Abstract
An extraordinary biodiversity of bacteria, fungi, viruses, and even small multicellular eukaryota inhabit the human skin. Genomic innovations have accelerated characterization of this biodiversity both at a species as well as the subspecies, or strain level, which further imparts a tremendous genetic diversity to an individual’s skin microbiome. In turn, these advances portend significant species- and strain-specificity in the skin microbiome’s functional impact on cutaneous immunity, barrier integrity, aging, and other skin physiologic processes. Future advances in defining strain diversity, spatial distribution, and metabolic diversity for major skin species will be foundational for understanding the microbiome’s essentiality to the skin ecosystem and for designing topical therapeutics that leverage or target the skin microbiome.
The skin harbors a diverse microbial community
Contrasted to the body’s other interfaces with the external environment, the human skin features a relatively low-nutrient barrier as its first line of defense against biotic and abiotic foreign matter. Despite a core function of ‘keep it out’, human skin is home to a diversity of microorganisms, including bacteria (primarily Cutibacterium, Staphylococcus, Corynebacterium, and Micrococcus spp. and other Actinobacteria, Proteobacteria, and Firmicutes, Figure 1A), fungi (primarily Malassezia sp.), viruses (both phage and human viruses, including papillomavirus and polyomavirus), and small eukaryotes such as mites (Demodex sp.)[1,2].
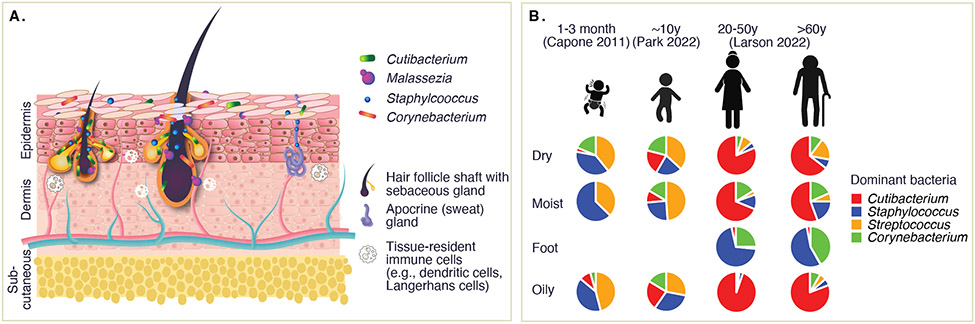
Figure 1. Overview of the skin microbiome.
A) The skin microbiome in healthy skin is considered to reside primarily in deeper structures such as pilosebaceous units (e.g., terminal or vellus hair follicles and sebaceous glands) and sweat glands, with relatively fewer microbes inhabiting the stratum corneum. Localization, density, and admixture of species within these structures remains little defined. These bacteria, fungi, and viruses have important roles in skin barrier homeostasis, cutaneous innate and adaptive immunity, and help condition the skin microenvironment. B) Representation of major skin microbiome changes over lifespan, where available. Piecharts are composite relative abundance data from the three indicated studies.
Considering the thick, scaly skin of the heel, the dry expanses of the forearm, the oily pores of the nose, or the hairy mucosal inner nares, the skin has marked physiologic differences over its breadth. In addition to differing numbers of pilosebaceous units, including different types of hair follicles such as terminal or vellus, sebaceous and apocrine glands, the thickness and composition of the skin’s major layers – the epidermis, the dermis, and the subcutaneous layer – vary based on location, features which collectively determine the skin’s numerous physiochemical variations in skin moisture, pH, salinity and oiliness[3]. Striking differences in the skin microbiome within a single individual reflect these physiologic differences, which dramatically remodel over lifespan (Figure 1B). In healthy individuals, the skin microbiome begins with seeding of the skin by the mother’s vaginal or skin microbiome during birth[4-6], then shifts towards a lipophile-dominated community following major hormonal changes during puberty, which increase skin oiliness[6,7]. Communities then remain relatively stable through adulthood[1], but will again remodel with age, diversifying in concurrence with major physiologic changes of thinning and drying skin[7]. Finally, the skin microbiome differs significantly between individuals and is influenced transiently or long-term by numerous additional intrinsic (e.g., genetics, immunocompetence, skin barrier status) and extrinsic factors (e.g., ethnicity, geography, hygiene and other personal habits, medication use, exposure to pathogens).
Different constituents of the skin microbiome have been linked to a wide range of cutaneous processes in health and disease, including modulation of innate and adaptive immunity through different stages of life[8-12], colonization resistance to pathogens[13,14], maintenance of the skin barrier and microenvironment[15,16], and wound healing[11,12,17], to name a few. Conversely, skin microbiome dysfunction and colonization of pathobionts like Staphylococcus aureus has been established to be associated with or to contribute to skin diseases such as atopic dermatitis[18-23], Netherton syndrome[24], and skin cancers[25]. As species-level interactions in skin health and disease have been reviewed excellently elsewhere [26-30], we will primarily discuss the contributions of two ubiquitous skin bacteria at sub-species level, which are the focus of the remainder of this review.
Deeper than species: host interactions are strain specific
Such studies have amply demonstrated the breadth of host processes that are regulated by different skin species. Adding to the challenges of investigating skin microbiome interactions is the extensive array of genetic diversity within each species. Indeed, microbial diversity and its phenotypic consequences are ultimately manifested at this finest taxonomic resolution, not necessarily at the species level. Infectious disease specialists are well-versed on the concept of this variation at the subspecies, or strain level – e.g., nosocomial vs. commensal, methicillin resistant vs. drug-susceptible S. aureus strains – thus a strain is defined as a genetic variant of a species, and “isolate”, a term often used interchangeably, is a heretofore uncharacterized strain. A lineage encompasses strains that are descended from a common ancestor, and strains can be very similar to their parent or differ significantly based on mutational rate or horizontal gene transfer introducing new genic elements.
Most studies to date have surveyed strain diversity in different individuals, or transmission from the environment in efforts to identify broad characteristic of disease-causing vs. healthy strains. However, two major recent efforts in skin, performing comparative genomics on libraries of Staphylococcus epidermidis and Cutibacterium acnes isolates, identified that striking strain diversity can exist within-individual; even within skin-site. In addition, these examples well exemplify that strain variation will take multiple manifestations depending on the species of interest.
Staphylococcal strain diversity
Zhou et al. surveyed 1,462 isolates of S. epidermidis isolated from 12 skin sites of 5 healthy individuals, targeting 10 isolates per skin sample[31]. While this depth is certainly not exhaustive, it was the first large-scale effort examining whether strains within a single skin site are clonal, or deriving from multiple lineages. Using a combination of tracing single nucleotide polymorphisms (SNPs) in the core genome (genic regions shared between all strains, which estimates evolutionary distance between genomes), and examining gene content differences (genic regions unique to only a subset of genomes, termed the accessory genome), they found that S. epidermidis strains within an individual and remarkably, even within a skin site, were far from clonal; nearly every isolate was a unique variant (Figure 2A).
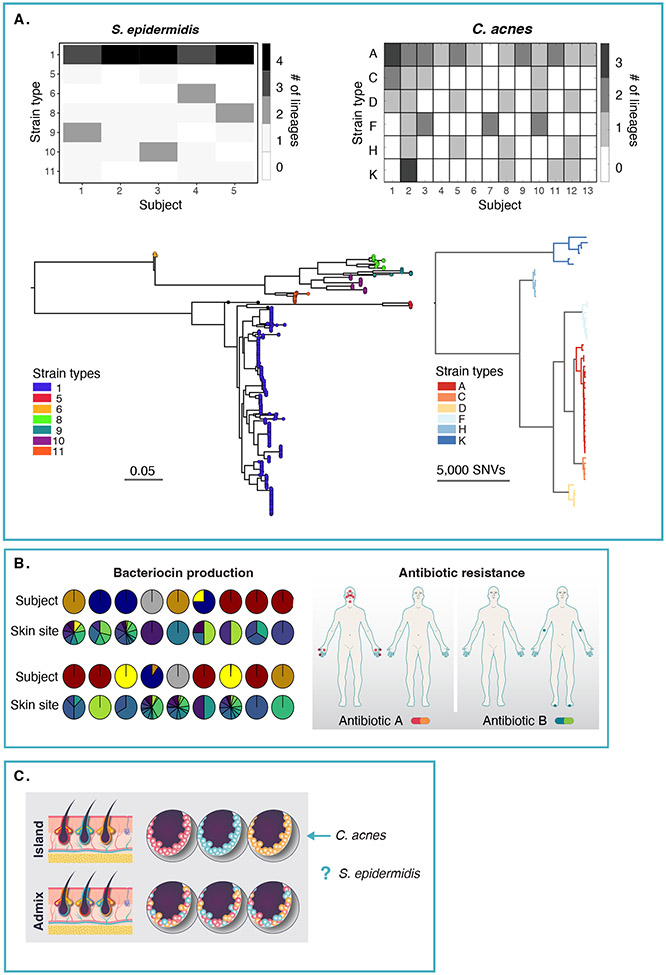
Figure 2. Extensive within-individual strain variation of two major skin species.
A) An individual simultaneously harbors multiple lineages of S. epidermidis (left) and C. acnes (right) across their skin. For S. epidermidis (left), the phylogenetic tree was inferred based on whole genome assemblies of 1477 isolates. Lineages were defined as groups of isolates (n>2) with genetic distances not exceeding 0.05. Similarly, isolates with genetic distances not exceeding 0.15 were annotated to be of the same "strain type", which are arbitrarily named. For C. acnes (figure adapted from Conwill et al., 2022), lineages were defined as sets of colonies separated by <100 mutations. The 53 lineages generated from 947 isolate genomes are shown in the tree, with strain types named by single locus sequence type. The distribution of lineage richness of each subject is visualized in the heatmaps for each species. B) For S. epidermidis, two examples of functional diversity arising from strain-level genetic diversity are shown. Distribution of 18 different types of predicted bacteriocins, and their distribution across subjects (top row) and skin site (bottom row) is shown. An example of antibiotic resistance gene reservoirs differing between individuals but disseminated across skin sites, with red or green dots showing presence in at least one isolate from that skin site. For both bacteriocin distribution and antibiotic resistance, most are individual-specific but distributed across multiple skin sites within that individual (figure adapted from Zhou et al., 2020). C) A key unresolved question is to what degree does genetically diverse strains co-exist at a microscale, in isolated skin structures? From Conwill et al., C. acnes follows the “island” hypothesis, but each species may differ in their spatial distributions (Figure adapted from Kong, Oh, 2022).
We note several findings of particular interest. First, a within-individual analysis of Bacteroides fragilis found that healthy individuals possess a single lineage that diversified within the gut of an individual over time[32], whereas S. epidermidis strains within an individual derived from multiple founder lineages, rather than a single colonizer. Populations were subsequently shaped by site adaptation, particularly in the thick scaly skin of the foot as well as through transmission events between high-touch sites such as the hands. Strikingly, horizontal gene transfer could occur on short evolutionary timeframes. Sister strains, zero SNP differences in the core genome – that is, the genome that is shared between all strains, could possess different gene contents, and very closely related strains with few SNP differences could differ by hundreds of genes. Such genes were often associated with mobile elements and, notably, plasmid-borne antibiotic resistance genes, whose function they verified experimentally. Given the striking co-occurrence of genetically diverse strains, what might be a potential role in skin health? By making admixtures of S. epidermidis strains observed within a skin site and examining transcriptional response to admixture, the presence of multiple diverse strains was shown to suppress expression of virulence factors and modulate metabolism on a population-level. Thus, strain diversity might be one mechanism to suppress S. epidermidis’ potential transition to pathogenicity, an area of interest to pursue in in vivo and bloodstream infection models.
C. acnes strain diversity
C. acnes differs from S. epidermidis in its relatively closed accessory genome, with ~10% of the genome estimated to vary between strain types, vs. 20% for the latter[33]. Conwill et al. surveyed 947 isolates of C. acnes obtained from 300 samples from 16 healthy adults, obtaining 1–15 colonies per sample[34]. The unique aspect of their design was that 145 of these samples were isolated pore samples from 5 individuals, allowing them to assess strain diversity at a much finer geographic scale allowable than a bulk skin swab. Indeed, they found that a pore was effectively a genetic island, with most isolates derived from the same pore having very few SNP level differences. On a broader scale, similar (but not clonal) strains could co-exist within an individual (Figure 2A), suggesting that pores are monocolonized at random, i.e., a population bottleneck. They purport that there is limited competition between co-existing C. acnes strains – one would suspect because there are relatively few genic differences between strains that would beget a significant functional advantage, although the authors observed differences in in vitro growth rate. Indeed, a compelling segue to this experiment would be to understand if these island-like population structures persist in C. acnes-associated diseases (like acne), or if certain strains possess gene-level differences that would overcome the neutral processes in healthy skin. It may very well be a combination of dispersion as well as a potential for genetic specialization (together with host immunity and intrinsic states) that mediates C. acnes’ contribution to acne.
Functional consequences of strain diversity
Now, how might we contextualize the findings of studies of S. epidermidis’ or C. acnes’ contribution to skin health and disease? Most studies have not – or are not – able to systematically study a wide breadth of strains for each species’ study. Yet there is ample precedent that that these strain-level differences are impactful. Continuing with the example of acne, while certain phylotypes (genetically similar groups of strains, of 6 identified in C. acnes[35]), e.g., “phylotype I” strains, have greater associations in acne formation than those of phylotype II, which are more closely related to healthy skin as well as, interestingly, deep tissue infections, potentially on account of the opportunistic nature of this phylotype[36]. In addition to the suppression of population-level virulence by S. epidermidis as discussed, there is ample evidence of phenotypic diversity of strains with significant health consequences (Figure 2B). Several examples include: in atopic dermatitis, patients with less severe skin flares had distinct S. epidermidis strain diversity (predicted by metagenomic sequencing), with a loss of genetic groups, or clades, typical of healthy controls[19,22]. This is further bolstered by evidence in mice, in which skin barrier disruption was observed with specific strains of S. epidermidis that produce excessive amounts of a damaging protease[37]. Other strains of S. epidermidis can produce metabolites that are protective against skin cancer in mice[38]. Numerous cases have identified different strains that produce different antimicrobials or possess antibiotic resistance genes[13,39], which may be a risk factor for transfer to other microbes in the environment[40]. A particularly intriguing study recently demonstrated that S. epidermidis can regulate wound healing in skin via recruitment of mucosa-associated invariant T (MAIT) cells in mice[11], with implications that different strains present in different regions might then modulate wound healing in a skin-site specific manner.
However, it is important to note that few ‘smoking gun’ characteristics of disease-causing strains have been identified, and that most strains lie on a continuum of commensal to virulent[33]. This is likely because of the complexity of genetic variants observed in a given strain - numerous genes/variants may endow plasticity in health vs. disease environments, or genetic background can modify gene essentiality[41] or virulence[42]. For example, while acne-associated phylotype I strains innately produce higher levels of porphyrins, this production can be regulated by the availability of vitamin B12, while other strains are non-producers and also non-responsive to B12[43]. S. epidermidis strains can produce widely varying amounts of the metabolite of the riboflavin biosynthesis pathway that mediates MAIT cell activation, dependent on environmental growth conditions (unpublished data).
Challenges and alternative approaches to resolving strain diversity
We note several challenges to bringing strain diversity into the mainstream vis-à-vis their mechanistic role in host-microbiome interactions. While there are numerous efforts now being taken to create patient-specific isolate collections[13,44-48], a significant challenge remains in systematically defining strain diversity genetically and phenotypically because of the scale and scope required. Isolation by cultivation followed by whole genome sequencing is the gold standard for generating sufficiently high-quality genomes for differentiation at the SNP level. However, cultivation and isolate sequencing is laborious, especially for investigating within-population diversity, and can be further limiting for low abundance microbes, in the absence of methods for enrichment. At the other extreme, algorithms to infer strain diversity from bulk metagenomic data require significant depth for each species interrogated, but still can dramatically underestimate within-population strain diversity, e.g., by identifying a dominant strain type based on SNP variation in a set of conserved marker genes[49-53]. Culture-assisted metagenomics, in which a limited number of cultivated isolates are used to track strains over time in additional metagenomic samples, may prove a useful intermediate, however; this has similarly limited ability to resolve population-level diversity, as an individual’s specific set of strains for each species of interest must be characterized a priori for most accurate tracking[53]. A very recent innovation in droplet-based, single-cell microbial metagenomics has promise in reconstructing, with extraordinary throughput, individual genomes; however, due to genome incompleteness, genomes obtained from multiple droplets had to be merged to make a composite genome, losing some information on strain variation[54].
Additional tools to examine the functional consequences of genetic variation across multiple strain types will be useful to probe the accessory genome. Transposon mutagenesis (e.g., Tn-seq[55]) knockout/knockdown or CRISPRi tools[41,56-59] are promising approaches that can be deployed in multiple strains as opposed to more laborious gene knockout approaches, and can be used to profile fitness effect of genes in different environmental conditions. However, these approaches are still predicated on genetic transformability, which remains a major challenge in primary isolates, which possess numerous restriction modification systems and other barriers to efficient genetic transformation. Overexpression of pangenome regions in a genetically tractable strain is another possibility to evaluate a phenotype of interest. Ultimately, genomic data should also be leveraged to identify genetically diverse strain sets worthy of phenotyping in low- mid-throughput assays, or to identify genes and variants of interest for synthesis and testing. For example, a clever deployment of genome-wide association studies (GWAS) with 415 strains of S. epidermidis identified 61 gene variants associated with infection vs. commensalism, further enabling 80% accuracy in a random forests machine learning model in classifying disease-causing vs. commensal strains[60]. High throughput screens, such as those searching for antimicrobial production[13], immunomodulatory ability, or other specific host interaction (i.e., production of molecules sensed by G-protein coupled receptors[61]), will also help to refine candidates for more laborious mechanistic follow-ups.
Outlook
Probing the microbiome’s role in the skin with not only species, but also strain-level resolution is a major emerging frontier in microbiome research, and we envision that these ecological principles will be similarly investigated in other prevalent skin species – e.g., the many additional staphylococcal species in the skin, Corynebacterium, Micrococcus and others. Characterizing strain diversity within and between individuals in states of health and disease will dramatically refine the genetic blueprint of the microbiome first established by metagenomic sequencing. Studies of strain diversity will be further complicated by a species’ unique evolutionary trajectory in the human body, as it is likely that many, if not most inferences on a species’ genetic diversity at the strain level will need to be reconstructed on a species-by-species basis. Given the already substantial species-level diversity of the human microbiota, systematic studies to reconstruct strain diversity on a greater scope will require technological and algorithmic innovations, including recent efforts in massively parallelized single cell approaches[54], integrated cultivation-based and metagenomic analyses, and algorithms that can delineate standing genetic variation. To complement genomic reconstructions, analogous high throughput efforts in strain phenotyping will be needed to translate the functional consequences of this genetic diversity. Recent examples have included screening for antibiotic resistance[31], production of useful antimicrobials[13,62], and production of immunomodulatory metabolites[63-66].
Finally, innovations in spatial resolution of both species and strains[67-69], perhaps drawing on recent technologies in spatial transcriptomics[70-72], are critically needed to understand to what degree strains (and species) actually co-exist in the different skin structures, and which are influencing a given host cell type, and what is the host response. For example, while C. acnes and S. epidermidis are ubiquitous in skin, some strains of C. acnes produce cutimycin, an antimicrobial that can reduce the presence of S. epidermidis in the same hair follicle[73]. This antagonistic relationship appears of increasing importance in relation to skin health, as altered Cutibacterium:Staphylococcus ratios have been observed in skin cancer[25] and aging[7]. However, the degree to which different S. epidermidis strains co-localize or compete on a microscale with other genetically diverse strains, C. acnes, or other skin microbiota remains unknown (Figure 2C). In addition, it would be of significant value to pinpoint host interactions that result from microbial colonization – for example, recent efforts using single cell transcriptomics characterized immune cell populations in the follicular environment that appeared to influence the resident skin microbiome composition[74] (and potentially, vice versa). Key questions remain: where do specific microbes reside in proximity to host cells and to each other? Which host cells are responding to which microbes, and what is their response? Such forays will continue to transform our understanding of how the microbiome shapes skin health.
Highlights.
The human skin microbiome differs between and within individuals
Ubiquitous bacteria S. epidermidis and C. acnes have major roles in cutaneous immunity and physiology
Strain-level differences can modulate interactions with the host
Knowledge gaps remain: strain diversity at a microscale and between species
Acknowledgements
JO is supported by the National Institutes of Health (1 R01 AR078634-01, DP2 GM126893- 01, 1 U19 AI142733, 1 R21 AR075174, 1U54NS105539).
Footnotes
Declaration of Interest
JO is on the scientific advisory board of Azitra, Inc., Dermbiont, Inc., and ELSI Skin Health, Inc.
References
- 1.Oh J, Byrd AL, Park M, Kong HH, Segre JA: Temporal Stability of the Human Skin Microbiome. Cell 2016, 165:854–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program, Kong HH, Segre JA: Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yousef H, Alhajj M, Sharma S: Anatomy, Skin (Integument), Epidermis. In StatPearls. . StatPearls Publishing; 2022. [PubMed] [Google Scholar]
- 4.Casterline BW, Paller AS: Early development of the skin microbiome: therapeutic opportunities. Pediatr Res 2021, 90:731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R: Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010, 107:11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capone KA, Dowd SE, Stamatas GN, Nikolovski J: Diversity of the Human Skin Microbiome Early in Life. Journal of Investigative Dermatology 2011, 131:2026–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Larson PJ, Zhou W, Santiago A, Driscoll S, Fleming E, Voigt AY, Chun O, Grady JJ, Kuchel GA, Robison JT, et al. : The microbiome of aging: instability, heterogeneity, and pathogenicity reservoirs in the skin, oral, and gut microbiome of older adults. Nature Aging [date unknown], In press. This paper describes the skin microbiome of older adults as a function of frailty, showing that the skin microbiome composition associates with frailty and the presence of pathogenicity reservoirs.
- 8.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. : Compartmentalized Control of Skin Immunity by Resident Commensals. Science 2012, 337:1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naik S, Bouladoux N, Linehan JL, Han S-J, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. : Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 2015, 520:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scharschmidt TC, Vasquez KS, Truong H-A, Gearty SV, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, et al. : A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity 2015, 43:1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linehan JL, Harrison OJ, Han S-J, Byrd AL, Vujkovic-Cvijin I, Villarino AV, Sen SK, Shaik J, Smelkinson M, Tamoutounour S, et al. : Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell [date unknown], doi: 10.1016/j.cell.2017.12.033. Here, they showed that some strains of S. epidermidis specifically activates CD8+ T cells able to express IL-17 and IFN-γ in mouse skin, and that these CD8+ T cells have a tissue repair signature and accelerate wound healing.
- 12. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han S-J, Chen YE, Li K, Farhat S, Weckel A, et al. : MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 2019, 366. Lack of colonization by microbes early in life results in deficiencies in mucosal-associated invariant T cells later in life that could not be rescued by microbial exposure late in life. In addition, they showed that different S. epidermidis cells could expand specific populations of cutaneous MAIT cells which promote wound healing – all in a mouse model.
- 13.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, et al. : Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Science Translational Medicine 2017, 9:eaah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakatsuji T, Hata TR, Tong Y, Cheng JY, Shafiq F, Butcher AM, Salem SS, Brinton SL, Rudman Spergel AK, Johnson K, et al. : Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med 2021, 27:700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng Y, Hunt RL, Villaruz AE, Fisher EL, Liu R, Liu Q, Cheung GYC, Li M, Otto M: Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host & Microbe 2022, 30:301–313.e9. Profiling 4 dozen different S. epidermidis isolates and additional skin commensal microbes, Zheng demonstrated in vitro and in vivo that S. epidermidis had a unique ability to produce a sphingomyelinase that generates ceramides, a key protective nutrient in skin barrier.
- 16. Uberoi A, Bartow-McKenney C, Zheng Q, Flowers L, Campbell A, Knight SAB, Chan N, Wei M, Lovins V, Bugayev J, et al. : Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host & Microbe 2021, 29:1235–1248.e8. Performing RNA-seq and skin barrier assessments in germ-free, antibiotic treated, or mice colonized with a cocktail of human commensal skin microbiota, Uberoi showed that commensal microbes mediate skin barrier homeostasis via the aryl hydrocarbon receptor signaling pathway.
- 17.Di Domizio J, Belkhodja C, Chenuet P, Fries A, Murray T, Mondéjar PM, Demaria O, Conrad C, Homey B, Werner S, et al. : The commensal skin microbiota triggers type I IFN–dependent innate repair responses in injured skin. Nat Immunol 2020, 21:1034–1045. [DOI] [PubMed] [Google Scholar]
- 18.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Murray PR, et al. : Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012, 22:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng W-I, Conlan S, Program NCS, Belkaid Y, Segre JA, Kong HH: Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Science Translational Medicine 2017, 9:eaal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, Shoji T, Takada S, Nakagawa S, Oguma R, et al. : Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Science Translational Medicine 2020, 12. Whole genome sequencing of S. aureus isolates from 268 infants showed that while strain diversity, presence of virulence factors, or quorum sensing (agr) type of S. aureus was not associated with predilection for developing atopic dermatitis, the lack of a functional agr system was associated with infants who did not develop AD. They found that agr is required for epidermal colonization and virulence and concluded that its retention is needed for disease progression.
- 21.Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, Cheng JY, Shafiq F, Higbee K, Hata TR, Horswill AR, et al. : Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J Allergy Clin Immunol 2020, doi: 10.1016/j.jaci.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chng KR, Tay ASL, Li C, Ng AHQ, Wang J, Suri BK, Matta SA, McGovern N, Janela B, Wong XFCC, et al. : Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nature Microbiology 2016, 1:16106. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, Kong HH, Amagai M, Nagao K: Dysbiosis and Staphyloccus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity 2015, 42:756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams MR, Cau L, Wang Y, Kaul D, Sanford JA, Zaramela LS, Khalil S, Butcher AM, Zengler K, Horswill AR, et al. : Interplay of Staphylococcal and Host Proteases Promotes Skin Barrier Disruption in Netherton Syndrome. Cell Reports 2020, 30:2923–2933.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voigt AY, Emiola A, Johnson JS, Fleming ES, Nguyen H, Zhou W, Tsai KY, Fink C, Oh J: Skin microbiome variation with cancer progression in human cutaneous squamous cell carcinoma. J Invest Dermatol 2022, doi: 10.1016/j.jid.2022.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris-Tryon TA, Grice EA: Microbiota and maintenance of skin barrier function. Science 2022, 376:940–945. [DOI] [PubMed] [Google Scholar]
- 27.Belkaid Y, Tamoutounour S: The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol 2016, 16:353–366. [DOI] [PubMed] [Google Scholar]
- 28.Chen YE, Fischbach MA, Belkaid Y: Skin microbiota–host interactions. Nature 2018, 553:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrd AL, Belkaid Y, Segre JA: The human skin microbiome. Nat Rev Microbiol 2018, 16:143–155. [DOI] [PubMed] [Google Scholar]
- 30.Dwyer LR, Scharschmidt TC: Early life host-microbe interactions in skin. Cell Host Microbe 2022, 30:684–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou W, Spoto M, Hardy R, Guan C, Fleming E, Larson PJ, Brown JS, Oh J: Host-Specific Evolutionary and Transmission Dynamics Shape the Functional Diversification of Staphylococcus epidermidis in Human Skin. Cell 2020, 180:454–470.e18. The first demonstration of within-individual S. epidermidis strain diversity, deeply sequencing multiple isolates from within a single individual.
- 32.Zhao S, Lieberman TD, Poyet M, Kauffman KM, Gibbons SM, Groussin M, Xavier RJ, Alm EJ: Adaptive Evolution within Gut Microbiomes of Healthy People. Cell Host Microbe 2019, doi: 10.1016/j.chom.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conlan S, Mijares LA, $author.lastName $author firstName, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, et al. : Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biology 2012, 13:R64. Conlan defined S. epidermidis strain diversity in a set of 28 genomes from commensal and nosocomial isolates. They demonstrated a strikingly open pangenome and considerable genetic diversity for S. epidermidis.
- 34. Conwill A, Kuan AC, Damerla R, Poret AJ, Baker JS, Tripp AD, Alm EJ, Lieberman TD: Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host Microbe 2022, 30:171–182. The first demonstration of within-individual C. acnes strain diversity, at the finest spatial resolution to date, at the level of individual pores, which were shown to be genetic islands.
- 35.Fitz-Gibbon S, Tomida S, Chiu B-H, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, et al. : Propionibacterium acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J Invest Dermatol 2013, 133:2152–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spittaels K-J, Ongena R, Zouboulis CC, Crabbé A, Coenye T: Cutibacterium acnes Phylotype I and II Strains Interact Differently With Human Skin Cells. Front Cell Infect Microbiol 2020, 10:575164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, Cheng JY, Shafiq F, Higbee K, Hata TR, Horswill AR, et al. : Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. Journal of Allergy and Clinical Immunology 2021, 147:955–966.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatsuji T, Chen TH, Butcher AM, Trzoss LL, Nam S-J, Shirakawa KT, Zhou W, Oh J, Otto M, Fenical W, et al. : A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Science Advances 2018, 4:eaao4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newstead LL, Varjonen K, Nuttall T, Paterson GK: Staphylococcal-Produced Bacteriocins and Antimicrobial Peptides: Their Potential as Alternative Treatments for Staphylococcus aureus Infections. Antibiotics 2020, 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jo J-H, Harkins CP, Schwardt NH, Portillo JA, NISC Comparative Sequencing Program, Zimmerman MD, Carter CL, Hossen MA, Peer CJ, Polley EC, et al. : Alterations of human skin microbiome and expansion of antimicrobial resistance after systemic antibiotics. Sci Transl Med 2021, 13:eabd8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rousset F, Cabezas-Caballero J, Piastra-Facon F, Fernández-Rodríguez J, Clermont O, Denamur E, Rocha EPC, Bikard D: The impact of genetic diversity on gene essentiality within the Escherichia coli species. Nature Microbiology 2021, doi: 10.1038/s41564-020-00839-y. An impressive demonstration of genome-wide fitness effects of CRISPRi-mediated gene knockdown in 18 genetically diverse strains of E. coli. They identified extensive variation in gene essentiality between strains and conditions.
- 42.van Opijnen T, Dedrick S, Bento J: Strain Dependent Genetic Networks for Antibiotic-Sensitivity in a Bacterial Pathogen with a Large Pan-Genome. PLOS Pathogens 2016, 12:e1005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnard E, Johnson T, Ngo T, Arora U, Leuterio G, McDowell A, Li H: Porphyrin Production and Regulation in Cutaneous Propionibacteria. mSphere 2020, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saheb Kashaf S, Proctor DM, Deming C, Saary P, Hölzer M, Taylor ME, Kong HH, Segre JA, Almeida A, Finn RD: Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions. Nat Microbiol 2021, doi: 10.1038/s41564-021-01011-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forster SC, Kumar N, Anonye BO, Almeida A, Viciani E, Stares MD, Dunn M, Mkandawire TT, Zhu A, Shao Y, et al. : A human gut bacterial genome and culture collection for improved metagenomic analyses. Nature Biotechnology 2019, 37:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI: Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. PNAS 2011, 108:6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagier J-C, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, et al. : Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 2016, 1:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Fleming E, Pabst V, Scholar Z, Xiong R, Voigt AY, Zhou W, Hoyt A, Hardy R, Peterson A, Beach R, et al. : Cultivation of common bacterial species and strains from human skin, oral, and gut microbiota. BMC Microbiol 2021, 21:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quince C, Delmont TO, Raguideau S, Alneberg J, Darling AE, Collins G, Eren AM: DESMAN: a new tool for de novo extraction of strains from metagenomes. Genome Biology 2017, 18:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, et al. : Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 2021, 10:e65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D: ConStrains identifies microbial strains in metagenomic datasets. Nat Biotech 2015, 33:1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olm MR, Crits-Christoph A, Bouma-Gregson K, Firek BA, Morowitz MJ, Banfield JF: inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol 2021, 39:727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aggarwala V, Mogno I, Li Z, Yang C, Britton GJ, Chen-Liaw A, Mitcham J, Bongers G, Gevers D, Clemente JC, et al. : Author Correction: Precise quantification of bacterial strains after fecal microbiota transplantation delineates long-term engraftment and explains outcomes. Nat Microbiol 2022, 7:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zheng W, Zhao S, Yin Y, Zhang H, Needham DM, Evans ED, Dai CL, Lu PJ, Alm EJ, Weitz DA: High-throughput, single-microbe genomics with strain resolution, applied to a human gut microbiome. Science 2022, 376:eabm1483. In a high throughput droplet-based, single sequencing approached applied to the gut microbiome, Zheng et al. were able to reconstruct >20K partial single cell genomes that were then co-assembled into species-level genomes.
- 55.van Opijnen T, Bodi KL, Camilli A: Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Meth 2009, 6:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui L, Vigouroux A, Rousset F, Varet H, Khanna V, Bikard D: A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nature Communications 2018, 9:1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spoto M, Puma JPR, Fleming E, Guan C, Nzutchi YO, Kim D, Oh J: Large-scale CRISPRi and transcriptomics of Staphylococcus epidermidis identify genetic factors implicated in commensal-pathogen lifestyle versatility. 2022, doi: 10.1101/2021.04.29.442003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spoto M, Guan C, Fleming E, Oh J: A Universal, Genomewide GuideFinder for CRISPR/Cas9 Targeting in Microbial Genomes. mSphere 2020, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo B-M, Marta E, et al. : A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell 2016, doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Méric G, Mageiros L, Pensar J, Laabei M, Yahara K, Pascoe B, Kittiwan N, Tadee P, Post V, Lamble S, et al. : Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis. Nat Commun 2018, 9:5034. A clever application of GWAS to 415 S. epidermidis isolates to identify genes (rather than isolates) associated with known pathogenicity traits.
- 61.Chen H, Nwe P-K, Yang Y, Rosen CE, Bielecka AA, Kuchroo M, Cline GW, Kruse AC, Ring AM, Crawford JM, et al. : A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell 2019, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma Y, Guo Z, Xia B, Zhang Y, Liu X, Yu Y, Tang N, Tong X, Wang M, Ye X, et al. : Identification of antimicrobial peptides from the human gut microbiome using deep learning. Nat Biotechnol 2022, 40:921–931. [DOI] [PubMed] [Google Scholar]
- 63.Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, et al. : Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tastan C, Karhan E, Zhou W, Fleming E, Voigt AY, Yao X, Wang L, Horne M, Placek L, Kozhaya L, et al. : Tuning of human MAIT cell activation by commensal bacteria species and MR1-dependent T-cell presentation. Mucosal Immunol 2018, doi: 10.1038/s41385-018-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Paik D, Yao L, Zhang Y, Bae S, D’Agostino GD, Zhang M, Kim E, Franzosa EA, Avila-Pacheco J, Bisanz JE, et al. : Human gut bacteria produce TH17-modulating bile acid metabolites. Nature 2022, 603:907–912. Paik performed a high throughput screen of 990 isolates the human gut to identify isolates that produced a Th17-cell modulating bile acid metabolite. The 238 identified were phylogenetically diverse and produced a range of amounts of the two key identified metabolites.
- 66.Han S, Van Treuren W, Fischer CR, Merrill BD, DeFelice BC, Sanchez JM, Higginbottom SK, Guthrie L, Fall LA, Dodd D, et al. : A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 2021, 595:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valm AM, Welch JLM, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG: Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. PNAS 2011, 108:4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, Borisy GG: Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A 2017, 114:E9105–E9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shi H, Shi Q, Grodner B, Lenz JS, Zipfel WR, Brito IL, De Vlaminck I: Highly multiplexed spatial mapping of microbial communities. Nature 2020, 588:676–681. This approach images microbial communities at single cell resolution with unprecedented scope. The combinatorial use of 10 fluorophores allow tagging of up to ~1000 unique cell types or species, given discriminatory probe design. Their proof of principle tracked 47 genera in the mouse gut microbiome.
- 70.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. : Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018, 361:eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, et al. : Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nature Protocols 2015, 10:442–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stickels RR, Murray E, Kumar P, Li J, Marshall JL, Di Bella DJ, Arlotta P, Macosko EZ, Chen F: Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol 2021, 39:313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Claesen J, Spagnolo JB, Ramos SF, Kurita KL, Byrd AL, Aksenov AA, Melnik AV, Wong WR, Wang S, Hernandez RD, et al. : A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Science Translational Medicine 2020, 12:eaay5445. Claesen computationally predicted, then experimentally verified the production of antimicrobials produced by select C. acnes strains, identifying cutimycin, which they then showed mediated an antagonistic relationship with Staphylococci, preventing its colonization in the hair follicle.
- 74.Kobayashi T, Voisin B, Kim DY, Kennedy EA, Jo J-H, Shih H-Y, Truong A, Doebel T, Sakamoto K, Cui C-Y, et al. : Homeostatic Control of Sebaceous Glands by Innate Lymphoid Cells Regulates Commensal Bacteria Equilibrium. Cell 2019, 176:982–997.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]