Summary
Electron microscopy is the gold standard to characterize centrosomal ultrastructure. However, production of significant morphometrical data is highly limited by acquisition time. We therefore developed a generalizable, semi-automated high-throughput electron tomography strategy to study centrosome aberrations in sparse patient-derived cancer cells at nanoscale. As proof of principle, we present electron tomography data on 455 centrioles of CD138pos plasma cells from one patient with relapsed/refractory multiple myeloma and CD138neg bone marrow mononuclear cells from three healthy donors as a control. Plasma cells from the myeloma patient displayed 122 over-elongated centrioles (48.8%). Particularly mother centrioles also harbored gross structural abnormalities, including fragmentation and disturbed microtubule cylinder formation, while control centrioles were phenotypically unremarkable. These data demonstrate the feasibility of our scalable high-throughput electron tomography strategy to study structural centrosome aberrations in primary tumor cells. Moreover, our electron tomography workflow and data provide a resource for the characterization of cell organelles beyond centrosomes.
Keywords: electron tomography, high-throughput electron microscopy, centriole over-elongation, centriole fragmentation, centrosome, plasma cell disorder, multiple myeloma, bone marrow
Graphical abstract
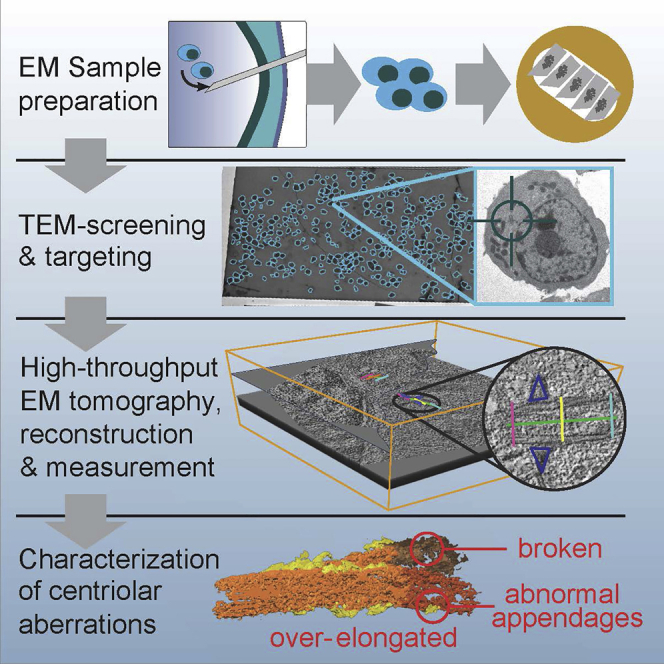
Highlights
-
•
High-throughput electron tomography reveals organelle structure in patient material
-
•
The workflow can be adapted to various cell types and organelles
-
•
We characterize 455 centrioles in human bone marrow cells at nanoscale
-
•
Myeloma cells contain over-elongated centrioles with gross structural aberrations
Motivation
Visualizing the organelle structure in patient-derived tissues at nanoscale can provide insights into numerous pathologies, but existing tools do not allow for analysis of sufficient numbers of cells at the required resolution. Here, by applying a semi-automated high-throughput electron tomography strategy, we quantify structural abnormalities of centrioles in human bone-marrow-derived cells at high numbers and share our data with the cell biological community to illustrate its utility as a tool for structural organelle analysis.
Dittrich et al. describe a semi-automated high-throughput electron tomography strategy to study organelle structure in patient-derived material at nanoscale. By applying their methodology to centrosomes, they show that plasma cells from a myeloma patient harbor over-elongated centrioles with gross structural abnormalities as the potential cause of chromosomal aberrations in multiple myeloma.
Introduction
Centrosomes are the major microtubule-organizing centers in mammalian cells and consist of a pair of centrioles embedded in pericentriolar material (Bettencourt-Dias and Glover, 2007; Cosenza et al., 2017; Ganem et al., 2009; Gönczy, 2015). Centrioles are microtubule-based cylinders with a physiological length of up to 500 nm and a diameter of approximately 200 nm (Bettencourt-Dias and Glover, 2007; Kong et al., 2020; Marteil et al., 2018; Sharma et al., 2021). They duplicate in S phase, with the formation of one daughter centriole next to each preexisting mother that subsequently elongates and matures until mitosis. Mature centrioles are decorated with two types of appendages, one set of nine distal appendages and a variable number of subdistal appendages, which can be located along the entire length of the centriole (Bettencourt-Dias and Glover, 2007; Hall and Hehnly, 2021). During mitosis, the two newly formed centrosomes of a cell migrate to opposite poles, contributing to bipolar spindle formation. To ensure accurate chromosome segregation, centriole number and structure are tightly controlled in non-transformed cells (Nigg and Holland, 2018).
Abnormalities in centrosome structure and number have been identified as drivers of genomic instability in various solid cancer entities and hematological malignancies (Chng et al., 2008; Cosenza et al., 2017; Gönczy, 2015; Krämer et al., 2003; Lopes et al., 2018; Maxwell et al., 2005). Centrosome aberrations have been detected in in situ carcinomas and low-grade tumors already and represent an early event in the evolution of malignant phenotypes in organotypic culture and animal models (Gönczy, 2015; Lopes et al., 2018). However, how centrosome aberrations develop remains unclear, with cancer-associated alterations in centrosomal genes being rare (Gönczy, 2015). Also, as most studies on centrosome aberrations in primary tumor tissues rely on single antigen immunostainings against pericentriolar matrix proteins at low resolution, virtually nothing is known about the detailed composition of centrosome aberrations in primary tumor cells at electron microscopy resolution.
The structural exploration of centrioles and other subcellular structures requires imaging techniques that go beyond the light microscopy diffraction limit of 200–300 nm. Significant advances have been made in super-resolution imaging methods over the last decade. Especially the recently introduced expansion microscopy allows for considerably improved resolution (Bowler et al., 2019; Chong et al., 2020; Gambarotto et al., 2019; Kong et al., 2020; Schermelleh et al., 2019; Vásquez-Limeta et al., 2022), and several centrosome-specific expansion microscopy protocols that allow for the analysis of centriole length (Kong et al., 2020) and structural features (Vásquez-Limeta et al., 2022), including appendages (Bowler et al., 2019), have been developed. These techniques are excellent for studying the function and structural contribution of individual proteins to the assembly of centrioles and centrosomes. Relying on the labeling of molecular components of the centriole, expansion microscopy can reveal protein complexes that are not necessarily visible by electron microscopy (Sahabandu et al., 2019). High-end deconvolution and computation of expansion microscopy images also allow for structural analysis of centrioles, although the resolution is still magnitudes apart from transmission electron microscopy images (Bowler et al., 2019; Gambarotto et al., 2019).
Plasma cell disorders comprise a range of increasingly malignant B cell neoplasias, spanning from premalignant monoclonal gammopathies of undetermined significance (MGUS) via asymptomatic smoldering myeloma to overt multiple myeloma and highly aggressive plasma cell leukemia (Manier et al., 2017; Maura et al., 2019). They arise from the malignant transformation of long-lived, terminally differentiated CD138pos bone marrow plasma cells (Halliley et al., 2015). In the absence of disease-defining mutational profiles, chromosome aberrations, including immunoglobulin heavy chain translocations and hyperdiploidy, are considered initiating events in plasma cell disorders, which are prevalent in early disease stages already (Manier et al., 2017; Maura et al., 2019).
Human plasma cells mainly reside within the bone marrow, which is only available by invasive bone marrow aspiration. Moreover, the physiological content of plasma cells in whole-bone-marrow aspirates is in the lower single-digit percentage range of mononuclear cells (Nombela-Arrieta and Manz, 2017). To understand the origin and evolution of centrosome aberrations in the spectrum of plasma cell disorders as a paradigm for malignant progression from a precursor lesion to aggressive malignancy, we aimed for the development of a method that allows for targeted imaging of centrioles as one specific subcellular structure at high resolution and high scale in sparce primary material.
Targeted imaging of specific subcellular morphology in human primary tissues by electron microscopy is challenging due to the absence of genetic or antigenic labels that could act as a guide to the feature(s) of interest. In 200-nm-thick sections of human plasma cells, the probability of including a centriole is well below 10% for any given cell, i.e., to find these features in statistically significant numbers, manual inspection of each cell is necessary. We enabled this screening by using computational detection of cell outlines in overview images and employing automated imaging of each cell at a magnification suitable for identifying centrioles by browsing through the generated image stack. This provides target coordinates that can be propagated to neighboring sections, enabling serial electron tomography of the target objects at high resolution (Schorb et al., 2019).
Results
A terabyte-scale electron tomography dataset of centrioles in primary human cells
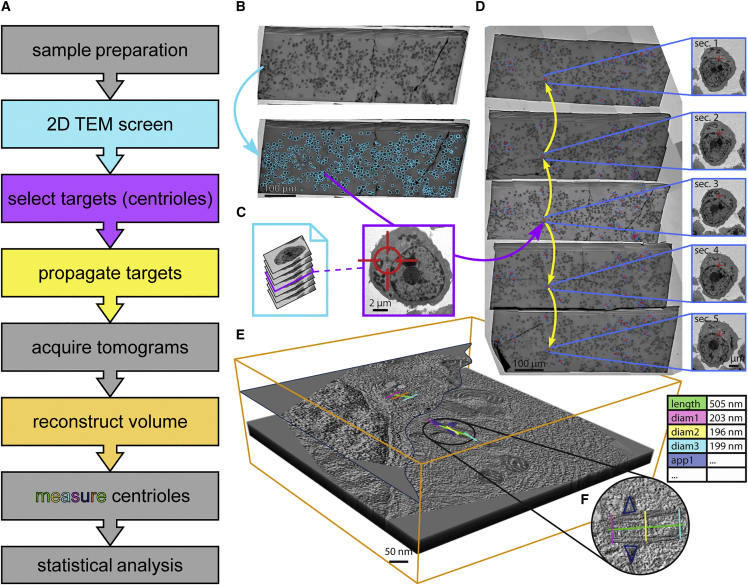
For ultrastructural analysis, we combined a transmission electron microscopy (TEM) screening procedure with targeted electron tomography (ET) of regions of interest (Figure 1). After magnetic-activated cell sorting (MACS)-based CD138 selection, preparation, and sectioning of both CD138pos and CD138neg fractions of bone marrow mononuclear cells for electron microscopy, a two-dimensional (2D) overview of the central section was obtained at 400× magnification (326.9 nm/px), and cells were semi-automatically detected and labeled for acquisition. Labeled cells were acquired at a magnification suitable for manual screening. For analysis of centrioles, we chose a 3,000× magnification with 42.75 nm/px, values that can be adapted according to the needs of other subcellular target features. The output was generated as an image stack that allowed for the identification of cells containing centrioles. Positions of selected cells containing target centrioles on the central section were then semi-automatically propagated, and the respective cells were re-identified on all adjacent sections to acquire the full length of the centrioles. Subsequently, single-axis ET (15,500× magnification, 1.55 nm/px; tilt range: −60° to 60°; increment: 1°) was performed at all targets on all sections. Next, corresponding tomograms from individual sections were reconstructed and joined to produce a final volume of at least 3.1 × 3.1 × 1 μm per target cell (X × Y × Z). Centrioles were measured and morphometrically analyzed within these volumes. Statistical analysis of centriole parameters was eventually performed using the exported measured parameters.
Figure 1.
Transmission electron microscopy and tomography workflow
(A) Overview of all relevant aspects of the workflow. Colors of the text boxes are matched with the respective procedure in (B)–(E). Steps not explicitly depicted are colored in gray.
(B) After sample preparation, a 2D overview of the central section is obtained at 400× magnification (326.9 nm/px), and cells are semi-automatically detected and labeled for acquisition (blue arrow).
(C) Labeled cells are acquired at a magnification suitable for manual screening (3,000×, 42.75 nm/px, in this study). Output is generated as an image stack (left) that allows for identification of cells containing subcellular target features (here: centrioles). An exemplary image of a plasma cell containing a centriole (red crosshair) at 3,000× magnification is depicted on the right.
(D) Selected cells (blue boxes) containing target centrioles (red crosses) on the central section are semi-automatically propagated and identified on all adjacent sections (yellow arrows). As an example, inlays on the right show the same cell containing target features (here: centrioles) on all five acquired neighboring sections. Subsequently, single-axis electron tomography (15,500× magnification, 1.55 nm/px; tilt range: −60° to 60°; increment: 1°) is performed at all targets on all sections.
(E) Corresponding tomograms from individual sections are reconstructed and joined to produce a final volume of at least 3.1 × 3.1 × 1 μm per target (X × Y × Z; orange bounding box).
(F) Target centrioles can be measured and morphometrically analyzed in these volumes. Statistical analysis of centriole parameters can eventually be performed using the exported measured parameters (exemplarily visualized as a table on the right). The volume shown in this panel can be interactively viewed in MoBIE as MMRR_06_Grid1_c442. For instructions, refer to the STAR Methods section of this paper. TEM, transmission electron microscopy.
Applying this methodology, a total of 6,504 cells were semi-automatically screened by TEM, and 455 completely pictured centrioles in 343 cells were examined, including 250 completely pictured centrioles in 221 bone-marrow-derived CD138pos plasma cells from a patient with relapsed/refractory multiple myeloma and 205 completely pictured centrioles in 138 CD138neg bone marrow mononuclear cells from three healthy donors. An online repository of the complete ET dataset (2.2 TB of raw data and 765 GB of tomography volume data) is publicly available for download and is also viewable in an interactive manner via MoBIE (Vergara et al., 2021) (see STAR Methods section for instructions).
Characterization of centriole parameters in CD138neg bone marrow mononuclear cells from healthy individuals
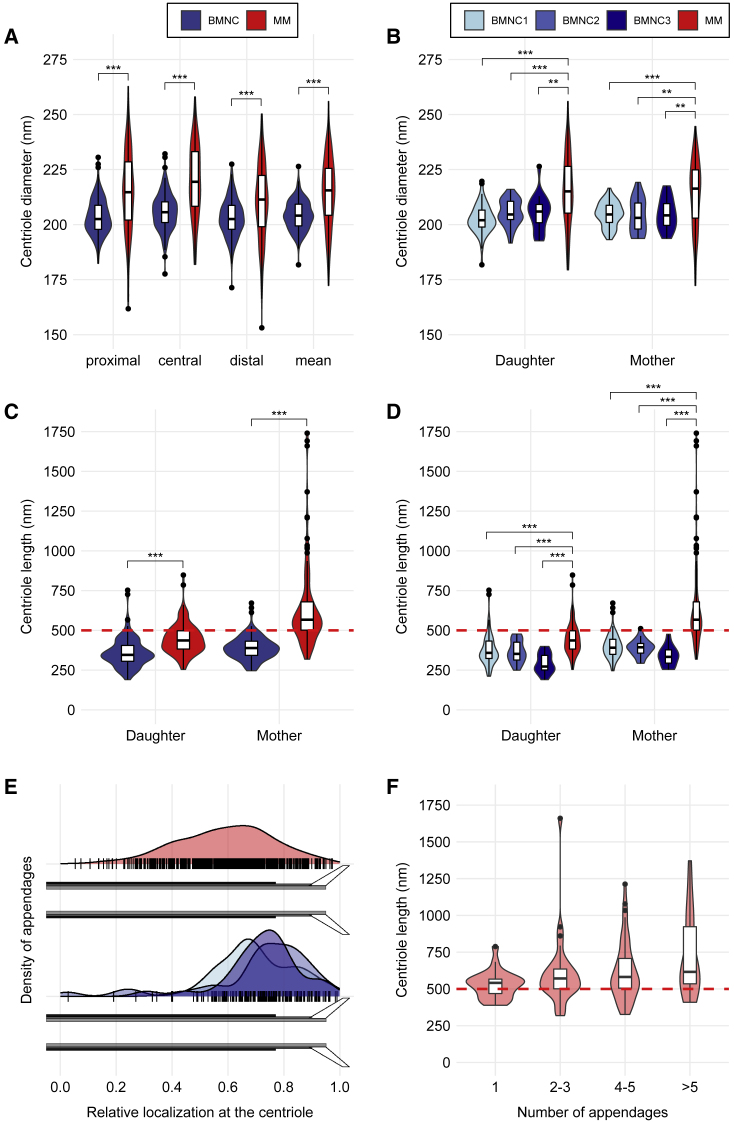
Both length and diameter of centrioles are tightly controlled and well conserved across evolution and usually do not exceed 500 and 250 nm in human cells, respectively (Bettencourt-Dias and Glover, 2007; Kohlmaier et al., 2009; Sharma et al., 2021). To determine length, diameter, and structural features of normal centrioles using the ET workflow described above, and to compare our results with those from previous analyses using conventional electron microscopy (Bettencourt-Dias and Glover, 2007; Chrétien et al., 1997; Kong et al., 2020; Marteil et al., 2018; Sharma et al., 2021), we first examined CD138neg bone marrow mononuclear cells from three healthy donors. In 138 cells, a total of 265 centrioles, among them 60 partially acquired and 205 completely pictured centrioles, were identified. Further analyses were restricted to complete centrioles. In line with the literature, the median length of centrioles (368 nm, range: 190–753 nm) was normal in these cells, whereas their median diameter (204 nm, range: 182–226 nm) was slightly reduced when compared with human cell lines (Chrétien et al., 1997; Sahabandu et al., 2019; Vásquez-Limeta et al., 2022) (Figures 2A–2D; Table 1). No structurally abnormal centrioles were found in CD138neg bone marrow mononuclear cells from healthy donors, and over-elongation was only detected in a very limited amount of these cells.
Figure 2.
Centriole characteristics
Completely acquired centrioles of CD138pos plasma cells from a patient with relapsed/refractory multiple myeloma (MM; 250 centrioles) compared with CD138neg bone marrow mononuclear cells (BMNCs) from three healthy donors (BMNC1, 125 centrioles, BMNC2, 47 centrioles, BMNC3, 33 centrioles). For BMNCs, measures of BMNC1–3 are pooled into one group. For a detailed description of how measures were performed, see the STAR Methods.
(A) Violin plots and integrated box plots showing the distribution of centriole diameters stratified by position of measurement. The diameter is measured at three different positions: proximal, central, and distal. Mean, the mean diameter of all three positions. For BMNCs, pooled data of BMNC1–3 are shown.
(B) Violin plots and integrated box plots showing the distribution of mean centriole diameters from proximal, central, and distal positions, stratified by mother and daughter centrioles. BMNC data from the three donors are displayed separately.
(C) Violin plots and integrated box plots showing the distribution of centriole lengths stratified by mother and daughter centrioles. The cutoff value for over-elongation of 500 nm is displayed as red dashed line. For BMNCs, pooled data of BMNC1–3 are shown.
(D) Violin plots and integrated box plots showing the distribution of centriole lengths stratified by mother and daughter centrioles. The cutoff value for over-elongation of 500 nm is displayed as red dashed line. BMNC data from the three donors are displayed separately.
(E) Distribution of relative appendage localizations along centrioles in MM (top) and the three BMNC donors (bottom). The relative localization ranging from 0 (proximal end) to 1 (distal end) is displayed on the x axis. Short black lines on the x axis correspond to individual relative localizations of appendages. Smoothed kernel density estimates of the relative appendage localizations are plotted on the y axis. For MM, 566 appendages, for BMNC1, 122 appendages, for BMNC2, 38 appendages, and for BMNC3, 31 appendages were examined.
(F) Violin plots and integrated box plots showing the distribution of the number of appendages in mother centrioles depending on centriole length in MM. The cutoff value for over-elongation of 500 nm is displayed as red dashed line. The number of appendages increases with centriole length (Kruskal-Wallis rank-sum test, p < 0.001). N (individual centrioles): “1” (23), “2–3” (31), “4–5” (45), “>5” (19). The Wilcoxon rank-sum test was used for pairwise comparisons in all panels. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figures S1 and S2 and Video S1.
Table 1.
Characteristics of centrioles
| N (including partially acquired) | MM |
CTRL1 |
CTRL2 |
CTRL3 |
CTRL1–3 |
|---|---|---|---|---|---|
| 409 | 160 | 60 | 45 | 265 | |
| Mother centrioles, N (%) | 223 (54.5) | 76 (47.5) | 24 (40) | 22 (48.9) | 122 (46) |
| Phenotypes of mother centrioles | |||||
| Broken, N (%) | 13 (5.8) | 0 | 0 | 0 | 0 |
| Incomplete, N (%) | 6 (2.7) | 0 | 0 | 0 | 0 |
| Asymmetric, N (%) | 18 (8.1) | 0 | 0 | 0 | 0 |
| Total abnormal, N (%) | 33 (14.8) | 0 | 0 | 0 | 0 |
| N (only completely acquired) | 250 | 125 | 47 | 33 | 205 |
| Appendages of mothers, N | 5 (1, 18) | 2 (1, 5) | 2 (1, 3) | 2 (2, 3) | 2 (1, 5) |
| Centriole diameter, mean (nm) | 215.6 (141.2, 256) | 203.6 (181.7, 219.7) | 204.1 (191.6, 219.2) | 205.7 (192.7, 226.4) | 204.1 (181.7, 226.4) |
| Centriole diameter, proximal (nm) | 214.7 (131.3, 262.8) | 201 (182.4, 227.5) | 204.1 (185.4, 230.6) | 204.1 (191.6, 226) | 202.5 (182.4, 230.6) |
| Centriole diameter, central (nm) | 219.4 (139.2, 258) | 205.6 (177.6, 225.9) | 207.2 (188.5, 230.6) | 208.7 (191.6, 232.1) | 205.6 (177.6, 232.1) |
| Centriole diameter, distal (nm) | 211.4 (131.6, 250.3) | 202.5 (171.4, 227.5) | 204.1 (188.5, 222.8) | 204.1 (185.4, 222.9) | 202.5 (171.4, 227.5) |
| Centriole length (nm) | 495.4 (253.8, 1740.2) | 377.1 (211.9, 752.6) | 377 (249.3, 511) | 328.9 (190.1, 475.3) | 367.6 (190.1, 752.6) |
| Centrioles >500 nm, N (%) | 122 (48.8) | 10 (8) | 1 (2.1) | 0 | 11 (5.4) |
| Centrioles <400 nm, N (%) | 56 (22.4) | 78 (62.4) | 31 (66) | 31 (93.9) | 140 (68.3) |
Data are shown as median (range) of the investigated centrioles or number of observed phenotypes (percentage), as appropriate. N, number; nm, nanometer; MM, centrioles of CD138pos plasma cells from a patient with multiple myeloma; CTRL, centrioles of CD138neg bone marrow mononuclear cells as a control; CTRL1–3, centrioles of CTRL1, CTRL2, and CTRL3 are pooled in one group. For a detailed description of the phenotypes, see the STAR Methods section morphometrical analysis of electron tomography data and Figure 3A. All phenotypes except for one asymmetric centriole were observed in mother centrioles. Abbreviations: total abnormal, presence of any of the phenotypes (broken, incomplete, or asymmetric); centriole diameter mean, mean of proximal, central, and distal diameters for each individual centriole. Analyses of centriole dimensions and total number of appendages (all lines below “N (only completely acquired)”) were restricted to completely acquired centrioles.
Characterization of centriole parameters in CD138pos plasma cells from a patient with relapsed/refractory multiple myeloma
The CD138pos plasma cells analyzed in this study were derived from the bone marrow of a 73-year-old male patient with relapsed/refractory multiple myeloma.
Immunofluorescence microscopy, using antibodies against centrin and pericentrin to label centrioles and pericentriolar material, respectively, revealed numerical centrosome aberrations in only 16 of 552 (2.9%) CD138pos plasma cells of the patient, a frequency within the range found in healthy cells of B lymphatic origin (Krämer et al., 2003) (Figure S1). On the contrary, supernumerary centrioles were frequent (55.7%) in U2OS cells constitutively overexpressing PLK4, the principal kinase regulating centrosome replication (Bettencourt-Dias and Glover, 2005; Habedanck et al., 2005; Cosenza et al., 2017), which were used as a positive control.
By application of the ET workflow described above to six independent sections, which were at least 3 μm apart from each other, we identified 410 individual centrioles in the bone marrow sample from the myeloma patient. As 160 centrioles were only partially acquired, the analysis of centriole dimensions was restricted to the remaining 250 complete centrioles. Manual analysis showed that 120 of these centrioles were decorated with appendages. Accordingly, 120 complete mother centrioles and 130 completely pictured daughter centrioles from the myeloma patient were available for further analysis, which revealed that centrosomal regions in CD138pos plasma cells from the myeloma patient contained a median of two centrioles (range: 1–4), thereby corroborating the immunofluorescence microscopy findings (Table 1). The median centriole diameter was 216 nm (range: 141–256 nm) and not significantly different in mother (216 nm, range: 172–245 nm) versus daughter centrioles (215 nm, range: 141–256 nm; p = 0.54) (Figures 2A and 2B; Table 1). Although significantly larger than in CD138neg bone marrow mononuclear cells from healthy donors, the median centriole diameter of CD138pos myeloma cells was still slightly reduced compared with human cell lines (Chrétien et al., 1997; Sahabandu et al., 2019; Vasquez-Limeta et al., 2022).
The median centriole length was 495 nm (range: 254–1740 nm), with 48.8% of centrioles being over-elongated when a cutoff of 500 nm was applied (Figures 2C and 2D). The median length of mother centrioles (567 nm, range: 319–1740 nm) was significantly longer than the median length of daughter centrioles (436 nm, range: 254–848 nm; p < 0.001), which translated into a higher fraction of over-elongated mother, compared with daughter centrioles (75.0% versus 24.6%). Compared with centrioles in CD138neg bone marrow mononuclear cells from three healthy donors, both mother and daughter centrioles in CD138pos plasma cells from the myeloma patient were significantly over-elongated (Figures 2C and 2D). In addition, subdistal appendages were often not located at the distal ends of the centrioles but frequently in their center (Figures 2E and S2). Also, the number of subdistal appendages increased with the length of centrioles in CD138pos plasma cells from the myeloma patient (Figure 2F).
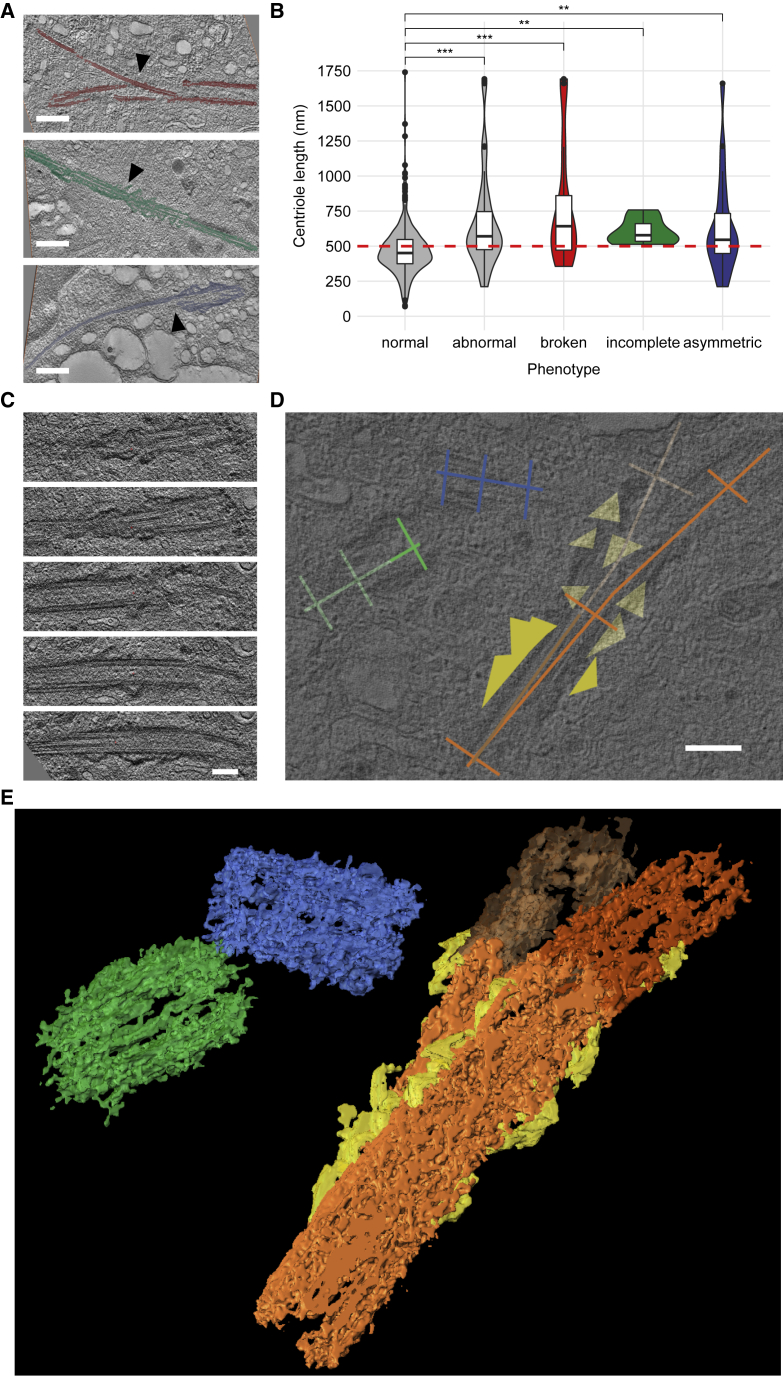
As additional structural abnormalities, 13 (5.8%), 6 (2.7%), and 18 (8.1%) of mother centrioles were broken, incomplete, and/or asymmetric at their elongated distal ends, respectively (Figure 3A; Table 1). All three phenotypes almost exclusively occurred in plasma cells with over-elongated centrioles, were restricted to mother centrioles, and were completely absent from CD138neg bone marrow mononuclear cells from healthy donors (Figure 3B; Table 1). A 3D visualization of ET data of a representative, structurally aberrant centrosome displaying over-elongation, a broken distal end, and supernumerary appendages is shown in Figures 3C–3E and Video S1.
Figure 3.
Over-elongated centrioles show additional structural abnormalities
(A) Segmented electron tomography data showing representative over-elongated centrioles with the phenotypes “broken” (top, red), “incomplete” (middle, green), and “asymmetric” (bottom, blue). The respective key feature of each phenotype is marked with a black triangle: breakage point with diverging triplets at the distal end (broken) in the top panel, multiple subdistal appendages on a longitudinal structure consisting of less than nine microtubule triplets (incomplete) in the middle panel, and an asymmetrically elongated distal end (asymmetric) in the bottom panel. Scale bars, 500 nm.
(B) Violin plots and integrated box plots showing the length distribution of normal versus abnormal centrioles in CD138pos plasma cells from a patient with relapsed/refractory MM. The abnormal group is subdivided and color coded into broken, incomplete, and asymmetric. The cutoff value for over-elongation of 500 nm is displayed as red dashed line. N (individual centrioles): normal (375), abnormal (33), broken (13), incomplete (6), asymmetric (18). The Wilcoxon rank-sum test was used for pairwise comparisons. ∗∗p < 0.01, ∗∗∗p < 0.001.
(C–E) Different views of electron tomography data, reconstructed and joined from five consecutive sections, of one representative structurally aberrant mother centriole and two adjacent structurally normal daughter centrioles.
(C) Different z-planes of the aberrant mother centriole as displayed in the original reconstructed electron tomography volume. Scale bar, 200 nm.
(D) Model view of the centrioles containing data used for measurements. For the mother centriole, length measurements for both parts of the broken centriole are displayed as two distinct vectors along the longitudinal axis. Appendages are marked with triangles. Scale bar, 200 nm.
(E) 3D visualization using Amira software. The mother centriole (orange) shows over-elongation (total length: 1692 nm) and a broken distal end (dark orange and light orange). Supernumerary subdistal appendages are marked in yellow. Two structurally normal daughter centrioles (lengths: 358 and 443 nm) are displayed in green and blue, respectively.
See also Figure S3.
Animated 3D visualization using Amira software of electron tomography data from one representative structurally aberrant mother centriole and two adjacent structurally normal daughter centrioles. The aberrant mother centriole (orange) shows over-elongation (total length: 1692 nm), a broken distal end (dark orange and light orange), and abundant, misplaced sets of appendages (yellow). Two non-over-elongated daughter centrioles (lengths: 358 and 443 nm) are displayed in green and blue, respectively. The depicted volume can be interactively viewed in MoBIE at https://github.com/mobie/centriole-tomo-examples.
Although the electron microscopy appearance of the over-elongated structures clearly resembled centrioles, we additionally performed immunostainings to ARL13B to exclude that these threads represent primary cilia rather than bona fide centrioles (Kong et al., 2020; Marteil et al., 2018). In contrast to serum-starved human BJ fibroblasts, which were used as a positive control, neither CD138neg bone marrow mononuclear cells from a healthy donor nor CD138pos plasma cells displaying over-elongated centrioles in electron microscopy contained ARL13Bpos primary cilia (Figure S3).
Discussion
High-throughput TEM as a morphology screening tool for primary tissues
The methodology we developed allows the screening of tens of thousands of cells from dozens of individual tissue donors. Manual inspection identified the cells to be targeted for ET acquisition. We propagated the target coordinates from the screened central section to the neighboring five consecutive 200-nm-thick sections in a semi-automated fashion.
This study systematically generates ET data of centrioles in human primary cells. Using the presented approach, it is possible to generate a statistically significant (n > 30) number of volume datasets depicting the subcellular target morphology of an individual tissue donor from bedside to data storage within 7 working days. As many of the acquisition steps are fully automated, data processing and analysis of already imaged data can be done in parallel. This enables a systematic characterization of organelles using volume electron microscopy with reasonable resource investment and within a decent experimental time frame. Due to the numbers of observations, a statistical correlation with clinical data is possible. A sample containing as little as 100,000 cells is sufficiently large for such an experiment. 150,000 cells even enable additional immunofluorescence imaging.
In principle, our workflow can be further improved and expedited by automated identification of regions of interest using machine-learning approaches. Automated analysis and recognition of different centriole morphologies, on the other hand, would require a significant amount of training data for machine learning.
Sharing the data through public repositories and providing means of on-the-fly visualization without the necessity of accessing or downloading huge datasets enables easy sharing, exploring, studying, and annotating volume electron microscopy data of primary (tumor) cells by a global research community.
Over-elongation and structural aberrations of centrioles in multiple myeloma
Recent data suggest a contribution of structural centriole/centrosome aberrations to several aspects of tumorigenesis including chromosomal instability (CIN) (Kohlmaier et al., 2009; Kong et al., 2020; Marteil et al., 2018). A recent screen identified centriole length deregulation as a recurrent structural aberration type in the NCI-60 panel of human cancer cell lines (Marteil et al., 2018). In vitro, over-elongated centrioles over-accumulated pericentriolar material, perturbing the symmetry of mitotic spindles. Also, they contributed to the generation of supernumerary centrosomes through fragmentation and generation of multiple procentrioles along their elongated walls, leading to subsequent multipolar and clustered, pseudo-bipolar spindle formation and CIN (Cosenza et al., 2017; Kohlmaier et al., 2009; Marteil et al., 2018). In addition, increased microtubule anchorage at supernumerary subdistal appendages of over-elongated centrioles might cause chromosome mis-segregation in asymmetric mitotic spindles.
The CD138pos plasma cells from the myeloma patient analyzed in this study exclusively harbored over-elongated and otherwise structurally aberrant centrioles but no extra centrosomes. In contrast, over-elongated centrioles were virtually absent from CD138neg healthy donor bone marrow mononuclear cells. Normal CD138pos plasma cells are terminally differentiated and, in contrast to the majority of other cell types of the bone marrow, survive for several decades in G1 phase arrest (Halliley et al., 2015). Similar to healthy plasma cells, the fraction of plasma cells in multiple myeloma that actively cycle is very low, whereas CD138neg bone marrow mononuclear cells mainly constitute fast-cycling immune cells and erythroid progenitors (Nombela-Arrieta and Manz, 2017). It has recently been shown that cell-cycle arrest leads to centriole over-elongation in different cell lines (Kong et al., 2020). Accordingly, our findings suggest that centriole over-elongation can occur in non-proliferating, quiescent cells, whereas numerical centrosome aberrations are subject to negative selection as they require cell-cycle progression for their development.
We also observed subdistal appendages to be often not located at the distal ends of the centrioles but frequently in their center. It has already been reported that aberrantly positioned subdistal appendages can occur on over-elongated centrioles in aortic endothelial cells (Bystrevskaya et al., 1992) and on over-elongated centrioles after CPAP overexpression (Kohlmaier et al., 2009).
Of note, our data demonstrate an unexpected variability in human centriole length. Apart from the large fraction of over-elongated centrioles in multiple myeloma cells, the mixture of CD138neg bone marrow mononuclear cell types from healthy donors revealed a fraction of centrioles that is shorter than the reported length of canonical human centrioles. Further analysis is warranted to evaluate the possibility that this variability in centriole length is specific for the bone marrow compartment.
Conclusion
Due to their small size, structural aberrations of centrosomes have been notoriously difficult to analyze by immunofluorescence microscopy. Using a semi-automated high-throughput workflow with cutting-edge ET, which allows for a resolution that is several magnitudes higher than the current limit of super-resolution microscopy (Chong et al., 2020; Gambarotto et al., 2019; Kong et al., 2020; Schermelleh et al., 2019; Vasquez-Limeta et al., 2022), we were able to determine the centriolar phenotype of healthy and rare, malignant bone marrow cells from patient material at the ultrastructural level on a high scale.
By assembling EM data requiring a total beam time of 273 h, of which more than 235 h were automated acquisition, we have analyzed the centrosomal phenotype of a primary cancer at the ultrastructural level on a high scale and describe over-elongated centrioles in multiple myeloma, a plasma cell malignancy that is characterized by a multitude of both numerical and structural chromosome aberrations.
Given the recent progress on the functional contribution of structural centriole aberrations in vitro, in-depth ultrastructural characterization of centriole phenotypes in primary cancer samples will be important to unravel the contribution of this aberration type to cancer development and evolution.
Limitations of study
The method we employ in this pilot study is generally applicable to various kinds of biological source material as long as a specimen for preparation compatible with conventional TEM is available (Cortese et al., 2020; Gomes Pereira et al., 2021). In our hands, a sample containing as little as 100,000 cells was sufficiently large to be handled for ET. Adaption of sample preparation to other types of specimens could allow for even smaller sample sizes. Our method is also applicable for targeting various different subcellular structures as long as they can be contrasted for and visualized by TEM. The use of open-source software facilitates the adaption and extension of our workflow to various experimental settings, which is an important prerequisite for further utilization and development of this imaging strategy by the scientific community.
Importantly, this workflow will always visualize only a fraction of all centrioles present in a given cell population and also potentially not all centrioles within single cells. For this reason, our EM methodology is not applicable for quantification of numeric centriole/centrosome aberrations at the cell population or single-cell level. Several immunofluorescence microscopy techniques have been developed within the past years to specifically address quantification of numerical centriole/centrosome aberrations. These techniques can be combined with the workflow described here to evaluate numeric and structural centriole/centrosome aberrations in parallel.
Relying on the labeling of molecular components of the centriole, expansion microscopy can reveal protein complexes that are not necessarily visible in electron microscopy (Sahabandu et al., 2019). Yet, even though expansion microscopy is performing at a throughput that surpasses electron microscopy even with our approach, it fails so far to reach the resolution of TEM tomography that gives access to subtriplet resolution. Only ultrastructure expansion microscopy (Gambarotto et al., 2019) is getting close but has only been achieved on purified centrioles so far and requires high-end deconvolution and computation. Our workflow therefore provides a valuable resource for high-throughput in situ characterization of subcellular structures in primary patient material.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-centrin | Merck Millipore | Cat.#04-1624, RRID:AB_10563501 |
| Rabbit anti-pericentrin | Abcam | Cat.#ab4448, RRID:AB_304461 |
| Rabbit anti-ARL13B | Proteintech | Cat.#17711-1-AP, RRID:AB_2060867 |
| Mouse anti-polyglutamylated Tubulin | AdipoGen | Cat.#AG-20B-0020B, RRID:AB_2490211 |
| Rabbit anti-MUM-1 (MRQ-43) | Cellmarque | Cat.#358R-74 |
| Mouse anti-Ki67 (MIB-1) | Agilent Dako | Cat.#GA626, RRID:AB_2687921 |
| Alexa Fluor 488 goat anti-mouse IgG | Molecular Probes Invitrogen | Cat.#A11017, RRID:AB_143160 |
| Alexa Fluor 488 goat anti-mouse IgG | Molecular Probes Invitrogen | Cat.#A11029, RRID:AB_2534088 |
| Alexa Fluor 568 goat anti-rabbit IgG | Molecular Probes Invitrogen | Cat.#A11036, RRID:AB_10563566 |
| Alexa Fluor 568 goat anti-rabbit IgG | Molecular Probes Invitrogen | Cat.#A11011, RRID:AB_143157 |
| Biological samples | ||
| Human bone marrow | This paper | N/A |
| Human peripheral blood | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Vectashield mounting medium | Vector Laboratories | Cat.#H-1000 |
| Vectashield mounting medium with DAPI | Vector Laboratories | Cat.#H-1200 |
| Puromycin | Life Technologies | Cat.#2600023 |
| Hygromycin | Life Technologies | Cat.#10687010 |
| Tetracyclin | SigmaAldrich | Cat.#T7660-5G |
| FicoLite-H separation medium | Linaris | Cat.#GTF1511KYA |
| Triton-X-100 | ThermoFisher Scientific | Cat.#HFH10 |
| Hoechst 33342 nucleic acid stain | Invitrogen | Cat.#H3570 |
| TRIzol RNA extraction reagent | Invitrogen | Cat.#15596026 |
| Critical commercial assays | ||
| RNeasy Mini Kit | Qiagen | Cat.#74104 |
| SV Total RNA Isolation System | Promega | Cat.#Z3101 |
| Small Sample Labeling Protocol | Affymetrix | N/A |
| GeneChip™ Human Genome U133 Plus 2.0 Array | Applied Biosystems | Cat.#900466 |
| Deposited data | ||
| Raw and analyzed data | This paper | EMPIAR DOI: 10.6019/EMPIAR-11243 |
| Code and exported workflows | This paper | Mendeley https://doi.org/10.17632/kf7fnbbbxp.1; Github: https://github.com/mobie/centriole-tomo-examples |
| Additional supplemental data | This paper | Mendeley, https://doi.org/10.17632/kf7fnbbbxp.1 |
| Gene expression profiling data | This paper; Weinhold et al. (2021); Seckinger et al. (2012) | ArrayExpress: E-MTAB-81/E-GEOD-2658 |
| Experimental models: Cell lines | ||
| U-2 OS | ATCC | Cat.#HTB-96, RRID:CVCL_0042 |
| U2OS-PLK4 | Konotop et al. (2016) | N/A |
| BJ | ATCC | Cat.#CRL-2522, RRID:CVCL_3653 |
| Software and algorithms | ||
| Konstanz Information Miner (KNIME) | Berthold et al. (2007) | https://www.knime.com/ |
| R 3.5.3 | R Core Team (2016) | https://cran.r-project.org/ |
| R ‘Survival‘ Package – 3.1-12 | Therneau and Grambsch (2000) | https://CRAN.R-project.org/package=survival |
| SerialEM | Mastronarde (2005); Schorb et al. (2019) | https://bio3d.colorado.edu/SerialEM/ |
| pyEM | Schorb et al. (2019) | https://git.embl.de/schorb/pyem |
| IMOD – 4.10.42 | Kremer et al. (1996); Mastronarde and Held (2017) | https://bio3d.colorado.edu/imod/ |
| AMIRA 2020.1 | ThermoFisher Scientific | https://thermofisher.com/amira-avizo |
| ImageJ (Fiji) – 1.53 | Schneider et al. (2012) | https://imagej.net/software/fiji/ |
| ZEN blue – 2.6 | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| NDP.view2 | Hamamatsu | https://www.hamamatsu.com/eu/en/product/life-science-and-medical-systems/digital-slide-scanner/U12388-01.html |
| MoBIE | Vergara et al. (2021) | https://github.com/mobie/mobie-viewer-fiji |
Resource availability
Lead contact
Further information and requests for resources, reagents, and scripts should be directed to and will be fulfilled by the lead contact, Alwin Krämer (a.kraemer@dkfz-heidelberg.de).
Materials availability
This study did not generate any new unique reagents. Epoxy resin blocks for electron microscopy experiments of the patient analyzed in this study were archived and are available upon request from the lead contact.
Experimental model and subject details
Plasma cell disorder patients and healthy donors
The investigated patient (73 year old male) and the healthy donors (Control 1: 62 year old female, control 2: 55 year old male, control 3: 56 year old female) gave written informed consent. The study was performed in accordance with the principles of the Declaration of Helsinki. Both the Ethics Committee of the University of Heidelberg as well as the European Molecular Biology Laboratory (EMBL) review board approved the study (Ethics Committee of the University of Heidelberg approval reference number: S-206/2011; EMBL BIAC application number: 2019-005).
Cell lines
U2OS (ATCC HTB-96, RRID:CVCL_0042) were obtained from ATCC and were kept in a humidified incubator with 37°C temperature and 5% CO2. The generation of U2OS-PLK4 cells was already described in (Konotop et al., 2016). U2OS cells were cultured in DMEM Glutamax medium (Life Technologies, Cat. No. 31966047) supplemented with 10% fetal bovine serum (FBS) (Clontech, Cat. No. 631106), 1.5 μg/mL puromycin (Life Technologies, Cat. No. 2600023), and 100 μg/mL hygromycin B (Life Technologies, Cat. No. 10687010). At 80% confluency, cells were passaged 1:10.
Method details
Plasma cell disorder and healthy donor samples
The analyzed bone marrow aspirate was acquired according to clinical standard operating procedures at the Department of Internal Medicine V, University of Heidelberg. Bone marrow mononuclear cells were isolated using Ficoll gradient centrifugation. Plasma cells were enriched by magnetic-activated cell sorting (MACS) for CD138 according to the manufacturer’s instructions, leading to a mean plasma cell purity of 88.9 ± 9.6% as checked by fluorescence-activated cell sorting (FACS).
Sample splitting
For comprehensive analysis, the sample was asymmetrically split: Up to 4 × 105 cells (depending on sample size) were used for immunofluorescence imaging, the remainder of the cells was fixed for electron microscopy and tomography.
Light microscopy and immunofluorescence imaging
After washing in PBS, approximately 105 cells were spun onto slides and fixed in 100% methanol at −20°C for 10 min.
Methanol-fixed cells were blocked with blocking buffer containing 10% goat serum in PBS at room temperature for 20 min and incubated with primary antibodies in blocking buffer at room temperature for additional 60 min. Samples were then washed three times with PBS and incubated with secondary antibodies in blocking buffer for 30 min at room temperature. After washing in PBS again, nuclei were stained using Hoechst (Invitrogen, Cat. No. H3570) in PBS for 5 min at room temperature. Samples were washed with PBS and de-mineralized purified water, incubated with 100% ethanol for 30 s, and then mounted onto coverslides using Vectashield mounting medium (Vector Laboratories, Cat. No. H-1000). Semi-automated immunofluorescence imaging with pre-defined regions of interest (ROI) was performed on the same or the following day by using ZEN blue 2.6 software (Carl Zeiss Microscopy) on a Zeiss Cell Observer equipped with a 40 × 1.3 Plan Apochromat objective.
Primary antibodies used for this study were: Centrosome staining: Mouse anti-centrin (Merck Millipore, 04–1624, RRID:AB_10563501) and rabbit anti-pericentrin (Abcam, Cat. No. ab4448, RRID:AB_304461). Alexa Fluor 488 (Molecular Probes Invitrogen, Cat. No. A11029, RRID:AB_2534088) and 568 (Molecular Probes Invitrogen, Cat. No. A11036, RRID:AB_10563566) conjugated with fluorescent dye were used as secondary antibodies.
Sample preparation for electron microscopy and tomography
A workflow scheme is depicted in Figure 1A. CD138pos plasma cells were fixed by adding freshly prepared electron microscopy (EM) fixative at 4°C (composition of fixative: 2.5% EM-grade glutaraldehyde and 2% EM-grade paraformaldehyde in 0.1 M Na-cacodylate buffer (pH 7.4)). After incubation in EM fixative at room temperature for 5 min, fixative was renewed, and samples were incubated in the fridge at 4°C overnight. After removal of fixative, cells were stained by incubation with Evans Blue (1 mg/mL in 0.1 M cacodylate buffer, pH 7.4) for 20 min at room temperature, followed by three washing steps with 0.1 M cacodylate buffer. For pre-embedding, cells were resuspended in 2% low-melting agarose and centrifuged (1000 g) for 10 min at 37°C to form a cell pellet. All subsequent embedding steps were performed using a temperature-controlled microwave at 24°C. For post-fixation and staining, cells were incubated in 1% osmium tetroxide in dH2O for 20 min. Cells were washed in dH2O for 1 min four times and stained with 1% uranyl acetate in dH2O for 14 min. Afterward, cells were rinsed in dH2O four times for 1 min each. Dehydration with an acetone series (50%, 70%, 90%, 2 × 100%) was performed for 45 s per step in the microwave. Cells were infiltrated with EPON epoxy resin 812 (hard formula) using increasing resin concentrations in 100% acetone (10%, 30%, 50%, 70%, 90%, 3 × 100% EPON in acetone). All infiltration steps were performed in the microwave for 3 min each at 24°C. Cell pellets in 100% resin were transferred into an embedding mould, incubated at room temperature overnight, and subsequently polymerized at 60°C for 2 days. Blocks were trimmed and serial sections (200 nm thickness each) were obtained using a Leica UC7 conventional ultramicrotome with a diamond knife (Diatome). At least five consecutive sections were collected on formvar-coated slot grids. Grids were post-stained with 2% uranyl acetate and lead citrate to enhance imaging contrast.
Electron microscopy and tomography
A high-throughput transmission electron microscopy workflow on a JEM 2100Plus electron microscope (JEOL Ltd., Akishima, Japan) equipped with a JEOL Matataki sCMOS camera was used to screen for centrosome-containing cells within the respective sections, as described previously (Schorb et al., 2019). After assessment of overall grid quality at 80× magnification, a 400× magnification montage image of the central section of the grid was obtained. Utilizing a KNIME (Konstanz Information Miner) (Berthold et al., 2007)- and pyEM-based software workflow, each cell on the grid was labeled and a virtual map of the cell at 1000× magnification was created by extrapolating data from the 400× magnification image. Using SerialEM’s advanced navigator functionality, we automatically acquired actual map images of each cell on the section at 1000× and 3000× magnification, respectively (Figure 1B). Output at 3000× magnification was generated as a gallery of TIFF images and manually evaluated for cells exposing centrosomal structures (Figure 1C).
After transferring the grid to a Tecnai F30 electron microscope (Thermo Fisher Scientific, Waltham, USA) equipped with a Gatan OneView camera (Gatan Inc., Pleasanton, USA), we semi-automatically targeted these cells on the remaining sections of the grid (Figure 1D). Map images were acquired at 2300× magnification for all cells of interest on all sections. Centrosome-containing regions were marked as points in SerialEM and labelled according to their location (i.e., cell and section). Single axis electron tomography was performed automatically at each point at 155,00× magnification (1.55 nm/px; tilt range: −60° to +60°; increment: 1°) using a custom SerialEM script.
Reconstruction of the acquired tilt series was achieved using an automated workflow based on the batch tomogram reconstruction feature of the IMOD software package (Kremer et al., 1996) on a high-performance computer cluster. The reconstructed tomograms of all sections were joined manually with etomo within the IMOD software package (Mastronarde, 2005; Mastronarde and Held, 2017) to produce full 3D volumes of each acquired centriole. The XY-dimensions for the final acquired 3D volumes are approximately 3.1 × 3.1 μm with at least 1 μm in Z (Figure 1E). Selected tomograms were manually segmented in Amira-Avizo software platform version 2020.1 (ThermoFisher Scientific), using the threshold-based brush segmentation tool. 3D visualizations of representative electron tomography data were generated using IMOD and Amira-Avizo, respectively (Figure 3; Video S1). Volume rendering and animations were computed and created in Amira-Avizo.
Multimodal big image sharing and exploration (MoBIE)
All acquired tomography data of this project can be visualized using the ImageJ (Schneider et al., 2012) plugin MoBIE (Vergara et al., 2021). We converted the reconstructed volumes into BDV-N5 format and determined the coordinate transformation to display the centrioles along their longitudinal axis from the model coordinates to produce insightful views to visualize the data.
General information on how to use and install MoBIE is accessible under the following link: https://github.com/mobie/mobie-viewer-fiji.
Visualizing the project data requires opening them as a MoBIE project. For this, open MoBIE in Fiji, and choose the plugin click ‘MoBIE - > Open MoBIE Project. Then enter the link to the dataset (https://github.com/mobie/centriole-tomo-examples). In the upcoming window, electron tomography data are listed in the ‘Tomograms’ dropdown menu (labeled according to their respective entity and patient number), where they can be visualized as reconstructed volumes. Alternatively, it is possible to display each centriole as a crop view along its longitudinal axis by selecting it from the ‘Centrioles’ dropdown menu in the same window.
All applicable metadata as well as KNIME, R, and, respectively, python scripts used to generate, visualize, and/or analyze the data of this project are made publicly available as well and can be found in the same Git repository and on Mendeley. DOIs are listed in the key resources table.
Quantification and statistical analysis
For all experiments, results with p values not larger than 5% were considered statistically significant. Performed statistical analyses of each experiment are described in the respective paragraph below and sample sizes are given in the in the figure legends.
Quantification and evaluation of numerical centrosome aberrations by immuno-fluorescence microscopy
After immunostaining, cells were evaluated manually in ZEN 2.6 blue edition (Carl Zeiss Microscopy, Germany) and classified as normal or amplified. Classification criteria were: normal, if four or less distinct signal maxima in the centrin channel, else amplified; normal, if two or less distinct signals in the pericentrin channel, else amplified. Cells were excluded from the evaluation if they (i) were not fully featured in 20 Z-stacks (out of focus), (ii) overlapped with other cells and thus, centrosomes could not be attributed to one or the other cell, or (iii) showed no maxima in the assumed centrosomal region in one of the two channels. Cells with visible amplification in at least one of both pericentrin and centrin signals were deemed amplified. At least 100 evaluable cells were analysed per sample.
Morphometrical analysis of electron tomography data
Using IMOD’s model feature, one electron microscopy expert marked the longitudinal axis, diameters (proximal, distal, and central), and appendages of each centriole. Structurally aberrant centriole phenotypes were tagged as well. A centriole was deemed (i) asymmetric, if it showed the typical, cylindrical 9 × 3 configuration on its one end, but only consisted of less than nine triplets on its other end; (ii) broken, if its 9 × 3 architecture was preserved, but showed clear breakage points (e.g. diverging triplets at the distal end); and (iii) incomplete, if it was clearly consisting of centriolar structures (e.g. microtubule triplets with or without appendages), but never displayed the typical 9 × 3 cylinder shape of a normally configurated centriole along its longitudinal axis. Sample images of phenotypically aberrant centrioles are depicted in Figure 3A. Centrioles carrying appendages and/or subdistal appendages were termed ‘mother’, else ‘daughter’.
Measurements of centriole dimensions were performed with the 3dmod tool within the IMOD software package. Using the “Slicer” window, a longitudinal slice through the center of the centriole at the largest possible longitudinal axis was generated, paying particular attention to the correct slicing angle to avoid incorrectness of diameter measures due to oblique cutting of the longitudinal axis. We measured centriole diameters at three different locations along the longitudinal axis: close to the proximal end, close to the distal end, and at the center. Branching microtubule blades as well as appendages were not included into the diameter measures. To measure diameters of fragmenting over-elongated centrioles, we only used the parts of the centriole that clearly showed no fragmentation. Fragmented areas which showed no 9 × 3 or 9 × 2 cylinder-shape were labeled, but neither used for measurements of length nor diameters.
Data extraction from IMOD-files and statistical analysis
Statistical analysis was done with R statistical environment 3.5.3 (R Core Team, 2016) on a ×86_64-w64-mingw32/×64 (64-bit) platform, together with ‘survival’ package (version 3.1-12) (Therneau et al., 2021; Therneau and Grambsch, 2000). A table containing coordinates and lengths for the longitudinal axis and the diameter marks as well as coordinates of individual appendages and phenotype tags was semi-automatically generated from the model files using a KNIME workflow (provided in the online resources). The Shapiro–Wilk test was used to test for normality and parametric or non-parametric statistics were applied for the analysis of centriole measures where applicable. Continuous data were described with median and range. If not stated otherwise, the Wilcoxon rank-sum test was used to test differences in continuous variables of two groups, the Kruskal-Wallis rank sum test was used to test differences in continuous variables of three or more groups, and Fisher exact test was used to test differences in categorical variables between groups. All statistical tests were two-sided.
Acknowledgments
We thank A. Baumann for sample preparation and C. Tischer and C. Pape for their support implementing data into MoBIE. This work was supported by funding from the Deutsche Forschungsgemeinschaft (DFG) to A.K. (KR 1981/4-1), S.O.S., and U.H. (Research Unit FOR 2969, projects HE 8472/1-1 and SCHO 1364/2-1) and the Dietmar Hopp Stiftung to M.S.R. T.D. received funding from the Medical Faculty of Heidelberg within the Physician Scientist Program.
Author contributions
A.K. conceived the project. A.K., Y.S., T.D., and S.K. designed the experiments. H.G., M.S.R., C.M.-T., U.H., and S.O.S. collected the samples. T.D., S.K., M.B., M.S., G.P., and I.H. performed experiments. A.K., Y.S., T.D., M.S., and S.K. analyzed data. A.K., Y.S., M.S., T.D., and S.K. wrote the paper. All co-authors approved the final manuscript.
Declaration of interests
H.G. has research funding from Amgen, BMS, Celgene, Chugai, Janssen, Incyte, Molecular Partners, Merck Sharp and Dohme, Sanofi, Mundipharma GmbH, Takeda, Dietmar-Hopp-Foundation, Johns Hopkins University, and Novartis; served as an advisor for Adaptive Biotechnology, Amgen, BMS, Celgene, Janssen, Sanofi, and Takeda; and received honoraria from Amgen, BMS, Celgene, Chugai, GlaxoSmithKline, Janssen, Novartis, and Sanofi. C.M.-T. served as a consultant for Janssen and received research funding from Bioline, Pfizer, and Janssen. M.S.R. received research funding from Amgen, Sanofi, Novartis, and Heidelberg Pharma and served as a consultant for Amgen, Sanofi, Novartis, BMS, Janssen, AbbVie, Regeneron, Takeda, and GSK. U.H. received travel grants from Janssen, Prothena, and Pfizer; served on advisory boards for Pfizer, Janssen, and Prothena; received honoraria from Janssen, Pfizer, Alnylam, and Akcea; and received research funding from Janssen and Prothena. A.K. received honoraria from Hoffmann-La Roche, Bayer, Daiichi Sankyo, and AbbVie and research funding from Bayer and Merck and is a paid consultant for Hoffmann-La Roche, Daiichi Sankyo, Bristol Myers Squibb, and AbbVie.
Published: November 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100322.
Contributor Information
Yannick Schwab, Email: yannick.schwab@embl.de.
Alwin Krämer, Email: a.kraemer@dkfz.de.
Supplemental information
Data and code availability
Data
Electron tomography data generated during this study have been deposited at EMPIAR (Iudin et al., 2016) and are publicly available using the accession code EMPIAR-11243 (https://doi.org/10.6019/EMPIAR-11243). Immunofluorescence and immunohistochemical data generated during this study will be shared by the lead contact upon request.
Code
All original code has been deposited at Github and Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T., Ohl P., Sieb C., Thiel K., Wiswedel B. Studies in Classification, Data Analysis, and Knowledge Organization (GfKL 2007) Springer; 2007. KNIME: the konstanz information miner. [Google Scholar]
- Bettencourt-Dias M., Glover D.M. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Bowler M., Kong D., Sun S., Nanjundappa R., Evans L., Farmer V., Holland A., Mahjoub M.R., Sui H., Loncarek J. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat. Commun. 2019;10:993. doi: 10.1038/s41467-018-08216-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrevskaya V.B., Lichkun V.V., Krushinsky A.V., Smirnov V.N. Centriole modification in human aortic endothelial cells. J. Struct. Biol. 1992;109:1–12. doi: 10.1016/1047-8477(92)90061-e. [DOI] [PubMed] [Google Scholar]
- Chng W.J., Braggio E., Mulligan G., Bryant B., Remstein E., Valdez R., Dogan A., Fonseca R. The centrosome index is a powerful prognostic marker in myeloma and identifies a cohort of patients that might benefit from aurora kinase inhibition. Blood. 2008;111:1603–1609. doi: 10.1182/blood-2007-06-097774. [DOI] [PubMed] [Google Scholar]
- Chong W.M., Wang W.-J., Lo C.-H., Chiu T.-Y., Chang T.-J., Liu Y.-P., Tanos B., Mazo G., Tsou M.-F.B., Jane W.-N., et al. Super-resolution microscopy reveals coupling between mammalian centriole subdistal appendages and distal appendages. Elife. 2020;9:e53580. doi: 10.7554/eLife.53580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D., Buendia B., Fuller S.D., Karsenti E. Reconstruction of the centrosome cycle from cryoelectron micrographs. J. Struct. Biol. 1997;120:117–133. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- Cortese M., Lee J.-Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosenza M.R., Cazzola A., Rossberg A., Schieber N.L., Konotop G., Bausch E., Slynko A., Holland-Letz T., Raab M.S., Dubash T., et al. Asymmetric centriole numbers at spindle Poles cause chromosome missegregation in cancer. Cell Rep. 2017;20:1906–1920. doi: 10.1016/j.celrep.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Gambarotto D., Zwettler F.U., Le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J.-G., Reuss M., Unser M., et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM) Nat. Methods. 2019;16:71–74. doi: 10.1038/s41592-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes Pereira S., Sousa A.L., Nabais C., Paixão T., Holmes A.J., Schorb M., Goshima G., Tranfield E.M., Becker J.D., Bettencourt-Dias M. The 3D architecture and molecular foundations of de novo centriole assembly via bicentrioles. Curr. Biol. 2021;31:4340–4353.e7. doi: 10.1016/j.cub.2021.07.063. [DOI] [PubMed] [Google Scholar]
- Gönczy P. Centrosomes and cancer: revisiting a long-standing relationship. Nat. Rev. Cancer. 2015;15:639–652. doi: 10.1038/nrc3995. [DOI] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., Nigg E.A. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hall N.A., Hehnly H. A centriole’s subdistal appendages: contributions to cell division, ciliogenesis and differentiation. Open Biol. 2021;11:200399. doi: 10.1098/rsob.200399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliley J.L., Tipton C.M., Liesveld J., Rosenberg A.F., Darce J., Gregoretti I.V., Popova L., Kaminiski D., Fucile C.F., Albizua I., et al. Long-lived plasma cells are contained within the CD19(-)CD38(hi)CD138(+) subset in human bone marrow. Immunity. 2015;43:132–145. doi: 10.1016/j.immuni.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iudin A., Korir P.K., Salavert-Torres J., Kleywegt G.J., Patwardhan A. EMPIAR: a public archive for raw electron microscopy image data. Nat. Methods. 2016;13:387–388. doi: 10.1038/nmeth.3806. [DOI] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B.F., Mogensen M.M., Spektor A., Dynlacht B.D., Khodjakov A., Gönczy P. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Sahabandu N., Sullenberger C., Vásquez-Limeta A., Luvsanjav D., Lukasik K., Loncarek J. Prolonged mitosis results in structurally aberrant and over-elongated centrioles. J. Cell Biol. 2020;219:e201910019. doi: 10.1083/jcb.201910019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konotop G., Bausch E., Nagai T., Turchinovich A., Becker N., Benner A., Boutros M., Mizuno K., Krämer A., Raab M.S. Pharmacological inhibition of centrosome clustering by slingshot-mediated cofilin activation and actin cortex destabilization. Cancer Res. 2016;76:6690–6700. doi: 10.1158/0008-5472.CAN-16-1144. [DOI] [PubMed] [Google Scholar]
- Krämer A., Schweizer S., Neben K., Giesecke C., Kalla J., Katzenberger T., Benner A., Müller-Hermelink H.K., Ho A.D., Ott G. Centrosome aberrations as a possible mechanism for chromosomal instability in non-Hodgkin’s lymphoma. Leukemia. 2003;17:2207–2213. doi: 10.1038/sj.leu.2403142. [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lopes C.A.M., Mesquita M., Cunha A.I., Cardoso J., Carapeta S., Laranjeira C., Pinto A.E., Pereira-Leal J.B., Dias-Pereira A., Bettencourt-Dias M., Chaves P. Centrosome amplification arises before neoplasia and increases upon p53 loss in tumorigenesis. J. Cell Biol. 2018;217:2353–2363. doi: 10.1083/jcb.201711191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manier S., Salem K.Z., Park J., Landau D.A., Getz G., Ghobrial I.M. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 2017;14:100–113. doi: 10.1038/nrclinonc.2016.122. [DOI] [PubMed] [Google Scholar]
- Marteil G., Guerrero A., Vieira A.F., de Almeida B.P., Machado P., Mendonça S., Mesquita M., Villarreal B., Fonseca I., Francia M.E., et al. Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nat. Commun. 2018;9:1258. doi: 10.1038/s41467-018-03641-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N., Held S.R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 2017;197:102–113. doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maura F., Bolli N., Angelopoulos N., Dawson K.J., Leongamornlert D., Martincorena I., Mitchell T.J., Fullam A., Gonzalez S., Szalat R., et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat. Commun. 2019;10:3835. doi: 10.1038/s41467-019-11680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell C.A., Keats J.J., Belch A.R., Pilarski L.M., Reiman T. Receptor for hyaluronan-mediated motility correlates with centrosome abnormalities in multiple myeloma and maintains mitotic integrity. Cancer Res. 2005;65:850–860. [PubMed] [Google Scholar]
- Nigg E.A., Holland A.J. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018;19:297–312. doi: 10.1038/nrm.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela-Arrieta C., Manz M.G. Quantification and three-dimensional microanatomical organization of the bone marrow. Blood Adv. 2017;1:407–416. doi: 10.1182/bloodadvances.2016003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2016. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Sahabandu N., Kong D., Magidson V., Nanjundappa R., Sullenberger C., Mahjoub M.R., Loncarek J. Expansion microscopy for the analysis of centrioles and cilia. J. Microsc. 2019;276:145–159. doi: 10.1111/jmi.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Ferrand A., Huser T., Eggeling C., Sauer M., Biehlmaier O., Drummen G.P.C. Super-resolution microscopy demystified. Nat. Cell Biol. 2019;21:72–84. doi: 10.1038/s41556-018-0251-8. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorb M., Haberbosch I., Hagen W.J.H., Schwab Y., Mastronarde D.N. Software tools for automated transmission electron microscopy. Nat. Methods. 2019;16:471–477. doi: 10.1038/s41592-019-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckinger A., Meissner T., Moreaux J., Depeweg D., Hillengass J., Hose K., Rème T., Rösen-Wolff A., Jauch A., Schnettler R., et al. Clinical and prognostic role of annexin A2 in multiple myeloma. Blood. 2012;120:1087–1094. doi: 10.1182/blood-2012-03-415588. [DOI] [PubMed] [Google Scholar]
- Sharma A., Olieric N., Steinmetz M.O. Centriole length control. Curr. Opin. Struct. Biol. 2021;66:89–95. doi: 10.1016/j.sbi.2020.10.011. [DOI] [PubMed] [Google Scholar]
- Therneau T.M., Grambsch P.M. Springer Science & Business Media; 2000. Modeling Survival Data: Extending the Cox Model. [Google Scholar]
- Therneau T.M., Thomas L., (original S.->R port and R. maintainer until 2009) Elizabeth A., Cynthia C. 2021. survival: Survival Analysis. [Google Scholar]
- Vásquez-Limeta A., Lukasik K., Kong D., Sullenberger C., Luvsanjav D., Sahabandu N., Chari R., Loncarek J. CPAP insufficiency leads to incomplete centrioles that duplicate but fragment. J. Cell Biol. 2022;221:e202108018. doi: 10.1083/jcb.202108018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara H.M., Pape C., Meechan K.I., Zinchenko V., Genoud C., Wanner A.A., Mutemi K.N., Titze B., Templin R.M., Bertucci P.Y., et al. Whole-body integration of gene expression and single-cell morphology. Cell. 2021;184:4819–4837.e22. doi: 10.1016/j.cell.2021.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold N., Salwender H.J., Cairns D.A., Raab M.S., Waldron G., Blau I.W., Bertsch U., Hielscher T., Morgan G.J., Jauch A., et al. Chromosome 1q21 abnormalities refine outcome prediction in patients with multiple myeloma - a meta-analysis of 2,596 trial patients. Haematologica. 2021;106:2754–2758. doi: 10.3324/haematol.2021.278888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animated 3D visualization using Amira software of electron tomography data from one representative structurally aberrant mother centriole and two adjacent structurally normal daughter centrioles. The aberrant mother centriole (orange) shows over-elongation (total length: 1692 nm), a broken distal end (dark orange and light orange), and abundant, misplaced sets of appendages (yellow). Two non-over-elongated daughter centrioles (lengths: 358 and 443 nm) are displayed in green and blue, respectively. The depicted volume can be interactively viewed in MoBIE at https://github.com/mobie/centriole-tomo-examples.
Data Availability Statement
Data
Electron tomography data generated during this study have been deposited at EMPIAR (Iudin et al., 2016) and are publicly available using the accession code EMPIAR-11243 (https://doi.org/10.6019/EMPIAR-11243). Immunofluorescence and immunohistochemical data generated during this study will be shared by the lead contact upon request.
Code
All original code has been deposited at Github and Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.