Summary
Naive human induced pluripotent stem cells (iPSCs) can be generated by reprogramming somatic cells with Sendai virus (SeV) vectors. However, only dermal fibroblasts have been successfully reprogrammed this way, and the process requires culture on feeder cells. Moreover, SeV vectors are highly persistent and inhibit subsequent differentiation of iPSCs. Here, we report a modified SeV vector system to generate transgene-free naive human iPSCs with superior differentiation potential. The modified method can be applied not only to fibroblasts but also to other somatic cell types. SeV vectors disappear quickly at early passages, and this approach enables the generation of naive iPSCs in a feeder-free culture. The naive iPSCs generated by this method show better differentiation to trilineage and extra-embryonic trophectoderm than those derived by conventional methods. This method can expand the application of iPSCs to research on early human development and regenerative medicine.
Keywords: Sendai virus vector, reprogramming, naive pluripotency, induced pluripotent stem cells, residual transgenes, temperature sensitivity, LMYC, hsa-microRNA-367, feeder-free culture, extra-embryonic trophectoderm
Graphical abstract
Highlights
-
•
A method for human iPSC generation that allows rapid removal of SeV vector
-
•
Naive iPSCs can be made from dermal fibroblasts or peripheral blood mononuclear cells
-
•
The method enables feeder-free naive iPSCs generation from dermal fibroblasts
-
•
The naive iPSCs generated by the method have significantly higher differentiation potency
Motivation
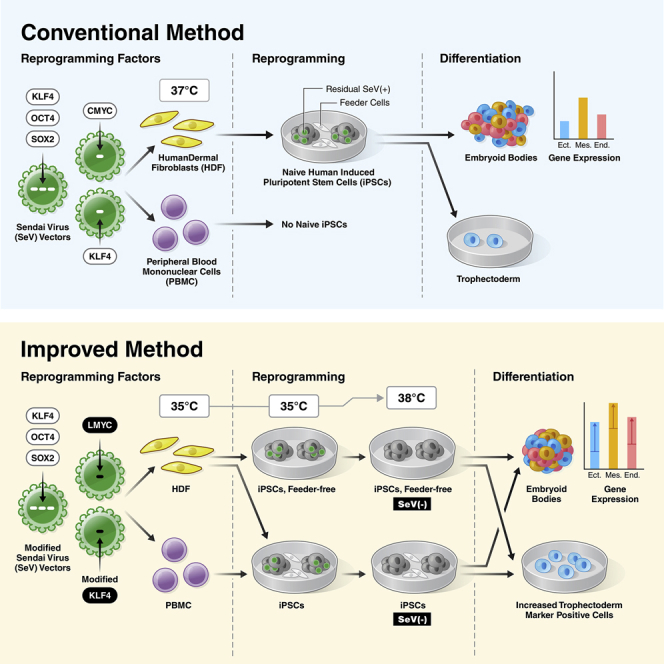
Although a method to establish naive human iPSCs directly from somatic cells using SeV vectors has been reported, it requires the use of dermal fibroblasts on feeder cells as a starting population. Furthermore, the SeV vector persists in the iPSCs after generation, limiting their differentiation potency. To solve these problems, we developed a SeV vector system that enables the generation of naive iPSCs from a variety of somatic cells, including under feeder-free conditions. Moreover, the method enables rapid removal of SeV vectors after iPSC generation, resulting in a superior differentiation potency.
Kunitomi et al. develop an improved SeV vector system to generate naive human iPSCs from various somatic cells by changing the structure and combination of SeV vectors. This method allows rapid removal of the SeV vectors, resulting in transgene-free naive iPSCs with superior differentiation potential.
Introduction
Human induced pluripotent stem cells (iPSCs) generated from somatic cells by conventional reprogramming methods possess “primed pluripotency” and resemble the post-implantation epiblast with restricted differential potency to extra-embryonic tissues. This is because fate determination of extra-embryonic tissues occurs prior to implantation. In contrast, naive human iPSCs resemble the pre-implantation epiblast and can differentiate into both embryonic and extra-embryonic lineages, characteristics known as “naive pluripotency” (Nichols and Smith, 2009; Weinberger et al., 2016). Many methods have been reported to generate naive human iPSCs from primed human iPSCs by introducing several pluripotency genes, cytokines, or chemicals (Guo et al., 2017; Takashima et al., 2014; Theunissen et al., 2014). However, these methods are time consuming and inefficient.
To solve this problem, several recent reports directly generated naive human iPSCs by introducing the four reprogramming factors—OCT4, SOX2, KLF4, and CMYC (OSKM)—into human dermal fibroblasts (HDFs) using Sendai virus (SeV) vectors (Kilens et al., 2018; Liu et al., 2017, 2021). SeV vectors have high infection efficiency, assert early and strong gene expression, and show no risk of genomic integration (Fusaki et al., 2009; Li et al., 2000). However, the SeV genome expression frequently persists at high levels in naive iPSCs after reprogramming, resulting in reduced trilineage differentiation potency compared with primed iPSCs (Liu et al., 2017). Moreover, continuous expression of the exogenous reprogramming factors may induce tumorigenicity, which is a serious hurdle in cell therapy (Ben-David and Benvenisty, 2011; Kono et al., 2019; Okita et al., 2007). SeV and other exogenous vectors can be removed by isolating single clones by manual colony picking (Takahashi et al., 2007), but this approach is difficult for naive human iPSCs because colonies are much smaller than those of primed iPSCs. Overall, no reliable methods to remove SeV vectors from naive human iPSCs have been reported. Thus, further optimization of naive iPSC reprogramming methods is essential to improve the study of early development and to enable their use in regenerative medicine.
In this study, we developed a system to significantly accelerate SeV removal by improving the structure of the SeV vector. This method enables removal of SeV vectors by simply changing the culture temperature after generating naive and primed human iPSCs. In addition, using OCT4, SOX2, KLF4, and LMYC as reprogramming factors, we were able to obtain naive iPSCs not only from dermal fibroblasts but also from peripheral blood mononuclear cells (PBMCs), and the resulting iPSCs possessed better differentiation propensity than those generated by conventional methods.
Results
We first transduced HDFs or PBMCs with the CytoTune-iPS 2.0 SeV reprogramming kit, containing three vectors, SeV(PM)KOS/TS12ΔF (SeV-KOS), SeV(HNL)CMYC/TS15ΔF (SeV-CMYC), and SeV18 + KLF4/TSΔF (SeV-KLF4), at 37°C, as previously reported (Kilens et al., 2018; Liu et al., 2017). As expected, we were able to generate naive iPSCs from HDFs (nCT2.0_1–3) but not from PBMCs (Figure S1A). We therefore substituted the proto-oncogene CMYC with LMYC (SeV-LMYC), which is reported to be more efficient in human iPSC reprogramming (Nakagawa et al., 2008, 2010). Importantly, using this modified CytoTune-iPS 2.0L, consisting of SeV-KOS, SeV-LMYC, and SeV-KLF4, we succeeded in deriving naive human iPSCs from both HDFs (nCT2.0L_1–2) and PBMCs (nCT2.0L_3–4) (Figure S1B).
Next, we collected cells at each passage and measured the amount of remaining SeV genome by real-time quantitative PCR (qPCR). We found that expression of the SeV genome persisted in all clones, and in some cell lines, the expression level was equal to or even greater than the housekeeping gene GAPDH (Figure 1A). We confirmed the existence of residual SeV in the established naive human iPSCs by immunocytochemistry (Figure S1C). All three SeV vectors persisted in all clones (Figure S1D).
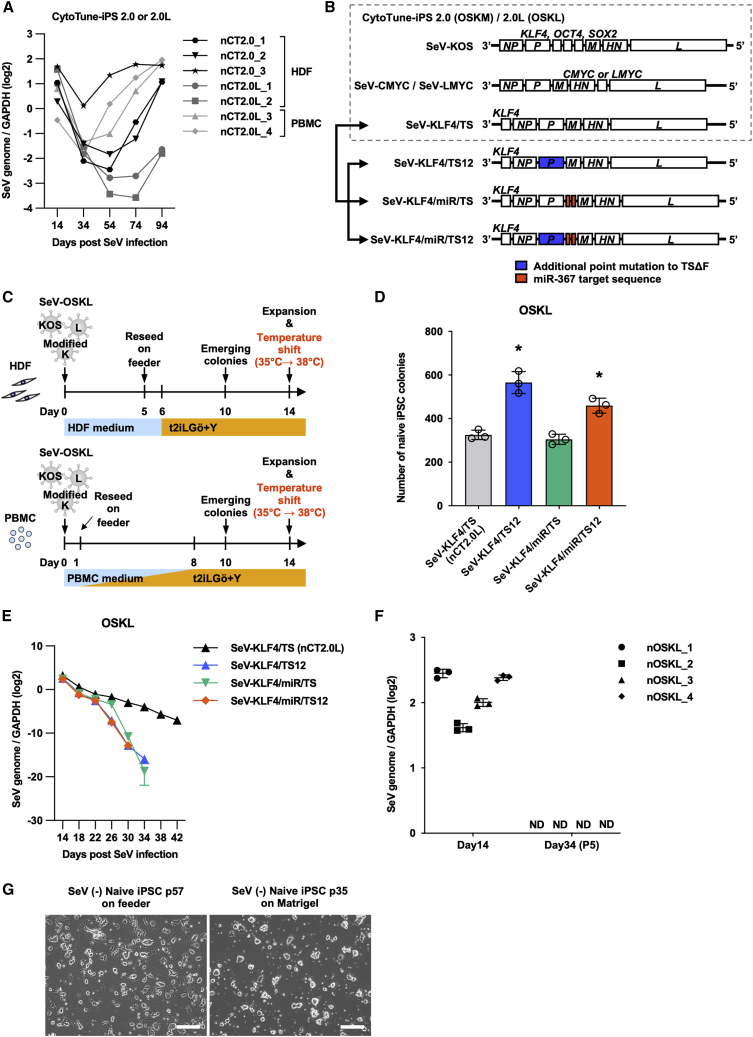
Figure 1.
Reprogramming HDFs and PBMCs into naive human iPSCs with a fast SeV vector removal system and modified SeV-KLF4 vectors
(A) qPCR analysis of SeV genome expression in naive iPSCs reprogrammed with the CytoTune 2.0 or 2.0L SeV reprogramming kit using TaqMan probes. Values are normalized to GAPDH expression and shown as the mean ± SD. n = 3 for each point.
(B) Schematic structure of the SeV vectors and the modified reprogramming cocktail. We changed the SeV18 + KLF4/TSΔF (KLF4/TS) vector in the CytoTune-2.0 or 2.0L reprogramming kit to one of three modified vectors: SeV18 + KLF4/TS12ΔF (KLF4/TS12), SeV18 + KLF4/PmiR367T2/TSΔF (KLF4/miR/TS), or SeV18 + KLF4/PmiR367T2/TS12ΔF (KLF4/miR/TS12). KLF4/miR/TS12 was developed for this study.
(C) Experimental design for the derivation of naive human iPSCs from HDFs or PBMCs using the modified SeV-OSKL cocktail and incubation temperature shift after the generation of naive human iPSCs.
(D) Number of naive human iPSC colonies generated from HDFs at day 14. Each of the SeV-KLF4/TS vectors was co-infected with SeV-KOS and SeV-LMYC. Data are shown as the mean ± SD. n = 3. ∗p < 0.05.
(E) qPCR analysis of SeV genome expression in naive iPSCs derived from HDFs reprogrammed with SeV-KOS, SeV-LMYC, and the respective SeV-KLF4 vectors using TaqMan probes. Data are shown as the mean ± SD. n = 3 of each point.
(F) qPCR analysis of SeV genome expression in HDF-derived naive iPSCs (nOSKL_1, 2) and PBMC-derived naive iPSCs (nOSKL_3, 4) reprogrammed with SeV-KOS, SeV-LMYC, and SeV-KLF4/miR/TS12 vectors at days 14 and 34 using TaqMan probes. Data are shown as the mean ± SD. n = 3.
(G) Representative phase-contrast images of naive iPSCs reprogrammed with SeV-KOS, SeV-LMYC, and SeV-KLF4/miR/TS12 vectors on iMEF feeder cells or in feeder-free conditions. Scale bars, 200 μm.
We hypothesized that the observed retention of SeV vectors may be attributable to the structure of SeV-TSΔF, which forms the backbone of the SeV-KLF4/TS vector. SeV-TSΔF has only one mutation in the viral polymerase (P) gene (Figure S1E) and possess very weak temperature sensitivity; therefore, it is difficult to remove from infected cells by raising the temperature, unlike the SeV-TS12ΔF and SeV-TS15ΔF vectors (Ban et al., 2011). Since the SeV vectors share polymerase and other viral proteins in mixed infection, persistent expression of the TSΔF vector may lead to sustained expression of the other two vectors.
To test this hypothesis, we generated three types of modified SeV-KLF4 vectors: SeV18 + KLF4/TS12ΔF (SeV-KLF4/TS12), SeV18 + KLF4/PmiR367T2/TSΔF (SeV-KLF4/miR/TS), and SeV18 + KLF4/PmiR367T2/TS12ΔF (SeV-KLF4/miR/TS12) (Figure 1B). The SeV-KLF4/TS12 vector is constructed on the SeV-TS12ΔF vector, which has extra point mutations in the polymerase gene to enhance temperature sensitivity (Figure 1B) (Ban et al., 2011). The SeV-KLF4/miR/TS vector has the hsa-microRNA-367 (miR-367) target sequence inserted in tandem on the 5′ side of the P gene of the SeV vector genome (Figure 1B). miR-367 has been shown to be specifically expressed in primed human PSCs (Zhang et al., 2015), and we confirmed that it is also expressed in naive human PSCs (Figure S1F). Thus, we expected that the addition of the miR-367 target sequence would facilitate more rapid vector removal after reprogramming to naive and primed PSCs because both express miR-367. In the SeV-KLF4/miR/TS12 vector, we incorporated both the TS12 backbone and the miR-367 target sequence to further facilitate vector removal (Figure 1B).
We compared the reprogramming efficiency of these modified vectors. It was previously reported that a high MOI was required to generate iPSCs from HDFs when all reprogramming factors were delivered by temperature-sensitive SeV vectors at 37°C, probably due to the loss of most vectors before completion of the reprogramming (Ban et al., 2011). Therefore, we cultured cells at 35°C from the time of infection until day 14, when we performed the first passage of naive iPSCs. Then, the culture temperature was raised to 38°C to remove the SeV vectors (Figure 1C). Reprogramming HDFs using SeV-KOS, SeV-LMYC, and the modified SeV-KLF4 vectors led to the appearance of naive iPSC colonies at around day 10, which is consistent with the conventional method using the SeV-KLF4/TS vector. We found that OSKL cocktails with SeV-KLF4/TS12 or SeV-KLF4/miR/TS12 showed significantly higher reprogramming efficiency than SeV-KL4/TS at day 14 under 35°C culture (Figure 1D).
We then assessed residual SeV vectors in naive iPSCs generated by the modified vectors using qPCR that could detect a single SeV-positive cell in 1 × 106 SeV-negative cells (Figure S1G). After the temperature shift at day 14, cells infected with the SeV-KLF4/TS12, SeV-KLF4/miR/TS, and SeV-KLF4/miR/TS12 vectors showed rapid clearance of the SeV genome, reaching the limit of detection limit at day 34 (Figure 1E). In contrast, SeV-KLF4/TS levels decreased more slowly and remained detectable even at day 42. Thus, the modified SeV vectors can be removed efficiently after reprogramming to naive iPSCs.
We then further characterized the reprogramming capacity of the SeV-KLF4/miR/TS12 vector. We were able to generate naive iPSC clones not only from HDFs (nOSKL_1, 2) but also from PBMCs (nOSKL_3, 4), in which SeV disappeared at day 34 (Figure 1F). We also succeeded in establishing and maintaining feeder-free naive iPSCs from HDFs (nOSKL_FF_1,2) using t2iLGö medium, conditioned by exposure to irradiated mouse embryonic fibroblasts (iMEFs), from the beginning of the reprogramming process (Figure 1G). Notably, this method could also be applied to the generation of primed iPSCs. Using protocols similar to that for naive iPSC generation, we were able to generate SeV-negative primed human iPSC colonies by the third passage (day 41) without colony picking for the selection of SeV-negative colonies (Figure S1H). These results demonstrate the utility of the SeV-KLF4/miR/TS12 vector in multiple applications.
Next, we validated the naive pluripotency of the generated nOSKL-iPSCs. For comparison, we sampled primed human iPSCs (OSKL_1–4) derived from the same somatic cells as nOSKL-iPSCs, primed WA09 H9 embryonic stem cells (ESCs; H9), primed 201B7 iPSCs (201B7), and reset naive H9 ESCs (nH9) (Takashima et al., 2014), which were converted from the primed to the naive state by overexpressing KLF2 and NANOG and maintained in the same conditions as nOSKL-iPSCs. We confirmed that nOSKL-iPSCs exhibited pluripotency marker expression (Figure 2A) and enhanced mitochondrial gene expression related to oxidative phosphorylation (Figure 2B) that were nearly identical to nH9 cells. Then, we compared the transcriptome of nOSKL-iPSCs with primed PSCs and previously published datasets from reset naive PSCs and the human inner cell mass (Takashima et al., 2014; Theunissen et al., 2014; Yan et al., 2013). A principal-component analysis (PCA) revealed that nOSKL-iPSCs were clearly distinguished from primed PSCs and shared almost identical characteristics with nH9 cells (Figure 2C). We also confirmed that nOSKL-iPSCs showed X chromosome reactivation (Figure 2D and 2E) (Balaton et al., 2015; Sahakyan et al., 2017) and a global DNA hypomethylation status compared with primed PSCs (Figure 2F and 2G) (Theunissen et al., 2016). These observations confirmed the naive status of nOSKL-iPSCs.
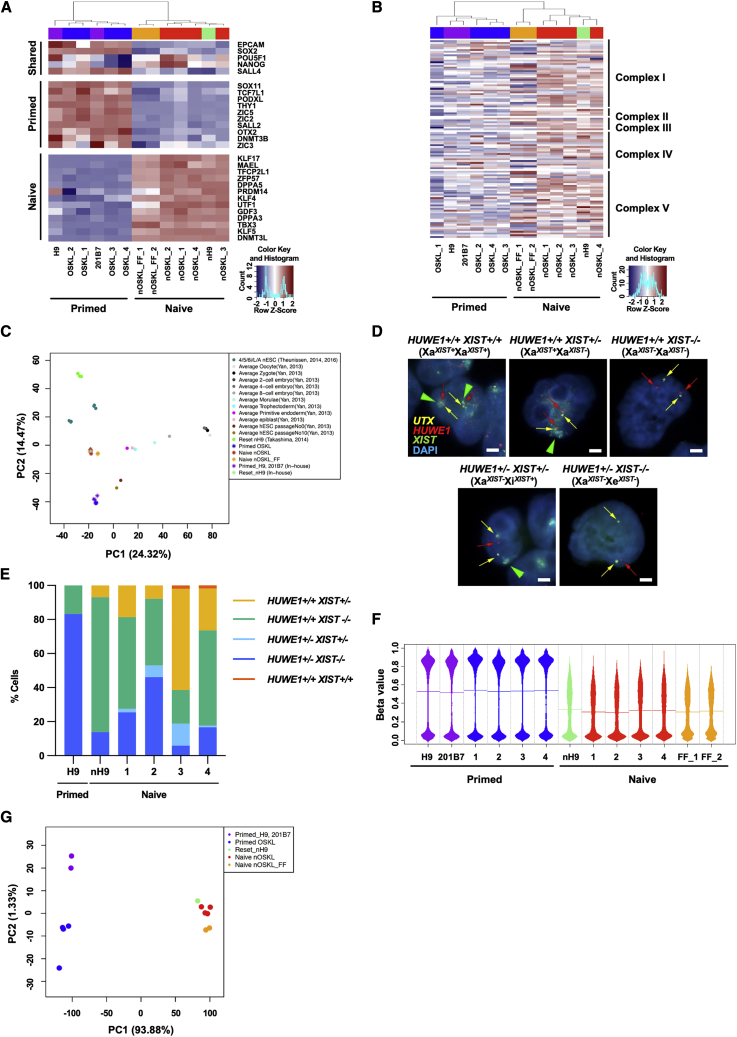
Figure 2.
Hallmarks of naive pluripotency in transcriptome, X chromosome reactivation, and global DNA methylation
(A) Heatmap of the RNA-seq data depicting expression levels of shared, primed, and naive pluripotency-associated marker genes in naive and primed human PSCs.
(B) Heatmap of genes encoding proteins of the electron transport chain located in the inner membrane of mitochondria. These genes reflect the activity of oxidative phosphorylation, are classified by the mitochondrion complex, and are hierarchically clustered.
(C) PCA of RNA-seq data from primed and naive human PSCs in this study compared with reset naive human iPSCs and pre-implantation embryo samples from Takashima et al., 2014, Theunissen et al., 2014, and Yan et al., 2013.
(D) Representative RNA-fluorescence in situ hybridization (FISH) images of naive and primed human iPSCs detecting X-linked genes UTX (escapes X chromosome inactivation) and HUWE1 and XIST (subject to X chromosome inactivation). UTX was analyzed to ensure only PSCs with normal X chromosome were included in the quantification. Scale bar, 20 μm.
(E) Quantification of the RNA-FISH patterns for XIST with HUWE1 in cells with biallelic UTX expression. 100 cells were analyzed in each cell line. Most naive human iPSCs showed X reactivation as indicated by the biallelic HUWE1 expression. Some cells in nOSKL_3 and 4 exhibited biallelic XIST expression, which is similar to the expression pattern of pre-implantation epiblast (Sahakyan et al., 2017; Theunissen et al., 2016).
(F) Beanplot of the global DNA methylation levels in primed and naive human PSCs analyzed using DNA methylation arrays. Horizontal lines in the beanplot represent mean methylation beta values.
(G) PCA of global DNA methylation in primed and naive human PSCs using DNA methylation arrays.
Previous reports demonstrated that naive human PSCs show various abnormal karyotypes more frequently than primed PSCs, and some cell lines show more than 50% abnormal karyotype even after fewer than 10 passages (Liu et al., 2017; Pastor et al., 2016; Theunissen et al., 2014). To examine the genome integrity of the naive iPSCs generated in this study, we performed SNP genotyping arrays of naive human iPSCs including feeder-free naive iPSCs by detecting copy-number variations (CNVs) in their genome. We also examined the source cells for each line to identify the genomic aberrations that occurred after reprogramming (Figures S2A and S2B). All naive PSCs except nOSKL_3 showed that various CNVs emerged after reprogramming to naive pluripotency, and those with a higher number of passages such as nH9 and nOSKL_1 tended to accumulate more abnormalities. Thus, our method did not appear to improve genome stability in either on-feeder or feeder-free naive iPSCs.
Next, to examine the trilineage differentiation potential of naive PSCs, we differentiated them to form embryoid bodies (EBs) (Figure 3A). All cell lines formed EBs, but their morphology differed between SeV (−) naive PSCs and SeV (+) naive iPSCs. The EBs generated from SeV (−) PSCs were often uniformly spherical in shape, whereas EBs generated from SeV (+) iPSCs were larger and tended to have irregular margins and lobes. We then quantified trilineage differentiation ability and residual pluripotent marker gene expression of these EBs using Scorecard assays, which generate an algorithmic score based on the expression of 96 genes analyzed by qPCR (Figure 3B) (Bock et al., 2011). The SeV (−) PSCs scored positively on trilineage potential, especially for mesoderm markers, and low on pluripotency, indicating that these EBs were well differentiated predominantly in the mesodermal lineage. In addition, the overall score profiles were nearly identical between nH9 ESCs and SeV (−) iPSCs. In contrast, the EBs generated from SeV (+) iPSCs showed lower trilineage scores and higher residual pluripotency scores than the SeV (−) PSCs. These results indicated that our method of removing the SeV vectors resulted in naive iPSCs with trilineage differentiation potential similar to naive ESCs and superior to conventionally derived naive iPSC lines.
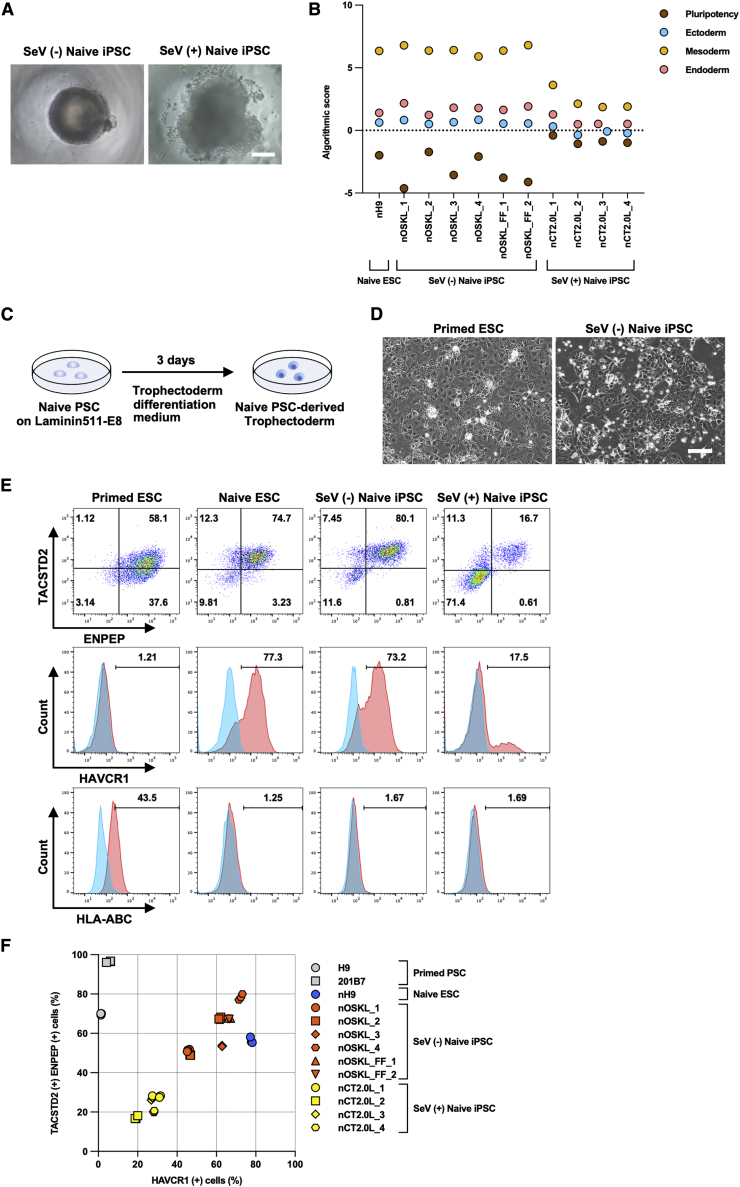
Figure 3.
Generation of naive PSC-derived EBs and trophectoderm
(A) Representative phase-contrast images of SeV (−) naive iPSC (nOSKL_1)- and SeV (+) naive iPSC (nCL2.0L_1)-derived EBs on day 7 after differentiation induction. Scale bars, 100 μm.
(B) Dot plot of algorithmic scores generated by Scorecard analysis based on 96 genes’ expression per sample. n = 1 of each point.
(C) Schematic representation of the trophectoderm differentiation protocol.
(D) Representative phase-contrast images of primed ESC (H9)- and SeV (−) naive iPSC (nOSKL_4)-derived cells on day 3 after trophectoderm induction. Scale bars, 400 μm.
(E) Quantification of TACSTD2, ENPEP, HAVCR1, and HLA-ABC expression in primed ESCs (H9), naive ESCs (nH9), SeV (−) naive iPSCs (nOSKL_4), and SeV (+) naive iPSCs (nCT2.0L_4) by flow cytometry.
(F) Percentage of TACSTD2 (+), ENPEP (+), and HAVCR1 (+) cells by flow cytometry. Each plot shows a single experiment.
Naive PSCs resemble pre-implantation epiblasts in that they have the potential to differentiate into extra-embryonic tissues, which distinguishes them from primed PSCs. We therefore examined the ability of nOSKL-iPSCs to differentiate into trophectoderm (TE) in a defined differentiation medium (Figure 3C) (Io et al., 2021). After inducing differentiation for 3 days, nOSKL-iPSC-derived TE showed identical cell morphology to that of the nH9-derived TE, which was clearly distinct from that of primed H9 ESC-derived differentiated cells (Figure 3D). We examined the expression of the TE-specific markers TACSTD2, ENPEP, and HAVCR1, as well as the negative marker HLA-ABC (Figure 3E and 3F). After differentiation, primed PSCs expressed TACSTD2 and ENPEP but not HAVCR1. They maintained expression of HLA-ABC. In great contrast, like nH9 cells, nOSKL-iPSCs generated by the modified system were positive for TACSTD2, ENPEP, and HAVCR1 and were negative for HLA-ABC. In some nOSKL-iPSC clones, we observed that higher proportions of cells expressed TACSTD2 and ENPEP than nH9-derived clones. Thus, nOSKL-iPSC clones generated with the SeV-KLF4/miR/TS12 vector are comparable to naive ESCs in TE differentiation potential.
Discussion
In this study, we developed an improved method of reprogramming human cells to a state of naive pluripotency. We used LMYC instead of CMYC and modified the SeV-KLF4 vector by introducing temperature-sensitive mutations in the viral polymerase gene and a target sequence for a miRNA specifically expressed in naive and primed pluripotent stem cells. We also revised a temperature setting during reprograming and maintenance of naive iPSCs. In the naive iPSCs generated by this method, SeV vectors rapidly disappeared, and the cells showed significantly improved potential to differentiate to trilineage and extra-embryonic lineages. Moreover, this method also generated more naive iPSC colonies than the conventional method. Although the mechanism for this improved yield was unclear, we observed more floating cells after infection with TS SeV than with TS12 SeV upon 35°C culture, which may indicate that TS12 is less toxic than TS in this condition, contributing to the difference in colony number. Indeed, our results indicate that the TS modification may have had a greater impact on the increased reprogramming efficiency than did the addition of miRNA target sequences.
nCT2.0L_1–4 iPSCs that were generated by the conventional method showed significantly lower differentiation potential in all clones than the nOSKL-iPSCs in our study. We found that the nCT2.0L-iPSC clone with the lowest residual expression of SeV genome on day 94 after infection maintained SeV expression similar to that of the housekeeping gene GAPDH. Our results suggest that even this low-level expression was sufficient to lead to persistent expression of pluripotency genes after differentiation induction, impairing differentiation potential.
The SNP genotyping array results showed that the naive iPSCs generated by our method had many genomic aberrations in both on-feeder and feeder-free iPSCs, similar to those previously reported in human naive PSCs (Liu et al., 2017). It has been shown that karyotypic abnormalities may be relatively suppressed by changing the components of naive PSC culture medium (Di Stefano et al., 2018). However, further optimized culture media and reprogramming methods are needed to avert this issue entirely.
Recently, naive human PSCs have been used not only for in-vitro-directed differentiation to extra-embryonic tissues but also for the study of human early development by generating blastoids (Liu et al., 2021; Yanagida et al., 2021; Yu et al., 2021). Our method may contribute to the development of these technologies. Furthermore, in regenerative medicine, naive iPSCs generated by this method will be useful in the study of organogenesis by blastocyst complementation (Kobayashi et al., 2010; Yamaguchi et al., 2017) due to their higher pluripotency.
The revised method can also be applied to the generation of primed iPSCs. Generally, primed iPSCs are generated through isolation of individual colonies, which is a labor-intensive process. One reason for this is to select iPSC clones without residual SeV or other exogenous genes. Our revised method results in elimination of SeV vectors without this cloning step, which should be useful in technologies such as autologous iPSC generation from a large number of donors, thus advancing various disease modeling, drug discovery, and cell therapy efforts (Madrid et al., 2021).
Finally, we successfully generated naive iPSCs from PBMCs, which are easier to collect than HDFs, by changing the combination of reprogramming factors to SeV-OSKL. Moreover, we established naive human iPSCs from HDFs in a feeder-free environment using iMEF-conditioned medium. These features should promote the generation of autologous iPSCs from as many donors as the primed iPSCs, and they will enable personalized clinical applications of naive human iPSCs, such as elucidation of infertility mechanisms by placental disease modeling and drug discovery.
Limitations of the study
There are several limitations in this study. We used various assays to evaluate naive pluripotency, but we used only one clone of reset naive PSCs as a control. It remains difficult to generate naive human iPSCs from PBMCs under feeder-free conditions. Even though HDFs have been successfully established in a feeder-free environment, a xeno-free environment has not been achieved, since we still used Matrigel as a substrate. As is the case with primed PSCs (Cahan and Daley, 2013; Kilpinen et al., 2017), we observed variations in quality among established naive cell clones. Therefore, further refinement of reprogramming methods is necessary, such as optimization of reprogramming factor combination.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Sendai Virus pAb (Polyclonal Antibody) | MBL International | Cat#PD029, RRID: AB_10597564 |
| Goat anti-Rabbit IgG Secondary Antibody Cyanine5 | Thermo Fisher Scientific | Cat#A10523, RRID: AB_2534032 |
| Pacific Blue anti-human HLA-A,B,C Antibody | BioLegend | Cat#311418, RRID: AB_493669 |
| Human TROP-2 (TACSTD2) Alexa Fluor 488-conjugated Antibody | R&D systems | Cat#FAB650G, RRID: not available |
| PE Mouse Anti-Human CD249 (ENPEP) | BD Biosciences | Cat#564533, RRID: AB_2738838 |
| TIM-1 (HAVCR1) Antibody, anti-human, Biotin, RE-Afinity | Miltenyi Biotec | Cat#130-106-023, RRID: AB_2654152 |
| APC Streptavidin | BioLegend | Cat#405207, RRID: not available |
| Bacterial and virus strains | ||
| CytoTune-iPS 2.0 | ID Pharma, Japan | Cat# 69000-41 |
| CytoTune-iPS 2.0L | ID Pharma, Japan | Cat# 69020-81 |
| CytoTuneEX-iPS (containing SeV18 + KLF4/PmiR367T2/TSΔF) | ID Pharma, Japan | Cat# 69060-61 |
| SeV18 + KLF4/TS12ΔF | ID Pharma, Japan | This paper |
| SeV18 + KLF4/PmiR367T2/TS12ΔF | ID Pharma, Japan | This paper |
| Chemicals, peptides, and recombinant proteins | ||
| CHIR99021 | Sigma-Aldrich | Cat#SML1046 |
| PD0325901 | Sigma-Aldrich | Cat#PZ0162 |
| Gö6983 | Sigma-Aldrich | Cat#G1918 |
| Recombinant Human LIF | Peprotech | Cat#300-05 |
| Y-27632 (hydrochloride) | Wako | Cat#036-24023 |
| DMEM/Ham’s F-12 | Sigma-Aldrich | Cat#D6421 |
| NDiff227 | Takara Bio | Cat#Y40002 |
| Matrigel ESC qualified Matrix | Corning | Cat#CLS354277 |
| iMatrix-511 (Laminin-E8) | Nippi, Japan | Cat#892012 |
| Stem-Cellbanker | Takara Bio | Cat#CB045 |
| Cell Banker 1 | Takara Bio | Cat#CB011 |
| Bovine Albumin Fraction V (7.5% solution) | Gibco | Cat#15260037 |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat#D2650 |
| Accutase | Innovative Cell Technologies | Cat#AT104 |
| A83-01 | Wako | Cat#039-24111 |
| JAK Inhibitor I | Sigma-Aldrich | Cat#420099 |
| Recombinant Human BMP-4 Protein | R&D systems | Cat#314-BP |
| 7-AAD Viability Staining Solution | BioLegend | Cat#420403 |
| StemFit AK02N | Takara Bio | Cat#AK02N |
| DMEM (4.5 g/L Glucose) with L-Gln, without Sodium Pyruvate, liquid | Nacalai Tesque | Cat#08459-64 |
| StemSpan ACF | STEMCELL Technologies | Cat#09860 |
| Recombinant Human SCF Protein | R&D systems | Cat#255-SC |
| Recombinant Human Thrombopoietin (NS0-expressed) Protein | R&D systems | Cat#288-TPN |
| Recombinant Human Flt-3 Ligand/FLT3L Protein | R&D systems | Cat#308-FK |
| Recombinant Human IL-6 Protein | R&D systems | Cat#206-IL |
| Recombinant Human IL-3 Protein | R&D systems | Cat#203-IL |
| 4%-Paraformaldehyde Phosphate Buffer Solution | Nacalai Tesque | Cat#09154-85 |
| Triton X-100 | Nacalai Tesque | Cat#35501-15 |
| Hydrochloric Acid | Wako | Cat#080-01066 |
| DMEM/F-12, GlutaMAX supplement | Gibco | 10565018 |
| 2-Mercaptoethanol | Gibco | 21985023 |
| MEM Non-Essential Amino Acids Solution (100X) | Gibco | 11140050 |
| Critical commercial assays | ||
| AllPrep DNA/RNA Mini Kit | QIAGEN | Cat#80284 |
| RNase-Free DNase Set | QIAGEN | Cat#79256 |
| NucleoSpin miRNA | MACHEREY-NAGEL | Cat#740971.50 |
| PrimeScript RT Master Mix | Takara Bio | Cat#RR036B |
| TaqMan Advanced miRNA Assays | Applied Biosystems | Cat#A25576 |
| Fast SYBR Green Master Mix | Applied Biosystems | Cat#4385612 |
| TaqMan Fast Advanced Master Mix | Applied Biosystems | Cat#4444557 |
| TaqMan hPSC Scorecard Kit, 384-well | Applied Biosystems | Cat#A15872 |
| TaqMan hPSC Scorecard Panel, Fast 96-well | Applied Biosystems | Cat#A15876 |
| TruSeq Stranded Total RNA Library Prep Gold | Illumina | Cat#20020599 |
| HiSeq PE Cluster Kit v4-cBot | Illumina | Cat#PE-401-4001 |
| HiSeq SBS Kit v4 | Illumina | Cat#FC-401-4003 |
| NovaSeq 6000 S1 Reagent Kit v1.5 | Illumina | Cat#20028318 |
| NextSeq 500/550 High Output Kit v2.5 | Illumina | Cat#FC-404-2005 |
| EZ DNA Methylation Kit | Zymo Research | Cat#D5001 |
| Infinium HumanMethylation450 BeadChip Kit | Illumina | Cat#WG-314-1003 |
| Infinium MethylationEPIC BeadChip Kit | Illumina | Cat#WG-317-1001 |
| Infinium OmniExpress24 v1.3 DNA Analysis Kit | Illumina | Cat#20024631 |
| Deposited data | ||
| RNA-seq, DNA methylation array and SNP genotyping array data | This paper | GEO: GSE179476 |
| Raw images and analyzed data | This paper | https://doi.org/10.5281/zenodo.7101941 |
| Systematic Identification of Defined Conditions for Induction and Maintenance of Naive Human Pluripotency | Theunissen et al. (2014) | GEO: GSE59435 |
| Molecular Criteria for Defining the Naive Human Pluripotent State | Theunissen et al. (2016) | GEO: GSE75868 |
| Tracing pluripotency of human early embryos and embryonic stem cells by single cell RNA-seq | Yan et al. (2013) | GEO: GSE36552 |
| RNA sequencing of conventional and reset human pluripotent stem cells | Takashima et al. (2014) | ArrayExpress: E-MTAB-2857 |
| Experimental models: Cell lines | ||
| Human embryonic stem cell line: H9 (WA09) | WiCell Research Institute | hPSCreg ID: WAe009-A |
| Human induced pluripotent stem cell line: 201B7 | Kyoto University | hPSCreg ID: KUIFMSi004-C |
| Human dermal fibroblast: TIG113 | JCRB Cell Bank | ID: JCRB0539 |
| Human dermal fibroblast: TIG120 | JCRB Cell Bank | ID: JCRB0542 |
| Human peripheral blood mononuclear cells (ID:HHU20120815) | Cellular Technology Ltd. | CTL-UP1 |
| Human peripheral blood mononuclear cells (ID:HHU20140120) | Cellular Technology Ltd. | CTL-UP1 |
| CF1 mouse embryonic fibroblasts, irradiated | ThermoFisher Scientific | A34181 |
| Oligonucleotides | ||
| SEV (Mr04269880_mr) | ThermoFisher Scientific | Cat#4331182 |
| GAPDH (Hs02786624_g1) | ThermoFisher Scientific | Cat#4331182 |
| hsa-miR-367-3p (478066_mir) | ThermoFisher Scientific | Cat#A25576 |
| hsa-miR-423-3p (478327_mir) | ThermoFisher Scientific | Cat#A25576 |
| Primer: SOX2 to M in SeV-KOS Forward: CTGCCCCTCTCACACATGT Reverse: GGGCTCCACAGTACCGTTAT |
This paper | N/A |
| Primer: LMYC to L in SeV-LMYC Forward: GAAAAGACAGCTCCGATGCC Reverse: CCTGCCCATCCATGACCTAG |
This paper | N/A |
| Primer: HN to CMYC in SeV-CMYC Forward: CTCTCAGTCTCTTACGTCTCTCA Reverse: CCTCCTCGTCGCAGTAGAAA |
This paper | N/A |
| Primer: Leader to KLF4 in SeV-KLF4 Forward: TTCCAGACCCTTTGCTTTGC Reverse: GCGAACGTGGAGAAAGATGG |
This paper | N/A |
| Primer: GAPDH Forward: TGCACCACCAACTGCTTAGC Reverse: TCTTCTGGGTGGCAGTGATG |
This paper | N/A |
| Software and algorithms | ||
| Graphpad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo software 10.6.1 | FlowJo, LCC | https://www.flowjo.com/ |
| bcl2fastq v2.17.1.14 | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| long-rna-seq-pipeline v2.3.4. | ENCODE DCC | https://github.com/ENCODE-DCC/long-rna-seq-pipeline/releases/tag/v2.3.4 |
| STAR 2.5.1b | Dobin et al. (2013) | https://github.com/alexdobin/STAR |
| RSEM 1.2.23 | Li and Dewey (2011) | https://github.com/deweylab/RSEM/releases/tag/v1.2.23 |
| Subread 1.5.1 | Liao et al. (2014) | https://sourceforge.net/projects/subread/files/subread-1.5.1/ |
| GenomeStudio V2011.1 | Illumina | https://support.illumina.com/downloads/genomestudio_software_20111.html |
| R 3.6.3 | The R Foundation for Statistical Computing | http://www.R-project.org/ |
| Minfi | Aryee et al. (2014) | https://bioconductor.org/packages/release/bioc/html/minfi.html |
| GenomeStudio v2.0.4 | Illumina | https://support.illumina.com/downloads/genomestudio-2-0.html |
| PennCNV 1.0.3 | Wang et al. (2007) | https://www.openbioinformatics.org/penncnv/penncnv_installation.html |
| Mosaic Alteration Detection-MAD (1.0.1) | González et al. (2011) | https://github.com/isglobal-brge/MAD/ |
| GWAS tools (1.16.1) | Gogarten et al. (2012) | https://www.bioconductor.org/packages/release/bioc/html/GWASTools.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Akira Kunitomi: akira.kunitomi@gladstone.ucsf.edu.
Materials availability
All unique/stable reagents generated in this study, including modified SeV-KLF4 vectors, are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
We obtained HDFs collected under informed consent from the Tokyo Metropolitan Institute of Gerontology (Kondo and Yonezawa, 1995). iMEFs were purchased from ThermoFischer Scientific and PBMCs were purchased from Cellular Technology Limited (CTL). Primed and naive ESC clones were obtained from WiCELL (Thomson et al., 1998) and Kyoto University (Takashima et al., 2014). All cells except naive PSCs were cultured in humidified incubators at 37°C in 5% CO2 and 20% O2, and naive PSCs were cultured under 5% O2 throughout. Recombinant DNA experiments in this study were carried out under the approval of Kyoto University and The J. David Gladstone Institutes.
Method details
Cell culture
HDFs and iMEFs were maintained in Dulbecco’s modified Eagle medium (DMEM, Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS, Japan Bio Serum). The day before naive PSCs were plated, iMEF cells were seeded in cell culture dishes at a concentration of 25,000 cells/cm2 and cultured overnight. The next day, the cells were washed twice with PBS(−) before plating. PBMCs were cultured in StemSpan ACF (STEMCELL) with 100 ng/mL human SCF (R&D), 100 ng/mL human TPO (R&D), 100 ng/mL human Flt3/Flk2 (R&D), 50 ng/mL human IL-6 (R&D), and 20 ng/mL human IL-3 (R&D) for 5 days before the SeV vector infection. Primed PSCs were maintained in StemFit AK02N medium (Ajinomoto) on laminin 511-E8 fragments (iMatrix-511, Nippi)-coated plates. Naive PSCs were cultured on iMEFs and maintained in t2iLGö (Takashima et al., 2014) medium composed of N2B27 medium (NDiff227, Takara Bio) with 1 μM CHIR99021 (Merck), 1 μM PD0325901 (Merck), 10 μg/mL human LIF (Peprotech), and 2.5 μM Gö6983 (Merck). Medium was changed every other day and 10 μM Y27632 (Wako) was added just before every medium change. Naive iPSCs were passaged every 3–4 days using Accutase (Innovative Cell Technologies).
Construction of modified SeV-KLF4 vectors
The insert sequence containing the open reading frame of human KLF4 gene was constructed by PCR from cDNAs using NotI-tagged gene-specific forward and reverse primers including SeV-specific transcriptional regulatory signal sequences. The point mutations on the TS and TS12 backbones are reported elsewhere (Schlaeger, 2018). The sequence of miR367T2, which is inserted after the P gene is TCACCATTGCTAAAGTGCAATTcgatTCACCATTGCTAAAGTGCAATT.
The method for constructing the plasmids that contain the F gene deficient SeV vector backbone is described elsewhere (Inoue et al., 2003), and in principle the same method was used to construct plasmids pSeV18 + KLF4/TS12ΔF, pSeV18 + KLF4/PmiR367T2/TSΔF and pSeV18 + KLF4/PmiR367T2/TS12ΔF (collectively, “SeV-KLF4 vector plasmids”), which contain the additional mutations and insert as described above.
The recovery and propagation of SeV-KLF4 vectors from the SeV-KLF4 vector plasmids were performed as previously described (Komuta et al., 2016).
Reprogramming of human somatic cells into naive or primed iPSCs
In the conventional method, HDFs or PBMCs were reprogrammed with the CytoTune-iPS 2.0 or 2.0L reprogramming kit. In this study, we replaced the KLF4/TS vector originally supplied with the kit, with one of the modified SeV-KLF4 vectors: KLF4/TS12, KLF4/PmiR367 T2/TS or KLF4/PmiR367T2/TS12. Cells were incubated at 35°C until the naive or primed iPSC colony generation. After the first passage of the generated iPSCs (usually day 14 for naive iPSCs and day 21 for primed iPSCs), the incubation temperature was changed to 38°C to remove the SeV vectors. After removal of the SeV vectors (day 34 when SeV-KLF4/PmiR367T2/TS12 was used), the temperature was changed to 37°C. In naive PSC reprogramming, the incubator was set to 5% O2 after the SeV vector infection.
HDFs were initially maintained in DMEM (Nacalai Tesque) supplemented with 10% FBS. At day 0, the cells were counted and infected with three SeV vectors: 1) polycistronic KLF4-OCT4-SOX2, 2) CMYC or LMYC, and 3) KLF4 at a multiplicity of infection of 5, respectively. From day 1, the medium was changed every other day. At day 5, the cells for naive reprogramming were reseeded on iMEF feeder cells, and the cells for primed reprogramming were reseeded on laminin 511-E8 fragment-coated plates. The next day the medium was changed to the respective PSC medium.
In PBMC reprogramming, PBMCs were cultured for 5 days in the StemSpan ACF based medium described in the cell culture section. At day 0, the cells were counted and infected with the three SeV vectors using the same reprogramming procedure as for HDFs. At day 1, the cells were counted and reseeded in the same conditions as for HDF reprogramming. Beginning on day 2 and then every other day, the same amount of PSC medium was added as day 1. At day 8, the medium was completely changed to PSC medium.
Preparation of iMEF conditioned t2iLGö medium for feeder-free naive iPSC culture
iMEF cells were seeded in cell culture dishes at a concentration of 25,000/cm2 cells and cultured in fibroblast medium (DMEM with 10% Fetal bovine serum) overnight. The next day, the cells were washed twice with PBS(−) and replaced with t2iLGö medium and cultured in a 37°C CO2 incubator for 24 h. The next day, the medium was collected using the MILLEX-HP 0.45 μm low protein binding filter (Merck) and used for feeder-free PSC culture.
Generation of feeder-free naive iPSCs from fibroblasts
Naive iPS cells were reprogrammed in on-feeder culture according to the protocol described above. At the first passage, iMEFs were removed by incubating naive PSCs for 2 h at 37°C on a gelatin-coated dish. This procedure was repeated in the next passage to completely remove the iMEFs. After collection, naive iPS cells were resuspended in iMEF conditioned medium plus 10 μM Y27632 (Wako) and seeded into culture dishes that had been incubated with Matrigel (hESC-qualified, Corning) at 37°C for 1 h before seeding. Following the seeding, the culture temperature was raised to 38°C to remove SeV vectors until SeV genome was not detected by qPCR.
RNA, DNA and miRNA extraction and real-time quantitative PCR (qPCR)
iMEFs were removed by incubating naive PSCs for 2 h at 37°C on a gelatin-coated dish before sampling for nucleic acid extraction. Total RNA and DNA were extracted from cell lysates using the AllPrep DNA/RNA Mini Kit (QIAGEN), and the RNA was incubated with RNase-Free DNase Set (QIAGEN) to remove genomic DNA. MicroRNA (miRNA) was extracted using NucleoSpin miRNA (MACHEREY-NAGEL). For qPCR, the reverse transcription reaction was performed with 1 μg of DNase-treated RNA using PrimeScript RT Master Mix (Takara) containing oligo dT primer and random 6 mers. cDNA was synthesized using TaqMan Advanced miRNA Assays (Applied Biosystems). qPCR including Scorecard analysis was performed on StepOne Plus (Applied Biosystems) or QuantStudio3 (Applied Biosystems) using TaqMan Fast Advanced Master Mix (Applied Biosystems) or Fast SYBR Green Master Mix (Applied Biosystems) according to the manufacturer’s protocol.
RNA sequencing and data analysis
RNA sequencing libraries were made from 100 ng of total RNA as starting materials with the TruSeq Stranded Total RNA Library Prep Gold (Illumina) following the manufacturer’s protocol. For Hiseq2500, clusters were generated with the HiSeq PE Cluster Kit v4-cBot (Illumina) using illumina cBot. Sequencing was performed with the HiSeq SBS Kit v4 using HiSeq2500 (2 × 126 PE mode). NovaSeq 6000 (2 × 101 PE mode) with the NovaSeq 6000 S1 Reagent Kit v1.5 (Illumina) and NextSeq 500 (76 SE mode) with the NextSeq 500/550 High Output Kit v2.5 was also used for sequencing. FASTQ files were generated from bcl files using bcl2fastq v2.17.1.14 (Illumina) and processed using ENCODE long-rna-seq-pipeline v2.3.4. Briefly, the sequenced reads were mapped to the human reference genome (GRCh38) using STAR 2.5.1b (Dobin et al., 2013) with GENCODE v24 gene annotations, the normalized gene expression data was calculated using RSEM 1.2.23 (Li and Dewey, 2011), and the gene count data was obtained using featureCounts bundled in Subread 1.5.1(Liao et al., 2014). For the characterization of our PSC lines, the datasets of GSE59435 (Theunissen et al., 2014) and GSE75868 (Theunissen et al., 2016) obtained from GEO and supplemental data in Yan et al. (2013) and Takashima et al. (2014) were used. The expression values of 4,720 genes included in all data were normalized by quantile among samples, and z-scores for each gene were used for the PCA. Log2-scaled, quantile normalized FPKM values were used for the expression heatmap of PSC markers and oxidative phosphorylation-related genes.
DNA methylation analysis
The bisulfite conversion of 500 ng genomic DNA was performed using the EZ DNA Methylation Kit (Zymo Research), and the global DNA methylation status was profiled using Infinium Human Methylation 450K or EPIC BeadChip Kit (illumina) according to the manufacturer’s protocols. After exporting the DNA methylation values using GenomeStudio V2011.1, data processing was conducted using the “minfi” package in R 3.6.3 (Aryee et al., 2014). In total, 424,444 probes common between 450K and EPIC and not located at known SNP sites were used for the PCA of PSC samples.
RNA-FISH
Dissociated naive iPSCs were incubated on a gelatin-coated dish for 2 h at 37°C to remove iMEF feeder cells. Then the cells were seeded on Matrigel-coated slides in PSC medium. The next day, the cells were fixed in 4% paraformaldehyde for 15 min at room temperature. The slides were treated with 0.2 M HCl for 20 min, permeabilized with 0.2% Triton X-100 for 10 min, digested with pepsin solution (0.005% in 0.1 M HCl) at 37°C for 2–6 min, and dehydrated. Bacterial artificial chromosomes (BACs) RP11-155O24 and RP11-256P2 were used to generate HUWE1 and UTX RNA FISH probes, respectively. BAC DNAs were labeled by nick-translation with Cy5-dUTP (RP11-155O24) and Cy3-dUTP (RP11-256P2). The labeled probes and the XIST RNA FISH probe (Chromosome Science Labo) were mixed with sonicated salmon sperm DNA and Cot-1 DNA in hybridization solution. The probes were denatured at 85°C for 10 min, applied to the pretreated slides, covered with cover slips, and hybridized at 37°C overnight. The slides were then washed with 50% formamide/2xSSC at 37°C for 20 min, 1xSSC for 15 min at room temperature, counterstained using DAPI, and mounted. The FISH images were captured with the CW4000 FISH application program (Leica Microsystems Imaging Solution) using a cooled CCD camera mounted on a Leica DMRA2 microscope.
In vitro differentiation
For embryoid body formation, 10,000 naive PSCs were plated per well of CELLSTAR 96 well V-bottom plates (Greiner) with 100 μL of differentiation media: DMEM/F12, GlutaMAX supplement (Gibco) with 20% fetal bovine serum supplemented with 55 μM 2-mercaptoethanol (2ME) (Gibco) and 100X MEM non-essential amino acids (NEAA) (Invitrogen). After 2 days, 100 μL of medium was added per well. Subsequently, the medium was changed every other day by half, i.e., 100 μL. The cells were harvested at day7 after differentiation induction, and the total RNA was extracted for Scorecard analysis.
Naive PSC-derived trophectoderm (TE) differentiation
For TE differentiation, naive PSCs were dissociated, and iMEF feeder cells were removed as described above. 500,000 cells were plated in laminin 511-E8 (0.15 μg/cm2 iMatrix 511; Nippi)-coated 6 wells with initial TE differentiation medium: Ndiff227, 2 μM A83-01 (Wako), 2 μM PD0325921 (Sigma) and 10 ng/mL BMP4 (R&D). The following day, the medium was changed to Ndiff227, 2 μM A83-01 (Wako), 2 μM PD0325921 (Sigma) and 1 μg/mL JAK inhibitor I (Sigma). The next day, the medium was changed again. At day 3, we harvested the cells using Accutase (Innovative Cell Technologies) for 30 min and analyzed TACSTD2, ENPEP, HAVCR1 and HLA-ABC expression by flow cytometry.
Flow cytometry
Dissociated cells were stained on ice for 20 min with Alexa Fluor 488-conjugated TROP-2 (TACSTD2), PE-conjugated CD249 (ENPEP), Tim-1 (HAVCR1) biotinylated antibody, and Pacific Blue-conjugated HLA-ABC antibody. After washing, APC Streptavidin was applied and incubated on ice for 20 min. Analyses were performed using the BD LSR Fortessa (BD Biosciences) and FACSAria II (BD Biosciences) flow cytometer equipped with FACS Diva software (BD biosciences). The data were analyzed using FlowJo software (LLC).
SNP genotyping array
Copy number variation (CNV) was evaluated with SNP genotyping array. Genomic DNA was hybridized onto the Infinium OmniExpress24 v1.3 DNA Analysis Kit (Illumina), and intensities were scanned by iScan (Illumina) following the manufacturer’s protocol. After exporting a final report using GenomeStudio (2.0.4) (Illumina), CNV analysis was performed with PennCNV (1.0.3) (Wang et al., 2007), Mosaic Alteration Detection-MAD (1.0.1) (González et al., 2011) and GWAS tools (1.16.1) (Gogarten et al., 2012). Only CNVs in test samples against control samples were reported. Log R Ratios and B-Allele Frequencies were visualized with GenomeStudio.
Quantification and statistical analysis
Exact n values are described in the relevant figure legends. Statistical significance was determined using a two-tailed unpaired Student’s t test and Prism software (GraphPad). p < 0.05 were considered significant and are indicated by asterisks in the figures. Error bars represent the mean ± s.d.
Acknowledgments
We are grateful to M. Iwasaki and K. Tomoda for sharing data and discussions prior to publication; M. Saito, A. Niwa, and M. Nakagawa for providing valuable experimental equipment; T. Okubo for technical assistance; and P. Karagiannis and K. Claiborn for critical reading of this manuscript. This work was supported by the Core Center for iPS Cell Research, Research Center Network for Realization of Regenerative Medicine, AMED under grant number JP21bm0104001; iPS Cell Research Fund; and JSPS KAKENHI under grant numbers 16K19429 and 18K15846. This work was also supported by funding from Mr. Hiroshi Mikitani, Mr. Marc Benioff, the L. K. Whittier Foundation, the Roddenberry Foundation, the Gladstone Institutes, the National Heart, Lung, and Blood Institute (NHLBI), and National Institutes of Health (NIH) (U01-HL100406, U01-HL098179, R01-HL130533, and R01-HL135358). Gladstone Institutes received support from National Center for Research Resources grant RR18928-01.
Author contributions
A.K. designed and conceived this study, performed most of the experiments, and analyzed the data. R.H. and V.A. cultured cells and performed differentiation assays. M.O. performed and supported flow cytometry. T.M.K. and M.N. analyzed the RNA sequencing (RNA-seq), DNA methylation array, and SNP genotyping array data. J.K., H.H., and K.K. developed and provided the SeV vectors. Y.T. and K.T. provided and instructed experimental techniques. K.F., N.T., and S.Y. supervised the project. A.K. and S.Y. wrote the manuscript.
Declaration of interests
A.K. and J.K. are co-inventors on a patent describing the method for producing naive human iPSCs from somatic cells. J.K. and H.H. are employees and K.K. is a board member of ID Pharma Co., Ltd., without compensation relating to this study. K.T. is on the scientific advisory board of I Peace, Inc., without salary. S.Y. is a scientific advisor to iPS Academia Japan without salary.
Inclusion and diversity
We worked to ensure diversity in experimental samples through the selection of the cell lines.
Published: October 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100317.
Supplemental information
Data and code availability
-
•
RNA-sequencing, DNA methylation array and SNP genotyping array data are accessible in the Gene Expression Omnibus database of the National Center for Biotechnology Information website. Accession numbers are listed in the key resources table. The original image data, numerical data and processed sequencing and array data that were not shown in the paper have been deposited at Zenodo and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaton B.P., Cotton A.M., Brown C.J. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differ. 2015;6:35. doi: 10.1186/s13293-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban H., Nishishita N., Fusaki N., Tabata T., Saeki K., Shikamura M., Takada N., Inoue M., Hasegawa M., Kawamata S., Nishikawa S.I. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U., Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H., et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B., Ueda M., Sabri S., Brumbaugh J., Huebner A.J., Sahakyan A., Clement K., Clowers K.J., Erickson A.R., Shioda K., et al. Reduced MNAÏVEhibition preserves genomic stability in naive human embryonic stem cells. Nat. Methods. 2018;15:732–740. doi: 10.1038/s41592-018-0104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten S.M., Bhangale T., Conomos M.P., Laurie C.A., McHugh C.P., Painter I., Zheng X., Crosslin D.R., Levine D., Lumley T., et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–3331. doi: 10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J.R., Rodríguez-Santiago B., Cáceres A., Pique-Regi R., Rothman N., Chanock S.J., Armengol L., Pérez-Jurado L.A. A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC Bioinf. 2011;12:166. doi: 10.1186/1471-2105-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., von Meyenn F., Rostovskaya M., Clarke J., Dietmann S., Baker D., Sahakyan A., Myers S., Bertone P., Reik W., et al. Epigenetic resetting of human pluripotency. Development. 2017;144:2748–2763. doi: 10.1242/dev.146811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M., Tokusumi Y., Ban H., Kanaya T., Tokusumi T., Nagai Y., Iida A., Hasegawa M. Nontransmissible virus-like particle formation by F-deficient sendai virus is temperature sensitive and reduced by mutations in M and HN proteins. J. Virol. 2003;77:3238–3246. doi: 10.1128/JVI.77.5.3238-3246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Io S., Kabata M., Iemura Y., Semi K., Morone N., Minagawa A., Wang B., Okamoto I., Nakamura T., Kojima Y., et al. Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell. 2021;28:1023–1039.e13. doi: 10.1016/j.stem.2021.03.013. [DOI] [PubMed] [Google Scholar]
- Kilens S., Meistermann D., Moreno D., Chariau C., Gaignerie A., Reignier A., Lelièvre Y., Casanova M., Vallot C., Nedellec S., et al. Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat. Commun. 2018;9:360. doi: 10.1038/s41467-017-02107-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F.P., Culley O.J., et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuta Y., Ishii T., Kaneda M., Ueda Y., Miyamoto K., Toyoda M., Umezawa A., Seko Y. In vitro transdifferentiation of human peripheral blood mononuclear cells to photoreceptor-like cells. Biol. Open. 2016;5:709–719. doi: 10.1242/bio.016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Yamaguchi T., Hamanaka S., Kato-Itoh M., Yamazaki Y., Ibata M., Sato H., Lee Y.S., Usui J.I., Knisely A.S., et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142:787–799. doi: 10.1016/j.cell.2010.07.039. [DOI] [PubMed] [Google Scholar]
- Kondo H., Yonezawa Y. Fetal-adult phenotype transition, in terms of the serum dependency and growth factor requirements, of human skin fibroblast migration. Exp. Cell Res. 1995;220:501–504. doi: 10.1006/excr.1995.1342. [DOI] [PubMed] [Google Scholar]
- Kono K., Sawada R., Kuroda T., Yasuda S., Matsuyama S., Matsuyama A., Mizuguchi H., Sato Y. Development of selective cytotoxic viral vectors for concentration of undifferentiated cells in cardiomyocytes derived from human induced pluripotent stem cells. Sci. Rep. 2019;9:3630. doi: 10.1038/s41598-018-36848-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.O., Zhu Y.F., Asakawa M., Kuma H., Hirata T., Ueda Y., Lee Y.S., Fukumura M., Iida A., Kato A., et al. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J. Virol. 2000;74:6564–6569. doi: 10.1128/jvi.74.14.6564-6569.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu X., Nefzger C.M., Rossello F.J., Chen J., Knaupp A.S., Firas J., Ford E., Pflueger J., Paynter J.M., Chy H.S., et al. Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat. Methods. 2017;14:1055–1062. doi: 10.1038/nmeth.4436. [DOI] [PubMed] [Google Scholar]
- Liu X., Tan J.P., Schröder J., Aberkane A., Ouyang J.F., Mohenska M., Lim S.M., Sun Y.B.Y., Chen J., Sun G., et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature. 2021;591:627–632. doi: 10.1038/s41586-021-03372-y. [DOI] [PubMed] [Google Scholar]
- Madrid M., Sumen C., Aivio S., Saklayen N. Autologous induced pluripotent stem cell-based cell therapies: promise, progress, and challenges. Curr. Protoc. 2021;1:e88. doi: 10.1002/cpz1.88. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Takizawa N., Narita M., Ichisaka T., Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pastor W.A., Chen D., Liu W., Kim R., Sahakyan A., Lukianchikov A., Plath K., Jacobsen S.E., Clark A.T. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell. 2016;18:323–329. doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., Kuoy E., Langerman J., Clark A.T., Jaenisch R., Plath K. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeger T.M. Nonintegrating human somatic cell reprogramming methods. Adv. Biochem. Eng. Biotechnol. 2018;163:1–21. doi: 10.1007/10_2017_29. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Okita K., Nakagawa M., Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W., et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Friedli M., He Y., Planet E., O'Neil R.C., Markoulaki S., Pontis J., Wang H., Iouranova A., Imbeault M., et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L., et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:524–526. doi: 10.1016/j.stem.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F.A., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Sato H., Kato-Itoh M., Goto T., Hara H., Sanbo M., Mizuno N., Kobayashi T., Yanagida A., Umino A., et al. Interspecies organogenesis generates autologous functional islets. Nature. 2017;542:191–196. doi: 10.1038/nature21070. [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yanagida A., Spindlow D., Nichols J., Dattani A., Smith A., Guo G. Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell. 2021;28:1016–1022.e4. doi: 10.1016/j.stem.2021.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Wei Y., Duan J., Schmitz D.A., Sakurai M., Wang L., Wang K., Zhao S., Hon G.C., Wu J. Blastocyst-like structures generated from human pluripotent stem cells. Nature. 2021;591:620–626. doi: 10.1038/s41586-021-03356-y. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hong Y., Xiang D., Zhu P., Wu E., Li W., Mosenson J., Wu W.S. MicroRNA-302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Rep. 2015;4:645–657. doi: 10.1016/j.stemcr.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-sequencing, DNA methylation array and SNP genotyping array data are accessible in the Gene Expression Omnibus database of the National Center for Biotechnology Information website. Accession numbers are listed in the key resources table. The original image data, numerical data and processed sequencing and array data that were not shown in the paper have been deposited at Zenodo and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.