Abstract
A major surface antigenic lipoprotein of Mycoplasma agalactiae, promptly recognized by the host's immune system, was characterized. The mature product, P48, showed significant similarity and shared conserved amino acid motifs with lipoproteins or predicted lipoproteins from Mycoplasma fermentans, Mycoplasma hyorhinis, relapsing fever Borrelia spp., Bacillus subtilis, and Treponema pallidum.
Mycoplasma agalactiae is the etiologic agent of contagious agalactia (CA) of sheep and goats, a disease involving acute mastitis, arthritis, keratoconjunctivitis, and abortion when first introduced in a susceptible population. In areas of endemicity, symptoms are usually reduced to a subacute, sometimes silent, mastitis with rare articular and ocular lesions. Once established in a flock, M. agalactiae colonizes several host tissues and can be recovered from apparently healthy animals even several years after the first outbreak of the disease and/or the last clinical episode. In the last few years, several authors have described mechanisms by which mycoplasmas may evade immune response. Of these, the best understood entails the variability of membrane lipoproteins (4, 5, 7, 17, 18, 25–27, 31, 33, 34, 39–42), while others involve the ability of membrane lipoproteins and lipopeptides to induce the expression of up- and down-regulating cytokines (10, 16, 19, 21, 25).
Very little is known about surface antigens of M. agalactiae and, although the closest related species—Mycoplasma bovis and Mycoplasma fermentans (99.8 and 95.0% 16S rRNA similarity, respectively) (23)—possess the above-mentioned features, it has not yet been possible to identify related genes in M. agalactiae. In previous studies, a 45- to 55-kDa major antigen of M. agalactiae, promptly recognized among total proteins by sera from naturally (36) or experimentally (8) infected sheep, was shown to be a membrane protein sensitive to trypsin treatment of whole cells (24, 36, 37). The aim of this study was to identify the gene encoding it and predict its possible role in the pathogenesis of CA.
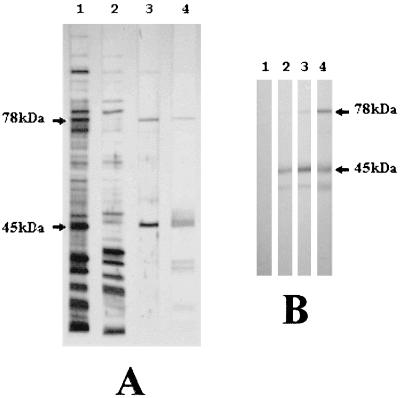
A Triton X-114 fraction (6, 24) of a field isolate from an outbreak of typical CA of sheep, MA7, was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting, with sera from naturally and experimentally infected sheep. Samples of whole organisms and aqueous and detergent phases were normalized to the original volumes and separated by SDS-PAGE, blotted on a nitrocellulose membrane, and visualized through India ink staining (11) (Fig. 1A, lanes 1 to 3). Western immunoblotting was performed with sera from naturally (Fig. 1A, lane 4) and experimentally infected sheep (Fig. 1B), as described elsewhere (8), in order to identify immunodominant membrane lipoproteins.
FIG. 1.
Characterization of the major surface antigens of M. agalactiae. (A) Total proteins (lane 1), soluble fraction (lane 2), and a Triton X-114 phase fraction (lane 3) were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and India ink stained (11). Detergent-phase membrane-bound proteins (lane 3) were also immunostained with serum of a symptomatic naturally infected sheep (lane 4). (B) Immunoblotting of Triton X-114 phase fractionated proteins from M. agalactiae with preimmune sheep serum (lane 1) and after 9 days (lane 2), 24 days (lane 3), and 57 days (lane 4) of experimental infection (8).
The Triton X-114 phase was resolved by SDS-PAGE and electroblotted on a polyvinylidene difluoride membrane (BioRad), and protein bands were visualized by Coomassie blue staining. The band of interest was cut and the N-terminal sequence was determined by Edman degradation by using an automated Applied Biosystems 477A gas-phase sequencer (Applied Biosystems Inc., Foster City, Calif.); PTH-derivative amino acids were identified by RP-high-performance liquid chromatography (Applied Biosystems 120A). A 12-amino-acid residue sequence was obtained from the N terminus of the 45- to 50-kDa membrane protein: AS(X)GDKYFKETE, in which X could be a modified C residue. We then synthesized a nondegenerate 24-residue oligonucleotide corresponding to the peptide sequence DKYFKETE and designed based on the preferred codon use of closely related mycoplasmas in order to avoid highly redundant nucleotide sequences. The probe sequence was 5′GATAAATATTTTAAAGAAACTGAA3′.
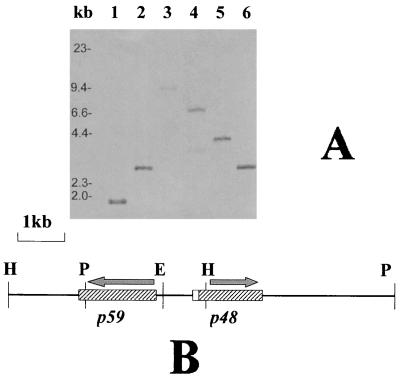
The oligonucleotide was 3′ tailing labeled with digoxigenin-dUTP (Boehringer Mannheim) by using terminal transferase. The tailing mixture contained dCTP in place of dATP in order to minimize the risk of nonspecific annealing to AT-rich regions, which are a common feature in mycoplasma DNA sequences. Genomic DNA was digested to completion with a panel of restriction enzymes, in various combinations (Fig. 2), resolved by 0.8% agarose gel electrophoresis and was capillary transferred to positively charged nylon membranes (Boehringer Mannheim). Hybridization, washing, and nonradioactive detection were done following the digoxigenin system user's guide (Boehringer Mannheim). A 4.1-kb HindIII fragment and a 6.6-kb PstI DNA fragment, identified by Southern analysis, were gel purified and ligated into digested and dephosphorylated pUC18 cloning vector (28). Positives clones were identified by standard colony hybridization and confirmed by slot blot hybridization with the same oligoprobe. Transformants were grown on a selective medium, and maxi-preps of plasmid DNA were cycle sequenced by using vector universal sequencing primers and sequence-generated primers on an ABI 373 DNA sequencer (Applied Biosystems) by the dideoxy chain termination method with fluorescence dye terminators (Perkin-Elmer). We partially sequenced the 4.1-kb HindIII fragment, with vector universal primers, and located a specific partial open reading frame (ORF), consistent with the results of N-terminal sequencing, 300 bp upstream of the 3′ terminus of the insert. The complete ORF was then sequenced in the 6.6-kb PstI fragment, which overlaps the former by 2 kb, with primers designed based on the previous sequence. A 4,521-bp sequence was obtained. Nucleotide and protein sequences were submitted to Orf-finder and BLAST sequence similarity searching (1, 2) at the National Center for Biotechnology Information (NCBI) web site and to the ExPasy web site facilities of the Swiss Institute of Bioinformatics (3). Conserved motifs identified by BLAST analysis were scanned in the SWISS-PROT database (43), and protein sequences of interest were aligned by using CLUSTAL W (14, 35); the multiple alignment was analyzed by GeneDoc (22).
FIG. 2.
Identification of the P48 gene in the M. agalactiae genome. (A) Typical results obtained by Southern analysis. Genomic DNA (5 μg/well) was digested with BglII and EcoRI (lane 1), EcoRI (lane 2), XbaI (lane 3), PstI (lane 4), HindIII (lane 5), and PstI and HindIII (lane 6). After electrophoresis and capillary transfer to a positively charged nylon membrane, digested DNA was hybridized with a digoxigenin-labeled oligoprobe derived from the N-terminal microsequencing of P48. Two overlapping fragments, namely the 4-kb HindIII and 6.6-kb PstI fragments, were gel purified, cloned in pUC18, and partially sequenced. (B) Physical map and partial characterization of coding regions corresponding to the 4-kb HindIII and 6.6-kb PstI overlapping fragments. The P48 gene is a malp (7) homolog and encodes a 22-residue leader peptide (white box), followed by the mature P48 lipoprotein (right dashed box). Another ORF (left dashed box) was found in opposite orientation and encodes a putative 59-kDa homolog of the P63 ABC transporter of M. fermentans. H, P, and E indicate HindIII, PstI, and EcoRI restriction sites, respectively.
The complete specific ORF nucleotide sequence corresponds to a lipoprotein-encoding gene. A 48-kDa mature lipoprotein (P48) derives from the cleavage of a typical leader peptide at the site VAASC immediately upstream of the cysteine residue, which is presumably the acylation site to which palmitate binds (32). Four UGA codons, as in all organisms belonging to the genus Mycoplasma, are translated into tryptophan. The termination of transcription or translation occurs at the level of three in-frame TAA stop codons followed by an imperfect inverted repeat sequence, which presumably forms a hairpin-like secondary structure. Two ORFs flank the P48 gene on the opposite strand, so that the hairpin terminator presumably functions even in the opposite orientation for the downstream ORF. The amino acid sequence was submitted to BLAST and BLAST 2.0 (1, 2) at the NCBI web site, and a significant similarity was found with two M. fermentans products and a Mycoplasma arginini product. The former products, P48 and Ag161, earlier described as human-derived tumor cell products, activating the differentiation of monocytes (19) and targeting homologue C′3 activation (20), respectively, are encoded by the same M. fermentans gene, malp, which also encodes a macrophage-activating lipopeptide (MALP-2) (7). The latter, originally described as an M. arginini metastasis-promoting factor (38), has recently been shown to be the P47 lipoprotein of Mycoplasma hyorhinis (7). Nucleotide sequencing of the P48 gene flanking regions confirmed the homology with malp. Indeed, although in opposite orientations, both lie upstream from a putative ABC transporter operon (7, 33), partially sequenced in this study. P48 also shows a lower degree of similarity (16% identical residue and 33% conserved substitutions) with the variable adherence-associated antigen, P50 adhesin, of Mycoplasma hominis (12, 13), but unlike P48, P50 is organized in repetitive blocks of amino acid sequences (41, 42). This organization is common to several surface lipoproteins of mycoplasmas and mammal pathogens and is consistent with a characteristic variability aimed at evading the immune response (4, 17, 25, 30, 31, 40). This study did not aim to identify variable expression of surface antigens, but posttranscriptional and posttranslational modification of P48 cannot be excluded a priori, as recently suggested for its closest homolog, MALP-404 of M. fermentans (7). On the basis of BLAST results, we identified two conserved motifs, SFNQS and IGVD-DQ. Both motifs were used to scan for a pattern in the SWISS-PROT database (43). Some of the protein sequences carrying both motifs were aligned by using CLUSTAL W (14, 35), and the multiple alignment was analyzed by GeneDoc (22). A longer version of the former motif, SLA (selective lipoprotein associated), distributed among selected lipoproteins, has been recently identified by Calcutt et al. (7). The use of two short motifs, SLA-1 (our shorter version of SLA) and SLA-2 (IGVD-DQ), led us to individuate a larger family of bacterial lipoproteins or putative products bearing them (Table 1). In particular, P48 homologs, not identified by SLA, were found in the complete genome sequences from Mycoplasma genitalium (9) and Mycoplasma pneumoniae (15) (MG040 and its M. pneumoniae homolog). On the other hand, two related products identified by Calcutt et al., namely the CD4+ T-cell-stimulating antigen (TCSA) from Listeria monocytogenes and the P20 hypothetical lipoprotein from Mycoplasma capricolum, were initially excluded because of the apparent absence of SLA-2. Actually, the TCSA published sequence is truncated immediately upstream of the site that might contain the SLA-2 motif by the vector cloning site (29). Nevertheless, it is noteworthy that, in the original paper (29), the CD4+ T-cell-stimulating activity of this product had been assigned to the published truncated version. On the contrary, the hypothetical P20 lipoprotein from M. capricolum was excluded even if an analysis of its encoding nucleotide sequence, retrieved from the GenBank database (accession no. Z33368.1), confirmed its homology with P48 and related products. In fact, (i) two partially overlapping additional ORFs downstream of the P20-encoding sequence should encode SLA-2, and (ii) an ABC transporter-encoding sequence is located immediately downstream from the above-mentioned malp homolog sequence.
TABLE 1.
Bacterial lipoproteins bearing SLAa motifs
| Species and product | Function(s) | SLA-1 motif (position) | SLA-2 motif (position) |
|---|---|---|---|
| M. agalactiae P48 | Surface antigen | DESFNQS (78–84) | FIIGVDADQ (318–326) |
| M. fermentans M161Ag or MALP-404 | Inducing cytokine production by human monocytes | DKSFNQS (75–81) | YVIGVDSDQ (290–298) |
| M. hyorhinis P47 (formerly M. arginini Ag243-5) | Metastasis-promoting activity | DKSFNQS (72–78) | YLIGVDTDQ (306–314) |
| M. genitaliumb MG040 | Putative membrane lipoprotein | DKSFSEM (54–60) | AIIGVDSAQ (358–366) |
| M. pneumoniaeb MG040 homolog | Putative membrane lipoprotein | DKSFSQM (54–60) | AVIGVDSAQ (359–367) |
| Treponema pallidum TMPC | Membrane lipoprotein C, 35-kDa antigen | DKSFNQQ (53–59) | WVIGVDRDQ (253–261) |
| Bacillus subtilis YUFN | Putative membrane lipoprotein | DKSFNQS (43–49) | WVIGVDKDQ (246–254) |
| Borrelia burgdorferibc BMPA | Basic membrane protein A, immunodominant antigen | DKSFNES (41–47) | YIIGVDEDQ (237–245) |
| B. burgdorferic BMPB | Basic membrane protein B, immunodominant antigen | DKSFNSS (40–46) | YVIGADQDQ (242–250) |
| L. monocytogenes TCSA | CD4+ TCSA | DRSFNQS (54–60) | —d |
SLA, selective lipoprotein-associated motifs (reference 7 and this work).
Not individuated by Calcutt et al. (7) by using a longer motif to scan database.
BMPA and BMPB of Borrelia garinii and Borrelia afzelii were omitted even if they bore both SLA motifs.
—, the TCSA published sequence is truncated immediately upstream of the site that might contain the SLA-2 motif by the vector cloning site (29).
Further research will be necessary to understand whether the SLA-1 and SLA-2 conserved amino acid motifs identified in this study will be helpful in assigning a more specific biological function to the proteins bearing them. Calcutt et al. (7), who also identified the longer version of SLA-1, speculate about its possible involvement in autoimmune disease or in targeting posttranslational proteolytic processing.
Establishing relations with biological and immunomodulatory features of M. fermentans and M. hyorhinis homologs would help to clarify the role of P48 in CA pathogenesis. Moreover, analogies between the arthritogenic properties of M. agalactiae and M. fermentans could lead to CA being considered an animal model of human mycoplasma arthritis.
Nucleotide sequence accession number.
The sequences reported in this paper were deposited in the EMBL database under accession no. AJ132423.
Acknowledgments
This work was supported by grants from MURST and Assessorato alla Programmazione, Regione Autonoma Sardegna, Progetto Biotecnologie.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel R D, Bairoch A, Hochstrasser D F. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 4.Behrens A, Heller M, Kirchhoff H, Yogev D, Rosengarten R. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect Immun. 1994;62:5075–5084. doi: 10.1128/iai.62.11.5075-5084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens A, Heller M, Rosenbusch R, Kirchhoff H. Immunoelectron microscopic localization of variable proteins on the surface of Mycoplasma bovis. Microbiology. 1996;142:1863–1871. doi: 10.1099/13500872-142-7-1863. [DOI] [PubMed] [Google Scholar]
- 6.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 7.Calcutt M J, Kim M F, Karpas A B, Muhlradt P F, Wise K S. Differential posttranslational processing confers intraspecies variation of a major surface lipoprotein and a macrophage-activating lipopeptide of Mycoplasma fermentans. Infect Immun. 1999;67:760–771. doi: 10.1128/iai.67.2.760-771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contini A, Pittau M, Cuccuru C, Marcello P, Briguglio P, Fadda M. Experimental infection of sheep with Mycoplasma agalactiae: studies of antibodies response. Note 1. Atti Soc Ital Sci Vet. 1989;43:1119–1123. [Google Scholar]
- 9.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J F, Dougherty B A, Bott K F, Hu P C, Lucier T S, Peterson S N, Smith H O, Hutchison III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 10.Hall R E, Agarwal S, Kestler D P, Cobb J A, Goldstein K M, Chang N-S. cDNA and genomic cloning and expression of the P48 monocytic differentiation/activation factor, a Mycoplasma fermentans gene product. Biochem J. 1996;319:919–927. doi: 10.1042/bj3190919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock K, Tsang V C W. India ink staining of proteins on nitrocellulose paper. Anal Biochem. 1983;133:157–162. doi: 10.1016/0003-2697(83)90237-3. [DOI] [PubMed] [Google Scholar]
- 12.Henrich B, Feldmann R-C, Hadding U. Cytoadhesins of Mycoplasma hominis. Infect Immun. 1993;61:2945–2951. doi: 10.1128/iai.61.7.2945-2951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henrich B, Kitzerov A, Feldmann R-C, Schaal H, Hadding U. Repetitive elements of the Mycoplasma hominis adhesin p50 can be differentiated by monoclonal antibodies. Infect Immun. 1996;64:4027–4034. doi: 10.1128/iai.64.10.4027-4034.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 15.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostyal D A, Butler G H, Beezhold D H. A 48-kilodalton Mycoplasma fermentans membrane protein induces cytokine secretion by human monocytes. Infect Immun. 1994;62:3793–3800. doi: 10.1128/iai.62.9.3793-3800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lysnyansky I, Rosengarten R, Yogev D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J Bacteriol. 1996;178:5395–5401. doi: 10.1128/jb.178.18.5395-5401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto M, Nishiguchi M, Kikkawa S, Nishimura H, Nagasawa S, Seya T. Structural and functional properties of complement-activating protein M161Ag, a Mycoplasma fermentans gene product that induces cytokine production by human monocytes. J Biol Chem. 1998;273:12407–12414. doi: 10.1074/jbc.273.20.12407. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto M, Takeda J, Inoue N, Hara T, Hatanaka M, Takahashi K, Nagasawa S, Akedo H, Seya T. A novel protein that participates in nonself discrimination of malignant cells by homologous complement. Nat Med. 1997;3:1266–1270. doi: 10.1038/nm1197-1266. [DOI] [PubMed] [Google Scholar]
- 21.Mühlradt P F, Kiess M, Meyer H, Süssmuth R, Jung G. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J Exp Med. 1997;185:1951–1958. doi: 10.1084/jem.185.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholas K B, Nicholas H B, Jr, Deerfield D W., II GeneDoc: analysis and visualization of genetic variation. EMBNEWNEWS. 1997;4:14. [Google Scholar]
- 23.Pettersson B, Uhlén M, Johansson K E. Phylogeny of some mycoplasmas from ruminants based on 16S rRNA sequences and definition of a new cluster within the hominis group. Int J Syst Bacteriol. 1996;46:1093–1098. doi: 10.1099/00207713-46-4-1093. [DOI] [PubMed] [Google Scholar]
- 24.Pittau M, Fadda M, Briguglio P, Farina S, Carboni A Q, Contini A. Triton X-114 phase fractionation of Mycoplasma agalactiae membrane proteins and affinity purification of specific antibodies. Atti Soc Ital Sci Vet. 1990;44:925–928. [Google Scholar]
- 25.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosengarten R, Behrens A, Stetefeld A, Heller M, Ahrens M, Sachse K, Yogev D, Kirchhoff H. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect Immun. 1994;62:5066–5074. doi: 10.1128/iai.62.11.5066-5074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosengarten R, Wise K S. The Vlp system of Mycoplasma hyorhinis: combinatorial expression of distinct variant lipoproteins generating high-frequency surface antigenic variation. J Bacteriol. 1991;173:4782–4793. doi: 10.1128/jb.173.15.4782-4793.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Sanderson S, Campbell D J, Shastri N. Identification of a CD4+ T cell-stimulating antigen of pathogenic bacteria by expression cloning. J Exp Med. 1995;182:1751–1757. doi: 10.1084/jem.182.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seifert H S, So M. Genetic mechanisms of bacterial antigenic variation. Microbiol Rev. 1988;52:327–336. doi: 10.1128/mr.52.3.327-336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons W I, Zuhua C, Glass J I, Simecka J W, Cassel G H, Watson H L. Sequence analysis of the chromosomal region around and within the V-1 encoding gene of Mycoplasma pulmonis: evidence for DNA inversion as a mechanism for V-1 variation. Infect Immun. 1996;64:472–479. doi: 10.1128/iai.64.2.472-479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutcliffe I C, Russell R R. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theiss P, Wise K S. Localized frameshift mutation generates selective, high-frequency phase variation of a surface lipoprotein encoded by a mycoplasma ABC transporter operon. J Bacteriol. 1997;179:4013–4022. doi: 10.1128/jb.179.12.4013-4022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Theiss P, Kim M F, Wise K S. Differential protein expression and surface presentation generate high-frequency antigenic variation in Mycoplasma fermentans. Infect Immun. 1993;61:5123–5128. doi: 10.1128/iai.61.12.5123-5128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tola S, Manunta D, Cocco M, Turrini F, Rocchigiani A M, Idini G, Angioi A, Leori G. Characterization of membrane surface proteins of Mycoplasma agalactiae during natural infection. FEMS Microbiol Lett. 1997;154:355–362. doi: 10.1111/j.1574-6968.1997.tb12667.x. [DOI] [PubMed] [Google Scholar]
- 37.Tola S, Idini G, Manunta D, Casciano I, Rocchigiani A M, Angioi A, Leori G. Comparison of Mycoplasma agalactiae isolates by pulsed field gel electrophoresis, SDS-PAGE and immunoblotting. FEMS Microbiol Lett. 1996;143:259–265. doi: 10.1111/j.1574-6968.1996.tb08490.x. [DOI] [PubMed] [Google Scholar]
- 38.Ushio S, Iwaki K, Taniai M, Otha T, Fukuda S, Sugimura K, Kurimoto M. Metastasis-promoting activity of a novel molecule, Ag 243-5, derived from mycoplasma, and the complete nucleotide sequence. Microbiol Immunol. 1995;39:393–400. doi: 10.1111/j.1348-0421.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 39.Yogev D, Menaker D, Strutzberg K, Levisohn S, Kirchhoff H, Hinz K H, Rosengarten R. A surface epitope undergoing high-frequency phase variation is shared by Mycoplasma gallisepticum and Mycoplasma bovis. Infect Immun. 1994;62:4962–4968. doi: 10.1128/iai.62.11.4962-4968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yogev D, Watson-McKown R, Rosengarten R, Im J, Wise K S. Increased structural and combinatorial diversity in an extended family of genes encoding Vlp surface proteins of Mycoplasma hyorhinis. J Bacteriol. 1995;177:5636–5643. doi: 10.1128/jb.177.19.5636-5643.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, Wise K S. Localized reversible frameshift mutation in an adhesin gene confers a phase-variable adherence phenotype in mycoplasma. Mol Microbiol. 1997;25:859–869. doi: 10.1111/j.1365-2958.1997.mmi509.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Wise K S. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect Immun. 1996;64:2737–2744. doi: 10.1128/iai.64.7.2737-2744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Schäffer A A, Miller W, Madden T L, Lipman D J, Koonin E V, Altschul S F. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]