Abstract
Background
Circular RNAs (circRNAs) dysregulation has been revealed to function in the pathological processes of cancers. Herein, the role and mechanisms of hsa_circ_0002082 in breast cancer (BC) progression were elucidated.
Methods
In vivo and in vitro functional experiments were conducted, and the interaction between miR‐508‐3p and hsa_circ_0002082 or Centromere Protein F (CENPF) was elucidated.
Results
Hsa_circ_0002082 expression was higher in BC tissues and cell lines. Functionally, knockdown of hsa_circ_0002082 induced apoptosis and suppressed proliferation and metastasis in BC cells in vitro. Mechanistically, hsa_circ_0002082 targeted miR‐508‐3p, which was confirmed to be decreased in BC. MiR‐508‐3p overexpression suppressed BC cell malignant phenotypes, moreover, inhibition of miR‐508‐3p attenuated the anticancer action of hsa_circ_0002082 silencing on BC cells. Besides that, miR‐508‐3p targeted CENPF, CENPF was highly expressed in BC, CENPF up‐regulation reversed the suppressive impacts of miR‐508‐3p on BC cell growth and metastasis. Besides, hsa_circ_0002082 silencing impeded BC growth in nude mice.
Conclusion
Knockdown of hsa_circ_0002082 suppresses breast cancer growth and metastasis by miR‐508‐3p/CENPF axis, suggesting that hsa_circ_0002082 may be a promising target for breast cancer treatment.
Keywords: breast cancer, CENPF, hsa_circ_0002082, miR‐508‐3p, tumorigenesis
Hsa_circ_0002082 promotes proliferation and metastasis but suppresses apoptosis of breast cancer cells by miR‐508‐3p/Centromere Protein F axis.
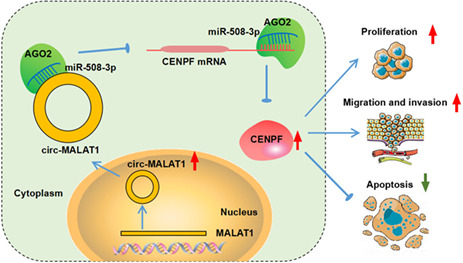
1. INTRODUCTION
Breast cancer (BC) has the highest incidence rate in women and is the second most common malignancy frequently occurring of newly diagnosed cancers. 1 Despite great improvements in screening, diagnosis and therapy, metastasis, recurrence, and drug resistance progression presently limit the successful management of BC. 2 Hence, ongoing investigations are necessary for ameliorating the outcome of BC patients.
Circular RNAs (circRNAs) are formed by back‐splicing and possess closed‐loop RNA structures. 3 More and more researchers uncovered that circRNAs are implicated in diverse biological and pathological processes. 4 , 5 Besides that, deregulated circRNAs are related to the tumorigenesis of BC. Increased circRNAs, such as circSEPT9 6 and circCDYL, 7 were found in BC, and acted as oncogenic genes to promote BC development by regulating cancer cell growth, metastasis or autophagy. In addition, circRNAs, such as circTADA2As 8 and hsa_circ_0025202, 9 possessed tumor‐suppressing capability in BC progression. Hsa_circ_0002082 is produced by Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) in chr11: 65,271,199–65,272,066 and finally forms a circular transcript of 867 bp. Chen et al. showed an increased circ‐MALAT1 (numbered hsa_circ_0002082) in cancer stem cells (CSCs) of hepatocellular carcinoma (HCC) and circ‐MALAT1 accelerated the self‐renewal of CSCs, implying the involvement of circ‐MALAT1 in the cancer. 10 By circRNA microarray, hsa_circ_0002082 was also found to show an elevated tendency in BC, and might mechanism as ceRNAs to play roles in BC. 11 Consistent this finding, we also exhibited an elevation of hsa_circ_0002082 in BC tissues via analyzing GSE182471, whereas the functions of hsa_circ_0002082 in BC remain unclear.
Herein, we speculated that hsa_circ_0002082 might act as an oncogenic to be implicated in the process of BC. Thus, the functions of hsa_circ_0002082 in BC cell growth and metastasis were investigated in this work. CircRNAs have been widely revealed to perform their functions via competitive endogenous RNA (ceRNA) mechanism, that is, act as sponges for specific microRNA (miRNA) and subsequently influence gene expression. 12 , 13 Preliminary bioinformatics analysis revealed that hsa_circ_0002082 and Centromere Protein F (CENPF) share the same microRNA response elements of miR‐508‐3p, thus, we further studied whether there was a ceRNA network among hsa_circ_0002082, miR‐508‐3p and CENPF in BC progression.
2. MATERIAL AND METHODS
2.1. Clinical tissue specimens
In total, 69 paired tumors were collected from BC patients newly diagnosed by pathological examination via surgery. None of them receive any preoperative therapy. Color Doppler ultrasound of BC was characterized by irregular shape, unclear boundary, burr‐like edge, uneven internal echo, aspect ratio > 1 and abundant blood flow signal; Immunohistochemistry (IHC) staining of BC displayed the positive of Ki67 and CK5/6, and the negative tumor suppressor P63 (Figure S1A,B). Besides, blood samples were collected from 69 BC patients. And 69 age‐ and gender‐matched healthy individuals were recruited and blood samples were obtained as the healthy control. The plasma was obtained by centrifugation at 1000 g at 4°C for 15 min. All samples were immediately stored at −80°C. This study was authorized by the Ethics Committee of Minda Hospital of Hubei Minzu University. Written informed consents had been sighed by all participators.
2.2. Cell culture
Normal MCF‐10A cells and BC cell lines (MCF‐7, MDA‐MB‐231, BT‐549, and T47D) were purchased from COBIOER. BC cell lines were cultured in the RPMI‐1640 medium (COBIOER) plus 1% penicillin–streptomycin (COBIOER) and 10% fetal bovine serum (FBS, COBIOER). MCF‐10A cells were grown in MEBM BulletKit. All medium was maintained at 37°Cwith 5% CO2.
2.3. IHC analysis
Paraffin‐embedded tissues were deparaffinized and rehydrated. Then, sections were incubated with anti‐CK5/6 (ab40, 1:1000), anti‐Ki67 antibody (ab16667, 1:400), anti‐CENPF (ab5, 1:500) and anti‐P63 (ab124762, 1:5000) (Abcam) for 12 h at 4°C, and HRP‐labeled anti‐rabbit or ‐mouse IgG for 30 min at 37°C. The sections were stained by diaminoaniline (DAB).
2.4. Quantitative real‐time PCR (qRT‐PCR)
Total RNA was prepared with the help of TRIzol reagent (Takara Biotech) and then used to generate cDNAs by reverse transcription, followed by qRT‐PCR analysis with specific primers (Table 1). β‐actin or U6 was used as the control.
TABLE 1.
Primers sequences used for qRT‐PCR
| Name | Primers for qRT‐PCR (5′–3′) | |
|---|---|---|
| circ‐MALAT1 (hsa_circ_0002082) | Forward | GTGTATTTTTAGAAACTTTGTCTG |
| Reverse | TTATTTAGAGGGCCTCTATTG | |
| CENPF | Forward | ACCTTCACAACGTGTTAGACAG |
| Reverse | CTGAGGCTCTCATATTCGGCA | |
| miR‐944 | Forward | CTCCGAGAAATTATTGTACATCG |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| miR‐508‐3p | Forward | GTCGCTGATTGTAGCCTTTTGG |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| U6 | Forward | CTTCGGCAGCACATATACT |
| Reverse | AAAATATGGAACGCTTCACG | |
| β‐actin | Forward | CTTCGCGGGCGACGAT |
| Reverse | CCACATAGGAATCCTTCTGACC | |
2.5. Cell transfection
The short hairpin RNA (shRNA) targeting hsa_circ_0002082 (sh‐hsa_circ_0002082), miR‐508‐3p mimics, inhibitors (anti‐miR‐508‐3p), pcDNA3.1 CENPF overexpression plasmids (oe‐CENPF) and the controls (nontarget shRNA [sh‐NC], NC mimic, NC inhibitor, vector) were procured from Geenseed. For stable transfection, the lentiviruses carrying sh‐hsa_circ_0002082 or sh‐NC were produced by Hanbio.
2.6. Cell counting Kit‐8 (CCK‐8) assay
After assigned transfection, BC cells were reacted with 10 μl CCK‐8 solution (Beyotime) (2 × 103/well) at 0, 1, 2, or 3 days for additional 2‐h incubation, followed by reading the absorbances by a microscope at 450 nm to assess cell proliferation.
2.7. 5‐Ethynyl‐2′‐deoxyuridine (EdU) assay
Transfected BC cells were incubated with 50 μM EdU reagent (RiboBio) for 3 h. Following dyeing with Apollo reaction mixture away from light for half an hour and Hoechst, the EdU‐positive cells were examined.
2.8. Flow cytometry
After assigned transfection, BC cells were harvested and dyed with fluorescein isothiocyanate (FITC)‐Annexin V and propidium iodide (PI) (BestBio) for 20 min away from light, and apoptotic cells were detected by the Flow Cytometer within 1 h.
2.9. Caspase‐3 activity analysis
The caspase3 activity was examined by a colorimetric assay kit (Abcam) after assigned transfection following the standard procedure. Later on, a microplate reader was applied to test the absorbance at 405 nm.
2.10. Transwell assay
A 24‐well chamber (8 μm pore size; Costar, Corning) precoated with or without matrigel was utilized to assess cell invasion and migration capacities. Transfected MCF‐7 and MDA‐MB‐231 cells with serum‐free medium were seeded into the top chamber. In the bottom chamber, complete medium was added as the chemoattractant. 24 h later, cells invaded or migrated to the bottom chamber were taken and counted by a light microscope after 0.1% crystal violet dyeing (Beyotime).
2.11. Western blotting
Total proteins were extracted using 100 μl of lysis buffer containing RIPA (99 μl) and protease inhibitor cocktail (1 μl). The proteins (about 50 μg) were resolved by 10% SDS‐poly acrylamide gel electrophoresis (SDS‐PAGE), and transferred to PVDF membranes (Millipore). Then primary antibodies including Vimentin (ab8978, 1:1000), E‐cadherin (ab1416, 1:500), N‐cadherin (ab76011, 1:5000), CENPF (ab5, 1:500) and β‐actin (ab6276, 1:1000) (Abcam) were applied to probe with the membranes all night at 4°C. After the secondary incubation at 37°C for 2 h, the ECL detection reagent (Beyotime) was employed for the detection of protein bands.
2.12. RNA pull‐down assay
BC cells were lysed and then incubated with specific probes including biotin‐labeled hsa_circ_0002082 probe (hsa_circ_0002082 probe) and control probe (Oligo probe) (Genepharma). Afterwards, the mixture was reacted with streptavidin‐coated magnetic beads. Finally, the enrichment of miRNAs was assayed.
2.13. RNA immunoprecipitation (RIP) assay
RIP assay was implemented as per the specification of the Magna RIP kit (Millipore) with antibodies of anti‐Argonaute 2 (Ago2) or anti‐IgG. Lastly, the enrichment of hsa_circ_0002082, miR‐508‐3p, and CENPF in isolated immunoprecipitated RNAs was assayed.
2.14. Dual‐luciferase reporter assay
The psiCHECK2 plasmids inserted with wild‐type (WT) fragments and the mutated (MUT) seed sequences of hsa_circ_0002082 and CENPF in miR‐508‐3p were constructed by Genepharma. BC cells were treated with a mixture of 200 ng recombinant psiCHECK2 plasmids and 50 nM of miRNA mimics or the controls, and the luciferase activity was assayed after 48‐h incubation.
2.15. Xenograft models
MCF‐7 cells (3 × 106) infected with lentiviruses carrying sh‐hsa_circ_0002082 or sh‐NC were subcutaneously vaccinated into the fourth mammary fat pad of nude mice (n = 6/group; 6 weeks; female; 18–22 g). The size of tumors was monitored every week. At days 28, the tumors were excised and weighed. All animal experiments were approved by the Animal Care Committee of Minda Hospital of Hubei Minzu University.
2.16. Statistical assay
Data are manifested as mean ± standard deviation (SD). The difference was evaluated by the Student's t test (two groups) and analysis of variance with hoc post Turkey test (multigroup). p < 0.05 means significant differences.
3. RESULTS
3.1. Hsa_circ_0002082 expression pattern
Hsa_circ_0002082 was one of the most up‐regulated circRNA, which was produced by MALAT1 in chr11: 65,271,199–65,272,066 (867 nT) (Figure 1A). Thereafter, the clinical samples were collected. Then we found a higher expression of hsa_circ_0002082 in BC tissues (n = 69) than those of adjacent normal tissues (Figure 1B). Meanwhile, hsa_circ_0002082 expression was also higher in the plasma samples of BC patients compared with the healthy control (Figure 1C). Furthermore, the association between hsa_circ_0002082 expression (both in tissues and plasma) and pathological characteristics was analyzed, Table 2 showed that higher hsa_circ_0002082 expression was closely related to advanced TNM stages and lymph node metastasis. Then, hsa_circ_0002082 expression was also increased in BC cell lines relative to the normal MCF‐10A cells (Figure 1D). Besides that, nuclear‐cytoplasmic fractionation assay suggested that hsa_circ_0002082 was mainly distributed in the cytoplasm of BC cells (Figure 1E,F).
FIGURE 1.
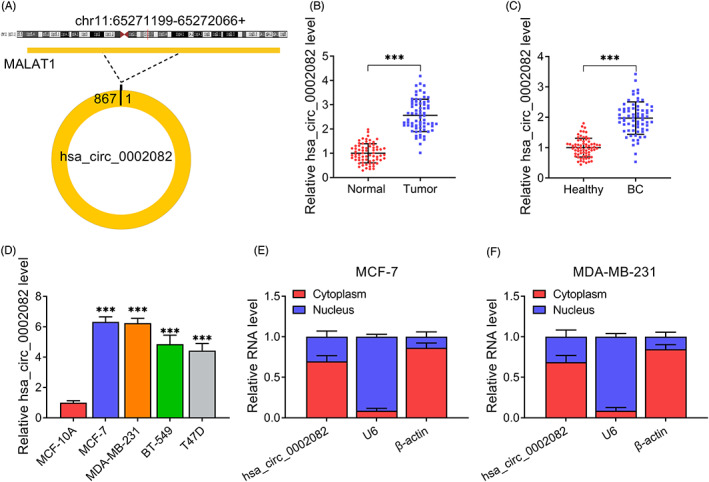
Hsa_circ_0002082 expression profile. (A) The formation of hsa_circ_0002082. (B–D) Increased hsa_circ_0002082 levels in BC tissues, plasma samples, and cell lines. (E, F) The subcellular localization of hsa_circ_0002082. ***p < 0.001
TABLE 2.
Correlation of the expression of hsa_circ_0002082 (tissues and plasma) with clinicopathologic features in BC patients
| Parameters | N = 69 | hsa_circ_0002082 | p‐Value | hsa_circ_0002082 | p‐Value | ||
|---|---|---|---|---|---|---|---|
| Level in tissues | Level in plasma | ||||||
| High N = 35 | Low N = 34 | High N = 35 | Low N = 34 | ||||
| Age, years | |||||||
| <50 | 30 | 14 | 16 | 0.554 | 12 | 18 | 0.118 |
| ≥50 | 39 | 21 | 18 | 23 | 16 | ||
| Tumor size | |||||||
| <2 | 24 | 10 | 14 | 0.272 | 9 | 15 | 0.109 |
| ≥2 | 45 | 25 | 20 | 26 | 19 | ||
| TNM stage | |||||||
| I–II | 37 | 13 | 24 | 0.005 | 11 | 26 | <0.001 |
| III | 32 | 22 | 10 | 24 | 8 | ||
| Lymph node metastasis | |||||||
| No | 35 | 10 | 25 | <0.001 | 8 | 27 | <0.001 |
| Yes | 34 | 25 | 9 | 27 | 7 | ||
3.2. Knockdown of hsa_circ_0002082 suppresses BC cell growth and metastasis
The clinical value of hsa_circ_0002082 on BC progression was then investigated. qRT‐PCR showed that the transfection of sh‐hsa_circ_0002082 markedly reduced hsa_circ_0002082 content in MCF‐7 and MDA‐MB‐231 cells compared with sh‐NC transfection (Figure S2A). Functionally, CCK‐8 and EdU assays exhibited that hsa_circ_0002082 silencing suppressed BC cell proliferation (Figure 2A–C). Flow cytomecry suggested that the apoptosis was enhanced after hsa_circ_0002082 silencing (Figure 2D). And the enhancement of apoptosis was also confirmed by the increased caspase3 activity in hsa_circ_0002082‐decreased BC cells (Figure 2E). Besides, transwell assay indicated that the migrated and invaded BC cells were reduced after the knockdown of hsa_circ_0002082 (Figure 2F,G). Western blotting suggested that hsa_circ_0002082 deficiency led to the decreases of Vimentin and N‐cadherin as well as an increase of E‐cadherin (Figure 2H,I), implying the impairment of epithelial‐mesenchymal transition (EMT) process.
FIGURE 2.
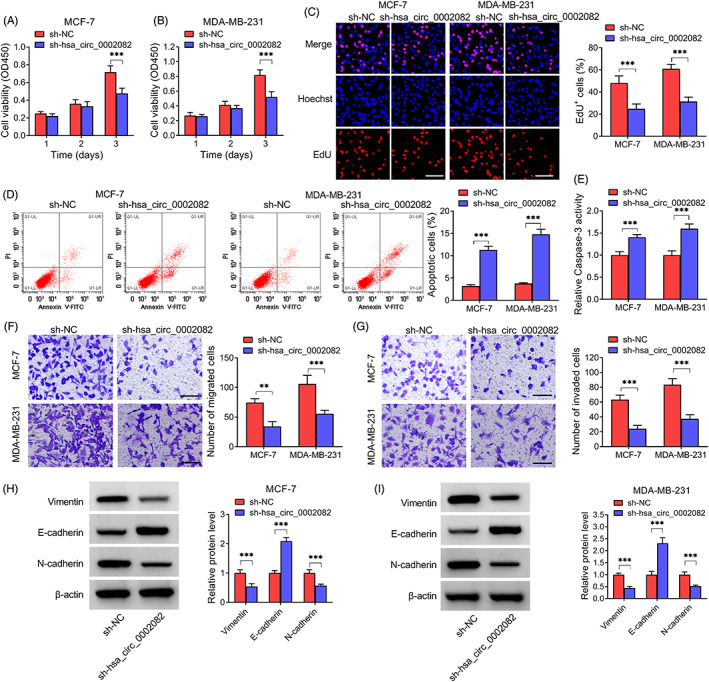
Knockdown of hsa_circ_0002082 suppresses BC cell growth and metastasis. After hsa_circ_0002082 knockdown, (A–C) cell proliferation detection. (D, E) Cell apoptosis analysis. (F, G) Measurement of cell migration and invasion. (H, I) Contents of EMT‐markers. **p < 0.01, ***p < 0.001
3.3. MiR‐508‐3p is targeted by hsa_circ_0002082
Given the cytoplasm distribution of hsa_circ_0002082, the potential targets of hsa_circ_0002082 were predicted using circinteractome, starbase, and circbank databases, and hsa_circ_0002082 was predicted to have putative conserved target sites for miR‐508‐3p and miR‐944 (Figure 3A). Then RNA pull‐down assay showed that miR‐508‐3p was significantly enriched by biotin‐labeled hsa_circ_0002082 probes compared with miR‐944 (Figure 3B,C). The Ago family of proteins is the core components of the RNA‐induced silencing complex (RISC) that is required for miRNA‐mediated gene silencing. We then analyzed if hsa_circ_0002082 and miR‐508‐3p contained the same RISC and performed RIP assay in BC cells, and found that miR‐508‐3p and hsa_circ_0002082 could be pulled down by anti‐Ago antibody relative to IgG antibody (Figure 3D,E). Thus, miR‐508‐3p might be a target of hsa_circ_0002082. Then, we constructed the mutated sequence in binding sites to verify this hypothesis (Figure 3F). After verifying the elevation efficiency of miR‐508‐3p mimic (Figure S2), dual‐luciferase reporter assay was executed, it was found that miR‐508‐3p overexpression overtly reduced the luciferase activities of the WT‐hsa_circ_0002082 group but not the mutated one (Figure 3G,H). Besides that, the content of miR‐508‐3p in BC tissues was decreased (Figure 3I), which was negatively correlated with hsa_circ_0002082 (Figure 3J). Also, miR‐508‐3p expression was lower in BC cell lines compared with the normal control (Figure 3K). Taken together, hsa_circ_0002082 directly targeted miR‐508‐3p in BC cells.
FIGURE 3.
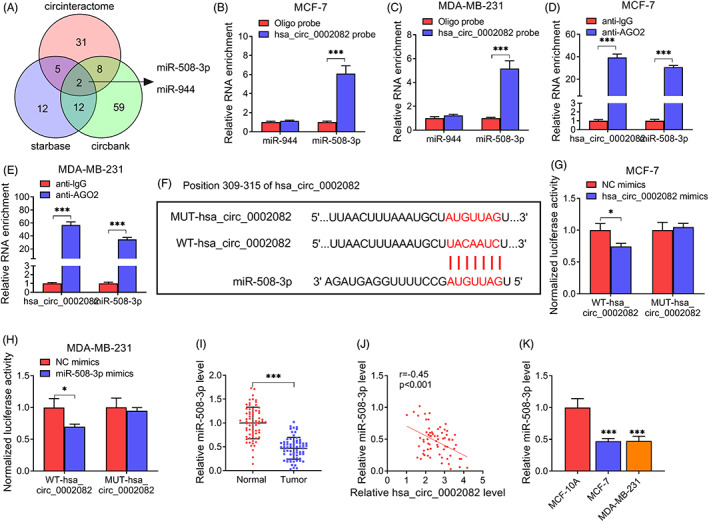
MiR‐508‐3p is targeted by hsa_circ_0002082. (A) The potential miRNAs interaction with hsa_circ_0002082. (B, C) RNA pull‐down assay. (D, E) RIP assay with anti‐AGO2 antibody. (F) Schematic illustration of the binding sequences. (G, H) Luciferase activity analysis. (I) Decreased miR‐508‐3p level in BC tissues. (J) Correlation analysis between hsa_circ_0002082 and miR‐508‐3p expression in BC tissues. (K) Detection of miR‐508‐3p level in BC cells and normal cells by qRT‐PCR. *p < 0.05, ***p < 0.001
3.4. CENPF is a target of miR‐508‐3p
According to the prediction of BRAC, starbase and targertscan databases, only CENPF was found to have the putative conserved target site for miR‐508‐3p (Figure 4A). Moreover, analysis from GEPIA showed much higher CENPF level in BC tissues (I = 1085) (Figure 4B). RIP assay showed that CENPF and miR‐508‐3p were enriched in Ago2‐containing microribonucleoproteins relative to control (Figure 4C,D). Then dual‐luciferase reporter assay was executed, the mutant version was displayed in Figure 4E. Thereafter, results showed the decline of luciferase activity in cells with the wild‐type CENPF reporter and miR‐508‐3p mimics (Figure 4F,G). Besides, the level of CENPF was increased in BC tissues, evidenced by qRT‐PCR and IHC analyses (Figure 4H,I). Also, an increased CENPF level in BC cell line was observed (Figure 4J,K). These data confirmed that CENPF was targeted by miR‐508‐3p.
FIGURE 4.
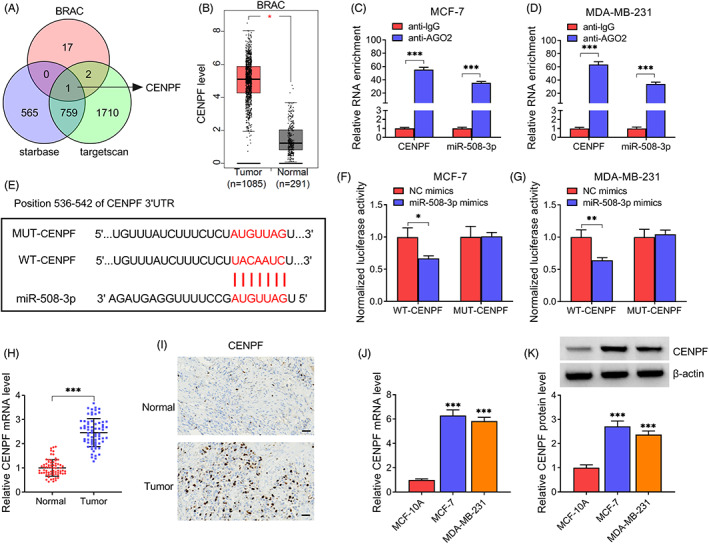
CENPF is targeted by miR‐508‐3p. (A) The potential targets interaction with miR‐508‐3p. (B) Analysis of CENPF expression profile based on GEPIA database. (C, D) RIP assay with anti‐AGO2 antibody. (E) Schematic illustration of the binding sequences. (F, G) Luciferase activity detection (H, I) Increased CENPF level BC tissues. (J, K) Increased CENPF level in BC cell lines. *p < 0.05, **p < 0.01, ***p < 0.001
3.5. MiR‐508‐3p suppresses BC cell growth and metastasis by CENPF
Next, rescue experiments were performed. Western blotting showed that CENPF overexpression plasmids (oe‐CENPF) markedly increased CENPF level in BC cells (Figure S2). Then, as expected, oe‐CENPF introduction rescued miR‐508‐3p mimics‐induced decrease of CENPF in BC cells (Figure 5A). Thereafter, miR‐508‐3p overexpression reduced the proliferation (Figure 5B–D) and induced apoptosis (Figure 5E,F), while these effects were attenuated by CENPF up‐regulation. Results of transwell assay implied that the migration and invasion of BC cells were enhanced after miR‐508‐3p mimics transfection and subsequently reduced in response to oe‐CENPF introduction (Figure 5G,H). Moreover, the inhibition of EMT process caused by miR‐508‐3p mimics was also abated by oe‐CENPF transfection (Figure 5I,J).
FIGURE 5.
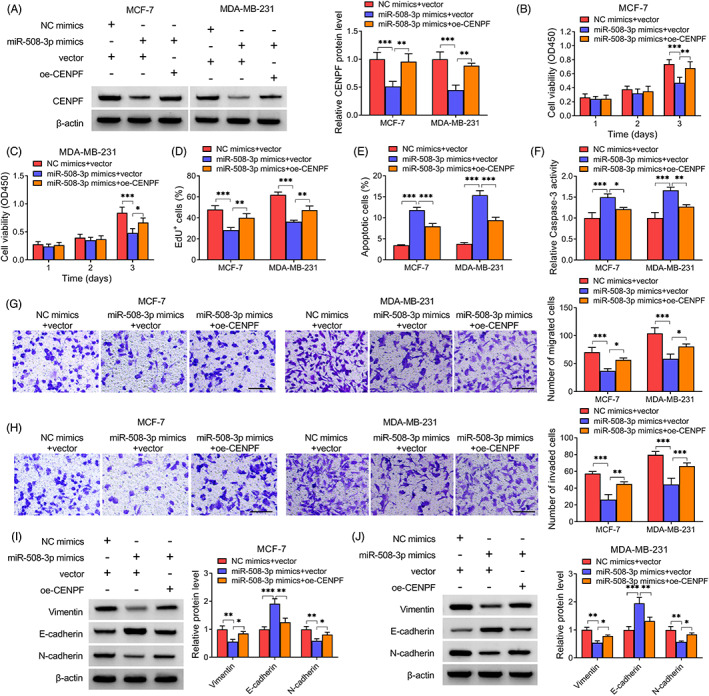
MiR‐508‐3p suppresses BC cell growth and metastasis by CENPF. After miR‐508‐3p mimics and oe‐CENPF co‐transfection. (A) Detection of CENPF level. (B–D) Cell proliferation analysis. (E, F) Cell apoptosis measurement. (G, H) Detection of cell migration and invasion. (I, J) V EMT‐markers contents. *p < 0.05, **p < 0.01, ***p < 0.001
3.6. Knockdown of hsa_circ_0002082 suppresses BC cell growth and metastasis by miR‐508‐3p
The knockdown efficiency of miR‐508‐3p inhibitors was first verified by qRT‐PCR (Figure S2). Then, we observed that hsa_circ_0002082 knockdown was accompanied with decreased CENPF expression, which was then rescued by subsequent miR‐508‐3p inhibition in cells (Figure 6A), suggesting the hsa_circ_0002082/miR‐508‐3p/CENPF axis. Thereafter, we studied whether hsa_circ_0002082 regulated BC tumorigenesis by absorbing miR‐508‐3p. It was found that miR‐508‐3p interference counteracted hsa_circ_0002082 knockdown mediated impairment of cell proliferation (Figure 6B–D) and expedition of cell apoptosis (Figure 6E,F). Additionally, the suppressive effects of hsa_circ_0002082 knockdown on cell migration and invasion abilities were also abated by miR‐508‐3p inhibitors (Figure 6G,H). Besides, miR‐508‐3p inhibitors reversed sh‐hsa_circ_0002082‐induced arrest of EMT process (Figure 6I,J).
FIGURE 6.
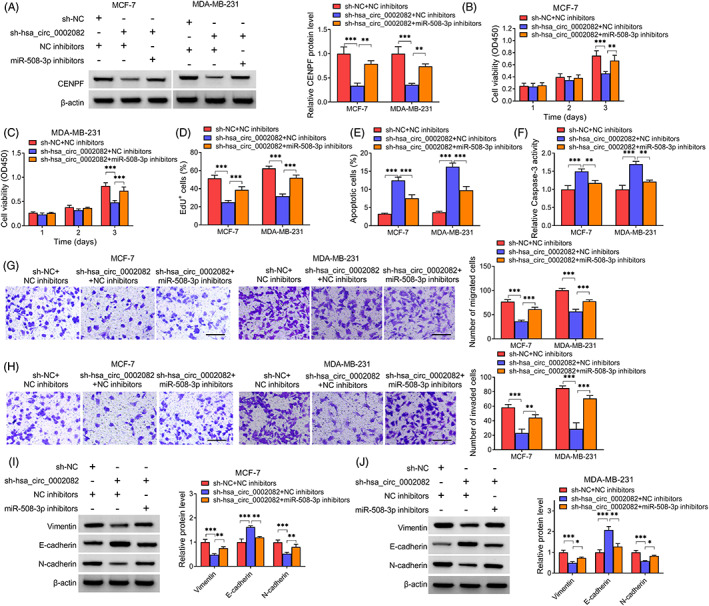
Knockdown of hsa_circ_0002082 suppresses BC cell growth and metastasis by miR‐508‐3p. After sh‐hsa_circ_0002082 and miR‐508‐3p inhibitors co‐transfection, (A) western blotting analysis of CENPF expression level. (B–D) Analysis of cell proliferation, (E, F) cell apoptosis, (G, H) cell migration and invasion. (I, J) Levels of EMT‐markers. *p < 0.05, **p < 0.01, ***p < 0.001
3.7. Knockdown of hsa_circ_0002082 impedes BC growth and EMT in vivo
Subsequently, in vivo assay was established with lentiviruses carrying sh‐hsa_circ_0002082 or sh‐NC. Hsa_circ_0002082 knockdown showed much smaller sizes and lower weights in tumors, indicating the inhibition of tumor growth (Figure 7A–C). Hsa_circ_0002082 expression in hsa_circ_0002082‐decreased tumor tissues was much lower (Figure 7D). Importantly, IHC staining revealed that CENPF, Vimentin and N‐cadherin contents were decreased, while E‐cadherin level was increased in xenograft tumor tissues derived from cell‐decreased hsa_circ_0002082 group (Figure 7E). Collectively, knockdown of hsa_circ_0002082 hindered BC growth and EMT in vivo.
FIGURE 7.
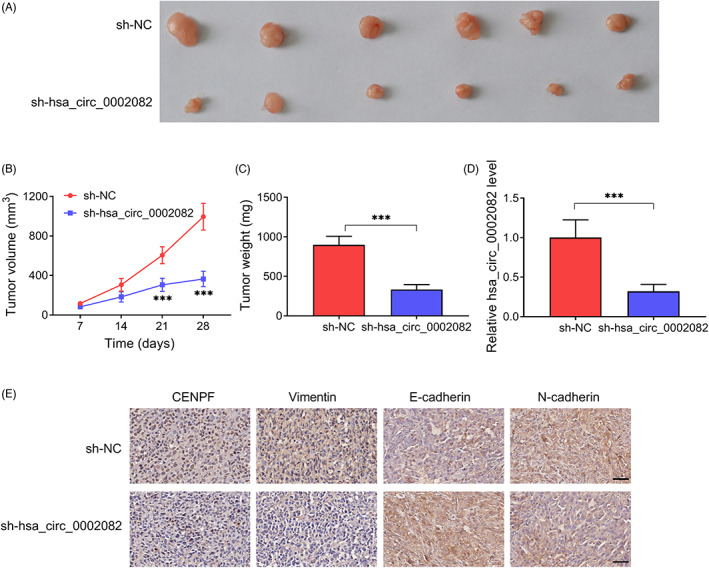
Knockdown of hsa_circ_0002082 impedes BC growth and EMT in vivo. (A) Representative images. (B) Growth curves. (C) Tumor weight was exhibited at the end point. (D) Level of hsa_circ_0002082 in xenograft tumors. (E) IHC staining in xenograft tumors. ***p < 0.001
4. DISCUSSION
Globally, BC is one of the most life‐threatening cancers in females ascribing to cancer distant metastasis and recurrence. 14 , 15 Our study first demonstrated that the hsa_circ_0002082/miR‐508‐3p/CENPF axis might contribute to the growth and metastasis of BC. CircRNAs have been identified to have functions in regulating complex human pathologies through controlling the alternation of cell proliferation, apoptosis, and metabolism. 16 , 17 For example, circCTDP1 up‐regulated Homeobox A10 through miR‐320b to expedite nasopharyngeal carcinoma tumorigenesis. 18 CircRNA‐5692 functioned as a tumor suppressor to repress HCC progression by miR‐328‐5p/disabled homolog 2‐interacting protein (DAB2IP) axis. 19 Besides, Wang et al. showed that circRNA‐002178 could promote lung adenocarcinoma progression by enhancing T‐cell exhaustion through miR‐34/programmed death‐ligand 1 (PD‐L1)/PD1 pathway. 20 Accordingly, circRNAs are considered to be the ideal biomarkers for cancer therapy. In the present work, an increased hsa_circ_0002082 was observed in BC tissues, which was consistent with the tendency of hsa_circ_0002082 in GSE182471 dataset. Thereafter, we confirmed that hsa_circ_0002082 deficiency could induce apoptosis and repress cell proliferative, migratory and invasive abilities and EMT process in vitro. Importantly, knockdown of hsa_circ_0002082 impeded BC growth and EMT in vivo.
Based on the ceRNA hypothesis, 21 , 22 we then probed the regulatory mechanism underlying the oncogenic action of hsa_circ_0002082, and discovered that hsa_circ_0002082 and CENPF shared the same microRNA response elements of miR‐508‐3p, and hsa_circ_0002082 sponged miR‐508‐3p to elevate CENPF expression, indicating the hsa_circ_0002082/miR‐508‐3p/CENPF axis in BC cells. MiRNAs are short segments of noncoding molecules with ~22 nucleotides, and have been reported to have pivotal roles in BC pathophysiology by modulating cellular biological processes. 23 , 24 As example, miR‐27a performed oncogenic action in BC by reinforcing cancer cell invasion and growth through blocking Glycogen Synthase Kinase 3 Beta. 25 MiR‐142‐3p induced apoptosis and impaired cancer stem cell properties in BC by targeting High Mobility Group A2 (HMGA2). 26 MiR‐508‐3p is a well‐identified tumor suppressor in many cancers, such as ovarian, 27 cervical 28 and renal cell carcinomas. 29 Also, miR‐508‐3p could impair BC cell EMT process and invasiveness through Zinc Finger E‐Box Binding Homeobox 1 in a targeted manner. 30 Consistently, we also proved the anticancer functions of miR‐508‐3p in BC, moreover, miR‐508‐3p was also able to stimulate apoptosis and suppress cell growth and metastasis in BC. What's more, we proved that miR‐508‐3p inhibition abated the anticancer action of hsa_circ_0002082 siRNA on BC.
CENPF, a transient kinetochore protein, is highly expressed in G2/M phase. 31 The up‐regulation of CENPF has been exhibited in many types of cancers, including BC. 32 , 33 High CENPF expression was linked with the bad outcome of BC, and accelerated the metastasis and proliferation of BC cells, indicating the oncogenic role of CENPF in BC development. 33 , 34 Thereafter, this study verified that CENPF up‐regulation attenuated the tumor‐suppressive functions of miR‐508‐3p in BC cells, implying the miR‐508‐3p/CENPF axis in BC cells.
Although some interesting results were found in this work, there are still some limitations. A larger cohort of sample sizes both in vitro and in vivo are essential to verify these conclusion. Besides, the function of hsa_circ_0002082 in other cancer types should be also examined in order to ensure its oncogenic action.
In all, we first confirmed that hsa_circ_0002082 functioned oncogene to promote BC progression via miR‐508‐3p/CENPF axis, filling the gap in knowledge for the molecular contribution of hsa_circ_0002082 in BC progression. Besides, these data also implied that the use of hsa_circ_0002082 siRNA might be a promising method for BC therapy.
CONFLICT OF INTEREST
The author declare that they have no conflicts of interest.
Supporting information
Figure S1
Figure S2
Data S1
Liu Y, Liu Y, Luo J, Zhao W, Hu C, Chen G. Hsa_circ_0002082 up‐regulates Centromere Protein F via abolishing miR‐508‐3p to promote breast cancer progression. J Clin Lab Anal. 2022;36:e24697. doi: 10.1002/jcla.24697
Yu Liu and Yun Liu contributed equally in this study.
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are available from the corresponding author upon request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Hong D, Fritz AJ, Zaidi SK, et al. Epithelial‐to‐mesenchymal transition and cancer stem cells contribute to breast cancer heterogeneity. J Cell Physiol. 2018;233(12):9136‐9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428‐442. [DOI] [PubMed] [Google Scholar]
- 4. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng X, Huang M, Xing L, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple‐negative breast cancer. Mol Cancer. 2020;19(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang G, Ling Y, Mehrpour M, et al. Autophagy‐associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR‐195‐5p to promote triple‐negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR‐182‐5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638‐1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen L, Kong R, Wu C, et al. Circ‐MALAT1 functions as both an mRNA translation brake and a microRNA sponge to promote self‐renewal of hepatocellular cancer stem cells. Adv Sci (Weinh). 2020;7(4):1900949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sang M, Wu M, Meng L, et al. Identification of epithelial‐mesenchymal transition‐related circRNA‐miRNA‐mRNA ceRNA regulatory network in breast cancer. Pathol Res Pract. 2020;216(9):153088. [DOI] [PubMed] [Google Scholar]
- 12. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67‐79. [DOI] [PubMed] [Google Scholar]
- 13. Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mayor S. A fifth of women with breast cancer have a recurrence, new UK figures show. BMJ. 2012;344:e4085. [DOI] [PubMed] [Google Scholar]
- 15. Peart O. Metastatic Breast Cancer. Radiol Technol. 2017;88(5):519m‐539m. [PubMed] [Google Scholar]
- 16. Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marques‐Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. Noncoding RNAs, cytokines, and inflammation‐related diseases. FASEB J. 2015;29(9):3595‐3611. [DOI] [PubMed] [Google Scholar]
- 18. Li H, You J, Xue H, Tan X, Chao C. CircCTDP1 promotes nasopharyngeal carcinoma progression via a microRNA‐320b/HOXA10/TGFβ2 pathway. Int J Mol Med. 2020;45(3):836‐846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Z, Yu Y, Huang Z, et al. CircRNA‐5692 inhibits the progression of hepatocellular carcinoma by sponging miR‐328‐5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J, Zhao X, Wang Y, et al. circRNA‐002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384‐388. [DOI] [PubMed] [Google Scholar]
- 22. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plantamura I, Cataldo A, Cosentino G, Iorio MV. miR‐205 in breast cancer: state of the art. Int J Mol Sci. 2020;22(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X, Zeng Z, Wang J, et al. MicroRNA‐9 and breast cancer. Biomed Pharmacother. 2020;122:109687. [DOI] [PubMed] [Google Scholar]
- 25. Chen H, Zhang Y, Cao X, Mou P. MiR‐27a facilitates breast cancer progression via GSK‐3β. Technol Cancer Res Treat. 2020;19:1533033820965576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mansoori B, Duijf PHG, Mohammadi A, et al. MiR‐142‐3p targets HMGA2 and suppresses breast cancer malignancy. Life Sci. 2021;276:119431. [DOI] [PubMed] [Google Scholar]
- 27. Guo F, Zhang K, Li M, et al. miR‐508‐3p suppresses the development of ovarian carcinoma by targeting CCNA2 and MMP7. Int J Oncol. 2020;57(1):264‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu P, Zhou G, Zhang X, Song G, Zhan L, Cao Y. Long non‐coding RNA Linc00483 accelerated tumorigenesis of cervical cancer by regulating miR‐508‐3p/RGS17 axis. Life Sci. 2019;234:116789. [DOI] [PubMed] [Google Scholar]
- 29. Han B, Shaolong E, Luan L, Li N, Liu X. CircHIPK3 promotes clear cell renal cell carcinoma (ccRCC) cells proliferation and metastasis via altering of miR‐508‐3p/CXCL13 signal. Onco Targets Ther. 2020;13:6051‐6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo SJ, Zeng HX, Huang P, Wang S, Xie CH, Li SJ. MiR‐508‐3p inhibits cell invasion and epithelial‐mesenchymal transition by targeting ZEB1 in triple‐negative breast cancer. Eur Rev Med Pharmacol Sci. 2018;22(19):6379‐6385. [DOI] [PubMed] [Google Scholar]
- 31. Shahid M, Lee MY, Piplani H, et al. Centromere protein F (CENPF), a microtubule binding protein, modulates cancer metabolism by regulating pyruvate kinase M2 phosphorylation signaling. Cell Cycle. 2018;17(24):2802‐2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim HE, Kim DG, Lee KJ, et al. Frequent amplification of CENPF, GMNN and CDK13 genes in hepatocellular carcinomas. PLoS One. 2012;7(8):e43223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun J, Huang J, Lan J, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer. Cancer Cell Int. 2019;19:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Q, Xu H, Zhu J, Feng K, Hu C. LncRNA MCM3AP‐AS1 promotes breast cancer progression via modulating miR‐28‐5p/CENPF axis. Biomed Pharmacother. 2020;128:110289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Data S1
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


