Abstract
CD26/dipeptidyl peptidase IV (DPPIV) is a cell surface ectoenzyme which participates in immune and inflammatory reactions. We found that CD26 was only partially expressed on human fibroblasts from periodontal tissues, whereas fibroblasts from lung and skin expressed CD26 constitutively as revealed by flow cytometry. We examined the possible upregulation of CD26 expression on human gingival fibroblasts in response to various stimulants. Interleukin-1α (IL-1α); tumor necrosis factor alpha; gamma interferon; lipopolysaccharide from Porphyromonas gingivalis, Prevotella intermedia, and Escherichia coli; and Prevotella glycoprotein augmented CD26 expression on gingival fibroblasts. Among the stimulants, IL-1α exhibited the most potent activity. Enzymatic activity of CD26 induced by IL-1α on fibroblasts was determined colorimetrically in terms of Gly-Pro hydrolysis of a synthetic chromogenic substrate, Gly-Pro p-nitroanilide. Among various inhibitors tested, diprotin A and phenylmethylsulfonyl fluoride inhibited the enzymatic activity, suggesting that the enzyme induced by IL-1α was DPPIV. The upregulation of CD26 mRNA expression upon stimulation with IL-1α was also revealed by a quantitative reverse transcription-PCR assay. In the kinetic experiment, 48 h and several days were required for maximum CD26 mRNA accumulation and CD26 molecule expression on the cell surface, respectively. The addition of cycloheximide at 2 h before IL-1α stimulation almost completely inhibited the accumulation of CD26 mRNA. These results suggested that induction of CD26 on human gingival fibroblasts is regulated at the transcriptional level and is also dependent on a de novo-synthesized protein factor(s).
Fibroblasts are the predominant cell type in the connective tissues of various organs. They are considered to function in the support of frameworks by synthesis of extracellular matrix such as collagens and in tissue turnover and repair. Recent studies have suggested that fibroblasts actively participate in inflammatory and immune responses. Fibroblasts produce various inflammatory cytokines such as interleukin-1 (IL-1) and IL-6 and chemokines such as IL-8, which in turn influence other cells (30, 31, 33, 34), and conversely, fibroblasts are regulated by inflammatory cytokines such as IL-1, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ), as well as bacterial components (8, 23, 26). Furthermore, fibroblasts are known to be heterogeneous in function and morphology (7).
Recently, it has been reported that cell surface ectoenzymes play an important role in inflammatory and immunological responses and in cell differentiation by extracellular degradation or modification of biologically active peptides, cytokines, and other cell surface proteins (16, 28). CD26 is a cell surface ectoenzyme, dipeptidylpeptidase IV (DPPIV) (EC 3.4.14.5), and is a highly glycosylated type II membrane sialoglycoprotein comprising two identical subunits of approximately 110 kDa. CD26/DPPIV has a unique specificity: it cleaves dipeptides from the N terminus of a polypeptide if proline is at the penultimate position. Peptides are also cleaved if alanine or hydroxyproline occupies the next (P1) position (28). Since N termini containing Xaa-Pro are not easily cleaved by other proteinases, the action of DPPIV is a rate-limiting step in the degradation of polypeptides. The only other proteinase with a similar specificity, DPPII (EC 3.4.14.2), has a lysosomal localization (6). CD26/DPPIV was originally characterized as a T-cell differentiation antigen and was reported to be expressed on various cell types, such as renal proximal tubules, intestinal epithelial cells, biliary caniliculae, alveolar pneumocytes, and skin fibroblasts (6, 28).
Many biologically active polypeptides have the sequence Xaa-Pro at the N terminus; therefore DPPIV may be essential for determining their biological half-lives. These substrates include substance P, chorionic gonadotropin, monomeric fibrin, and several releasing hormones which could regulate the biological reactions (40). Recently, it was reported that CD26/DPPIV was able to induce the N-terminal truncation of several chemokines, including RANTES (for regulated on activation, normal T-cell expressed and secreted) and stromal cell-derived factor-1 (SDF-1). CD26-mediated processing of these chemokines resulted in the alteration of chemokine receptor specificity (19) or abrogation of chemotactic activities (27). Furthermore, truncated RANTES inhibited infection of mononuclear cells by a macrophage-tropic human immunodeficiency virus type 1 (HIV-1) strain (19, 21). In contrast to truncated RANTES, truncated SDF-1 diminished the potency to inhibit HIV-1 infection (27). These findings suggest that CD26/ DPPIV regulates inflammatory and immunological responses.
In this study, we compared CD26/DPPIV expression in various human fibroblasts from different tissues. We found that fibroblasts from periodontal tissues only partially expressed CD26/DPPIV on their surfaces, in contrast to those from skin and lung, which expressed this molecule constitutively. We then investigated possible upregulation of CD26/DPPIV expression on human gingival fibroblasts (HGF) in response to inflammatory cytokines and cell surface components from periodontal disease-associated bacteria. We also investigated the characteristics of this enzyme and the regulatory mechanisms, and we discuss possible involvement of the enzyme in the pathogenesis of periodontal diseases.
MATERIALS AND METHODS
Reagents.
Glycyl-prolyl p-nitroanilide (Gly-Pro-pNA), p-nitroanilide (pNA), diprotin A, phorbol 12-myristate 13-acetate (PMA), phenylmethylsulfonyl fluoride (PMSF), leupeptin, pepstatin A, bestatin, aprotinin, 1,10-phenanthroline, bovine serum albumin (BSA), and cycloheximide (CHX) were obtained from Sigma Chemical Co. (St. Louis, Mo.). α-Minimum essential medium (α-MEM) and 0.25% trypsin–1 mM EDTA were from Gibco BRL (Rockville, Md.). Fetal calf serum (FCS) was obtained from Flow Laboratories (McLean, Va.). Anti-CD26 monoclonal antibody (MAb) (M-A261; mouse immunoglobulin G1 [IgG1]) was purchased from Pharmingen (San Diego, Calif.). Anti-CD14 MAb (MEM-18; mouse IgG1) was purchased from Monosan (Uden, The Netherlands). Isotype control antibody (Ab) (679.1Mc7; mouse IgG1) was purchased from Coulter (Miami, Fla.). Human natural IFN-γ (antiviral activity, 8.0 × 106 IU/mg of protein) was kindly provided by the Hayashibara Bioscience Institute (Okayama, Japan). Human recombinant IL-1α, anti-human IL-1α rabbit serum, and human recombinant TNF-α were obtained from Dainippon Pharmaceutical Co. Ltd. (Osaka, Japan).
Cells and cell culture.
HGF were prepared from the explants of normal gingival tissues of 8- to 25-year-old patients, with informed consent, as described previously (31). Human periodontal ligament (PDL) fibroblasts were prepared from the root surfaces of healthy human erupted third molars. Ligamental tissues were obtained by scraping the root surface. Both explants were cut into pieces and cultured in 100-mm-diameter tissue culture dishes (Falcon; Becton Dickinson Labware, Lincoln Park, N.J.) in α-MEM supplemented with 10% FCS with a medium change every 3 days for 10 to 15 days until confluent cell monolayers were formed. The cells were detached with 0.25% trypsin–1 mM EDTA, washed with phosphate-buffered saline (PBS), and subcultured in plastic flasks (Corning Coster, Acton, Mass.). After three or four subcultures by trypsinization, homogeneous, slim, spindle-shaped cells grown in characteristic swirls were obtained. The cells were used as confluent monolayers at subculture levels 5 through 15. Human skin fibroblasts (SF-MA) and human lung fibroblasts (WI-38, MRC-5, and IMR-90) were obtained from the Japanese Cancer Research Resources Bank (Tokyo, Japan). Human skin fibroblasts (FS-4) were generously supplied by M. Kohase, National Institute of Infectious Diseases (Tokyo, Japan). These fibroblasts were maintained in α-MEM supplemented with 10% FCS.
Preparations of bacterial components.
Lipopolysaccharide (LPS) was prepared from Porphyromonas gingivalis 381 and Prevotella intermedia ATCC 25611 by the hot phenol-water extraction method as described previously (14, 32). LPS was also extracted from P. intermedia ATCC 25611 by use of a phenol-chloroform-petroleum ether mixture (PCP) (14). Hot phenol-water-extracted LPS from Escherichia coli (O127:B8) was purchased from Sigma Chemical Co. Prevotella glycoprotein (PGP), which has been reported to show stimulatory activity on HGF and to possess no Limulus activity, was prepared as described previously (14). Fimbriae prepared from P. gingivalis 381 (18) were generously supplied by T. Ogawa (Asahi University Dental School, Gifu, Japan).
Stimulation of fibroblasts.
Fibroblasts were cultured in 96-well multiplates for enzyme assay, in 24-well multiplates for flow cytometry, and in 6-well multiplates for reverse transcription-PCR (RT-PCR). The final volumes were 200 μl, 1 ml, and 5 ml of 10% FCS–α-MEM, respectively. When the cultured cells were almost confluent, the culture media were renewed and various stimulants were added and then incubated for the indicated times.
Immunostaining.
Fibroblasts were collected by trypsinization, washed with PBS (pH 7.2), and used for staining. To determine the expression of CD26 and CD14, 105 fibroblasts were incubated at 4°C for 30 min with 1 μg of anti-CD26 MAb (M-A261) and anti-CD14 MAb (MEM-18) diluted in 0.1% NaN3–0.1% BSA in PBS. After being washed twice with 0.1% NaN3–0.1% BSA in PBS, the cells were stained with fluorescein-conjugated goat anti-mouse Igs (Biosource International, Camarillo, Calif.) at 4°C for a further 30 min and then washed twice more. Staining was analyzed on a FACScan fluorescence-activated cell analyzer (Becton Dickinson, Mountain View, Calif.). Data were collected for 5,000 events, which were stored in list mode and then analyzed with Lysis II software (Becton Dickinson).
Assay for DPPIV activity.
DPPIV activity was assayed by using a chromogenic substrate, Gly-Pro-pNA, as described previously (22). HGF (104) were seeded into 96-well flat-bottomed plates (Nunc, Roskilde, Denmark) in 200 μl of α-MEM with 10% FCS until confluent cell monolayers formed. Confluent HGF were stimulated with various cytokines and bacterial components for 6 days before being washed with PBS three times and used for enzyme assay. Proteolytic activity was determined by measurement of the amount of pNA formed in the supernatant at 405 nm. The confluent monolayer in the 96-well flat-bottomed plate was incubated at 37°C for the indicated times with 2 mM Gly-Pro-pNA in 100 mM HEPES buffer (pH 7.6) containing 0.12 M NaCl, 5 mM KCl, 1.2 mM MgSO4, 8 mM glucose, and 10 mg of BSA per ml. The plate was covered with an adhesive plate cover during the culture, and the final volume of the incubation mixture was 100 μl. Tests were run in triplicate; cell-free and substrate-free blanks were run in parallel. The results were expressed as nanomoles of pNA formed per hour per confluent-monolayer cell at 37°C. To examine the effects of potential inhibitors, cells were preincubated with inhibitors for 15 min at 37°C before addition of the substrate to the incubation mixture. To examine the effect of anti-CD14 MAb on DPPIV induction, the HGF monolayer in the 96-well plate was incubated with dialyzed anti-CD14 MAb (MY4, mouse IgG2b; Coulter) at 10 μg/ml or with dialyzed isotype control mouse IgG2b (Becton Dickinson) at 10 μg/ml at 37°C for 30 min. Antibody-treated cells were stimulated with 10 μg of E. coli LPS per ml for 6 days. To examine the effect of anti-IL-1α Ab on DPPIV induction by LPS, the HGF monolayer in the 96-well plate was incubated with 10 μg of E. coli LPS per ml for 6 days with addition of anti-IL-1α Ab at a 1:100 dilution at days 0, 1, and 2.
Real-time quantitative PCR.
The RNAgents Total RNA Isolation System (Promega Corporation, Madison, Wis.) was used for the extraction of total RNA from cultured fibroblasts according to the manufacturer's instructions. The application volume for RT-PCR was determined by electrophoresis. RT and real-time quantitative PCR were performed (9, 13) with use of EZ RT-PCR Core reagents (PE Applied Biosystems, Foster City, Calif.). Briefly, 100 ng of RNA was subjected to quantitative RT-PCR. The primers used for PCR had the following sequences: forward primer, 5′-GCTTGTCACCATCATCACCGT-3′, and reverse primer, 5′-AGTGTAAGTTTTGCGACTGTCAGC-3′. The reaction produced an 85-bp PCR product. The RT-PCR mixture (50 μl) contained Taq Man buffer A, 12.5 U of Tth DNA polymerase, a 300 nM concentration of each primer, 1.25 U of AmpliTaq Gold DNA polymerase, 5.5 mM MgCl2, 300 μM dATP, 300 μM dCTP, 300 μM dGTP, and 600 μM dUTP. The reaction mixture contained the following detection probe (200 nM): 5′-(FAM)TCTGCTGAACAAAGGCACAGATGATGCTAC(TAMRA)-3′, where FAM is 6-carboxyfluorescein and its emission spectrum is quenched by the second fluorescent dye, TAMRA (6-carboxy-tetramethylrhodamine). The nuclease degradation of the hybridization probe releases the quenching of the FAM fluorescent emission, resulting in an increase in peak fluorescent emission at 518 nm. All reactions were performed in an ABI Prism 7700 sequence detector (PE Applied Biosystems), which allows measurement of the fluorescent spectra of the 96 wells of the thermal cycler continuously during PCR amplification. The thermal cycler conditions were as follows: 35 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 20 s, and extension at 72°C for 30 s. Reaction conditions were programmed on a Mac Power PC 7200/120 linked directly to the model 7700 sequence detector. Analysis of data was performed with ABI Prism 7200/7700 sequence detection system software version 1.6.3. Briefly, ΔRn (reporter dye emission/quencher dye emission) was plotted on the y axis, and the PCR cycle number was plotted on the x axis. The threshold was determined from the data points collected from the baseline of the amplification plot. The point at which the amplification plot crosses the threshold was defined as the Ct value (cycle number at this point). Ct can be used as a quantitative measurement of the input target number with 105-order linearity. Because PCR products theoretically double in number every cycle, the difference between the target number can be calculated to be 2x in the case that the difference of the Ct value is x (9, 13).
Statistical analysis.
Most of the experiments were carried out in triplicate assays. The statistical significance between two means was analyzed by Student's unpaired t test. All experiments in this study were repeated to test the reproducibility of the results.
RESULTS
Expression of CD26/DPPIV on fibroblasts derived from various organs.
CD26 expression on fibroblasts derived from various organs was investigated by flow cytometry (Table 1). The number of cells expressing CD26 among fibroblasts from lung and skin was very high, with the averages being 94.7 and 84.1%, respectively. These results were consistent with previous reports (22) which stated that most dermal fibroblasts (more than 70%) expressed CD26. However, few fibroblasts from periodontal tissue (gingiva and periodontal ligaments) expressed CD26, with the averages being 20.8 and 17.5%, respectively. The mean fluorescence values were also correlated to the percent positivity of CD26. It has been reported that the phenotypic characteristics of fibroblasts depend on the organs or tissues from which the fibroblasts are derived (1, 2, 29, 31). These results suggested organ specificity in respect to CD26 expression on fibroblasts.
TABLE 1.
Expression of CD26 (DPPIV) and CD14 on human fibroblasts of various organsa
| Fibroblasts | Origin | % Positive cells (mean
fluorescence value)
|
|
|---|---|---|---|
| CD26 | CD14 | ||
| MH-12 | Gingiva | 32.5 (15.0) | 56.1 |
| KEK | Gingiva | 32.9 (11.1) | 42.3 |
| NIK | Gingiva | 5.2 (5.6) | 61.3 |
| YS-G | Gingiva | 22.6 (5.7) | 69.9 |
| AK-G | Gingiva | 5.2 (4.2) | 58.6 |
| FE | Gingiva | 26.1 (8.4) | NDb |
| AK | Periodontal ligament | 19.7 (3.8) | 24.9 |
| TT | Periodontal ligament | 7.7 (5.2) | ND |
| YS | Periodontal ligament | 11.1 (7.9) | 14.6 |
| KS | Periodontal ligament | 18.1 (4.5) | 10.9 |
| IMR-90 | Lung | 96.9 (51.9) | 1.1 |
| WI-38 | Lung | 88.1 (94.5) | 3.9 |
| MRC-5 | Lung | 99.2 (95.4) | 0.5 |
| FS-4 | Skin | 87.2 (88.4) | 0.7 |
| SF-MA | Skin | 80.9 (96.8) | 1.9 |
Fibroblasts derived from various organs were maintained in α-MEM supplemented with 10% FCS. Cells were collected from confluent monolayers with trypsin-EDTA, and CD26 and CD14 antigens on the cell surface were assessed by flow cytometry with indirect staining as described in Materials and Methods. Isotype-matched Abs were used as negative controls.
ND, not done.
Induction of CD26 expression on the surface of HGF in response to cytokines and bacterial components.
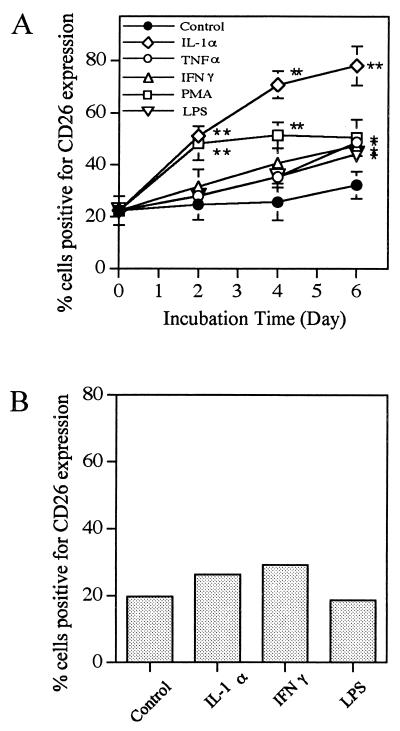
The difference in CD26 expression on fibroblasts between periodontal tissues and lung or skin moved us to investigate whether CD26 expression was inducible on periodontal tissue fibroblasts upon stimulation with various materials. HGF and PDL fibroblasts were stimulated with IL-1α, PMA, IFN-γ, TNF-α, and E. coli LPS for up to 6 days and then stained with anti-CD26 MAb for flow cytometry. Figure 1A shows that a marked increase of CD26 expression on HGF was observed upon stimulation with IL-1α compared with controls at day 6 (78.1% expression after stimulation versus 31.2% for the control), and the other stimulants, i.e., PMA, IFN-γ, TNF-α, and E. coli LPS, also upregulated the CD26 expression on HGF significantly (P < 0.05) compared with the control at day 6 (50.5, 47.4, 48.7, and 44.2% expression, respectively). In regard to the kinetics of CD26 expression upon stimulation, the upregulation was a slow response, with expression increasing gradually upon each stimulation and reaching a plateau at around day 6 after stimulation. On the other hand, as shown in Fig. 1B, only slight induction of CD26 expression on PDL fibroblasts was observed upon stimulation with IL-1α and IFN-γ. To represent the amount of CD26 expression more accurately, the mean fluorescence channel values of CD26 before stimulation and after stimulation for 6 days are shown in Table 2. The mean fluorescence value and percent positivity of CD26 were almost correlated. E. coli LPS did not exhibit activity on PDL fibroblasts. It must be noted here that HGF expressed CD14 on the cell surface at a high level, whereas PDL fibroblasts expressed it at a low level (Table 1). These findings suggested the heterogeneity of fibroblasts even within periodontal tissues. In the following studies, HGF were used because of their high responsiveness.
FIG. 1.
Induction of CD26 on cell surfaces of HGF and PDL fibroblasts in response to various stimulants. Confluent HGF (donor, YS-G) (A) and PDL fibroblasts (donor, AK) (B) were stimulated with IL-1α (10 ng/ml), PMA (100 ng/ml), IFN-γ (200 U/ml), TNF-α (40 ng/ml), or E. coli LPS (10 μg/ml) for the indicated times (A) and for 6 days (B) in α-MEM supplemented with 10% FCS. After being harvested by trypsinization, cells were stained with anti-CD26 MAb and analyzed by fluorescence-activated cell sorting. The results in panels A and B are representative of four different experiments with four different donors (YS-G, NIK, AK-G, and KEK) and three different experiments with three different donors (AK, TT, and YS), respectively. Triplicate (A) and single (B) assays were carried out, and differences from the control were significant at P < 0.01 (∗∗) and P < 0.05 (∗).
TABLE 2.
Mean channel fluorescence of CD26 before and after stimulationa
| Cells | Mean channel fluorescence value (SD)
|
||||||
|---|---|---|---|---|---|---|---|
| Day 0 (control) | Day 6
|
||||||
| Control | IL-1α | TNF-α | IFN-γ | PMA | LPS | ||
| HGF | 5.66 (0.42) | 9.29 (0.47) | 26.36 (5.35) | 15.77 (0.8) | 14.67 (0.92) | 17.70 (1.71) | 12.94 (1.91) |
| PDL fibroblasts | 5.30 | 5.58 | 6.33 | NDb | 9.35 | ND | 4.07 |
Induction of CD26 on the cell surfaces of HGF and PDL fibroblasts in response to various stimulants is shown. Confluent HGF (donor, YS-G) and PDL fibroblasts (donor, AK) were stimulated with IL-1α (10 ng/ml), TNF-α (40 ng/ml), IFN-γ (200 U/ml), PMA (100 ng/ml), or E. coli LPS (10 μg/ml) for 6 days in α-MEM supplemented with 10% FCS. After being harvested by trypsinization, cells were stained with anti-CD26 MAb and analyzed by fluorescence-activated cell sorting. The results are representative of four different experiments with four different donors (YS-G, NIK, AK-G, and KEK) for HGF and three different experiments with three different donors (AK, TT, and YS) for PDL fibroblasts. Triplicate (HGF) and single (PDL fibroblasts) assays were carried out.
ND, not done.
CD26-associated DPPIV activity on HGF stimulated with cytokines and bacterial components.
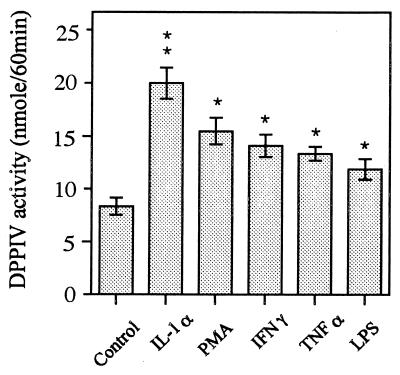
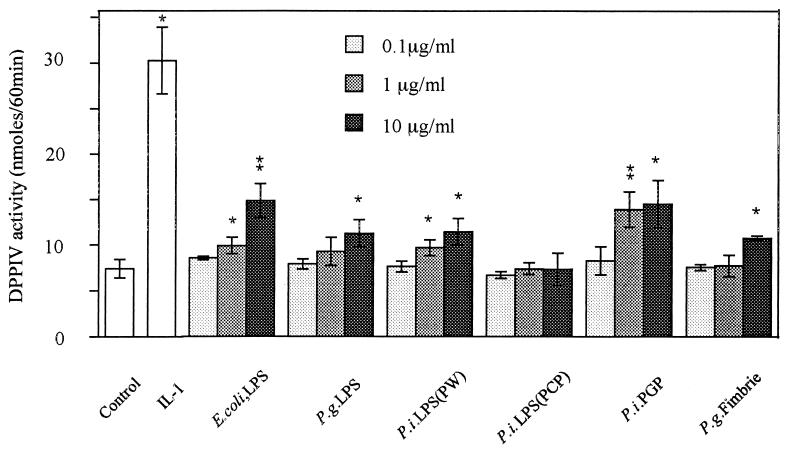
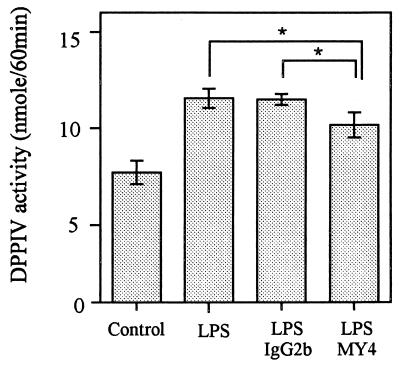
HGF were cultured for 6 days with or without stimulants, and confluent-monolayer cells were subjected to DPPIV assay with Gly-Pro-pNA as a substrate. Figure 2 shows that all stimulants induced the enzymatic activities on HGF significantly (P < 0.01) compared with unstimulated HGF. Among the stimulants, IL-1α had the most potent effect on the induction of DPPIV activity on HGF. Since CD26 is an ectoenzyme, it is natural that this observation was consistent with that of CD26 expression analyzed by flow cytometry (percent positivity and mean fluorescence value of CD26) (Fig. 1). It has been reported that LPS from oral black-pigmented bacteria (BPB) exhibits atypical biological activities, unlike the common LPS from Enterobacteriaceae (11, 39). We examined the response of HGF to various cell surface components from BPB in respect to DPPIV activity induction. Figure 3 shows that the phenol-water-extracted LPS from P. intermedia and P. gingivalis induced the DPPIV activity with dose dependency and that the activities were slightly weaker than those of E. coli LPS. In contrast, the PCP-extracted LPS from P. intermedia was scarcely active in induction of DPPIV activity, whereas PGP exhibited a definite ability to induce DPPIV activity. The activity of PGP was the strongest among the bacterial components so far examined. P. gingivalis fimbriae, another possible HGF activator, exhibited only weak activity at the highest concentration (10 μg/ml). Gingival fibroblasts release IL-8 in response to LPS in a CD14-dependent manner (31). We examined whether the mechanism by which CD26/DPPIV is induced on gingival fibroblasts after stimulation by LPS could depend on membrane CD14. Figure 4 shows that anti-CD14 MAb (MY4) only slightly inhibited the induction of CD26/DPPIV stimulated by LPS. These results indicated that induction of CD26/DPPIV by LPS was in part CD14 dependent.
FIG. 2.
Induction of DPPIV activity in HGF stimulated with various stimulants. Confluent HGF (donor, MH-12) were stimulated with IL-1α (10 ng/ml), PMA (100 ng/ml), IFN-γ (200 U/ml), TNF-α (40 ng/ml), or E. coli LPS (10 μg/ml) for 6 days in α-MEM supplemented with 10% FCS in 96-well plates and then washed with PBS three times, and confluent-monolayer cells in the plates were examined by DPPIV assay with Gly-Pro-pNA as a substrate. The results are representative of four different experiments with four different donors (MH-12, NIK, FE, and YS-G). Differences from the control were significant at P < 0.001 (∗∗) and P < 0.01 (∗).
FIG. 3.
Induction of DPPIV activity in HGF stimulated with various bacterial components. Confluent HGF (donor, NIK) were stimulated with various concentrations of bacterial components for 6 days in α-MEM supplemented with 10% FCS in 96-well plates and then washed with PBS three times, and confluent-monolayer cells in 96-well plates were examined by DPPIV assay with Gly-Pro-pNA as a substrate. E. coli LPS extracted with phenol-water, P. gingivalis (P.g.) LPS extracted with phenol-water, P. intermedia (P.i.) LPS extracted with phenol-water (PW), P. intermedia LPS extracted with PCP, PGP, and P. gingivalis fimbriae were used. The results are representative of two different experiments with two different donors (NIK and AK-G). Differences from the control were significant at P < 0.01 (∗∗) and P < 0.05 (∗).
FIG. 4.
Partial inhibition of LPS-induced CD26/DPPIV by anti-CD14 MAb. Confluent HGF (donor, NIK) were stimulated with E. coli LPS (10 μg/ml) for 6 days along with anti-CD14 MAb (MY4; 10 μg/ml) or isotype control mouse IgG2b (10 μg/ml), and then DPPIV assay was performed. The results are representative of two different experiments with two different donors (NIK and YS-G). Differences were significant at P < 0.05 (∗).
Characterization of CD26-associated DPPIV activity.
DPPIV was reported to be a serine protease (22), and diprotin A (Ile-Pro-Ile) is specifically used to inhibit DPPIV activity (37). We examined whether the enzyme expressed on HGF stimulated with IL-1α was sensitive to such inhibitors. As shown in Table 3, diprotin A strongly inhibited Gly-Pro-pNA hydrolysis (88.2% inhibition). A serine protease inhibitor, PMSF, inhibited Gly-Pro-pNA hydrolysis partially. By flow cytometry, we also confirmed that 1 μM PMSF had no effect on CD26 expression on gingival fibroblasts during 1 h of culture (data not shown). The other inhibitors, bestatin (aminopeptidase inhibitor), aprotinin (trypsin inhibitor), pepstatin A (aspartic and acid protease inhibitor), leupeptin (serine and thiol protease inhibitor), 1,10-phenanthroline (metalloprotease inhibitor), and EDTA (metalloprotease inhibitor), had no effects on Gly-Pro-pNA hydrolysis. Although DPPIV activity on HGF was low without stimulation, the activity on unstimulated HGF had a sensitivity to inhibitors similar to that of IL-1-stimulated HGF (data not shown). These results indicated that DPPIV activity was induced in HGF in correlation with CD26 expression.
TABLE 3.
Effect of protease inhibitors on DPPIV activity on IL-1-stimulated fibroblastsa
| Inhibitor | Concn | % Inhibitionb |
|---|---|---|
| Diprotin A | 1 mM | 88.2 |
| PMSF | 1 mM | 42.9 |
| 0.5 mM | 26.9 | |
| Bestatin | 1 mM | 5.1 |
| Aprotinin | 10 μg/ml | <1 |
| Pepstatin | 10 μg/ml | <1 |
| Leupeptin | 20 μg/ml | <1 |
| 1,10-Phenanthroline | 1 mM | <1 |
| EDTA | 5 mM | 7.6 |
Confluent HGF (donor, FE) were stimulated with 10 ng of IL-1α per ml for 6 days in 96-well plates and then washed with PBS three times, and confluent-monolayer cells were examined by DPPIV assay in the presence or absence of the indicated inhibitors.
Percent inhibition was calculated as [(DPPIV activity on IL-1-stimulated HGF in the absence of inhibitor) − (DPPIV activity on IL-1-stimulated HGF in the presence of inhibitor)]/(DPPIV activity on IL-1-stimulated HGF in the absence of inhibitor) × 100. DPPIV activities of fibroblasts stimulated with or without IL-1α were 33.4 and 9.3 nmol/60 min/culture, respectively. The results are representative of three different experiments with three different donors (FE, NIK, and MH-12).
Cell surface DPPIV activity on HGF induced by IL-1α.
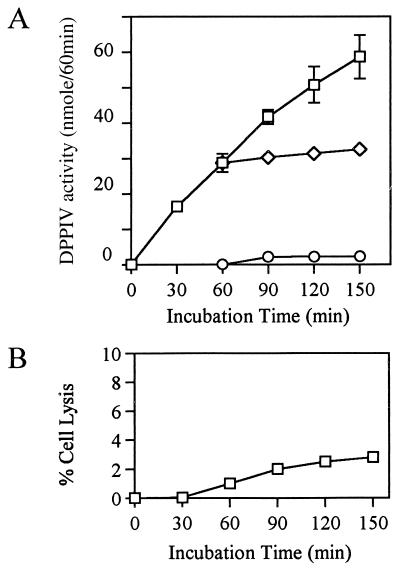
We further examined whether the degradation of the substrate by the cell suspension resulted from enzymatic activity of DPPIV on the cell surface. It is possible that incorporation of a substrate by HGF followed by intracellular degradation and release of pNA could occur or that intracellular enzymes were secreted. We carried out the following experiments. IL-1α-stimulated HGF were incubated at 37°C for 1 h with the substrate, and then the supernatants were harvested from monolayer cells. The harvested supernatants were incubated for an additional 90 min without addition of any reagents, and monolayer cells in enzyme assay buffer without substrate were incubated for an additional 90 min. As shown in Fig. 5A, during this 90 min, the control group (without separation of cells from substrate) exhibited an enzymatic response that was linear and time dependent. On the other hand, neither additional degradation of the substrate by the supernatant nor pNA release from the cell suspension was observed during 90 min after the separation of cells from substrate. These results excluded the possibility that the secretion of intracellular enzyme or degradation of the substrate inside cells was responsible for the degradation of the substrate in the cell suspension. Furthermore, the possibility that enzymatic activity could be released by lysed cells could be ruled out, since fewer than 3% of the cells died during the experiment (Fig. 5B).
FIG. 5.
Cell surface enzyme is responsible for the degradation of substrates by cells. (A) Confluent HGF (donor, MH-12) were stimulated with IL-1α (10 ng/ml) for 6 days in α-MEM supplemented with 10% FCS in 96-well plates and then washed with PBS three times, and confluent-monolayer cells were examined by DPPIV assay. Monolayer cells were incubated with 2 mM substrate for DPPIV assay as described in Materials and Methods. After a 60-min incubation, the supernatant and the corresponding monolayer cells, which were resuspended in peptidase medium, were incubated for an additional 90 min without addition of the substrate. □, pNA formed by intact fibroblasts; ◊, pNA formed by supernatant resulting from 60 min of preincubation of cells with the substrate; ○, pNA released by cells preincubated for 60 min with the substrate. (B) The percentage of lysed fibroblasts was determined by trypan blue exclusion at each time point. The results are representative of three different experiments with three different donors (MH-12, KEK, and NIK).
Induction of CD26/DPPIV mRNA expression in HGF in response to IL-1α.
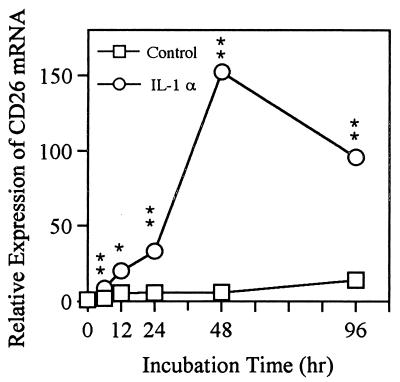
To determine whether the induction of CD26 by IL-1α was due to a change in gene expression, as distinct from translation, posttranslational degradation, or modulation of the protein, the level of CD26 mRNA was assessed by real-time quantitative PCR. Total RNA was extracted from IL-1α-stimulated HGF at 0, 6, 12, 24, 48, and 96 h of culture. Table 4 shows the Ct value (see Materials and Methods) at each point. Evaluation of the Ct value for the measurement makes it possible to quantify at the range of 105, compared with evaluation of the end point, which has a linear range of two orders of magnitude (9, 13). Figure 6 shows the relative expression of CD26 mRNA in HGF for control cells and upon IL-1 stimulation. An increase in CD26-specific mRNA was observed at 6 h (P < 0.001) after addition of IL-1α to the culture, reached a maximum at 48 h, and decreased thereafter. Quantitative analysis of CD26 mRNA levels revealed an approximately 150-fold increase of CD26 mRNA after 48 h of culture with IL-1α compared with the control. The relative expression of CD26 mRNA was approximately 90-fold greater than that of the control at 96 h. In the control culture without stimulation, CD26 mRNA accumulation was scarcely observed during the 96 h. Thus, the changes in relative mRNA levels analyzed by real-time quantitative PCR were consistent with the changes in protein levels analyzed by flow cytometry.
TABLE 4.
Induction of CD26 mRNA expression upon stimulation with IL-1αa
| Sample | Mean Ct value (SD) at h:
|
|||||
|---|---|---|---|---|---|---|
| 0 | 6 | 12 | 24 | 48 | 96 | |
| IL-1α | 31.75 (0.08) | 28.69** (0.13) | 27.43* (0.41) | 26.71** (0.12) | 24.50** (0.11) | 25.17** (0.09) |
| Control | 31.75 (0.08) | 30.78 (0.12) | 29.33 (0.74) | 29.27 (0.25) | 29.24 (0.25) | 27.95 (0.11) |
Confluent HGF (donor, NIK) were stimulated with or without IL-1α (10 ng/ml) for the indicated times, and then the total RNA was extracted for real-time quantitative PCR. The Ct value is defined in Materials and Methods. The results are representative of two different experiments with two different donors (NIK and FB). Differences from the respective controls were significant at P < 0.001 (∗∗) and P < 0.02 (∗).
FIG. 6.
Induction of CD26/DPPIV mRNA expression in response to IL-1α in HGF. HGF (donor, NIK) were stimulated with or without IL-1α (10 ng/ml) for the indicated times, and then total RNA was extracted for real-time quantitative PCR. The Ct value is defined in Materials and Methods. The relative difference in CD26 mRNA expression was calculated according to the Ct value (Table 3). The relative value of mRNA expression at day 0 was converted to 1. The results are representative of two different experiments with two different donors (NIK and FB). Differences from the respective controls were significant at P < 0.001 (∗∗) and P < 0.02 (∗).
Induction of CD26 mRNA requires de novo protein synthesis.
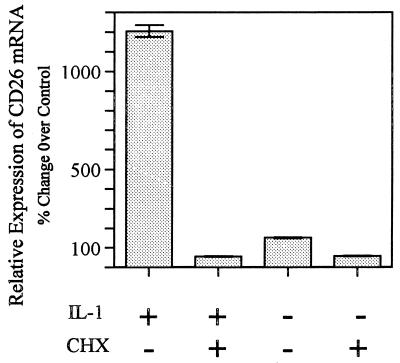
When HGF were stimulated with IL-1α, the induction of CD26 at both the mRNA and protein levels was slow (Fig. 1 and 6). These observations raise the question of whether the CD26 mRNA requires de novo protein synthesis for induction. We examined the effect of the protein synthesis inhibitor CHX on IL-1-induced accumulation of CD26 mRNA in HGF. Figure 7 shows that CHX (10 μg/ml) added at 2 h before IL-1α blocked the induction of CD26 mRNA completely, indicating that de novo protein synthesis was required for CD26 mRNA induction.
FIG. 7.
Inhibition of CD26 mRNA accumulation in HGF by CHX. HGF (donor, NIK) were preincubated with or without CHX (10 μg/ml) for 2 h and stimulated with or without IL-1α (10 ng/ml) for 24 h, and then the total RNA was extracted for real-time quantitative PCR. The relative difference in CD26 mRNA expression was calculated according to the Ct value as described in Materials and Methods. The relative value of mRNA expression at day 0 was converted to 100%.
DISCUSSION
Since CD26 was identified as DPPIV in T cells, its biological function has been investigated. In T cells, CD26 is capable of transmitting signals to activate cytotoxicity, IL-2 secretion, and proliferation (6). CD45 is comodulated with CD26 and is coprecipitated in T-cell lysates (36). Furthermore, CD26 was reported to bind to HIV-1 Tat protein (10). However, the enzymatic activity is not absolutely required for such functions of CD26. On the other hand, several functions which require the enzymatic activity have been postulated. For example, the proliferation of T cells stimulated with mitogen was inhibited with a DPPIV-specific inhibitor (25), as was IL-2 and IFN-γ production from T cells (24). Other studies showed that substance P, chorionic gonadotropin, monomeric fibrin, and several releasing hormones were hydrolyzed by DPPIV (40). Recently, several chemokines, including RANTES and SDF-1, which attract inflammatory cells to the inflamed lesion, have been revealed to be substrates for CD26/DPPIV (19, 27), and the specificity of chemokine receptors was altered after degradation of chemokines by CD26/DPPIV, resulting in regulation of inflammatory responses. Furthermore, alteration of the specificity of truncated RANTES for chemokine receptor causes the inhibition of HIV infection (19, 21). On the other hand, truncated SDF-1 had diminished potency to inhibit HIV-1 infection (27). These results suggested that CD26/ DPPIV actively participates in inflammatory and immunological responses.
In this study, we first investigated whether CD26 was expressed on fibroblasts from various tissues and the regulation of CD26 expression on fibroblasts. Recent studies suggested that fibroblasts play an important role in the regulation of inflammatory and immunological responses as well as support frameworks for other cells (31, 33, 34); i.e., fibroblasts are able to function as antigen-presenting cells in immunological reactions (8, 26), to produce cytokines (such as IL-1, IL-6, TNF-α, and granulocyte-macrophage colony-stimulating factor) and chemokines (such as IL-8, RANTES, eotaxin, and monocyte chemoattractant protein-1) in inflammatory responses (20, 30, 33, 34), and to express various adhesion molecules upon activation (4). Here, we found that CD26 was induced on gingival fibroblasts upon stimulation both at the protein level as determined by flow cytometry (Fig. 1) and at the mRNA level as determined by real-time quantitative RT-PCR (Fig. 6). In local infection with periodontal disease-associated bacteria, HGF should be exposed to inflammatory cytokines and various bacterial components. We used cytokines (IL-1α, IFN-γ, and TNF-α), a protein kinase C activator (PMA), and bacterial components such as LPS fractions, PGP, and fimbriae for activation of fibroblasts. All were able to induce CD26/DPPIV on the cell surface. However, IL-1α had the most potent effect for induction of CD26/DPPIV on the cell surface (Fig. 1A and 2).
Although bacterial components had less of an effect than IL-1α on the induction, at a high concentration (10 μg/ml), bacterial components significantly enhanced the induction. It has been reported that the activities of LPS differed among sources, probably because of differences in chemical structures (11, 39). In fact, P. intermedia LPS (PCP extract) had practically no ability to induce DPPIV, whereas the PGP fraction exhibited strong activity. The phenol-water-extracted LPSs tested here apparently exhibited similar activity with regard to the induction of DPPIV activity. It is possible, however, that the activity of phenol-water-extracted LPSs from P. gingivalis and P. intermedia in this study was derived mainly from PGP in the LPS fraction, as suggested by Iki et al. (14). If so, the activity of the BPB LPS by itself was considerably weaker than that of E. coli LPS. The HGF used in this study generally expressed membrane CD14 (mCD14) on the cell surface at high levels. A recent report by Sugawara et al. (31) demonstrated that E. coli LPS activated HGF to release IL-8 in an mCD14-dependent manner. In this study, E. coli LPS activated HGF to induce CD26/DPPIV in an mCD14-dependent manner in part (Fig. 4). This is also supported by the fact that PDL fibroblasts expressing low levels of CD14 on the cell surface did not respond to E. coli LPS for CD26 induction (Fig. 1B). Given the previous report by Takada et al. (33) demonstrating that BPB LPS induced cell-associated IL-1α in gingival fibroblasts in culture, CD26/DPPIV may be induced by LPS in an autocrine manner. We set up an experiment using anti-IL-1α Ab, which blocked the CD26/DPPIV induction stimulated by 1 ng of IL-1α per ml. Anti-IL-1α Ab had no effect on the CD26/DPPIV induction stimulated by 10 μg of E. coli LPS per ml when added on days 0, 1, and 2 (data not shown), suggesting that induction of CD26/DPPIV by LPS is not via IL-1α in an autocrine manner.
We also showed that induction of CD26/DPPIV upon stimulation occurred on the cell surfaces of fibroblasts (Fig. 5) by demonstrating that the degradation of the substrate resulted from DPPIV activity on the cell surface and not from activity inside the cells. Furthermore, we clearly identified the enzyme induced on fibroblasts as DPPIV by experiments using various inhibitors, among which diprotin A (Ile-Pro-Ile) and PMSF (a serine protease inhibitor) inhibited the enzymatic activity (Table 3). DPPIV was reported to be a serine protease; thus, PMSF inhibited DPPIV activity partially (17). Diprotin A is specifically used to inhibit DPPIV activity (37), since diprotin A is a tripeptide which cannot be hydrolyzed by DPPIV and which competes with the substrate for the active site of DPPIV. These characteristics were consistent with our results (Table 3). Interestingly, while CD26/DPPIV was only partially expressed on HGF and PDL fibroblasts, the fibroblasts from lung and skin expressed CD26/DPPIV on the cell surface constitutively (Table 1). The CD26/DPPIV expression in HGF was upregulated by various stimulants, whereas that in PDL fibroblasts was only slightly upregulated (Fig. 1A). These findings are supported by the evidence that fibroblasts differ in morphology and function within or between tissues and/or organs (7). The findings in this study further suggested that CD26/DPPIV on fibroblasts plays important roles in inflammatory reactions and that these roles differ between organs.
Several days were required for the expression of CD26 on the cell surface following stimulation (Fig. 1A). This slow response was also demonstrated at the transcriptional level (a maximum at around 48 h) by real-time quantitative RT-PCR, an extremely sensitive technique for quantifying mRNA; unlike other quantitative PCR methods, whose linear dynamic range is approximately two orders of magnitude, this method has a very large dynamic range of starting target molecule determination (at least five orders of magnitude) (9, 13). We demonstrated that the induction of CD26 mRNA was completely blocked when cells were pretreated with CHX at 2 h before addition of the inducer. This result suggested that the requirement of de novo protein synthesis can be attributed to the slow response for induction of CD26 on the cell surface. It is of interest to identify the de novo-synthesized protein which induces CD26 on the cell surface directly, and investigations to find the protein are under way. This slow induction of CD26 suggested that CD26 on fibroblasts in periodontal tissue might function at a late stage of inflammation or to repair the damaged tissues.
Different patterns of CD26/DPPIV expression on fibroblasts have been reported for several diseases, such as rheumatoid arthritis, psoriasis, lichen planus, and systemic sclerosis (2, 22). CD26/DPPIV is downregulated on skin fibroblasts in vivo and in vitro in systemic sclerosis (scleroderma), which is an autoimmune disease characterized as fibroblast activation and endothelial cell damage (2, 29), leading to the overproduction of extracellular matrix and the development of fibrotic lesions. On the other hand, CD26/DPPIV is upregulated on fibroblasts in psoriasis and rheumatoid arthritis (22), both of which are associated with inflammatory and immunological characteristics in which extracellular matrix breakdown is an important part of the disease process (12, 38). Specific DPPIV inhibitors suppress collagen-induced and alkyldiamine-induced arthritis in vivo (35). In periodontal tissue, CD26/DPPIV is localized in gingival tissue from chronic periodontitis patients by immunohistochemical analysis (15). The CD26/DPPIV level in gingival crevicular fluid correlates positively with clinical indications of disease severity in untreated patients and is reduced after periodontal treatment (3, 5).
These findings suggest that CD26/DPPIV on fibroblasts is associated with the regulation of inflammatory and immunological responses by regulation of chemokines and biologically active peptides or by binding to the ligands on other cells and that it is possibly associated with the onset and course of dermatological, rheumatic, and periodontal diseases.
ACKNOWLEDGMENTS
This work was supported in part by Grants-in-Aid for Scientific Research (no. 10307053, 10470378, 09671843, and 09671943) and for Encouragement of Young Scientists (no. 10771212) from the Ministry of Education, Science, Sports and Culture, Japan.
We thank C. Hirata, A. Sugiyama, and J. Hatakeyama for providing cultured human fibroblasts. We also thank D. Mrozek (Medical English Service, Kyoto, Japan) for reviewing the paper.
REFERENCES
- 1.Agarwal S, Chandra C S, Piesco N P, Langkamp H H, Bowen L, Baran C. Regulation of periodontal ligament cell functions by interleukin-1β. Infect Immun. 1998;66:932–937. doi: 10.1128/iai.66.3.932-937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bou-Gharios G, Osman J, Amanda A, Monoghan P, Vancheeswaran R, Black C, Olsen I. Expression of ectopeptidase in scleroderma. Ann Rheum Dis. 1995;54:111–116. doi: 10.1136/ard.54.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox S W, Eley B M. Cathepsin B/L-, elastase-, tryptase-, trypsin- and dipeptidylpeptidase IV-like activities in gingival crevicular fluid: a comparison of levels before and after basic periodontal treatment of chronic periodontitis patients. J Clin Periodontol. 1992;19:333–339. doi: 10.1111/j.1600-051x.1992.tb00655.x. [DOI] [PubMed] [Google Scholar]
- 4.Dustin M L, Rothlein R, Bhan A K, Dinarello C A, Springer T A. Induction by IL1 and interferon-γ: tissue distribution, biochemistry, and function of a natural adherence molecule (ICAM-1) J Immunol. 1986;137:245–254. [PubMed] [Google Scholar]
- 5.Eley B M, Cox S W. Cathepsin B/L-, elastase-, tryptase-, trypsin- and dipeptidylpeptidase IV-like activities in gingival crevicular fluid: correlation with clinical parameters in untreated chronic periodontitis patients. J Periodont Res. 1992;27:62–69. doi: 10.1111/j.1600-0765.1992.tb02087.x. [DOI] [PubMed] [Google Scholar]
- 6.Fleischer B. CD26: a surface protease involved in T-cell activation. Immunol Today. 1994;15:180–184. doi: 10.1016/0167-5699(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 7.Fries K M, Blieden T, Looney R J, Sempowski G D, Silvera M R, Willis R A, Phipps R P. Evidence of fibroblast heterogeneity and the role of fibroblast subpopulations in fibrosis. Clin Immunol Immunopathol. 1994;72:283–292. doi: 10.1006/clin.1994.1144. [DOI] [PubMed] [Google Scholar]
- 8.Geppert T D, Lipsky P E. Antigen presentation by interferon-γ-treated endothelial cells and fibroblasts: differential ability to function as antigen presenting despite comparable la expression. J Immunol. 1985;135:3750–3762. [PubMed] [Google Scholar]
- 9.Gibson U E M, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 10.Gutheil W G, Subramanyam M, Flentke G R, Sanford D G, Munoz E, Huber B T, Bachovchin W W. Human immunodeficiency virus 1 Tat binds to dipeptidylaminopeptidase IV (CD26): a possible mechanism for Tat's immunosuppressive activity. Proc Natl Acad Sci USA. 1994;91:6594–6598. doi: 10.1073/pnas.91.14.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamada S, Takada H, Ogawa T, Fujiwara T, Mihara J. Lipopolysaccharides of oral anaerobes associated with chronic inflammation: chemical and immunomodulating properties. Int Rev Immunol. 1990;6:247–261. doi: 10.3109/08830189009056635. [DOI] [PubMed] [Google Scholar]
- 12.Harris E D. Rheumatoid arthritis. Pathophysiology and implications for therapy. N Engl J Med. 1990;322:1277–1289. doi: 10.1056/NEJM199005033221805. [DOI] [PubMed] [Google Scholar]
- 13.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 14.Iki K, Kawahara K, Sawamura S, Arakaki R, Sakuta T, Sugiyama A, Tamura H, Sueda T, Hamada S, Takada H. A novel component different from endotoxin extracted from Prevotella intermediaATCC 25611 activates lymphoid cells from C3H/HeJ mice and gingival fibroblasts from humans. Infect Immun. 1997;65:4531–4538. doi: 10.1128/iai.65.11.4531-4538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennett C N, Cox S W, Eley B M. Histochemical and immunocytochemical localization of dipeptidyl peptidases II and IV in human gingiva. J Periodontol. 1996;67:846–852. doi: 10.1902/jop.1996.67.9.846. [DOI] [PubMed] [Google Scholar]
- 16.Kenny A J, O'Hare M J, Gusterson B A. Cell-surface peptidases as modulators of growth and differentiation. Lancet. 1989;30:785–787. doi: 10.1016/s0140-6736(89)90841-6. [DOI] [PubMed] [Google Scholar]
- 17.McDonald J K, Schwabe C. Intracellular exopeptidases. In: Barrett A J, editor. Proteinases in mammalian cells and tissues. Amsterdam, The Netherlands: Elsevier/North-Holland Biomedical Press; 1977. pp. 311–391. [Google Scholar]
- 18.Ogawa T, Shimauchi H, Hamada S. Mucosal and systemic immune responses in BALB/c mice to Bacteroides gingivalisfimbriae administered orally. Infect Immun. 1989;57:3466–3471. doi: 10.1128/iai.57.11.3466-3471.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oravecz T, Pall M, Roderiquez G, Gorrell M D, Ditto M, Nguyen N Y, Boykins R, Unsworth E, Norcross M A. Regulation of the receptor specificity and function of the chemokine RANTES (Regulated on Activation, Normal T Cell Expressed and Secreted) by dipeptidyl peptidase IV (CD26)-mediated cleavage. J Exp Med. 1997;186:1865–1872. doi: 10.1084/jem.186.11.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patil R R, Borch R F. Granulocyte-macrophage colony-stimulating factor expression by human fibroblasts is both upregulated and subsequently downregulated by interleukin-1. Blood. 1995;85:80–86. [PubMed] [Google Scholar]
- 21.Proost P, Meester I D, Schols D, Struyf S, Lambeir A M, Wuyts A, Opdenakker G, Clercq E D, Scharpe S, Damme J V. Amino-terminal truncation of chemokines by CD26/dipeptidyl-peptidase IV. J Biol Chem. 1998;273:7222–7227. doi: 10.1074/jbc.273.13.7222. [DOI] [PubMed] [Google Scholar]
- 22.Raynaud F, Bauvois B, Gerbaud P, Evain-Brion D. Characterization of specific proteases associated with the surface of human skin fibroblasts, and their moduration in pathology. J Cell Physiol. 1992;151:378–385. doi: 10.1002/jcp.1041510219. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt J A, Mizel S B, Cohen D, Green I. Interleukin 1, a potential regulator of fibroblast proliferation. J Immunol. 1982;128:2177–2182. [PubMed] [Google Scholar]
- 24.Schon E, Demuth H U, Eichmann E, Horst H J, Korner I J, Kopp J, Mattern T, Neubert K, Noll F, Ulmer A J, Barth A, Ansorge S. Dipeptidyl peptidase IV in human T lymphocytes. Impaired induction of interleukin 2 and γ interferon due to specific inhibition of dipeptidyl peptidase. Scand J Immunol. 1989;29:127–132. doi: 10.1111/j.1365-3083.1989.tb01108.x. [DOI] [PubMed] [Google Scholar]
- 25.Schon E, Jahn S, Kiessig S T, Demuth H U, Neubert K, Barth A, Von Baehr R, Ansorge S. The role of dipeptidyl peptidase IV in human T lymphocyte activation. Inhibition and antibodies against dipeptidyl peptidase IV suppress lymphocyte proliferation. Eur J Immunol. 1987;17:1821–1826. doi: 10.1002/eji.1830171222. [DOI] [PubMed] [Google Scholar]
- 26.Shimabukuro Y, Murakami S, Okada H. Antigen-presenting-cell function of interferon γ-treated human gingival fibroblasts. J Periodontal Res. 1996;31:217–228. doi: 10.1111/j.1600-0765.1996.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 27.Shioda T, Kato H, Tashiro Y, Ikegawa M, Nakayama E E, Hu H, Kato A, Sakai Y, Liu H, Honjo T, Nomoto A, Iwamoto A, Morimoto C, Nagai Y. Anti-HIV-1 and chemotactic activities of human stromal cell-derived factor 1 α (SDF-1 α) and SDF-1 β are abolished by CD26/dipeptidyl peptidase IV-mediated cleavage. Proc Natl Acad Sci USA. 1998;95:6331–6336. doi: 10.1073/pnas.95.11.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shipp M A, Look A T. Hematopoietic differentiation antigens that are membrane-associated enzymes: cutting is the key! Blood. 1993;82:1052–1070. [PubMed] [Google Scholar]
- 29.Shiwen X, Vancheeswaran R, Bou-Gharios G, O'Hare M J, Olsen I, Abraham D, Black C. Scleroderma-derived human fibroblasts retain abnormal phenotypic and functional characteristics following retroviral transduction with the SV40 tsT antigen. Exp Cell Res. 1995;220:407–414. doi: 10.1006/excr.1995.1332. [DOI] [PubMed] [Google Scholar]
- 30.Smith R S, Smith T J, Blieden T M, Phipps R P. Fibroblasts as sentinel cells: synthesis of chemokines and regulation of inflammation. J Immunol. 1997;151:317–322. [PMC free article] [PubMed] [Google Scholar]
- 31.Sugawara S, Sugiyama A, Nemoto E, Rikiishi H, Takada H. Heterogeneous expression and release of CD14 by human gingival fibroblasts: characterization and CD14-mediated interleukin-8 secretion in response to lipopolysaccharide. Infect Immun. 1998;66:3043–3049. doi: 10.1128/iai.66.7.3043-3049.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takada H, Hirai H, Fujiwara T, Koga T, Ogawa T, Hamada S. Bacteroideslipopolysaccharides (LPS) induce anaphylactoid and lethal reactions in LPS-responsive and -nonresponsive mice primed muramyl dipeptide. J Infect Dis. 1990;162:428–434. doi: 10.1093/infdis/162.2.428. [DOI] [PubMed] [Google Scholar]
- 33.Takada H, Mihara J, Morisaki I, Hamada S. Induction of interleukin-1 and -6 in human gingival fibroblast cultures stimulated with Bacteroideslipopolysaccharides. Infect Immun. 1991;59:295–301. doi: 10.1128/iai.59.1.295-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura M, Tokuda M, Nagaoka S, Takada H. Lipopolysaccharides of Bacteroides intermedius (Prevotella intermedia) and Bacteroides (Porphyromonas) gingivalisinduce interleukin-8 gene expression in human gingival fibroblast cultures. Infect Immun. 1992;60:4932–4937. doi: 10.1128/iai.60.11.4932-4937.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka S, Murakami T, Sugiura H, Kawashima K, Sugita T. Suppression of arthritis by the inhibitors of dipeptidyl peptidase IV. Int J Immunopharmacol. 1997;19:15–24. doi: 10.1016/s0192-0561(97)00004-0. [DOI] [PubMed] [Google Scholar]
- 36.Torimoto Y, Dang N H, Vivier E, Tanaka T, Schlossman S F, Morimoto C. Coassociation of CD26 (dipeptidyl peptidase IV) with CD45 on the surface of human T lymphocytes. J Immunol. 1991;147:2514–2517. [PubMed] [Google Scholar]
- 37.Umezawa H, Aoyagi T, Ogawa K, Naganawa H, Hamada M, Takeuchi T. Diprotin A and B, inhibitors of dipeptidyl aminopeptidase IV, produced by bacteria. J Antibiot (Tokyo) 1984;37:422–425. doi: 10.7164/antibiotics.37.422. [DOI] [PubMed] [Google Scholar]
- 38.Valdimarsson H, Baker B S, Jonsdohir I, Fry L. Psoriasis: a disease of abnormal keratinocyte proliferation induced by T-lymphocytes. Immunol Today. 1986;7:256–259. doi: 10.1016/0167-5699(86)90005-8. [DOI] [PubMed] [Google Scholar]
- 39.Wilson M. Biological activities of lipopolysaccharide and endotoxin. In: Shah H N, Mayrand D, Genco R J, editors. Biology of the species Porphyromonas gingivalis. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 171–198. [Google Scholar]
- 40.Yaron A, Naider F. Proline-dependent structural and biological properties of peptides and proteins. Crit Rev Biochem Mol Biol. 1993;28:31–81. doi: 10.3109/10409239309082572. [DOI] [PubMed] [Google Scholar]