Summary
Researching the murine epigenome in disease models has been hampered by the lack of appropriate and cost-effective DNA methylation arrays. Here we perform a comprehensive, comparative analysis between the Mouse Methylation BeadChip (MMB) and reduced-representation bisulfite sequencing (RRBS) in two murine models of colorectal carcinogenesis. We evaluate the coverage, variability, and ability to identify differential DNA methylation of RRBS and MMB. We show that MMB is an effective tool for profiling the murine methylome that performs comparably with RRBS, identifying similar differentially methylated pathways. Although choice of technology is experiment dependent and will be predicated on the underlying biology being probed, these analyses provide insights into the relative strengths and weaknesses of each approach.
Keywords: DNA methylation, microarray, epigenetics, colorectal, epigenome, methylome, genomics, gastroenterology, bioinformatics, murine models
Graphical abstract

Highlights
-
•
The Illumina Mouse Methylation array allows precise measurements of DNA methylation
-
•
RRBS and the Mouse Methylation array identify similar aberrantly methylated pathways
-
•
Choice of platform should be guided by the underlying biology of the experimental model
Motivation
Array-based platforms have been used extensively to evaluate human DNA methylation, but DNA methylation profiling of model systems has been limited to next-generation sequencing-based approaches. Commercial release of a murine DNA methylation array is of interest to the epigenetics community, but the performance of this platform versus existing sequencing-based methodologies has not been evaluated previously. In this resource article, we attempt to provide insight into the performance of MMB in a real-world setting.
Fennell et al. comprehensively compare reduced-representation bisulfite sequencing and the Mouse Methylation BeadChip. We assess the ability of each platform to identify differentially methylated CpGs and pathways, their coverage of different genomic elements, variability and precision of methylation measurements, and other important factors that influence choice of experimental platform.
Introduction
DNA methylation is the covalent modification of DNA to include a methyl group (CH3). This is a common epigenetic alteration that can govern chromatin accessibility, transcription factor activity, gene regulation, and transcript expression. DNA methylation is deposited by DNA methyltransferase (DNMT) enzymes and removed by ten-eleven-twelve (TET) enzymes. Most DNA methylation occurs in the context of CG dinucleotides (CpG [cytosine-phosphate-guanine]), with the cytosine nucleotide becoming methylated. Deregulation of the DNA methylation landscape is linked to several diseases. The CpG island methylator phenotype, which describes the widespread accumulation of DNA methylation at CpG islands, occurs in several forms of cancer, including colorectal and gastric cancers and glioma (Fennell et al., 2019; Zouridis et al., 2012; Noushmehr et al., 2010; Liu et al., 2019). DNA methylation dysregulation also occurs in heart disease (Navas-Acien et al., 2021; Serra-Juhé et al., 2015), Alzheimer’s disease (Levine et al., 2015; Mastroeni et al., 2010), and rheumatoid arthritis (Nakano et al., 2013).
These associations have been borne out by studies utilizing increasingly accessible genome-wide DNA methylation technologies. In the late 1990s and early 2000s, PCR-based methodologies were the primary modality of choice for investigating DNA methylation alterations. These approaches are limited in throughput and, thus, stymie discovery-based investigations. In 2006, Illumina released the GoldenGate BeadArray methylation assay, a microarray-based approach that allowed simultaneous assessment of the DNA methylation state of ∼1,500 CpG sites located in the proximal promoters of ∼370 genes (Bibikova and Fan, 2009). The BeadArray platform has evolved over the past 15 years to include more than 850,000 CpG sites in its latest rendition, the EPIC array (Pidsley et al., 2016). The EPIC array and its predecessor, the 450K array, have been used extensively to characterize the DNA methylation landscape of normal and pathological states. This has led to several paradigm-shifting publications (Wockner et al., 2014; Hinoue et al., 2012; Horvath 2013; Nazor et al., 2012). These technologies are limited to human samples, and, thus, experimental biologists seeking to understand the mechanisms behind these associations using animal models must resort to other methodologies.
Transgenic murine models are an important tool for understanding the effects of certain genes on DNA methylation. A typical experiment might include knocking out a gene of interest, observing a phenotype, and assessing the underlying DNA methylation profile of a given tissue. To achieve the latter, sequencing-based approaches, such as whole-genome bisulfite sequencing (WGBS) (Lister et al., 2009) or reduced-representation bisulfite sequencing (RRBS), are usually employed (Gu et al., 2011). WGBS, when performed with sufficient depth, cover more than 99% of CpGs in the murine methylome but currently costs several thousands of dollars per sample. RRBS involves enzymatic digestion by MspI, which cuts DNA at CCGG motifs (Gu et al., 2011) and, thus, generates smaller fragments that map to CpG-dense regions of the genome, reducing sequencing library size and, ultimately, sequencing costs at the expense of coverage of CpGs in other regions (Gu et al., 2011). Inclusion of an enzyme digestion and size selection step also creates technical variability in the specific CpGs captured in each library preparation.
Illumina recently developed a mouse DNA methylation microarray based on the BeadArray technology. This array captures more than 285,000 CpG sites. These sites are curated to include proximal promoter regions and other regulatory regions of the murine methylome. In this study, we generate DNA methylation data from transgenic Kras and Braf mutant animals using RRBS and the Mouse Methylation BeadChip (MMB). In human colorectal cancer, BRAF and KRAS mutation are associated with DNA methylation dysregulation (Fennell et al., 2019; Hinoue et al., 2012; Weisenberger et al., 2006). Therefore, transgenic murine models recapitulating these mutations are ideal models for examining DNA methylation profiling technologies. We performed a comprehensive comparative analysis to critically appraise this new platform and provide insights into the benefits and limitations of each technology.
Results
Coverage of MMB and RRBS
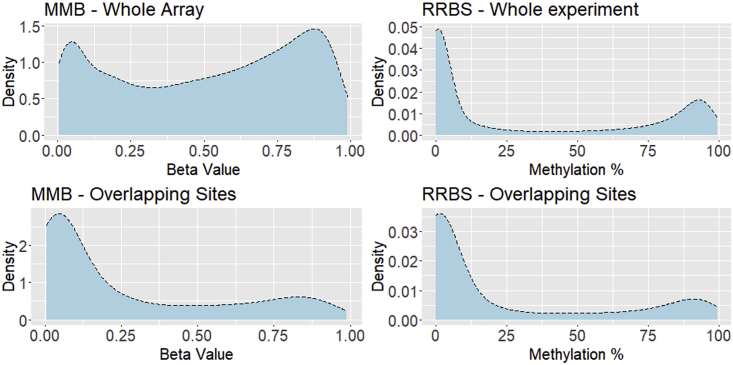
We first sought to examine the coverage of both platforms. After filtering, 264,145 CpGs were captured by MMB, of which 207,468 were captured in all samples (no sample-level filtering). For RRBS, at our threshold of 10× coverage, we captured 1,487,242 individual CpG sites. We also examined the number of CpGs covered at sequencing depths from 5× to 40× (Figure S1). As expected, there was a decrease in the number of CpGs captured with increasing sequencing depth filtering. These data indicate that the depth to which the library is sequenced is an important factor in RRBS experimental design. For MMB and RRBS, DNA methylation profiles followed the bimodal distribution expected in DNA methylation data. MMB data contained a greater proportion of intermediately methylated CpG sites (Figure 1). When we compared only sites that were covered by both platforms, the global DNA methylation patterns were consistent between MMB and RRBS (Figure 1).
Figure 1.
Global methylation profiles of the cohort as captured by MMB and RRBS
Methylation values from single CpG sites were collapsed to a mean value across all samples. The height of the density plot represents a function depicting the number of CpGs with a given level of DNA methylation. Entire datasets are presented at the top and only sites that are covered by MMB and RRBS at the bottom.
Next we examined the coverage of CpG islands (CGIs) by each technology. In this study, we adopted the UCSC definition of a CGI, which describes islands as having more than 50% GC content, a length of more than 200 bp, and a ratio of observed to expected CG dinucleotides of greater than 0.6. Using these criteria, we identify 17,017 CGIs in the murine genome. RRBS covered 13,778 CGIs (∼80% of all annotated islands) with at least one CpG site. The median number of CpG sites covered per CGI was 41. MMB covered a similar number of CGIs (13,365), but the median number of CpGs per CpG island on the array was only 2. CpGs covered in RRBS were enriched for CGIs compared with those on MMB, with 48.9% of RRBS CpGs residing in CGIs versus 11.5% on MMB. These data indicate that the breadth of the array, at the level of CGIs, is similar to RRBS, but the intra-island coverage is substantially lower.
We also compared the genomic distribution of CpGs on MMB with RRBS. Although RRBS had a comparative enrichment for 5 untranslated regions (UTRs), transcription start sites (TSSs), and exonic regions, the array had greater coverage of intronic regions (p < 0.0001; Figure S2A). In overlapping sites, we saw a similar distribution of DNA methylation in these regions (Figure S3). When considering both technologies in totality, although patterns of DNA methylation in each genomic region were similar, RRBS tended to have a greater proportion of completely hypomethylated TSSs and 5′ UTRs. The breadth of DNA methylation in intergenic sites was higher in MMB, which is likely a result of the curated nature of the probe locations on the array.
Both technologies covered repetitive elements such as long and short interspersed nuclear elements (LINEs and SINEs, respectively) and long terminal repeats (LTRs). 36,405 and 252,752 CpGs mapped to annotated repetitive elements for MMB and RRBS, respectively. Mean methylation at repetitive elements was similar for both platforms, especially in CpGs that are common to both platforms (Figure S2B).
To examine the breadth of coverage of MMB and RRBS in regulatory regions, we downloaded candidate cis-regulatory elements (CREs) from the murine ENCODE project. These elements are subdivided into promoter-like, proximal enhancer-like, distal enhancer-like, CTCF-only, and DNase-H3K4me3. The murine ENCODE project describes 23,762 promoter like signatures (PLSs), 72,964 proximal enhancer like signatures (pELSs), 209040 distal enhancer like signatures (dELSs), 23836 CTCF-only, and 10,383 DNase-H3K4me3 regions.
MMB has probes that cover 2.4% of PLS (571 regions), 6.8% of dELS (4927 regions), 0.73% of pELS (1542 regions), 0.88% of DNase-H3K4me3 regions (211 regions), and 5.12% of CTCF-only sites (532 regions). In total, MMB has probes that cover 7,783 independent candidate cis-regulatory regions.
By comparison, at 10× coverage, the RRBS consensus dataset contained CpGs that mapped to 57,068 unique candidate cis-regulatory elements, including 60.9% of PLS (14,480 PLS regions), 29.4% of pELS (21,450 regions), and 8.8% of dELS (18,356 regions).
Although RRBS covered a greater number of cCREs, there was a significant bias toward coverage of proximal sites compared with the genomic distribution of cCREs (p < 0.0001 for all proximal elements versus genome; Figure S2C). In contrast, representation of cCREs on the array was more reflective of the genome, with a small enrichment for dELS and depletion for pELS elements (p = 0.0091 and p = 0.0034, respectively; Figure S2C).
Appropriate subsampling of CpGs in CGIs can capture most intra-CGI variability
RRBS captures significantly more CpG sites per CGI. We sought to determine whether capturing these CpG sites yielded additional insight into the DNA methylation profile of the CGI or whether they were methylated similarly and provided redundant information. To better understand the variability within CGIs, we first calculated the variance of methylation across CGIs in each sample and then calculated the mean variance at each CGI across the entire dataset (Figures 2 and S5). The positive skew in the distribution of mean variances indicates that most CGIs are relatively stable, with little variability in DNA methylation between CpGs residing in the same island. Figures 2B–2I depict CGIs with increasing intra-island variability. We hypothesized that CGI length and ratio of CGI length to number of CpG sites may influence intra-island variability, with longer CGIs and those represented by fewer CpGs being more variable. To test this, we regressed the variance of each island against the length (base pairs) and the ratio of CGI length to number of CpG sites (CpG:CGI length ratio). Although CGI length and CpG:CGI length ratio were highly significantly associated with increased intra-island variability (1.75 × 10−6 and 2 × 10−16, respectively), the R2 was low (R2 ≈ 0.01), indicating that most of the variability is ascribed to other factors.
Figure 2.
Intra-CGI variability differs between CGIs
(A) Variance of DNA methylation across CGIs containing multiple CpG sites. The histogram represents the distribution of the log10(variance) between CpG sites in all CGIs with more than 1 CpG. DNA methylation was measured by RRBS. Each dot and line represents methylation in an individual sample.
(B–H) Representative DNA methylation profiles of CGIs with low variance (B) through CGIs with extreme intra-island variability (H). Each line represents a single sample across an entire CGI, and each dot represents a single CpG site. Line and dot colors represent mouse age groups.
Next we examined the effect of DNA methylation level on the stability of DNA methylation across the island. DNA methylation was most consistent across CGIs when islands were nearly entirely methylated or demethylated (Figure S4). We quantified this statistically by calculating the absolute difference between average methylation across the island and hemimethylation (50%). As distance from hemimethylation increased (toward 0% or 100% methylation), variability across the island significantly decreases (p = 2 × 10−16, R2 ≈ 0.76).
To identify whether most of the variability between CpGs in an island could be captured by a smaller subset of CpGs, we performed variable clustering. Variable clustering captures subsets of highly correlated variables, and identifies if most of the variance within a cluster can be explained by a single variable. The average CGI in our RRBS dataset contained 52.59 CpG sites, and 13.4 variable clusters. We calculated how much of the total variability within a CGI could be explained by capturing only data from representative CpGs from variable clusters within an island. Using data from only the most representative CpGs of variable clusters within a CGI, we can recapitulate an average of 64.6% of the variability within a CGI and reduce the size of the total dataset pertaining to CGIs from 702,978 CpGs to 179,166 CpGs.
We observed that CGIs with low variability have a lower percentage of variance explained by variable clusters (Figure S4) and require a greater number of clusters to explain the variance across the CGI. This is likely due to the small variance, most of which is produced by noise rather than biological signal. In contrast, highly variable CGIs, which reflect biological variability across the island, can be well explained by capturing several CpGs (Figure S4). These data indicate that highly variable CGIs can be accurately summarized by profiling a subset of carefully selected CpGs within the CGI.
Base-resolution methylation profiling via MMB versus RRBS
We sought to assess the correlation between methylation values attained at base resolution using MMB and RRBS. After coverage filtering (>10 reads per CpG), our RRBS experiment covered 1.48 × 106 CpG sites. 25,548 filter-passing CpGs were covered by MMB and RRBS, representing 12.3% and 2.44% of CpGs covered by MMB and RRBS, respectively. We extracted methylation values for these CpG sites from both platforms and computed a sample-wise correlation analysis. In CpGs covered by both assays, we observed high concordance between both platforms (Pearson’s R2 > 0.95 in all samples; Figure 3). The average absolute difference between measurements obtained on MMB and RRBS was small, with DNA methylation measurements of more than 98% of overlapping probes with an absolute methylation difference of less than 20% between platforms (Figure 3C).
Figure 3.
RRBS and MMB DNA methylation measurements are highly correlated
Comparative analyses revealed significant genome-wide concordance of DNA methylation between MMB and RRBS.
(A) Representative DNA methylation measurements as captured by the MMB and RRBS in CpG sites that were common to both platforms.
(B) Pearson’s R2 values for same-sample, whole-methylome comparisons on RRBS and MMB.
(C) Distribution of the difference between the mean MMB beta value and the mean RRBS methylation percentage at individual CpG sites.
(D) Correlation of DNA methylation between MMB and RRBS (Rsq) versus the average beta value for the same CpG. CpGs were binned in increments of 0.05.
Next we examined correlations between methylation values obtained on MMB and RRBS at individual CpG sites. There was substantial inter-CpG variability in the correlation between MMB and RRBS (Pearson’s R2 range, 0–0.98). We hypothesized that the correlation between readouts obtained on each platform may be weakest at measurement extremes (fully [un]methylated). To examine this hypothesis, we calculated the average beta value obtained by MMB for each probe and binned them in beta value increments of 0.05. We observed that correlations between MMB and RRBS followed a unimodal distribution centered around an average beta value of ∼0.4 (Figure 3D).
Differential DNA methylation analysis of common CpGs
Most studies attempt to identify differential DNA methylation between experimental groups. We next sought to examine the ability of both technologies to identify differential methylation with age and by sample group. It is well established that methylation at specific loci shifts in accordance with advancing age (Fennell et al., 2021; Horvath 2013; Teschendorff et al., 2013). For an age-associated differential DNA methylation analysis, we regressed the beta values (or percentage of methylated reads) against age (days) for CpGs that were assayed with both technologies. The Illumina methylation array vastly outperformed RRBS in this respect, identifying statistically significant age-associated DNA methylation alterations in 3,274 of 25,396 overlapping CpGs (Figures 4A and 4B). In contrast, RRBS identified a mere 278 of 25,396 CpGs. 92.4% of CpGs that were identified as changing with age by RRBS were also detected by MMB. These data indicate that, although both approaches can detect differential methylation with age, MMB is more sensitive and able to detect more subtle age-associated DNA methylation events.
Figure 4.
MMB detects significantly more differential DNA methylation with age
(A) Overlap between genes common to both platforms and identified as significantly age associated in wild-type samples by RRBS, MMB, or both.
(B) Significant differential methylation with age detected by RRBS and MMB.
We also performed differential DNA methylation analysis between Kras or Braf mutant animals and their wild-type counterparts at 8 and 14 months. In keeping with our aging analysis, MMB detected significantly more statistically differentially methylated loci (Table 1). Most of the differences detected by MMB were small. By applying the common |Δ| 0.2 beta value threshold for calling differential methylation, the discrepancies in differential methylation calling on the two platforms shrank substantially (Table 1), indicating that MMB may be more suited for detecting more subtle DNA methylation alterations.
Table 1.
Number of differentially methylated CpG sites between 8- and 14-month-old Braf and Kras mutant animals versus wild-type animals, as assessed by RRBS and MMB, in CpG sites that were common to both techniques
| Group | 8 months (FDR) |
8 months (FDR and |Δ| β > 0.2) |
||||
|---|---|---|---|---|---|---|
| RRBS | MMB | p value | RRBS | MMB | p value | |
| KRAS | 260 | 4,949 | <0.0001 | 246 | 401 | <0.0001 |
| BRAF | 577 | 5,154 | <0.0001 | 577 | 841 | <0.0001 |
| Group | 14 months (FDR) |
14 months (FDR and |Δ| β > 0.2) |
||||
|---|---|---|---|---|---|---|
| RRBS | MMB | p Value | RRBS | MMB | p Value | |
| KRAS | 1,046 | 6,921 | <0.0001 | 947 | 870 | 0.07 |
| BRAF | 1,620 | 6,887 | <0.0001 | 1,467 | 1,447 | 0.72 |
MMB is more precise than RRBS
We hypothesized that the within-group variation in detected methylation may be smaller on the MMB platform compared with RRBS, improving our ability to detect more subtle age and genotype-associated changes. To explore this, we analyzed within-group variance on each platform at single-CpG resolution. For each experimental group and for most CpG sites (Figure 5), we observed substantially less variability between samples on MMB. For Braf and Kras mutant samples at 8 months, variability was significantly greater on the RRBS platform compared with MMB, with 3,277 (Braf mutant animals) and 2,815 CpGs (Kras mutant animals), having a greater intra-group variability on RRBS compared with MMB. In contrast, the RRBS was less variable than MMB at 74 and 75 CpGs, respectively. The same trend was observed at 14 months (Figure 5). These data support the hypothesis that RRBS-generated DNA methylation data are more variable than MMB and, thus, are limited in detecting smaller DNA methylation changes.
Figure 5.
MMB-derived DNA methylation data are less variable than RRBS
Shown is the log(variance ratio) of CpG sites common to both platforms by experimental group. CpGs with a log(variance ratio) of less than 0 have lower within-group variability on the MMB and more than 0 on RRBS. For all experimental groups, most CpGs have lower within-group variability on MMB. Log(variance ratio) was calculated by F tests.
To investigate whether sequencing coverage was influencing variability on RRBS, we regressed the mean coverage of each CpG site in the consensus dataset against the within-group variance for each experimental group (Figure S6). For all experimental groups, we observed that, as mean sequencing coverage increased, there was a congruent decrease in the within-group variance. These data indicate that the variability is most likely technical rather than biological.
Detection of common pathway alterations
Gene promoter and pathways analysis is commonly used to identify processes that are driving phenotypes of interest. We sought to assess whether each technology could independently identify differentially methylated promoters and pathways in our Braf and Kras mutant murine models. We performed differential methylation analysis on each platform, using the entire quality-filtered dataset (rather than just CpGs shared by each technology) in each case, to emulate the situation where the investigator has chosen a particular technology from the outset. In keeping with earlier analyses, we considered CpGs to be differentially methylated when the false discovery rate (FDR) was less than 0.05 and there was an absolute difference in DNA methylation of 0.2 (MMB) or 20% (RRBS) (Table S1). Both technologies detected a large number of significant DNA methylation alterations under each condition versus wild-type animals. Because of the higher number of CpGs, RRBS detected more differentially methylated CpG sites than MMB in most comparisons (Table 2). The proportion of differential methylation compared with the total number of CpGs captured by each technique was between 1% and 3%.
Table 2.
Differential DNA methylation in Braf and Kras mutated intestine as detected by MMB and RRBS
| Mutation | Direction | 8 months |
14 months |
||
|---|---|---|---|---|---|
| RRBS | MMB | RRBS | MMB | ||
| Braf | hypermethylated | 11,253 (0.76%) | 4,423 (1.7%) | 46,757 (3.14%) | 6,276 (2.41%) |
| hypomethylated | 13,908 (0.93%) | 9,423 (3.61%) | 16,399 (1.1%) | 7,808 (2.99%) | |
| Kras | hypermethylated | 5,288 (0.36%) | 1,962 (0.75%) | 30,625 (2.06%) | 3,729 (1.43%) |
| hypomethylated | 2,127 (0.14%) | 2,263 (0.87%) | 5,597 (0.38%) | 3,312 (1.27%) | |
The entire probe set was included in the analysis of MMB and all CpGs with more than 10× coverage for RRBS. Data represent the number of differentially methylated CpGs and the proportion of differentially methylated CpGs compared with all CpGs covered.
Because these models represent intestinal oncogenes that are usually associated with the CGI methylator phenotype, we next segregated the data by whether the CpG resided in a CGI. As outlined earlier, 727,509 (48.9%) and 30,385 (11.6%) CpGs reside in CGIs in our RRBS and MMB datasets, respectively. On both platforms, we observed the expected pattern of distribution of hyper(hypo)methylation events (Figure S7), with hypermethylation concentrated in CGIs and hypomethylation in non-island-associated genomic regions.
Next we performed Gene Ontology analysis on hypermethylated promoters in 14-month-old Braf and Kras mutant mice on MMB and RRBS. For each comparison, we observed an enrichment for hypermethylation at promoters encoding genes involved in differentiation and development (Figure 6), consistent with our prior work. We observed substantial overlap in the Gene Ontology processes identified by MMB and RRBS) (64.4% and 51% for Braf and Kras, respectively), and ∼65% of the top 50 enriched terms for both models were identified by RRBS and MMB (Figure 6). These data indicate that both technologies can reliably detect differentially methylated pathways that are relevant to the disease model.
Figure 6.
RRBS and MMB detect similar pathway-level differential methylation alterations
Top: Gene Ontology enrichment analysis for differentially hypermethylated promoters detected in Braf or Kras mutant 14-month-old mice versus wild-type littermates as captured by RRBS and MMB. Hypergeometric test for overrepresentation was used to generate p values, which were subsequently adjusted using the FDR method. Bottom: overlap of Gene Ontology terms determined by analysis of MMB and RRBS data.
Discussion
DNA methylation is an important epigenetic modification that can govern chromatin state and gene transcriptional programs. Several diseases alter the DNA methylation landscape (Robertson 2005), and understanding the basis of these modifications is important for understanding disease etiology. Murine models are ideal for investigating disease, but genome-scale DNA methylation analyses have been limited to bisulfite-based sequencing approaches, such as WGBS, which is prohibitively expensive, and RRBS, which has a coverage bias for CGI-associated CpGs. Here we compared a new array-based DNA methylation analysis tool, MMB, with RRBS. We report that MMB performs comparably with RRBS. MMB produces precise measurements of DNA methylation and is able to detect disease-specific DNA methylation alterations.
Both techniques capture a similar number of CGIs, but MMB has significantly fewer CpGs per island. There is some evidence that suggests a high level of correlation between the DNA methylation of CpG sites within a CGI, especially when the CGI is small (<400 bp) (Zhang et al., 2015). However, most of this research is confined to humans, and it is not clear whether this was also true in murine models. Here we sought to assess whether increased intra-CGI coverage by RRBS might contain biologically relevant DNA methylation or whether additional CpG coverage within an island is redundant, with DNA methylation of a few CpGs being sufficient to infer DNA methylation of the entire island. This is an important distinction, given that the CGIs are typically only represented by one or two probes on MMB. Here we report that most CGIs had relatively low intra-island methylation variability. As the mean methylation of a CGI approached 50%, we observed significant variability in DNA methylation between CpGs within the island, and, thus, it is important to consider the validity of generalizing MMB data generated from a small number of probes to the entire CGI at intermediate levels of DNA methylation.
In sequencing experiments, investigators are often faced with the coverage versus cost trade-off, where increased coverage and sequencing depth are associated with readouts that are more reliable but can dramatically increase the overall cost of the experiment. In this study, we considered 10× coverage to be an appropriate cutoff for calling DNA methylation fractions, allowing us to profile ∼1.5 million CpG sites. As we increase our coverage stringency, we obtain more accurate methylation fractions that converge on the methylation readout obtained for the same sites on MMB. If the CpG sites of interest are covered by MMB, this technology may offer a more precise readout that is more cost effective than deep sequencing of bisulfite sequencing libraries. As little as 100 ng is sufficient for generating RRBS libraries. In contrast, MMB requires more than 250ng. Investigators should keep these factors in mind when considering which technology to use. Ultimately, the underlying biological processes being probed and the resources of the laboratory will guide these decisions.
One common application is differential methylation analysis. Here the question is whether there is a statistically significant difference in DNA methylation at a given site between two experimental groups. Small, consistent differences can often yield significant p values but have little biological relevance. We and others also apply a Δ methylation cutoff, usually 0.2 for methylation arrays or 20% for methylation sequencing . One of our key findings here is that, although MMB can detect substantially more differential methylation between experimental groups at a purely statistical level, this difference is negligible when we apply an absolute methylation change cutoff. For most animal experiments, investigators will be far more interested in large changes in DNA methylation at a given locus; thus, these data should provide confidence that both methods, when covering the locus of interest, will detect these alterations. For more niche applications where subtle changes in DNA methylation are of interest (for example, when profiling a locus that is methylated in a subpopulation of cells in bulk samples), MMB may be more appropriate. Epigenetic age is another common analysis. Although we did not assess epigenetic age in this study, we did identify many age-associated loci on MMB. The low technical variability and the sheer number of age-associated loci in our experiment indicate that MMB data will be well suited for this application. MMB consistently captures the same loci and does not suffer from variable dropout, which affects RRBS-based epigenetic age predictions.
The expertise and availability of bioinformatics infrastructure and support should be considered when choosing an approach. RRBS is a sequencing-based approach and, therefore, will generate millions of sequencing reads that must be preprocessed, aligned, and methylation called. This process is computationally intensive and requires specialized bioinformatics expertise. MMB data can be processed entirely in the R environment, with several well described R packages facilitating array processing (Xu et al., 2016; Aryee et al., 2014; Müller et al., 2019). In contrast, RRBS data, as with all large sequencing experiments, must first be processed using command-line tools, presenting an additional technical barrier for inexperienced users. It is not possible to process these data on standard computers, and access to compute infrastructure is required. MMB generates a comparably smaller amount of data and can be processed on high-end desktop computers. Basic array analysis and processing are more accessible to non-bioinformatically trained scientists and can also be performed on platforms with graphical interfaces (i.e., Galaxy). The availability of specialized expertise and infrastructure is an important element that should be considered when choosing a technology.
In our final analysis, we performed a side-by-side differential methylation and pathways analysis on both platforms to confirm that similar biological signatures would emerge with both techniques. We have shown previously that Braf mutation in humans and mice results in widespread accumulation of DNA methylation at CGIs (Fennell et al., 2019, 2021; Bond et al., 2018). We have also shown that Kras mutation can generate a similar but distinct methylator phenotype. Using RRBS and MMB, we recapitulated these signatures, validating the animal models and the approaches used to assess DNA methylation.
Here we comprehensively evaluated the performance of the Illumina Mouse Methylation Array in comparison with RRBS in two models of murine colorectal carcinogenesis using matched samples. In CpG sites that are covered by both platforms, we report high correlation between DNA methylation profiles. MMB could detect a greater number of statistically differentially methylated sites than RRBS; MMB was able to detect smaller methylation alterations because of the increased precision of DNA methylation readouts, especially in regions that are poorly covered by RRBS. However, this difference was not significant when we imposed a minimum methylation change threshold, as is common in the literature. MMB is a valuable addition to the toolbox of experimental epigeneticists that performs comparably with sequencing-based methodologies. MMB is highly suited to applications requiring highly precise methylation calls and detection of small methylation differences between experimental groups.
Limitations of the study
Our approach in this study does bear some limitations. We compared RRBS with the new MMB. RRBS predominantly assesses DNA methylation in CGIs, and therefore we did not cover many of the non-island-associated CpGs on the MMB and were not able to make direct comparisons at these loci. WGBS assesses the whole methylome for DNA methylation, and future work should evaluate how MMB performs in comparison with WGBS.
We also used the beta value difference threshold of 0.2, which has been established empirically for human methylation arrays. Appropriate thresholds are likely to vary in mouse models and will vary depending on the specific biology being assessed. For example, in our study, we assessed non-neoplastic tissue, which is relatively homogeneous and has little within-group variability, providing additional confidence when calling differential methylation at smaller beta value thresholds. In contrast, cancerous tissues are likely to be more heterogeneous and therefore may warrant a higher threshold.
We also did not assess the effect of SNPs on MMB probe behavior, but Zhou et al. (2022) published work describing the design and annotation of MMB. Probes on the array were designed to exclude any that harbored a SNP within 10 bp of the 3′ end of the probe sequence in any of the 15 inbred mouse strains selected for assessment.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| AllPrep DNA/RNA/Protein Mini Kit | Qiagen | 80004 |
| Ovation® RRBS Methyl-Seq | Tecan Life Sciences | 0553–32 |
| Infinium Mouse Methylation BeadChip | Illumina Inc. | 20041559 |
| Zymo EZ-96 DNA Methylation kit | Zymo | D5001 |
| Deposited data | ||
| Reduced Representation Bisulphite Sequencing Data | ArrayExpress and Zenodo | E-MTAB-12214 https://doi.org/10.5281/zenodo.7102827 |
| Mouse Methylation BeadChip Data | ArrayExpress and Zenodo | E-MTAB-11985 https://doi.org/10.5281/zenodo.7102827 |
| Experimental models: Organisms/strains | ||
| model organism: Villin-CreERT2: Tg(Vil1-cre/ERT2)23Syr mice | The Jackson Laboratory | JAX stock #020282 |
| model organism: BRAF V637: Braftm1Mmcm mice | The Jackson Laboratory | JAX stock #017837 |
| model organism: KRASG12D−LSL: Krastm4Tyj mice | The Jackson Laboratory | JAX stock #008179 |
| Other | ||
| Code | Zenodo | https://doi.org/10.5281/zenodo.7102827 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lochlan Fennell (lochlan.fennell@monash.edu).
Materials availability
This study did not generate any unique reagents.
Experimental model and subject details
Animal models
The BrafV637 CA (FBV background) and KRASG12D-LSL (C57/BL6J background) murine models were used for this study. The BrafV637 CA model is a cre-recombinase dependent conditionally activated model that facilitates the expression of the oncogenic Braf V637E allele. The V637E allele is analogous to the human V600E mutation.
The KRASG12D-LSL conditionally activated model similarly expresses the oncogenic allele upon exposure to cre-recombinase. Both models were independently crossed with animals bearing the Villin-CreERT2 transgene (C57BL6J background). Villin-CreERT2 animals express the CreERT2 fusion gene under the guise of the Villin promoter, which is only active in the lower gastrointestinal tract. A single IP injection (75mg/kg) of tamoxifen facilitates the translocation of CreERT2 to the nucleus, where it induces recombination of the targeted alleles. We injected animals with tamoxifen at weaning and sacrificed them at two, eight and fourteen months of age. We also sacrificed age-matched littermates as wild type controls. We did not actively select animal sex, and animals were house in standard conditions. The Samples investigated in the study table is a description of the animals used throughout the study. All animal experiments were approved by the QIMR Berghofer Animal Ethics Committee (P2178).
Method details
Sample collection and processing
Non-neoplastic tissue from the proximal small intestine was dissected at necropsy and cryopreserved in liquid nitrogen. DNA and RNA was extracted from fresh-frozen tissue samples using the AllPrep DNA/RNA/Protein minikit (Qiagen, USA) as per the manufacturer’s instructions. The same DNA extraction was used for both RRBS and MMB.
Samples investigated in the study
| Sample Group | Age (months) | n (MMB) | n (RRBS) |
|---|---|---|---|
| Wild-type | 2 | 4 | 4 |
| Wild-type | 8 | 6 | 6 |
| Wild-type | 14 | 6 | 6 |
| Kras mutant | 2 | 3 | 3 |
| Kras mutant | 8 | 3 | 3 |
| Kras mutant | 14 | 3 | 3 |
| Braf mutant | 2 | 3 | 3 |
| Braf mutant | 8 | 3 | 3 |
| Braf mutant | 14 | 3 | 3 |
Reduced representation bisulphite sequencing and data processing
We generated single base resolution DNA methylation data using the Ovation RRBS Methyl-Seq System 1–16 (Tecan Life Sciences) with 100ng of input DNA. This approach employs the MspI restriction enzyme to digest DNA at the CCGG motif, which is highly enriched in CpG dense regions of the genome. This generates a library of small (>300bp) fragments that are rich in CpG content. Sequencing libraries are generated from these fragments, bisulphite converted and sequenced. For this study, we sequenced these libraries to a target depth of 30 million single end 100bp reads per sample on an Illumina NovaSeq instrument.
Data were processed as per Fennell et al. (2021). Briefly, sequencing reads were inspected for quality using FastQC (v0.11.7). Reads were trimmed to remove sequencing adaptors and poor quality bases using TrimGalore (v0.6.6, (Krueger et al., 2021)). Reads were then aligned to the murine methylome (mm10) using Bismark (v0.20.0) and methylation calculated from alignments using the methylation_extractor and bismark2bedGraph functions of Bismark (v0.20.0, (Krueger and Andrews, 2011)). Data were imported into the R environment using the methylKit package (v1.14.2, (Akalin et al., 2012)). Using methylKit, CpG sites covered by < 10 reads in any sample were discarded to generate a consensus dataset. We also performed analyses at various levels of coverage to determine how many CpGs are lost when coverage requirements increase.
Mouse Methylation BeadChip
Genome-wide DNA methylation was performed using the Illumina Infinium Mouse Methylation BeadChip (Illumina, San Diego, CA, USA) following the standard manufacturer’s protocol. 500ng of high-quality genomic DNA was bisulfite converted using the Zymo EZ-96 DNA Methylation kit (Zymo Research, Irine, CA, USA). Bisulfite converted samples were then amplified, fragmented, purified and hybridized onto the Mouse Methylation BeadChip according to the manufacturer’s standard protocol. The arrays were washed and scanned using the Illumina iScan System. Mouse Methylation BeadChips were processed at Australian Genome Research Facility (AGRF), Melbourne. For analysis, idat files were imported into R using EnMix (v1.25.1, (Xu et al., 2016)). Data was normalized using the preprocessENmix function, which models background noise using out of band intensities on Infinium type I probes and corrects for probe type dye biases using the RELIC method. Data was subsequently filtered by detection P and for sex chromosomes. For detection P filtering, we masked values that had a detection p > 0.05, and removed the probe entirely if >50% of samples had a detection p > 0.05.
Data annotations
To annotate CpGs with respect to whether they reside in CpG islands, we downloaded the mm10 cpgIslandExt table from the UCSC table browser. The cpgIslandExt table contains annotations of CpG islands, where a genomic region is a CpG island if it meets the following criteria: having >50% GC content, a length of >200bp and a ratio of observed to expected CG dinucleotides of >0.6. We assigned each CpG island a unique identifier, and examined overlaps between CpGs on MMB and RRBS using the intersect function of bedtools (v2.29.0, (Quinlan and Hall, 2010)).
Annotations of candidate regulatory elements were downloaded from the murine ENCODE project web portal (Mouse ENCODE Consortium et al., 2012). These annotation tracks were generated by the ENCODE consortia through evaluation of ChIP-Seq data pertaining to histone modifications, and DNase-hypersensitivity sequencing. These annotations included promoter-like (PLS; within 200bp of TSS, ++ DNase accessibility and H3K4me3 signal), proximal enhancer-like (pELS; within 2000bp of TSS, ++ DNase accessibility and H3K27ac signal, low H3K4me3 if < 200bp to TSS), distal enhancer-like (dELS; >2000bp from TSS, ++ DNase accessibility and H3K27ac signal), CTCF-only (high DNase and CTCF, low H3K27ac and H3K4me3) and DNase-H3K4me3 (>200bp from TSS, ++ DNase accessibility and H3K4me3 signal). We examined overlaps between CpGs and candidate regulatory elements using the intersect function of bedtools (v2.29.0).
Repetitive elements were annotated using RepeatMasker. Briefly, pre-computed annotations for the mm10 genome were downloaded from the RepeatMasker web interface (RepeatMasker open-4.0.5 - Repeat Library, 20140131, Dec 2011 annotations). Bedtools was used to find the overlap between these repetitive elements and CpGs in the RRBS and MMB datasets.
Gene promoter annotations were generated using the annotatePeaks function of the HOMER tool (v4.8, (Heinz et al., 2010)) and the mm10 reference genome. CpGs that were subsequently annotated as promoter and ascribed to a gene were used for downstream gene promoter analysis.
Quantification and statistical analyses
To identify a subsets of similarly methylated CpGs in each CpG island we used the Cluster Variables procedure in JMP (v16, SAS Institute, Care NC, USA). The algorithm starts with a set of variables and splits them into clusters comprising highly correlated variables, and identifies a single representative variable which explains the largest proportion of the variance in the cluster. To evaluate the variability of DNA methylation at single CpG sites within experimental groups we used the F-test method as employed in the matrixStats R Package (v0.61.1). This method tests whether the variances across two groups are equal. Data is presented as the log(variance) ratio, with <0 being representing less within group variability on MMB and >0 representing greater within group variability on MMB.
To assess the degree of correlation between MMB and RRBS we first generated a consensus dataset that contained DNA methylation measurements of the same CpG on both platforms. For each sample, we then performed linear regression analysis on the entire consensus data set, comparing measurements on MMB with measurements on RRBS. For differential methylation analysis we used the Limma R package (v3.14, (Ritchie et al., 2015)), considering only samples assayed with both approaches. For gene ontology enrichment analysis we used the ClusterProfiler R package (v3.16.1, (Wu et al., 2021)). In comparing the number of differentially methylated CpGs or gene ontology terms, we the X2 method with Yates correction. Statistical methods are referred to in figure legends, table captions or in text as appropriate.
Acknowledgments
We acknowledge funding support from Pathology Queensland, Australian Rotary Health, Tour De Cure, and Illumina Inc.
Author contributions
Conceptualization, L.J.F. and V.L.J.W.; methodology, L.J.F., G.H., and A.-M.P.; investigation, D.M.M., C.E.B., and A.K.; software, L.J.F.; formal analysis, L.J.F. and G.H.; funding acquisition, L.J.F. and V.L.J.W.; supervision, B.A.L. and V.L.J.W.; resources, D.M.M., C.E.B., and A.K.; writing – original draft, L.J.F.; writing – reviewing and editing, G.H., D.M.M., C.E.B., A.K., B.A.L., A.-M.P., and V.L.J.W.
Declaration of interests
Illumina Inc. contributed to reagent expenses but was not involved in conducting the study, analyzing data, or deciding to publish.
Published: November 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100323.
Supplemental information
Data and code availability
-
•
All raw sequencing and microarray data has been deposited to ArrayExpress and will be publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw and processed data has also been deposited to Zenodo and the corresponding DOI is listed in the key resources table.
-
•
Code to process sequencing and microarray data has been uploaded to Zenodo. The corresponding DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.
References
- Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Fan J.-B. In: DNA Methylation. Tost J., editor. Vol. 507. Humana Press; 2009. GoldenGate® assay for DNA methylation profiling; pp. 149–163.http://link.springer.com/10.1007/978-1-59745-522-0_12 (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- Bond C.E., Liu C., Kawamata F., McKeone D.M., Fernando W., Jamieson S., Pearson S.-A., Kane A., Woods S.L., Lannagan T.R.M., et al. Oncogenic BRAF mutation induces DNA methylation changes in a murine model for human serrated colorectal neoplasia. Epigenetics. 2018;13:40–48. doi: 10.1080/15592294.2017.1411446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell L., Dumenil T., Wockner L., Hartel G., Nones K., Bond C., Borowsky J., Liu C., McKeone D., Bowdler L., et al. Integrative genome-scale DNA methylation analysis of a large and unselected cohort reveals 5 distinct subtypes of colorectal adenocarcinomas. Cell. Mol. Gastroenterol. Hepatol. 2019;8:269–290. doi: 10.1016/j.jcmgh.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell L., Kane A., Liu C., McKeone D., Hartel G., Su C., Bond C., Bettington M., Leggett B., Whitehall V. Braf mutation induces rapid neoplastic transformation in the aged and aberrantly methylated intestinal epithelium. Gut. 2021;71:1127–1140. doi: 10.1136/gutjnl-2020-322166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Smith Z.D., Bock C., Boyle P., Gnirke A., Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T., Weisenberger D.J., Lange C.P.E., Shen H., Byun H.-M., Van Den Berg D., Malik S., Pan F., Noushmehr H., van Dijk C.M., et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;16:96. doi: 10.1186/s13059-015-0649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., James F., Ewels P., Afyounian E., Schuster-Boeckler B. FelixKrueger/TrimGalore: v0.6.7. Zenodo. 2021 https://zenodo.org/record/5127899 [Google Scholar]
- Levine M.E., Lu A.T., Bennett D.A., Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging. 2015;7:1198–1211. doi: 10.18632/aging.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.-M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Fennell L.J., Bettington M.L., Walker N.I., Dwine J., Leggett B.A., Whitehall V.L.J. DNA methylation changes that precede onset of dysplasia in advanced sessile serrated adenomas. Clin. Epigenetics. 2019;11:90. doi: 10.1186/s13148-019-0691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D., Grover A., Delvaux E., Whiteside C., Coleman P.D., Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol. Aging. 2010;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse ENCODE Consortium. Stamatoyannopoulos J.A., Snyder M., Hardison R., Ren B., Gingeras T., Gilbert D.M., Groudine M., Bender M., Kaul R., et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Scherer M., Assenov Y., Lutsik P., Walter J., Lengauer T., Bock C. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol. 2019;20:55. doi: 10.1186/s13059-019-1664-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Boyle D.L., Firestein G.S. Regulation of DNA methylation in rheumatoid arthritis synoviocytes. J. Immunol. 2013;190:1297–1303. doi: 10.4049/jimmunol.1202572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Acien A., Domingo-Relloso A., Subedi P., Riffo-Campos A.L., Xia R., Gomez L., Haack K., Goldsmith J., Howard B.V., Best L.G., et al. Blood DNA methylation and incident coronary heart disease: evidence from the strong heart study. JAMA Cardiol. 2021;6:1237–1246. doi: 10.1001/jamacardio.2021.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazor K.L., Altun G., Lynch C., Tran H., Harness J.V., Slavin I., Garitaonandia I., Müller F.J., Wang Y.-C., Boscolo F.S., et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P., Pan F., Pelloski C.E., Sulman E.P., Bhat K.P., et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R., Zotenko E., Peters T.J., Lawrence M.G., Risbridger G.P., Molloy P., Van Djik S., Muhlhausler B., Stirzaker C., Clark S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17:208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Serra-Juhé C., Cuscó I., Homs A., Flores R., Torán N., Pérez-Jurado L.A. DNA methylation abnormalities in congenital heart disease. Epigenetics. 2015;10:167–177. doi: 10.1080/15592294.2014.998536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A.E., West J., Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Hum. Mol. Genet. 2013;22:R7–R15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Wockner L.F., Noble E.P., Lawford B.R., Young R.M., Morris C.P., Whitehall V.L.J., Voisey J. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl. Psychiatry. 2014;4:e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Niu L., Li L., Taylor J.A. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44:e20. doi: 10.1093/nar/gkv907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Spector T.D., Deloukas P., Bell J.T., Engelhardt B.E. Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome Biol. 2015;16:14. doi: 10.1186/s13059-015-0581-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Hinoue T., Barnes B., Mitchell O., Iqbal W., Lee S.M., Foy K.K., Lee K.-H., Moyer E.J., VanderArk A., et al. DNA methylation dynamics and dysregulation delineated by high-throughput profiling in the mouse. Cell Genom. 2022;2:100144. doi: 10.1016/j.xgen.2022.100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouridis H., Deng N., Ivanova T., Zhu Y., Wong B., Huang D., Wu Y.H., Wu Y., Tan I.B., Liem N., et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci. Transl. Med. 2012;4:156ra140. doi: 10.1126/scitranslmed.3004504. https://www.science.org/ [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw sequencing and microarray data has been deposited to ArrayExpress and will be publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw and processed data has also been deposited to Zenodo and the corresponding DOI is listed in the key resources table.
-
•
Code to process sequencing and microarray data has been uploaded to Zenodo. The corresponding DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.