Abstract
The CRISPR/Cas9 system is a versatile genome editing platform in biotechnology and therapeutics. However, the requirement of protospacer adjacent motifs (PAMs) limits the genome targeting scope. To expand this repertoire, we revisited and engineered a compact Cas9 orthologue derived from Neisseria cinerea (NcCas9) for efficient genome editing in mammal cells. We demonstrated that NcCas9 generates genome editing at target sites with N4GYAT (Y = T/C) PAM which cannot be recognized by existing Cas9s. By optimizing the NcCas9 architecture and its spacer length, editing efficacy of NcCas9 was further improved in human cells. In addition, the NcCas9-derived Base editors can efficiently generate base conversions. Six anti-CRISPR (Acr) proteins were identified as off-switches for NcCas9. Moreover, NcCas9 successfully generated efficient editing of mouse embryos by microinjection of NcCas9 mRNA and the corresponding sgRNA. Thus, the NcCas9 holds the potential to broaden the CRISPR/Cas9 toolsets for efficient gene modifications and therapeutic applications.
Subject terms: CRISPR-Cas9 genome editing, Transfection
A Neisseria cinerea Cas9 variant is characterised in PAM preference, editing efficiency (for both nuclease and base editing) and anti-CRISPR protein blocking, with a PAM of (N)4GYAT reported alongside a mouse zygote assay.
Introduction
CRISPR-Cas9 system and derived genomic manipulation tools have transformed biotechnology and medicine1,2. They are composed of the Cas9 nucleases and a single guide RNA (sgRNA). Cas9 binds to the ~20 nucleotide (nt) spacer sequence and scaffold of sgRNA, and typically cleaves target DNA about 3 bp upstream of the PAM3,4. The CRISPR-guided DNA base editors (BEs), comprising deaminases fused to the N-terminus of Cas9 nickase (nCas9), enable targeted C-to-T (CBE) or A-to-G (ABE) conversions in genomic DNA5,6. The BEs, which can precisely install targeted point mutations without requiring DSBs or donor DNA templates, have exhibited powerful genome manipulation capability in various organisms7,8.
To date, a series of natural Cas9 orthologs and artificially-designed Cas9 variants with various PAM requirements have been developed, such as SaCas9 (NNGRRT)9, Nme1Cas9 (NNNNGATT)10, Nme2Cas9 (NNNNCC)11, St1Cas9 (NNRGAA)12, SpaCas9 (NNGYRA)13, Cje1Cas9 (NNNVRYAC)14, Cje3Cas9 (NNNNCYA)15, SauriCas9 (NNGG)16, SchCas9 (NNGR)17, FrCas9 (NNTA)18, ScCas9 (NNG)19, SpCas9-VQR (NGA)20, SpCas9-NG (NG)21, SpCas9-NRNH (NRNH)22, SpG (NG)23, SpRY (NR > NY)23, etc. These Cas9 nucleases expand the PAM compatibility of Cas9 orthologs and improve the flexibility of genome editing applications24. However, the NGG PAM requirement of conventional SpCas9 and relevant BEs, together with the complicated PAM requirements of small Cas enzymes, restrict the genome targeting scope. Therefore, it is necessary to explore new Cas9 nucleases with distinctive PAMs to expand the nuclease repertoire.
In this study, we revisited the attributes of Neisseria cinerea Cas9 (NcCas9), a small Cas9 orthologue, which were discovered yet has not been fully examined and utilized for genome editing in human cells9. NcCas9 was reported to be inefficient and recognized a restrictive N4GTA PAM, limiting its application in genome editing9,25.
Here, we indicated that the engineered NcCas9 system enables efficient genome editing and base editing at endogenous sites with a distinct N4GYAT PAM, offering an alternative tool for both basic research and clinical applications.
Results
PAM identification of NcCas9 by PAM-DOSE
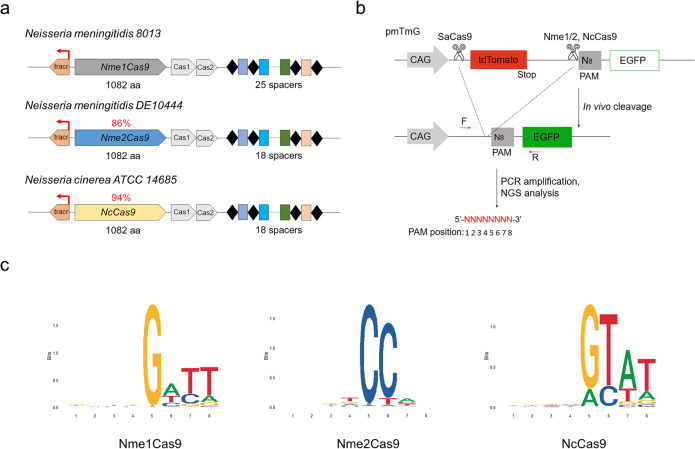
The type II-C Cas9 orthologue NcCas9 is of small size (1082 aa) and is closely related to conventional Neisseria meningitidis Cas9 (NmeCas9)11 (Fig. 1a). The NcCas9 is 94% identical to Nme1Cas9, and the divergences lie mainly in the C-terminal PAM-interacting domain (PID) (Fig. 1a and S1). The NcCas9 locus also contains an array composed of 36-bp direct repeats (DRs) interspaced by 30-bp spacers in the proximity of the Cas genes operon, similar to that of Nme1/2Cas9 locus (Fig. 1a). Crystal structures of Nme1Cas9 and Nme2Cas9 have indicated that key residues of H1024/N1029 or D1028/R1033 determine the N4GATT or N4CC PAM specificity, respectively26. Interestingly, we noticed that the NcCas9 harbors PAM-interacting residues N1024 and A1029, different from that of Nme1/2Cas9 orthologs by protein sequence alignment (Fig. S1). Therefore, we speculated that the NcCas9 may recognize distinct PAM sequences from Nme1/2Cas9. The PAM of NcCas9 was identified as NNNNGTA using in vitro cleavage of a degenerate 7-bp sequence in the previous report9. However, many II-C Cas9 orthologues recognize an 8-nt PAM sequence (such as Nme1Cas9 (N4GATT) and CjeCas9 (N3VRYAC)) that cannot be fully characterized by the 7-bp library11,14. In addition, many Cas9 orthologues identified by library cleavage in vitro or in bacteria often do not function well in mammalian cells9,27.
Fig. 1. PAM identification by PAM-DOSE.
a Schematic showing the CRISPR/Cas loci of the strains encoding the Nme1Cas9, Nme2Cas9, and NcCas9 orthologs. Red arrows denote tracrRNA transcription initiation sites. Percent identities of Nme2Cas9 and NcCas9 with Nme1Cas9 are 86% and 94%, respectively. b Schematic showing the PAM-DOSE assay for characterization of PAM sequences recognized by Cas9s. An 8 N library of PAM sequences were introduced into the constructs. In the presence of a functional PAM, cleavage-mediated tdTomato cassette excision was performed, which led to the expression of EGFP. After Cas9 cleavage, cleaved products were PCR amplified and subjected to deep sequencing to identify PAM sequences. c The sequence logo showing the PAM specificity of Nme1Cas9, Nme2Cas9, and NcCas9 was obtained using deep-sequencing data.
Therefore, we fully identified the functional PAMs of NcCas9 in human cells using a widely-used positive screening system designated as PAM-DOSE (PAM Definition by Observable Sequence Excision)28,29. Briefly, a library of pmTmG plasmids containing a randomized 8-bp sequence (5′-NNNNNNNN-3′) was generated. After the cleavage of Nme1Cas9, Nme2Cas9 or NcCas9, the tdTomato cassette was excised, and the CAG promoter drove the expression of the EGFP gene28. Cleaved products were polymerase chain reaction (PCR) amplified and subjected to deep sequencing to identify PAM sequences recognized by these Cas9 nucleases (Fig. 1b). As expected, the conventional Nme1Cas9 and Nme2Cas9 recognized N4GATT and N4CC as optimal PAMs, respectively (Fig. 1c). In addition, a relaxed N4RYWT (R = A/G, Y = C/T, W = A/T) was recognized by NcCas9, which is different from that of Nme1Cas9, consistent with the previous speculation based on the divergent PAM-interacting residues (Fig. 1c). Importantly, the NcCas9 mainly recognized an N4GTAT PAM and showed an obvious preference for T in the eighth base, demonstrating that the previously identified N4GTA PAM was inaccurate (Fig. 1c).
Detailed determination of efficient PAM sequences recognized by NcCas9
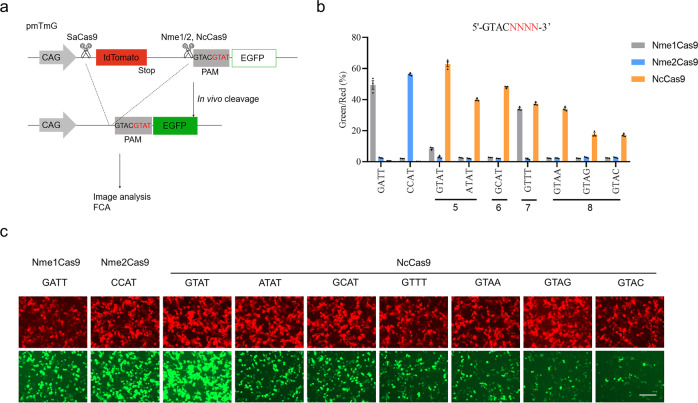
To further verify the PAM specificity of NcCas9, we performed a quantitative comparison by co-transfecting plasmids encoding Cas9, corresponding sgRNA, and pmTmG reporter plasmids containing variable PAM sequences based on an optimal N4GATT (Nme1Cas9), N4CC (Nme2Cas9), and N4GTAT (NcCas9) PAM (Fig. 2a). The editing efficiency of different PAMs was quantified by calculating the ratio of EGFP-positive cells to tdTomato-positive cells by flow cytometry analysis (Fig. 2a). As a result, Nme1Cas9 efficiently cleaved optimal N4GATT PAM and suboptimal N4GTTT PAM; Nme2Cas9 only potently recognized N4CC PAM (Fig. 2b, c). In contrast, reporter assays showed NcCas9 efficiently cleaved target sites containing optimal N4GTAT PAM and suboptimal N4GCAT PAM (Fig. 2b, c). In addition, the NcCas9 can also accept A or T at the fifth (N4ATAT) or seventh (N4GTTT) base of the PAM, respectively (Fig. 2b, c). At the eighth base, a strong preference for a T of NcCas9 was observed, and all other three bases significantly reduced editing efficiency (Fig. 2b, c). Importantly, despite the high sequence conservation, Nme1/2Cas9 could not cleave at primary NcCas9 PAMs (N4GYAT, where Y is T or C), highlighting distinct PAM specificity of the NcCas9 (Fig. 2b). These quantitative comparison results were consistent with the previous observations of PAM identification from PAM-DOSE. Taken together, these data demonstrated that NcCas9 can efficiently recognize distinct N4GYAT and suboptimal N4ATAT/N4GTTT PAMs, enriching Cas9 nucleases toolsets.
Fig. 2. Detailed PAM sequence analysis of NcCas9.
a Schematic showing the pmTmG reporter assay for comparing different PAMs of Nme1Cas9, Nme2Cas9, and NcCas9 orthologs. Variable PAM sequences were introduced into the constructs. In the presence of a functional PAM, cleavage-mediated tdTomato cassette excision was performed, which led to the expression of EGFP, thereby allowing live visualization under a fluorescence microscope or detection via flow cytometry analysis (FCA). b Comparison of Nme1Cas9, Nme2Cas9, and NcCas9 cleavage efficiency of different PAMs by FCA. The different PAMs were designed based on the optimal GTACGTAT PAM sequence. Red or green represents the tdTomato or EGFP signal, respectively. c Representative fluorescence microscopy images of PAM analysis. Scale bar, 200 μm. Error bars indicate the s.e.m. (n = 3).
Efficient genome editing in human cells by optimized NcCas9 system
In addition to previously-identified incomplete PAM sequences, the relatively low activity is what hinders NcCas9 from being widely used in the past9,25. Therefore, we attempted to improve its activity by optimizing the NcCas9 architecture and its sgRNA. First, the human codon-optimized ORF of NcCas9 was refined using codon usages from GenScript and flanked with bipartite nuclear localization signal (bpNLS), which have been proved to substantially improve the expression levels of BEs30 (Fig. S2a). As a result, engineered NcCas9 showed a significant increase in editing efficiency compared to the previously-reported NcCas9 system in both pmTmG reporter assay and endogenous sites (Fig. S2b and S2c). In addition, we identified the sgRNA scaffold sequence used by NcCas9 and attempted to explore the optimal spacer length requirement (Fig. S3a and S3b). A series of sgRNAs targeting site5 with variable spacer lengths (20–27 nt) were tested. An additional guanine was added to the 5′-terminal sequence of each sgRNA to ensure proper transcription. We proved that 24 nt (G + 23) is the ideal spacer length, and activity decreased upon other lengths, indicating that the 20-nt spacer length of NcCas9 as utilized previously affected its editing activity9,25 (Fig. S3c).
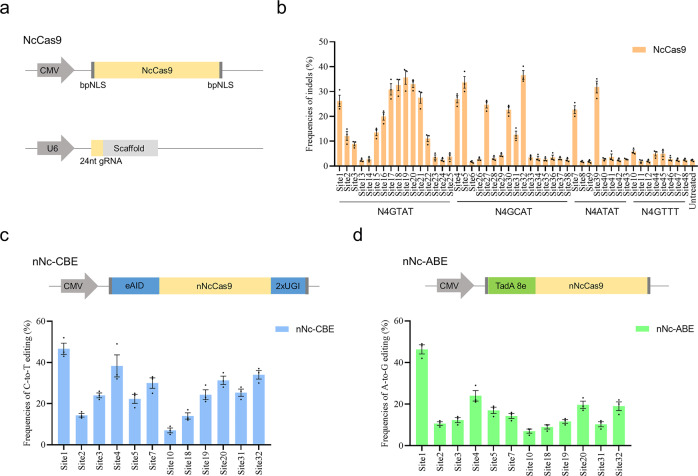
To thoroughly attest to the efficacy of NcCas9 for genome editing, we co-transfected the optimized NcCas9 system and corresponding sgRNAs with optimal 24-nt spacer length into HEK293T cells (Fig. 3a). A panel of 48 endogenous sites with functional PAMs including N4GTAT, N4GCAT, N4ATAT, and N4GTTT were tested (Fig. 3b and Table S2). As a result, obvious indels were observed at 25 out of 48 endogenous sites with varied editing efficiencies from 4.9% to 36.6% (Fig. 3b). Notably, NcCas9 generated efficient indels at target sites with N4GYAT (Y = T/C) PAMs, while showed lower efficiency at target sites with N4ATAT and N4GTTT PAMs (Fig. 3b). These endogenous gene editing results were consistent with the results of pmTmG reporter assays. In addition, we used the classical GUIDE-seq method to assess the on-target and off-target frequencies of NcCas9 targeting two different loci, Site1 and Site531,32. As a result, no off-target cleavage was detected in the two sites (Fig. S4). Collectively, these results validated that the optimized NcCas9 system is a potent genome editing platform in human cells.
Fig. 3. Genome editing of NcCas9 and its base editors in human cells.
a Schematic of optimized NcCas9 and its sgRNA architecture. b Genome editing of NcCas9 with N4GYAT, N4ATAT and N4GTTT PAMs in 48 endogenous loci. c Top: Schematic of the nNc-CBE. Bottom: Efficient C-to-T conversions in 12 endogenous loci by four nNc-CBE. d Top: Schematic of the nNc-ABE, TadA 8e was used. Bottom: Efficient A-to-G conversions in 12 endogenous loci by nNc-ABE. The highest C-to-T/A-to-G editing frequency within the editing window represents the base editing frequency at each site. Error bars indicate s.e.m. (n = 3).
Efficient base editing in human cells by NcCas9-derived BEs
In addition to Cas9-mediated gene knockout, base editing is a revolutionary technology that can expediently induce targeted base conversions in desired sites without donor templates7. However, BEs are highly dependent on a proper PAM adjacent to the target base, limiting the targeting range7,33. Although many Cas9 variants have been used for base editing, more Cas9 nucleases with distinct PAMs are needed to enrich the BE tools. To test whether NcCas9 could be employed for base editing, we replaced the SpCas9 nickase in our previously-designed eAID-CBE system with NcCas9 nickase (D16A) and to create nNc-CBE (Fig. 3c). The eAID-CBE system has exhibited high editing activity and excellent Cas9 compatibility in the previous report13,34,35. Notably, the nNc-CBE showed efficient C-to-T editing in all 12 sites with average frequencies from 7.0% to 46.7% (Figs. 3c and S5). Encouraged by the results of CBE, we further generated nNc-ABE system with TadA 8e, the current optimal version of ABE13,36 (Fig. 3d). As a result, the nNc-ABE also mediated the efficient conversion of A-to-G in genomic DNA with average frequencies from 7.0% to 46.3% (Figs. 3d and S6). Taken together, these data indicated that NcCas9 can be used to expand the targeting range of BEs.
Identification of efficient Acr inhibitors for NcCas9
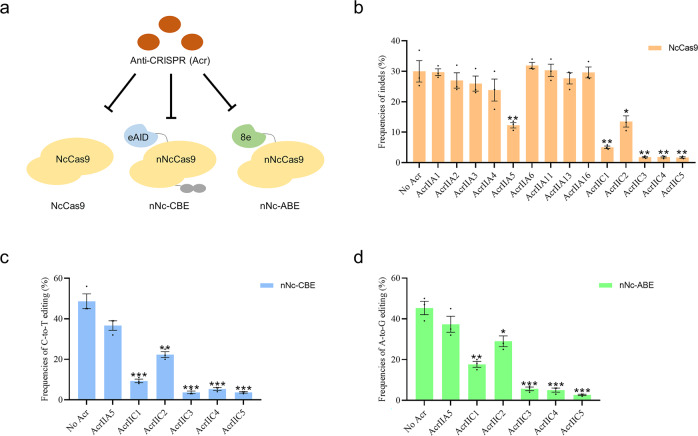
Bacterial CRISPR–Cas systems utilize sequence-specific RNA-guided nucleases to defend against bacteriophage infection37. In response to the bacterial war on phage infection, numerous phages evolved anti-CRISPR (Acr) proteins to block the function of CRISPR-Cas systems37,38. As a natural brake for CRISPR/Cas nucleases, Acr has been widely used to manipulate genome editing in mammalian cells and organisms38. To test Acr inhibition of NcCas9, we performed NcCas9 and corresponding sgRNA transfections (targeting site5) in HEK293T cells in the presence or absence of 14 representatives Acr proteins (AcrIIA1-A6, A11, A13, A16, and AcrIIC1-C5). Consequently, AcrIIC3-C5 showed strong inhibition of NcCas9 genome editing, followed by AcrIIC1, C2, and A5, while others showed no obvious effect (Fig. 4a, b). Similar inhibitory effects of the six Acrs were observed in both nNc-CBE and nNc-ABE systems, indicating that Acrs can also be used to regulate the activity of nNc-BEs (Fig. 4c, d). Notably, the most robust inhibition was presented by AcrIIC3-C5 in both NcCas9 and its BEs, suggesting that they have a high potential fight against NcCas9 (Fig. 4b–d). These results suggested that the six Acr proteins can be used as off switches for NcCas9 applications.
Fig. 4. NcCas9 is inhibited by Acr proteins in human cells.
a Schematic of Acr proteins is used to inhibit the activities of NcCas9 and its base editors. b Genome editing of NcCas9 in the presence or absence of the 14 previously described Acr families. Plasmids expressing NcCas9, sgRNA targeting site5, and each Acr (1:1:1) were co-transfected into HEK293T cells. c Base editing of nNc-CBE in the presence or absence of six Acrs at the site1. d Base editing of nNc-ABE in the presence or absence of six Acrs at site1. Error bars indicate the s.e.m. (n = 3).
Genome editing in mouse embryos by NcCas9
To evaluate the feasibility of NcCas9 genome editing in the mouse embryo, we selected four target sites with N4GYAT PAMs from Tyr and Pcsk9 gene. Genome editing was performed in mouse zygotes by microinjection of NcCas9-encoding mRNA and the appropriate sgRNA, as previously reported39,40. After injection, mouse zygotes were cultured in vitro to blastocysts and then genotyped. Notably, two target sites with N4GTAT PAM showed efficient editing efficiency of 25% (Tyr-1) and 80% (Pcsk9-1) (Table 1). In addition, the editing frequencies in single embryo ranged from 26.1% to 30.7% in Tyr-1 and from 21.2% to 98.6% in Pcsk9-1 (Table 1). However, no obvious editing events were observed in two target sites with a suboptimal N4GCAT PAM (Table 1). These results demonstrated efficient genome editing in mouse embryos by NcCas9, underscoring the potential of the NcCas9 system in generating disease models in animals.
Table 1.
Generation of targeted editing in mouse embryos by NcCas9.
| PAM | Target site | No. of zygotes | No. of blastocysts | No. of editing (%) | Frequencies of editingin single embryo (%) |
|---|---|---|---|---|---|
| N4GTAT | Tyr-1 | 20 | 8 | 2 (25) | (26.1–30.7) |
| Pcsk9-1 | 20 | 10 | 8 (80) | (21.2–98.6) | |
| N4GCAT | Tyr-2 | 20 | 8 | 0 (0) | (0) |
| Pcsk9-2 | 20 | 9 | 0 (0) | (0) |
Discussion
In this study, we identified a Cas9 orthologue derived from N. cinerea that can be utilized for efficient genome editing. Importantly, we re-identified that NcCas9 recognizes a relaxed N4GYAT PAM, demonstrating that the previously identified N4GTA PAM was inaccurate. The editing efficiency of the NcCas9 system was improved through rational optimization of NcCas9 architecture and spacer length. We further validated that NcCas9 and NcCas9-derived BEs can potently perform gene editing in both human cells and mouse embryos. Efficient Acr inhibitors of NcCas9 were also developed for precise regulation and applications. Our study comprehensively characterized NcCas9 as a versatile and robust genome editing tool.
In addition, the compact size of NcCas9 (1082 aa) is compatible with the all-in-one-AAV vector for in vivo delivery. Although several small Cas9 nucleases have been used for genome editing via single-AAV delivery, the range of targetable sequences remains limited because of the PAM requirements. We demonstrated that NcCas9 recognized a novel PAM of N4GYAT, which is different from those of currently used small Cas9s, including SaCas9 (NNGRRT)9, Nme1Cas9 (NNNNGATT)10, Nme2Cas9 (NNNNCC)11, CjeCas9 (NNNVRYAC)14, St1Cas9 (NNRGAA)12 and SpaCas9 (NNGYRA)13. Therefore, NcCas9 holds the potential to further expand the PAM compatibility of small Cas9 orthologues and improve the flexibility of therapeutic applications in the future.
We have validated that NcCas9-mediated CBE and ABE can induce robust C-to-T and A-to-G base editing. Recently, some new precise editing tools were generated, including glycosylase BEs that can generate C-to-G transversions41–43, dual BEs that can simultaneously catalyze both cytosine and adenine base conversions44–47, and prime editors that can directly write new genetic information into a specified site48. The NcCas9 could be used as a new scaffold to expand the versatility of these precise genome editing systems in the future.
In summary, we developed a Cas9 ortholog, NcCas9, with a distinct N4GYAT PAM (N4GTAT > N4GCAT), that can induce efficient genome editing frequency of up to 36.6% in human cells. In addition, the NcCas9-derived CBE or ABE systems can efficiently generate targeted C-to-T or A-to-G base conversions of up to 46.7% or 46.3%, respectively. We also presented that AcrIIC3–C5 are robust inhibitors of both NcCas9 and its BEs, having the potential to be off-switches for NcCas9 applications. Moreover, the NcCas9 also successfully generated efficient editing in mouse embryos. Thus, the NcCas9 system is a promising tool for both basic research and therapeutic applications.
Methods
Animals
C57/BL6 mice were obtained from the Laboratory Animal Center of Jilin University (Changchun, China). Superovulated C57BL/6J mice (6–8 week old females) were mated with C57BL/6J males. All mice and experimental protocols used in this project have been approved by the Institutional Animal Care and Use Committee of Jilin University (SY202111300).
Plasmid construction
The previous NcCas9, Nme1Cas9, and Nme2Cas9 was obtained from Addgene (#68331, #115694 and #119923). The pmTmG reporter plasmid was a kind gift from Feng Gu28. The human codon-optimized NcCas9 and its sgRNA scaffold were synthesized by Genscript Biotech (China). Plasmid site-directed mutagenesis was performed using the Fast Site-Directed Mutagenesis Kit (Tiangen, China). The site-directed mutation primers are listed in Table S1. The sequences of plasmids are listed in Supplementary Note 1.
Cell culture and transfection
HEK293T cell lines (obtained from ATCC and preserved by our laboratory) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (HyClone, China) and incubated at 37 °C in an atmosphere of 5% CO2. The cells were seeded in 24-well plates and transfected using Hieff TransTM Liposomal Transfection Reagent (Yeasen, China) according to the manufacturer’s instructions. The puromycin (Meilunbio, China) was added at the final concentration of 3 μg/mL to enrich the positively transfected cells at 24 h after transfection. After 72 h, the cells were collected and used for genotyping by TIDE49 or EditR50. All target sites and primers used for genotyping are listed in Tables S2 and S3.
PAM characterization using PAM-DOSE
The PAM-DOSE assay was carried out as previously reported28. Briefly, HEK293T cells were co-transfected with Nme1Cas9/Nme2Cas9/NcCas9, corresponding sgRNA, SaCas9, and its sgRNA, and pmTmG library plasmids (1:1:1:1:1) using Hieff TransTM Liposomal Transfection Reagent (Yeasen, China). PCR is used to amplify the sequences flanking the Cas9 target locus, and products were subjected to deep sequencing (Sangon Biotech, China). PAM regions without indel within three bases near PAM were extracted. PAMs were counted and used to generate sequence logos.
pmTmG reporter assay
HEK293T cells were co-transfected with Nme1Cas9/Nme2Cas9/NcCas9, corresponding sgRNA and pmTmG reporter plasmids (1:1:1) using Hieff TransTM Liposomal Transfection Reagent. After 72 h of incubation, the fluorescent images were imaged with a microscope (ts100; Nikon, Tokyo, Japan). Cells were harvested for editing quantification by flow cytometry. Quantification was based on the relative fluorescent frequencies of green/red.
GUIDE-seq analysis
GUIDE-seq was constructed to assess the on-target and off-target frequencies of NcCas9 targeting Site1 and Site5 as previous studies reported18,31,32. GUIDE-seq and data analysis were completed by GeneRulor Company Bio-X Lab (Guangzhou, Guangdong, China). Briefly, 5 μl dsODN (100 μM) and 10 μg NcCas9-sgRNA plasmids were co-transfected into HEK293T cell line. After 72 h, cells were harvested for DNA extraction, followed by dsODN-PCR verification of effective cleavage. Then, GUIDE-seq libraries were constructed. The DNA went through shearing, adding Y adapters and two rounds of PCR, and was finally sequenced using MGISEQ-2000RS sequencer (PE150, paired-end) with customized settings for 16 bp UMI. Data were demultiplexed using in-house python scripts and then analyzed using guideseq v1.151. No SpCas9 positive controls have been included to compare off-target reads in this study.
mRNA and gRNA preparation
The NcCas9, nNc-CBE, and nNc-ABE plasmids were linearized with NotI and transcribed in vitro using the HiScribe™ T7 ARCA mRNA kit (NEB). The mRNA was purified using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. The sgRNA oligos were annealed into the pUC57-Nc sgRNA expression vector containing a T7 promoter. The sgRNAs were then amplified and transcribed in vitro using the MAXIscript T7 kit (Ambion) and purified using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol.
Microinjection of mouse zygotes and genotyping
Briefly, a mixture of mRNA (50 ng/µl) and sgRNA (30 ng/µl) was co-injected into the cytoplasm of pronuclear-stage zygotes. Each group was injected with an average of approximately 20 zygotes to test the base editing efficiency. The injected zygotes were transferred to KSOM medium for culture at 37 °C, 5% CO2, and 100% humidity. Then, the injected single zygote was collected at the blastocyst stage. Genomic DNA was extracted in embryo lysis buffer (1% NP40) at 56 °C for 60 minutes and then at 95 °C for 10 minutes in a Bio-Rad PCR Amplifier. Then, the extracted products were amplified by PCR (95 °C, 5 min for predegeneration, 42 cycles of (95 °C, 30 s, 58 °C, 30 s, 72 °C, 30 s), 72 °C, 5 min for extension) and determined by Sanger sequencing. The genomic DNA of newborn mice was extracted from ear clips and analyzed by PCR genotyping. All target sites and primers used for genotyping are listed in Tables S2 and S3.
Statistics and reproducibility
All data are expressed as mean ± s.e.m. of at least three individual determinations for all experiments. Data were analyzed by student’s t-test via GraphPad prism software 8.0.1. The probability value that was smaller than 0.05 (p < 0.05) was statistically significant. *p < 0.05, **p < 0.01, ***p < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This study was financially supported by the National Key Research and Development Program (Grant no. 2018YFB1404205) of China Stem Cell and Translational Research (Grant no. 2017YFA0105101) and the National Natural Science Foundation of China (Grant nos. 32170543 and 31970574).
Author contributions
Z.Q.L., L.X.L., and Z.J.L. conceived and designed the experiments. Z.Q.L., S.Y.C., W.H.X., and H.Y. performed the experiments and analyzed the data. Z.Q.L. and S.Y.C. wrote the paper. All authors read and approved the final paper.
Peer review
Peer review information
Communications Biology thanks Junho Hur, Pranam Chatterjee and Vijay Tripathi for their contribution to the peer review of this work. Primary Handling Editor: Eve Rogers. Peer reviewer reports are available.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary files. Deep sequencing data of the PAM-DOSE assay have been deposited at Sequence Read Archive under accession code SRP404972. Raw sequencing data of GUIDE-seq have been deposited at the Sequence Read Archive under accession code SRP398785. The constructed plasmids are available from addgene (accession IDs: 194093 to 194095). The source data are listed in Supplementary Data 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhiquan Liu, Siyu Chen.
Contributor Information
Liangxue Lai, Email: lai_liangxue@gibh.ac.cn.
Zhanjun Li, Email: lizj_1998@jlu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-04258-z.
References
- 1.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Q, et al. Fusing T5 exonuclease with Cas9 and Cas12a increases the frequency and size of deletion at target sites. Sci. China Life Sci. 2020;63:1918–1927. doi: 10.1007/s11427-020-1671-6. [DOI] [PubMed] [Google Scholar]
- 5.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudelli NM, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 9.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibraheim R, et al. All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol. 2018;19:137. doi: 10.1186/s13059-018-1515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edraki A, et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell. 2019;73:714–726.e714. doi: 10.1016/j.molcel.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agudelo D, et al. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 2020;30:107–117. doi: 10.1101/gr.255414.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, et al. Versatile and efficient in vivo genome editing with compact Streptococcus pasteurianus Cas9. Mol. Therap. 2022;30:256–267. doi: 10.1016/j.ymthe.2021.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim E, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, et al. Compact Cje3Cas9 for efficient in vivo genome editing and adenine base editing. CRISPR J. 2022;5:472–486. doi: 10.1089/crispr.2021.0143. [DOI] [PubMed] [Google Scholar]
- 16.Hu Z, et al. A compact Cas9 ortholog from Staphylococcus auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 2020;18:e3000686. doi: 10.1371/journal.pbio.3000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, et al. Compact SchCas9 recognizes the simple NNGR. Pam. Adv. Sci. 2022;9:e2104789. doi: 10.1002/advs.202104789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui Z, et al. FrCas9 is a CRISPR/Cas9 system with high editing efficiency and fidelity. Nat. Commun. 2022;13:1425. doi: 10.1038/s41467-022-29089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018;4:eaau0766. doi: 10.1126/sciadv.aau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimasu H, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller SM, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020;38:471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collias D, Beisel CL. CRISPR technologies and the search for the PAM-free nuclease. Nat. Commun. 2021;12:555. doi: 10.1038/s41467-020-20633-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen F, et al. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat. Commun. 2017;8:14958. doi: 10.1038/ncomms14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun W, et al. Structures of Neisseria meningitidis Cas9 complexes in catalytically poised and Anti-CRISPR-inhibited states. Mol. Cell. 2019;76:938–952.e935. doi: 10.1016/j.molcel.2019.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano S, et al. Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9. Nat. Commun. 2019;10:1968. doi: 10.1038/s41467-019-09741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang L, et al. Efficient cleavage resolves PAM preferences of CRISPR-Cas in human cells. Cell Regeneration. 2019;8:44–50. doi: 10.1016/j.cr.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye J, et al. Can SpRY recognize any PAM in human cells? J. Zhejiang Univ. Sci. B. 2022;23:382–391. doi: 10.1631/jzus.B2100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koblan LW, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malinin NL, et al. Defining genome-wide CRISPR-Cas genome-editing nuclease activity with GUIDE-seq. Nat. Protoc. 2021;16:5592–5615. doi: 10.1038/s41596-021-00626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, et al. Highly efficient base editing with expanded targeting scope using SpCas9-NG in rabbits. FASEB J. 2020;34:588–596. doi: 10.1096/fj.201901587R. [DOI] [PubMed] [Google Scholar]
- 34.Liu, Z. et al. Improved base editor for efficient editing in GC contexts in rabbits with an optimized AID-Cas9 fusion. FASEB J. fj201900476RR (2019). [DOI] [PubMed]
- 35.Liu Z, et al. Efficient and high-fidelity base editor with expanded PAM compatibility for cytidine dinucleotide. Sci. China Life Sci. 2021;64:1355–1367. doi: 10.1007/s11427-020-1775-2. [DOI] [PubMed] [Google Scholar]
- 36.Richter MF, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2022;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauch BJ, et al. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell. 2017;168:150–158.e110. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marino ND, Pinilla-Redondo R, Csörgő B, Bondy-Denomy J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods. 2020;17:471–479. doi: 10.1038/s41592-020-0771-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang D, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J. Mol. Cell Biol. 2014;6:97–99. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, et al. Highly efficient RNA-guided base editing in rabbit. Nat. Commun. 2018;9:2717. doi: 10.1038/s41467-018-05232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao D, et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021;39:35–40. doi: 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 42.Kurt IC, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021;39:41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, et al. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. commun. 2021;12:1384. doi: 10.1038/s41467-021-21559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grünewald J, et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 2020;38:861–864. doi: 10.1038/s41587-020-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakata RC, et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020;38:865–869. doi: 10.1038/s41587-020-0509-0. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 2020;38:856–860. doi: 10.1038/s41587-020-0527-y. [DOI] [PubMed] [Google Scholar]
- 47.Xie J, et al. ACBE, a new base editor for simultaneous C-to-T and A-to-G substitutions in mammalian systems. BMC Biol. 2020;18:131. doi: 10.1186/s12915-020-00866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anzalone AV, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brinkman EK, Chen T, Amendola M, Van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kluesner MG, et al. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai SQ, Topkar VV, Joung JK, Aryee MJ. Open-source guideseq software for analysis of GUIDE-seq data. Nat. Biotechnol. 2016;34:483. doi: 10.1038/nbt.3534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary files. Deep sequencing data of the PAM-DOSE assay have been deposited at Sequence Read Archive under accession code SRP404972. Raw sequencing data of GUIDE-seq have been deposited at the Sequence Read Archive under accession code SRP398785. The constructed plasmids are available from addgene (accession IDs: 194093 to 194095). The source data are listed in Supplementary Data 1.