Abstract
The molecular mechanisms that underlie oncogene-induced genomic damage are still poorly understood. To understand how oncogenes affect chromatin architecture, it is important to visualize fundamental processes such as DNA replication and transcription in intact nuclei and quantify the alterations of their spatiotemporal organization induced by oncogenes. Here, we apply superresolution microscopy in combination with image cross correlation spectroscopy to the U937-PR9 cell line, an in vitro model of acute promyelocytic leukemia that allows us to activate the expression of the PML-RARα oncogene and analyze its effects on the spatiotemporal organization of functional nuclear processes. More specifically, we perform Tau-stimulated emission depletion imaging, a superresolution technique based on the concept of separation of photons by lifetime tuning. Tau-stimulated emission depletion imaging is combined with a robust image analysis protocol that quickly produces a value of colocalization fraction on several hundreds of single cells and allows observation of cell-to-cell variability. Upon activation of the oncogene, we detect a significant increase in the fraction of transcription sites colocalized with PML/PML-RARα. This increase of colocalization can be ascribed to oncogene-induced disruption of physiological PML bodies and the abnormal occurrence of a relatively large number of PML-RARα microspeckles. We also detect a significant cell-to-cell variability of this increase of colocalization, which can be ascribed, at least in part, to a heterogeneous response of the cells to the activation of the oncogene. These results prove that our method efficiently reveals oncogene-induced alterations in the spatial organization of nuclear processes and suggest that the abnormal localization of PML-RARα could interfere with the transcription machinery, potentially leading to DNA damage and genomic instability.
Significance
The molecular mechanisms that underlie oncogene-induced genomic damage are still poorly understood. Here, we combine image cross correlation spectroscopy with state-of-the-art superresolution microscopy to quantify alterations induced by the PML-RARα oncogene in an in vitro model of acute promyelocytic leukemia. We find that activation of the oncogene induces a significant increase in the fraction of transcription sites colocalized with PML/PML-RARα due to disruption of physiological PML bodies into a large number of PML-RARα microspeckles. We also detect a heterogeneous response of the cells to the activation of the oncogene. These results suggest that our image cross correlation spectroscopy-based approach can be useful for characterizing global alterations in the spatial organization of chromatin, in single cells, in response to oncogene activation, or in other triggering events.
Introduction
The genome is the template for a cell’s fundamental processes, including DNA replication, DNA repair, epigenetic modification, and transcription. Maintaining this vast information requires the combined action of diverse cellular processes such as cell cycle, DNA repair, and cellular metabolism (1). Many actors are involved in the tight regulation of these mechanisms, and most of these activities occur in the context of chromatin, thus needing a large number of proteins controlling chromatin dynamics (2). The collapse of the mechanisms involved in the spatial organization and regulation of the genome is often linked to various human diseases, including cancer (3). It has been proposed that activation of oncogenes triggers the molecular events responsible for the induction of genomic damage early in tumorigenesis. However, despite their potential importance in cancer development, the molecular mechanisms that underlie oncogene-induced genomic damage are still poorly understood. Activated oncogenes may induce genomic damage through alterations of the dynamics of DNA replication, collectively known as “replication stress” (4), that can be generated by several mechanisms, including dysregulation of gene transcription (5) or interferences between DNA replication and transcription (6).
Acute promyelocytic leukemia (APL) represents a highly malignant form of leukemia with a fatal course of only a few weeks (7) and accounts for approximately 10% of all acute myeloid leukemia (8). APL’s main diagnostic feature is an aberrant chromosomal translocation that juxtaposes the promyelocytic leukemia protein (PML) and the retinoic acid receptor RARα genes, respectively, on chromosomes 15 and 17 (9), with PML and RARα domains retaining their wild-type functions. In healthy cells, PML is localized in PML nuclear bodies (NBs) (10,11,12), discrete subnuclear structures of which PML is a crucial component (13,14,15). Indeed, PML promotes NB biogenesis and maintains its integrity and is involved in the recruitment and localization of about 100 proteins into this complex (e.g., SUMO-1, CBP, DAXX, BLM) that are involved in major cellular processes such as stem cell self-renewal, cell death, and transcription (16). Liquid-liquid phase separation, an emerging principle of compartmentalization inside cells, may also participate in the multistep biogenesis of PML NBs (17,18). In APL, chromosomal translocation generates the PML-RARα oncogene. The resulting fusion protein PML-RARα, in vitro, has an affinity and binding specificity to retinoids equal to the wild-type RARα and can regulate RARα target genes (19).
The U937-PR9 cell line is a well-established in vitro model of APL. U937 is a hematopoietic cell line derived from a lymphoid malignancy of a 37-year-old patient suffering from histiocytic lymphoma. However, these cells retain myeloid features rather than lymphoid. U937 cells have been engineered by transduction with a vector carrying PML-RARα cDNA under the control of the inducible mouse metallothionein 1 promoter. Upon infection and selection, the PR9 clone expressed high levels of PML-RARα upon induction by ZnSO4 (zinc) already after 24 h (20). More importantly, PML-RARα levels were comparable with those expressed in fresh APL blasts collected from patients.
Here, we apply advanced optical microscopy to the U937-PR9 model to investigate alterations of nuclear organization induced by the PML-RARα oncogene. Our main goal is to set up a method for the extraction of quantitative parameters related to the spatial organization of functional sites in the nucleus (e.g., colocalization between PML and transcription sites) in a large number of single cells. On the image acquisition side, we take advantage of stimulated emission depletion (STED) superresolution microscopy (21) and, more specifically, of a STED-based method called separation of photons by lifetime tuning (SPLIT) (22,23,24), previously shown to provide good performances on U937-PR9 cells (25). On the image analysis side, we evaluate the spatial distribution of nuclear sites using a colocalization algorithm based on image cross correlation spectroscopy (ICCS) (26,27). More specifically, we introduce a robust image acquisition and analysis protocol that quickly produces a value of colocalization fraction on several hundreds of single cells and allows observation of cell-to-cell variability. Thanks to this approach, we monitor the colocalization between PML and sites of active transcription in single cells and quantify the alterations induced by the PML-RARα oncogene. Following activation of the oncogene, we detect a significant increase in the fraction of transcription sites colocalized with PML/PML-RARα. This increase of colocalization can be ascribed to the abnormal occurrence of a relatively large number of oncogene-induced PML-RARα microspeckles compared with the physiological PML bodies. The analysis also reveals the cell-to-cell variability of this colocalization value, with activated cells showing higher variability compared with the control sample. This variability can be ascribed, at least in part, to a heterogeneous response of the cells to the activation of the oncogene.
These results show that the proposed method efficiently reveals oncogene-induced alterations in the spatial organization of chromatin and discloses by optical superresolved microscopy that the abnormal nuclear localization of the PML-RARα oncoprotein corresponds to an increased degree of colocalization with the transcription machinery.
Materials and methods
Cell culture and treatments
U937-PR9 cells were cultured in RPMI-1640 medium (Sigma-Aldrich R7388, Burlington, MA, USA) supplemented with 1% penicillin/streptomycin (Sigma-Aldrich P4333) and 10% fetal bovine serum (Sigma-Aldrich F9665) and maintained at 37°C and 5% CO2. To induce the expression of PML-RARα, U937-PR9 cells were incubated with 100 μM ZnSO4 solution and left growing for 8 or 24 h. Before experiments, cells were seeded on Cell-Tak (Corning 354240, Corning, NY, USA)-coated glass coverslips.
Immunofluorescence
Primary antibodies used in this work are α-PML (mouse, sc-966 Santa Cruz Biotechnology, Dallas, TX, USA), α-H3K9me2 (mouse, ab1220 Abcam, Waltham, MA, USA), α-H3K9me3 (rabbit, ab8898 Abcam), and α-Pol2 (RNA polymerase II CTD repeat [phospho S2] rabbit, ab5095 Abcam). Secondary antibodies used in this work are goat α-mouse Alexa Fluor 488 (ab150113 Abcam), goat α-rabbit Atto 594 (77671 Sigma-Aldrich), goat α-mouse Atto 594 (76085 Sigma-Aldrich), goat α-rabbit Atto 647N (40839 Sigma-Aldrich), and goat α-mouse Atto 647N (50185 Sigma-Aldrich). DNA dyes used in this study are DAPI (62,248 Thermo Fisher Scientific, Waltham, MA, USA) and Picogreen (P7581 Sigma-Aldrich).
Indirect immunofluorescence experiments were performed as follows. Cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde (w/v) for 10 min at room temperature, and permeabilized with 0.5% (v/v) Triton X-100 in PBS for 20 min. Cells were then blocked with 3% BSA in PBS and incubated in a wet chamber with primary antibody diluted in incubation buffer overnight at 4°C. Cells were then extensively washed with washing buffer 3 × 15 min and incubated with secondary antibody diluted in incubation buffer for 1 h at room temperature, followed by the same washing procedure with washing buffer. Finally, cells were extensively washed with PBS, incubated with DNA dye dilutions, and then mounted on glass slides with ProLong Diamond Antifade Mountant (Invitrogen P36961, Waltham, MA, USA). For Tau-STED experiments, cells have been blocked with 10% normal goat serum (ab7481 Abcam) in PBS.
Image acquisition
Confocal images were acquired on a Leica TCS SP8 confocal microscope, using an HCX PL APO CS2 63× 1.40 numerical aperture oil immersion objective lens (Leica Microsystems, Mannheim, Germany). Excitation wavelengths/emission bandwidths were the following: DAPI (405/410–483), Alexa 488 (488/500–550), and Atto 594 (561/589–643). The pinhole size was set to a value corresponding to 0.8 Airy units at a wavelength of 580 nm. 2,048 × 2,048 pixel images were acquired with a pixel size of 45 nm.
STED images were acquired on a Leica Stellaris 8 Tau-STED microscope using an HC PL APO CS2 100×/1.40 oil immersion objective lens (Leica Microsystems). Tau-STED is a commercial implementation of a method called SPLIT, which exploits the additional information encoded into additional channels of the microscope to generate superresolution images with low background (22,23,24). Emission depletion was accomplished with a 775 nm STED laser. A white-light laser provided excitation at the desired wavelength for each sample. Excitation wavelengths/emission bandwidths were the following: Picogreen (488/500–510), Alexa 594 (590/595–641 using a hybrid detector), and Atto647N (646/651–720). 2,048 × 2,048 pixel images were acquired with a pixel size of 45 nm. Tau-STED parameters were set as follows: Tau-strength value 100, denoise value 50, and background suppression checked.
Image correlation spectroscopy and ICCS analysis
The image correlation spectroscopy (ICS) and ICCS analyses were performed in MATLAB (MathWorks, Natick, MA, USA) using a modified version of the ICCS algorithm (27) (https://github.com/llanzano/ICCS), adapted for the analysis of large images containing many cells. All images were preprocessed in ImageJ before ICCS analysis. To generate the nuclei selection mask, the image in the DNA channel was converted into a binary image using the function “threshold” of ImageJ, then single nuclei were identified and listed as objects using “analyze particles,” and the image of the “count masks” was saved. The background was subtracted from the images prior to ICCS analysis using the function “subtract Background.”
The nuclei selection mask was used to sequentially define a region of interest corresponding to a single-cell nucleus and perform the analysis. The two-dimensional (2D) image correlation function was calculated as
| (1) |
where I1(x,y) and I2(x,y) are the images in the first and the second channels, respectively, and the angle brackets indicate averaging over all the selected pixels of the image (28,29). The two auto-correlation functions were obtained by setting i = j = 1 and i = j = 2, respectively, whereas the cross correlation function was obtained by setting i = 1 and j = 2. The numerator in Eq. 1 was calculated by a 2D fast Fourier-transform algorithm (26,27). We assume that the 2D auto- and cross correlation functions have circular symmetry (Fig. S1). The 2D correlation functions were then converted into radial 1D correlation functions Gij(δr) by performing an angular mean, as described previously (30) (Fig. S1). The resulting radial auto-correlation functions (ACFs) were then fitted to a Gaussian model:
| (2) |
where Gii(0) represents the amplitude and G(∞) represents an offset value. The width parameter wii represents the 1/e2 radius of a Gaussian function corresponding to the convolution of the microscope point spread function and the object function (31). The corresponding full width at half maximum parameter, R = (2ln2)1/2wii, represents the average apparent size of the structures in the region of interest extracted by ICS.
The amplitude of the cross correlation function (CCF), G12(0), was determined as the value of the CCF at spatial lag zero. The amplitude parameters were used to calculate the coefficients of colocalizations fch1 and fch2 and f:
| (3) |
In cross correlation spectroscopy, the cross correlation fractions (Eq. 3) are a measure of the overlap between intensity fluctuations in the two channels. In the context of fluorescence cross-correlation spectroscopy, where intensity fluctuations are generated by labeled molecules diffusing through the observation volume, the cross correlation fractions represent the fractions of interacting molecules (32,33). In our ICCS analysis, the colocalization fractions (Eq. 3) are a measure of the overlap between spatial fluctuations in the two channels. Specifically, the coefficient fch1 (fch2) represents the fraction of signal in channel 1 (channel 2), which is correlated with the signal in channel 2 (channel 1).
We note that a well-established parameter for evaluation of the cross-correlation is the Pearson correlation coefficient (PCC), defined as (34)
| (4) |
where Ii,av = < Ii(x,y) >. We can calculate a similar parameter in our ICCS analysis:
| (5) |
where the sign + (−) stands for positive (negative) correlation. In the absence of shot noise, PCC = PCCICCS. More generally, the denominator in Eq. 4 (definition of PCC) will contain an additional variance due to shot noise that can affect the measured value of PCC (34) In contrast, the auto-correlation amplitudes Gii(0) used in Eq. 5, evaluated from fitting, represent extrapolated values at zero lag and do not contain the contribution of the shot noise. Thus, in general, values calculated with Eqs. 4 and 5 could be different, PCC ≠ PCCICCS.
The analysis shown in Fig. S2 was performed in Matlab using the quality by ICS algorithm (25,35), freely available at https://github.com/llanzano/QuICS. The quality by ICS algorithm calculates the image correlation function and extracts three parameters (R, B, N) related to the quality of the image: R is the full width at half maximum width calculated from the auto-correlation function, related to the spatial resolution (smaller values of R corresponding to higher spatial resolution and/or smaller size of the objects); B is the brightness, related to the image contrast (higher values of B corresponding to higher contrast); and N is the relative noise variance, related to the signal-to-noise ratio of the image (smaller values of N corresponding to higher signal-to-noise ratios).
Object-based analysis
The object-based analysis of the number and size of PML NBs/PML-RARα microspeckles was performed on binary images. The image in the PML channel was converted into a binary image using the function “threshold” of ImageJ, then single spots were identified and listed as objects using “analyze particle,” and the image of the “count masks” was saved. For this MATLAB analysis, both “count masks” identifying nuclei and spots were used. The area A of each spot was evaluated counting the number of pixels forming the spot. The size (diameter) of the spot was then calculated as Robj2(A/π)1/2.
Results
Optimization of ICCS for the semiautomated analysis of large numbers of cells
We evaluated the spatial distribution of nuclear sites using a colocalization algorithm based on ICCS (26,27). ICCS is a pixel-based colocalization method that does not require a presegmentation of the image into objects. We have recently shown that the combination of superresolution STED and ICCS is a suitable tool for analyzing the nanoscale distribution of functional nuclear sites (27). ICCS has also been combined with other superresolution techniques such as structured illumination microscopy (36,37) and stochastic optical reconstruction microscopy (38). In this work, we optimize ICCS for the analysis of PML-RARα oncogene-induced alterations in the U937-PR9 model.
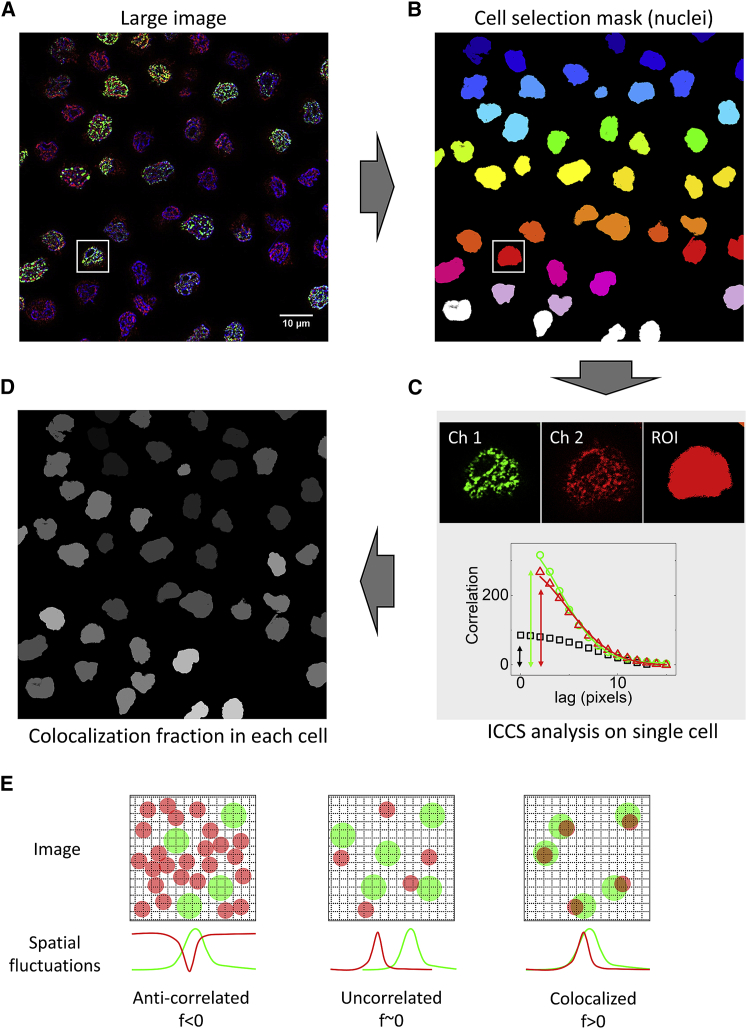
In order to be able to analyze a representative sample of the whole cell population, we acquired, by confocal or superresolution microscopy, multiple large images of the sample, as depicted in Fig. 1 A. Large images covered approximately 10,000 μm2 of the sample, and, for each experiment, approximately 10 images were acquired. The acquisition time for each image was approximately 1 min (depending on the number of lines averaging), and the number of U937-PR9 cells in the field of view was on the order of 30–50 cells (variable depending on the level of cell confluency). Using the DNA counterstain signal, we identified each cell nucleus as an object, and we created a nuclei selection mask so that every single cell from the large field of view was automatically selected and analyzed sequentially (Fig. 1 B). Nuclei of cells in mitosis were identified manually and excluded from the selection mask.
Figure 1.
Large-image acquisition and analysis protocol. Schematic of the image cross correlation spectroscopy (ICCS) protocol adapted for the analysis of multiple cells in large images. (A) Large images are acquired (typically 2,048 × 2,048 pixels with a pixel size of 45 or 60 nm) containing several cells. (B) Nuclei are selected based on the nuclear stain signal creating a cell selection mask. (C) Regions of interest for each nucleus are defined and analyzed with ICCS algorithm, which extracts the value of the colocalization rate from the correlation functions. (D) A value of colocalization is associated with each cell. (E) Schematic representation of the spatial intensity fluctuations generated by the fluorescent particles in a dual-color image and the calculation of the colocalization fraction f by ICCS. To see this figure in color, go online.
We then applied a modified version of the ICCS algorithm (27) that was adapted to analyze large images. The modified algorithm processes all the acquired images together with the corresponding nuclei selection masks to perform the ICCS analysis sequentially for each cell of the sample. The ICCS algorithm calculated and fitted the spatial ACF for each channel of the acquired images and the CCF of the dual-color image (Fig. 1 C). Finally, the ICCS algorithm provided a value of the colocalization fraction for each cell (Eq. 3), normalizing the amplitude of the CCF to that of the ACFs (Fig. 1 D).
In our ICCS analysis, the colocalization fractions should be interpreted as a measure of the overlap between spatial intensity fluctuations in the two channels (Fig. 1 E). Specifically, the colocalization fraction fch1 (fch2), defined by Eq. 3, represents the degree of overlap of spatial fluctuations in channel 1 (channel 2) with spatial fluctuations in the other channel. These spatial fluctuations originate from the convolution of the fluorescent particles with the microscope point spread function (31). A value of 0 indicates uncorrelated particles, whereas negative values indicate anti-correlated particles. Positive values indicate some degree of colocalization. A value of colocalization of 1 is obtained when all the spatial fluctuations in one channel are perfectly overlapped to spatial fluctuations in the other channel. In addition to the distribution of the particles, other factors, including the size of the particles and of the point spread function in the two channels, may affect the measured value of colocalization.
Validation of the large-image ICCS protocol
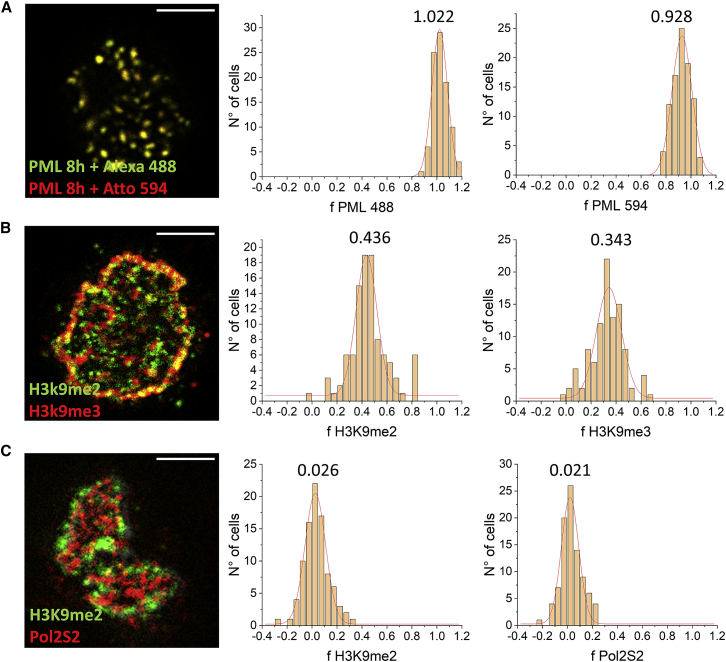
We validated the analysis method through ICCS analysis of a cell’s population labeled against multiple nuclear proteins. As a positive control, we labeled the PML-RARα fusion protein, 8 h after zinc activation, with a primary antibody against the PML protein and two secondary antibodies carrying different fluorophores, namely Alexa 488 and Atto 594. We obtained a Gaussian fit of the distribution of the colocalization fraction f, measured through the ICCS analysis (Fig. 2 A), with a value close to 1, as expected (fPML488 = 1.022 ± 0.002, fPML594 = 0.928 ± 0.002, n = 176, mean ± SE). Furthermore, as a negative control, we analyzed large images of PR9 cells labeled for factors generally associated with separate nuclear compartments, namely the dimethylated lysine 9 of histone H3, mainly present in the heterochromatin region, and the elongating form of RNA polymerase 2, mainly related with more accessible and euchromatic nuclear regions (Fig. 2 C). After ICCS analysis, we measured the Gaussian fit’s peak of the distribution of the colocalization fraction close to 0, as expected (fH3K9me2 = 0.026 ± 0.003, fPol2S2 = 0.021 ± 0.004, n = 91). In order to also take into account an intermediate situation of partial colocalization, we performed the ICCS analysis on large images of PR9 cells labeled with two primary antibodies directed against two posttranslational modifications, namely di- and trimethylation of H3K9 (Fig. 2 B). The result obtained is a value of the Gaussian fit’s peak of fH3K9me2 = 0.436 ± 0.006 and fH3K9me3 = 0.343 ± 0.008 (n = 210).
Figure 2.
Validation of the image analysis protocol. (A) Positive control: PML/PML-RARα labeled with two secondary antibodies in U937-PR9 cells, 8 h after PML-RARα activation. (B) Intermediate colocalization: H3K9me2 + α-mouse Alexa Fluor 488 and H3K9me3 + α-rabbit Atto 594. (C) Negative control: H3K9me2 + α-mouse Alexa Fluor 488 and Pol2S2 + α-rabbit Atto 594. Shown are, from left to right, representative confocal images of U937-PR9 cells and the histograms showing the distribution of the colocalization fraction. Red lines are Gaussian fits. Indicated numbers represent the peak values. Scale bar, 3 μm. To see this figure in color, go online.
Activation of the PML-RARα oncogene in the U937-PR9 model
Next, we monitored the expression of the oncoprotein in the U937-PR9 model in response to activation of the oncogene. The indirect immunofluorescences against PML/PML-RARα were performed using a primary antibody against the PML protein and a secondary antibody coupled to a fluorophore. Thus, the fluorescent signal we obtained is a reporter of the localization of both the PML and PML-RARα proteins.
To improve the visualization of PML-RARα micropeckles in the activated sample, we used superresolution microscopy (even though confocal microscopy is sufficient to visualize PML NBs in the control sample). Specifically, we used SPLIT, a STED-based method that exploits the additional information encoded into additional channels of the microscope to generate superresolution images with low background (22,23,24). The commercially available Tau-STED microscope is essentially based on the same principle. We have recently evaluated, in U937-PR9 cells, the quality of images generated by different SPLIT approaches (including the commercially available Tau-STED) versus conventional STED and found that the SPLIT images had higher resolution and nonreduced contrast and noise levels than the STED images (25). In the samples of the present work, Tau-STED imaging provides an improvement of spatial resolution of about 110 nm, in the Alexa 594 channel, with respect to confocal (Fig. S2).
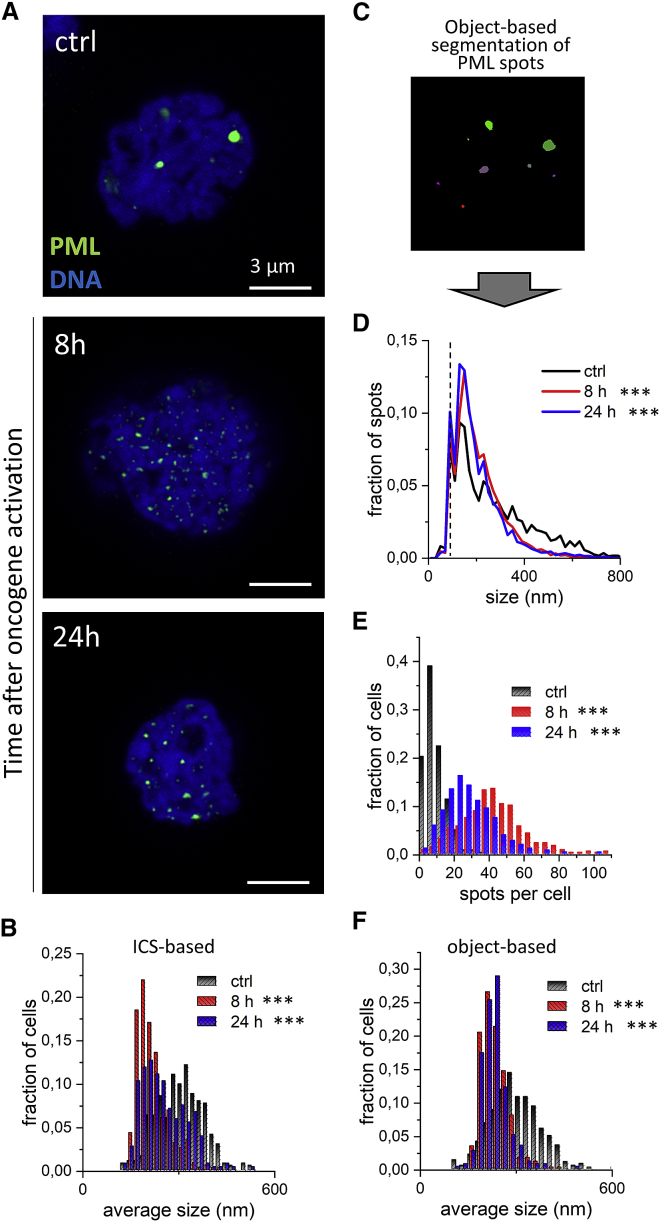
Representative Tau-STED images of PML/PML-RARα in U937-PR9 cells are shown in Fig. 3 A. In our control condition (Fig. 3 A, top row), before induction of the expression of PML-RARα, we could identify the PML NB structures. The induction of the expression of PML-RARα fusion protein was obtained through a zinc treatment (see materials and methods). The induction effect is maximum after 8 h and then decreases with time. At its maximum (Fig. 3 A, middle row), we observed the disruption of PML NBs into nuclear microspeckles, as previously reported (39,40).
Figure 3.
Activation of the PML-RARα oncogene in the U937-PR9 model. (A) Representative Tau-STED images of PML in U937-PR9 cells before (control [ctrl]) and 8/24 h after PML-RARα expression induction following ZnSO4 treatment. Nuclear DNA is counterstained with Picogreen. Scale bar, 3 μm. (B) Histograms show the distribution of the average size of PML/PML-RARα spots per cell, before and after oncogene induction. Values represent the average apparent size of the structures extracted by ICS. (C) Example of the spot selection mask used for the object-based analysis of size and number of PML/PML-RARα spots. (D) Histograms show the distribution of the size of PML/PML-RARα spots in all cells, before and after oncogene induction, measured through object-based segmentation. (E) Histograms show the distribution of the number of PML/PML-RARα spots per cell, before and after oncogene induction, measured through object-based segmentation. (F) Histograms show the distribution of the average size of PML/PML-RARα spots per cell, before and after oncogene induction, measured through object-based segmentation. Significance: ctrl versus 8 h ∗∗∗p ≤ 0.001; ctrl versus 24 h ∗∗∗p ≤ 0.001. To see this figure in color, go online.
The average size of PML spots (representing PML NBs/speckles) per cell, measured by ICS, decreases after induction of the oncogene (Fig. 3 B). We complemented this analysis using object-based segmentation (Fig. 3 C) and measuring the number and size of single PML spots in the images before and after induction of the oncogene (Fig. 3 D–F). The observed distribution of size of PML spots in control cells (Fig. 3 B, top) is in keeping with previously reported values for PML bodies ranging from 200 nm to 1 μm (41). As expected, induction of PML-RARα disrupts PML bodies and reduces the fraction of detected spots with a size larger than 300 nm (Fig. 3 D). Moreover, the number of detected PML spots per cell significantly increases after induction of the oncogene with an average of 9.6 ± 0.3 spots/cell (mean ± SE) for the control condition and 41.5 ± 1 spots/cell 8 h after PML-RARα induction (Fig. 3 E). The average size of PML spots in each cell, determined by object-based analysis, decreases (Fig. 3 F), in keeping with the ICS-based measurement of size.
PML/PML-RARα colocalization with the transcription machinery evaluated by ICCS
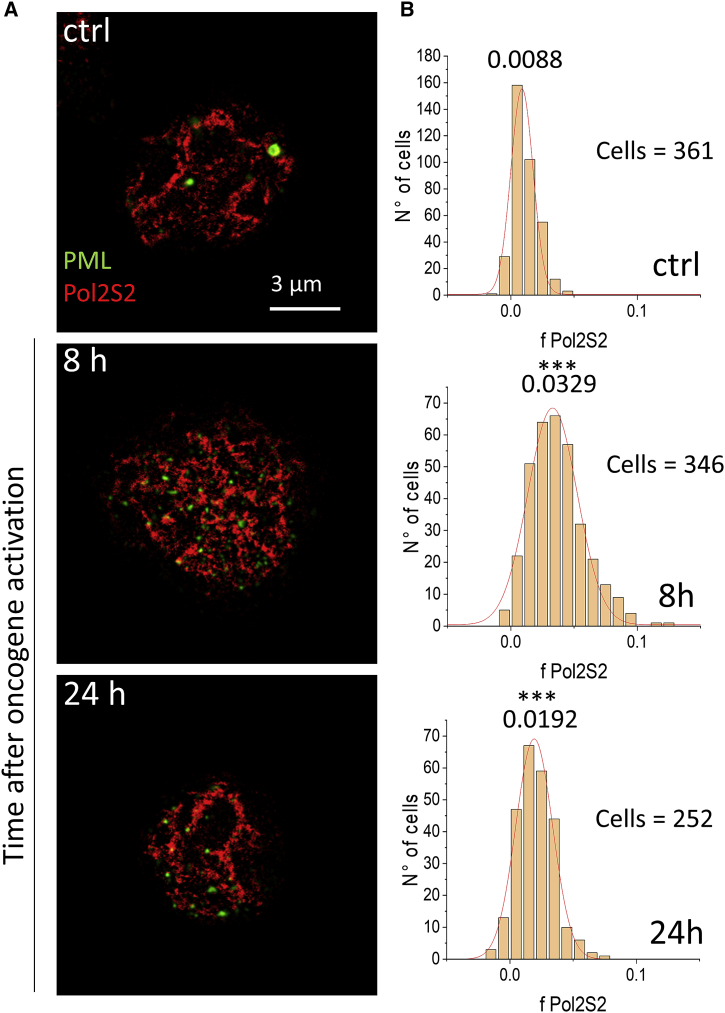
Next, we measured the colocalization rate of PML/PML-RARα with the transcription machinery that we visualized through indirect immunofluorescence against the elongating form of RNA polymerase 2 (hereafter Pol2S2) through a primary antibody against the phosphorylated Ser 2 of the RNA polymerase 2 C-terminal domain. We verified that, also in the Atto 647N channel, Tau-STED imaging provided better resolution and contrast compared with gated-STED (time gating at 1 ns) or conventional STED (integrated intensity) (Fig. S3). Then, we took advantage of two-color superresolution microscopy in order to be able to better resolve transcription foci and PML-RARα microspeckles and to improve the accuracy of our colocalization analysis. Representative two-color Tau-STED images of PML and Pol2S2 are shown in Fig. 4 A.
Figure 4.
Colocalization analysis of superresolved signals of PML and elongating Pol2. (A) Representative two-color Tau-STED images of PML/PML-RARα (green) and Pol2S2 (red) in U937-PR9 cells before (ctrl) and after PML-RARα expression induction, at the specified time point. Scale bar, 3 μm. (B) Histograms show the distribution of the colocalization fraction fPol2S2, representing the fraction of Pol2S2 molecules colocalized with PML/PML-RARα molecules. Red lines are Gaussian fits. Numbers represent the peak values. Significance: ctrl versus 8 h ∗∗∗p ≤ 0.001; ctrl versus 24 h ∗∗∗p ≤ 0.001. To see this figure in color, go online.
We quantified the fraction of Pol2S2 colocalized with PML. Upon induction of PML-RARα expression, the fraction of Pol2S2 colocalized with PML increases from fPol2S2 = 0.0088 ± 0.0005 (Fig. 4 B, control, n = 361 cells) to the value fPol2S2 = 0.0329 ± 0.0006 after 8 h (Fig. 4, 8 h, n = 346 cells, ∗∗∗p ≤ 0.001). At 24 h after PML-RARα expression induction (Fig. 4, bottom row), the colocalization fraction decreases to the value fPol2S2 = 0.0192 ± 0.0004 (n = 252 cells, control versus 24 h ∗∗∗p ≤ 0.001), intermediate between the control and the 8 h condition.
These data indicate that, following induction of PML-RARα oncogene, an increasing fraction of Pol2S2 colocalizes with PML/PML-RARα molecules at the resolution of our imaging system. To verify that this difference was due to the presence of oncogene-induced PML-RARα microspeckles, we combined the ICCS data with the PML spot analysis performed in Fig. 2.
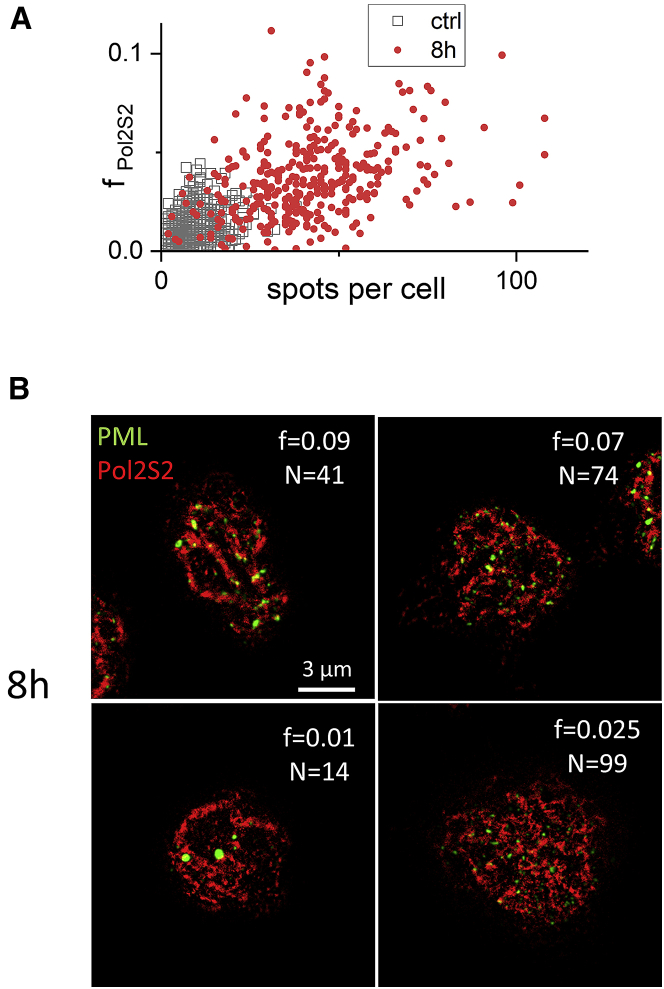
Fig. 5A shows the value of colocalization fraction fPol2S2 measured in each cell as a function of the number of PML spots per cell. Data from cells at 8 h postactivation are compared with control cells. The plot suggests that the increase in the value of fPol2S2 (fraction of Pol2S2 molecules colocalized with PML/PML-RARα molecules) is caused by the large number of small PML-RARα microspeckles that “spread” the PML close to a larger portion of the Pol2S2 pool of the nucleus. This analysis also shows the cell-to-cell variability in the colocalization value fPol2S2 and the number of PML spots N, with activated cells showing higher variability compared with the control sample. Fig. 5 B shows representative images of activated cells, corresponding to different values of fPol2S2 and N. In a fraction of activated cells, the values fPol2S2 and N are similar to control, indicating a heterogeneous response of the sample to the activation of the oncogene (see, for instance, the cell in the lower left corner of Fig. 5 B).
Figure 5.
Cell-to-cell variability of oncoprotein expression and distribution. (A) Scatter plot of the colocalization fraction fPol2S2 as a function of the number of PML/PML-RARα spots per cell. (B) Examples of Tau-STED images of U937-PR9 cells at 8 h after PML-RARα induction. For each cell are reported the corresponding values of the colocalization fraction fPol2S2 (f) and number of PML spots (N). Scale bar, 3 μm. To see this figure in color, go online.
Discussion
In this work, we have applied ICCS to the U937-PR9 cell line, an in vitro model for studying the effects of the PML-RARα oncogene. On the image acquisition side, we have exploited a recently introduced, STED-based, superresolution imaging technique. The performances of STED can be significantly improved by fluorescence lifetime detection and phasor plot analysis, as demonstrated by the SPLIT (22) and subsequent works (23,24,42,43,44). The Tau-STED technique is one of the first commercial implementations of this strategy, and we had previously demonstrated that, in U937-PR9 cells, the SPLIT or Tau-STED images were of better quality compared with conventional STED images (25). On the image analysis side, we have adapted our previously reported ICCS algorithm to the semiautomated analysis of a large number of cells. This new protocol, which simply requires the acquisition of images with a large field of view and a counterstain signal to select single cell nuclei, has been fundamental to increase the statistical power of our analysis and to detect heterogeneity in single-cell response.
An advantage of ICCS-based colocalization is that it does not require presegmentation of the image into objects and can be applied to images (including confocal images) where object segmentation would not be accurate. For a comparison, we performed an analysis similar to that reported in Fig. 4 using confocal microscopy (Fig. S4). The absolute values of colocalization measured by confocal-ICCS are different (the peak value at 8 h is about 2× higher in confocal-ICCS compared with STED-ICCS) (Fig. S4 B). This can be explained by a larger degree of overlap between spatial fluctuations in the confocal images. Nevertheless, the confocal-ICCS analysis (Fig. S4 B) shows that PML-RARα activation induces an increase in the fraction of Pol2S2 colocalized with PML/PML-RARα, in keeping with the STED-ICCS analysis (Fig. 4). This suggests that, at least for the results in Fig. 4, a confocal-ICCS analysis would have been sufficient. This confirms the versatility of ICCS as a method that can be combined with different imaging techniques. On the other hand, superresolution increases our capability of resolving target objects in the images. In our case, we exploited the higher spatial resolution provided by STED microscopy to better resolve single PML spots and characterize their heterogeneous distribution. The apparent size of the PML-RARα microspeckles in the confocal images is about 260 nm (Fig. S2 A), which is in the order of the diffraction limit. The apparent size of the PML-RARα microspeckles in the STED images decreases to about 150 nm (Fig. S2 B). The combination of object- and ICCS-based types of analysis (Fig. 5) is useful to investigate and interpret cell-to-cell variability of the extracted parameters.
Our results show that the ICCS, complemented with object-based analysis, can quantify oncogene-induced alterations in the spatial organization of chromatin. As a representative application, we have analyzed the variation in the colocalization between PML and Pol2S2 induced by the PML-RARα oncogene. PML-RARα activation induces an increase in the fraction of Pol2S2 colocalized with PML/PML-RARα. This increase can be ascribed to disruption of PML bodies and the formation of a large number of smaller PML-RARα speckles. This analysis suggests that the abnormal localization of PML-RARα could interfere with fundamental processes such as DNA replication and transcription, which are tightly coordinated in space and in time. In general, we expect that our ICCS-based approach can be useful for characterizing global alterations in the spatial organization of chromatin, in single cells, in response to specific triggering events.
Author contributions
E.C., A.D., M.F., G.I.D., and L.L. designed the study, conceived the experiments, and wrote the manuscript. E.C., I.C., and M.D. prepared samples. E.C. and L.L. collected data. L.L. wrote the software. E.C., G.I.D., M.F., P.G.P., A.D., and L.L. analyzed data and discussed results. All authors critically reviewed the manuscript.
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC) through MFAG (My First AIRC Grant) grant ID 21931 and from the University of Catania under the program Programma Ricerca di Ateneo UNICT 2020-2022-linea 2 and linea Open Access. Part of this work has been developed under the PRIN 20177XJCHX_003. The authors gratefully acknowledge the Bio-Nanotech Research and Innovation Tower (BRIT; PON project financed by the Italian Ministry for Education, University, and Research MIUR).
Declaration of interests
The authors declare no competing interests.
Editor: Dylan Myers Owen.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.10.003.
Contributor Information
Gaetano Ivan Dellino, Email: gaetano.dellino@ieo.it.
Luca Lanzanò, Email: luca.lanzano@unict.it.
Supporting material
Data availability
All source data generated and/or analyzed during this study are included in this article or are available from the corresponding authors upon reasonable request.
References
- 1.Bansbach C.E., Cortez D. Defining genome maintenance pathways using functional genomic approaches. Crit. Rev. Biochem. Mol. Biol. 2011;46:327–341. doi: 10.3109/10409238.2011.588938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos E.I., Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 3.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability--an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 4.Gaillard H., García-Muse T., Aguilera A. Replication stress and cancer. Nat. Rev. Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 5.Kotsantis P., Silva L.M., et al. Petermann E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016;7:13087. doi: 10.1038/ncomms13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helmrich A., Ballarino M., et al. Tora L. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z.Y., Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- 8.Jing Y. The PML-RARalpha fusion protein and targeted therapy for acute promyelocytic leukemia. Leuk. Lymphoma. 2004;45:639–648. doi: 10.1080/10428190310001609933. [DOI] [PubMed] [Google Scholar]
- 9.Kakizuka A., Miller W.H., Jr., et al. Evans R.M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor. PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 10.Kastner P., Perez A., et al. Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen K., Shiels C., Freemont P.S. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 12.Kentsis A., Gordon R.E., Borden K.L. Self-assembly properties of a model RING domain. Proc. Natl. Acad. Sci. USA. 2002;99:667–672. doi: 10.1073/pnas.012317299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z.G., Delva L., et al. Pandolfi P.P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 14.Ishov A.M., Sotnikov A.G., et al. Maul G.G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong S., Hu P., et al. Pandolfi P.P. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- 16.Rabellino A., Scaglioni P.P. PML degradation: multiple ways to eliminate PML. Front. Oncol. 2013;3:60. doi: 10.3389/fonc.2013.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corpet A., Kleijwegt C., et al. Lomonte P. PML nuclear bodies and chromatin dynamics: catch me if you can. Nucleic Acids Res. 2020;48:11890–11912. doi: 10.1093/nar/gkaa828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banani S.F., Rice A.M., et al. Rosen M.K. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nervi C., Poindexter E.C., et al. Jetten A.M. Characterization of the PML-RAR alpha chimeric product of the acute promyelocytic leukemia-specific t(15;17) translocation. Cancer Res. 1992;52:3687–3692. [PubMed] [Google Scholar]
- 20.Grignani F., Ferrucci P.F., et al. Nicoletti I. The acute promyelocytic leukemia-specific PML-RAR alpha fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 21.Diaspro A., Bianchini P. Optical nanoscopy. La Rivista del Nuovo Cimento. 2020;43:385–455. [Google Scholar]
- 22.Lanzano L., Coto Hernandez I., et al. Vicidomini G. Encoding and decoding spatio-temporal information for super-resolution microscopy. Nat. Commun. 2015;6:6701. doi: 10.1038/ncomms7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelicci S., Tortarolo G., et al. Lanzanò L. Improving SPLIT-STED super-resolution imaging with tunable depletion and excitation power. J. Phys. Appl. Phys. 2020;53:234003. [Google Scholar]
- 24.Sarmento M.J., Oneto M., et al. Lanzano L. Exploiting the tunability of stimulated emission depletion microscopy for super-resolution imaging of nuclear structures. Nat. Commun. 2018;9:3415. doi: 10.1038/s41467-018-05963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerutti E., D’Amico M., et al. Lanzanò L. Evaluation of sted super-resolution image quality by image correlation spectroscopy (QuICS) Sci. Rep. 2021;11:20782. doi: 10.1038/s41598-021-00301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comeau J.W., Kolin D.L., Wiseman P.W. Accurate measurements of protein interactions in cells via improved spatial image cross-correlation spectroscopy. Mol. Biosyst. 2008;4:672–685. doi: 10.1039/b719826d. [DOI] [PubMed] [Google Scholar]
- 27.Oneto M., Scipioni L., et al. Lanzano L. Nanoscale distribution of nuclear sites by super-resolved image cross-correlation spectroscopy. Biophys. J. 2019;117:2054–2065. doi: 10.1016/j.bpj.2019.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrix J., Dekens T., et al. Lamb D.C. Arbitrary-region raster image correlation spectroscopy. Biophys. J. 2016;111:1785–1796. doi: 10.1016/j.bpj.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leutenegger M., Lasser T., et al. Robelek R. Imaging of G protein-coupled receptors in solid-supported planar lipid membranes. Biointerphases. 2008;3:FA136. doi: 10.1116/1.3054189. [DOI] [PubMed] [Google Scholar]
- 30.Scipioni L., Gratton E., et al. Lanzanò L. Phasor analysis of local ICS detects heterogeneity in size and number of intracellular vesicles. Biophys. J. 2016;111:619–629. doi: 10.1016/j.bpj.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiseman P.W., Capani F., et al. Martone M.E. Counting dendritic spines in brain tissue slices by image correlation spectroscopy analysis. J. Microsc. 2002;205:177–186. doi: 10.1046/j.0022-2720.2001.00985.x. [DOI] [PubMed] [Google Scholar]
- 32.Bacia K., Majoul I.V., Schwille P. Probing the endocytic pathway in live cells using dual-color fluorescence cross-correlation analysis. Biophys. J. 2002;83:1184–1193. doi: 10.1016/S0006-3495(02)75242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foldes-Papp Z., Rigler R. Quantitative two-color fluorescence cross-correlation spectroscopy in the analysis of polymerase chain reaction. Biol. Chem. 2001;382:473–478. doi: 10.1515/BC.2001.057. [DOI] [PubMed] [Google Scholar]
- 34.Adler J., Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry. 2010;77:733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- 35.D'Amico M., Di Franco E., et al. Lanzano L. A phasor-based approach to improve optical sectioning in any confocal microscope with a tunable pinhole. Microsc. Res. Tech. 2022;85:3207–3216. doi: 10.1002/jemt.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cainero I., Cerutti E., et al. Lanzanò L. Chromatin investigation in the nucleus using a phasor approach to structured illumination microscopy. Biophys. J. 2021 doi: 10.1016/j.bpj.2021.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cainero I., Cerutti E., et al. Lanzano L. Measuring nanoscale distances by structured illumination microscopy and image cross-correlation spectroscopy (SIM-ICCS) Sensors. 2021;21 doi: 10.3390/s21062010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelicci S., Furia L., et al. Faretta M. Novel tools to measure single molecules colocalization in fluorescence nanoscopy by image cross correlation spectroscopy. Nanomaterials. 2022;12:686. doi: 10.3390/nano12040686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dyck J.A., Maul G.G., et al. Evans R.M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 40.Koken M.H., Puvion-Dutilleul F., et al. Chomienne C. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernardi R., Pandolfi P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 42.Wang L., Chen B., et al. Qu J. Resolution improvement in STED super-resolution microscopy at low power using a phasor plot approach. Nanoscale. 2018;10:16252–16260. doi: 10.1039/c8nr03584a. [DOI] [PubMed] [Google Scholar]
- 43.Coto Hernandez I., Castello M., et al. Vicidomini G. Efficient two-photon excitation stimulated emission depletion nanoscope exploiting spatiotemporal information. Neurophotonics. 2019;6:045004. doi: 10.1117/1.NPh.6.4.045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tortarolo G., Sun Y., et al. Vicidomini G. Photon-separation to enhance the spatial resolution of pulsed STED microscopy. Nanoscale. 2019;11:1754–1761. doi: 10.1039/c8nr07485b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data generated and/or analyzed during this study are included in this article or are available from the corresponding authors upon reasonable request.