue of ALI mice, which were downregulated after LPS induction but were significantly reversed by DM treatment (Fig. 12D–F). It was suggested that DM might have a therapeutic effect on ALI by activating PI3K/Akt.
Fig. 12.
Cell proliferation and apoptosis detection of LPS-induced ALI. A Immunohistochemistry staining of Bcl-2 and Bax proteins (magnification 200 ×). The expression of Bcl-2 (B) and Bax (C) in lung tissue. D Immunohistochemistry staining of p-PI3K and p-Akt proteins (magnification 200 ×). The expression of p-PI3K (E) and p-Akt (F) in lung tissue. Data are displayed as Mean ± SD. #P < 0.05, ##P < 0.01 vs. control group, *P < 0.05, **P < 0.01 vs. LPS group
To further verify whether DM could regulate apoptosis in LPS-induced ALI mice, we detected the levels of Bcl-2 and Bax in the lung tissue of ALI mice by western blotting. After LPS stimulation, the expression of Bcl-2 was downregulated, and Bax was significantly upregulated. After DM treatment, the level of Bcl-2 was significantly increased, and Bax was significantly decreased (Fig. 13A–C). The results revealed that DM could inhibit LPS-induced apoptosis. Furthermore, we also detected the expression of PI3K/Akt in lung tissue. LPS caused a decrease in the phosphorylation levels of PI3K and Akt, whereas DM treatment significantly increased their expression (Fig. 13D–F). These data confirm that DM could effectively ameliorate LPS-induced ALI by activating PI3K/Akt.
Fig. 13.
The proliferation and apoptosis of ALI cells were detected by western blotting. A Images of Bcl-2 and Bax proteins. The expression of Bcl-2 (B) and Bax (C) in lung tissue. D Images of p-PI3K and p-Akt proteins. The expression of p-PI3K E and p-Akt F in lung tissue. Data are displayed as the mean ± SD. #P < 0.05, ##P < 0.01 vs. control group and *P < 0.05, **P < 0.01 vs. LPS group
Discussion
Nauclea officinalis is involved in the regulation of the inflammatory response, oxidative stress, immune regulation, and cell proliferation [29]. These activities are closely related to the occurrence and development of ALI. Previous studies have demonstrated that DM could inhibit LPS-induced secretion of NO, TNF-α, IL-1β, and IL-6 and inhibit the phosphorylation levels of NF-κ B, p38, ERK, and JNK, showing outstanding anti-inflammatory activity [30]. In this study, we found that DM could maintain the structural integrity of lung tissue in ALI mice, specifically by inhibiting the infiltration of inflammatory cells and improving alveolar wall thickening, alveolar collapse, and hemorrhagic characteristics. These results reveal that DM could effectively improve lung injury.
In the metabolomic analysis, samples of diseased organs are typically collected for subsequent analysis. From a physiological point of view, the lung is the main diseased organ, and the blood is rich in small molecule metabolites. Changes in blood components can reflect the physiological and pathological changes of the organ. When the body is diseased, the inflammatory mediators in the blood are transferred to the lungs and attack lung tissue. Hence, this experiment chose blood and lung tissues as ideal specimens for metabolic analysis. We employed UPLC-QTOF-MS/MS to characterize and analyze endogenous metabolites in a DM-treated mouse model of LPS-induced ALI and found 16 candidate metabolites in plasma and 19 candidate metabolites in lung tissue. These metabolites are mainly related to metabolic pathways such as linoleic acid metabolism, arachidonic acid metabolism, glycerophospholipid metabolism, sphingolipid metabolism, purine metabolism, and arginine and proline metabolism.
Fatty acid metabolism is an essential component of biofilms and can supply energy for the body and play a prominent role in inhibiting inflammation, resisting bacterial and viral infections, and immune regulation [31]. Linoleic acid (LA) cannot be synthesized in the body and is an essential fatty acid. Moreover, LA produces arachidonic acid (AA), which can produce prostaglandins (PGs) and leukotrienes (LTs) under the action of cyclooxygenase (COX). Studies have shown that increasing LA intake may decrease the risk of pneumonia [32]. Moreover, AA has activity against bacteria, fungi, and influenza viruses, whereas alveolar macrophages can release AA and other unsaturated fatty acids into alveolar fluid, thereby exerting their antibacterial impact and protecting the lungs from various microbial infections [33]. The content of unsaturated fatty acids in ALI may change with the pathological state. One study found that in the LPS-induced ALI mouse model, LA had different trend changes in serum, lung, BALF and spleen [14]. Similarly, our study also demonstrated that the contents of LA and AA in the plasma of the ALI model group were increased, whereas the contents of PGA2 and LTC4 in the lung tissues were decreased. The conversion of excess LA to AA indicated that ALI mice were in an inflammatory state, and downregulation of PGA2 and LTC4 in lung tissues may be associated with changes in COX activity in disease states. Although PGs are proinflammatory factors, they have both pro- and anti-inflammatory effects. PGs may trigger the inflammatory process in the early stage of the disease. However, when the concentration of PGs reaches a sufficient level and the inflammatory process is at an optimal level, the anti-inflammatory process is initiated by increasing the synthesis of active anti-inflammatory lipids [34, 35]. In addition, other fatty acids, such as pantothenic acid and oleic acid, were observed to have increased in the mouse plasma samples of the model group. Pantothenic acid has anti-lipid peroxidation effects [36], whereas oleic acid plays a role in the inflammatory response [37]. According to reports, fatty acids can protect the body from disease by regulating various inflammatory cells, and cytokines can promote the production of fatty acids [38]. Some fatty acids can also inhibit apoptosis and reduce oxidative stress [39]. Additionally, we also found that the specific amount and type of glycerophospholipid changes in LPS-induced ALI mice differed in plasma and lung tissue samples [15]. Disturbances in glycerophospholipid metabolism are mainly affected by the activity of phospholipase A2, which in turn is associated with inflammation [40].
Sphingomyelin is involved in numerous biological processes such as cell growth and differentiation, regulation of membrane stability, apoptosis, and angiogenesis. Furthermore, studies have indicated that sphingomyelin degradation products are involved in the pathophysiological process of ALI [41]. Sphingomyelin regulates PI3K/Akt to improve vascular endothelial permeability, reduce vascular inflammation, and maintain vascular endothelial integrity [42]. Sphingosine is an essential mediator of pulmonary edema and alveolar damage, and it maintains alveolar stability by altering lung-damaging surfactants [31]. Our results showed that the levels of sphingosine in ALI mouse plasma were decreased, but the sphingosine levels were reversed after DM treatment. These data suggested that DM could activate sphingosine in LPS-induced ALI. Furthermore, arginine is involved in tissue repair, inflammation, and the secretion of various hormones [43]. It is a precursor for synthesizing nitric oxide, urea, polyamines, proline, glutamate, and creatine [44]. Elevation of proline in LPS-induced ALI results in increased neutrophil recruitment and protein leakage, suggesting that it mediates lung inflammation in ALI [45]. In our study, the L-proline levels in ALI mouse lung tissue were increased, and the expression was downregulated after treatment with DM, indicating that DM could alleviate ALI by inhibiting the increase in neutrophil recruitment and protein leakage during lung inflammation.
Our study identified 31 potentially active components of DM against ALI using network pharmacology analysis, among which strictosamide is the most abundant indole alkaloid in DM [8]. Strictosamide has excellent anti-inflammatory and analgesic activities, and its anti-inflammatory effect is related to the inhibition of the NF-κ B and MAPK signaling pathways [46]. Our previous study also indicated that strictosamide has favorable proliferation activity and angiogenesis effects on HUVECs. In this study, we found 20 core genes for DM treatment of ALI, which were mainly involved in cell growth, proliferation, differentiation and apoptosis; angiogenesis; and inflammatory responses. STAT3 can stimulate various growth factors, such as IFN, EGF, IL5, and IL6, and play a role in cell growth and apoptosis. However, this gene is also involved in coronavirus biology, immune response, and antiviral activity. Previous studies have shown that LPS-induced ALI can be improved by regulating the MAPK and IL-6/STAT3 signaling pathways [47, 48]. The MAPK kinase family is involved in various cellular processes such as cell proliferation, differentiation, and cell cycle progression. Studies have also shown that regulation of caspase-3, Bax/Bcl-2 and MAPK signaling could attenuate microvascular endothelial cell injury and alleviate inflammation and pulmonary edema in ALI mice while maintaining the integrity of alveolar structures [49]. Moreover, PI3K/Akt is involved in multifarious biological processes, including the regulation of cell proliferation, apoptosis, and the release of anti-inflammatory cytokines [50]. Our preceding studies have also confirmed that DM might repair the endothelial cell barrier of lung injury by activating the PI3K/Akt signaling pathway, inhibiting the inflammatory response, improving intercellular connections, and promoting angiogenesis. Therefore, it was determined that PI3K/Akt is the main influencing pathway of DM in the treatment of ALI.
The integrated analysis of metabolomics and network pharmacology found 10 key targets (PTGS2, PLA2G1B, CYP1A2, ALOX15, PTGS1, ALOX5, ALOX12, CYP1A1, CYP1B1, and CYP19A1) and 3 related pathways (arachidonic acid metabolism, linoleic acid metabolism, and glycerophospholipid metabolism). This analytical strategy is also suitable for screening metabolites and targets in other natural compounds. PTGS2, also known as cyclooxygenase, can catalyze AA to generate PGs, which are mainly involved in inflammation, cell proliferation and apoptosis. In addition, PTGS1 can regulate angiogenesis in endothelial cells [51]. PLA2G1B is a phospholipase A2 that catalyzes membrane glycerophospholipids to release AA and Lys phospholipids, which are involved in cell contraction and proliferation. The hydrolysis of surfactant phospholipids (PL) by phospholipase A2 results in surfactant damage in ALI/ARDS [52]. ALOX15, ALOX5, and ALOX12 are all members of the lipoxygenase protein family that can act on AA and LA and affect inflammation, immunity, cell proliferation, apoptosis, and angiogenesis [53]. CYP1A1, CYP1A2, CYP1B1, and CYP19A1 are members of the cytochrome P450 family and have a therapeutic effect on hyperoxia-induced lung injury [54]. After LPS stimulation, the levels of the AA-related genes CBR2, CYP4F18 and CYP2E1 were downregulated, whereas ALOX12, PTGES and PTGES2 were upregulated. Studies have shown that LPS-induced ALI can be alleviated by regulating AA, anti-apoptotic and anti-inflammatory effects [33]. In brief, integrated network pharmacology and metabolomic studies revealed that the effect of DM on reversing ALI is mainly mediated by regulating arachidonic acid metabolism, linoleic acid metabolism, and glycerophospholipid metabolism. Furthermore, these actions might be closely related to the regulation of the inflammatory response, cell proliferation and apoptosis.
Apoptosis of lung cells plays a pivotal role in ALI/ARDS development [55, 56]. Therefore, it is urgent to solve the problem of apoptosis in ALI/ARDS. Apoptosis of lung endothelial and epithelial cells might initiate or contribute to the progression of many lung diseases [57]. The balance of survival and death of lung endothelial cells is essential for angiogenesis, degeneration, and barrier function. Lung endothelial cells can be damaged by hypoxia or LPS stimulation. However, VEGF is abundant in endothelial cells and can promote endothelial cell survival and maintain normal alveolar structure. [1]. Our previous study also demonstrated that DM could promote the expression of VEGF in lung tissue, thereby protecting lung endothelial cells from apoptosis. The Bcl-2 protein family is an essential regulator of apoptotic cells [58, 59]. Bax is a crucial executor of mitochondrial regulation of cell death, and its dysregulation guides abnormal cell death [60]. PI3K/Akt plays a vital role in regulating anti-inflammatory cytokines, immune regulation, extracellular matrix degradation, proliferation, and apoptosis [61]. Studies have confirmed that the alleviation of lung injury is related to activation of the PI3K/Akt pathway, downregulation of proinflammatory cytokines, and regulation of apoptosis-related proteins (downregulation of caspase-3, caspase-9 and Bax and upregulation of Bcl-2) [62]. In our study, DM preconditioning also downregulated Bax, upregulated Bcl-2 and activated the PI3K/Akt signaling pathway. Therefore, it was revealed that DM could significantly alleviate lung injury by activating PI3K/Akt, promoting proliferation and regulating apoptosis. These findings lay the foundation for further in-depth exploration of the pathological mechanisms of and interventional strategies for lung injury.
Conclusion
In this study, we first investigated the possible mechanism of DM in treating ALI through a comprehensive strategy combining metabolomics, network pharmacology and molecular biology experiments. The integrated analysis of metabolomics and network pharmacology predicted that DM could treat ALI mainly by improving the inflammatory response and regulating cell apoptosis. The accuracy of the strategy and predictive results were preliminarily validated by biological experiments. The results confirmed that integrated metabolomics and network pharmacology analysis to predict the potential mechanism of DM treatment of ALI is accurate and feasible. This comprehensive strategy provides a more precise direction for exploring the treatment of ALI with natural medicines.
Supplementary Information
Additional file 1: Fig. S1. The stem of Nauclea officinalis Pierre ex Pitard (Danmu in Chinese, DM). Fig. S2. HPLC analysis of DM.
Additional file 2: Fig. S3. The representative total ion chromatograms (TICs) of QC samples on positive and negative ion mode in plasma and lung tissue. PCA score plots and RSD% distribution of QC samples for metabolomic validation. Fig. S4. The representative total ion chromatograms (TICs) of plasma and lung tissue samples in positive and negative ion mode. Fig. S5. PLS-DA analysis of plasma and lung tissue in mice. Fig. S6. S-plot analysis of plasma and lung tissue samples in positive and negative ion mode. Table S1. Parameters of PLS-DA model and OPLS-DA model for prediction. Table S2. Enrichment analysis of potential metabolic pathways in mouse plasma and lung tissue.
Additional file 3: Table S3. Summary of chemical constituents in DM. Table S4. Active components of DM in the treatment of ALI.
Additional file 4: Targets information of DM.
Additional file 5: Fig. S7. The core genes for the integrated analysis of metabolomics and network pharmacology.
Acknowledgements
Not applicable.
Abbreviations
- ALI
Acute lung injury
- DM
Nauclea officinalis (Danmu in Chinese, DM)
- LPS
Lipopolysaccharide
- BALF
Bronchoalveolar lavage fluid
- VEGF
Vascular endothelial growth factor
- PCA
Principal component analysis
- PLS-DA
Partial least squares discriminant analysis
- OPLS-DA
Orthogonal projections to latent structures- discriminant analysis
- LA
Linoleic acid
- AA
Arachidonic acid
- PGs
Prostaglandins
- LTs
Leukotrienes
- COX
Cyclooxygenase
- PEs
Phosphatidylethanolamines
- PCs
Phosphatidylcholines
- PIs
Phosphatidylinositols
- PAs
Phosphatidic acids
- LPCs
Lyso-phosphatidylcholines
- LPEs
Lyso-phosphatidylethanolamines
Author contributions
XH designed the study and performed the experiments. XH, XSC analyzed the data and wrote the manuscript. LLY, WYH, MSY and XYQ participated in the process of animal experimental modeling and sample collection. TYF, LAL and XFM revised the manuscript. HXN in charge of funding. LYH designed and funded experiments. All authors have read and approved the final manuscript.
Funding
This work was supported by Hainan Provincial Natural Science Foundation of China (No.2019RC207 and 820RC768), National Natural Science Foundation of China (No.82160826) and Innovative Research Project for postgraduates of Hainan Province (No. Hys2020-367).
Availability of data and materials
The data used and/or investigated during the present study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The animal experiment project was conducted in strict accordance with the guidelines for the care and use of laboratory animals and approved by the Ethics Committee of Hainan Medical University (No. HYLL-2022-019).
Consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoning He, Email: hexiaoningvv@aliyun.com.
Yonghui Li, Email: lyhssl@126.com.
References
- 1.Cen M, Ouyang W, Zhang W, Yang L, Lin X, Dai M, et al. MitoQ protects against hyperpermeability of endothelium barrier in acute lung injury via a Nrf2-dependent mechanism. Redox Biol. 2021;41:101936. doi: 10.1016/j.redox.2021.101936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W, Long L, Yang X, Tong Z, Southwood M, King R, et al. Circulating BMP9 protects the pulmonary endothelium during inflammation-induced lung injury in mice. Am J Respir Crit Care Med. 2021;203(11):1419–1430. doi: 10.1164/rccm.202005-1761OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McVey MJ, Steinberg BE, Goldenberg NM. Inflammasome activation in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2021;320(2):L165–l178. doi: 10.1152/ajplung.00303.2020. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Cui Y, Zhou Z, Ding Y, Nie H. Alveolar type 2 epithelial cells as potential therapeutics for acute lung injury/acute respiratory distress syndrome. Curr Pharm Des. 2019;25(46):4877–4882. doi: 10.2174/1381612825666191204092456. [DOI] [PubMed] [Google Scholar]
- 5.Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315(8):788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 6.He YQ, Zhou CC, Yu LY, Wang L, Deng JL, Tao YL, et al. Natural product derived phytochemicals in managing acute lung injury by multiple mechanisms. Pharmacol Res. 2021;163:105224. doi: 10.1016/j.phrs.2020.105224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu WY, Gao CX, Zhang HH, Wu YG, Yu CH. Herbal active ingredients: potential for the prevention and treatment of acute lung injury. Biomed Res Int. 2021;2021:5543185. doi: 10.1155/2021/5543185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li N, Cao L, Cheng Y, Meng ZQ, Tang ZH, Liu WJ, et al. In vivo anti-inflammatory and analgesic activities of strictosamide from Nauclea officinalis. Pharm Biol. 2014;52(11):1445–1450. doi: 10.3109/13880209.2014.895910. [DOI] [PubMed] [Google Scholar]
- 9.Song LL, Mu YL, Zhang HC, Wu GY, Sun JY. A new indole alkaloid with anti-inflammatory from the branches of Nauclea officinalis. Nat Prod Res. 2020;34(16):2283–2288. doi: 10.1080/14786419.2018.1536130. [DOI] [PubMed] [Google Scholar]
- 10.Song S, Liu P, Wang L, Li D, Fan H, Chen D, et al. In vitro anti-inflammatory activities of naucleoffieine H as a natural alkaloid from Nauclea officinalis Pierrc ex Pitard, through inhibition of the iNOS pathway in LPS-activated RAW 264.7 macrophages. Nat Prod Res. 2020;34(18):2694–2697. doi: 10.1080/14786419.2018.1550765. [DOI] [PubMed] [Google Scholar]
- 11.Mai SY, Li YH, Zhang XG, Wang YR, Zhang JQ, Jia A. A new indole alkaloid with HUVEC proliferation activities from Nauclea officinalis. Nat Prod Res. 2021;35(18):3049–3055. doi: 10.1080/14786419.2019.1689497. [DOI] [PubMed] [Google Scholar]
- 12.Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, Jarmusch AK, Dorrestein PC. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol. 2022;20(3):143–160. doi: 10.1038/s41579-021-00621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchev AS, Vasileva LV, Amirova KM, Savova MS, Balcheva-Sivenova ZP, Georgiev MI. Metabolomics and health: from nutritional crops and plant-based pharmaceuticals to profiling of human biofluids. Cell Mol Life Sci CMLS. 2021;78(19–20):6487–6503. doi: 10.1007/s00018-021-03918-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Lin S, Liu R, Li H, Liu Z, Zhang X, et al. Metabolomic profile perturbations of serum, lung, bronchoalveolar lavage fluid, spleen and feces in LPS-induced acute lung injury rats based on HPLC-ESI-QTOF-MS. Anal Bioanal Chem. 2020;412(5):1215–1234. doi: 10.1007/s00216-019-02357-1. [DOI] [PubMed] [Google Scholar]
- 15.Ding Z, Zhong R, Yang Y, Xia T, Wang W, Wang Y, et al. Systems pharmacology reveals the mechanism of activity of Ge-Gen-Qin-Lian decoction against LPS-induced acute lung injury: a novel strategy for exploring active components and effective mechanism of TCM formulae. Pharmacol Res. 2020;156:104759. doi: 10.1016/j.phrs.2020.104759. [DOI] [PubMed] [Google Scholar]
- 16.Fu C, Wu Q, Zhang Z, Xia Z, Ji H, Lu H, et al. UPLC-ESI-IT-TOF-MS metabolomic study of the therapeutic effect of Xuefu Zhuyu decoction on rats with traumatic brain injury. J Ethnopharmacol. 2019;245:112149. doi: 10.1016/j.jep.2019.112149. [DOI] [PubMed] [Google Scholar]
- 17.Wang ZY, Wang X, Zhang DY, Hu YJ, Li S. [Traditional Chinese medicine network pharmacology: development in new era under guidance of network pharmacology evaluation method guidance] Zhongguo Zhong Yao Za Zhi. 2022;47(1):7–17. doi: 10.19540/j.cnki.cjcmm.20210914.702. [DOI] [PubMed] [Google Scholar]
- 18.Lai X, Wang X, Hu Y, Su S, Li W, Li S. Editorial: network pharmacology and traditional medicine. Front Pharmacol. 2020;11:1194. doi: 10.3389/fphar.2020.01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, Si Y, Cheng J, Ding J, Zhao H, Liu W, et al. Metabolic and network pharmacological analyses of the therapeutic effect of Grona styracifolia on calcium oxalate-induced renal injury. Front Pharmacol. 2021;12:652989. doi: 10.3389/fphar.2021.652989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Q, Zhu W, Diao Y, Xu G, Wang L, Qu S, et al. Elucidation of the mechanism of action of ginseng against acute lung injury/acute respiratory distress syndrome by a network pharmacology-based strategy. Front Pharmacol. 2020;11:611794. doi: 10.3389/fphar.2020.611794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia L, Zhou H, Li W, Lv Z. Network pharmacology integrated molecular docking revealed the mechanism of Jianpi Yiqi Taohua decoction against ulcerative colitis. Med Sci Monit. 2022;28:e933537. doi: 10.12659/MSM.933537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu L, Wang Y, Sun H, Xiong Y, Zhong L, Wu Z, et al. An untargeted metabolomics approach to investigate the wine-processed mechanism of Scutellariae radix in acute lung injury. J Ethnopharmacol. 2020;253:112665. doi: 10.1016/j.jep.2020.112665. [DOI] [PubMed] [Google Scholar]
- 23.Cui Y, Liu S, Zhang X, Ding X, Duan X, Zhu Z, et al. Metabolomic analysis of the effects of adipose-derived mesenchymal stem cell treatment on rats with sepsis-induced acute lung injury. Front Pharmacol. 2020;11:902. doi: 10.3389/fphar.2020.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang N, Lu F, Li Z, Zhao H, Pang M, Ye T, et al. Effects of Radix Scrophulariae on Hyperthyroidism Assessed by Metabonomics and Network Pharmacology. Front Pharmacol. 2021;12:727735. [DOI] [PMC free article] [PubMed]
- 25.Guo W, Liu S, Zheng X, Xiao Z, Chen H, Sun L, et al. Network pharmacology/metabolomics-based validation of AMPK and PI3K/AKT signaling pathway as a central role of Shengqi Fuzheng injection regulation of mitochondrial dysfunction in cancer-related fatigue. Oxid Med Cell Longev. 2021;2021:5556212. doi: 10.1155/2021/5556212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T, Zhang W, Hu E, Sun Z, Li P, Yu Z, et al. Integrated metabolomics and network pharmacology to reveal the mechanisms of hydroxysafflor yellow A against acute traumatic brain injury. Comput Struct Biotechnol J. 2021;19:1002–1013. doi: 10.1016/j.csbj.2021.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Z, Wang Y, Zhong R, Feng Y, Xia T, Xie Y, et al. Systems pharmacology reveals the mechanism of activity of Physalis alkekengi L. var. franchetii against lipopolysaccharide-induced acute lung injury. J Cell Mol Med. 2020;24(9):5039–5056. doi: 10.1111/jcmm.15126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Zhang Z, Xie Z, Lv S, Shi Y, Zhai C, Li X, et al. Integrated metabolomics and network pharmacology study on the mechanism of Kangfuxiaoyan suppository for treating chronic pelvic inflammatory disease. Front Pharmacol. 2022;13:812587. doi: 10.3389/fphar.2022.812587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan X, Wu J, Yang H, Yan L, Wang S. Paeoniflorin blocks the proliferation of vascular smooth muscle cells induced by platelet-derived growth factor-BB through ROS mediated ERK1/2 and p38 signaling pathways. Mol Med Rep. 2018;17(1):1676–1682. doi: 10.3892/mmr.2017.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai XT, Zhang ZY, Jiang CH, Chen JQ, Ye JQ, Jia XB, et al. Nauclea officinalis inhibits inflammation in LPS-mediated RAW 264.7 macrophages by suppressing the NF-κB signaling pathway. J Ethnopharmacol. 2016;183:159–165. doi: 10.1016/j.jep.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Das UN. Arachidonic acid and other unsaturated fatty acids and some of their metabolites function as endogenous antimicrobial molecules: a review. J Adv Res. 2018;11:57–66. doi: 10.1016/j.jare.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Wang Z, Chen W, Cao B, Zhang M, Gu Q, et al. An integrative metabolomic and network pharmacology study revealing the regulating properties of xihuang pill that improves anlotinib effects in lung cancer. Front Oncol. 2021;11:697247. doi: 10.3389/fonc.2021.697247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruan Y, Fan Y, Xie Y, Ma C, Mo B, Lai Y, et al. Modified Xiaoqinglong decoction alleviates lipopolysaccharide-induced acute lung injury in mice by regulating arachidonic acid metabolism and exerting anti-apoptotic and anti-inflammatory effects. Anat Rec (Hoboken). 2021;305(7):1672–1681. doi: 10.1002/ar.24822. [DOI] [PubMed] [Google Scholar]
- 34.Elajami TK, Colas RA, Dalli J, Chiang N, Serhan CN, Welty FK. Specialized proresolving lipid mediators in patients with coronary artery disease and their potential for clot remodeling. FASEB J . 2016;30(8):2792–2801. doi: 10.1096/fj.201500155R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norris PC, Arnardottir H, Sanger JM, Fichtner D, Keyes GS, Serhan CN. Resolvin D3 multi-level proresolving actions are host protective during infection. Prostaglandins Leukot Essent Fatty Acids. 2018;138:81–89. doi: 10.1016/j.plefa.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann A, Brunssen C, Morawietz H. Contribution of lectin-like oxidized low-density lipoprotein receptor-1 and LOX-1 modulating compounds to vascular diseases. Vasc Pharmacol. 2017;S1537-1891(17):30171–30174. doi: 10.1016/j.vph.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Liu YJ, Li H, Tian Y, Han J, Wang XY, Li XY, et al. PCTR1 ameliorates lipopolysaccharide-induced acute inflammation and multiple organ damage via regulation of linoleic acid metabolism by promoting FADS1/FASDS2/ELOV2 expression and reducing PLA2 expression. Lab Invest. 2020;100(7):904–915. doi: 10.1038/s41374-020-0412-9. [DOI] [PubMed] [Google Scholar]
- 38.Wu B, Walker J, Spur B, Rodriguez A, Yin K. Effects of lipoxin A4 on antimicrobial actions of neutrophils in sepsis. Prostaglandins Leukot Essent Fatty Acids. 2015;94:55–64. doi: 10.1016/j.plefa.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Das U. HLA-DR expression, cytokines and bioactive lipids in sepsis. Arch Med Sci AMS. 2014;10(2):325–335. doi: 10.5114/aoms.2014.42586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong X, Yao S, Wu W, Cao J, Sun L, Li H, et al. Gas explosion-induced acute blast lung injury assessment and biomarker identification by a LC-MS-based serum metabolomics analysis. Hum Exp Toxicol. 2021;40(4):608–621. doi: 10.1177/0960327120960761. [DOI] [PubMed] [Google Scholar]
- 41.Navarrete A, Rupérez FJ, Mendes TO, Pérez-Rial S, Girón-Martínez A, Terrón-Expósito R, et al. A metabolomic approach shows sphingosine 1-phosphate and lysophospholipids as mediators of the therapeutic effect of liver growth factor in emphysema. J Pharm Biomed Anal. 2017;139:238–246. doi: 10.1016/j.jpba.2017.02.045. [DOI] [PubMed] [Google Scholar]
- 42.Zhou X, Wang R, Zhang T, Liu F, Zhang W, Wang G, et al. Identification of lysophosphatidylcholines and sphingolipids as potential biomarkers for acute aortic dissection via serum metabolomics. Eur J Vasc Endovasc Surg. 2019;57(3):434–441. doi: 10.1016/j.ejvs.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Suchner U, Heyland DK, Peter K. Immune-modulatory actions of arginine in the critically ill. Br J Nutr. 2002;87(Suppl 1):S121–132. doi: 10.1079/BJN2001465. [DOI] [PubMed] [Google Scholar]
- 44.Wu G, Bazer FW, Satterfield MC, Gilbreath KR, Posey EA, Sun Y. L-Arginine nutrition and metabolism in ruminants. Adv Exp Med Biol. 2022;1354:177–206. doi: 10.1007/978-3-030-85686-1_10. [DOI] [PubMed] [Google Scholar]
- 45.Sharma NS, Lal CV, Li JD, Lou XY, Viera L, Abdallah T, et al. The neutrophil chemoattractant peptide proline-glycine-proline is associated with acute respiratory distress syndrome. Am J Physiol Lung Cell Mol Physiol. 2018;315(5):L653–l661. doi: 10.1152/ajplung.00308.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li D, Chen J, Ye J, Zhai X, Song J, Jiang C, et al. Anti-inflammatory effect of the six compounds isolated from Nauclea officinalis Pierrc ex Pitard, and molecular mechanism of strictosamide via suppressing the NF-κB and MAPK signaling pathway in LPS-induced RAW 264.7 macrophages. J Ethnopharmacol. 2017;196:66–74. doi: 10.1016/j.jep.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 47.Chen IC, Wang SC, Chen YT, Tseng HH, Liu PL, Lin TC, et al. Corylin ameliorates LPS-induced acute lung injury via suppressing the MAPKs and IL-6/STAT3 signaling pathways. Pharmaceuticals. 2021;14(10):1046. doi: 10.3390/ph14101046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong F, Sun Y, Song W, Zhou Y, Zhu S. MiR-216a alleviates LPS-induced acute lung injury via regulating JAK2/STAT3 and NF-κB signaling. Hum Cell. 2020;33(1):67–78. doi: 10.1007/s13577-019-00289-7. [DOI] [PubMed] [Google Scholar]
- 49.Xu J, Xu D, Yu Z, Fu Z, Lv Z, Meng L, et al. Exosomal miR-150 partially attenuated acute lung injury by mediating microvascular endothelial cells and MAPK pathway. Biosci Rep. 2022;42(1):BSR20203363. doi: 10.1042/BSR20203363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao YR, Wang D, Liu Y, Shan L, Zhou JL. The PI3K/Akt, p38MAPK, and JAK2/STAT3 signaling pathways mediate the protection of SO2 against acute lung injury induced by limb ischemia/reperfusion in rats. J Physiol Sci JPS. 2016;66(3):229–239. doi: 10.1007/s12576-015-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelin LD, Jin Y, Chen B, Liu Y, Rogers LK, Reese J. Cyclooxygenase-2 deficiency attenuates lipopolysaccharide-induced inflammation, apoptosis, and acute lung injury in adult mice. Am J Physiol Regul Integr Comp Physiol. 2022;322(2):R126–r135. doi: 10.1152/ajpregu.00140.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hite RD, Grier BL, Waite BM, Veldhuizen RA, Possmayer F, Yao LJ, et al. Surfactant protein B inhibits secretory phospholipase A2 hydrolysis of surfactant phospholipids. Am J Physiol Lung Cell Mol Physiol. 2012;302(2):L257–265. doi: 10.1152/ajplung.00054.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu C, Weng QY, Zhou LR, Cao C, Li F, Wu YF, et al. Homeostatic and early-recruited CD101(-) eosinophils suppress endotoxin-induced acute lung injury. Eur Respir J. 2020;56(5):1902354. doi: 10.1183/13993003.02354-2019. [DOI] [PubMed] [Google Scholar]
- 54.Lingappan K, Maity S, Jiang W, Wang L, Couroucli X, Veith A, et al. Role of cytochrome P450 (CYP)1A in hyperoxic lung injury: analysis of the transcriptome and proteome. Sci Rep. 2017;7(1):642. doi: 10.1038/s41598-017-00516-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan D, Wang D, Zhu L. Protective role of scutellarin on LPS induced—Acute lung injury and regulation of apoptosis, oxidative stress and reduction of mitochondrial dysfunction. Saudi J Biol Sci. 2022;29(1):371–378. doi: 10.1016/j.sjbs.2021.08.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan S, He J, Yang Y, Wang D. Intermedin reduces oxidative stress and apoptosis in ventilator-induced lung injury via JAK2/STAT3. Front Pharmacol. 2021;12:817874. doi: 10.3389/fphar.2021.817874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Liu L. Anagliptin alleviates lipopolysaccharide-induced inflammation, apoptosis and endothelial dysfunction of lung microvascular endothelial cells. Exp Ther Med. 2021;22(6):1472. doi: 10.3892/etm.2021.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arulananda S, Lee EF, Fairlie WD, John T. The role of BCL-2 family proteins and therapeutic potential of BH3-mimetics in malignant pleural mesothelioma. Expert Rev Anticancer Ther. 2021;21(4):413–424. doi: 10.1080/14737140.2021.1856660. [DOI] [PubMed] [Google Scholar]
- 59.Zhang J, Zhang S, Shi Q, Allen TD, You F, Yang D. The anti-apoptotic proteins Bcl-2 and Bcl-xL suppress Beclin 1/Atg6-mediated lethal autophagy in polyploid cells. Exp Cell Res. 2020;394(1):112112. doi: 10.1016/j.yexcr.2020.112112. [DOI] [PubMed] [Google Scholar]
- 60.Spitz AZ, Gavathiotis E. Physiological and pharmacological modulation of BAX. Trends Pharmacol Sci. 2022;43(3):206–220. doi: 10.1016/j.tips.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abd El-Ghafar OAM, Hassanein EHM, Sayed AM, Rashwan EK, Shalkami AS, Mahmoud AM. Acetovanillone prevents cyclophosphamide-induced acute lung injury by modulating PI3K/Akt/mTOR and Nrf2 signaling in rats. Phytother Res PTR. 2021;35(8):4499–4510. doi: 10.1002/ptr.7153. [DOI] [PubMed] [Google Scholar]
- 62.Zhong R, Xia T, Wang Y, Ding Z, Li W, Chen Y, et al. Physalin B ameliorates inflammatory responses in lipopolysaccharide-induced acute lung injury mice by inhibiting NF-κB and NLRP3 via the activation of the PI3K/Akt pathway. J Ethnopharmacol. 2022;284:114777. doi: 10.1016/j.jep.2021.114777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. The stem of Nauclea officinalis Pierre ex Pitard (Danmu in Chinese, DM). Fig. S2. HPLC analysis of DM.
Additional file 2: Fig. S3. The representative total ion chromatograms (TICs) of QC samples on positive and negative ion mode in plasma and lung tissue. PCA score plots and RSD% distribution of QC samples for metabolomic validation. Fig. S4. The representative total ion chromatograms (TICs) of plasma and lung tissue samples in positive and negative ion mode. Fig. S5. PLS-DA analysis of plasma and lung tissue in mice. Fig. S6. S-plot analysis of plasma and lung tissue samples in positive and negative ion mode. Table S1. Parameters of PLS-DA model and OPLS-DA model for prediction. Table S2. Enrichment analysis of potential metabolic pathways in mouse plasma and lung tissue.
Additional file 3: Table S3. Summary of chemical constituents in DM. Table S4. Active components of DM in the treatment of ALI.
Additional file 4: Targets information of DM.
Additional file 5: Fig. S7. The core genes for the integrated analysis of metabolomics and network pharmacology.
Data Availability Statement
The data used and/or investigated during the present study are available from the corresponding author upon reasonable request.