Abstract
Plasmid DNA (pDNA) nanoparticles synthesized by complexation with linear polyethylenimine (lPEI) are one of the most effective non-viral gene delivery vehicles. However, the lack of scalable and reproducible production methods and the high toxicity have hindered their clinical translation. Previously, we have developed a scalable flash nanocomplexation (FNC) technique to formulate pDNA/lPEI nanoparticles using a continuous flow process. Here we report a tangential flow filtration (TFF)-based scalable purification method to reduce the uncomplexed lPEI concentration in the nanoparticle formulation and improve its biocompatibility. The optimized procedures achieved a 60% reduction of the uncomplexed lPEI with preservation of the nanoparticle size and morphology. Both in vitro and in vivo studies showed the purified nanoparticles significantly reduced toxicity while maintaining transfection efficiency. TFF also allows for gradual exchange of solvent to isotonic solution and further concentrating the nanoparticles for injection. Combining FNC production and TFF purification, we validated the purified pDNA/lPEI nanoparticles for future clinical translation of this gene nanomedicine.
Keywords: DNA nanoparticles, linear PEI, tangential flow filtration, gene delivery, biocompatibility
Graphical Abstract
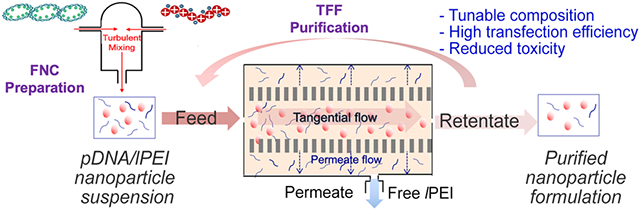
Introduction:
Plasmid DNA (pDNA)/polycation nanoparticles are one of the most promising non-viral vehicles for the delivery of the gene of interest to treat a variety of inherited and acquired diseases such as cancer, immunodeficiency, and metabolic disorders.1,2 Among cationic polymers developed for gene therapy applications, polyethylenimine (PEI) is a gold standard with a high transfection efficiency in vivo.3-5 A commercially available reagent, in vivo-jetPEI®, provides an optimized linear PEI (lPEI) carrier with a defined structure and uniform charge density, prepared under good manufacturing practice (GMP)-compliant conditions. lPEI-based nanoparticles have been tested in several preclinical and clinical studies.6-9 However, although pDNA/lPEI nanoparticles offer high transfection efficiency, they cause significant toxicity in vivo, especially when delivered systemically through intravenous (i.v.) injection. Previous studies have shown that lPEI changes the gene expression pattern of endothelial cells10 and induces apoptosis, stress responses, and oncogenesis.11 In order to achieve efficient condensation of pDNAs into nanoparticles and thus high transfection efficiency, pDNA/lPEI nanoparticles are usually generated with a molar ratio of nitrogen on lPEI to phosphate on pDNA (N/P ratio) greater than 6. The manufacturer of in vivo-jetPEI® recommends an N/P ratio of 6 to 8 for complexing pDNA. This condition results in a considerable amount of uncomplexed free lPEI in the final nanoparticle suspension.12 It should be noted that toxicity has been correlated with high N/P ratios, as well as with high concentrations of free lPEI.13,14 Besides lPEI, a high N/P ratio, ranging from 8 to 20, is often recommended for many other polycation carriers,15-18 where similar toxicity concerns arise from the free/uncomplexed polycations that are in great excess. This dilemma in choosing between higher N/P ratios for high efficiency of complexation/condensation and high transfection efficiency, and lower N/P ratios for lower free lPEI and better biocompatibility is a long-standing challenge for polycation carriers. It remains a critical issue to address towards a formulation with better efficacy and safety profile for clinical translation of nanoparticle-based gene medicine.
A direct and feasible solution is to purify pDNA/lPEI nanoparticles by partially removing the free lPEI to reduce the total lPEI amount dosed with the nanoparticles. The most common methods, size exclusion chromatography,14 conventional ultrafiltration, and ultracentrifugation,19 suffer from low efficiency and low yield due to entrapment of nanoparticles. These batch-mode purification processes have substantial difficulties in scale-up production. In addition, the centrifugation process likely results in substantial nanoparticle loss due to caking and particle aggregation.20,21 Nanoparticles purified by ultracentrifugation also requires further purification to adjust the excess amount of sucrose for the purpose of achieving isotonic condition for in vivo studies.21-24 Dialysis, another commonly used method for laboratory scale purification,25 is time consuming and difficult to scale up. Among current purification methods, tangential flow filtration (TFF) is particularly suitable for purification of nanoparticles, allowing the removal of impurities (i.e., surfactant and excess materials).19,26 As the direction of filtration is perpendicular to the direction of the bulk flow, there is minimal blockage of the filters by nanoparticles. To date, reported applications of TFF technique on nanomedicine have been primarily focused on poly(lactic-co-glycolic acid) and lipid nanoparticles.27 Its utility for purifying polyelectrolyte complex nanoparticles with nucleic acid therapeutics including pDNA has not been reported previously. Due to the highly charged nature of pDNA/lPEI nanoparticles and the excess lPEI polycation, it remains a great challenge to effectively remove excess lPEI without compromising the integrity of the nanoparticles and maintaining sufficient colloidal stability, i.e. preventing dissociation or aggregation of the intact nanoparticles during the purification process. In this study, we tested the hypotheses that TFF with optimized parameters permits efficient purification of pDNA/lPEI nanoparticles by maximal removal of free lPEI molecules and retaining stability of the purified nanoparticles in a continuous manner, and the purified pDNA/lPEI nanoparticles can yield higher level of biocompatibility while retaining high efficiency of gene delivery.
We have previously developed a scalable flash nanocomplexation (FNC) technique28 to formulate pDNA/lPEI nanoparticles with controlled size, uniformity and high transfection efficiency. In this study we applied the TFF method to reduce the concentration of free lPEI in the formulation at an N/P ratio of 6. The technical parameters of TFF were screened and optimized to achieve the best efficiency of lPEI removal, whilst minimizing the loss of nanoparticles and keeping the characteristics of nanoparticles stable in the suspensions, including size and morphology. We conducted in vitro and in vivo experiments in two mouse models to determine the effect of reducing free lPEI in the nanoparticle formulation on transfection efficiency and toxicity. We also confirmed that TFF is effective to replace the original solvent of the nanoparticles with an isotonic solution (9.5% w/w trehalose) and to concentrate the nanoparticle suspension to a target concentration. With FNC plus TFF as a scalable synthesis and purification process, purified pDNA/lPEI nanoparticles are poised for future testing for systemic delivery of pDNA in clinic.
Materials and Methods:
1. Materials
in vivo-jet PEI®, 150 mM was purchased from Polyplus (Illkirch, France). gWiz-Luc plasmid was purchased from Aldevron (Fargo, ND, USA) and dissolved at 1 mg/mL in DI water. Dulbecco’s Modification of Eagle’s Medium (DMEM), RPMI 1640, penicillin-streptomycin, and fetal bovine serum were purchased from Invitrogen (Carlsbad, CA, USA). Precision Red Advanced Protein Assay (PRAPA) reagent was from Cytoskeleton (Denver, CO, USA). Modified Polyethersulfone (mPES/300 kD) filters were purchased from Spectrum® Lab (Rancho Dominguez, CA, USA). Unless otherwise stated, all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
2. Nanoparticle preparation
FNC device setup.
The confined impinging jet (CIJ) mixer as well as the FNC process were previously developed by our lab.29 pDNA/lPEI complex nanoparticles were prepared in a CIJ mixer equipped with two streams. The two streams were independently loaded with lPEI and pDNA solutions, and complex nanoparticles were formed in a CIJ mixer before being collected. For the CIJ device, the two inlets connected to gas-tight plastic syringes (5, 10, 20, or 50 mL) by PTFE tubing with an ID of 0.75 mm are introducing the solutions of in vivo-jet PEI® and DNA. The CIJ device was manufactured by Johns Hopkins University Whiting School of Engineering machine shop based on a CIJ design reported previously.29 The same flow rate (20 mL/min) was used to inject both solutions into the CIJ mixer, controlled by a programmable syringe pump (New Era Pump System, model NE-4000). The complex nanoparticles were collected in scintillation vials through a PTFE tubing (ID = 0.75 mm) connected to the mixing chamber.
pDNA/lPEI complex nanoparticles formulation.
The plasmid DNA was diluted with an appropriate amount of DI water to give an input concentration of 400 μg/mL. The in vivo-jet PEI was used as received and diluted by ultrapure water to desired concentrations corresponding to N/P ratio 6. The pH of the lPEI solution was adjusted to 3.5 by NaOH to keep a consistent charge density on lPEI molecules across all experiments. The nanoparticles were formulated by injection the two working solutions into confined impinging jet (CIJ) chamber with flow rate 20 mL/min.
3. Nanoparticle purification by TFF method.
KrosFlo RESEARCH IIi TFF SYSTEM and modified polyethersulfone (mPES/300 kDa and 500 kDa) filters were from Spectrum® Lab. The purification of nanoparticles was performed with the setup shown in Figure 1a. The diafiltration mode was firstly used to purify the pDNA/lPEI nanoparticles at a DNA concentration of 40 μg/mL. In the process, the purified retentate or concentrate was directed to a separate retentate collector, and the feed reservoir was diluted by a phosphate-buffered saline (PBS) at the same rate as filtrate or permeate was being generated, hence maintaining a constant volume. The filtrate was taken at designated times. At the end, the retentate was collected and its free polymer and DNA content were determined. A 9.5% trehalose replaced the washing solution of PBS after the purification step. After full replacement, the system was switched to the concentration mode to concentrate the nanoparticles. Between purification of batches, the TFF system was cleaned by 1-h continuous circulation of a large volume of D.I. water. The system was stored in 30% ethanol.
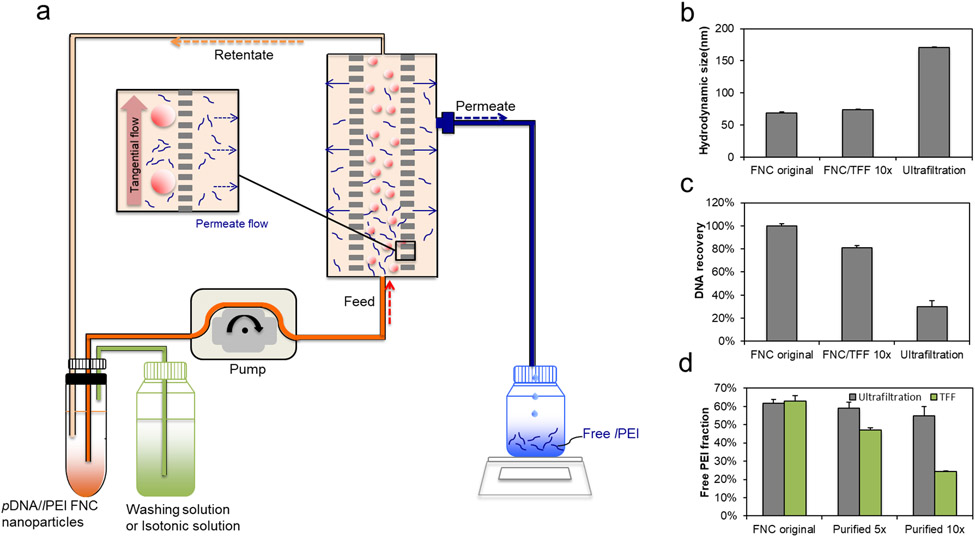
Figure 1.
(a) Schematic diagram of pDNA/lPEI nanoparticle purification by TFF. Comparison of (b) the z-average hydrodynamic size (c) DNA recovery and (d) free lPEI fraction of the nanoparticles before purification (FNC original nanoparticles) vs. following purification by TFF and ultrafiltration methods using the 5-volume washing (5×) and 10-volume washing (10×) protocols, respectively (n = 3). The 5× and 10× washing refer to 5 and 10 times of the volume of the solvent used, respectively, in reference to the volume of nanoparticles. This is measured by the volume of the permeate collected during the TFF process.
4. Transmission electron microscopy (TEM) of nanoparticles.
TEM imaging samples of pDNA/lPEI nanoparticles were prepared by incubating 10 μL of pDNA/lPEI nanoparticle solution on an ionized nickel grid covered with a carbon film. After 10 min, the solution was removed, and a 6-μL drop of 2% uranyl acetate was added to the grid. After 20 sec, the staining solution was removed, and the grid was dried at room temperature. The samples were imaged with a Tecnai FEI-12 electron microscope (Johns Hopkins University, Integrated Imaging Center) at 100 kV. All images were taken by a Megaview III wide-angle camera.
5. Characterization of pDNA/lPEI nanoparticles
Particle size and polydispersity index (PDI) were measured by Zetasizer Nano ZS90 (Malvern Instruments, Southborough, MA). Size measurement was performed at 25°C using a 90° scattering angle. The mean hydrodynamic diameter was determined by cumulative analysis. The z-average hydrodynamic diameter was obtained and used as the size of the pDNA/lPEI nanoparticles for all the analysis in this study. Three independent measurements were conducted. DNA concentration was measured by NanoDrop Spectrophotometer (Thermo Scientific ND-1000). An aliquot of 2 μL standard nanoparticle suspension with five pDNA concentrations (25, 50, 100, 150, and 200 μg/mL) was prepared, with the absorbance at 260 nm measured to generate the standard curve. Three independent measurements were conducted. The absorbance of an aliquot of 2 μL of pDNA/lPEI nanoparticles were measured at the same wavelength. By using the generated standard curve, the corresponding pDNA concentration for the absorbance of pDNA/lPEI nanoparticles could be determined. To evaluate the pDNA integrity of the TFF-purified nanoparticles, gel retardation assay was performed. Nanoparticle samples were incubated for 30 min with 1 mg/mL heparin sulfate to release pDNA, after which 0.5 μg pDNA dose was added to each well and analyzed by electrophoresis at 80 V for 45 min on a 0.8% (w/v) agarose gel. The pDNA bands were visualized under a UV transilluminator.
6. Free polymer measurement of pDNA/lPEI nanoparticles.
The Precision Red Advanced Protein Assay (PRAPA) was used to determine of the concentration and the fraction of uncomplexed in vivo-jet® PEI. The method was adopted from a previous report.14 The pDNA concentration of nanoparticles was diluted to 40 μg/mL and 500 μL of the diluted nanoparticle suspension was added in each Vivaspin column (PES, MWCO of 100,000, Sartorius) in their appropriate microcentrifuge tubes in order to isolate uncomplexed in vivo-jet® PEI from nanoparticle suspensions. The standard in vivo-jet® PEI solutions were prepared in the concentration within 0.25 mM to 0.025 mM to generate the absorbance-to-concentration standard curve. Three Vivaspin columns were prepared for one sample and centrifuged at 7200 RPM at room temperature for 60 seconds. The flow-through was collected for the measurement. An aliquot of 60 μL of the flow-through solution and the standard in vivo-jet® PEI solution was separately added to a 96-well plate, followed by addition of 200 μL of PRAPA reagent working solution into each well. The plate was incubated at room temperature for 10 min. The absorbance at a wavelength of 590 nm was assessed and the concentrations were calculated against a standard curve.
7. In vitro transfection of pDNA/lPEI nanoparticles.
The nanoparticles prepared were evaluated for their transfection efficiency in vitro using a luciferase reporter pDNA in two separate cell lines: PC3 prostate cancer cell line and NCI-H1299 lung cancer cell line. PC3 cell line was maintained in Dulbecco’s Modified Eagle’s Medium (DMEM). NCI-H1299 cell line was maintained in RPMI-1640 medium. All media were supplemented with 10% fetal bovine serum (FBS) and 100 U/mL Penicillin and 100 μg/mL Streptomycin. Cells were cultured at 37°C and 5% CO2.
The in vitro transfection efficiency of the nanoparticle followed different incubation times, expression times, and doses were screened as following. Cells were seeded in 24-well plates at a density of 5 × 104 cells/well 24 h prior to the transfection experiments. Nanoparticle suspensions equivalent to 0.2, 0.4, 0.6, 0.8, and 1.0 μg of DNA dose/well were added to the cells and incubated for 1, 2, 3 or 4 h, followed which the mixture was aspirated. The cells were then washed by 1× PBS (pH 7.4) twice and immediately placed in fresh medium for another 6, 12, or 24 h incubation to express the luciferase. One hundred μL of reporter lysis buffer (Promega, Madison, WI) was added to each well and cells were subjected to two freeze-thaw cycles. Twenty μL of cell lysate from each well was assayed using a luciferase assay kit (Promega, Madison, WI) on a luminometer (20/20n, Turner BioSystems, Sunnyvale, CA). The luciferase activity was converted to the amount of luciferase expressed using a recombinant luciferase protein (Promega, US) as the standard. Total protein content in the lysate was measured by BCA assay (Pierce BCA reagents, Thermo Scientific, US), and luciferase expressed was normalized against total protein content. For all tests, n = 4 wells for each group were tested.
8. In vitro cytotoxicity of pDNA/lPEI nanoparticles.
The Thermo Scientific Alamar Blue® reagent Assay was performed to determine the in vitro cytotoxicity. PC3 cells were seeded in 96-well plates at a density of 1 × 104 cells/well at 24 h prior to the cytotoxicity experiments. The nanoparticle suspensions equivalent to 0.05 μg, 0.1 μg, 0.25 μg, 0.5 μg, 0.75 μg, and 1 μg of DNA dose/well were added to the cells and incubated for 4 h, followed which the mixture was aspirated. The cells were then washed by 1× PBS (pH 7.4) twice and immediately placed in fresh medium. The cells were incubated for another 24 h with 10 μL of AlamarBlue® reagent to each well and the plates are incubated at 37°C overnight to allow cells to convert resazurin to resorufin completely, and then the cell viability was measured using AlarmaBlue® assay according to the manufacturer’s protocol. The absorbance of the mixture at 570 nm while using 600 nm as a reference wavelength was measured. For all tests, n = 4 wells for each group were tested.
9. In vivo transfection of pDNA/lPEI nanoparticles.
All in vivo procedures were approved by the Johns Hopkins University Institutional Animal Care and Use Committee. Balb/c mice (7-week old, female, n = 5 per group, Charles River Laboratories, Frederick, MD) and CD-1 mice (12-week old, female, n = 5 per group, Charles River Laboratories, Frederick, MD) were injected through lateral tail vein with a dose 40 μg DNA of nanoparticles per mouse. pDNA/lPEI nanoparticle formulations (N/P = 6) were prepared as control group for all in vivo experiments according to the manufacturer’s protocol (Polyplus Transfection, France). All the nanoparticle groups were suspended in 9.5% trehalose solution to formulate isotonic for systemic injection. Bioluminescence imaging was performed by using the IVIS® Spectrum (PerkinElmer, Waltham, MA) at 24 h and 48 h time points after nanoparticle injection. Mice were anesthetized with 3% isoflurane and injected i.p. with 100 μL 30 mg/mL D-luciferin solution (Gold Biotechnology, St. Louis. MO). After 5 min injection of D-luciferin, bioluminescence images were obtained; and the images were processed with Living Image Software (PerkinElmer).
10. In vivo toxicity of pDNA/lPEI nanoparticles.
After the bioluminescence imaging analyses were performed at 24 h and 48 h time points following nanoparticle injection, mice were euthanized by isoflurane. The liver and the blood sample of each mouse were collected immediately. The whole liver was fixed in 10% neutral buffered formalin for 24 h and embedded in paraffin followed by cutting into 5 μm sections. The hematoxylin-eosin (H&E) staining was conducted on sectioned tissue samples. The stained slides were visualized with an Olympus IX 50 microscope. Quantification of positive staining was performed using NIH Image J program. For assessing the necrosis area of the liver, scores were evaluated by visual assessment of the percentage of the necrotic area over normal area involvement of the staining section by both low (10X) and high (20X) magnification microscopic evaluation on at least 10 different fields of views on each slide. Biochemical analyses of liver enzymes, including alanine transaminase (ALT) and aspartate transaminase (AST), were carried out for each blood sample using Spotchem EZ chemistry analyzer (Arkray USA, Edina, MN). The averages of 5 samples were plotted for different groups.
Results:
The free lPEI in the pDNA/lPEI nanoparticle suspension was reduced by TFF (Figure 1a). When washing solution, i.e. diluted phosphate-buffered saline (PBS), was supplemented into the system, free lPEI was gradually washed out, thus purifying the nanoparticles. Nanoparticles prepared at N/P = 6 were washed by TFF with either 5 or 10 times of washing volume, i.e. 5 or 10 times of the volume of the solvent that was filtered out as the permeate compared to the volume of nanoparticle suspension. To be more specific in washed times, if the initial sample volume is 5 mL (5 g), the total mass of permeate is set to 50 g for 10 times washing. In later experiments, TFF was used to replace the diluted PBS of the purified nanoparticle suspension with an isotonic solution and subsequently to concentrate the nanoparticles to a sufficiently high DNA concentration (> 100 μg/mL). In the liquid replacement mode, the washing solution was changed to 9.5% w/w trehalose; while in the concentration mode, water was filtered out in a continuous and gradual manner.
We conducted comparison with other purification methods, such as conventional ultrafiltration. However, the average diameter of the purified nanoparticles increased significantly. The average hydrodynamic size increased from 68 nm to 171 nm after purified by ultrafiltration; in contrast, nanoparticles purified under the “FNC/TFF 10×” (TFF with 10 times washing volume that was filtered out as the permeate) condition maintained a similar size as the unpurified nanoparticles (Figure 1b). More importantly, the majority of the nanoparticles were lost likely due to membrane adhesion and aggregation on to the membrane, leading to low recovery yield and low purification efficiency. As shown in Figure 1c and 1d, the ultrafiltration method provided a yield of only 30% of nanoparticles (vs. 80% for FNC/TFF 10× method). With a ten-volume of washing procedure, ultrafiltration only removed 7% free lPEI, whereas TFF yielded a 61.9% reduction of lPEI content.
We tested the effect of the ion concentration (Figure S2a) and pH (Figure S2b) of the washing solutions on the efficiency of lPEI removal. The PBS was diluted to 0.07×, 0.08× and 0.1× for the washing buffer solutions. Higher PBS concentrations were not tested as these caused nanoparticle aggregations (Figure S3a). On the other hand, PBS concentrations lower than 0.07× led to low lPEI removal efficiency. The results (Figure S2a) suggest that with higher PBS concentrations and hence higher ionic strength the purification efficiency was increased. We also optimized the pH of the washing solution for lPEI polymer removal efficiency (Figure S2b). While maintaining weak acidic pH in the nanoparticle suspension (e.g. pH 6.5) is important for retaining good solubility of lPEI, reducing pH below 6.5 decreased the efficiency of free lPEI removal. At the same ionic strength (0.08× PBS), a solution pH of 6.5 reduced the concentration of free lPEI by 18% compared with that achieved at pH 5.5. We further optimized the molecular weight cut off (MWCO) of the filter (Figure S4a and S4b), and the TFF flow rate (shear rate) (Figure S4c and S4d). The results showed that 300 kD MWCO and a flow rate of 15.5 mL/min were optimal for TFF purification of nanoparticles to achieve higher DNA recovery and greater removal of free lPEI among tested conditions.
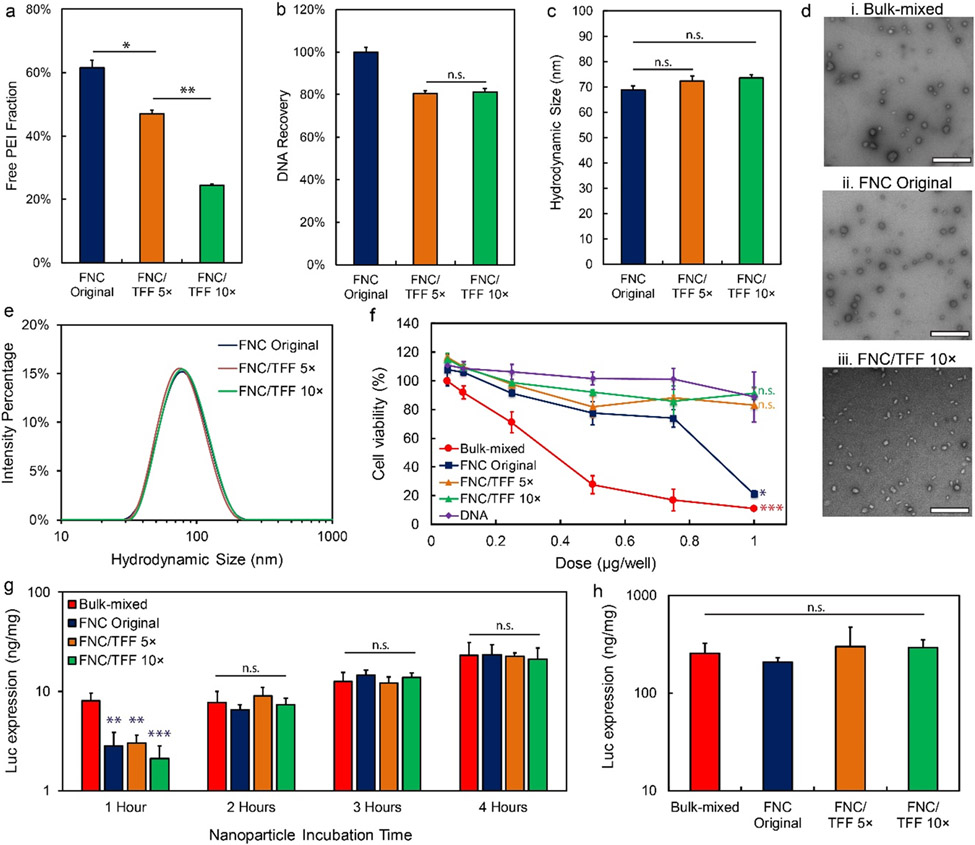
We then optimized the output volume of the TFF wash needed for nanoparticle purification. With only 5× volume washing, the free lPEI remained at a high level of 47% of the total dose added. After 10× volume washing, the free lPEI concentration was reduced to 24% (Figure 2a) with 80% nanoparticle retention in the purified suspension as measured by DNA recovery (Figure 2b). To evaluate the DNA compaction ability of the TFF-purified nanoparticles, gel retardation assay was performed. The results confirmed that DNA integrity did not change after TFF purification (Figure S5). We characterized the pDNA/lPEI nanoparticles purified with our optimized TFF conditions (washing buffer in pH 6.5, 0.1 × PBS, 300 kDa MWCO filter, flow rate 15.5 mL/min, 10× washing volume). Nanoparticle size was well preserved with a similar average size and size distribution before and after TFF purification under these conditions (Figure 2c, 2d, 2e). Representative TEM images of the nanoparticle preparations indicated that FNC nanoparticles were smaller and had slightly more uniform size distribution compared to those of bulk-mixed nanoparticles prepared at a small batch size (200 μL). The polydispersity index (PDI) of nanoparticles prepared by FNC, FNC/TFF 5×, FNC/TFF 10× were below 0.2 (Figure S6). The PDI of nanoparticles prepared by bulk-mixing method is above 0.25 with the higher heterogeneity from pipet mixing (Figure S6).
Figure 2.
The pDNA/lPEI nanoparticle characteristics and in vitro transfection activities after TFF purification with optimized conditions. (a) Percentage of the free lPEI, (b) DNA recovery, and (c) z-average size of the nanoparticles before and after TFF purification; FNC/TFF 5× and FNC/TFF 10× (see Fig. 1 for group description). (d) TEM images of the nanoparticles prepared by (i) bulk-mixed: Polyplus protocol (ii) FNC original (unpurified), and (iii) FNC/TFF 10×. Scale bar is 500 nm. (e) The intensity average size distribution of the nanoparticles before and after TFF. (f) In vitro cellular viability of PC3 cell line in bulk-mixed: Polyplus protocol, FNC original, FNC/TFF 5×, FNC/TFF 10× nanoparticles and naked DNA with different DNA concentrations (nanoparticle concentration was titrated to match the DNA concentration for comparison purpose). For statistical analysis, the comparison is against DNA group and bulk-mixed group, respectively. (g) In vitro transfection efficiency with different incubation time of bulk-mixed: Polyplus protocol, FNC original, FNC/TFF 5×, FNC/TFF 10× purified pDNA/lPEI nanoparticles by PC3 cell line. (h) In vitro transfection efficiency of bulk-mixed: Polyplus protocol, FNC original, FNC/TFF 5×, FNC/TFF 10× purified nanoparticles by NCI-H1299 cell line. For statistical analysis, n.s. denotes no statistical significance with p > 0.05, *p < 0.05, **p < 0.01, and ***p < 0.001 from one-way ANOVA test.
To determine the in vitro toxicity of the pDNA/lPEI nanoparticles, AlamarBlue® assay was performed as an indicator of metabolic cell function following treatment with nanoparticles (Figure 2f) over the dose range of 0.05 to 1 μg pDNA per 1 × 104 cells (the corresponding free lPEI concentration was 0 to 0.5 mM in the cell culture medium). Both bulk-mixed nanoparticles and unpurified FNC nanoparticles showed higher cytotoxicity at higher doses: bulk-mixed nanoparticles caused a significant decrease in cell viability at doses higher than 0.25 μg per well (1 × 104 cells); while FNC nanoparticles showed much lower toxicity at DNA doses lower than 0.75 μg/well, but a similarly high cytotoxicity at 1 μg per well. On the other hand, purified nanoparticles by TFF for 10 times maintained 80% of the original cell viability over the entire dose range tested (0.05–1 μg/well), which was comparable with treating with uncomplexed plasmid DNA.
The transfection of bulk-mixed and purified nanoparticles was conducted in the PC3 prostate cancer cell line in serum-containing medium using gWiz-Luc as a reporter plasmid; and the transfection efficiency was evaluated by comparing the expression of luciferase enzyme and normalized against the total protein content in the cell lysate. The following parameters were varied: the duration of incubation with the nanoparticles (Figure 2g), the time after removal of the nanoparticles and medium change (Figure S7a), and different doses of the nanoparticles (Figure S7b). When these parameters were not the variable being tested, the incubation time was kept at 4 h, the expression time was kept at 24 h, and the dose was kept at 0.4 μg per 104 cells. Transfection outcomes with different nanoparticle incubation times (Figure 2g) suggested that with a 1 h incubation time, bulk-mixed nanoparticles transfected cells more efficiently than FNC original, FNC/TFF 5×, or FNC/TFF 10× nanoparticles, probably because the larger size of bulk-mixed nanoparticles resulted in faster sedimentation to the cell monolayer.30,31 At longer incubation times, the in vitro transfection efficiency was unaffected by the concentration of free lPEI. Similarly, when expression time was varied (Figure S7a), while the DNA dose was fixed at 0.4 μg per 104 cells and the incubation time with the nanoparticles was fixed at 4 h, there were no significant differences among groups dosed with nanoparticles with different concentrations of free lPEI. In addition, we studied DNA doses ranging from 0.2 to 1 μg per 104 cells (Figure S7b). The result showed that at lower doses (0.2 or 0.4 μg per 104 cells) transfection efficiency was higher for nanoparticle formulations with higher proportions of free lPEI for both bulk mixed and unpurified FNC original nanoparticles. However, when the dose was increased to 0.6 μg per 104 cells, transfection efficiency was similar at all levels of free lPEI tested. With higher doses (0.8 or 1 μg per 104 cells) the TFF purified nanoparticles had significantly higher transfection efficiency compared to non-purified nanoparticles, which may be attributed to the lower cytotoxicity of TFF nanoparticles at the higher dose (Figure 2f). The transfection efficiency in the NCI-H1299 lung cancer cells (Figure 2h) showed a similar trend as that observed in PC3 cells described above. When the DNA dose was fixed at 0.4 μg per 104 cells and the incubation time was 4 h, there were no significant differences among groups dosed with nanoparticles with different concentrations of free lPEI. The transfection efficiencies were similar for nanoparticles with different levels of free lPEI.
We evaluated the cytotoxicity and transfection efficiency for the purified nanoparticles supplemented with different levels of free lPEI; and compared them with unpurified nanoparticles. After free lPEI is added to TFF-purified pDNA/lPEI nanoparticles at intermediate (47%, FNC/TFF 10×+) and high (61.5%, similar to the unpurified FNC original nanoparticles, FNC/TFF 10×++) levels, the average transfection efficiency was similar to all purified and unpurified nanoparticles (Figure S8a). In contrast, the purified nanoparticles FNC/TFF 10× and FNC/TFF 5× significantly reduced cytotoxicity of the nanoparticles; and the lPEI-supplemented nanoparticles showed even higher levels of cytotoxicity than the unpurified nanoparticles (Figure S8b). These results indicate that while removal of uncomplexed free lPEI substantially reduced the cytotoxicity, the supplemented lPEI appeared to induce a higher level of cell toxicity.
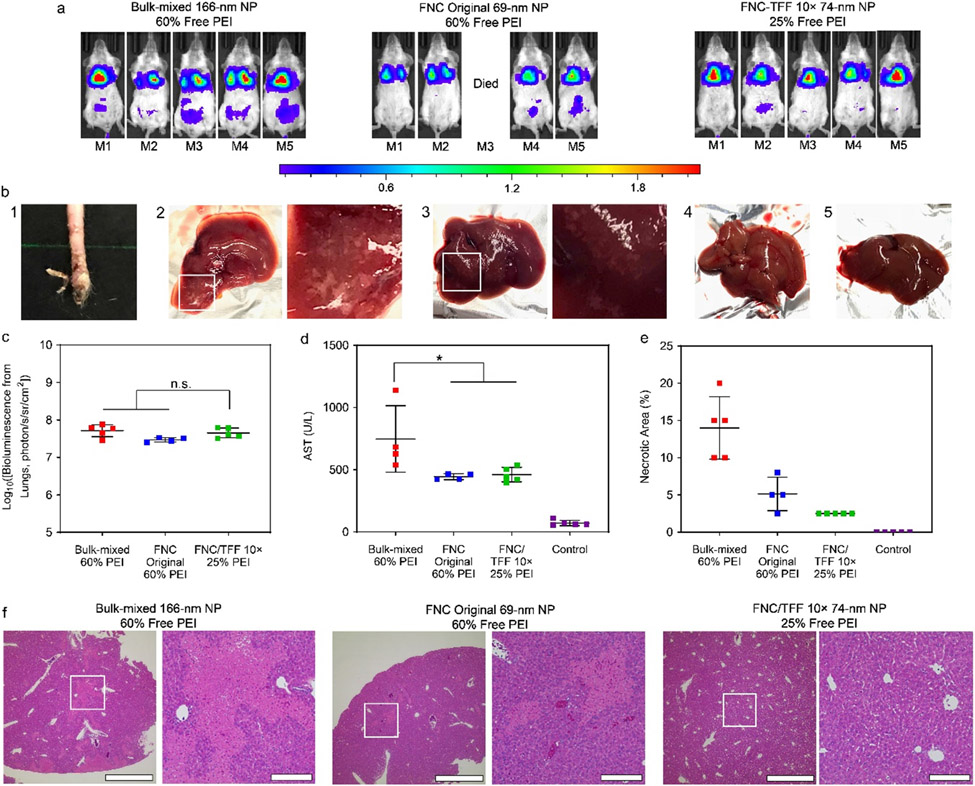
The in vivo transfection efficiency in mice dosed with purified nanoparticles was investigated in two immunocompetent models, Balb/c mice (inbred strain) and CD-1 mice (outbred strain). The dose administered was 40 μg of gWiz-Luc plasmid per mouse. The IVIS whole-body images of Balb/c mice and the IVIS ROI quantitative analysis data are shown (Figures 3a and 3c). The results showed that there was no significant difference in transfection efficiency in the lung between bulk-mixed nanoparticles, unpurified FNC original nanoparticles or TFF-purified nanoparticles, in Balb/c mice at either 24 h (Figures 3a and 3c) or 48 h (Figures S10a and S10c) time points. This shows that the presence of the additional free lPEI (~35% removed by TFF) was not essential to achieving high transfection efficiency in the lung.
Figure 3.
(a) In vivo transfection efficiency of the injected Polyplus bulk-mixed, FNC original and FNC/TFF 10× pDNA/lPEI nanoparticles, respectively in Balb/c mice. The injection dose is 40 μg DNA/mice. IVIS whole-body images were taken at 24 h post injection of particles. Scale bar: local radiance with unit of 106 photon/s/sr/cm2 (b) 1: The necrosis at the tail injection site of the unpurified nanoparticles. The images of harvested livers in each group. 2: Polyplus bulk-mixed, 3: FNC original, 4: FNC/TFF 10× pDNA/lPEI nanoparticles, 5: Naked DNA. (c) The IVIS ROI quantitative analysis results at 24 h time point from Balb/c mice. Each bar represents mean ± standard deviation (n = 5). (d) AST assessments of Balb/c mice injected with Polyplus bulk-mixed, FNC original, and FNC/TFF 10× pDNA/lPEI nanoparticles, respectively. (e) Percentage of necrotic area in liver of Balb/c mice injected with Polyplus bulk-mixed, FNC original, and FNC/TFF 10× pDNA/lPEI nanoparticles, respectively. (f) H&E-stained liver tissue sections of Balb/c mice injected with Polyplus bulk-mixed, FNC original, and FNC/TFF 10× pDNA/lPEI nanoparticles, respectively. Scale bar 1000 μm (Left) and 200 μm (Right). For statistical analysis, n.s. denotes no statistical significance with p > 0.05 and *p < 0.05 from one-way ANOVA test.
In these experiments, we also observed that one out of five mice died after the injection of unpurified nanoparticles within one hour, and there was visible necrosis at the injection site (Figure 3b-1). In contrast, the bulk-mixed nanoparticles and TFF-purified nanoparticles did not show signs of shock and acute toxicity. Nonetheless, macroscopic examination showed that both bulk-mixed (Figure 3b-2) and unpurified FNC original nanoparticles (Figure 3b-3) resulted in necrotic regions on the liver. No appreciable damage was observed in the livers of mice dosed with TFF-purified nanoparticles (Figure 3b-4) or pDNA alone (Figure 3b-5).
Liver damage can be assessed by the levels of aspartate transaminase (AST) and alanine transaminase (ALT) enzymes detected in the serum. Bulk-mixed nanoparticles elicited substantially elevated levels of AST and ALT following i.v. administration of the nanoparticles, whereas mice that received original and TFF-purified nanoparticles only showed a mild increase of AST and ALT levels (Figures 3d and S11a). Although the “FNC original 60%-lPEI” group and “bulk-mixed 60%-lPEI” group had the same level of free lPEI, the hydrodynamic size for “FNC original 60%-lPEI” group was 68 nm, which was much smaller than that of the “bulk-mixed 60%-lPEI” group (161 nm, Figure S12a). The intensity size distribution measurements shown in Figure S12b confirmed that the bulk-mixed nanoparticles had a wider and less reproducible size distribution.
Among all the groups tested, TFF-purified nanoparticles resulted in the lowest extent of necrosis measured by the percentage of necrotic area. The histological assessments confirmed that significant levels of inflammation and necrosis were observed in the livers of mice treated with bulk-mixed nanoparticles or FNC original nanoparticles (Figures 3e and 3f).
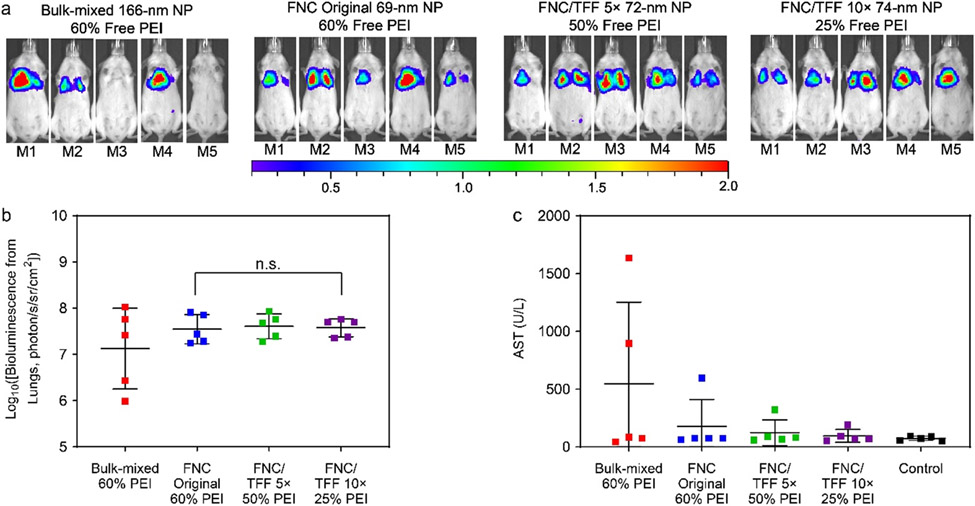
In a different strain of mice, CD-1, all tested nanoparticle preparations, including bulk-mixed nanoparticles (60% free lPEI), unpurified FNC original nanoparticles (60% free lPEI), FNC/TFF 5× or 10×-purified nanoparticles to 50% or 25% free lPEI levels, showed comparable levels of transfection efficiency at both 24-h (Figures 4a and 4b) and 48-h time points (Figures S10b and S10d). No significant level of acute or subacute toxicity was detected in any of these groups, in particular between unpurified FNC original nanoparticles and FNC/TFF 10× (25% free lPEI) nanoparticles for AST (p = 0.46) and ALT (p = 0.61) enzyme levels. Comparison between bulk-mixed and FNC original nanoparticles also did not yield significant difference in AST (p = 0.30) and ALT (p = 0.27) levels (Figures 4c and S11b). Our results indicate that Balb/c mice were more susceptible to 35% difference of free lPEI of the pDNA/lPEI nanoparticles than CD-1 mice.
Figure 4.
(a) In vivo transfection efficiency of CD-1 mice. The dose is 40 μg DNA/mice. IVIS whole-body images were taken at 24 h post injection of particles. Scale bar: local radiance with unit of 106 photon/s/sr/cm2 (b) The IVIS ROI quantitative analysis results at 24-h time point from CD-1 mice. Each bar represents mean ± standard deviation (n = 5). (c) AST assessments of CD-1 mice injected with Polyplus bulk-mixed, FNC original, FNC/TFF 5×, and FNC/TFF 10× pDNA/lPEI nanoparticles, respectively. For statistical analysis, n.s. denotes no statistical significance with p > 0.05 from one-way ANOVA test.
Discussion:
The production of highly uniform pDNA/lPEI complex nanoparticles at large scale with high reproducibility is a key challenge to successful translation of these nanoparticles from the bench to the clinic. In conventional nanoparticle preparation methods, such as that recommended by Polyplus using in vivo-jet PEI®, the lPEI and pDNA solutions are mixed in vials in a batch mode at an N/P ratio of 6 to 8. The mixing volume is typically several hundred microliters, and batch-to-batch variations are high. It is rather difficult to scale up such a batch-mixing preparation process while maintaining the uniformity of the nanoparticles prepared.29 We reported the use of a turbulent mixing process to generate pDNA/lPEI nanoparticles with high uniformity, high reproducibility and the ability to scale up. When the flow rates were higher than 20 mL/min, reaching towards a turbulent mixing process, the average sizes of nanoparticles could be controlled to smaller than those of the bulk-mixed nanoparticles. We followed the manufacturer’s recommendation by adopting an N/P = 6 to formulate the nanoparticles. With this N/P ratio, there would be ~60% of the total amount of lPEI left uncomplexed (free lPEI) in the nanoparticle suspension (see Materials and Methods section for details).
The free lPEI in the pDNA/lPEI nanoparticle suspension was successfully reduced by TFF (Figure 1a). Compared with the traditional purification by ultrafiltration, TFF is a better method to remove free lPEI with the ease of scalability (Figure S1), while reducing nanoparticle aggregation and maintaining high yield and purification efficiency. With the increasing nanoparticle preparation batch size from 1 mL to 50 mL, TFF purification efficiency was maintained at the same level following ten cycles of solvent exchange (based on the volume of solvent used for purification). Larger scale of purification could be further achieved by changing the hollow fiber cartridge with a larger inner lumen diameter, a longer length, and a larger number of hollow fibers. In addition, TFF could control the level of free lPEI more precisely by adjusting the experimental parameters for the purification step. To obtain the highest nanoparticle recovery and purification efficiency, we optimized the essential parameters including the washing solution, washing times (the replacement solvent mass), the pore size of filter and the shear rate. Initial experiments were conducted on lPEI polymer solution alone as a model sample in Figure S2. The results (Figure S2a) suggest that with higher PBS concentrations and hence higher ionic strength the purification efficiency was increased. The reason for lower purification efficiency in deionized (DI) water (Figure S2a) may be attributed to a higher degree of binding affinity between lPEI and pDNA/lPEI nanoparticles than that in PBS, which provides charge screening and facilitates release of excess lPEI from the complexes. In addition, lPEI assumes more rigid rod chain confirmation with a higher hydrodynamic volume in DI water, slowing down its effective filtration through the TFF membrane (30 kDa MWCO). Therefore, the efficiency of purification efficiency would be significantly reduced in DI water. Diluted PBS washing solution was then selected for our TFF purification system. The reduction in free lPEI removal at lower pH (Figure S2b) may have been due to a higher charge density on lPEI, which extended the polymer chains, resulting in a larger radius of gyration, and thus made it more difficult for the lPEI to diffuse through the membrane pores. To guide the screening experiments, the stability of unpurified nanoparticles in different concentrations of PBS was determined (Figure S3a). As described above, due to the charge screening effect to the net positive charges on nanoparticle surfaces, the nanoparticles aggregated in buffer with a concentration higher than 0.4× PBS. The optimized TFF washing solution was then identified as 0.1× PBS (pH 6.5). We then tested the stability of the nanoparticles under this condition (Figure S3b) to confirm that the nanoparticles would not aggregate during the TFF washing process (3 h).
After the purification with the optimized TFF conditions, the free lPEI concentration was reduced significantly to 24% (Figure 2a) with 80% nanoparticle retention (Figure 2b). TFF purification with the optimized conditions was effective in maintaining the physiochemical properties of the nanoparticles (Figures 2c, 2d, and 2e). With consistent size, size distribution, and morphology, the cytotoxicity was studied on different levels of free lPEI. We found the observed cytotoxicity could be attributed primarily to the free lPEI content of the nanoparticle preparation rather than the nanoparticles per se (Figure 2f). These results suggest that TFF purified nanoparticles can maintain the same level of in vitro transfection efficiency with reduced cytotoxicity (Figures 2f, 2g, and 2h).
Another advantage of using TFF over traditional ultrafiltration method is that TFF can easily exchange the solvent of nanoparticles solution by simply shifting the washing buffer. After the purification step, nanoparticles were suspended in 0.1× PBS solution. The solvent was then adjusted to 9.5% w/w trehalose solution to have trehalose as a cryoprotectant for the purpose of generating lyophilized formulation and to render the formulation isotonic for systemic injection. This step was tested by either bulk-mixing method, in which 19% w/w trehalose was mixed with the nanoparticle suspension in a 1:1 volume ratio (Figure S9a), or TFF (Figure S9b). First, we directly bulk-mixed nanoparticles (400 μg/mL DNA) with 19% trehalose solution at 1:1 ratio. The size distribution results are shown (Figure S9a). By bulk mixing with 19% trehalose, the nanoparticles aggregated within 5 min since the mixing rate cannot be controlled. This implied that these particles were not stable for further applications. In contrast, when the solvent of the TFF nanoparticles was gradually replaced with 9.5% trehalose using the TFF process, nanoparticle size and distribution were maintained without measurable aggregation (Figures S9b and S9c). To identify the washing volume to fully replace the solvent from PBS solution to 9.5% trehalose, we used both PBS solution (no nanoparticles) and nanoparticle solution as the original feed solution in TFF and then washed with 9.5% trehalose. Samples were lyophilized and weighed to determine the sugar concentration. The results showed that after washing with 5× volume of the 9.5% trehalose solution, solvent exchange was completed; and the size distribution of the nanoparticle preparation in 9.5% trehalose was maintained (Figure S9b). This was not achievable by the bulk mixing of trehalose solution with the nanoparticle suspension. The nanoparticle suspension was further concentrated by 5-fold (Figures S9b, S9c, and S9d). During the concentration step, no washing solution was added. The results suggest that after nanoparticles have been washed for 5 times with 9.5% trehalose and then concentrated 5-fold, their average size and size distributions were preserved.
These results highlighted the advantages of TFF for purifying pDNA/lPEI nanoparticles. TFF has the ability to scale up the purification while minimizing the risk of particle aggregation. It affords high recovery yield of nanoparticles, control for the level of free polymer, and ease of exchange of solvent or medium. These features of the optimized TFF purification process can be easily extended to other nanoparticle systems during scaleup production.
The in vivo study results demonstrate that TFF purification is capable of reducing the amount of free lPEI in nanoparticle formulations without significantly reducing the level of transfection efficiency below that of unpurified nanoparticles in both Balb/c mice and CD-1 mice. The in vivo toxicity responses were minimized by our TFF purification protocol, although the inflammation and liver toxicity appeared to be animal strain-dependent. The in vivo toxicity responses were also reported in another study14 in which Balb/c mice receiving nanoparticles with excess free polymer showed signs of shock, i.e. ruffled fur and reduced activity after injection. Here we found that there was a high degree of infiltration by mononuclear cells in the liver of mice treated with bulk-mixed nanoparticles (Figure 3f). These cells were found predominantly perivascular and included larger, blast like, immature monocytes, and were accompanied by extramedullary hematopoiesis with large, possibly atypical cells found around perivascular and intravascular regions. In contrast, liver damage was absent from the liver tissue collected from the purified FNC/TFF 10× nanoparticles treated group; and the sinusoidal structure of the liver remained normal with minimum infiltration of monocytes. Collectively, these serum biochemical analysis and pathological assessment confirmed that the hepatic injury observed in Balb/c mice was induced by the excess amount of free lPEI in nanoparticle formulations and the larger size of the nanoparticles following i.v. injection. The FNC/TFF purified nanoparticles significantly reduced the toxicity without compromising the transfection efficiency in the lung.
Conclusion:
The pDNA/lPEI nanoparticles produced by the FNC method was effectively purified by the optimized TFF protocol. TFF purification was able to reduce free lPEI from nanoparticle suspension by 60% (from 60% to 24% in a nanoparticle formulation prepared at an N/P ratio of 6) with a satisfactory yield and preserved physical characteristics, including size, size distribution, and morphology. We also developed a TFF protocol to replace the carrier liquid of the nanoparticle suspension with an isotonic solution, 9.5% w/w trehalose, and to increase the concentration of the nanoparticles without compromising their stabilities. The purified pDNA/lPEI nanoparticles with reduced free lPEI content (24% of the input lPEI) delivered lower toxicities and similarly high level of transfection efficiencies both in vitro and in vivo, in comparison with unpurified nanoparticles. This TFF technique coupled with the FNC production process enabled the continuous and scalable production of purified nanoparticles at a tailored concentration. This method can be easily extended to other nanoparticle formulations and has high translational potential for the development of nanoparticle gene medicine.
Supplementary Material
Acknowledgements:
This study is partially supported by a research contract from Cancer Targeting Systems, Inc. and an NIH grant R01EB018358 (HQM), P41EB024495 (MGP), and P50CA058236 (MGP).
Footnotes
Supporting Information. Detailed experimental procedures and data for TFF optimization, and Supplementary Figures S1 to S12 for the in vitro and in vivo studies.
References:
- (1).Ginn SL; Alexander IE; Edelstein ML; Abedi MR; Wixon J Gene Therapy Clinical Trials Worldwide to 2012 - an Update. Journal of Gene Medicine. 2013, 15 (2), 65–77. [DOI] [PubMed] [Google Scholar]
- (2).Peer D; Karp JM; Hong S; Farokhzad OC; Margalit R; Langer R Nanocarriers as an Emerging Platform for Cancer Therapy. Nat. Nanotechnol 2007, 2, 751–760. [DOI] [PubMed] [Google Scholar]
- (3).Patnaik S; Gupta KC Novel Polyethylenimine-Derived Nanoparticles for in Vivo Gene Delivery. Expert Opin. Drug Deliv 2013, 10 (2), 215–228. [DOI] [PubMed] [Google Scholar]
- (4).Jere D; Jiang HL; Arote R; Kim YK; Choi YJ; Cho MH; Akaike T; Cho CS Degradable Polyethylenimines as DNA and Small Interfering RNA Carriers. Expert Opin. Drug Deliv 2009, 6 (8), 827–834. [DOI] [PubMed] [Google Scholar]
- (5).Wightman L; Kircheis R; Rossler V; Carotta S; Ruzicka R; Kursa M; Wagner E Different Behavior of Branched and Linear Polyethylenimine for Gene Deliveryin Vitro Andin Vivo. J. Gene Med 2001, 3 (4), 362–372. [DOI] [PubMed] [Google Scholar]
- (6).Ami SA; Patricia O; Shalva B; Moshe S; H. RJ; Donald L; Avraham H; Ilan L Phase I/II Marker Lesion Study of Intravesical BC-819 DNA Plasmid in H19 Over Expressing Superficial Bladder Cancer Refractory to Bacillus Calmette-Guerin. J. Urol 2008, 180 (6), 2379–2383. [DOI] [PubMed] [Google Scholar]
- (7).Davies LA; McLachlan G; Sumner-Jones SG; Ferguson D; Baker A; Tennant P; Gordon C; Vrettou C; Baker E; Zhu J; Alton EWFW; Collie DDS; Porteous DJ; Hyde SC; Gill DR Enhanced Lung Gene Expression After Aerosol Delivery of Concentrated PDNA/PEI Complexes. Mol. Ther 2008, 16 (7), 1283–1290. [DOI] [PubMed] [Google Scholar]
- (8).Lisziewicz J; Gabrilovich DI; Varga G; Xu J; Greenberg PD; Arya SK; Bosch M; Behr J-P; Lori F Induction of Potent Human Immunodeficiency Virus Type 1-Specific T-Cell-Restricted Immunity by Genetically Modified Dendritic Cells. J. Virol 2001, 75 (16), 7621–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Buscail L; Bournet B; Vernejoul F; Cambois G; Lulka H; Hanoun N; Dufresne M; Meulle A; Vignolle-Vidoni A; Ligat L; Saint-Laurent N; Pont F; Dejean S; Gayral M; Martins F; Torrisani J; Barbey O; Gross F; Guimbaud R; Otal P; Lopez F; Tiraby G; Cordelier P First-in-Man Phase 1 Clinical Trial of Gene Therapy for Advanced Pancreatic Cancer: Safety, Biodistribution, and Preliminary Clinical Findings. Mol. Ther 2015, 23 (4), 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Godbey WT; Wu KK; Mikos AG Poly(Ethylenimine)-Mediated Gene Delivery Affects Endothelial Cell Function and Viability. Biomaterials 2001, 22 (5), 471–480. [DOI] [PubMed] [Google Scholar]
- (11).Regnström K; Ragnarsson EGE; Köping-Höggård M; Torstensson E; Nyblom H; Artursson P PEI – a Potent, but Not Harmless, Mucosal Immuno-Stimulator of Mixed T-Helper Cell Response and FasL-Mediated Cell Death in Mice. Gene Ther. 2003, 10, 1575–1583. [DOI] [PubMed] [Google Scholar]
- (12).Yue Y; Jin F; Deng R; Cai J; Dai Z; Lin MCM; Kung H-F; Mattebjerg MA; Andresen TL; Wu C Revisit Complexation between DNA and Polyethylenimine — Effect of Length of Free Polycationic Chains on Gene Transfection. J. Control. Release 2011, 152 (1), 143–151. [DOI] [PubMed] [Google Scholar]
- (13).Morimoto K; Nishikawa M; Kawakami S; Nakano T; Hattori Y; Fumoto S; Yamashita F; Hashida M Molecular Weight-Dependent Gene Transfection Activity of Unmodified and Galactosylated Polyethyleneimine on Hepatoma Cells and Mouse Liver. Mol. Ther 2003, 7 (2), 254–261. [DOI] [PubMed] [Google Scholar]
- (14).Boeckle S; von Gersdorff K; van der Piepen S; Culmsee C; Wagner E; Ogris M Purification of Polyethylenimine Polyplexes Highlights the Role of Free Polycations in Gene Transfer. J. Gene Med 2004, 6 (10), 1102–1111. [DOI] [PubMed] [Google Scholar]
- (15).Köping-Höggård M; Tubulekas I; Guan H; Edwards K; Nilsson M; Vårum KM; Artursson P Chitosan as a Nonviral Gene Delivery System. Structure–Property Relationships and Characteristics Compared with Polyethylenimine in Vitro and after Lung Administration in Vivo. Gene Ther. 2001, 8, 1108–1121. [DOI] [PubMed] [Google Scholar]
- (16).Lavertu M; Méthot S; Tran-Khanh N; Buschmann MD High Efficiency Gene Transfer Using Chitosan/DNA Nanoparticles with Specific Combinations of Molecular Weight and Degree of Deacetylation. Biomaterials 2006, 27 (27), 4815–4824. [DOI] [PubMed] [Google Scholar]
- (17).Peng S-F; Tseng MT; Ho Y-C; Wei M-C; Liao Z-X; Sung H-W Mechanisms of Cellular Uptake and Intracellular Trafficking with Chitosan/DNA/Poly(γ-Glutamic Acid) Complexes as a Gene Delivery Vector. Biomaterials 2011, 32 (1), 239–248. [DOI] [PubMed] [Google Scholar]
- (18).Thibault M; Astolfi M; Tran-Khanh N; Lavertu M; Darras V; Merzouki A; Buschmann MD Excess Polycation Mediates Efficient Chitosan-Based Gene Transfer by Promoting Lysosomal Release of the Polyplexes. Biomaterials 2011, 32 (20), 4639–4646. [DOI] [PubMed] [Google Scholar]
- (19).Musumeci T; Leonardi A; Bonaccorso A; Puglisi R and P. G Tangential Flow Filtration Technique: An Overview on Nanomedicine Applications. Pharmaceutical Nanotechnology. 2018, 6 (1), 48–60. [DOI] [PubMed] [Google Scholar]
- (20).Dalwadi G; Benson HAE; Chen Y Comparison of Diafiltration and Tangential Flow Filtration for Purification of Nanoparticle Suspensions. Pharm. Res 2005, 22 (12), 2152–2162. [DOI] [PubMed] [Google Scholar]
- (21).Robertson JD; Rizzello L; Avila-Olias M; Gaitzsch J; Contini C; Magoń MS; Renshaw SA; Battaglia G Purification of Nanoparticles by Size and Shape. Sci. Rep 2016, 6, 27494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jasinski DL; Schwartz CT; Haque F; Guo P Large Scale Purification of RNA Nanoparticles by Preparative Ultracentrifugation BT- RNA Nanotechnology and Therapeutics: Methods and Protocols. 2015, 1297, 67–82. [DOI] [PubMed] [Google Scholar]
- (23).Mao H-Q; Roy K; Troung-Le VL; Janes KA; Lin KY; Wang Y; August JT; Leong KW Chitosan-DNA Nanoparticles as Gene Carriers: Synthesis, Characterization and Transfection Efficiency. J. Control. Release 2001, 70 (3), 399–421. [DOI] [PubMed] [Google Scholar]
- (24).Johnson ME; Hanna SK; Montoro Bustos AR; Sims CM; Elliott LCC; Lingayat A; Johnston AC; Nikoobakht B; Elliott JT; Holbrook RD; Scott KCK; Murphy KE; Petersen EJ; Yu LL; Nelson BC Separation, Sizing, and Quantitation of Engineered Nanoparticles in an Organism Model Using Inductively Coupled Plasma Mass Spectrometry and Image Analysis. ACS Nano. 2017, 11 (1), 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Fornaguera C; Solans C Analytical Methods to Characterize and Purify Polymeric Nanoparticles. International Journal of Polymer Science. 2018, 2018, 1–10. [Google Scholar]
- (26).Pansare VJ; Tien D; Thoniyot P; Prud’homme RK Ultrafiltration of Nanoparticle Colloids. J. Memb. Sci 2017, 538, 41–49. [Google Scholar]
- (27).He Z; Hu Y; Nie T; Tang H; Zhu J; Chen K; Liu L; Leong KW; Chen Y; Mao H-Q Size-Controlled Lipid Nanoparticle Production Using Turbulent Mixing to Enhance Oral DNA Delivery. Acta Biomater. 2018, 81, 195–207. [DOI] [PubMed] [Google Scholar]
- (28).Hu Y; He Z; Hao Y; Gong L; Pang M; Howard GP; Ahn H-H; Brummet M; Chen K; Liu H; Ke X; Zhu J; Anderson CF; Cui H; Ullman CG; Carrington CA; Pomper MG; Seo J-H; Mittal R; Minn I; Mao H-Q Kinetic Control in Assembly of Plasmid DNA/Polycation Complex Nanoparticles. ACS Nano. 2019, 13 (9), 10161–10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Santos JL; Ren Y; Vandermark J; Archang MM; Williford J-M; Liu H-W; Lee J; Wang T-H; Mao H-Q Continuous Production of Discrete Plasmid DNA-Polycation Nanoparticles Using Flash Nanocomplexation. Small. 2016, 12 (45), 6214–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cho EC; Zhang Q; Xia Y The Effect of Sedimentation and Diffusion on Cellular Uptake of Gold Nanoparticles. Nat. Nanotechnol 2011, 6, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Pezzoli D; Giupponi E; Mantovani D; Candiani G Size Matters for in Vitro Gene Delivery: Investigating the Relationships among Complexation Protocol, Transfection Medium, Size and Sedimentation. Sci. Rep 2017, 7, 44134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.