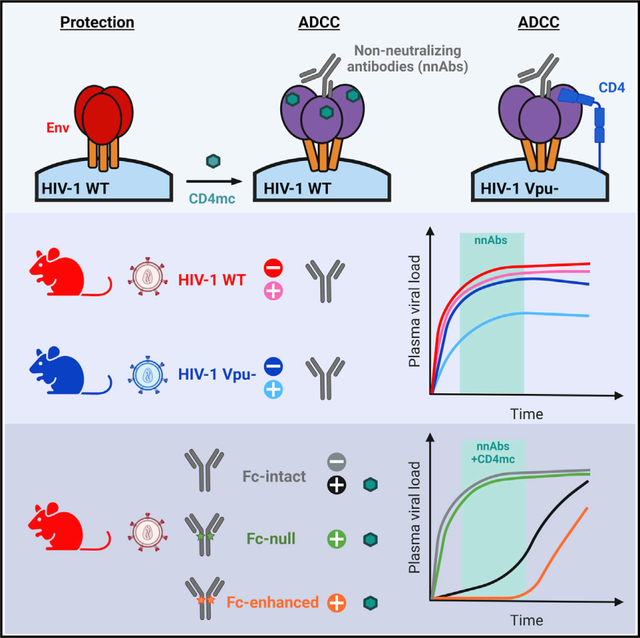
SUMMARY
Non-neutralizing antibodies (nnAbs) can eliminate HIV-1-infected cells via antibody-dependent cellular cytotoxicity (ADCC) and were identified as a correlate of protection in the RV144 vaccine trial. Fc-mediated effector functions of nnAbs were recently shown to alter the course of HIV-1 infection in vivo using a vpu-defective virus. Since Vpu is known to downregulate cell-surface CD4, which triggers conformational changes in the viral envelope glycoprotein (Env), we ask whether the lack of Vpu expression was linked to the observed nnAbs activity. We find that restoring Vpu expression greatly reduces nnAb recognition of infected cells, rendering them resistant to ADCC. Moreover, administration of nnAbs in humanized mice reduces viral loads only in animals infected with a vpu-defective but not with a wild-type virus. CD4-mimetics administration, known to “open” Env and expose nnAb epitopes, renders wild-type viruses sensitive to nnAbs Fc-effector functions. This work highlights the importance of Vpu-mediated evasion of humoral responses.
In brief
Prévost et al. demonstrate that the HIV-1 accessory protein Vpu protects infected cells from Fc-effector functions both in vitro and in vivo. Furthermore, they show that non-neutralizing antibodies (nnAb) fail to alter HIV-1 replication in humanized mice unless small CD4-mimetics are used to “open up” Env and enable nnAb interaction.
Graphical Abstract
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins (Env) mediate viral entry into target cells. Env is synthesized as a trimeric gp160 precursor, which is further cleaved into two subunits linked by non-covalent bonds: the exterior gp120 and the transmembrane gp41 subunits (Earl et al., 1990; Freed et al., 1989; McCune et al., 1988). During the entry process, the gp120 subunit sequentially interacts with the CD4 surface molecule as well as one of its co-receptors (CCR5 or CXCR4) (Kwong et al., 1998; Shaik et al., 2019; Wu et al., 2010a), allowing the gp41 subunit to mediate fusion between the viral and target cell lipid membranes (Chan et al., 1997; Weissenhorn et al., 1997). Since Env is the sole viral antigen exposed at the surface of virions and infected cells, it represents the main target for humoral responses. Fusion-competent Env trimers can sample different conformations. Broadly neutralizing antibodies (bNAbs) mainly recognize the pre-fusion “closed” conformation (Li et al., 2020; Lu et al., 2019; Munro et al., 2014; Stadtmueller et al., 2018), while non-neutralizing antibodies (nnAbs) mainly bind to “open” CD4-bound conformations (Alsahafi et al., 2019; Jette et al., 2021; Munro et al., 2014; Yang et al., 2019). Primary difficult-to-neutralize HIV-1 isolates favor the pre-fusion “closed” conformation, thus exposing Env regions that are heavily glycan shielded (Doores, 2015; Lee et al., 2016; Li et al., 2020).
Since antiretroviral therapy (ART) is unable to eradicate HIV-1 reservoirs, monotherapy, or combination of bNAbs targeting the CD4-binding site (3BNC117, VRC01, VRC07-523), the V3 glycan supersite (10–1074, PGT121), and the V2 apex (PGDM1400) are currently under investigation in multiple clinical trials as therapeutic agents to reduce or eliminate cellular reservoirs through Fc-mediated effector functions (NCT02140255, NCT03837756, NCT04319367, NCT03721510). Thus far, results have shown that bNAbs can control HIV-1 viremia and delay viral rebound upon treatment interruption in HIV-1-infected individuals (Bar et al., 2016; Caskey et al., 2015,2017; Lynch et al., 2015; Scheid et al., 2016). Similar outcomes were also observed in non-human primates (NHPs) and humanized mice (Barouch et al., 2013; Bolton et al., 2016; Freund et al., 2017; Halper-Stromberg et al., 2014; Hessell et al., 2016; Horwitz et al., 2013; Klein et al., 2012; Nishimura et al., 2017; Parsons et al., 2019; Schommers et al., 2020; Schoofs et al., 2019; Shingai et al., 2013). In vivo studies in animal models have demonstrated that Fc-mediated effector functions are required for the optimal therapeutic activity of bNAbs (Asokan et al., 2020; Bournazos et al., 2014; Halper-Stromberg et al., 2014; Lu et al., 2016; Wang et al., 2020). However, bNAbs rarely arise during natural infection and have yet to be consistently elicited by vaccination (Landais and Moore, 2018; Pauthner et al., 2019).
Given the difficulty of eliciting bNAbs in vivo, nnAbs have been evaluated as a potential alternative. nnAbs represent the majority of antibodies (Abs) in the plasma of HIV-1-infected individuals and are easily elicited by vaccination (Beaudoin-Bussières et al., 2020; Davis et al., 2009; Decker et al., 2005; Madani et al., 2016; Madani et al., 2018; Tomaras and Haynes, 2009; Tomaras et al., 2008; Visciano et al., 2019). Despite poor neutralization capacity, nnAbs can mediate other functions, such as the elimination of HIV-1-infected cells by Ab-dependent cellular cytotoxicity (ADCC) or Ab-dependent cellular phagocytosis (ADCP). Among these functions, ADCC was associated with the protection observed in the RV144 vaccine trial (Haynes et al., 2012a; Rerks-Ngarm et al., 2009). Thus, several studies have examined the antiviral effects of nnAbs in NHPs and humanized mice. These included prophylactic administration of nnAbs targeting the CD4-binding site (b6), the V3 loop (KD-247, 2219), the V1V2 region (830A, 2158), the gp120 cluster A (A32), and the gp41 immunodominant region (246D, 4B3, F240, 7B2). The results from these studies showed a reduction in the number of transmitted/founder (T/F) viruses and/or plasma viral loads after challenge with lab-adapted tier 1 viruses (Burton et al., 2011; Eda et al., 2006; Hessell et al., 2018; Hioe et al., 2022; Moog et al., 2014; Santra et al., 2015). Finally, therapeutic administration of large quantities of the monoclonal anti-gp41 246D nnAb to humanized mice infected with a vpu-deleted tier 2 HIV-1 molecular clone (HIV-1NL4/3YU2) led to the elimination of infected cells and selected for escape mutations that stabilized the Env “closed” conformation in an Fc-dependent manner, suggesting a protective effect (Horwitz et al., 2017). However, other studies administrating a cocktail of nnAbs (A32 and 17b) to humanized mice infected with a wild-type (WT) tier 2 HIV-1 strain (JR-CSF) had no impact on viral replication, except when combined with a small CD4 mimetic compound (CD4mc) that “open up” the trimer and expose these otherwise occluded epitopes (Rajashekar et al., 2021).
Differences between the various nnAb studies could be attributed to the specificity of the Abs (anti-gp41 versus anti-gp120), the humanized mouse model used or specific viral determinants. In particular, certain studies conducted using an infectious molecular clone (IMC) of HIV-1 that expressed a tier 2 Env (YU2) lacked a functional Vpu (HIV-1NL4/3YU2). However, Vpu plays an important role in downregulating cell-surface CD4, which can bind and trigger conformational changes in the viral Env, therefore exposing vulnerable epitopes (Prévost et al., 2022; Veillette et al., 2014, 2015). We thus asked whether the lack of a functional Vpu was responsible for the observed nnAbs activity by comparing isogenic viruses that differed solely in Vpu expression. Importantly, we tested the influence of Vpu expression on nnAb function not only in vitro but also in humanized mice. We found that anti-gp41 246D nnAb mediates potent ADCC responses against HIV-1NL4/3YU2, but not its Vpu-positive (Vpu+) counterpart. Accordingly, 246D was found to alter viral replication in vivo only in the absence of Vpu. These data thus provide conclusive evidence that Vpu allows HIV-1 to evade humoral responses in vivo and emphasizes the need to use fully functional IMCs to assess nnAb Fc-mediated effector functions.
RESULTS
Elicitation of anti-gp41 nnAbs following HIV-1 infection
First, we characterized the susceptibility of the HIV-1NL4/3YU2 IMC to nnAbs-mediated Fc-effector responses to confirm previous observations (Horwitz et al., 2017). To do so, we used plasma samples from a cross-sectional cohort of 50 HIV-1-infected individuals (HIV+ plasma), which were grouped according to the inferred time post infection and ART treatment (Table S1). The nnAbs present in the HIV+ plasma from this cohort were previously shown to mediate potent ADCC responses against infected cells presenting Env in the “open” CD4-bound conformation (Ding et al., 2016a; Richard et al., 2015; Veillette et al., 2015). We infected primary CD4+ T cells with the HIV-1NL4/3YU2 IMC and evaluated the ability of HIV+ plasma to recognize and eliminate infected cells. Consistent with its susceptibility to nnAbs, HIV-1NL4/3YU2-infected primary CD4+ T cells were efficiently recognized (Figure 1A) and susceptible to ADCC (Figure 1B) mediated by all the tested plasma samples, with a significant increase of activity starting 6 months post infection. Similar levels of activity were also present in plasma from ART-treated individuals (Figures 1A and 1B).
Figure 1. Cells infected with HIV-1NL4/3YU2 are susceptible to nnAb-mediated ADCC responses.
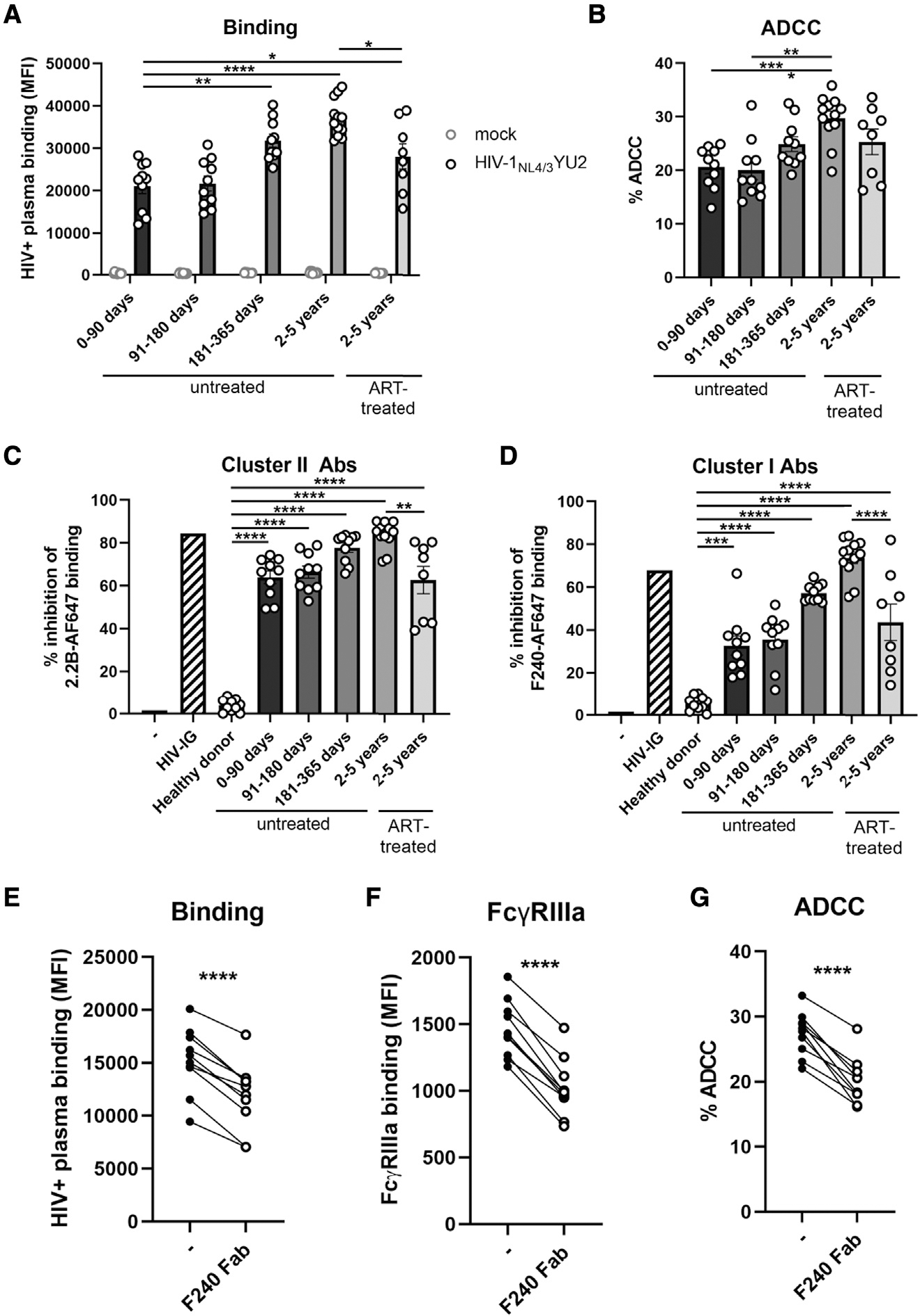
(A and B) Primary CD4+ T cells were either mock infected (gray symbols) or infected with the HIV-1NL4/3YU2 virus (black symbols). (A) Cell-surface staining and (B) ADCC responses mediated by plasma from 50 different HIV-1-infected individuals categorized by the time post infection and the antiretroviral therapy (ART) treatment status. (A) The graph shows the mean fluorescence intensities (MFIs) on the whole cell population (mock) or infected p24+ cell population (HIV-1NL4/3YU2).
(C and D) The binding of Alexa Fluor 647 (AF647)-precoupled (C) anti-gp41 cluster II 2.2B mAb or (D) anti-gp41 cluster I F240 mAb was evaluated in presence of unlabeled human plasma from healthy controls or HIV-1-infected individuals. A pool of purified immunoglobulins from HIV-1-infected individuals (HIV-IG) was used as a positive control.
(E–G) (E) Plasma binding, (F) FcγRIIIa binding, and (G) ADCC responses mediated by plasma from 10 untreated HIV-1-infected individuals (2–5 years) were evaluated in the presence of competing antigp41 cluster I F240 Fab fragment. Error bars indicate mean ± standard error of the means (SEM). Statistical significance was tested using (A–D) one-way ANOVA with a Holm-Sidak post test or (E–G) a paired t test (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, nonsignificant).
Since the HIV-1NL4/3YU2 IMC has been reported to be sensitive to anti-gp41 Fc-mediated antiviral activity in vivo (Horwitz et al., 2017), we evaluated the contribution of anti-gp41 nnAbs present in HIV+ plasma to infected-cell recognition and ADCC activity by performing binding competition experiments (Figures 1C and 1D). We focused on two main classes of antigp41 nnAbs based on observations from previous studies showing potent ADCC responses (Ding et al., 2016a; Gohain et al., 2016; Moog et al., 2014; Santra et al., 2015; Sojar et al., 2019; von Bredow et al., 2016; Williams et al., 2019; Yang et al., 2018): the anti-cluster II Abs targeting the heptad repeat region 2 (HR2) (Frey et al., 2010) and anti-cluster I Abs targeting the disulfide loop region (C-C loop) (Gohain et al., 2016; Santra et al., 2015) (Figure S1A). Anti-gp41 cluster II nnAbs, inferred from binding competition experiments using the prototypic anti-cluster II 2.2B nnAb, were elicited in the acute phase of the infection (within 90 days) (Figure 1C). Elicitation of anti-gp41 cluster I nnAbs appears to take more time, as revealed in binding competition experiments using the prototypic F240 anti-cluster I nnAb. While some competition with F240 binding was observed within the first 3 months post infection, it culminates in the chronic phase of the infection (more than 2 years) (Figure 1D). In agreement with previous studies showing potent ADCC activity by anticluster I gp41 monoclonal Abs (Ding et al., 2016a; Gohain et al., 2016; Moog et al., 2014; Santra et al., 2015; von Bredow et al., 2016; Williams et al., 2019), we observed that blockade with anti-cluster I gp41 F240 Fab fragment significantly decreased plasma binding, FcγRIIIa engagement, and ADCC responses against HIV-1NL4/3YU2-infected cells (Figures 1E–1G).
Vpu protects HIV-1-infected cells from recognition and Fc-effector functions mediated by anti-Env Abs in vitro and ex vivo
Since the HIV-1NL4/3YU2 does not express the accessory protein Vpu due to a mutation in the start codon of the vpu gene (Horwitz et al., 2017), we asked whether the efficient recognition and ADCC-mediated elimination of HIV-1NL4/3YU2 infected primary CD4+ T cells by nnAbs was linked to the lack of Vpu expression by this IMC. We restored the vpu open reading frame (ORF; Figure 2A) and infected primary CD4+ T cells using both Vpu-negative (Vpu−) and Vpu+ constructs. Using a recently developed fluorescence-activated cell sorting (FACS)-based intracellular staining method (Prévost et al., 2022), we confirmed Vpu expression upon restoration of the vpu ORF (Figure 2B). We also confirmed by intracellular staining the equivalent expression of Nef in both IMCs (Figure 2B). Vpu efficiently downregulated cell-surface CD4 and BST-2 (Figure 2C). We also measured cell-surface expression of NTB-A and PVR, which were shown to modulate ADCC responses against HIV-1-infected cells (Prévost et al., 2019) and are downregulated by Vpu (Matusali et al., 2012; Shah et al., 2010). As expected, Vpu expression significantly downregulated their expression from the surface of primary CD4+ T cells from five different healthy individuals (Figures 2D and 2E).
Figure 2. Reversion of Vpu open reading frame in the HIV-1NL4/3YU2 construct.
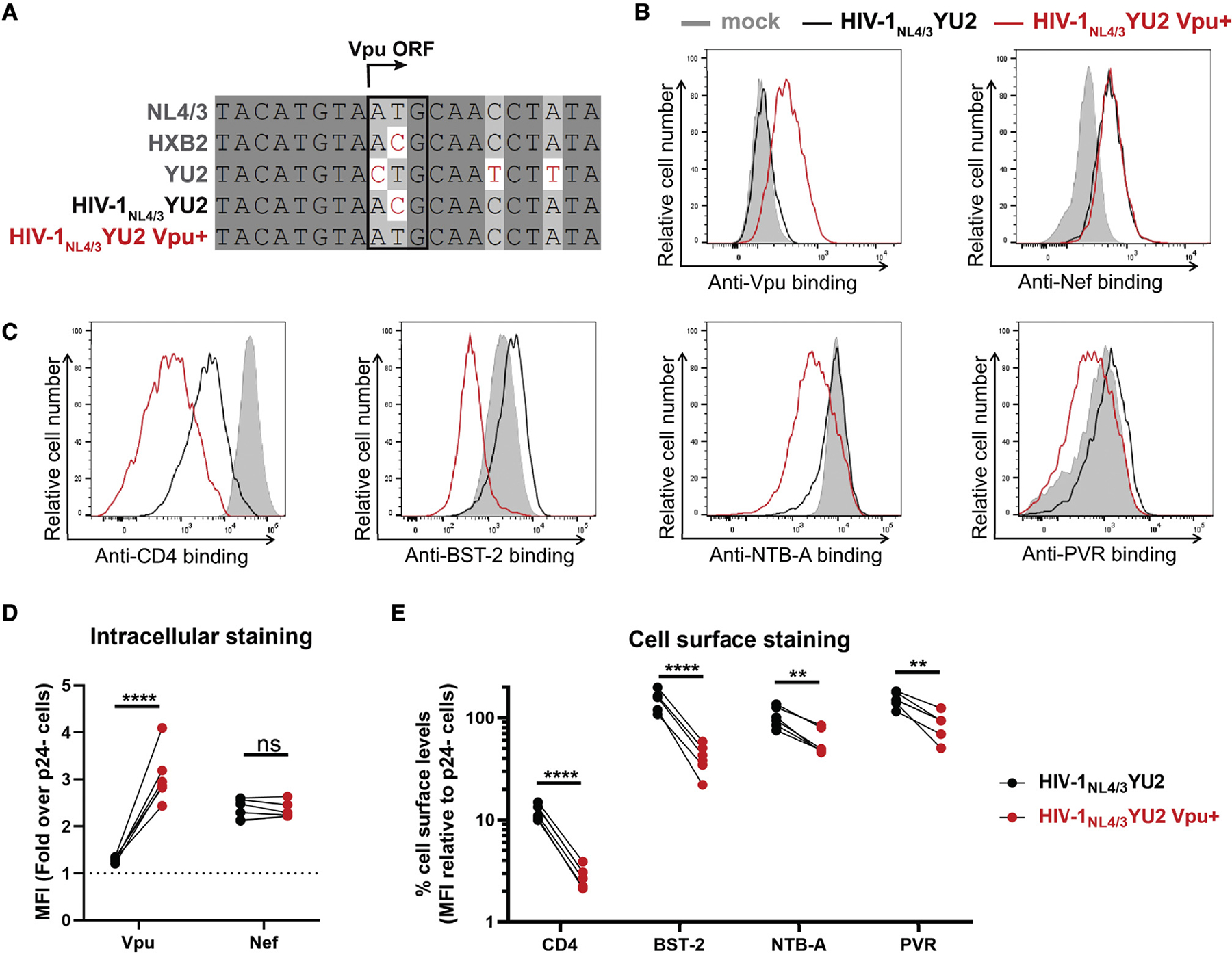
(A) Sequence alignment of the Vpu open reading frame (ORF) region of selected HIV-1 reference isolates (NL4/3, HXB2, YU2) with the HIV-1NL4/3YU2 construct and its Vpu+ counterpart. Identical nucleotides are shaded in dark gray, conserved nucleotides in light gray, and non-identical nucleotides are highlighted in red. The vpu start codon is highlighted by a black box.
(B–E) Primary CD4+ T cells either mock infected or infected with the HIV-1NL4/3YU2 virus or its Vpu+ counterpart were stained for intracellular detection of (B and D) viral proteins Vpu and Nef and cell-surface detection of (C and E) cellular proteins CD4, BST-2, NTB-A, and PVR.
(B and C) Histograms depicting representative stainings for (B) viral proteins and (C) their target substrates.
(D and E) The graphs show the MFIs obtained from five independent experiments using primary cells from five different healthy donors.
(D) Viral protein levels were reported as a fold increase in the signal detected on infected p24+ cells compared with uninfected p24− cells.
(E) Cellular protein levels were reported as a percentage of detection at the surface of infected p24+ cells compared with uninfected p24− cells. Error bars indicate means ± SEM. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (**p < 0.01; ****p < 0.0001; ns, nonsignificant).
We next evaluated the effect of Vpu on the recognition and elimination of infected cells by nnAbs. Consistent with the observed downmodulation of CD4 and BST-2, Vpu expression strongly reduced the recognition of infected cells by monoclonal Abs (mAbs) targeting the anti-gp41 cluster I (246D), the gp120 cluster A (A32), or by polyclonal HIV+ plasma (Figure 3A). We extended these results to a panel of 27 nnAbs and 10 HIV+ plasma, which yielded the same results (Figures 3B and 3C). As a measure of this panel of nnAbs to mediate Fc-effector functions, we examined their ability to interact with a soluble dimeric FcγRIIIa protein. This recombinant protein is used as a surrogate of FcγR clustering, which is required to trigger Fceffector functions (Anand et al., 2019; Wines et al., 2016, 2017). Consistent with a significant reduction in the recognition of infected cells by nnAbs, we observed that Vpu expression diminished the ability of all mAbs and HIV+ plasma to engage with FcγRIIIa (Figures 3D and 3E) and to mediate ADCC (Figures 3F and 3G). We noted that nnAbs targeting the gp41 were more potent at engaging soluble dimeric FcγRIIIa and mediating ADCC against cells infected with the vpu-defective IMC than the panel of anti-gp120 nnAbs tested (Figures S2A–S2C).
Figure 3. Vpu expression impairs Env recognition and Fc-effector functions mediated by anti-Env nnAbs.
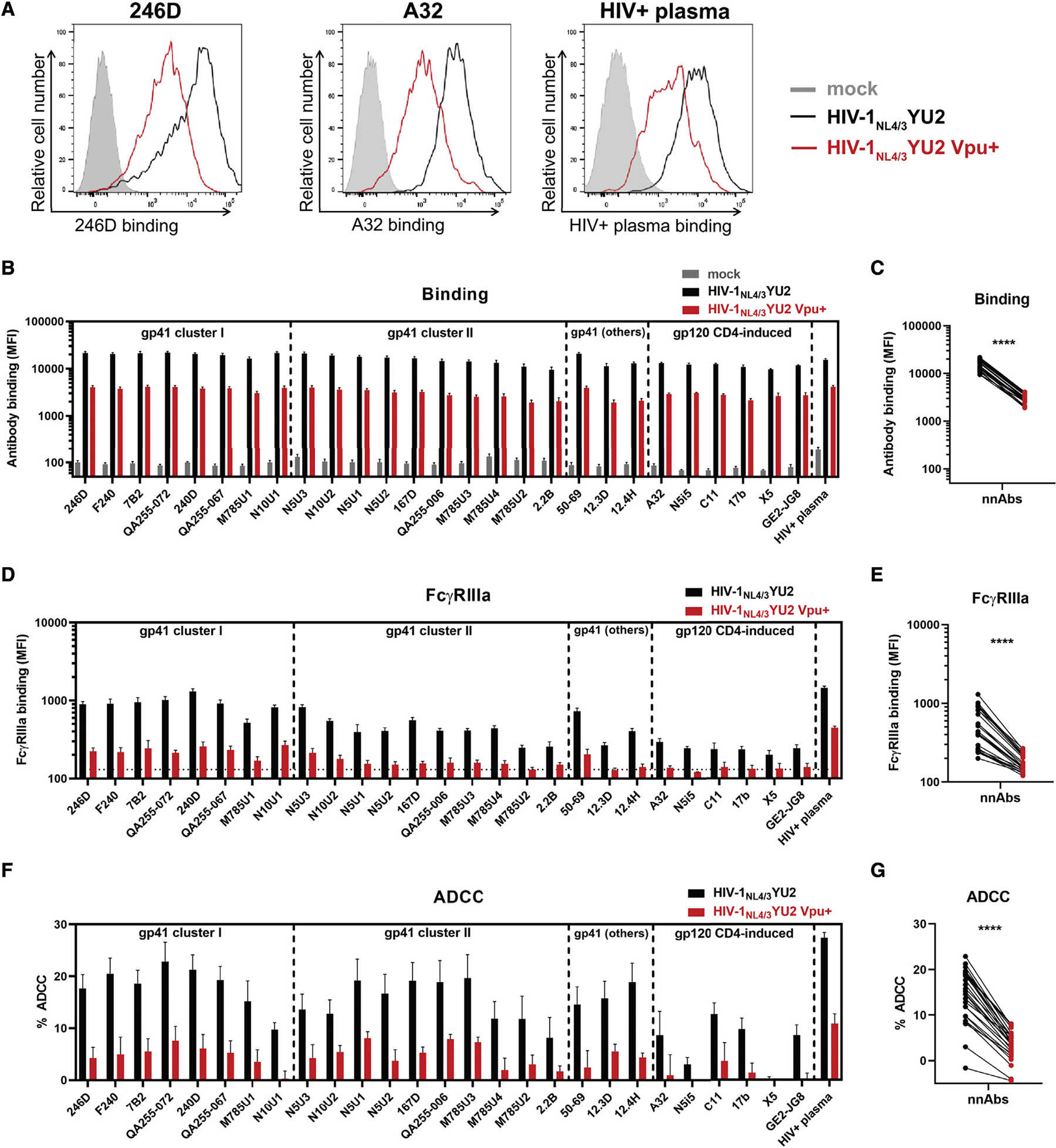
Primary CD4+ T cells were either mock infected or infected with the HIV-1NL4/3YU2 virus or its Vpu+ counterpart.
(A–E) Cell-surface staining performed using anti-gp41 nnAbs targeting the gp41 cluster I, cluster II, or other gp41 regions as well as anti-gp120 CD4-induced Abs and plasma from 10 HIV-1-infected individuals (HIV+ plasma). Ab binding was detected either by using (A–C) Alexa Fluor 647-conjugated anti-human secondary Abs or (D and E) by using biotinylated recombinant soluble dimeric FcγRIIIa followed by the addition of Alexa Fluor 647-conjugated streptavidin.
(A) Histograms depicting representative stainings using anti-gp41 cluster I 246D, mAb, anti-gp120 cluster A A32 mAb and HIV+ plasma.
(B and D) The graphs represent the MFI obtained from the infected p24+ cell population using cells from five different healthy donors. (D) The horizontal dotted line represents the signal obtained in absence of mAb.
(F and G) Primary CD4+ T cells infected with HIV-1NL4/3YU2 viruses were used as target cells. Autologous peripheral blood mononuclear cells (PBMCs) were used as effector cells in a FACS-based ADCC assay.
(F) The graph represents the percentages of ADCC obtained in the presence of the respective Abs using cells from five different healthy donors. Error bars indicate means ± SEM.
(C, E, and G) Statistical significance was tested using a paired t test (****, p < 0.0001).
Vpu facilitates viral release by downregulating the restriction factor BST-2 (Neil et al., 2008; Van Damme et al., 2008). Multiple studies have shown that this activity decreases the overall amount of Env at the cell surface, consequently decreasing the susceptibility of infected cells to ADCC (Alvarez et al., 2014; Arias et al., 2014; Pham et al., 2016; Richard et al., 2017; Veillette et al., 2014). Since the vpu-defective HIV-1NL4/3YU2 was used to evaluate the in vivo activity of several bNAbs (Bournazos et al., 2014,2016; Diskin et al., 2013; Freund et al., 2015, 2017; Halper-Stromberg et al., 2014; Horwitz et al., 2013; Klein et al., 2012,2014; Lu et al., 2016; Schommers et al., 2020; Schoofs et al., 2019; Vanshylla et al., 2021), we tested whether their binding to infected cells was also influenced by Vpu expression. Consistent with decreased Env expression on cells infected with a Vpu+ virus, we observed a significant reduction in CD4-binding site (3BNC117), V3 glycan (10–1074), and V2-apex (PG16) bNAb binding (Figure 4A). The phenotype was validated using a panel of 35 bNAbs targeting different epitopes, indicative of an overall reduction of cell-surface Env expression in the presence of a functional Vpu (Figures 4B and 4C). Accordingly, infected cells were significantly less susceptible to bNAbs-mediated Fc-effector functions (Figures 4D–4G). Among the different classes of bNAbs, Abs targeting the V3 glycan supersite, the CD4-binding site, and the V2 apex demonstrated stronger ADCC-mediated killing of cells infected with the vpu-defective HIV-1NL4/3YU2, compared with Abs known to interact with the gp120 silent face, the gp120-gp41 interface, or the gp41 membrane-proximal external region (MPER), despite similar levels of Env recognition (Figures S2D–S2F). These findings support previous observations indicating that, in addition to Env recognition, the angle of approach of the Ab is important to mediate ADCC as it modulates the exposure of the Fc region required to activate effector cells (Acharya et al., 2014; Tolbert et al., 2020).
Figure 4. Vpu expression decreases Env recognition and Fc-effector functions mediated by anti-Env bNAbs.
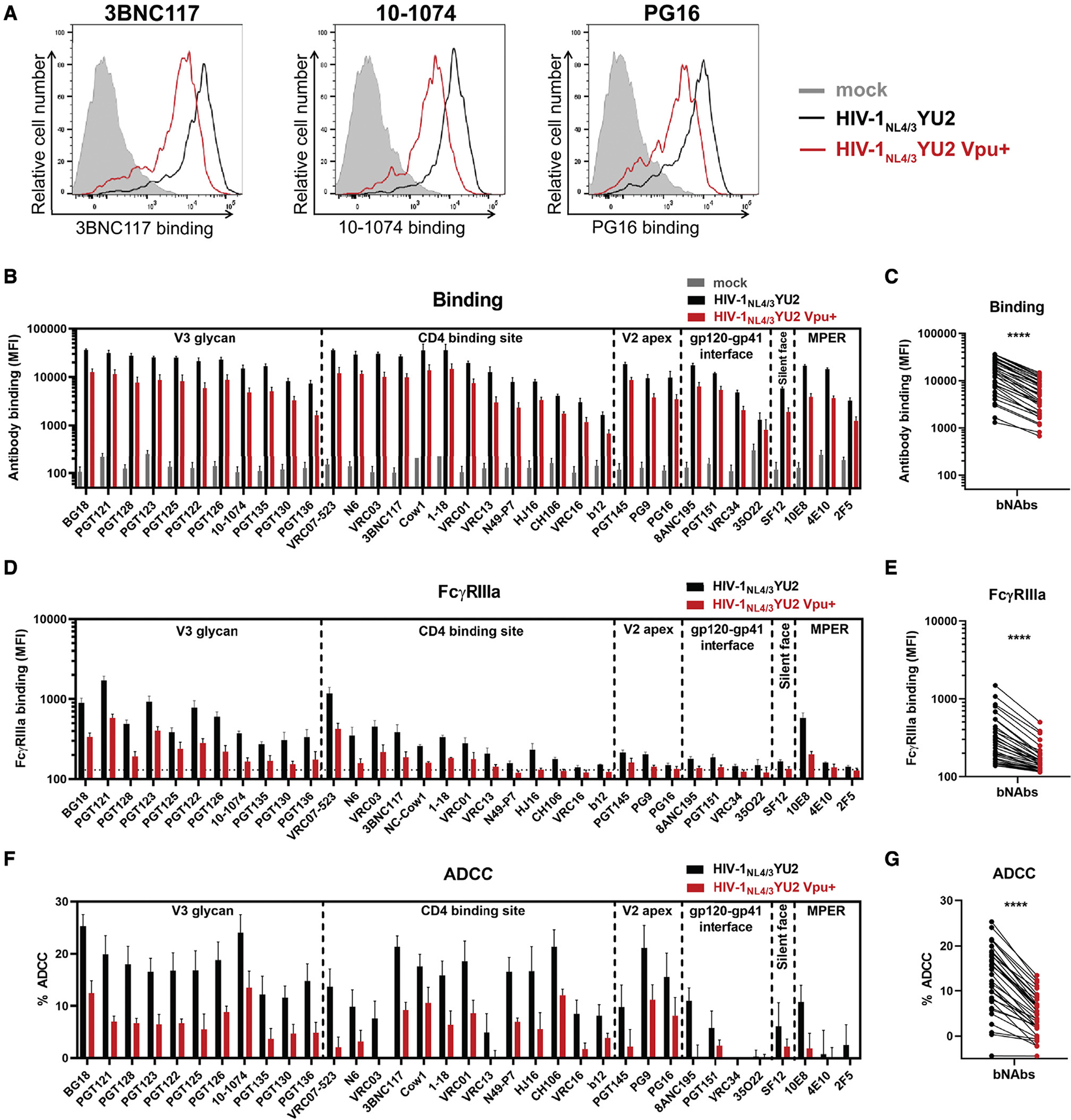
Primary CD4+ T cells were either mock infected or infected with the HIV-1NL4/3YU2 virus or its Vpu+ counterpart.
(A–E) Cell-surface staining performed using broadly neutralizing Abs targeting the gp120 V3 glycan supersite, CD4 binding site, V2 apex and silent face, the gp120-gp41 interface, and the gp41 MPER. Ab binding was detected either by using (A–C) Alexa Fluor 647-conjugated anti-human secondary Abs or (D and E) by using biotinylated recombinant soluble dimeric FcγRIIIa followed by the addition of Alexa Fluor 647-conjugated streptavidin.
(A) Histograms depicting representative stainings using anti-CD4 binding site 3BNC117, anti-V3 glycan 10–1074, and anti-V2 apex PG16 mAbs.
(B and D) The graphs represent the MFI obtained from the infected p24+ cell population using cells from five different healthy donors.
(D) The horizontal dotted line represents the signal obtained in the absence of mAb.
(F and G) Primary CD4+ T cells infected with HIV-1NL4/3YU2 viruses were used as target cells. Autologous PBMCs were used as effector cells in a FACS-based ADCC assay.
(F) The graph represents the percentages of ADCC obtained in the presence of the respective Abs using cells from five different healthy donors. Error bars indicate mean ± SEM.
(C, E, and G) Statistical significance was tested using a paired t test (****p < 0.0001).
Since all coding regions of the HIV-1NL4/3YU2 IMC other than the env gene are derived from a lab-adapted proviral backbone, we wished to test unmodified HIV-1 primary isolates. Primary CD4+ T cells were infected with a panel of 13 IMCs coding for Vpu or defective for its expression. This panel includes transmitted/founder (T/F) viruses, molecular clones derived during chronic infection, and IMCs from lab-adapted strains as controls (Figures 5A and 5B). Vpu expression consistently reduced nnAbs (246D and M785U3) and bNAbs (3BNC117 and 10–1074) recognition of infected cells, which in turn protected infected cells from ADCC responses mediated by all Abs tested (Figures 5A and 5B). Env polymorphisms present in CH167 (E460), REJO (N334), and CH293 (T332, N334) viruses abrogated the capacity of bNAbs 3BNC117 or 10–1074 to recognize infected cells. Of note, while Vpu expression has a profound effect in the recognition of infected cells by all tested anti-Env Abs, it did not affect their neutralization profile (Figures S1B–S1C).
Figure 5. The ability of Vpu to limit anti-Env ADCC responses is conserved among different HIV-1 strains.
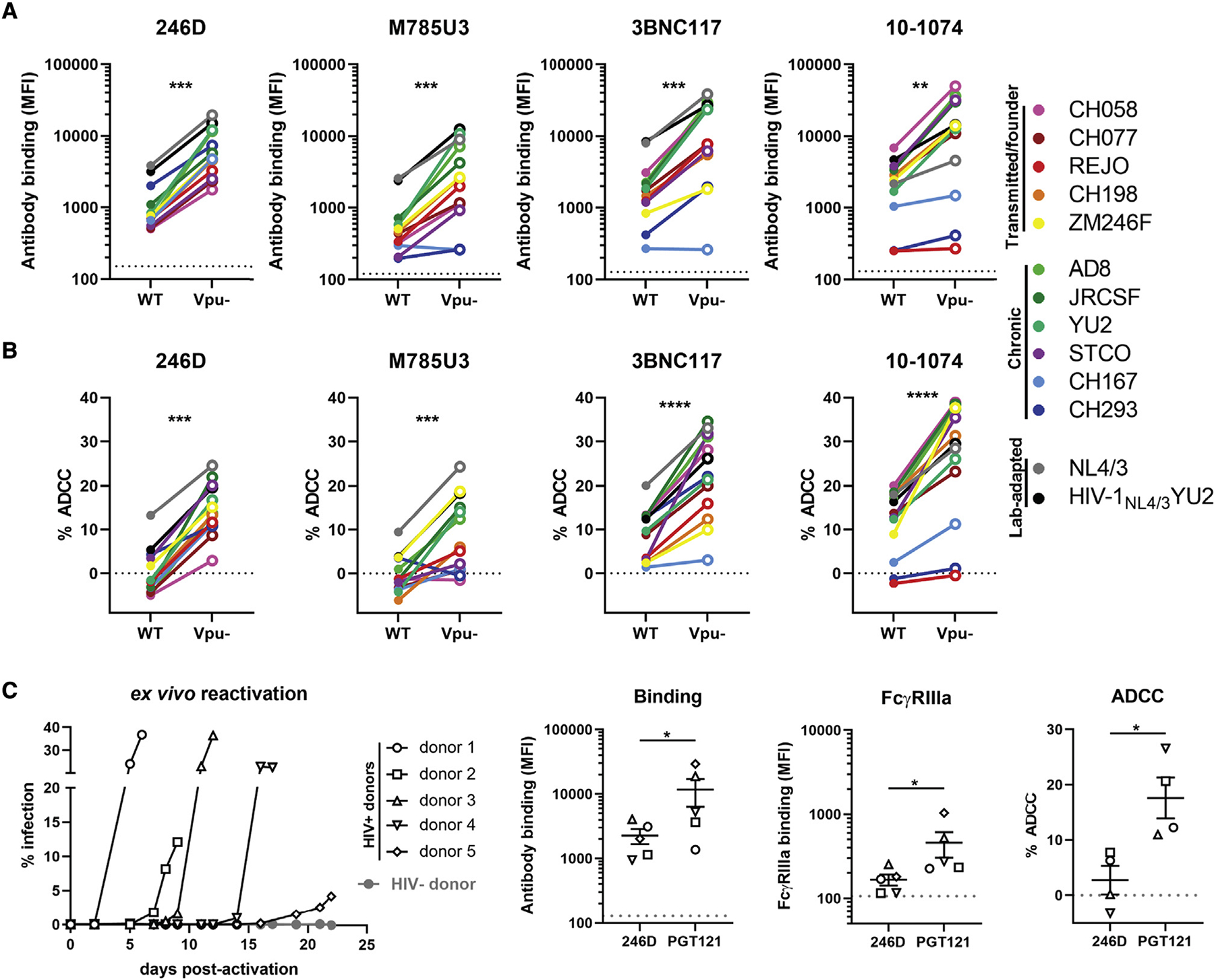
(A and B) Primary CD4+ T cells were infected with HIV-1 clade B and clade C transmitted/founder (CH058, CH077, REJO, CH198, ZM246F), chronic (AD8, JR-CSF, YU2, STCO, CH167, CH293), and lab-adapted (NL4/3, HIV-1NL4/3YU2) wild-type (WT) strains or their Vpu – counterpart. (A) Cell-surface staining performed using anti-gp41 nnAbs 246D and M785U3, as well as bNAbs 3BNC117 and 10–1074. Ab binding was detected by using Alexa Fluor 647-conjugated anti-human secondary Abs. (B) Infected primary CD4+ T cells were used as target cells and autologous PBMCs were used as effector cells in a FACS-based ADCC assay.
(C) Primary CD4+ T cells from five different ART-treated HIV-1-infected individuals were isolated and activated with PHA-L/IL-2 to expand the endogenous virus. Cell-surface staining and ADCC experiments were performed upon reactivation. Ab binding was detected using Alexa Fluor 647-conjugated anti-human secondary Abs or biotinylated recombinant soluble dimeric FcγRIIIa followed by the addition of Alexa Fluor 647-conjugated streptavidin. Ex vivo-expanded infected primary CD4+ T cells from four HIV-1-infected individuals were used as target cells and autologous PBMCs were used as effector cells in a FACS-based ADCC assay. ADCC susceptibility was only measured when the percentage of infection (p24+ cells) was higher than 10%.
(A and C) The horizontal dotted lines represent the signal obtained in absence of mAb. The Ab binding and FcγRIIIa graphs represent the MFI obtained from the infected p24 + cell population. The ADCC graphs represent the percentages of ADCC obtained in the presence of the respective Abs. Error bars indicate means ± SEM. Statistical significance was tested using (A and B) a paired t test or Wilcoxon signed-rank test based on statistical normality or (C) a Mann-Whitney U test (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, nonsignificant).
To extend our observations to a more physiological model, we expanded patient-derived infected CD4+ T cells. Briefly, we isolated and activated primary CD4+ T cells from five ART-treated HIV-1-infected individuals. Viral replication upon reactivation was monitored by intracellular p24 staining and flow cytometry (Figure 5C). These endogenously infected cells were protected from Fc-mediated effector functions mediated by nnAb 246D but remained susceptible to bNAb PGT121 (Figure 5C), in agreement with our in vitro results generated using Vpu+ IMCs (Figures 5A and 5B).
Vpu expression limits the antiviral activity of the 246D Ab in HIV-1-infected humanized mice
To determine the impact of Vpu expression on 246D antiviral activities in vivo, we infected SRG-15 (SIRPAh/m Rag2−/− Il2rg−/− IL15h/m) humanized mice with HIV-1NL4/3YU2 or its Vpu-competent counterpart. Similar to a previously used humanized-mouse model (Horwitz et al., 2017), this humanized-mouse model supports HIV-1 replication and Fc-effector functions in vivo (Herndler-Brandstetter et al., 2017; Rajashekar et al., 2021). Immunodeficient mice engrafted with human peripheral blood lymphocytes (hu-PBLs) were infected intraperitoneally (i.p.) with 30,000 plaque-forming units (PFU) of either virus (Figure 6A). Half of each cohort received subcutaneous (s.c.) injections of the 246D nnAb at days 2, 4, and 6 post infection. Infected humanized mice were then monitored for plasma viral loads (PVLs). Both viruses replicated efficiently in SRG-15 hu-PBL mice, reaching on average 1 × 107 viral RNA copies/mL of plasma at 3 days post infection (Figure 6B). While PVLs in mice infected with HIV-1NL4/3YU2 stabilized after day 3, infection with the Vpu+ variant further increased PVLs to 3.5 × 107 copies/mL by day 10. A single administration of the 246D nnAb at day 2 resulted in a significant reduction in PVLs (~36-fold decrease) in humanized mice infected with the vpu-defective virus but not significantly in those infected with the vpu-competent virus (Figure 6B). The 246D maximal inhibitory effect (~85-fold reduction) was reached 10 days post infection with the vpu-defective virus. At the end of the experiment (day 11), 246D treatment induced on average a 41-fold decrease in PVLs in mice infected with the vpu-defective virus but no difference in humanized mice infected with the Vpu+ virus. These results highlight the role played by Vpu in promoting viral replication in presence of nnAbs.
Figure 6. Vpu promotes HIV-1 replication in humanized mice treated with nnAb 246D.
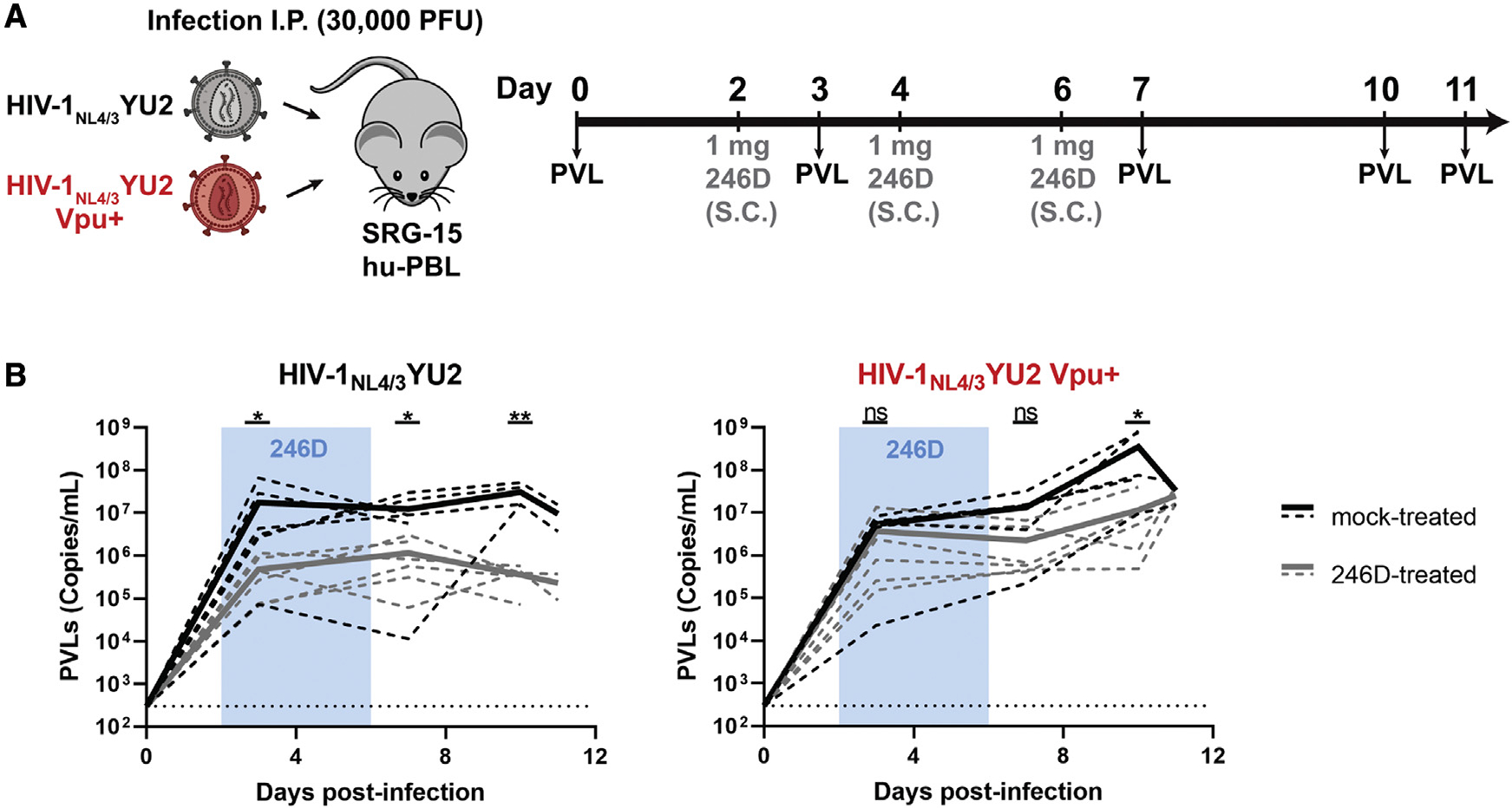
(A) Experimental outline. SRG-15-Hu-PBL mice were infected with HIV-1NL4/3YU2 or its Vpu+ counterpart intraperitoneally. At day 2, 4, and 6 post infection, mice were administered 1 mg of 246D mAb subcutaneously (s.c.). Mice were bled routinely for plasma viral load (PVL) quantification.
(B) PVL levels were measured by quantitative real-time PCR (limit of detection = 300 copies/mL, dotted line). Twelve mice were infected using each virus; six of them were mock treated (black lines) and the other six were treated with 246D mAb (gray lines). PVL measurements for individual mice are shown as dashed lines and mean values for each regimen are shown as solid lines. Statistical significance was tested using an unpaired t test or a Mann-Whitney U test based on statistical normality (*p < 0.05; **p < 0.01; ns, nonsignificant).
Harnessing nnAb Fc-effector functions by improving epitope exposure and FcγRIIIa interaction
Enhancing the affinity of the Fc fragment of Abs for FcγRs was shown to increase Fc-effector functions of bNAbs in HIV-1-infected humanized mice (Bournazos et al., 2014; Wang et al., 2020). To evaluate whether this strategy could apply to nnAbs, we introduced well-characterized IgG1 Fc mutations in the 246D heavy chain to modulate its interaction with FcγRs. The GRLR mutations (G236R/L328R) and the GASDALIE mutations (G236A/S239D/A330L/I332E) are respectively known to decrease and increase the affinity for activating FcγRs (Bournazos et al., 2014; Horton et al., 2010; Smith et al., 2012). To characterize these Fc variants, primary CD4+ T cells were infected with the HIV-1NL4/3YU2 constructs expressing Vpu or not. As expected, Fc modifications did not alter the ability of 246D to recognize infected cells, but it modulated the interaction with the soluble dimeric FcγR probe, with the GRLR mutations abrogating FcγRIIIa binding and the GASDALIE mutations improving it (Figures 7A and 7B). Introduction of the GASDALIE mutations enhanced ADCC against cells infected with the vpu-defective virus. Interestingly, it also allowed 246D to mediate ADCC against cells infected with the Vpu+ IMC, while the unaltered native 246D failed to do so (Figure 7C). To evaluate whether the 246D GASDALIE was able to mediate ADCC against cells infected with a primary isolate, we infected primary CD4+ T cells with the transmitted/founder virus CH058, a strain shown to be resistant to 246D-mediated ADCC responses (Figure 5B). CH058-infected cells were poorly recognized by 246D and resistant to ADCC mediated by all 246D Fc variants tested, including 246D GASDALIE (Figure S4). Consistent with the role of Nef and Vpu in preventing the exposure of epitopes recognized by nnAbs, disruption of the expression of both accessory proteins enhanced recognition and ADCC susceptibility of infected cells by 246D (Figure S4). These results agree with the requirement of Env-CD4 interaction to expose the gp41 cluster I region. The 246D recognizes with picomolar affinity a highly conserved linear peptide (596WGCSGKLICTT606), which corresponds to the gp41 C-C loop, located between HR1 and HR2 helices (Figures S3A–S3C). This epitope is occluded in the closed Env trimer structure but can be exposed upon CD4-triggered conformational changes (Figures S3D–S3E).
Figure 7. CD4 mimetics and Fc engineering enhance the antiviral activity of anti-gp41 nnAbs in vivo.
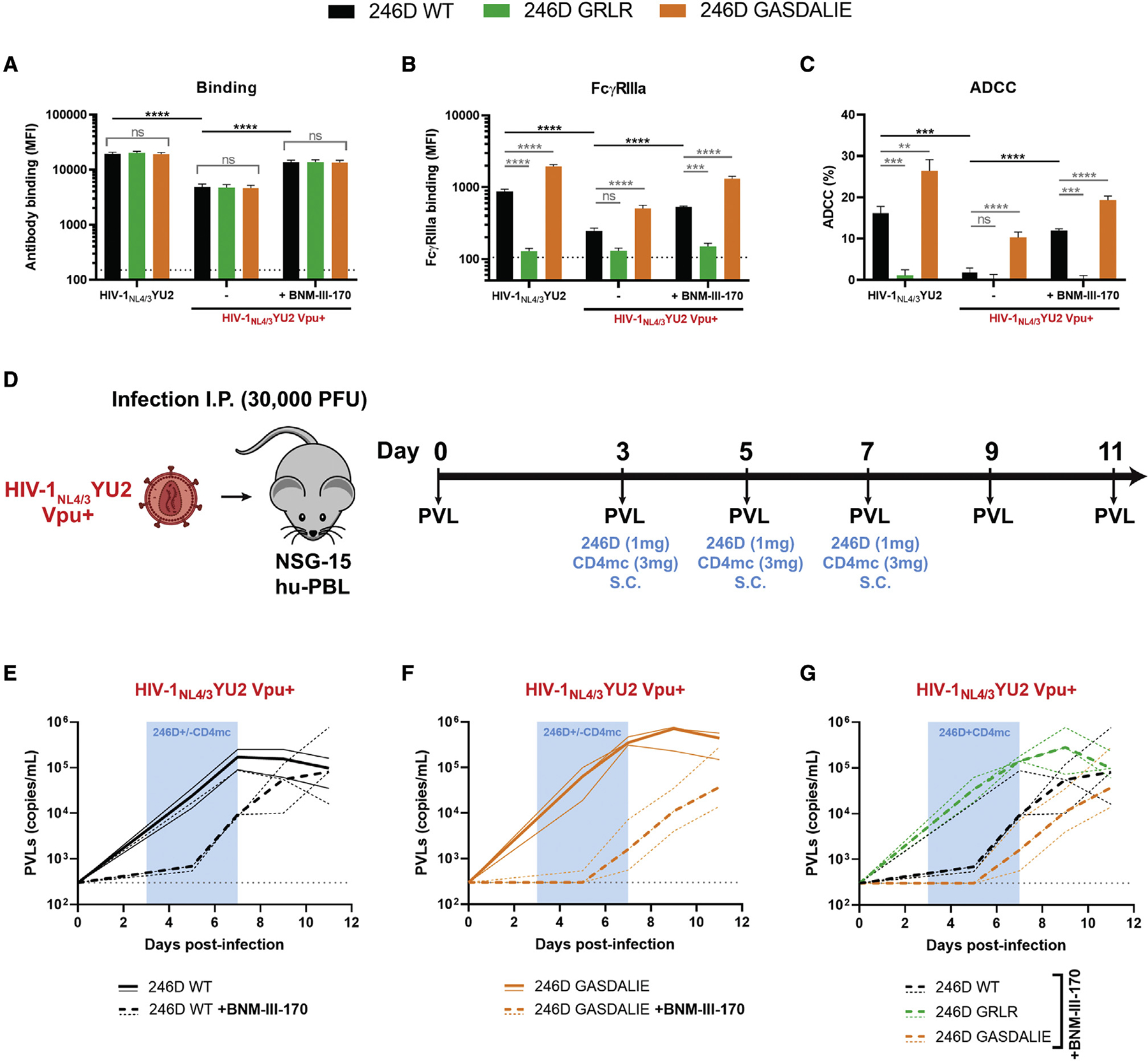
(A–C) Primary CD4+ T cells were either infected with HIV-1NL4/3YU2 virus or its Vpu+ counterpart. Forty-eight hours post infection, cell-surface staining and ADCC responses were measured in the presence of the anti-gp41 nnAb 246D WT or its Fc-mutated variants to impair (G236R/L328R; GRLR) or to enhance (G236A/S239D/A330L/I332E; GASDALIE) Fc-effector functions. Alternatively, cells infected with HIV-1NL4/3YU2 Vpu+ were treated with CD4mc BNM-III-170 during staining and ADCC experiments. Ab binding was detected using (A) Alexa Fluor 647-conjugated anti-human secondary Abs or (B) by using biotinylated recombinant soluble dimeric FcγRIIIa followed by the addition of Alexa Fluor 647-conjugated streptavidin.
(A and B) The graphs represent the MFI obtained from the infected p24+ cell population using cells from five different healthy donors. The horizontal dotted lines represent the signal obtained in absence of mAb.
(C) Infected primary CD4+ T cells were used as target cells and autologous PBMCs were used as effector cells in a FACS-based ADCC assay. The graph represents the percentages of ADCC obtained in the presence of the respective Abs using cells from five different healthy donors. Error bars indicate means ± SEM.
(A–C) Statistical significance was tested using a one-way ANOVA with a Holm-Sidak post test or a Kruskal-Wallis test with a Dunn’s post test when comparing between the different 246D Fc variants and an unpaired t test or a Mann-Whitney U test when comparing between viruses and treatment. Appropriate statistical test (parametric or nonparametric) was applied based on dataset distribution normality (**p < 0.01; ***p < 0.001; ****p < 0.0001; ns, nonsignificant).
(D) Experimental outline. NSG-15-Hu-PBL mice were infected with HIV-1NL4/3YU2 Vpu+ intraperitoneally. At day 3, 5, and 7 post infection, mice were administered 1 mg of 246D mAb s.c. alongside or without of 3 mg of CD4mc BNM-III-170 (n = 2–3 per cohort).
(E–G) PVLs were measured by quantitative real-time PCR (limit of detection = 300 copies/mL, dotted gray line) upon treatment with (E) 246D WT ± CD4mc BNM-III-170, (F) 246D GASDALIE ± CD4mc BNM-III-170, or (G) 246D WT, GRLR, or GASDALIE in presence of CD4mc BNM-III-170. PVL measurements for individual mice are shown as thin lines and median values for each cohort as thick lines.
While it is becoming increasingly clear that HIV-1 successfully evades nnAbs responses by keeping its Env in a “closed” conformation (Bruel et al., 2017; Dufloo et al., 2020; Veillette et al., 2014, 2015; von Bredow et al., 2016), new strategies are currently being tested to harness their potential antiviral activity. Small CD4 mimetic compounds have been optimized to “open up” Env trimers, therefore exposing otherwise occluded epitopes recognized by nnAbs (Ding et al., 2019b; Fritschi et al., 2021; Jette et al., 2021; Laumaea et al., 2020; Melillo et al., 2016). Using this strategy, CD4mc were shown to synergize with monoclonal CD4i Abs or nnAbs found in plasma from infected individuals to eliminate HIV-1-infected cells in vitro, ex vivo, and in vivo in humanized mice (Alsahafi et al., 2019; Anand et al., 2019; Ding et al., 2016b; Lee et al., 2015; Madani et al., 2018; Prévost et al., 2020b; Rajashekar et al., 2021; Richard et al., 2015, 2016, 2017). Since 246D was unable to efficiently recognize cells infected with a Vpu+ virus, we combined it with the CD4mc BNM-III-170, which greatly improved its capacity to bind to cells infected with HIV-1NL4/3YU2 Vpu+ or the primary isolate CH058 (Figures 7A and S4A). Accordingly, FcγRIIIa engagement and ADCC responses against WT-infected cells were significantly enhanced upon CD4mc addition (Figures 7B, 7C, S4B, and S4C). Given these observations, we investigated whether CD4mc and Fc modifications could improve the ability of 246D to alter HIV-1 WT replication in vivo. To do so, NSG-15 humanized mice engrafted with hu-PBL were infected with HIV-1NL4/3YU2 Vpu+ and were administered with 246D nnAb alongside or without CD4mc BNM-III-170 at day 3, 5, and 7 post infection (Figure 7D). In line with the data obtained in vitro (Figures 7A–7C), we observed that co-administration of BNM-III-170 with 246D WT or GASDALIE considerably reduced PVLs at day 7 (18-fold and 219-fold, respectively), compared with the Ab alone (Figures 7E and 7F). In both cases, treatment interruption led to a significant increase of PVLs in the following days. To evaluate the contribution of Fc-effector functions to this phenotype, infected humanized mice were co-administered with BNM-III-170 and 246D Fc variants (WT, GRLR, or GASDALIE). At day 7 post infection, PVLs were 15-fold higher in presence of 246D GRLR and 6-fold lower in presence of 246D GASDALIE, compared with 246D WT, confirming the primordial role of Fceffector functions in limiting HIV-1 replication in presence of nnAbs and CD4mc (Figure 7G). Altogether, our results indicate that Fc-effector functions mediated by nnAbs in vivo are limited by the occluded nature of their epitopes.
DISCUSSION
The HIV-1 accessory protein Vpu is a multi-functional protein that promotes viral replication by interfering with the intracellular trafficking of various host proteins (Dubét al., 2010). CD4 and BST-2 downregulation by Vpu was shown to increase viral release in cell culture systems (Bour et al., 1999; Neil et al., 2008; Van Damme et al., 2008). Humanized mouse models of acute HIV-1 infection using HIV-1-infected humanized mice confirmed the role of Vpu in promoting viral replication in the initial phase of infection (Dave et al., 2013; Sato et al., 2012; Yamada et al., 2018). Similarly, chronic infection of NHPs with SHIV constructs encoding a defective or mutated vpu gene were found to be less pathogenic and exhibit lower viral loads (Hout et al., 2005; Shingai et al., 2011). In these in vivo studies, elevated viral loads were linked to Vpu-mediated CD4 and BST-2 downregulation, but the potential contributions of nnAbs to PVLs were not addressed. Beyond its effect on viral release, Vpu protects HIV-1-infected cells from nnAbs-mediated ADCC responses by limiting the presence of Env-CD4 complexes at the plasma membrane (Prévost et al., 2022; Veillette et al., 2014, 2015). Here, we show that Vpu expression enhances HIV-1 viral replication in vivo by limiting nnAbs recognition of infected cells and therefore their capacity to mediate Fc-effector functions. Beyond infected-cell elimination, the absence of Vpu could also affect the level of circulating infectious viral particles. While we did not observe any changes in the neutralization by antigp41 nnAbs against virions produced in 293T cells in vitro (Figures S1B and –S1C), one could speculate that the capacity of nnAbs to neutralize viral particles originating from primary cells could also be altered in the absence of Vpu. Indeed, CD4 incorporation into virions has been shown to sensitize them to neutralization by various CD4i mAbs and HIV-IG (Ding et al., 2019a). While this was observed with nef-defective viruses, CD4 incorporation is also modulated by Vpu expression, and this could apply to vpu-defective viruses (Levesque et al., 2003). Our results highlight the importance of carefully selecting viruses for in vitro and in vivo studies, since several widely used HIV-1 strains are defective for Vpu expression (e.g., HXB2, YU2, ADA) (Li et al., 1991; Shaw et al., 1984; Theodore et al., 1996).
Different approaches aimed at generating bNAbs by vaccination are being investigated using germline-targeting Env immunogens (Dosenovic et al., 2015; Jardine et al., 2016; Steichen et al., 2016) followed by sequential Env trimer immunization (Escolano et al., 2016; Haynes et al., 2012b; Saunders et al., 2019; Williams et al., 2017) to guide Ab maturation. However, none of these vaccine studies have yet resulted in the consistent elicitation of bNAbs. Facing the difficulty in eliciting bNAbs by vaccination, nnAbs have been studied as an alternative for vaccine development. If ADCC activity contributes to vaccine protection, as suggested in the RV144 trial (Haynes et al., 2012a), our results suggest that elicitation of nnAbs are unlikely to confer protection in vaccine settings unless strategies to “open” the Env trimer are in place. Supporting this, NHPs immunized with monomeric gp120 were completely protected from a heterologous SHIV infection if a CD4mc was combined with the challenge viral stock (Madani et al., 2018). This study showed that nnAbs elicited by the gp120 immunogen did not protect in the absence of CD4mc. In addition, it was suggested that the modest protection observed in the RV144 trial could be linked to the circulating strains in Thailand (clade CRF01_AE), which harbor Env in a more “open” conformation, while a similar vaccine trial (HVTN 702) was unsuccessful when performed in South Africa, where Env from the circulating strains (clade C) samples a more “closed” conformation (Gray et al., 2021; Prévost et al., 2017).
In this study, we selected the SRG-15 and NSG-15 humanized mouse models over other humanized mouse models because of their endogenous expression of human interleukin (IL)-15, which allows the development of a functional natural killer (NK) cell compartment (Brehm et al., 2018; Herndler-Brandstetter et al., 2017). NK cells play a central role in the ADCC responses in vitro and in vivo, and their specific depletion has been shown to abrogate the elimination of HIV-1 reservoirs by a combination of CD4mc and nnAbs in SRG-15 humanized mice (Rajashekar et al., 2021). Human IL-15 expression likely also increases HIV-1 replication by regulating the susceptibility of CD4+ T cells to infection (Manganaro et al., 2018). This could explain the higher viral load peak (~107 copies/mL) (Figure 6B) compared with those achieved in the NRG humanized mice using the same HIV-1NL4/3YU2 IMC (~105 copies/mL) in previous studies (Halper-Stromberg et al., 2014; Horwitz et al., 2017). We also used humanized mice as a model of HIV-1 infection since it allows the use of clinically relevant HIV-1 isolates and does not require Env adaptation to replicate.
In addition to accessory proteins, Env conformation is a critical factor when studying nnAbs Fc-effector functions (Alsahafi et al., 2015; Ding et al., 2016a; Dufloo et al., 2020; Prévost et al., 2017, 2018a, 2018b, 2021; Richard et al., 2015; Veillette et al., 2014, 2015). Multiple studies reported significant effects of nnAbs in limiting HIV-1/SHIV replication in vivo using viruses coding for easy-to-neutralize tier 1 Env from lab-adapted strains, which are readily recognized by nnAbs (Burton et al., 2011; Eda et al., 2006; Hessell et al., 2018; Hioe et al., 2022; Moog et al., 2014; Santra et al., 2015). A recent study showed that tier 2 primary virus can be affected in vivo by nnAbs but only when combined with CD4mc (Rajashekar et al., 2021). The combination of nnAbs or HIV+ plasma with CD4mc significantly reduced PVLs and the size of the viral reservoir in an Fc-effector-function- and NK-cell-dependent manner (Rajashekar et al., 2021). These results emphasize the need to utilize fully functional viruses in ADCC assays to preclude Vpu-related artifacts. However, one could speculate that the development of broad-spectrum Vpu inhibitors may enhance the efficacy of nnAbs to eliminate HIV-1-infected cells (Robinson et al., 2022). Overall, our study indicates that it is unlikely for nnAbs-based immunotherapies to alter HIV-1 viral replication in vivo in the absence of strategies aimed at exposing the vulnerable epitopes they recognize.
Limitations of the study
The humanized mice experiments were performed with only one nnAb (246D) and using a chimeric virus (HIV-1NL4/3YU2). Further in vivo experiments with additional nnAbs of different epitope specificities and using clinically relevant HIV-1 primary isolates are needed to confirm our observations. Moreover, our study mainly focuses on the role of the accessory protein Vpu to evade humoral responses in vivo, but Nef is also known to affect Env conformation at the surface of HIV-1-infected cells. Nef plays a major role in CD4 downregulation from the cell surface and we cannot rule out the possibility that Nef could also contribute to HIV-1 evasion of nnAb responses in vivo, especially when using primary viruses.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andrés Finzi (andres.finzi@umontreal.ca).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
All data generated and reported in this paper are available from the lead contact (andres.finzi@umontreal.ca) upon request.
This paper does not report original code
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Ethics statement
Written informed consent was obtained from all study participants and research adhered to the ethical guidelines of CRCHUM and was reviewed and approved by the CRCHUM institutional review board (ethics committee, approval number CE 16.164 - CA). Research adhered to the standards indicated by the Declaration of Helsinki. All participants were adult and provided informed written consent prior to enrollment in accordance with Institutional Review Board approval.
Cell lines and primary cells
293T human embryonic kidney cells (obtained from ATCC) and TZM-bl cells (NIH AIDS Reagent Program) were maintained at 37°C under 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) (Wisent, St. Bruno, QC, Canada), supplemented with 5% fetal bovine serum (FBS) (VWR, Radnor, PA, USA) and 100 U/mL penicillin/streptomycin (Wisent). 293T cells were derived from 293 cells, into which the simian virus 40 T-antigen was inserted. TZM-bl cells were derived from HeLa cells and were engineered to stably express high levels of human CD4 and CCR5 and to contain the firefly luciferase reporter gene under the control of the HIV-1 promoter (Platt et al., 1998). Human peripheral blood mononuclear cells (PBMCs) from ten HIV-negative individuals (8 males and 2 females, unknown age) and five antiretroviral therapy (ART)-treated HIV-positive individuals (all males) obtained by leukapheresis and Ficoll-Paque density gradient isolation were cryopreserved in liquid nitrogen until further use. CD4+ T lymphocytes were purified from resting PBMCs by negative selection using immunomagnetic beads per the manufacturer’s instructions (StemCell Technologies, Vancouver, BC) and were activated with phytohemagglutinin-L (10 μg/mL) for 48 h and then maintained in RPMI 1640 (Thermo Fisher Scientific, Waltham, MA, USA) complete medium supplemented with rIL-2 (100 U/mL).
Human plasma samples
The FRQS-AIDS and Infectious Diseases Network supports a representative cohort of newly-HIV-infected subjects with clinical indication of primary infection [the Montreal Primary HIV Infection Cohort (Fontaine et al., 2011; Fontaine et al., 2009)]. Cross-sectional plasma samples from 50 HIV-1-infected individuals were segregated in five groups based on infection duration and treatment with antiretroviral therapy (ART). Plasma samples were obtained from treatment-naive subjects during the acute phase of infection (first 3 months after HIV acquisition), the early phase of infection (3–6 and 6–12 months after acquisition) and during the chronic phase of infection (>2 years after acquisition). Another group of chronically-infected individuals (>2 years after acquisition) received ART treatment. A summary of demographic parameters for all HIV-1-infected subjects is included in Table S1. Plasma samples were also obtained from ten age- and sex-matched HIV-negative healthy volunteers.
Experimental animal models
SRG-15 mice encoding human SIRPA and IL-15 in a 129xBALB/c (N3) genetic background were originally generated in the laboratory of Dr Richard Flavell (Yale University) (Herndler-Brandstetter et al., 2017). NSG-15 mice with expression of the human IL15 gene in the NOD/ShiLtJ background were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) (Brehm et al., 2018). The mice were bred and maintained under specific pathogen-free conditions. All animal studies were performed with authorization from Institutional Animal Care and Use Committees (IACUC) of Yale University. The mice were a mix of male and female randomly distributed between groups. SRG-15-Hu-PBL and NSG-15-Hu-PBL mice were engrafted as described (Rajashekar et al., 2021). Briefly, 1 × 107 PBMCs, purified by Ficoll density gradient centrifugation of healthy donor blood buffy coats (two anonymous donors, obtained from the New York Blood Bank) were injected IP in a 200-μL volume into 6- to 8-week-old SRG-15 or NSG-15 mice, using a 1-cm3 syringe and 25-gauge needle. Cell engraftment was tested 15 days post-transplant. 100 μL of blood was collected by retroorbital bleeding. PBMCs were isolated by Ficoll density gradient centrifugation; stained with fluorescently-labelled anti-human CD45, CD3, CD4, CD8 and CD56 antibodies and analyzed by flow cytometry to confirm engraftment. Humanized mice were intraperitoneally challenged with 30,000 PFU of HIV-1NL4/3YU2 Vpu− or Vpu+. Infection profile was analyzed for plasma viral load (PVL) analysis of serially collected plasma samples.
METHOD DETAILS
Plasmids and proviral constructs
The HIV-1NL4/3YU2 proviral construct has been previously reported (Horwitz et al., 2017). A mutation was introduced in the putative vpu start codon (ACG → ATG) to restore the vpu open reading frame (ORF) in the HIV-1NL4/3YU2 IMC using the QuikChange II XL site-directed mutagenesis protocol (Agilent Technologies, Santa Clara, CA). Transmitted/Founder (T/F) and chronic infectious molecular clones (IMCs) of patients CH058, CH077, CH198, ZM246F, CH167, CH293, REJO, STCO were inferred and constructed as previously described (Ochsenbauer et al., 2012; Parrish et al., 2013; Parrish et al., 2012; Salazar-Gonzalez et al., 2009). The generation of vpu-defective IMCs was previously described (Kmiec et al., 2016; Langer et al., 2015; Sauter et al., 2015; Yamada et al., 2018) and consists in the introduction of premature stop codons in the vpu reading frame using the QuikChange II XL site-directed mutagenesis protocol. CH058 IMCs defective for Vpu and/or Nef expression were previously described (Heigele et al., 2016; Kmiec et al., 2016). The IMCs encoding for HIV-1 reference strains NL4/3, AD8, YU2 and JR-CSF were described elsewhere (Adachi et al., 1986; Koyanagi et al., 1987; Krapp et al., 2016; Li et al., 1991; Theodore et al., 1996). To generate vpu-defective JR-CSF IMC, a stop-codon was introduced directly after the start-codon of vpu using the QuikChange II XL site-directed mutagenesis protocol. The plasmids encoding for the heavy and light chains of the 246D antibody are available through the NIH AIDS Reagent Program. Site-directed mutagenesis was performed on the plasmid expressing 246D antibody heavy chain to introduce the GRLR mutations (G236R/L328R) or the GASDALIE mutations (G236A/S239D/A330L/I332E) using the QuikChange II XL site-directed mutagenesis protocol. The presence of the desired mutations was determined by automated DNA sequencing. The vesicular stomatitis virus G (VSV-G)-encoding plasmid was previously described (Emi et al., 1991).
Viral production, infections and ex vivo amplification
For in vitro infection, vesicular stomatitis virus G (VSV-G)-pseudotyped HIV-1 viruses were produced by co-transfection of 293T cells with an HIV-1 proviral construct and a VSV-G-encoding vector using the calcium phosphate method. Two days post-transfection, cell supernatants were harvested, clarified by low-speed centrifugation (300 × g for 5 min), and concentrated by ultracentrifugation at 4°C (100,605 × g for 1 h) over a 20% sucrose cushion. Pellets were resuspended in fresh RPMI, and aliquots were stored at −80°C until use. Viruses were then used to infect activated primary CD4+ T cells from healthy HIV-1 negative donors by spin infection at 800 × g for 1 h in 96-well plates at 25°C. Viral preparations were titrated directly on primary CD4+ T cells to achieve similar levels of infection among the different IMCs tested (around 10% of p24 + cells). To expand endogenously infected CD4+ T cells, primary CD4+ T cells obtained from ART-treated HIV-1-infected individuals were isolated from PBMCs by negative selection. Purified CD4+ T cells were activated with PHA-L at 10 μg/mL for 48 h and then cultured for at least 6 days in RPMI 1640 complete medium supplemented with rIL-2 (100 U/mL) to reach greater than 10% infection for the ADCC assay.
Antibodies
The following Abs were used to assess cell-surface Env staining: anti-gp41 nnAbs QA255-006, QA255-067, QA255-072 (kindly provided by Julie Overbaugh), 7B2, 2.2B, 12.3D, 12.4H (kindly provided by James Robinson), F240 (NIH AIDS Reagent Program), M785U1, M785U2, M785U3, M785U4, N10U1, N10U2, N5U1, N5U2, N5U3 (kindly provided by George Lewis), 246D, 240D, 167D, and 50–69; anti-cluster A A32 (NIH AIDS Reagent Program), C11 (kindly provided by James Robinson) and N5i5; anti-co-receptor binding site 17b (NIH AIDS Reagent Program) and X5; anti-V3 loop GE2-JG8 (kindly provided by Gunilla Karlsson Hedestam); anti-CD4 binding site VRC01, VRC03, VRC07-523, VRC13, VRC16 (kindly provided by John Mascola), N6 (kindly provided by Mark Connors), 1–18 (kindly provided by Florian Klein), HJ16, CH106 (NIH AIDS Reagent Program), NC-Cow1, b12, 3BNC117 and N49-P7; anti-V3 glycan PGT121, PGT122, PGT123, PGT125, PGT126, PGT128, PGT130, PGT135, PGT136 (IAVI), BG18 and 10–1074; anti-V2 apex PGT145, PG9 and PG16 (IAVI); anti-gp120-gp41 interface PGT151 (IAVI), 35O22 (kindly provided by Mark Connors), VRC34 (kindly provided by John Mascola) and 8ANC195; anti-silent face SF12; anti-MPER 10E8 (kindly provided by Mark Connors), 4E10 and 2F5 (NIH AIDS Reagent Program). The HIV-IG polyclonal antibody consists of anti-HIV immunoglobulins purified from a pool of plasma from HIV + asymptomatic donors (NIH AIDS Reagent Program). Mouse anti-human CD4 (clone OKT4; Thermo Fisher Scientific), mouse anti-human BST-2 (clone RS38E, PE-Cy7-conjugated; Biolegend, San Diego, CA, USA), mouse anti-human NTB-A (clone NT-7, Biolegend) and mouse anti-PVR (clone SKII.4, Biolegend) were also used as primary antibodies for cell-surface staining. Goat anti-mouse IgG (H + L), goat anti-human IgG (H + L) (Thermo Fisher Scientific) and mouse anti-human IgG Fc (Biolegend) antibodies pre-coupled to Alexa Fluor 647 were used as secondary antibodies in flow cytometry experiments. Rabbit antisera raised against Nef (NIH AIDS Reagent Program) or Vpu (Prévost et al., 2020a) were used as primary antibodies in intracellular staining. BrillantViolet 421 (BV421)-conjugated donkey anti-rabbit antibodies (Biolegend) were used as secondary antibodies to detect Nef and Vpu antisera binding by flow cytometry. Goat anti-human IgG Fc antibodies conjugated to horseradish peroxidase (HRP; Thermo Fisher Scientific) were used as secondary antibodies in ELISA experiments.
Small CD4-mimetics
The small-molecule CD4-mimetic compound (CD4mc) BNM-III-170 was synthesized as described previously (Chen et al., 2019). The compounds were dissolved in dimethyl sulfoxide (DMSO) at a stock concentration of 10 mM and diluted to 50 μM in phosphate-buffered saline (PBS) for cell-surface staining or in RPMI-1640 complete medium for ADCC assays.
Protein production of recombinant proteins
FreeStyle 293F cells (Thermo Fisher Scientific) were grown in FreeStyle 293F medium (Thermo Fisher Scientific) to a density of 1 × 106 cells/mL at 37°C with 8% CO2 with regular agitation (150 rpm). Cells were transfected with plasmids expressing the light and heavy chains of a given antibody using ExpiFectamine 293 transfection reagent, as directed by the manufacturer (Thermo Fisher Scientific). One week later, the cells were pelleted and discarded. The supernatants were filtered (0.22-μm-pore-size filter), and antibodies were purified by protein A affinity columns, as directed by the manufacturer (Cytiva, Marlborough, MA, USA). The recombinant protein preparations were dialyzed against phosphate-buffered saline (PBS) and stored in aliquots at −80°C. To assess purity, recombinant proteins were loaded on SDS-PAGE polyacrylamide gels in the presence or absence of β-mercaptoethanol and stained with Coomassie blue. Purified F240 and 2.2B IgG were conjugated with Alexa Fluor 647 dye according to the manufacturer’s protocol (Thermo Fisher Scientific). The F240 Fab fragments were prepared from purified IgG (10 mg/mL) by proteolytic digestion with immobilized papain (Pierce, Rockford, IL) and purified using protein A, followed by gel filtration chromatography on a Superdex 200 16/60 column (Cytiva). The biotin-tagged dimeric recombinant soluble FcγRIIIa (V158) protein was produced and characterized as described (Wines et al., 2016) with additional purification step using a Superdex 200 10/300 GL column (Cytiva).
Flow cytometry analysis of cell-surface staining
Cell surface staining was performed at 48h post-infection. Mock-infected or HIV-1-infected primary CD4+ T cells were incubated for 30 min at 37°C with anti-CD4 (0.5 μg/mL), anti-BST-2 (2 μg/mL), anti-NTB-A (5 μg/mL), anti-PVR (10 μg/mL), anti-Env mAbs (5 μg/mL), HIV-IG (50 μg/mL) or plasma (1:1000 dilution). Cells were then washed once with PBS and stained with the appropriate Alexa Fluor 647-conjugated secondary antibody (2 μg/mL), when needed, for 20 min at room temperature. After one more PBS wash, cells were fixed in a 2% PBS-formaldehyde solution. Alternatively, the binding of anti-Env Abs was detected using a biotin-tagged dimeric recombinant soluble FcγRIIIa (0.2 μg/mL) followed by the addition of Alexa Fluor 647-conjugated streptavidin (Thermo Fisher Scientific; 2 μg/mL). Anti-human IgG Fc secondary antibodies (3 μg/mL) were used when cell surface binding was performed in presence of F240 Fab blockade. Infected cells were then permeabilized using the Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences, Mississauga, ON, Canada) and stained intracellularly using PE-conjugated mouse anti-p24 mAb (clone KC57; Beckman Coulter, Brea, CA, USA; 1:100 dilution) in combination with Nef or Vpu rabbit antisera (1:1000 dilution). The percentage of infected cells (p24+) was determined by gating on the living cell population according to a viability dye staining (Aqua Vivid; Thermo Fisher Scientific). Alternatively, cells were stained intracellularly with rabbit antisera raised against Nef or Vpu (1:1000) followed by BV421-conjugated anti-rabbit secondary antibody. Samples were acquired on an LSR II cytometer (BD Biosciences), and data analysis was performed using FlowJo v10.5.3 (Tree Star, Ashland, OR, USA).
Antibody-dependent cellular cytotoxicity (ADCC) assay
Measurement of ADCC using a fluorescence-activated cell sorting (FACS)-based infected cell elimination (ICE) assay was performed at 48 h post-infection. Briefly, HIV-1-infected primary CD4+ T cells were stained with AquaVivid viability dye and cell proliferation dye eFluor670 (Thermo Fisher Scientific) and used as target cells. Cryopreserved autologous PBMC effectors cells, stained with cell proliferation dye eFluor450 (Thermo Fisher Scientific), were added at an effector: target ratio of 10:1 in 96-well V-bottom plates (Corning, Corning, NY). A 1:1000 final dilution of plasma or 5 μg/mL of anti-Env mAbs was added to appropriate wells and cells were incubated for 5 min at room temperature. The plates were subsequently centrifuged for 1 min at 300 × g, and incubated at 37°C, 5% CO2 for 5 h before being fixed in a 2% PBS-formaldehyde solution. Infected cells were identified by intracellular p24 staining as described above. Samples were acquired on an LSR II cytometer (BD Biosciences) and data analysis was performed using FlowJo v10.5.3 (Tree Star). The percentage of ADCC was calculated with the following formula: [(% of p24 + cells in Targets plus Effectors) − (% of p24 + cells in Targets plus Effectors plus plasma)/(% of p24 + cells in Targets) × 100] by gating on infected lived target cells.
Virus neutralization assay
TZM-bl target cells were seeded at a density of 2 × 104 cells/well in 96-well luminometer-compatible tissue culture plates (PerkinElmer, Waltham, MA, USA) 24 h before infection. HIV-1NL4/3YU2 Vpu− or Vpu+ (in a final volume of 50 μL) was pre-incubated with the indicated amounts of mAbs for 1 h at 37°C before adding to the target cells. Following an incubation of 24 h at 37°C, 100 μL of media was added to each well. Two days later, the medium was removed from each well, and the cells were lysed by the addition of 30 μL of passive lysis buffer (Promega, Madison, WI, USA) followed by one freeze-thaw cycle. An LB 942 TriStar luminometer (Berthold Technologies, Bad Wildbad, Germany) was used to measure the luciferase activity in each well after the addition of 100 mL of Luciferin buffer (15 mM MgSO4, 15 mM KH2PO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μL of 1 mM Firefly D-luciferin (Prolume, Pinetop, AZ, USA).
Plasma viral load measurements
Quantification of PVLs was done using the method described previously (Gibellini et al., 2004). Briefly, 100 μL of blood was collected from mice at each time point by retroorbital bleed. Plasma viral RNA was extracted using the QIAamp viral RNA mini kit (QIAGEN, Hilden Germany) following the manufacturer’s protocol. SYBR green real-time PCR assay was carried out in a 20-μL PCR mixture volume consisting of 10 μL of 2× Quantitect SYBR green RT-PCR Master Mix (QIAGEN) containing HotStarTaq DNA polymerase, 0.5 μL of 500 nM each oligonucleotide primer, 0.2 μL of 100× QuantiTect RT Mix (containing Omniscript and Sensiscript RTs), and 8 μL of RNA extracted from plasma samples or standard HIV-1 RNA (from 5 × 105 to 5 copies per 1 mL). Highly conserved sequences on the gag region of HIV-1 were chosen, and specific HIV-1 gag primers were selected. The sequences of HIV-1 gag primers are 5′ TGCTATGTCAGTTCCCCTTGGTTCTCT-3′ and 5′AGTTGGAGGACATCAAGCAGCCATGCAAAT-3′. Amplification was done in an Applied Biosystems 7500 real-time PCR system, and it involved activation at 45°C for 15 min and 95°C for 15 min followed by 40 amplification cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s. For the detection and quantification of viral RNA, the real-time PCR of each sample was compared with threshold cycle value of a standard curve.
Sequence analysis
The LOGO plot (Crooks et al., 2004) was created using the Analyze Align tool at the Los Alamos National Laboratory - HIV database which is based on the WebLogo program (https://www.hiv.lanl.gov/content/sequence/ANALYZEALIGN/analyze_align.html) and the HIV-1 database global curated and filtered 2019 alignment, including 6,223 individual Env protein sequences. The relative height of each letter within individual stack represents the frequency of the indicated amino acid at that position. The numbering of Env amino acid sequences is based on the HXB2 reference strain of HIV-1, where 1 is the initial methionine.
gp41 peptide ELISA
Peptides corresponding to the gp41 C-C loop region (residues 583–618) were either previously described (Gohain et al., 2016) or purchased from Genscript (Piscataway, NJ, USA). A peptide corresponding to the SARS-CoV-2 Spike S2 stem helix (residues 1153–1163) was used as a negative control and was previously reported (Li et al., 2021). Peptides were prepared in PBS at a concentration of 1 μg/mL and were adsorbed to white 96-well plates (MaxiSorp Nunc) overnight at 4°C. Coated wells were subsequently blocked with blocking buffer (Tris-buffered saline [TBS] containing 0.1% Tween 20 and 2% BSA) for 1 h at room temperature. Wells were then washed four times with washing buffer (TBS containing 0.1% Tween 20). Anti-gp41 246D and F240 or anti-gp120 A32 (50 ng/mL) were prepared in a diluted solution of blocking buffer (0.1% BSA) and incubated with the peptide-coated wells for 90 min at room temperature. Plates were washed four times with washing buffer followed by incubation with horseradish peroxidase (HRP)-conjugated anti-human IgG secondary Ab (0.3 μg/mL in a diluted solution of blocking buffer [0.4% BSA]) for 1 h at room temperature, followed by four washes. HRP enzyme activity was determined after the addition of a 1:1 mix of Western Lightning Plus-ECL oxidizing and luminol reagents (PerkinElmer Life Sciences). Light emission was measured with an LB942 TriStar luminometer (Berthold Technologies).
Surface plasmon resonance (SPR)
Surface plasmon resonance assays were performed on a Biacore 3000 (Cytiva) with a running buffer of 10 mM HEPES pH 7.5 and 150 mM NaCl, supplemented with 0.05% Tween 20 at 25°C. The binding kinetics between the gp41 C-C loop and the 246D antibody were obtained in a format where 246D IgG was immobilized onto a Protein A sensor chip (Cytiva) with ~300 response units (RU) and serial dilutions of gp41 (583–618) synthetic peptide were injected with concentrations ranging from 0.488 to 31.25 nM. The protein A chip was regenerated with a wash step of 0.1 M glycine pH 2.0 and reloaded with IgG after each cycle. Kinetic constants were determined using a 1:1 Langmuir model in bimolecular interaction analysis (BIA) evaluation software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics were analyzed using GraphPad Prism version 9.3.1 (GraphPad, San Diego, CA, USA). Every data set was tested for statistical normality and this information was used to apply the appropriate (parametric or nonparametric) statistical test. Statistical details of experiments are indicated in the figure legends. p values < 0.05 were considered significant; significance values are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Monoclonal anti-Env gp41 246D | NIH AIDS Reagent Program (Xu et al., 1991) | Cat# 1245 |
| Monoclonal anti-Env gp41 246D GRLR | This paper | N/A |
| Monoclonal anti-Env gp41 246D GASDALIE | This paper | N/A |
| Monoclonal anti-Env gp41 F240 | NIH AIDS Reagent Program (Cavacini et al., 1998) | Cat# 7623 |
| Anti-Env gp41 F240 Fab fragment | (Gohain et al., 2016) | N/A |
| Monoclonal anti-Env gp41 240D | NIH AIDS Reagent Program (Xu et al., 1991) | Cat# 1242 |
| Monoclonal anti-Env gp41 167D | NIH AIDS Reagent Program (Xu et al., 1991) | Cat# 11681 |
| Monoclonal anti-Env gp41 50–69 | NIH AIDS Reagent Program (Gorny et al., 1989) | Cat# 531 |
| Monoclonal anti-Env gp41 QA255-006 | Dr Julie Overbaugh (Williams et al., 2019) | N/A |
| Monoclonal anti-Env gp41 QA255-067 | Dr Julie Overbaugh (Williams et al., 2019) | N/A |
| Monoclonal anti-Env gp41 QA255-072 | Dr Julie Overbaugh (Williams et al., 2019) | N/A |
| Monoclonal anti-Env gp41 7B2 | NIH AIDS Reagent Program | Cat# 12556 |
| Monoclonal anti-Env gp41 2.2B | Dr James E. Robinson | N/A |
| Monoclonal anti-Env gp41 12.3D | Dr James E. Robinson | N/A |
| Monoclonal anti-Env gp41 12.4H | Dr James E. Robinson | N/A |
| Monoclonal anti-Env gp41 M785U1 | Dr George Lewis (Ding et al., 2016a) | N/A |
| Monoclonal anti-Env gp41 M785U2 | Dr George Lewis | N/A |
| Monoclonal anti-Env gp41 M785U3 | Dr George Lewis | N/A |
| Monoclonal anti-Env gp41 M785U4 | Dr George Lewis | N/A |
| Monoclonal anti-Env gp41 N10U1 | Dr George Lewis (Ding et al., 2016a) | N/A |
| Monoclonal anti-Env gp41 N10U2 | Dr George Lewis | N/A |
| Monoclonal anti-Env gp41 N5U1 | Dr George Lewis (Ding et al., 2016a) | N/A |
| Monoclonal anti-Env gp41 N5U2 | Dr George Lewis | N/A |
| Monoclonal anti-Env gp41 N5U3 | Dr George Lewis (Ding et al., 2016a) | N/A |
| Monoclonal anti-Env cluster A A32 | NIH AIDS Reagent Program | Cat# 11438 |
| Monoclonal anti-Env cluster A N5i5 | (Guan et al., 2013) | N/A |
| Monoclonal anti-Env cluster A C11 | Dr James E. Robinson (Robinson JE et al., 1992) | N/A |
| Monoclonal anti-Env co-receptor binding site 17b | NIH AIDS Reagent Program | Cat# 4091; RRID:AB_2905603 |
| Monoclonal anti-Env co-receptor binding site X5 | (Huang et al., 2004) | N/A |
| Monoclonal anti-Env V3 loop GE2-JG8 | Dr Gunilla Karlsson Hedestam (Phad etal., 2015) | N/A |
| Monoclonal anti-Env CD4 binding site 3BNC117 | (Scheid et al., 2011) | RRID:AB_2491033 |
| Monoclonal anti-Env CD4 binding site N49-P7 | (Sajadi et al., 2018) | N/A |
| Monoclonal anti-Env CD4 binding site VRC01 | NIH AIDS Reagent Program (Wu etal., 2010b) | Cat# 12033; RRID:AB_2491019 |
| Monoclonal anti-Env CD4 binding site VRC03 | NIH AIDS Reagent Program (Wu etal., 2010b) | Cat# 12032; RRID:AB_2491021 |
| Monoclonal anti-Env CD4 binding site VRC07-523 | Dr John Mascola (Rudicell et al., 2014) | N/A |
| Monoclonal anti-Env CD4 binding site VRC13 | Dr John Mascola (Zhou etal., 2015) | N/A |
| Monoclonal anti-Env CD4 binding site VRC16 | Dr John Mascola (Zhou etal., 2015) | N/A |
| Monoclonal anti-Env CD4 binding site N6 | NIH AIDS Reagent Program (Huang et al., 2016) | Cat# 12968 |
| Monoclonal anti-Env CD4 binding site NC-Cow1 | (Sok etal., 2017) | RRID:AB_2687423 |
| Monoclonal anti-Env CD4 binding site b12 | NIH AIDS Reagent Program (Burton et al., 1994) | Cat# 2640; RRID:AB_2491069 |
| Monoclonal anti-Env CD4 binding site 1–18 | Dr Florian Klein (Schommers et al., 2020) | N/A |
| Monoclonal anti-Env CD4 binding site HJ16 | NIH AIDS Reagent Program (Corti et al., 2010) | Cat# 12138; RRID:AB_2491032 |
| Monoclonal anti-Env CD4 binding site CH106 | NIH AIDS Reagent Program (Liao et al., 2013) | Cat# 12566 |
| Monoclonal anti-Env V3 glycan PGT121 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491041 |
| Monoclonal anti-Env V3 glycan PGT122 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491042 |
| Monoclonal anti-Env V3 glycan PGT123 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491043 |
| Monoclonal anti-Env V3 glycan PGT125 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491044 |
| Monoclonal anti-Env V3 glycan PGT126 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491045 |
| Monoclonal anti-Env V3 glycan PGT128 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491047 |
| Monoclonal anti-Env V3 glycan PGT130 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491048 |
| Monoclonal anti-Env V3 glycan PGT135 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491050 |
| Monoclonal anti-Env V3 glycan PGT136 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491060 |
| Monoclonal anti-Env V3 glycan BG18 | (Freund et al., 2017) | N/A |
| Monoclonal anti-Env V3 glycan 10-1074 | (Mouquet et al., 201H2) | RRID:AB_2491062 |
| Monoclonal anti-Env V2 apex PGT145 | International AIDS Vaccine Initiative (Walker et al., 2011) | RRID:AB_2491054 |
| Monoclonal anti-Env V2 apex PG9 | International AIDS Vaccine Initiative (Walker et al., 2009) | RRID:AB_2491030 |
| Monoclonal anti-Env V2 apex PG16 | International AIDS Vaccine Initiative (Walker et al., 2009) | RRID:AB_2491031 |
| Anti-Env silent face SF12 monoclonal Ab | (Schoofs et al., 2019) | N/A |
| Monoclonal anti-Env gp120-gp41 interface PGT151 | International AIDS Vaccine Initiative (Falkowska et al., 2014) | N/A |
| Monoclonal anti-Env gp120-gp41 interface 8ANC195 | (Scheid et al., 2011) | RRID:AB_2491037 |
| Monoclonal anti-Env gp120-gp41 interface 35O22 | Dr Mark Connors (Huang et al., 2014) | N/A |
| Monoclonal anti-Env gp120-gp41 interface VRC34 | Dr John Mascola (Kong et al., 2016) | RRID:AB_2819225 |
| Monoclonal anti-Env gp41 MPER 10E8 | NIH AIDS Reagent Program (Huang et al., 2012) | Cat# 12294; RRID:AB_2491067 |
| Monoclonal anti-Env gp41 MPER4E10 | NIH AIDS Reagent Program (Stiegler et al., 2001) | Cat# 10091; RRID:AB_2491029 |
| Monoclonal anti-Env gp41 MPER 2F5 | NIH AIDS Reagent Program (Buchacher et al., 1994) | Cat# 1475; RRID:AB_2491015 |
| Polyclonal Anti-HIV Immune Globulin, Pooled Inactivated Human Sera (HIV-IG) | NIH AIDS Reagent Program | Cat# 3957; RRID:AB_2890264 |
| Mouse anti-Human CD4 clone OKT4 | Thermo Fisher Scientific | Cat# 14-0048-82; RRID:AB_467075 |
| PE-Cy7 Mouse anti-Human CD317 (BST-2) clone RS38E | Biolegend | Cat# 348416; RRID:AB_2716221 |
| Mouse anti-human CD155 (PVR) clone SKII.4 | Biolegend | Cat# 337602; RRID:AB_2300508 |
| Mouse anti-human CD352 (NTB-A) clone NT-7 | Biolegend | Cat# 317202; RRID:AB_571931 |
| Rabbit Anti-HIV-1 Nef Polyclonal Antibody | NIH AIDS Reagent Program | Cat# 2949 |
| Rabbit Anti-HIV-1 Vpu Polyclonal Antibody | (Prévost et al., 2020a) | N/A |
| Goat anti-Human IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A21445; RRID:AB_2535862 |
| Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A21235; RRID:AB_2535804 |
| Alexa Fluor 647 anti-Human IgG Fc Antibody | Biolegend | Cat# 409319; RRID:AB_2563329 |
| Brilliant Violet 421 Donkey anti-Rabbit IgG (minimal x-reactivity) Antibody | Biolegend | Cat# Poly4064; RRID:AB_10643424 |
| Goat anti-Human IgG Fc Cross-Adsorbed Secondary Antibody, HRP | Thermo Fisher Scientific | Cat# A18823; RRID:AB_2535600 |
| Streptavidin, Alexa Fluor 647 conjugate | Thermo Fisher Scientific | Cat# S32357; RRID:AB_2336066 |
| PE Mouse anti-HIV-1 p24 clone KC57 | Beckman Coulter | Cat# 6604667; RRID:AB_1575989 |
| APC Mouse Anti-Human CD45 Clone HI30 | BD Biosciences | Cat# 561864; RRID:AB_11153499 |
| FITC Mouse Anti-Human CD3 Clone HIT3a | BD Biosciences | Cat# 555339; RRID:AB_395745 |
| PE Mouse Anti-Human CD8 Clone RPA-T8 | Biolegend | Cat# 555367; RRID:AB_395770 |
| PerCP Mouse Anti-human CD4 Clone RPA-T4 | Biolegend | Cat# 300527; RRID:AB_893327 |
|
| ||
| Biological samples | ||
|
| ||
| Human PBMCs from HIV-1-infected and uninfected individuals | FRQS AIDS network and New York Blood Bank | N/A |
| Plasma form HIV-1-infected individuals | FRQS AIDS network | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Dulbecco’s modified Eagle’s medium (DMEM) | Wisent | Cat# 319-005-CL |
| Gibco™ Roswell Park Memorial Institute 1640 medium (RPMI) | Thermo Fisher Scientific | Cat# 11875-093 |
| Fetal bovine serum (FBS) | VWR | Cat# 97068-085 |
| Penicillin/streptomycin | Wisent | Cat# 450-200-EL |
| Tris-buffered saline (TBS) | Thermo Fisher Scientific | Cat# BP24711 |
| BSA | BioShop | Cat# ALB001.100 |
| Tween 20 | Thermo Fisher Scientific | Cat# BP337-500 |
| Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate | Perkin Elmer Life Sciences | Cat# NEL105001EA |
| Phosphate buffered saline (PBS) | Wisent | Cat# 311-010-CL |
| Lymphocyte separation medium | Wisent | Cat# 305-010-CL |
| FreeStyle 293F expression medium | ThermoFisher Scientific | Cat# 12338002 |
| ExpiFectamine 293 transfection reagent | ThermoFisher Scientific | Cat# A14525 |
| Passive lysis buffer | Promega | Cat# E1941 |
| Firefly D-Luciferin Free Acid | Prolume | Cat# 306 |
| eBioscience™ Cell proliferation dye eFluor670 | Thermo Fisher Scientific | Cat# 65-0840-90 |
| eBioscience™ Cell proliferation dye eFluor450 | Thermo Fisher Scientific | Cat# 65-0842-85 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain | Thermo Fisher Scientific | Cat# L34966 |
| Formaldehyde 37% | Thermo Fisher Scientific | Cat# F79-500 |
| Biotininylated recombinant soluble dimeric FcγRIIIa (V158) protein | (Wines et al., 2016) | N/A |
| CD4 mimetic BNM-III-170 | Dr Amos B. Smith III (Chen etal., 2019) | N/A |
| Dimethyl sulfoxide (DMSO) | Thermo Fisher Scientific | Cat# D1391 |
| Phytohemagglutinin-L (PHA-L) | Sigma | Cat# L2769 |
| Recombinant IL-2 (rIL-2) | NIH AIDS Reagent Program | Cat# 136 |
| Protein A Sepharose CL-4B | Cytiva | Cat # 17096303 |
| Papain-agarose resin | Thermo Fisher Scientific | Cat # 20341 |
| β-mercaptoethanol | Bio-Rad | Cat# #1610710 |
| Acrylamide/Bis-Acrylamide | Thermo Fisher Scientific | Cat# BP1410-1 |
| Sodium dodecyl sulfate (SDS) | Thermo Fisher Scientific | Cat # BP166-500 |
| Ammonium Persulfate | Bio-Rad | Cat# 1610700 |
| TEMED | Bio-Rad | Cat# 1610801 |
| Coomassie Brilliant Blue R-250 | Thermo Fisher Scientific | Cat# BP101-50 |
| Magnesium phosphate (MgSO4) | Bioshop | Cat# MAG511.500 |
| Potassium phosphate monobasic (KH2PO4) | Thermo Fisher Scientific | Cat# P285-500 |
| Adenosine 5-triphosphate disodium salt hydrate (ATP) | Sigma | Cat# A3377-10G |
| Dithiothreitol (DTT) | Thermo Fisher Scientific | Cat# BP172-5 |
| HIV-1 Env gp41 peptide (583–618) | (Gohain et al., 2016) | N/A |
| HIV-1 Env gp41 peptide (587–597) | Genscript | N/A |
| HIV-1 Env gp41 peptide (596–606) | Genscript | N/A |
| SARS-CoV-2 Spike S2 peptide (1153–1163) | (Li et al., 2021) | N/A |
|
| ||
| Critical commercial assays | ||
|
| ||
| QuikChange II XL Site-Directed Mutagenesis Kit | Agilent Technologies | Cat # #200522 |
| Alexa Fluor 647 Protein Labeling Kit | Thermo Fisher Scientific | Cat# A20173 |
| EasySep human CD4+ T cell enrichment kit | StemCell Technologies | Cat# 19052 |
| Cytofix/Cytoperm Fixation/Permeabilization Kit | BD Biosciences | Cat# 554714 |
| QIAamp viral RNA mini kit | QIAGEN | Cat# 52906 |
| Quantitect SYBR green RT-PCR Master Mix | QIAGEN | Cat# 204245 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| 293T human embryonic kidney cells | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| TZM-bl cells | NIH AIDS Reagent Program | Cat# 8129; RRID:CVCL_B478 |
| FreeStyle 293F cells | Thermo Fisher Scientific | Cat# R79007; RRID: CVCL_D603 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| SIRPAh/m Rag2−/− Il2rg−/− IL15h/m (SRG-15) mice | (Herndler-Brandstetter et al., 2017) | N/A |
| NOD.Cg-Prkdcscid IL2rg−/− Tg(Hu-IL15) (NSG-15) mice | Jackson Laboratory | Cat# 030890; RRID:IMSR_JAX:030890 |
| Oligonucleotides | ||
| 246D heavy chain G236R FWD: 5′-CCTGAACTCCTGCGGGGACCGTCAGTC-3′ | Integrated DNA Technologies | N/A |
| 246D heavy chain G236R REV: 5′-GACTGACGGTCCCCGCAGGAGTTCAGG-3′ | Integrated DNA Technologies | N/A |
| 246D heavy chain L328R FWD: 5′-CCAACAAAGCCCGCCCAGCCCCCATC-3′ | Integrated DNA Technologies | N/A |
| 246D heavy chain L328R REV: 5′-GATGGGGGCTGGGCGGGCTTTGTTGG-3′ | Integrated DNA Technologies | N/A |
| 246D heavy chain G236A/S239D FWD: 5′-CTCCTGGCGGGACCGGATGTCTTCCTCTTC-3′ | Integrated DNA Technologies | N/A |
| 246D heavy chain G236A/S239D REV: 5′-GAAGAGGAAGACATCCGGTCCCGCCAGGAG-3′ | Integrated DNA Technologies | N/A |
| 246D heavy chain A330L/I332E FWD: 5′-GCCCTCCCACTCCCCGAAGAGAAAACCATC-3′ | Integrated DNA Technologies | N/A |
| 246D heavy chain A330L/I332E REV: 5′-GATGGTTTTCTCTTCGGGGAGTGGGAGGGC-3′ | Integrated DNA Technologies | N/A |
| HIV-1 IMC JR-CSF Vpu-FWD: 5′ -GTGCATGT AATGTAACCTTTACAAATATTAGC-3′ | Integrated DNA Technologies | N/A |
| HIV-1 IMC JR-CSF Vpu-REV: 5′-GCTAATATTTGTAAAGGTTACATTACATGCAC-3′ | Integrated DNA Technologies | N/A |
| HIV-1NL4/3YU2 Vpu + FWD: 5′-GTAAGTAGTACATGTAATGCAACCTATACC-3′ | Integrated DNA Technologies | N/A |
| HIV-1NL4/3YU2Vpu + REV: 5′-GGTATAGGTTGCATTACATGTACTACTTAC-3′ | Integrated DNA Technologies | N/A |
| HIV-1 gag primer FWD: 5′-TGCTATGTCAGTTCCCCTTGGTTCTCT-3′ | (Rajashekar et al., 2021) | N/A |
| HIV-1 gag primer REV: 5′-AGTTGGAGGACATCAAGCAGCCATGCAAAT-3′ | (Rajashekar et al., 2021) | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| 246D mAb Light Chain Expression Vector | NIH AIDS Reagent Program | Cat# 13742 |
| 246D mAb Heavy Chain Expression Vector | NIH AIDS Reagent Program | Cat# 13741 |
| 246D GRLR mAb Heavy Chain Expression Vector | This paper | N/A |
| 246D GASDALIE mAb Heavy Chain Expression Vector | This paper | N/A |
| HIV-1NL4/3YU2 | (Horwitz et al., 2017) | N/A |
| HIV-1NL4/3YU2 Vpu+ | This paper | N/A |
| HIV-1 IMC JR-CSF | NIH AIDS Reagent Program (Koyanagi et al., 1987) | Cat# 2708 |
| HIV-1 IMC JR-CSF Vpu− | This paper | N/A |
| HIV-1 IMC NL4/3 | NIH AIDS Reagent Program (Adachi et al., 1986) | Cat# 114 |
| HIV-1 IMC NL4/3 Vpu− | (Neil et al., 2006) | N/A |
| HIV-1 IMCYU2 | (Li et al., 1991) | N/A |
| HIV-1 IMC YU2 Vpu+ | (Krapp et al., 2016) | N/A |
| HIV-1 IMCAD8 | (Theodore et al., 1996) | N/A |
| HIV-1 IMC AD8 Vpu+ | (Krapp et al., 2016) | N/A |
| HIV-1 IMC CH058 | (Ochsenbauer et al., 2012) | N/A |
| HIV-1 IMC CH058 Vpu− | (Kmiec et al., 2016) | N/A |
| HIV-1 IMC CH058 Nef− | (Heigele et al., 2016) | N/A |
| HIV-1 IMC CH058 Nef− Vpu− | (Heigele et al., 2016) | N/A |
| HIV-1 IMC CH077 | (Ochsenbauer et al., 2012) | N/A |
| HIV-1 IMC CH077 Vpu− | (Kmiec et al., 2016) | N/A |
| HIV-1 IMC REJO | (Ochsenbauer et al., 2012) | N/A |
| HIV-1 IMC REJO Vpu− | (Yamada et al., 2018) | N/A |
| HIV-1 IMC STCO | (Parrish et al., 2013) | N/A |
| HIV-1 IMC STCO Vpu− | (Kmiec et al., 2016) | N/A |
| HIV-1 IMCCH198 | (Parrish et al., 2013) | N/A |
| HIV-1 IMC CH198 Vpu− | (Sauter et al., 2015) | N/A |
| HIV-1 IMC ZM246F | (Parrish et al., 2012) | N/A |
| HIV-1 IMC ZM246F Vpu− | (Langer et al., 2015) | N/A |
| HIV-1 IMCCH167 | (Parrish et al., 2013) | N/A |
| HIV-1 IMC CH167 Vpu− | (Kmiec et al., 2016) | N/A |
| HIV-1 IMC CH293 | (Parrish et al., 2013) | N/A |
| HIV-1 IMC CH293 Vpu− | (Sauter et al., 2015) | N/A |
| VSV G glycoprotein expression vector | (Emi et al., 1991) | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| BD FACSDiva v9.0 | BD Biosciences | RRID: SCR_001456 |
| FlowJo v10.5.3 | Tree Star | https://www.flowjo.com/; RRID:SCR_008520 |
| GraphPad Prism v9.3.1 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
|
| ||
| Other | ||
|
| ||
| BD LSR II Flow Cytometer | BD Biosciences | N/A |
| TriStar LB 942 Microplate Reader | Berthold Technologies | N/A |
| Applied Biosystems 7500 real-time PCR system | Thermo Fisher Scientific | N/A |
| Biacore 3000 | Cytiva | RRID: SCR_019954 |
| Protein A sensor chip | Cytiva | Cat #29127558 |
| Superdex 200 16/60 column | Cytiva | Cat# 17-1069-01 |
| Superdex 200 10/300 GL column | Cytiva | Cat# 17-5175-01 |
| Clear V-bottom 96-well plates (cell culture-treated) | Corning | Cat# 0720096 |
| White flat-bottom 96-well plates (cell culture-treated) | Corning | Cat# 0877126 |
| White Maxisorp™ Nunc™ 96-well plates | Thermo Fisher Scientific | Cat# 437796 |
Highlights.
Vpu limits HIV-1 Env recognition and ADCC responses mediated by nnAbs and bNAbs
Vpu deletion impairs HIV-1 replication in humanized mice in presence of nnAbs
CD4 mimetics and Fc engineering boost nnAbs antiviral activities in vivo
ACKNOWLEDGMENTS
The authors thank the CRCHUM BSL3 and flow cytometry platforms for technical assistance and Mario Legault from the Fonds de recherche du Québec-Santé (FRQS) AIDS and Infectious Diseases network for cohort coordination and clinical samples. We also thank David T. Evans (University of Wisconsin) for helpful discussions. We thank the following collaborators for kindly providing Abs: Julie Overbaugh (Fred Hutchinson Cancer Research Center) for QA255-006, QA255-067, and QA255-072; James Robinson (Tulane University) for 7B2, 2.2B, 12.3D, 12.4H, A32, C11, and 17b; George Lewis (University of Maryland) for M785U1, M785U2, M785U3, M785U4, N5U1, N5U2, N5U3, N10U1, and N10U2; Gunilla Karlsson Hedestam (Karolinska Institutet) for GE2-JG8; John Mascola (Vaccine Research Center, NIAID) for VRC01, VRC03, VRC07-523, VRC13, VRC16, and VRC34; Mark Connors (NIAID) for 10E8, N6, and 35O22; Florian Klein (University of Cologne) for 1–18; and the International AIDS Vaccine Initiative (IAVI) for PG9, PG16, PGT121, PGT122, PGT123, PGT125, PGT126, PGT128, PGT130, PGT135, PGT136, PGT145, and PGT151. We thank P. Mark Hogarth (Burnet Institute) for kindly providing recombinant dimeric FcγRIIIa. The graphical abstract and Figures 6 and 7 were prepared using illustrations from BioRender.com. This study was supported by a Canadian Institutes of Health Research (CIHR) foundation grant #352417 to A.F. Funds were also provided by a CIHR team grant #422148 to P.K. and A.F.; a Canada Foundation for Innovation (CFI) grant #41027 to A.F.; and by the National Institutes of Health to A.F. (R01 AI148379 and R01 AI150322), to M.P. and A.F. (R01 AI129769), M.P. (AI116274), and to P.K. (R01 AI145164, R33 AI122384, and P50 AI150464 [CHEETAH]). Support for this work was also provided by P01 GM56550/AI150471 to A.B.S. and A.F. This work was partially supported by 1UM1AI164562-01; co-funded by National Heart, Lung, and Blood Institute; National Institute of Diabetes and Digestive and Kidney Diseases; National Institute of Neurological Disorders and Stroke; National Institute on Drug Abuse, and the National Institute of Allergy and Infectious Diseases to A.F. A.F. is the recipient of a Canada Research Chair on Retroviral Entry #RCHS0235 950-232424. F.K. is supported by the German Research Foundation (DFG CRC 1279 and SPP1923) and the Baden-Württemberg Foundation (BWST-ISF2018-032). J.P. and S.P.A. are recipients of CIHR doctoral fellowships. M.W.G. is a recipient of the Gruber Science Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The opinions and assertions expressed herein are those of the author(s) and do not necessarily reflect the official policy or position of the Uniformed Services University or the Department of Defense.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111624.
REFERENCES
- Acharya P, Tolbert WD, Gohain N, Wu X, Yu L, Liu T, Huang W, Huang CC, Kwon YD, Louder RK, et al. (2014). Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J. Virol. 88, 12895–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, and Martin MA (1986). Production of acquired immunodeficiency syndromeassociated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsahafi N, Ding S, Richard J, Markle T, Brassard N, Walker B, Lewis GK, Kaufmann DE, Brockman MA, and Finzi A (2015). Nef proteins from HIV-1 elite controllers are inefficient at preventing antibody-dependent cellular cytotoxicity. J. Virol. 90, 2993–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding S, Bhattacharyya S, Das D, Anand SP, Prévost J, Tolbert WD, et al. (2019). An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25, 578–587.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, and Chen BK (2014). HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J. Virol. 88, 6031–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand SP, Prévost J, Baril S, Richard J, Medjahed H, Chapleau JP, Tolbert WD, Kirk S, Smith AB III, Wines BD, et al. (2019). Two families of env antibodies efficiently engage Fc-gamma receptors and eliminate HIV-1-Infected cells. J. Virol. 93. e01823–e01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias JF, Heyer LN, von Bredow B, Weisgrau KL, Moldt B, Burton DR, Rakasz EG, and Evans DT (2014). Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc. Natl. Acad. Sci. USA 111, 6425–6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan M, Dias J, Liu C, Maximova A, Ernste K, Pegu A, McKee K, Shi W, Chen X, Almasri C, et al. (2020). Fc-mediated effector function contributes to the in vivo antiviral effect of an HIV neutralizing antibody. Proc. Natl. Acad. Sci. USA 117, 18754–18763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW, et al. (2016). Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N. Engl. J. Med. 375, 2037–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, et al. (2013). Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin-Bussières G, Prévost J, Gendron-Lepage G, Melillo B, Chen J, Smith Iii AB, Pazgier M, and Finzi A (2020). Elicitation of cluster A and Co-receptor binding site antibodies are required to eliminate HIV-1 infected cells. Microorganisms 8, E710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton DL, Pegu A, Wang K, McGinnis K, Nason M, Foulds K, Letukas V, Schmidt SD, Chen X, Todd JP, et al. (2016). Human immunodeficiency virus type 1 monoclonal antibodies suppress acute simian-human immunodeficiency virus viremia and limit seeding of cell-associated viral reservoirs. J. Virol. 90, 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S, Perrin C, and Strebel K (1999). Cell surface CD4 inhibits HIV-1 particle release by interfering with Vpu activity. J. Biol. Chem. 274, 33800–33806. [DOI] [PubMed] [Google Scholar]
- Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, and Ravetch JV (2014). Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158, 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, Gazumyan A, Seaman MS, Nussenzweig MC, and Ravetch JV (2016). Bispecific anti-HIV-1 antibodies with enhanced breadth and potency. Cell 165, 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm MA, Aryee K-E, Bruzenksi L, Greiner DL, Shultz LD, and Keck J (2018). Transgenic expression of human IL15 in NOD-scid IL2rgnull (NSG) mice enhances the development and survival of functional human NK cells. J. Immunol. 200, 103.120. [Google Scholar]
- Bruel T, Guivel-Benhassine F, Lorin V, Lortat-Jacob H, Baleux F, Bourdic K, Noël N, Lambotte O, Mouquet H, and Schwartz O (2017). Lack of ADCC breadth of human non-neutralizing anti-HIV-1 antibodies. J. Virol 91, e02440–e02516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. (1994). Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10, 359–369. [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. (1994). Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027. [DOI] [PubMed] [Google Scholar]
- Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA, Moldt B, Dunlop DC, Poignard P, Doyle LA, Cavacini L, et al. (2011). Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc. Natl. Acad. Sci. USA 108, 11181–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP Jr., Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, et al. (2015). Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, et al. (2017). Antibody 10–1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 23, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavacini LA, Emes CL, Wisnewski AV, Power J, Lewis G, Montefiori D, and Posner MR (1998). Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res. Hum. Retroviruses 14, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, and Kim PS (1997). Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273. [DOI] [PubMed] [Google Scholar]
- Chen J, Park J, Kirk SM, Chen HC, Li X, Lippincott DJ, Melillo B, and Smith AB III. (2019). Development of an effective scalable enantioselective synthesis of the HIV-1 entry inhibitor BNM-III-170 as the Bis-trifluoroacetate salt. Org. Process Res. Dev. 23, 2464–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Langedijk JPM, Hinz A, Seaman MS, Vanzetta F, FernandezRodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, et al. (2010). Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5, e8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave VP, Hajjar F, Dieng MM, Haddad É, and Cohen ÉA (2013). Efficient BST2 antagonism by Vpu is critical for early HIV-1 dissemination in humanized mice. Retrovirology 10, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, Montefiori DC, Graham BS, Keefer MC, Pinter A, Morris L, et al. (2009). High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387, 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, et al. (2005). Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201, 1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Veillette M, Coutu M, Prévost J, Scharf L, Bjorkman PJ, Ferrari G, Robinson JE, Stürzel C, Hahn BH, et al. (2016a). A highly conserved residue of the HIV-1 gp120 inner domain is important for antibody-dependent cellular cytotoxicity responses mediated by anti-cluster A antibodies. J. Virol. 90, 2127–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Verly MM, Princiotto A, Melillo B, Moody AM, Bradley T, Easterhoff D, Roger M, Hahn BH, Madani N, et al. (2016b). Small molecule CD4-mimetics sensitize HIV-1-infected cells to ADCC by antibodies elicited by multiple envelope glycoprotein immunogens in non-human primates. AIDS Res. Hum. Retroviruses 33, 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Gasser R, Gendron-Lepage G, Medjahed H, Tolbert WD, Sodroski J, Pazgier M, and Finzi A (2019a). CD4 incorporation into HIV-1 viral particles exposes envelope epitopes recognized by CD4-induced antibodies. J. Virol. 93. e01403–e01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Grenier MC, Tolbert WD, Vézina D, Sherburn R, Richard J, Prévost J, Chapleau JP, Gendron-Lepage G, Medjahed H, et al. (2019b). A new family of small-molecule CD4-mimetic compounds contacts highly conserved aspartic acid 368 of HIV-1 gp120 and mediates antibody-dependent cellular cytotoxicity. J. Virol. 93, e01325–e01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Klein F, Horwitz JA, Halper-Stromberg A, Sather DN, Marcovecchio PM, Lee T, West AP Jr., Gao H, Seaman MS, et al. (2013). Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. J. Exp. Med. 210, 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ (2015). The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 282, 4679–4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. (2015). Immunization for HIV-1 broadly neutralizing antibodies in human ig knockin mice. Cell 161, 1505–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé M, Bego MG, Paquay C, and Cohen ÉA (2010). Modulation of HIV1-host interaction: role of the Vpu accessory protein. Retrovirology 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufloo J, Guivel-Benhassine F, Buchrieser J, Lorin V, Grzelak L, Dupouy E, Mestrallet G, Bourdic K, Lambotte O, Mouquet H, et al. (2020). Anti-HIV-1 antibodies trigger non-lytic complement deposition on infected cells. EMBO Rep. 21, e49351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Doms RW, and Moss B (1990). Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda Y, Murakami T, Ami Y, Nakasone T, Takizawa M, Someya K, Kaizu M, Izumi Y, Yoshino N, Matsushita S, et al. (2006). Anti-V3 humanized antibody KD-247 effectively suppresses ex vivo generation of human immunodeficiency virus type 1 and affords sterile protection of monkeys against a heterologous simian/human immunodeficiency virus infection. J. Virol. 80, 5563–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi N, Friedmann T, and Yee JK (1991). Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J. Virol. 65, 1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, et al. (2016). Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in ig knockin mice. Cell 166, 1445–1458.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. (2014). Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine J, Coutlée F, Tremblay C, Routy JP, Poudrier J, and Roger M; Montreal Primary HIV Infection and Long-Term Nonprogressor Study Groups (2009). HIV infection affects blood myeloid dendritic cells after successful therapy and despite nonprogressing clinical disease. J. Infect. Dis. 199, 1007–1018. [DOI] [PubMed] [Google Scholar]
- Fontaine J, Chagnon-Choquet J, Valcke HS, Poudrier J, and Roger M; Montreal Primary HIV Infection and Long-Term Non-Progressor Study Groups (2011). High expression levels of B lymphocyte stimulator (BLyS) by dendritic cells correlate with HIV-related B-cell disease progression in humans. Blood 117, 145–155. [DOI] [PubMed] [Google Scholar]
- Freed EO, Myers DJ, and Risser R (1989). Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J. Virol. 63, 4670–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund NT, Horwitz JA, Nogueira L, Sievers SA, Scharf L, Scheid JF, Gazumyan A, Liu C, Velinzon K, Goldenthal A, et al. (2015). A new glycan-dependent CD4-binding site neutralizing antibody exerts pressure on HIV-1 in vivo. PLoS Pathog. 11. e1005238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund NT, Wang H, Scharf L, Nogueira L, Horwitz JA, Bar-On Y, Golijanin J, Sievers SA, Sok D, Cai H, et al. (2017). Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med. 9. eaal2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey G, Chen J, Rits-Volloch S, Freeman MM, Zolla-Pazner S, and Chen B (2010). Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat. Struct. Mol. Biol. 17, 1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschi CJ, Liang S, Mohammadi M, Anang S, Moraca F, Chen J, Madani N, Sodroski JG, Abrams CF, Hendrickson WA, and Smith AB 3rd. (2021). Identification of gp120 residue His105 as a novel target for HIV-1 neutralization by small-molecule CD4-mimics. ACS Med. Chem. Lett. 12, 1824–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini D, Vitone F, Gori E, La Placa M, and Re MC (2004). Quantitative detection of human immunodeficiency virus type 1 (HIV-1) viral load by SYBR green real-time RT-PCR technique in HIV-1 seropositive patients. J. Virol. Methods 115, 183–189. [DOI] [PubMed] [Google Scholar]
- Gohain N, Tolbert WD, Orlandi C, Richard J, Ding S, Chen X, Bonsor DA, Sundberg EJ, Lu W, Ray K, et al. (2016). Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240. Sci. Rep. 6, 36685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Gianakakos V, Sharpe S, and Zolla-Pazner S (1989). Generation of human monoclonal antibodies to human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86, 1624–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GE, Bekker LG, Laher F, Malahleha M, Allen M, Moodie Z, Grunenberg N, Huang Y, Grove D, Prigmore B, et al. (2021). Vaccine efficacy of ALVAC-HIV and bivalent subtype C gp120-MF59 in adults. N. Engl. J. Med. 384, 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Pazgier M, Sajadi MM, Kamin-Lewis R, Al-Darmarki S, Flinko R, Lovo E, Wu X, Robinson JE, Seaman MS, et al. (2013). Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc. Natl. Acad. Sci. USA 110, E69–E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, Nogueira L, Eisenreich TR, Liu C, Gazumyan A, Schaefer U, et al. (2014). Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158, 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. (2012a). Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366, 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, and Kepler TB (2012b). B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 30, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Stürzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, et al. (2016). The potency of nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe 20, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndler-Brandstetter D, Shan L, Yao Y, Stecher C, Plajer V, Lietzenmayer M, Strowig T, de Zoete MR, Palm NW, Chen J, et al. (2017). Humanized mouse model supports development, function, and tissue residency of human natural killer cells. Proc. Natl. Acad. Sci. USA 114, E9626–E9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell AJ, Jaworski JP, Epson E, Matsuda K, Pandey S, Kahl C, Reed J, Sutton WF, Hammond KB, Cheever TA, et al. (2016). Early short-term treatment with neutralizing human monoclonal antibodies halts SHIV infection in infant macaques. Nat. Med. 22, 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell AJ, Shapiro MB, Powell R, Malherbe DC, McBurney SP, Pandey S, Cheever T, Sutton WF, Kahl C, Park B, et al. (2018). Reduced cell-associated DNA and improved viral control in macaques following passive transfer of a single anti-V2 monoclonal antibody and repeated simian/human immunodeficiency virus challenges. J. Virol. 92. e02198–e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioe CE, Li G, Liu X, Tsahouridis O, He X, Funaki M, Klingler J, Tang AF, Feyznezhad R, Heindel DW, et al. (2022). Non-neutralizing antibodies targeting the immunogenic regions of HIV-1 envelope reduce mucosal infection and virus burden in humanized mice. PLoS Pathog 18. e1010183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton HM, Bernett MJ, Peipp M, Pong E, Karki S, Chu SY, Richards JO, Chen H, Repp R, Desjarlais JR, and Zhukovsky EA (2010). Fc-engineered anti-CD40 antibody enhances multiple effector functions and exhibits potent in vitro and in vivo antitumor activity against hematologic malignancies. Blood 116, 3004–3012. [DOI] [PubMed] [Google Scholar]
- Horwitz JA, Halper-Stromberg A, Mouquet H, Gitlin AD, Tretiakova A, Eisenreich TR, Malbec M, Gravemann S, Billerbeck E, Dorner M, et al. (2013). HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc. Natl. Acad. Sci. USA 110, 16538–16543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz JA, Bar-On Y, Lu CL, Fera D, Lockhart AAK, Lorenzi JCC, Nogueira L, Golijanin J, Scheid JF, Seaman MS, et al. (2017). Nonneutralizing antibodies alter the course of HIV-1 infection in vivo. Cell 170, 637–648.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hout DR, Gomez ML, Pacyniak E, Gomez LM, Inbody SH, Mulcahy ER, Culley N, Pinson DM, Powers MF, Wong SW, and Stephens EB (2005). Scrambling of the amino acids within the transmembrane domain of Vpu results in a simian-human immunodeficiency virus (SHIVTM) that is less pathogenic for pig-tailed macaques. Virology 339, 56–69. [DOI] [PubMed] [Google Scholar]
- Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, et al. (2004). Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. USA 101, 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. (2012). Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, et al. (2014). Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515, 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. (2016). Identification of a CD4-binding-site antibody to HIV that evolved near-Pan neutralization breadth. Immunity 45, 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereño-Orbea J, Kalyuzhniy O, Deresa I, et al. (2016). HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette CA, Barnes CO, Kirk SM, Melillo B, Smith AB 3rd, and Bjorkman PJ (2021). Cryo-EM structures of HIV-1 trimer bound to CD4-mimetics BNM-III-170 and M48U1 adopt a CD4-bound open conformation. Nat. Commun. 12, 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, et al. (2012). HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492, 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Nogueira L, Nishimura Y, Phad G, West AP Jr., Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, et al. (2014). Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J. Exp. Med. 211, 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec D, Iyer SS, Stürzel CM, Sauter D, Hahn BH, and Kirchhoff F (2016). Vpu-mediated counteraction of tetherin is a major determinant of HIV-1 interferon resistance. mBio 7. e09344–e01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, et al. (2016). Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, and Chen IS (1987). Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236, 819–822. [DOI] [PubMed] [Google Scholar]
- Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Stürzel CM, Mack K, Reith E, Engelhart S, Ciuffi A, et al. (2016). Guanylate binding protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe 19, 504–514. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, and Hendrickson WA (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais E, and Moore PL (2018). Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers. Retrovirology 15, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer SM, Hopfensperger K, Iyer SS, Kreider EF, Learn GH, Lee LH, Hahn BH, and Sauter D (2015). A naturally occurring rev1-vpu fusion gene does not confer a fitness advantage to HIV-1. PLoS One 10. e0142118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumaea A, Smith AB 3rd, Sodroski J, and Finzi A (2020). Opening the HIV envelope: potential of CD4 mimics as multifunctional HIV entry inhibitors. Curr. Opin. HIV AIDS 15, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WS, Richard J, Lichtfuss M, Smith AB III, Park J, Courter JR, Melillo BN, Sodroski JG, Kaufmann DE, Finzi A, et al. (2015). Antibody-dependent cellular cytotoxicity against reactivated HIV-1-Infected cells. J. Virol. 90, 2021–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, and Ward AB (2016). Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque K, Zhao YS, and Cohen EA (2003). Vpu exerts a positive effect on HIV-1 infectivity by down-modulating CD4 receptor molecules at the surface of HIV-1-producing cells. J. Biol. Chem. 278, 28346–28353. [DOI] [PubMed] [Google Scholar]
- Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, and Hahn BH (1991). Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65, 3973–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li W, Lu M, Bess J Jr., Chao CW, Gorman J, Terry DS, Zhang B, Zhou T, Blanchard SC, et al. (2020). Subnanometer structures of HIV-1 envelope trimers on aldrithiol-2-inactivated virus particles. Nat. Struct. Mol. Biol. 27, 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen Y, Prévost J, Ullah I, Lu M, Gong SY, Tauzin A, Gasser R, Vézina D, Anand SP, et al. (2021). Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern. Cell Rep. 38, 110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. (2013). Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CL, Murakowski DK, Bournazos S, Schoofs T, Sarkar D, HalperStromberg A, Horwitz JA, Nogueira L, Golijanin J, Gazumyan A, et al. (2016). Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science 352, 1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, et al. (2019). Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, et al. (2015). Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 7. 319ra206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N, Princiotto AM, Easterhoff D, Bradley T, Luo K, Williams WB, Liao HX, Moody MA, Phad GE, Vázquez Bernat N, et al. (2016). Antibodies elicited by multiple envelope glycoprotein immunogens in primates neutralize primary human immunodeficiency viruses (HIV-1) sensitized by CD4-mimetic compounds. J. Virol. 90, 5031–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N, Princiotto AM, Mach L, Ding S, Prevost J, Richard J, Hora B, Sutherland L, Zhao CA, Conn BP, et al. (2018). A CD4-mimetic compound enhances vaccine efficacy against stringent immunodeficiency virus challenge. Nat. Commun. 9, 2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganaro L, Hong P, Hernandez MM, Argyle D, Mulder LCF, Potla U, Diaz-Griffero F, Lee B, Fernandez-Sesma A, and Simon V (2018). IL-15 regulates susceptibility of CD4(+) T cells to HIV infection. Proc. Natl. Acad. Sci. USA 115, E9659–E9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusali G, Potestà M, Santoni A, Cerboni C, and Doria M (2012). The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J. Virol. 86, 4496–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, and Weissman IL (1988). Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53, 55–67. [DOI] [PubMed] [Google Scholar]
- Melillo B, Liang S, Park J, Schön A, Courter JR, LaLonde JM, Wendler DJ, Princiotto AM, Seaman MS, Freire E, et al. (2016). Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med. Chem. Lett. 7, 330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog C, Dereuddre-Bosquet N, Teillaud JL, Biedma ME, Holl V, Van Ham G, Heyndrickx L, Van Dorsselaer A, Katinger D, Vcelar B, et al. (2014). Protective effect of vaginal application of neutralizing and nonneutralizing inhibitory antibodies against vaginal SHIV challenge in macaques. Mucosal Immunol. 7, 46–56. [DOI] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, et al. (2012). Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA 109, E3268–E3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, et al. (2014). Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJD, Eastman SW, Jouvenet N, and Bieniasz PD (2006). HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJD, Zang T, and Bieniasz PD (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Gautam R, Chun TW, Sadjadpour R, Foulds KE, Shingai M, Klein F, Gazumyan A, Golijanin J, Donaldson M, et al. (2017). Early antibody therapy can induce long-lasting immunity to SHIV. Nature 543, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, et al. (2012). Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 86, 2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, et al. (2012). Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4-beta7. PLoS Pathog 8. e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, et al. (2013). Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. USA 110, 6626–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MS, Lee WS, Kristensen AB, Amarasena T, Khoury G, Wheatley AK, Reynaldi A, Wines BD, Hogarth PM, Davenport MP, and Kent SJ (2019). Fc-dependent functions are redundant to efficacy of anti-HIV antibody PGT121 in macaques. J. Clin. Invest. 129, 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prévost J, Nedellec R, von Bredow B, Abbink P, et al. (2019). Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 50, 241–252.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phad GE, Vázquez Bernat N, Feng Y, Ingale J, Martinez Murillo PA, O’Dell S, Li Y, Mascola JR, Sundling C, Wyatt RT, and Karlsson Hedestam GB (2015). Diverse antibody genetic and recognition properties revealed following HIV-1 envelope glycoprotein immunization. J. Immunol. 194, 5903–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TNQ, Lukhele S, Dallaire F, Perron G, and Cohen ÉA (2016). Enhancing virion tethering by BST2 sensitizes productively and latently HIV-infected T cells to ADCC mediated by broadly neutralizing antibodies. Sci. Rep. 6, 37225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, and Kabat D (1998). Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72, 2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J, Zoubchenok D, Richard J, Veillette M, Pacheco B, Coutu M, Brassard N, Parsons MS, Ruxrungtham K, Bunupuradah T, et al. (2017). Influence of the envelope gp120 phe 43 cavity on HIV-1 sensitivity to antibody-dependent cell-mediated cytotoxicity responses. J. Virol. 91. e02452–e02516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J, Richard J, Ding S, Pacheco B, Charlebois R, Hahn BH, Kaufmann DE, and Finzi A (2018a). Envelope glycoproteins sampling states 2/3 are susceptible to ADCC by sera from HIV-1-infected individuals. Virology 515, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J, Richard J, Medjahed H, Alexander A, Jones J, Kappes JC, Ochsenbauer C, and Finzi A (2018b). Incomplete downregulation of CD4 expression affects HIV-1 env conformation and antibody-dependent cellular cytotoxicity responses. J. Virol. 92. e04844–e0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J, Pickering S, Mumby MJ, Medjahed H, Gendron-Lepage G, Delgado GG, Dirk BS, Dikeakos JD, Stürzel CM, Sauter D, et al. (2019). Upregulation of BST-2 by type I interferons reduces the capacity of vpu to protect HIV-1-Infected cells from NK cell responses. mBio 10. e01113–e01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J, Edgar CR, Richard J, Trothen SM, Jacob RA, Mumby MJ, Pickering S, Dubé M, Kaufmann DE, Kirchhoff F, et al. (2020a). HIV-1 vpu downregulates tim-3 from the surface of infected CD4(+) T cells. J. Virol. 94. e019999–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J, Tolbert WD, Medjahed H, Sherburn RT, Madani N, Zoubchenok D, Gendron-Lepage G, Gaffney AE, Grenier MC, Kirk S, et al. (2020b). The HIV-1 env gp120 inner domain shapes the Phe43 cavity and the CD4 binding site. mBio 11. e02800–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J, Medjahed H, Vezina D, Chen HC, Hahn BH, Smith AB 3rd, and Finzi A (2021). HIV-1 envelope glycoproteins proteolytic cleavage protects infected cells from ADCC mediated by plasma from infected individuals. Viruses 13. 10.3390/v13112236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J, Richard J, Gasser R, Medjahed H, Kirchhoff F, Hahn BH, Kappes JC, Ochsenbauer C, Duerr R, and Finzi A (2022). Detection of the HIV-1 accessory proteins Nef and Vpu by flow cytometry represents a new tool to study their functional interplay within a single infected CD4+ T cell. J. Virol jvi0192921. 10.1128/jvi.01929-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajashekar JK, Richard J, Beloor J, Prévost J, Anand SP, Beaudoin-Bussières G, Shan L, Herndler-Brandstetter D, Gendron-Lepage G, Medjahed H, et al. (2021). Modulating HIV-1 envelope glycoprotein conformation to decrease the HIV-1 reservoir. Cell Host Microbe 29, 904–916.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. (2009). Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361, 2209–2220. [DOI] [PubMed] [Google Scholar]
- Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, Pazgier M, Schön A, Freire E, Routy JP, et al. (2015). CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc. Natl. Acad. Sci. USA 112, E2687–E2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J, Pacheco B, Gohain N, Veillette M, Ding S, Alsahafi N, Tolbert WD, Prévost J, Chapleau JP, Coutu M, et al. (2016). Co-Receptor binding site antibodies enable CD4-mimetics to expose conserved anti-cluster A ADCC epitopes on HIV-1 envelope glycoproteins. EBioMedicine 12, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J, Prévost J, von Bredow B, Ding S, Brassard N, Medjahed H, Coutu M, Melillo B, Bibollet-Ruche F, Hahn BH, et al. (2017). BST-2 expression modulates small CD4-mimetic sensitization of HIV-1-Infected cells to antibody-dependent cellular cytotoxicity. J. Virol. 91. e02199–e0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CA, Lyddon TD, Gil HM, Evans DT, Kuzmichev YV, Richard J, Finzi A, Welbourn S, Rasmussen L, Nebane NM, et al. (2022). Novel compound inhibitors of HIV-1NL4–3 vpu. Viruses 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JE YH, Holton D, Elliott S, and Ho DD (1992). Distinct antigenic sites on HIV gp120 identified by a panel of human monoclonal antibodies. J. Cell. Biochem, Q449. [Google Scholar]
- Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, et al. (2014). Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 88, 12669–12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajadi MM, Dashti A, Rikhtegaran Tehrani Z, Tolbert WD, Seaman MS, Ouyang X, Gohain N, Pazgier M, Kim D, Cavet G, et al. (2018). Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell 173, 1783–1795.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, et al. (2009). Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206, 1273–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra S, Tomaras GD, Warrier R, Nicely NI, Liao HX, Pollara J, Liu P, Alam SM, Zhang R, Cocklin SL, et al. (2015). Human non-neutralizing HIV-1 envelope monoclonal antibodies limit the number of founder viruses during SHIV mucosal infection in rhesus macaques. PLoS Pathog 11. e1005042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Misawa N, Fukuhara M, Iwami S, An DS, Ito M, and Koyanagi Y (2012). Vpu augments the initial burst phase of HIV-1 propagation and downregulates BST2 and CD4 in humanized mice. J. Virol. 86, 5000–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders KO, Wiehe K, Tian M, Acharya P, Bradley T, Alam SM, Go EP, Scearce R, Sutherland L, Henderson R, et al. (2019). Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science 366. eaay7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Hotter D, Van Driessche B, Stürzel CM, Kluge SF, Wildum S, Yu H, Baumann B, Wirth T, Plantier JC, et al. (2015). Differential regulation of NF-kappaB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 10, 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. (2011). Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu CL, Lorenzi JCC, Feldmann A, Braunschweig M, Nogueira L, Oliveira T, et al. (2016). HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommers P, Gruell H, Abernathy ME, Tran MK, Dingens AS, Gristick HB, Barnes CO, Schoofs T, Schlotz M, Vanshylla K, et al. (2020). Restriction of HIV-1 escape by a highly broad and potent neutralizing antibody. Cell 180, 471–489.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs T, Barnes CO, Suh-Toma N, Golijanin J, Schommers P, Gruell H, West AP Jr., Bach F, Lee YE, Nogueira L, et al. (2019). Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity 50, 1513–1529.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AH, Sowrirajan B, Davis ZB, Ward JP, Campbell EM, Planelles V, and Barker E (2010). Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe 8, 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik MM, Peng H, Lu J, Rits-Volloch S, Xu C, Liao M, and Chen B (2019). Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature 565, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, Hahn BH, Arya SK, Groopman JE, Gallo RC, and Wong-Staal F (1984). Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science 226, 1165–1171. [DOI] [PubMed] [Google Scholar]
- Shingai M, Yoshida T, Martin MA, and Strebel K (2011). Some human immunodeficiency virus type 1 Vpu proteins are able to antagonize macaque BST-2 in vitro and in vivo: vpu-negative simian-human immunodeficiency viruses are attenuated in vivo. J. Virol. 85, 9708–9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M Jr., Lifson JD, et al. (2013). Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503, 277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, DiLillo DJ, Bournazos S, Li F, and Ravetch JV (2012). Mouse model recapitulating human Fcgamma receptor structural and functional diversity. Proc. Natl. Acad. Sci. USA 109, 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojar H, Baron S, Sullivan JT, Garrett M, van Haaren MM, Hoffman J, Overbaugh J, Doranz BJ, and Hicar MD (2019). Monoclonal antibody 2C6 targets a cross-clade conformational epitope in gp41 with highly active antibody-dependent cell cytotoxicity. J. Virol. 93. e07722–e0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Le KM, Vadnais M, Saye-Francisco KL, Jardine JG, Torres JL, Berndsen ZT, Kong L, Stanfield R, Ruiz J, et al. (2017). Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature 548, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmueller BM, Bridges MD, Dam KM, Lerch MT, Huey-Tubman KE, Hubbell WL, and Bjorkman PJ (2018). DEER spectroscopy measurements reveal multiple conformations of HIV-1 SOSIP envelopes that show similarities with envelopes on native virions. Immunity 49, 235–246.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, et al. (2016). HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, and Katinger H (2001). A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17, 1757–1765. [DOI] [PubMed] [Google Scholar]
- Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, and Peden KW (1996). Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 12, 191–194. [DOI] [PubMed] [Google Scholar]
- Tolbert WD, Sherburn R, Gohain N, Ding S, Flinko R, Orlandi C, Ray K, Finzi A, Lewis GK, and Pazgier M (2020). Defining rules governing recognition and Fc-mediated effector functions to the HIV-1 co-receptor binding site. BMC Biol. 18, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, and Haynes BF (2009). HIV-1-specific antibody responses during acute and chronic HIV-1 infection. Curr. Opin. HIV AIDS 4, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, et al. (2008). Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82, 12449–12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, and Guatelli J (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanshylla K, Held K, Eser TM, Gruell H, Kleipass F, Stumpf R, Jain K, Weiland D, Munch J, Gruttner B, et al. (2021). CD34T+ humanized mouse model to study mucosal HIV-1 transmission and prevention. Vaccines (Basel) 9. 10.3390/vaccines9030198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette M, Désormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, et al. (2014). Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J. Virol. 88, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette M, Coutu M, Richard J, Batraville LA, Dagher O, Bernard N, Tremblay C, Kaufmann DE, Roger M, and Finzi A (2015). The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-Infected individuals. J. Virol. 89, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visciano ML, Gohain N, Sherburn R, Orlandi C, Flinko R, Dashti A, Lewis GK, Tolbert WD, and Pazgier M (2019). Induction of Fc-mediated effector functions against a stabilized inner domain of HIV-1 gp120 designed to selectively harbor the A32 epitope region. Front. Immunol. 10, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bredow B, Arias JF, Heyer LN, Moldt B, Le K, Robinson JE, Zolla-Pazner S, Burton DR, and Evans DT (2016). Comparison of antibody-dependent cell-mediated cytotoxicity and virus neutralization by HIV-1 env-specific monoclonal antibodies. J. Virol. 90, 6127–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. (2009). Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. (2011). Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Gajjar MR, Yu J, Padte NN, Gettie A, Blanchard JL, RussellLodrigue K, Liao LE, Perelson AS, Huang Y, and Ho DD (2020). Quantifying the contribution of Fc-mediated effector functions to the antiviral activity of anti-HIV-1 IgG1 antibodies in vivo. Proc. Natl. Acad. Sci. USA 117, 18002–18009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, and Wiley DC (1997). Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430. [DOI] [PubMed] [Google Scholar]
- Williams WB, Zhang J, Jiang C, Nicely NI, Fera D, Luo K, Moody MA, Liao HX, Alam SM, Kepler TB, et al. (2017). Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Nat. Commun. 8, 1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KL, Stumpf M, Naiman NE, Ding S, Garrett M, Gobillot T, Vézina D, Dusenbury K, Ramadoss NS, Basom R, et al. (2019). Identification of HIV gp41-specific antibodies that mediate killing of infected cells. PLoS Pathog 15. e1007572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wines BD, Vanderven HA, Esparon SE, Kristensen AB, Kent SJ, and Hogarth PM (2016). Dimeric FcgammaR ectodomains as probes of the Fc receptor function of anti-influenza virus IgG. J. Immunol. 197, 1507–1516. [DOI] [PubMed] [Google Scholar]
- Wines BD, Billings H, McLean MR, Kent SJ, and Hogarth PM (2017). Antibody functional assays as measures of Fc receptor-mediated immunity to HIV - new Technologies and their impact on the HIV vaccine field. Curr. HIV Res. 15, 202–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. (2010a). Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010b). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JY, Gorny MK, Palker T, Karwowska S, and Zolla-Pazner S (1991). Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 65, 4832–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada E, Nakaoka S, Klein L, Reith E, Langer S, Hopfensperger K, Iwami S, Schreiber G, Kirchhoff F, Koyanagi Y, et al. (2018). Human-specific adaptations in vpu conferring anti-tetherin activity are critical for efficient early HIV-1 replication in vivo. Cell Host Microbe 23, 110–120.e7. [DOI] [PubMed] [Google Scholar]
- Yang Z, Liu X, Sun Z, Li J, Tan W, Yu W, and Zhang M (2018). Identification of a HIV gp41-specific human monoclonal antibody with potent antibody-dependent cellular cytotoxicity. Front. Immunol. 9, 2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wang H, Liu AZ, Gristick HB, and Bjorkman PJ (2019). Asymmetric opening of HIV-1 Env bound to CD4 and a coreceptor-mimicking antibody. Nat. Struct. Mol. Biol. 26, 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. (2015). Structural repertoire of HIV-1-Neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and reported in this paper are available from the lead contact (andres.finzi@umontreal.ca) upon request.
This paper does not report original code
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.


