Abstract
Use of molecular data in human and ecological health risk assessments of industrial chemicals and agrochemicals has been anticipated by the scientific community for many years; however, these data are rarely used for risk assessment. Here, a logic framework is proposed to explore the feasibility and future development of transcriptomic methods to refine and replace the current apical endpoint-based regulatory toxicity testing paradigm. Four foundational principles are outlined and discussed that would need to be accepted by stakeholders prior to this transformative vision being realized. Well-supported by current knowledge, the first principle is that transcriptomics is a reliable tool for detecting alterations in gene expression that result from endogenous or exogenous influences on the test organism. The second principle states that alterations in gene expression are indicators of adverse or adaptive biological responses to stressors in an organism. Principle 3 is that transcriptomics can be employed to establish a benchmark dose-based point of departure (POD) from short-term, in vivo studies at a dose level below which a concerted molecular change (CMC) is not expected. Finally, Principle 4 states that the use of a transcriptomic POD (set at the CMC dose level) will support a human health-protective risk assessment. If all four principles are substantiated, this vision is expected to transform aspects of the industrial chemical and agrochemical risk assessment process that are focused on establishing safe exposure levels for mammals across numerous toxicological contexts resulting in a significant reduction in animal use while providing equal or greater protection of human health. Importantly, these principles and approaches are also generally applicable for ecological safety assessment.
Keywords: risk assessment, toxicogenomics, point of departure, transcriptomics
For almost a century, animal studies have been used to determine the potential for adverse consequences of exposure to chemicals and to inform human health risk assessments (Daniel Krewski et al., 2010; Wax, 1995). These whole animal studies are designed to identify a dose at which no adverse effects occur and to enable an assessment of the potential for risk to humans. Risks are then managed using reference doses, reference concentrations, tolerable daily intakes, and other safety standards for chemicals. These safety standards have been extrapolated from the no observed adverse effect level (NOAEL) or mathematically derived benchmark dose (BMD) that are used as the point of departure (POD) for risk assessment (Haber et al., 2018). BMDs are most often used to describe experimentally observed dose-responsive tissue effects, clinical pathology changes, adverse responses, and apical endpoints. This approach is founded on the premise that humans and ecological species will be sufficiently protected by setting tolerable chemical exposure to a level below which a defined observable endpoint is expected. When capital and human resources are available to conduct and evaluate these studies, this traditional toxicity testing approach has been generally predictive of human toxicity/safety and therefore protective of human health (Monticello et al., 2017).
However, the traditional experimental identification of a POD for risk assessment requires the use of extensive, low-throughput, and resource-intensive animal testing. For example, agrochemical registration requires a battery of tests in multiple species that cover all life stages (Sewell et al., 2021). These traditional testing approaches run counter to current socioethical calls for a reduction in animal use and limitations on high-dose testing (Tan et al., 2021; US EPA, n.d.). Furthermore, the time- and resource-intensive nature of current testing practices puts significant limitations on our collective ability to generate data on environmental chemicals needed to protect human health. The potential for genomic tools to aid in addressing many of these challenges has been discussed for more than 2 decades (Inoue and Pennie, 2003; National Research Council, 2005; Yauk et al., 2020). A combination of these drivers for change has gained the attention of the risk assessment community including the need for data to address a greater number of chemicals of regulatory interest, the broad availability of high-throughput transcriptomic tools that allow for relatively comprehensive characterization of molecular alterations associated with chemical effects, and a series of case studies demonstrating the utility and reliability of molecular analysis for hazard and risk assessment. In addition, the reduced costs and time needed to complete these studies would allow for the screening and regulatory assessment of industrial chemicals that may not otherwise be tested for toxicity. There is now more support to shift away from decades of risk assessment based exclusively on apical endpoints to a paradigm centered on comprehensive molecular analyses (eg, omics). The goal is to identify the most sensitive effects at the molecular level to determine levels of risk and a POD based on concerted molecular change (CMC) (Krewski et al., 2020).
It is understood that the current requirement of traditional risk assessment is to determine the critical effect occurring at the lowest dose level in order to establish a POD. The vision is to transition from this historical approach to a paradigm that does not rely on the identification of specific critical effects from animal studies and instead establishes a POD that is protective of all potential critical effects. While dissimilar from the current toxicity testing practices of characterizing various apical outcomes, this paradigm is expected to be equivalent in the level of human health protection. This vision serves as a starting point which could be adapted and built upon to meet different regulatory and programmatic needs (Stucki et al., 2022).
Although the specifics related to study design, analysis, and application context vary and other molecular data could be used, one area of focus supporting the paradigm shift to novel methods is the use of transcriptomic data to set a POD without linkage to a concurrent/specific apical endpoint. Specifically, the derived PODs are agnostic to the nature of the molecular level change (such as the specific pathway or gene set associated with the POD) (Farmahin et al., 2017; Johnson et al., 2020; LaRocca et al., 2020; Pagé-Larivière et al., 2019). A transcriptome POD value is derived from BMD analysis of transcriptome change and represents a dose below which there is no CMC. The value of this approach is based on the premise that a transcriptomic POD will be health protective if it is anchored by a BMD below a concentration/exposure necessary to initiate causal events linked at a molecular level change (ie, a molecular initiating event) to a potential adverse outcome (similar in principle to an NOAEL). This allows these methods to be generically related to the most sensitive mechanistic/molecular processes altered by chemical exposure (ie, a CMC) and at the same time are agnostic to the nature of the biological processes and their relationship to any specific apical outcomes (ie, hazard) that may be associated with the CMC. Simply put, these approaches aim to identify the lowest dose at which biological alterations are expected to occur in response to a treatment and not the adverse effect(s) that could occur. Retrospectively evaluating available datasets has demonstrated good concordance (typically within 10-fold) between transcriptomic PODs derived from in vivo short term-studies and those established by longer term, apical endpoint focused guideline toxicity studies (Bhat et al., 2013; Farmahin et al., 2017; Gwinn et al., 2020; Johnson et al., 2020; Pagé-Larivière et al., 2019; Thomas et al., 2013).
In a precedent-setting decision that aligns principally with what is proposed here, the United States Environmental Protection Agency (US EPA) has used a POD based upon rat in vivo gene expression in a human health risk assessment (US Environmental Protection Agency, 2016). In rats orally exposed to the herbicide halauxifen-methyl, liver is the target organ. Mode-of-action studies demonstrated that aryl hydrocarbon receptor activation was the cause of liver toxicity, and liver Cyp1a1 gene expression was used by the US EPA as a biomarker for activation of this mode-of-action. Rat subchronic toxicity studies were performed, but no rat chronic toxicity or carcinogenicity studies were conducted with halauxifen-methyl. To set the chronic dietary reference dose, the US EPA used the nonadverse endpoint of liver Cyp1a1 gene expression from a subchronic (90 days) rat toxicity study. As stated in the US EPA Human Health Risk Assessment, the Cyp1a1 gene expression POD value “was selected to be protective of potential liver effects resulting from chronic dietary exposure to halauxifen-methyl.” Our vision extends this precedent-setting regulatory decision by seeking to identify a human health-protective POD value for any repeat dose critical effect based upon a POD derived from a comprehensive analysis of the in vivo transcriptome.
The approach discussed here diverges from the use of PODs linked to defined transcriptomic biomarkers (eg, molecular pathways or signatures) or adverse outcome pathways because it creates the near-term opportunity to employ transcriptomics at scale for risk assessment. If successful, this novel use of transcriptomic PODs is expected to reduce the size and duration of animal studies by eliminating the need for medium- and long-term rodent studies, yet still be human health protective against chronic toxicities. The immediate implementation of this approach alone for regulatory decision-making is limited because of insufficient numbers of case studies covering a broad enough chemical and biological space and temporal scope as well as the absence of consensus methods and a sufficient number of case studies to demonstrate the validity of the approach.
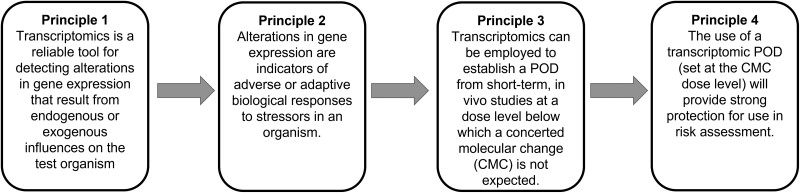
A logic framework (Figure 1) is proposed that explores the feasibility and future development of transcriptomic methods to refine and replace the current apical endpoint-based regulatory toxicity testing and risk assessment paradigm. It is important to note that the discussions were focused on risk assessment applications, not screening and prioritization, where some of these methods are already in use (Harrill et al., 2019; Krewski et al., 2020; Reardon et al., 2021; Wang et al., 2018). The threshold for establishing confidence and fitness for purpose is lower for the application of this approach in screening and prioritization as compared with risk assessment (Parish et al., 2020). However, lessons learned from tiered testing and chemical screening applications are highly informative and thus far also broadly support the logic framework presented.
Figure 1.
Proposed logic framework for the implementation of pathway agnostic transcriptomic-derived PODs in risk assessment. Strong evidence base and community consensus of acceptable practices will be required to support robust, reproducible application of these methods for risk assessment. Abbreviation: POD, point of departure.
The logic framework is established as a series of foundational principles that must all be realized for these methods to be adopted for human health risk assessment at scale. Principle 2 is dogma from decades of research in molecular biology and toxicology. Principles 1, 3, and 4 have robust supporting data with established standards of practice, are comprised of evolving science, and provide ripe opportunities for new research and consensus building. In identifying these points, the authors considered both gaps in methodological development and data interpretation as well as the ways in which this approach challenges and requires clarification of expectations for the goals of chemical risk assessment.
Principle 1: Transcriptomics is a reliable tool for detecting alterations in gene expression that result from endogenous or exogenous influences on the test organism. This principle is well supported by publication and practice. Transcriptomics is broadly defined as the measurement of gene expression profiles (mRNA), from all or part of the genome, via platforms such as genomic microarrays, RNAseq, etc. Transcriptomic tools are widely accessible and routinely used to detect and characterize transcriptional activity in a cell, tissue, or organism prior to the manifestation of a phenotype. A detailed review of these methods and their biological outputs is beyond the scope of this article. Although bioinformatic pipelines vary, standardized approaches and widely accepted tools have been published and made publicly available to identify differentially expressed transcripts from these various platforms. Best practices to enhance the reproducibility of these data have been published (eg, microarray quality control consortium) and continue to evolve (FDA, n.d.; Verheijen et al., 2020). Progress has been made toward frameworks for reporting transcriptomic data by the Organisation for Economic Co-operation and Development (OECD) although standards and technologies also continue to evolve (Harrill et al., 2021). Finally, methods for dose-response analysis of genomic data have been formulated and peer-reviewed and open-source software that enables the implementation of an analysis pipeline is available (National Toxicology Program, 2018; Phillips et al., 2019).
Principle 2: Alterations in gene expression are indicators of adverse or adaptive biological responses to stressors in an organism. The principle that (given sufficient exposure) biological stressors can cause cells or tissues to deviate from homeostasis is a well-established scientific observation (Calabrese et al., 2007; Kültz, 2020). After an exposure threshold is reached, cells respond by attempting to re-establish homeostasis, and toxicity occurs when this adaptive response is insufficient or, in some cases, when the response is inappropriate thereby inducing additional stress. The establishment of a mechanism-agnostic transcriptomic POD for chemical safety will require consensus across sectors and scientific communities that all physiologically manifested toxicological outcomes are accompanied by a change in an organism’s gene expression. That is, given sufficient exposure, cells will upregulate or downregulate the amount of mRNA that they produce in a gene-specific manner, which in turn alters the amount of proteins that are translated to perform their specific functions related to maintaining homeostasis (eg, clearing or neutralizing foreign chemicals, repairing damage, increasing energy production, etc.). Indeed, this phenomenon is supported by decades of experimental evidence linking chemically induced effects to transcriptomic responses (Davis et al., 2021). Recent transcriptomic studies with relatively large chemical sets have also clearly shown a high correlation between the emergence of transcriptomic responses (ie, a transcriptomic POD) with the emergence of traditional apical adverse effects (Gwinn et al., 2020; Johnson et al., 2020). In these studies, it is notable that the degree of response at the transcriptomic level can vary significantly based on the mode of action, with nuclear receptor activators showing very strong responses and agents that act through other mechanisms (eg, G-protein coupled receptors, ion channels) or have a primary target tissue (eg, brain) that has not been queried in the above studies often producing less pronounced effects in sentinel organs (eg, liver and kidney; Gwinn et al., 2020). Hence, it will be critical to explore additional chemicals that act through mechanisms that have more subtle transcriptional effects to calibrate analysis pipelines to confidently identify the molecular POD of these types of agents. Collectively, the evidence for Principle 2 is strong; however, further studies of agents that may challenge this principle will be critical for refining study protocols (eg, broader tissue sampling) and analysis methods that will establish greater certainty in Principle 2.
Principle 3: Transcriptomics can be employed to establish a POD from short-term, in vivo studies at a dose level below which a CMC is not expected. This principle requires substantive additional data and scientific discussion. While the theoretical premise may be sound, pragmatic implementation will require substantial future effort. Scientific consensus around the term concerted molecular change in the context of transcriptomic dose-response must be developed both conceptually and methodologically. Broadly speaking, we understand a CMC to be the dose-dependent inflection point in gene expression where the frequency or breadth of change exceeds the stochastic gene variations of a control. Foundational to the identification of a CMC is the establishment of normal variation, as the intent is to identify a POD where there is a clear departure from a normal range. However, the experimental and informatic methodology to identify this inflection point prospectively in a rigorous, reproducible manner that can be used to extrapolate across species and from acute to chronic settings remains undefined. For example, a key challenge associated with signal detection is characterizing background noise in order to distinguish true and false positives. High false positive rates are a well-documented concern in ‘omics experiments due to the massive number of individual datapoints and subsequent statistical tests involved in any single experiment (Storey and Tibshirani, 2003). Empirical studies are required to characterize the occurrence of spurious dose-response behavior across the transcriptome and determine the most suitable filtering criteria to minimize any impacts on the final transcriptomic POD. The specific methods used to derive the POD are of critical importance. These will have to be calibrated to be protective of toxicity while balancing sensitivity and specificity. Further work is required to determine the methodology that yields a single highly repeatable and protective POD that provides adequate health protection. One of the significant benefits of the vision will be to identify a transcriptomic POD estimating a chronic apical POD from a study of much shorter duration. There is a growing body of evidence to support this idea, but the optimal exposure duration most protective of a chronic apical POD remains to be determined (Bianchi et al., 2021; Gwinn et al., 2020; LaRocca et al., 2020). Realization of Principle 3 will require further methodological development and discussion to build an accepted weight of evidence around reproducible methods and appropriate study design.
Principle 4: The use of a transcriptomic POD (set at the CMC dose level) will provide strong protection for use in risk assessment. If broad scientific consensus for Principles 1–3 can be established, it follows that Principle 4 will be well supported and robust. In the transformative risk assessment paradigm envisioned here, a POD based upon an adverse outcome (current method) would be replaced by a POD built on a transcriptome CMC (Principle 3). In current practice, the human or ecological chemical risk assessment process identifies a POD for the most sensitive adverse outcome (hazard), and this POD is compared with chemical exposure levels to determine appropriate chemical use. Adverse outcomes are derived from apical endpoints such as organ histopathology or high-level functional changes in the organism. Due to the hierarchical nature of organisms, all apical endpoints require prior change at the molecular level (Ankley et al., 2010; Krewski et al., 2020; Meek et al., 2014). Therefore, a transcriptomic POD based upon CMC would be protective of any adverse effect. Indeed, experimental studies examining the concordance of transcriptomic and apical POD values in various species show remarkable similarity between these 2 POD values (Bhat et al., 2013; Bianchi et al., 2021; Farmahin et al., 2017; Gwinn et al., 2020; Pagé-Larivière et al., 2019; Thomas et al., 2013). In cases where there is a potential risk, a tiered testing approach using cell-based, in vitro, or other assays could be used to obtain additional information about the toxicity profile.
Despite the promising experimental data cited here, there are outstanding questions that remain to be addressed before a transcriptomic POD can be used as the sole basis to derive a POD for risk assessment. For example, is the transcriptomic POD too conservative (much lower than a traditional adverse endpoint POD)? The published data suggest this is not the case, but more experimental work in this area is needed. Identification of a specific hazard with confidence is rarely achieved by examination of transcriptomic data; hence, amending what is an acceptable metric of change to base a POD on will be necessary. Another question relates to the breadth of toxicities that are captured by transcriptomics. Much of the published literature highlighted here has focused on chronic toxicity and carcinogenicity, but will this POD also be protective for developmental and reproductive toxicity? In some cases, chronic toxicity and carcinogenicity studies yield the most sensitive (lowest dose) POD. If the molecular landscape is comprehensively examined to derive a POD, when would mechanistic information to characterize the specific hazard still be required to protect human or ecological health? Answering this last question is perhaps the biggest, and most challenging, hurdle to the implementation of a transcriptomic POD in risk assessment.
CONCLUSION
We acknowledge that to significantly change the current toxicology testing paradigm there are major hurdles beyond the science, such as regulatory reform, which may be more challenging to overcome. The focus of this study is the presentation of a scientific framework and knowledge needed to determine the plausibility of the proposed approach to identify a chemical-specific, health-protective POD that will be acceptable for use in risk assessment. Challenges beyond the scientific plausibility of what is proposed will need to be addressed through different fora.
Furthermore, it is important to recognize that during the paradigm shift there may be the need for additional steps in the risk assessment process for substances where exposure levels exceed those deemed to be protective of human health; specifically, those substances identified to pose a risk to human health based on the transcriptomics POD. In such cases, refinements to the risk assessment parameters using more traditional testing approaches may be necessary to meet legislative requirements or to further inform risk management actions (ie, nature of the toxicity, severity, reversibility, and dose-related incidence), which is not characterized in the approach outlined here. As confidence is gained in the application of transcriptomics for the characterization of the vast landscape of hazard outcomes, it is envisioned that the need for additional toxicity studies will progressively be reduced.
The social psychologist, Dr Kurt Lewin, might well have been considering the challenges described here when he wrote that “If you want to truly understand something, try to change it.” We acknowledge that shifting away from an apical endpoint-driven regulatory toxicology paradigm to one anchored in short-term transcriptomic PODs will require the collective deepening and harmonization of our understanding of the interpretation of transcriptional signaling, specifically how dose-related changes in molecular processes as manifest in the transcriptome inform risk decisions that support public protection goals through chemical risk assessment. What is proposed here is intended to be a health protective and practical first step to the implementation of transcriptomic PODs in risk assessment that has significant potential for advancement and refinement as our biological and toxicological understanding of the transcriptome evolves. This logic framework will serve to organize and, hopefully, spur action toward this goal.
ACKNOWLEDGMENTS
We would like to thank John Bucher, Warren Casey, David Geter, Andrey Boyadzhiev, Matthew Meier, Jay Harriman, Nina Hallmark, and Doug Wolf for critical reading of the article. We also thank Members of the Health and Environmental Sciences Institute (HESI) eSTAR Committee. HESI is a nonprofit organization whose mission is to collaboratively create science-based solutions to various challenges affecting human health and the environment. HESI’s activities are organized around scientific committees comprised scientists from academia, NGOs, government, and industry.
FUNDING
This research was supported, in part, by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS). The HESI scientific initiative is primarily supported by in-kind contributions (from public and private sector participants) of time, expertise, and experimental effort. These contributions are supplemented by direct funding (that largely supports program infrastructure and management) that was provided by HESI’s corporate sponsors. A list of supporting organizations is available at www.hesiglobal.org (last accessed October 6, 2022).
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Disclaimer: Some of the information in this study has been funded by the U.S. Environmental Protection Agency. It has been subjected to review by the Center for Computational Toxicology and Exposure and approved for publication. Approval neither signifies that the content reflects the views of the Agency nor mentions trade names or commercial products that constitute endorsement or recommendation for use. The opinions expressed in this article do not reflect the views or policies of Health Canada, the Division of the National Toxicology Program, or the National Institute of Environmental Health Sciences.
Contributor Information
Kamin J Johnson, Predictive Safety Center, Corteva Agriscience, Indianapolis, Indiana 46268, USA.
Scott S Auerbach, Predictive Toxicology Branch, Division of the Translational Toxicology, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina 27709, USA.
Tina Stevens, Drug Safety and Drug Metabolism and Pharmacokinetics, Boehringer-Ingelheim, Athens, Georgia 30601, USA.
Tara S Barton-Maclaren, Healthy Environments and Consumer Safety Branch, Health Canada, Ottawa, Ontario K1P 5J9, Canada.
Eduardo Costa, Predictive Safety Center, Corteva Agriscience, Indianapolis, Indiana 46268, USA.
Richard A Currie, Product Safety, Syngenta Jealotts Hill International Research Center, Bracknell RG42 6EY, UK.
Deidre Dalmas Wilk, Investigative Safety, In vitro In Vivo Translation, GlaxoSmithKline, Collegeville, Pennsylvania 19426, USA.
Saddef Haq, Health and Environmental Sciences Institute, Washington, District of Columbia 20005, USA.
Julia E Rager, The Department of Environmental Sciences and Engineering, Gillings School of Global Public Health, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
Anthony J F Reardon, Healthy Environments and Consumer Safety Branch, Health Canada, Ottawa, Ontario K1P 5J9, Canada.
Leah Wehmas, U.S. Environmental Protection Agency, Office of Research and Development, Center for Computational Toxicology and Exposure, Research Triangle Park, North Carolina 27711, USA.
Andrew Williams, Healthy Environments and Consumer Safety Branch, Health Canada, Ottawa, Ontario K1P 5J9, Canada.
Jason O’Brien, Wildlife Toxicology Research Section, Environment and Climate Change Canada, Ottawa, Ontario K1A 0H3, Canada.
Carole Yauk, Department of Biology, University of Ottawa, Ottawa, Ontario K1N 6N5, Canada.
Jessica L LaRocca, Predictive Safety Center, Corteva Agriscience, Indianapolis, Indiana 46268, USA.
Syril Pettit, Health and Environmental Sciences Institute, Washington, District of Columbia 20005, USA.
REFERENCES
- Ankley G. T., Bennett R. S., Erickson R. J., Hoff D. J., Hornung M. W., Johnson R. D., Mount D. R., Nichols J. W., Russom C. L., Schmieder P. K., et al. (2010). Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Bhat V. S., Hester S. D., Nesnow S., Eastmond D. A. (2013). Concordance of transcriptional and apical benchmark dose levels for conazole-induced liver effects in mice. Toxicol. Sci. 136, 205–215. [DOI] [PubMed] [Google Scholar]
- Bianchi E., Costa E., Yan Z. J., Murphy L., Howell J., Anderson D., Mukerji P., Venkatraman A., Terry C., Johnson K. J. (2021). A rat subchronic study transcriptional point of departure estimates a carcinogenicity study apical point of departure. Food Chem. Toxicol. 147, 111869. [DOI] [PubMed] [Google Scholar]
- Calabrese E. J., Bachmann K. A., Bailer A. J., Bolger P. M., Borak J., Cai L., Cedergreen N., Cherian M. G., Chiueh C. C., Clarkson T. W., et al. (2007). Biological stress response terminology: Integrating the concepts of adaptive response and preconditioning stress within a hormetic dose–response framework. Toxicol. Appl. Pharmacol. 222, 122–128. [DOI] [PubMed] [Google Scholar]
- Davis A. P., Grondin C. J., Johnson R. J., Sciaky D., Wiegers J., Wiegers T. C., Mattingly C. J. (2021). Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 49, D1138–D1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmahin R., Williams A., Kuo B., Chepelev N. L., Thomas R. S., Barton-Maclaren T. S., Curran I. H., Nong A., Wade M. G., Yauk C. L. (2017). Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch. Toxicol. 91, 2045–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (n.d.). MicroArray/Sequencing Quality Control (MAQC/SEQC). Available at: https://www.fda.gov/science-research/bioinformatics-tools/microarraysequencing-quality-control-maqcseqc. Accessed February 15, 2022.
- Gwinn W. M., Auerbach S. S., Parham F., Stout M. D., Waidyanatha S., Mutlu E., Collins B., Paules R. S., Merrick B. A., Ferguson S., et al. (2020). Evaluation of 5-day in vivo rat liver and kidney with high-throughput transcriptomics for estimating benchmark doses of apical outcomes. Toxicol. Sci. 176, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber L. T., Dourson M. L., Allen B. C., Hertzberg R. C., Parker A., Vincent M. J., Maier A., Boobis A. R. (2018). Benchmark dose (BMD) modeling: Current practice, issues, and challenges. Crit. Rev. Toxicol. 48, 387–415. [DOI] [PubMed] [Google Scholar]
- Harrill J., Shah I., Setzer R. W., Haggard D., Auerbach S., Judson R., Thomas R. S. (2019). Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions. Curr. Opin. Toxicol. 15, 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J. A., Viant M. R., Yauk C. L., Sachana M., Gant T. W., Auerbach S. S., Beger R. D., Bouhifd M., O’Brien J., Burgoon L., et al. (2021). Progress towards an OECD reporting framework for transcriptomics and metabolomics in regulatory toxicology. Regul. Toxicol. Pharmacol. 125, 105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Pennie W. (2003). Toxicogenomics (Inoue T., Pennie W. D., Eds.). Springer, Tokyo, Japan. doi: 10.1007/978-4-431-66999-9. [DOI] [Google Scholar]
- Johnson K. J., Auerbach S. S., Costa E. (2020). A rat liver transcriptomic point of departure predicts a prospective liver or non-liver apical point of departure. Toxicol. Sci. 176, 86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewski D., Acosta D., Andersen M., Anderson H., Bailar J. C., Boekelheide K., Brent R., Charnley G., Cheung V. G., Green S., et al. (2010). Toxicity testing in the 21st century: A vision and a strategy. J. Toxicol. Environ. Health. B Crit. Rev. 13, 51–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewski D., Andersen M. E., Tyshenko M. G., Krishnan K., Hartung T., Boekelheide K., Wambaugh J. F., Jones D., Whelan M., Thomas R., et al. (2020). Toxicity testing in the 21st century: Progress in the past decade and future perspectives. Arch. Toxicol. 94, 1–58. [DOI] [PubMed] [Google Scholar]
- Kültz D. (2020). Defining biological stress and stress responses based on principles of physics. J. Exp. Zool. A Ecol. Integr. Physiol. 333, 350–358. [DOI] [PubMed] [Google Scholar]
- LaRocca J., Costa E., Sriram S., Hannas B. R., Johnson K. J. (2020). Short-term toxicogenomics as an alternative approach to chronic in vivo studies for derivation of points of departure: A case study in the rat with a triazole fungicide. Regul. Toxicol. Pharmacol. 113, 104655. [DOI] [PubMed] [Google Scholar]
- Meek M. E., Boobis A., Cote I., Dellarco V., Fotakis G., Munn S., Seed J., Vickers C. (2014). New developments in the evolution and application of the WHO/IPCS framework on mode of action/species concordance analysis. J. Appl. Toxicol. 34, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticello T. M., Jones T. W., Dambach D. M., Potter D. M., Bolt M. W., Liu M., Keller D. A., Hart T. K., Kadambi V. J. (2017). Current nonclinical testing paradigm enables safe entry to First-In-Human clinical trials: The IQ consortium nonclinical to clinical translational database. Toxicol. Appl. Pharmacol. 334, 100–109. [DOI] [PubMed] [Google Scholar]
- National Research Council (2005). Toxicogenomic Technologies and Risk Assessment of Environmental Carcinogens: A Workshop Summary Committee on How Toxicogenomics Could Inform Critical Issues in Carcinogenic Risk Assessment of Environmental Chemicals, Committee on Emerging Issues and Data. National Academies Press. Available at: http://www.nap.edu/catalog/11335.html. Accessed October 6, 2022. [Google Scholar]
- National Toxicology Program (2018). NTP Research Report on National Toxicology Program Approach to Genomic Dose-Response Modeling: Research Report 5. [PubMed]
- Pagé-Larivière F., Crump D., O'Brien J. M. (2019). Transcriptomic points-of-departure from short-term exposure studies are protective of chronic effects for fish exposed to estrogenic chemicals. Toxicol. Appl. Pharmacol. 378, 114634. [DOI] [PubMed] [Google Scholar]
- Parish S. T., Aschner M., Casey W., Corvaro M., Embry M. R., Fitzpatrick S., Kidd D., Kleinstreuer N. C., Lima B. S., Settivari R. S., et al. (2020). An evaluation framework for new approach methodologies (NAMs) for human health safety assessment. Regul. Toxicol. Pharmacol. 112, 104592. [DOI] [PubMed] [Google Scholar]
- Phillips J. R., Svoboda D. L., Tandon A., Patel S., Sedykh A., Mav D., Kuo B., Yauk C. L., Yang L., Thomas R. S., et al. (2019). Bmdexpress 2: Enhanced transcriptomic doseresponse analysis workflow. Bioinformatics 35, 1780–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon A. J. F., Rowan-Carroll A., Ferguson S. S., Leingartner K., Gagne R., Kuo B., Williams A., Lorusso L., Bourdon-Lacombe J. A., Carrier R., et al. (2021). Potency ranking of per- and polyfluoroalkyl substances using high-throughput transcriptomic analysis of human liver spheroids. Toxicol. Sci. 184, 154–169. [DOI] [PubMed] [Google Scholar]
- Sewell F., Lewis D., Mehta J., Terry C., Kimber I. (2021). Rethinking agrochemical safety assessment: A perspective. Regul. Toxicol. Pharmacol. 127, 105068. [DOI] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U S A 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki A. O., Barton-Maclaren T. S., Bhuller Y., Henriquez J. E., Henry T. R., Hirn C., Miller-Holt J., Nagy E. G., Perron M. M., Ratzlaff D. E., et al. (2022). Use of new approach methodologies (NAMs) to meet regulatory requirements for the assessment of industrial chemicals and pesticides for effects on human health. Front. Toxicol. 4, 964553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y. M., Barton H. A., Boobis A., Brunner R., Clewell H., Cope R., Dawson J., Domoradzki J., Egeghy P., Gulati P., et al. (2021). Opportunities and challenges related to saturation of toxicokinetic processes: Implications for risk assessment. Regul. Toxicol. Pharmacol. 127, 105070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. R., Wesselkamper S. C., Wang N. C. Y., Zhao J. J., Petersen D. D., Lambert J. C., Cote I., Yang L., Healy E., Black M. B., et al. (2013). Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol. Sci. 134, 180–194. [DOI] [PubMed] [Google Scholar]
- US Environmental Protection Agency (2016). Halauxifen-Methyl—New Active Ingredient Human Health Risk Assessment for Proposed Uses on Cereal Grains (Barley, Wheat, and Triticale). Available at: https://www.regulations.gov/document/EPA-HQ-OPP-2012-0919-0005. Accessed October 6, 2022.
- US EPA (n.d.). Administrator Memo Prioritizing Efforts to Reduce Animal Testing, September 10, 2019. Available at: https://www.epa.gov/research/administrator-memo-prioritizing-efforts-reduce-animal-testing-september-10-2019. Accessed January 29, 2022.
- Verheijen M., Tong W., Shi L., Gant T. W., Seligman B., Caiment F. (2020). Towards the development of an omics data analysis framework. Regul. Toxicol. Pharmacol. 112, 104621. [DOI] [PubMed] [Google Scholar]
- Wang P., Xia P., Yang J., Wang Z., Peng Y., Shi W., Villeneuve D. L., Yu H., Zhang X., Baird D. J. (2018). Reduced transcriptomic approach for screening and prediction of chemical toxicity. Chem. Res. Toxicol. 31, 532–533. [DOI] [PubMed] [Google Scholar]
- Wax P. M. (1995). Elixirs, diluents, and the passage of the 1938 federal food, drug and cosmetic act. Ann. Intern. Med. 122, 456–461. [DOI] [PubMed] [Google Scholar]
- Yauk C. L., Harrill A. H., Ellinger-Ziegelbauer H., van der Laan J. W., Moggs J., Froetschl R., Sistare F., Pettit S. (2020). A cross-sector call to improve carcinogenicity risk assessment through use of genomic methodologies. Regul. Toxicol. Pharmacol. 110, 104526. [DOI] [PMC free article] [PubMed] [Google Scholar]