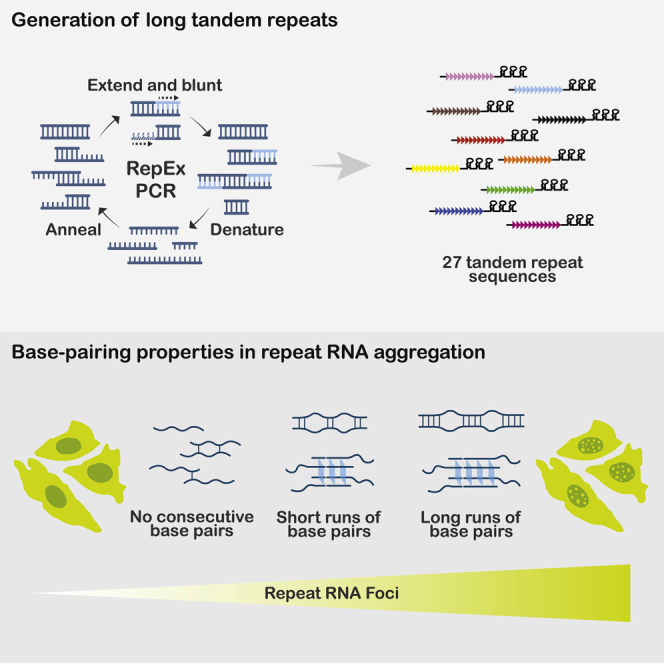
Summary
A common pathological feature of RNAs containing expanded repeat sequences is their propensity to aggregate in cells. While some repeat RNA aggregates have been shown to cause toxicity by sequestering RNA-binding proteins, the molecular mechanism of aggregation remains unclear. Here, we devised an efficient method to generate long tandem repeat DNAs de novo and applied it to systematically determine the sequence features underlying RNA aggregation. Live-cell imaging of repeat RNAs indicated that aggregation was promoted by multivalent RNA-RNA interactions via either canonical or noncanonical base pairs. While multiple runs of two consecutive base pairs were sufficient, longer runs of base pairs such as those formed by GGGGCC hexanucleotide repeats further enhanced aggregation. In summary, our study provides a unifying model for the molecular basis of repeat RNA aggregation and a generalizable approach for identifying the sequence and structural determinants underlying the distinct properties of repeat DNAs and RNAs.
Keywords: short tandem repeat, repeat expansion disorder, RNA aggregation, multivalent interactions, RNA base-pairing, RNA imaging, microsatellite, RNA structure, G-quadruplex
Graphical abstract
Highlights
-
•
A generalizable and efficient method, RepEx-PCR, generates tandem repeat DNAs de novo
-
•
Multivalent RNA-RNA base-pairing promotes repeat RNA aggregation
-
•
Canonical and noncanonical consecutive base pairs could promote RNA aggregation
-
•
Longer runs of consecutive base pairs further enhanced repeat RNA aggregation
Motivation
Expansion of short tandem repeats (STRs) in the human genome underlies over 50 genetic disorders, some of which involve RNA gain-of-function pathophysiological mechanisms. Compared with physiological RNAs, RNAs containing repeat expansion sequences exhibit many distinct properties such as a high propensity to form aggregates in cells, although the causes of these distinct properties remain largely unclear. We set out to develop an unbiased approach to determine the sequence features of repeat RNAs that promote their aggregation in cells, with potentially broad applications in understanding the pathological basis of repeat expansions.
By developing and applying RepEx-PCR, an efficient method to generate tandem repeat DNAs de novo, Isiktas et al. find that repeat RNA aggregation is generally promoted by consecutive runs of either canonical or noncanonical RNA-RNA base pairs. Long runs of base pairs such as those formed by C9orf72 hexanucleotide repeats further enhance aggregation.
Introduction
An increasing number of human diseases, especially neurological disorders, have been shown to arise from STR expansion mutations. While some of them are recessive mutations (e.g., fragile X syndrome), many repeat expansion mutations are inherited in a dominant manner, suggesting possible involvement of gain-of-function mechanisms (Rodriguez and Todd, 2019; Schwartz et al., 2021). One such mechanism that has been well established in myotonic dystrophy type 1 and 2 (DM1/2) is mediated by RNA transcripts containing the expanded repeat sequences, which form aggregates also known as repeat RNA foci (Ranum and Cooper, 2006; Wojciechowska and Krzyzosiak, 2011). The aggregated CUG and CCUG repeat RNAs in DM1 and DM2, respectively, sequester several RNA-binding proteins (RBPs) including the muscleblind-like (MBNL) family of splicing regulators, thereby causing widespread dysregulation in pre-mRNA splicing (Scotti and Swanson, 2016; Wang et al., 2012). In addition to DM1/2, repeat RNA foci have been widely observed in other repeat expansion disorders such as Huntington disease (CAG repeats) (Didiot et al., 2018) and C9orf72-associated amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (GGGGCC repeats) (DeJesus-Hernandez et al., 2011; Zu et al., 2013). However, the pathophysiological significance of repeat RNA foci in these diseases is less well understood.
By imaging CAG/CUG and GGGGCC repeat RNA foci assembly and disassembly in cells, a previous study has shown that these RNA foci are liquid-like, dynamic structures that share similarities with phase-separated protein condensates formed by multivalent interactions (Jain and Vale, 2017). The same study has further proposed that repeat RNAs may assemble by directly base-pairing with one another, a model reminiscent of cytoplasmic stress granule assembly (Van Treeck et al., 2018). While this model is plausible and further supported by in silico simulation (Nguyen et al., 2022), direct evidence of a critical role of RNA-RNA base-pairing is currently lacking. Alternative mechanisms of repeat RNA aggregation may involve, for example, RBPs that contain low-complexity domains that are prone to multimerize via either homotypic or heterotypic interactions (Kato et al., 2012). Once recruited by repeat RNAs, such RBPs may facilitate condensate formation, which may in turn recruit additional repeat RNAs in an indirect manner.
For understanding the mechanism of repeat RNA aggregation, it is important to identify the sequence features that promote this process. However, previous studies of repeat RNA foci have been largely restricted to disease-associated expanded repeats, which represent only a small fraction of all possible repeat sequences. In addition, most dominantly pathogenic repeats have been shown to form RNA foci to some extent, with few counterexamples available. Furthermore, each endogenous repeat region is expressed in a distinct genomic context and cellular environment, thereby prohibiting a direct comparison between repeat sequences.
To circumvent these confounding factors, we developed a generalizable approach to efficiently generate long tandem repeat DNA fragments of any desired sequence and length, which can be subsequently cloned in expression constructs for functional assays. By using this approach, we monitored the formation of RNA foci for a variety of repeats, most of which have not been previously studied. Results from this unbiased analysis strongly supported RNA-RNA base-pairing as a general cause and further revealed the characteristics of RNA base pairs that promote repeat RNA aggregate formation.
Results
Generation of long tandem repeats by RepEx-PCR
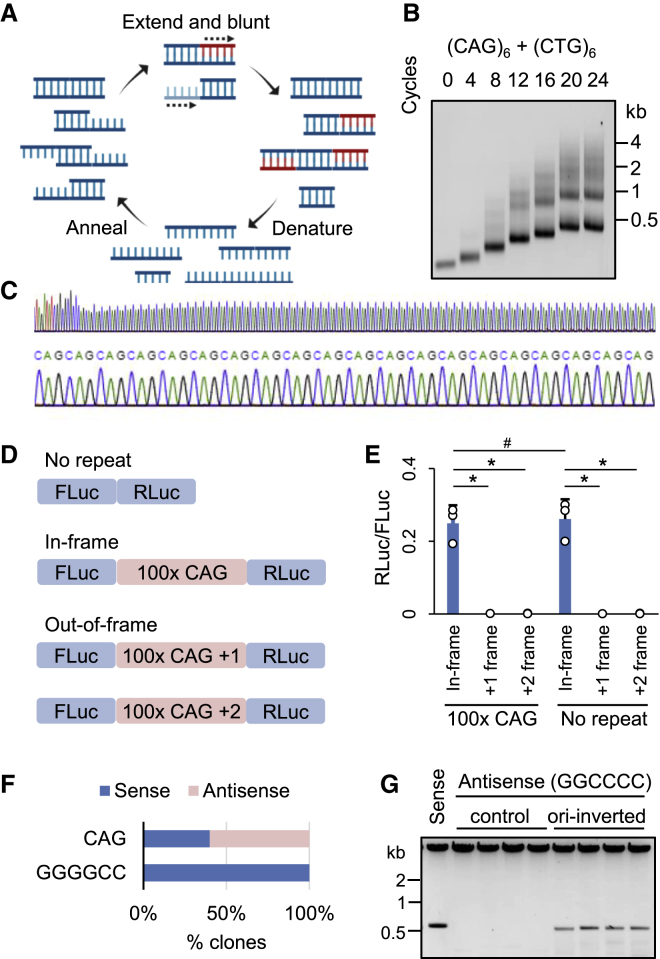
To investigate the sequence determinants of repeat RNA aggregation, we needed an efficient and generalizable method to synthesize long tandem repeats of any desired sequence. While a previously reported method based on type IIS restriction endonucleases allows sequential elongation of any repeat (Scior et al., 2011) (hereafter referred to as the sequential method), it requires multiple rounds of cloning to create long repeats in the pathological range. We took inspiration from the seminal work by Khorana and colleagues on generating di- and trinucleotide repeat DNA polymers by using E. coli DNA polymerase I (Khorana et al., 1965), and improved upon a previously described (Ordway and Detloff, 1996) PCR-based method to generate long DNA repeats from short DNA oligos, which we termed repeat-extension PCR (RepEx-PCR) (Figure 1A). RepEx-PCR is initiated by annealing two complimentary single-stranded DNA oligos (typically 16–20 nt and 5ʹ phosphorylated) containing two or more repeats of the desired sequence. While most oligos would form perfect duplexes, some would anneal in an offset manner, yielding either 5ʹ or 3ʹ overhangs. Next, Taq DNA polymerases fill in the 5ʹ overhangs with additional repeat units before all duplexes are denatured for the next cycle of annealing and extension. In addition to filling 5ʹ overhangs, Taq polymerases with 3ʹ-to-5ʹ proofreading exonuclease activity remove 3ʹ overhangs, thereby blunting both ends of the double-stranded repeat DNA products (Figure 1A).
Figure 1.
Generation of long tandem repeats by RepEx-PCR
(A) Schematic illustration of RepEx-PCR.
(B) Length distribution of RepEx-PCR products.
(C) Sanger sequencing results of a ∼100x CAG construct, showing the junction between 5ʹ flanking region and repeat insert (top) and a magnified view of CAG repeats (bottom).
(D) Schematic illustration of a dual-luciferase reporter for detecting frameshifted CAG repeats.
(E) Relative RLuc/FLuc activities of out-of-frame 0x or 100x CAG constructs. Data are shown as mean ± SD, #p > 0.1; ∗p < 0.05, two-tailed Student’s t test.
(F) Percentage of clones with CAG or GGGGCC repeats inserted in the sense and antisense direction.
(G) Restriction enzyme digestion of clones containing antisense (GGCCCC) repeats inserted in the original or ori-inverted vector.
As expected, increasing numbers of RepEx-PCR cycles generated longer repeat DNAs spanning a wide range in length. For example, RepEx-PCR starting from two oligos containing six units (6x) of CAG and CTG repeats, respectively, generated CAG:CTG repeat DNA products ranging from tens to thousands of copies (Figure 1B), well into the pathological range associated with Huntington disease and DM1. Size-selected, 5ʹ phosphorylated repeat DNA products were ligated to linearized vectors (typically non-phosphorylated PCR products), cloned and amplified in E. coli, and verified by sequencing (Figure 1C). In principle, RepEx-PCR products should consist only of full repeats. To test whether incomplete repeats caused by small indels may arise during either RepEx-PCR or subsequent cloning, we generated a series of dual-luciferase reporter constructs containing CAG repeats (Figure 1D). In these reporters, ∼100x CAG repeats were in-frame with the upstream firefly luciferase (FLuc) coding sequencing, while a downstream Renilla luciferase (RLuc) coding sequencing was in either 0- (in-frame), +1-, or +2-frame (out-of-frame) relative to FLuc. If RepEx-PCR or subsequent cloning generated incomplete repeats, we would observe decreased RLuc activity from the in-frame reporter and increased RLuc activity from +1 or +2-frame reporters. After transfection in HEK293T cells, all four tested clones of out-of-frame reporters yielded background levels of RLuc activity, similar to the negative control reporters with no repeats inserted (Figure 1E), suggesting that most if not all RepEx-PCR products are pure tandem repeats with no incomplete repeat units.
Blunt-end ligation of RepEx-PCR products should in principle result in repeats being inserted in either the sense or antisense orientation. Indeed, we obtained similar numbers of clones containing either CAG or CTG repeats (Figure 1E). When we applied RepEx-PCR to generate other repeats, however, some repeats showed strong orientation bias. For example, RepEx-PCR for the hexanucleotide GGGGCC repeats only yielded clones in sense orientation but not in the antisense (GGCCCC) orientation (Figure 1F). Attempts to reverse the G-rich repeats by using restriction enzymes resulted in the contraction of repeats (Figure 1G), indicating that the observed orientation bias was largely due to repeat instability instead of ligation bias. Similar strand-specific repeat instability has been previously attributed to the position of repeats relative to the origin of replication (ori) (Kang et al., 1995). Indeed, inverting the ori prior to repeat insertion substantially stabilized the repeats in the antisense orientation (Figure 1G).
Sequence determinants of triplet repeat RNA aggregation
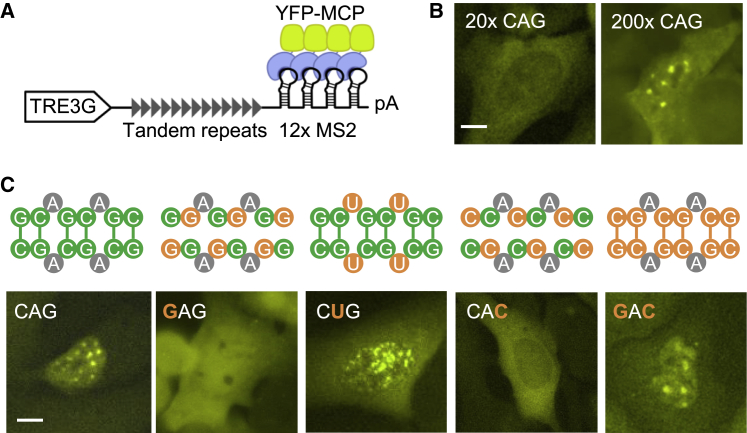
An MS2 stem-loop/MS2 coat protein (MCP)-based RNA imaging system has previously been used to monitor repeat RNA aggregation in living cells (Jain and Vale, 2017) (Figure 2A). After transfecting 12x MS2 stem-loop-tagged repeat RNA expression constructs in U2OS cells that stably express MCP-YFP and reverse tetracycline-controlled trans-activator (rtTA), we induced repeat RNA expression by adding doxycycline and imaged the formation of RNA foci. By comparing 20x and ∼200x CAG repeats, we first confirmed the previous observation that repeat RNA aggregation was strongly dependent on repeat length (Jain and Vale, 2017) (Figure 2B). To determine the sequence features that promote CAG repeat RNA foci formation, we applied RepEx-PCR to generate variants of CAG repeats with similar length (∼200x) (Figure S1), each containing substitutions of one of the three nucleotides. Substituting C1 with G (GAG repeats) or G3 with C (CAC repeats) abolished the formation of RNA foci, whereas A2U substitution (CUG repeats) had little effect (Figure 2C). Similar to a previous study on endogenous CUG repeat RNA foci in DM1 myoblasts (Dansithong et al., 2005), knocking down MBNL1 and, to a lesser extent, MBNL2, reduced CUG repeat foci in U2OS cells (Figure S2). To further assess the role of base-pairing, we generated the compensatory double mutant C1G/G3C (GAC repeats), in which consecutive C:G pairs were restored. Indeed, GAC repeat RNAs readily formed foci in U2OS cells (Figure 2C). Together, our results were consistent with the previously proposed model (Jain and Vale, 2017), in which repeat RNAs form multivalent interactions via base-pairing.
Figure 2.
Sequence determinants of CAG repeat RNA aggregation
(A) Schematic illustration of the 12x MS2-tagged doxycycline-inducible repeat RNA imaging construct.
(B) Representative images of cells expressing 20x (left) and ∼200x (right) CAG repeat RNAs. Scale bar, 10 μm.
(C) Potential base-pairing patterns (top) and representative images (bottom) of CAG sequence variants. Substituted nucleotides are highlighted in orange. See Figure 3C for quantification. Scale bar, 10 μm.
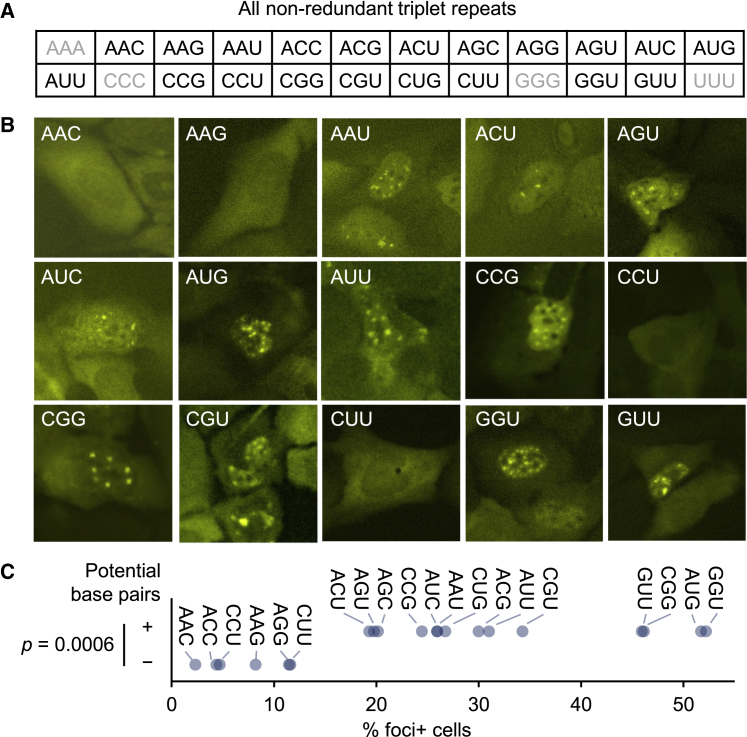
To more systematically determine the sequence characteristics that promote repeat RNA aggregation, we sought to generate all possible triplet repeat sequences. Except for the four homopolymers (polyA, polyC, polyG, and polyT), we generated all of the remaining 20 non-redundant triplet repeats (Figure 3A). Consistent with the analysis on CAG variants, we restricted the length at ∼600 base pairs or ∼200x. Because the percentage of foci-positive cells was highly correlated with the average RNA foci area per cell (Figure S3), we used the former as our primary measurement.
Figure 3.
Expanded analysis of trinucleotide repeat RNA aggregation
(A) List of all of 20 non-redundant triplet repeats. Homopolymers are indicated in gray.
(B) Representative images of cells expressing each of 15 triplet repeats not included in Figure 2. Scale bar, 10 μm.
(C) Relationship between predicted base pairs and foci forming ability. p value, two-tailed Mann-Whitney U test.
Similar to CAG variants, the 20 tested triplet repeat RNAs exhibited a wide range of aggregation propensity (Figure 3B), which was not explained by the differences between their expression levels (Figure S4). To further rule out the potential effect of differential expression levels between repeats, we measured repeat RNA expression levels and foci formation at each of a series of doxycycline concentrations, and then interpolated the percentage of foci-positive cells at a fixed expression level (50% of GADPH expression level measured by RT-qPCR), which yielded highly consistent results with those from experiments using a constant doxycycline concentration (Figure S5).
The 20 triplet repeat sequences allowed us to assess more broadly the role of base-pairing in repeat RNA aggregation. Consistent with our CAG variant analysis, binary categorization of triplet repeats based on their potential to form Watson−Crick or G:U wobble base pairs was highly predictive of foci formation (Figure 3C), accounting for more than half of the variance (r2 = 0.59). These results strongly supported a general role of RNA-RNA base-pairing in mediating repeat RNA aggregation.
Base-pairing properties in repeat RNA aggregation
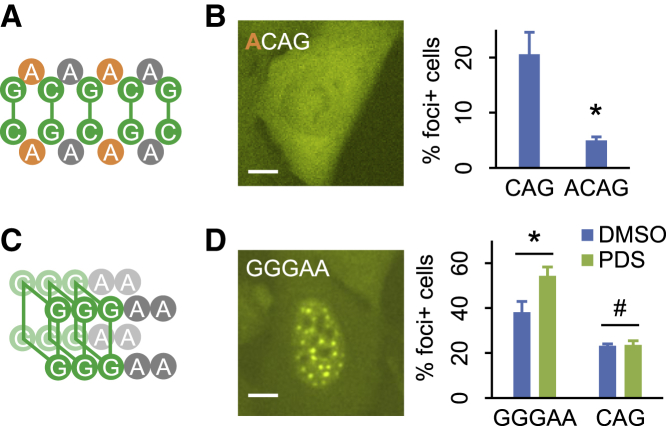
We next sought to further characterize the type and strength of base-pairing required for repeat RNA aggregation. For triplet repeats, base pairs were invariably in runs of two (Figure 2C). To test whether multiple runs of two consecutive base pairs were the minimal interactions, we generated ∼150x ACAG repeats by RepEx-PCR, which could potentially form non-consecutive C:G pairs (Figure 4A). In contrast to 200x CAG repeats, 150x ACAG repeat RNA did not form foci (Figure 4B), suggesting that runs of as few as two consecutive base pairs were the minimal requirement for RNA aggregation.
Figure 4.
Base-pairing properties in repeat RNA aggregation
(A) Potential base-pairing pattern of ACAG repeats. The added A nucleotides are highlighted in orange.
(B) Representative image (left) and quantification (right) of cells expressing 150x ACAG repeat RNA. Scale bar, 10 μm. Quantification data are shown as mean ± SD, ∗p < 0.05, two-tailed Student’s t test.
(C) Predicted G-quadruplex structure of GGGAA repeats.
(D) Representative image (left) and quantification (right) of cells expressing 120x GGGAA or 200x CAG repeat RNAs, treated with DMSO or PDS. Scale bar, 10 μm. Quantification data are shown as mean ± SD, #p > 0.1; ∗p < 0.05, two-tailed Student’s t test.
Previous studies have proposed that repeat RNAs containing multiple consecutive G nucleotides may aggregate via the formation of G-quadruplex (Fay et al., 2017; Jain and Vale, 2017), a four-stranded RNA structure involving noncanonical G:G base pairs. Of the three tested triplet repeats containing GG dinucleotides, we observed RNA foci formation for CGG and UGG, but not AGG (Figures 3B and 3C). However, CGG and UGG foci could also be due to C:G and G:U base pairs, respectively, whereas the scarcity of AGG foci might be due to RBPs and helicases that normally prevented RNA G-quadruplex formation in cells (Guo and Bartel, 2016). To test whether repeat RNA aggregation could be mediated by stronger G-quadruplex interactions, we generated 120x repeats of AAGGG, a well-established G-quadruplex motif with no potential to form canonical base pairs (Guo and Bartel, 2016) (Figure 4C). Not only 120x AAGGG repeat RNAs strongly aggregated (Figure 4D), treating cells with pyridostatin (PDS), a G-quadruplex stabilizer (Biffi et al., 2014), further increased AAGGG but not CAG foci (Figure 4D). These results suggested that aside from runs of two (or more) consecutive canonical base pairs, multiple runs of three (or more) consecutive G nucleotides could also mediate repeat RNA aggregation through noncanonical base pairs and G-quadruplex assembly.
Aggregation of ALS/FTD-associated GGGGCC repeat RNAs
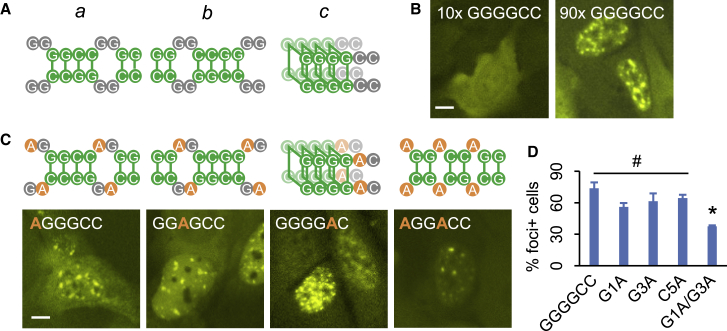
Expansion of hexanucleotide GGGGCC repeats within the first intron of C9orf72 gene is the most common genetic cause of ALS and FTD (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Similar to other pathogenic repeats, GGGGCC repeat as well as its antisense GGCCCC repeat RNAs are both known to form foci in patient tissues (DeJesus-Hernandez et al., 2011; McEachin et al., 2020; Zu et al., 2013). In keeping with the notion that runs of consecutive base pairs mediate RNA aggregation, GGGGCC repeats could form runs of four consecutive base pairs in multiple configurations (Figure 5A), two of which involving canonical C:G pairs between C5/C6 and either G3/G4 (a) or G1/G2 (b). GGGGCC repeats could also form four-layer G-quadruplexes (c), as have been previously shown in vitro (Fay et al., 2017; Fratta et al., 2012; Jain and Vale, 2017; Reddy et al., 2013), as well as a variety of configurations with shorter runs of base pairs (e.g., G2/G3 pairing to C5/C6 (Wang et al., 2019a; Wang et al., 2019b)). Consistent with previous studies, we observed strong aggregation propensity of GGGGCC repeat RNAs in a repeat length-dependent manner (Figure 5B), with 60% to 80% transfected cells showing clear RNA foci upon induction. To test whether the strong base-pairing potential of GGGGCC repeat RNA may promote foci formation, we generated three mutant sequences, each containing substitutions that disrupted two of the three possible “run-of-four” configurations (Figure 5C). Specifically, AGGGCC (G1A) repeats could still form configuration a, as well as a three-layer G-quadruplex (not shown), whereas GGAGCC (G3A) and GGGGAC (C5A) repeats could form configurations b and c, respectively. Indeed, with the remaining ability to form runs of four consecutive base pairs, each of the three mutants still strongly aggregated (Figures 5C and 5D). In contrast, foci formation of a G1A/G3A double mutant, which could only form runs of two consecutive C:G pairs (Figure 5C), was significantly decreased to a level comparable to other GC-rich triplet repeats (Figure 5D). These results suggested that longer runs of consecutive base pairs enhanced GGGGCC repeat RNA aggregation, further supporting the role of base-pairing in promoting repeat RNA foci formation.
Figure 5.
Aggregation of ALS/FTD-associated GGGGCC repeat RNAs
(A) Potential base-pairing patterns of GGGGCC repeats.
(B) Representative images of cells expressing 10x (left) or 90x (right) GGGGCC repeat RNAs. Scale bar, 10 μm.
(C) Potential base-pairing patterns (top) and representative images (bottom) of cells expressing each GGGGCC variant. Substituted nucleotides are highlighted in orange. Scale bar, 10 μm.
(D) Quantification of foci + cells expressing each GGGGCC variant. Quantification data are shown as mean ± SD, #p > 0.1; ∗p < 0.05, two-tailed Student’s t test.
Discussion
Our PCR-based repeat extension provides an efficient and generalizable approach to generate tandem repeats of any desired sequence spanning a wide range of lengths. Compared with a previous type IIS restriction endonuclease-based method (the sequential method) (Scior et al., 2011), RepEx-PCR has two clear advantages: First, the sequential method relies on a pair of type IIS restriction sites flanking the repeats, which may interfere with certain applications. In contrast, blunt-end ligation of RepEx-PCR products does not require any specific flanking sequence. Second, the sequential method starts with a relatively short synthetic repeat and doubles the repeat length after each round of cloning. Therefore, multiple rounds of cloning are required to obtain more than a few dozen repeats. On the contrary, RepEx-PCR can generate long repeats in a single round of cloning, with the minor trade-off being that multiple bacterial colonies need to be screened in order to identify those with the desired length and orientation. Nonetheless, the sequential method, due to its PCR-free nature, provides a complementary strategy that may be more suited for assembling multiple repeat sequences in the same construct.
RepEx-PCR enabled us to generate a large variety of repeat RNAs and delineate the sequence features that promote RNA foci formation. First, mutation analysis of CAG repeats showed that disrupting the predicted C1:G3 base pairs abolished RNA aggregation (Figure 2), whereas the compensatory double mutations that restored base-pairing also restored RNA aggregation. These observations were corroborated by our expanded analysis that surveyed all 20 non-redundant, non-homopolymeric triplet repeats (Figure 3), in which binary categorization of base-pairing potential could effectively predict foci formation, accounting for more than half of the variance. Finally, to test whether runs of consecutive base pairs were required for RNA aggregation, we interrupted the GC and GGCC/CCGG motifs in CAG (Figure 4) and GGGGCC repeats (Figure 5), respectively, both causing significant reductions in RNA foci. Collectively, these results supported and expanded the previously proposed model in which RNA-RNA base pairs mediate repeat RNA aggregation (Jain and Vale, 2017; Nguyen et al., 2022).
Our analysis revealed important contributions of noncanonical RNA-RNA interactions in repeat RNA aggregation. G-rich RNAs have long been known to form highly stable G-quadruplex structure, in which each of four adjacent runs of consecutive G nucleotides (G-runs) pair to two other G-runs on both Watson-Crick and Hoogsteen faces. While many endogenous RNAs contain regions that can form G-quadruplexes in vitro, these regions are predominantly unfolded in eukaryotic cells by RNA helicases and RBPs (Guo and Bartel, 2016), presumably to prevent their potential negative impact on mRNA translation and degradation. In contrast to physiological mRNAs, some repeat RNAs such as those in C9orf72-associated ALS/FTD and SCA36 (UGGGCC repeats) contain hundreds or thousands of adjacent G-runs and therefore have extraordinarily high propensity to form intra- or intermolecular G-quadruplexes (Conlon et al., 2016; Fratta et al., 2012; Reddy et al., 2013). Indeed, our analyses of GGGAA and GGGGCC repeats provided evidence that expanded repeat RNAs with runs of three or more G nucleotides could readily aggregate via G-quadruplexes, suggesting that unlike those in physiological RNAs, G-quadruplexes within repeat RNA aggregates might become less accessible to the cellular remodeling machinery. These results also raised the intriguing possibility that G-rich repeat RNA foci might further sequester G-quadruplex-remodeling factors and thereby stabilize otherwise transient G-quadruplexes formed within physiological RNAs (Yang et al., 2018).
On one hand, our model that multiple runs of two or more consecutive base pairs were sufficient to promote RNA aggregation explains why RNAs containing expanded repeats, with their high density of base-pairing motifs, were particularly prone to aggregation. On the other hand, this model also suggests that RNA aggregates may not consist purely of RNAs containing expanded repeats but also of other endogenous RNAs with the potential to base-pair extensively to repeat RNAs, such as pre-mRNAs with similar albeit non-expanded repeat sequences, potentially causing the misprocessing or retention of these secondarily recruited RNAs. Considering that previous studies have largely focused on the RBPs sequestered within repeat RNA foci, we suggest that investigating their secondary RNA components may shed new light on the pathophysiological significance of repeat RNA aggregates.
While the base-pairing potential explained more than half of the variance in RNA foci formation between repeat sequences, some sequence features associated with foci formation were not readily explained by base-pairing. For example, while G:U wobble pairs are in general weaker than C:G pairs, GGU and GUU repeats were more prone to aggregation than GGC and GCC repeats, respectively (Figure 3C), suggesting that G/U-rich sequences may promote RNA aggregation in ways additional to base-pairing, possibly due to their interactions with certain RBPs. Consistent with a previous study in DM1 myoblasts (Dansithong et al., 2005), MBNL1/2 knock down also reduced but did not completely abolish CUG repeat RNA foci in our system, suggesting that RNA-RNA and RNA-RBP interactions may co-exist and both contribute to aggregate formation. At high RNA abundance (e.g., ectopically expressed repeats), intermolecular RNA-RNA interactions may be favored, whereas at low RNA abundance (e.g., endogenous C9orf72 intronic repeats), interactions with abundant RBPs may initiate aggregation and promote subsequent RBP-RNA and RNA-RNA interactions (Van Treeck and Parker, 2018). While RBP-RNA interactions may interfere with base-pairing locally, by bridging multiple RNAs they may cause more base pairs to form elsewhere. Determining the phase diagrams of RNA foci formation, in which RNA and RBP abundance can be independently titrated, will be useful to assess the relative roles of RNA-RNA versus RNA-RBP interactions at different concentrations.
Our systematic approach of identifying sequence and structural features in repeat RNAs is not limited to the studies of RNA foci, but generalizable for studying other properties of repeat sequences, such as their impact on pre-mRNA processing (Sznajder et al., 2018), non-AUG translation (Zu et al., 2011), antisense RNA expression (Zu et al., 2013), cytoplasmic stress granules (Fay et al., 2017), and cell viability (Sun et al., 2020). Therefore, RepEx-PCR represents a valuable addition to the repertoire of methods enabling deeper investigations into the physiology and pathophysiology of tandem repeats and repeat expansion disorders.
Limitations of the study
While RepEx-PCR can generate tandem repeat DNAs tens of kilobases in length, blunt-end ligation is much less efficient for these longer inserts. Furthermore, longer repeats are substantially less stable during E. coli culture and more likely to undergo partial or complete contraction. Future modifications of the cloning procedure including using alternative host organisms may enhance the cloning efficiency and/or stability of ultralong repeats to better recapitulate those observed in certain repeat expansion diseases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-MBNL1 | Developmental Studies Hybridoma Bank | Cat# MB1a(4A8); RRID: AB_2618248 |
| Mouse monoclonal anti-MBNL2 | Developmental Studies Hybridoma Bank | Cat# MB2A(3B4); RRID: AB_2618250 |
| Mouse monoclonal anti-GAPDH | Proteus Biosciences Inc | Cat# 40-1246; Clone 1D4 |
| Bacterial and virus strains | ||
| NEB Stable Competent E. coli | New England Biolabs | Cat# C3040H |
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin sodium salt | Sigma-Aldrich | Cat# A9518 |
| Doxycycline Hydrochloride, Ready Made Solution | Sigma-Aldrich | Cat# D3072 |
| Puromycin Dihydrochloride | Gibco | Cat# A1113803 |
| Pyridostatin hydrochloride | Sigma-Aldrich | Cat# SML2690 |
| AscI | New England Biolabs | Cat# R0558S |
| NotI-HF | New England Biolabs | Cat# R3189S |
| Critical commercial assays | ||
| Q5 Hot Start High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0493S |
| NEBuilder HiFi DNA Assembly | New England Biolabs | Cat# E5520 |
| KAPA HiFi PCR Kit | Roche | Cat# KK2103; 07,958,854,001 |
| Monarch DNA Gel Extraction Kit | New England Biolabs | Cat# T1020S |
| Blunt/TA Ligase Master Mix | New England Biolabs | M0367S |
| Instant Sticky-end Ligase Master Mix | New England Biolabs | Cat# M0370S |
| QIAprep Spin Miniprep Kit | Qiagen | Cat# 27106 |
| Dual-Luciferase Reporter Assay System | Promega | Cat# E1910 |
| Monarch Total RNA Miniprep Kit | New England Biolabs | Cat# T2010S |
| Luna Universal One-Step RT-qPCR Kit | New England Biolabs | Cat# E3005S |
| TURBO DNA-free™ Kit | Invitrogen | Cat# AM1907 |
| Experimental models: Cell lines | ||
| Human: U2OS cells | Jain and Vale (2017) | N/A |
| Human: HEK293T cells | ATCC | ATCC CRL-3216 |
| Oligonucleotides | ||
| Primer: pTRE-MS2x12 RT-qPCR_Fwd: AGATCTGCGCGCGATCG | Integrated DNA Technologies | N/A |
| Primer: pTRE-MS2x12 RT-qPCR_Rev: AGCCAGAAGTCAGATGCTCAAG | Integrated DNA Technologies | N/A |
| Primers used to generate lentiCRISPRv2_sgRNAs | This paper | Table S2 |
| Primers used to generate repeat sequences | This paper | Table S3 |
| Recombinant DNA | ||
| pmirGLO Dual-Luciferase miRNA Target Expression Vector | Promega | Cat# E1330 |
| Tet-On 3G Inducible Expression System | TaKaRa-Clontech | Cat# 631168 |
| pTRE3G_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_CAGx20_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_CAGx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AACx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AAGx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AAUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_ACCx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_ACGx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_ACUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AGGx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AGUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AUCx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AUGx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AUUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_CCGx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_CCUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_CGGx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_CGUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_CUGx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_CUUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_GGUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_GUUx200_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_ACAGx150_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_GGGAAx120_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_GGGGCCx10_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_GGGGCCx90_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AGGGCCx90_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_GGAGCCx90_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_GGGGACx90_MS2x12 | This paper | Subcloned from Tet-On 3G |
| pTRE3G_AGGACCx90_MS2x12 | This paper | Subcloned from Tet-On 3G |
| lentiCRISPRv2 | PMCID: PMC4486245 | Addgene Plasmid #52961 |
| pMDLg/pRRE | PMCID: PMC110254 | Addgene Plasmid #12251 |
| pRSV-Rev | PMCID: PMC110254 | Addgene Plasmid #12253 |
| VSV.G | PMID: 12717450 | Addgene Plasmid #14888 |
| LentiCRISPRv2_sgRNA1_MBNL1 | This paper | Subcloned from lentiCRISPRv2 |
| LentiCRISPRv2_sgRNA2_MBNL1 | This paper | Subcloned from lentiCRISPRv2 |
| LentiCRISPRv2_sgRNA1_MBNL2 | This paper | Subcloned from lentiCRISPRv2 |
| LentiCRISPRv2_sgRNA2_MBNL2 | This paper | Subcloned from lentiCRISPRv2 |
| LentiCRISPRv2_sgRNA1_NT | This paper | Subcloned from lentiCRISPRv2 |
| LentiCRISPRv2_sgRNA2_NT | This paper | Subcloned from lentiCRISPRv2 |
| Software and algorithms | ||
| ImageJ | PMCID: PMC5554542 | https://imagej.nih.gov/ij/ |
| Other | ||
| 24 Well glass bottom plates | Cellvis | P24-1.5H-N |
| DMEM, high glucose | Gibco | Cat# 11965084 |
| Fetal Bovine Serum, qualified, heat inactivated, USDA-approved regions | Gibco | Cat# 10438034 |
| Penicillin-streptomycin | Gibco | Cat# 15140122 |
| LB Broth | Gibco | Cat# 10855001 |
| GloMax 20/20 Luminometer | Promega | Cat# E5311 |
| Lionheart FX Automated Microscope | Agilent | N/A |
| Lenti-X Concentrator | TaKaRa-Clontech | Cat# 631232 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Junjie Guo (junjie.guo@yale.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
U2OS and HEK293T cell lines
The U2OS cell line stably expressing rtTA and MCP-YFP was a gift from A. Jain and R. Vale. Both U2OS and HEK293T (ATCC CRL-3216) cell lines were maintained in DMEM high-glucose medium (Gibco) with 10% (v/v) heat inactivated fetal bovine serum (Gibco) and penicillin–streptomycin (100 U/mL, Gibco) at 37°C with 5% CO2.
Method details
Generation of tandem repeats by RepEx-PCR
RepEx-PCR was performed by using 0.5 μM each of the two 5ʹ phosphorylated single-stranded DNA oligos (Integrated DNA Technologies) containing 5-7x sense and antisense repeats, respectively. For most triplet repeats, Q5 hot-start high-fidelity DNA polymerase (New England Biolabs) was used. For GC-rich repeats such as GGGGCC, KAPA HiFi DNA polymerase (Roche) was used with GC buffer and/or GC enhancer. RepEx-PCR products were size-selected and purified from a 0.8% agarose gel. Linear vectors were typically generated by PCR using non-phosphorylated primers. Vectors linearized by restriction enzymes would need to be blunted and dephosphorylated to prevent self-ligation. After blunt-end ligation, 1 μL ligation product was transformed in NEB Stable competent cells (New England Biolabs). After overnight incubation, individual bacterial colonies were amplified in liquid cultures before plasmid DNA was extracted. Longer repeats tend to be more unstable and therefore have lower positive rates. For ∼600 bp repeats, typically 4–16 colonies were screened for the correct repeat insert size and orientation, which were determined by restriction enzyme digestion and DNA sequencing, respectively. After sequencing validation, the repeat insert was further subcloned to a doxycycline-inducible (Tet-On, Clontech) expression vector containing 12x MS2 hairpins.
Luciferase assays
RepEx-PCR-generated ∼100x CAG repeats were inserted into a dual-luciferase reporter plasmid modified from pmirGLO (Promega) between firefly luciferase (FLuc) coding sequence in the 0 frame and Renilla luciferase (RLuc) coding sequence in the 0, +1, or +2 frame. Reporter plasmid DNAs as well as control reporter plasmid DNAs with no repeats inserted were transfected in HEK293T cells in 24-well plates using Lipofectamine 2000 (Invitrogen). 16-24 h after transfection, cells were lysed in Glo Lysis Buffer (Promega) at room temperature for 5 min. FLuc and RLuc activities in lysates were sequentially measured by using Dual-Glo luciferase assay reagents and a GloMax 20/20 luminometer (Promega) according to the manufacturer’s instructions.
Live-cell imaging of repeat RNA foci
U2OS cells plated in glass-bottom 24-well plates (Cellvis) were transfected with 320 ng 12x MS2-tagged repeat expression construct and 80 ng CMV-BFP plasmid DNA (for labeling transfected cells) mixed with 1.6 μL FuGENE HD (Promega) according to the manufacturer’s instructions. 4-6 h after transfection, the medium was replaced with fresh DMEM supplemented with 10% FBS and 1 μg/mL doxycycline (Sigma). 16-24 h after induction, cells were imaged at 37°C by using a Lionheart FX automated microscope (Agilent) with 20x and 60× objectives. Typically, 50-100 BFP-positive cells were imaged in each well for RNA foci quantification.
RT-qPCR
After live-cell imaging, total RNA from U2OS cells were extracted by using Monarch Total RNA Miniprep kit (New England Biolabs) and treated with TURBO DNase (Invitrogen). RT-qPCR was performed by using Luna Universal One-Step RT-qPCR reagents (New England Biolabs), following the manufacturer’s instructions. Primers targeting the MS2 stem-loop regions were used for quantifying repeat RNA expression levels, which were normalized by GAPDH expression levels.
Lentivirus preparation and transduction
Single guide RNA (sgRNA) sequences (Table S1) were cloned into lentiCRISPRv2 (Addgene, #52961) by using NEBuilder HiFi DNA assembly reagents (New England Biolabs). For lentivirus production, 60-70% confluent HEK293T cells were transfected with sgRNA containing lentiCRISPRv2, pMDLg/pRRE (Addgene, #12251), pRSV-Rev (Addgene, #12253) and VSV.G (Addgene, #14888) plasmids by using Lipofectamine 2000 (Invitrogen). 72 h after transfection, media was collected and centrifuged at 3,000 rpm for 10 min at 4°C to pellet the cell debris. The supernatant was filtered through a 0.45 μm low-protein-binding membrane. Viral particles were further concentrated by adding Lenti-XTM concentrator (Clontech) to the supernatant at a 1:3 ratio. The mixture was incubated for 2 h at 4°C and centrifuged at 1,500 g for 45 min at 4°C. After removing the supernatant, the pellet containing lentiviruses was resuspended in 1 mL DMEM, aliquoted, and stored at −80°C.
For lentivirus transduction, 2 × 105 U2OS cells were seeded in each well of 24-well plates. One aliquot of viral particles was added to each well and mixed. After 48 h, fresh media containing 1 μg/mL puromycin were added for selection. After two rounds of 48-h selection, puromycin-resistant cells were harvested and expanded for downstream analyses.
Quantification and statistical analysis
For comparisons of RNA aggregates between two repeat sequences, each with multiple biological replicates, two-tailed Student’s t tests were used unless indicated otherwise. For the comparison of RNA aggregates between two groups of repeat sequences (Figure 3C), two-tailed Mann−Whitney U test was used.
Acknowledgments
We thank D. Bartel for the suggestion of using PCR for repeat synthesis, A. Jain and R. Vale for providing the U2OS cell line stably expressing rtTA and MCP-YFP, and members of the Guo lab for helpful discussions. This work is supported by an NIH New Innovator Award (DP2 GM132930) and the Muscular Dystrophy Association (MDA602934). J.U.G. is a Klingenstein-Simons Fellow in Neuroscience and a New York Stem Cell Foundation-Robertson Investigator.
Author contributions
Conceptualization, A.U.I. and J.U.G.; methodology, A.U.I., A.E., and J.U.G.; investigation, A.U.I., A.E., S.Y., and J.U.G.; writing – original draft, J.U.G.; writing – review & editing, A.U.I., A.E., S.Y., and J.U.G.; funding acquisition and supervision, J.U.G.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 9, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100334.
Supplemental information
Data and code availability
-
•
Any data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Biffi G., Di Antonio M., Tannahill D., Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014;6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife. 2016;5:e17820. doi: 10.7554/eLife.17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansithong W., Paul S., Comai L., Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot M.C., Ferguson C.M., Ly S., Coles A.H., Smith A.O., Bicknell A.A., Hall L.M., Sapp E., Echeverria D., Pai A.A., et al. Nuclear localization of huntingtin mRNA is specific to cells of neuronal origin. Cell Rep. 2018;24:2553–2560.e5. doi: 10.1016/j.celrep.2018.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay M.M., Anderson P.J., Ivanov P. ALS/FTD-Associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 2017;21:3573–3584. doi: 10.1016/j.celrep.2017.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M., Parkinson G., Isaacs A.M. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U., Bartel D.P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016;353:aaf5371. doi: 10.1126/science.aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Vale R.D. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Jaworski A., Ohshima K., Wells R.D. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorana H.G., Jacob T.M., Moon M.W., Narang S.A., Ohtsuka E. Studies on Polynucleotides.Xlii. The synthesis of deoxyribopolynucleotides containing repeating nucleotide sequences. Introduction and general considerations. J. Am. Chem. Soc. 1965;87:2954–2956. doi: 10.1021/ja01091a027. [DOI] [PubMed] [Google Scholar]
- McEachin Z.T., Parameswaran J., Raj N., Bassell G.J., Jiang J. RNA-mediated toxicity in C9orf72 ALS and FTD. Neurobiol. Dis. 2020;145:105055. doi: 10.1016/j.nbd.2020.105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.T., Hori N., Thirumalai D. Condensates in RNA repeat sequences are heterogeneously organized and exhibit reptation dynamics. Nat. Chem. 2022;14:775–785. doi: 10.1038/s41557-022-00934-z. [DOI] [PubMed] [Google Scholar]
- Ordway J.M., Detloff P.J. In vitro synthesis and cloning of long CAG repeats. Biotechniques. 1996;21:609–610. doi: 10.2144/96214bm08. [DOI] [PubMed] [Google Scholar]
- Ranum L.P., Cooper T.A. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Reddy K., Zamiri B., Stanley S.Y.R., Macgregor R.B., Jr., Pearson C.E. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez C.M., Todd P.K. New pathologic mechanisms in nucleotide repeat expansion disorders. Neurobiol. Dis. 2019;130:104515. doi: 10.1016/j.nbd.2019.104515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J.L., Jones K.L., Yeo G.W. Repeat RNA expansion disorders of the nervous system: post-transcriptional mechanisms and therapeutic strategies. Crit. Rev. Biochem. Mol. Biol. 2021;56:31–53. doi: 10.1080/10409238.2020.1841726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scior A., Preissler S., Koch M., Deuerling E. Directed PCR-free engineering of highly repetitive DNA sequences. BMC Biotechnol. 2011;11:87. doi: 10.1186/1472-6750-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Eshov A., Zhou J., Isiktas A.U., Guo J.U. C9orf72 arginine-rich dipeptide repeats inhibit UPF1-mediated RNA decay via translational repression. Nat. Commun. 2020;11:3354. doi: 10.1038/s41467-020-17129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznajder L.J., Thomas J.D., Carrell E.M., Reid T., McFarland K.N., Cleary J.D., Oliveira R., Nutter C.A., Bhatt K., Sobczak K., et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl. Acad. Sci. USA. 2018;115:4234–4239. doi: 10.1073/pnas.1716617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B., Parker R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell. 2018;174:791–802. doi: 10.1016/j.cell.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B., Protter D.S.W., Matheny T., Khong A., Link C.D., Parker R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. USA. 2018;115:2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.T., Cody N.A., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S., Schroth G.P., Housman D.E., Reddy S., et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Goodrich K.J., Conlon E.G., Gao J., Erbse A.H., Manley J.L., Cech T.R. C9orf72 and triplet repeat disorder RNAs: G-quadruplex formation, binding to PRC2 and implications for disease mechanisms. RNA. 2019;25:935–947. doi: 10.1261/rna.071191.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.F., Ursu A., Childs-Disney J.L., Guertler R., Yang W.Y., Bernat V., Rzuczek S.G., Fuerst R., Zhang Y.J., Gendron T.F., et al. The hairpin form of r(G4C2)(exp) in c9ALS/FTD is repeat-associated non-ATG translated and a target for bioactive small molecules. Cell Chem. Biol. 2019;26:179–190.e12. doi: 10.1016/j.chembiol.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowska M., Krzyzosiak W.J. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.Y., Lejault P., Chevrier S., Boidot R., Robertson A.G., Wong J.M.Y., Monchaud D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018;9:4730. doi: 10.1038/s41467-018-07224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A., et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T., Liu Y., Banez-Coronel M., Reid T., Pletnikova O., Lewis J., Miller T.M., Harms M.B., Falchook A.E., Subramony S.H., et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Any data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.