Abstract
Introduction:
Toxicity data are unavailable for many thousands of chemicals in commerce and the environment. Therefore, risk assessors need to rapidly screen these chemicals for potential risk to public health. High-throughput screening (HTS) for in vitro bioactivity, when used with high-throughput toxicokinetic (HTTK) data and models, allows characterization of these thousands of chemicals.
Areas covered:
This review covers generic physiologically-based toxicokinetic (PBTK) models and high-throughput PBTK modeling for in vitro-in vivo extrapolation (IVIVE) of HTS data. We focus on “httk”, a public, open-source set of computational modeling tools and in vitro toxicokinetic (TK) data.
Expert opinion:
HTTK benefits chemical risk assessors with its ability to support rapid chemical screening/prioritization, perform IVIVE, and provide provisional TK modeling for large numbers of chemicals using only limited chemical-specific data. Although generic TK model design can increase prediction uncertainty, these models provide offsetting benefits by increasing model implementation accuracy. Also, public distribution of the models and data enhances reproducibility. For the httk package, the modular and open-source design can enable the tool to be used and continuously improved by a broad user community in support of the critical need for high-throughput chemical prioritization and rapid dose estimation to facilitate rapid hazard assessments.
Keywords: Generic Physiologically Based Toxicokinetic Models, High-Throughput, In Vitro to In Vivo Extrapolation, Modeling Software Tools, Open Source Tools, Physiologically-based toxicokinetics, Toxicokinetics
Introduction
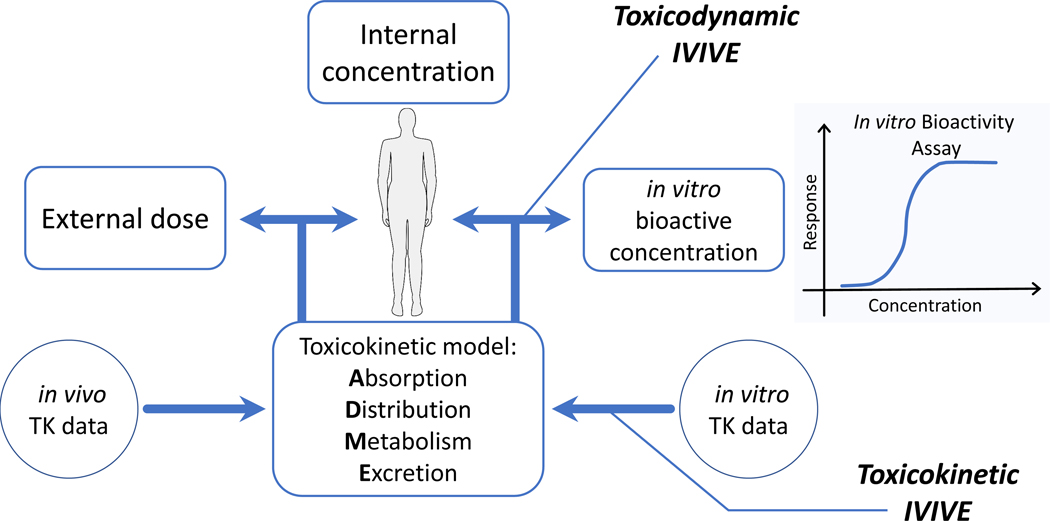
There are thousands of chemicals present in our environment, many of which are routinely detected in environmental and human blood samples [1,2]. There is a need to rapidly assess potential risk to human health for these chemicals, but the necessary toxicity data are unavailable for most [3–5]. In vivo toxicity testing of such a large number of chemicals is not feasible [6,7]. As an alternative, in vitro high-throughput screening (HTS) assays are performed, in which a large number of chemicals may be examined by a battery of in vitro tests for biological activity across a variety of different biologically-based endpoints [8–10]. HTS typically characterizes the concentration at which a chemical causes bioactivity in vitro. To use in vitro HTS to assess in vivo potential risk to humans, the in vitro bioactive concentration, representing an internal dose or threshold, must be extrapolated to an equivalent in vivo external dose or exposure (in vitro-in vivo extrapolation, or IVIVE, Figure 1) [11]. This equivalent external dose can then be compared to estimates of exposure to assess potential risk [12].
Figure 1. In vitro to in vivo extrapolation (IVIVE).
We perform that IVIVE using toxicokinetic (TK) modeling. TK models relate external dose to internal body concentration by describing “what the body does to the chemical”: absorption, distribution, metabolism, and excretion (ADME). For IVIVE of in vitro bioactive concentrations, we assume that bioactivity would occur in the body at a concentration equal to an in vitro bioactive concentration, and use TK modeling in reverse (that is, reverse dosimetry) to find the “equivalent dose” – an external dose that would produce the specified body concentration. While IVIVE broadly includes any use of in vitro data to predict phenomena in vivo, it is useful to distinguish between TK IVIVE (that is, the use of in vitro data to predict ADME) and toxicodynamic (TD) IVIVE, which includes the use of in vitro data to predict toxic effects in vivo.
A model of toxicokinetics (TK) quantitatively describes the body’s absorption, distribution, metabolism, and excretion (ADME) of a chemical or substance [13]. (Different terms for this concept are preferred in different fields, including “toxicokinetics”, “pharmacokinetics”, “biokinetics”, and simply “kinetics”; in this review, we use “TK”). TK models are used to estimate internal dose following an external exposure. For IVIVE of toxicological bioactivity data, an administered equivalent dose (AED) is determined using reverse dosimetry by solving the TK model in reverse, deriving the external dose of chemical (that is TK model input) that produces a specified internal concentration (that is TK model output) (Figures 1 and 2) [14]. While reverse dosimetry has been applied one chemical at a time [15–17], applying reverse dosimetry to HTS data requires rapid approaches amenable to thousands of chemicals. Therefore, a high-throughput toxicokinetics (HTTK) approach is needed [18]. HTTK methods use generic TK models that can be parameterized rapidly for large numbers of chemicals, using in vitro measurements and/or in silico predictions of chemical-specific TK properties like metabolism [7]. Thus, HTTK relies on TK IVIVE to perform toxicological IVIVE of HTS data.
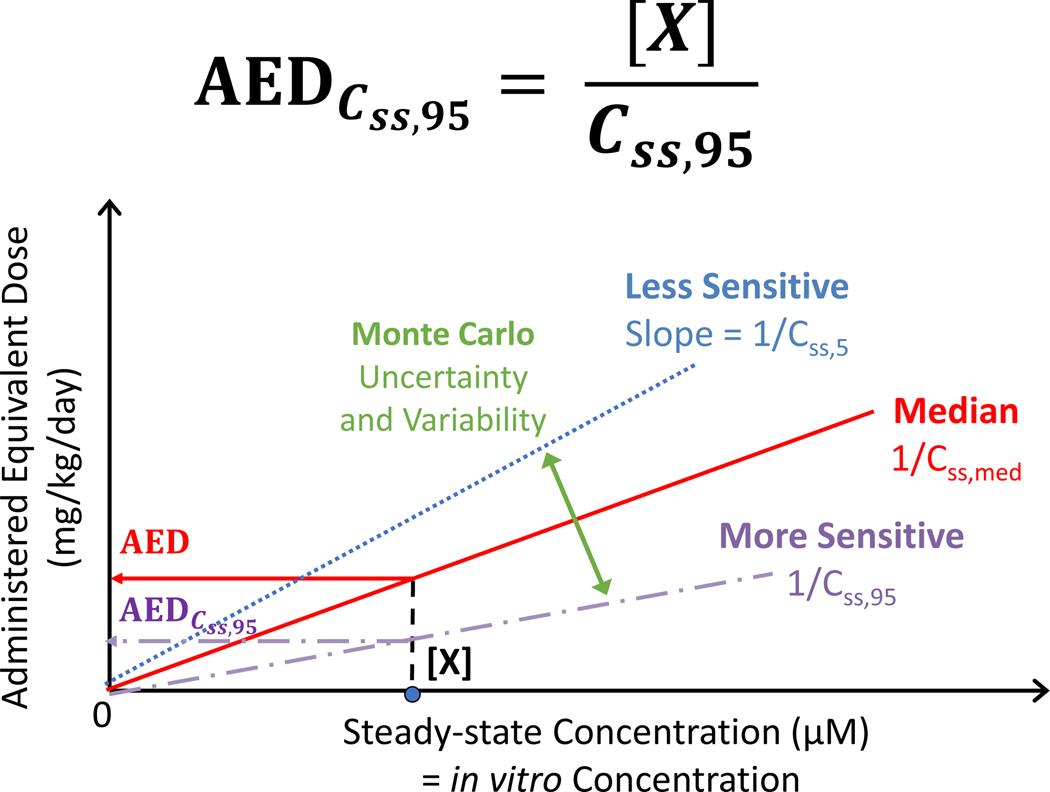
Figure 2. Reverse Dosimetry Toxicodynamic IVIVE.
Equivalent external dose is determined by solving the TK model in reverse by deriving the external dose (that is, TK model input) that produces a specified internal concentration (that is, TK model output). Reverse dosimetry and IVIVE using HTTK relies on the linearity of the models. We calculate a scaling factor to relate in vitro concentrations (μM) to administered equivalent doses (AED). The scaling factor is the inverse of the steady state plasma concentration (Css) predicted for a 1 mg/kg/day exposure dose rate. We use Monte Carlo to simulate variability and propagate uncertainty to calculate an upper 95th percentile Css,95 for individuals who get higher plasma concentrations from the same exposure.
While IVIVE broadly includes any use of in vitro data to predict phenomena in vivo it is useful to distinguish between TK IVIVE (that is, the use of in vitro data to predict ADME) and toxicodynamic (TD) IVIVE, which includes the use of in vitro data to predict toxic effects in vivo (Figure 1). Both TK IVIVE and in vitro disposition modeling (for example, the Armitage model [19]) are useful for enhancing TD IVIVE [20], but the specific in vitro measurements and IVIVE methods used to inform TK IVIVE may be very different from those for TD IVIVE. For example, TK IVIVE often uses the clearance of hepatocyte suspensions and the binding of plasma proteins along with a TK model to predict ADME [21,22]. Those ADME predictions might in turn be used to scale concentrations for TD IVIVE in which the in vitro bioactivity data are statistically correlated with in vivo toxic endpoints observed in animals [23]. Both the data and the methods (for example, mechanistic ADME vs. statistical modeling) can vary significantly between TK IVIVE and TD IVIVE.
HTTK methods have been used by the pharmaceutical industry to determine the range of efficacious doses, to prospectively evaluate success of planned clinical trials, and to minimize side effects and drug-drug interactions. The TK IVIVE methods initially developed for pharmaceutical HTTK have also been applied to environmental and industrial chemicals, to rapidly assess potential human health risks for chemicals which (rightly) cannot ethically be tested in humans, but to which humans are nonetheless exposed. Because of the demonstrated utility of HTTK methods for environmental and industrial chemicals, the U.S. Environmental Protection Agency (U.S. EPA) provides an implementation of HTTK methods through the publicly-available software package called “httk” (https://CRAN.R-project.org/package=httk). Here we distinguish the scientific methodology of HTTK from the R package httk via capitalization. The httk package was developed to achieve two main goals: IVIVE of in vitro HTS bioactive concentrations to predict human dose context [6,28], and providing open source data and models for evaluations and applications by the broader scientific community [31]. The use of HTTK methodology and the httk package has been recently advocated by the Health Canada, as they envision using such an approach for future screening level assessments under the Canadian Environmental Protection Act [32]. This review provides a description of HTTK models, with a focus on the httk package, its capabilities, and applications. However, comparison between other HTTK implementations is also provided.
1. Generic Physiologically-Based Toxicokinetic Models
TK models describe the ADME of a chemical by representing the body as one or more connected compartments among which the substance can enter (absorption), move (distribution), and leave (metabolism and elimination) [33,34]. Mathematically, a TK model describes the time-dependent amount of a substance in each compartment using a set of mass-balance ordinary differential equations [35]. The parameters of the model describe the size of each compartment and the rates at which the substance moves into, between, and out of compartments. The structure of the model defines the number of compartments and how they are connected.
Physiologically-based toxicokinetic (PBTK) models are designed such that the structure and parameters of the model have physiologically meaningful interpretations, as opposed to “empirical” non-physiological compartmental TK models [35]. Compartments in PBTK models typically represent individual organs or tissues and/or groups of physiologically similar organs or tissues [36–38]. Instead of simply applying uncertainty factors or extrapolating from non-physiological TK models to assess potential risk, PBTK models can estimate internal dose metrics (that is the biologically effective dose) by predicting internal dose of target organs to replace the externally administered dose (or exposure), as the observed effects are expected to be more directly related to the target tissue dose than the administered dose [37,39].
PBTK model parameters are based on anatomical, physiological, and biochemical properties [37]. Parameters may represent either physiological quantities (for example, tissue blood flow, tissue volume) or chemical-specific quantities describing the interactions between the chemical and the body (for example, tissue:plasma equilibrium partition coefficients) [36,37]. Model parameters are estimated using data obtained from known exposure scenarios. The models can be used to make predictions of hypothetical exposure scenarios (exposure conditions of interest) and simulate the chemical TK in potentially untestable situations. These extrapolations can be not only from in vitro to in vivo conditions [11,40], but also across exposure routes [41,42]; between species [41–43]; between chemicals [44,45]; across populations [46–49]; and across life-stages [50,51]. The model parameters are modified to represent differences among the extrapolation situations, with no need to collect new chemical-specific or in vivo data [39].
PBTK models are traditionally developed for individual chemicals, with both the parameters and structure of the models tailored to make the most accurate predictions possible for each substance [52]. Different physiological processes may be included or omitted depending on their importance to the chemical under consideration [37], for example saturable resorption of ethylene glycol by the kidney [53] or extrahepatic metabolism of bisphenol-A and naloxone [54,55]. These “bespoke” (chemical-specific or study-specific) PBTK models require large amounts of information regarding the behavior and partitioning of a chemical in the body. Because of the challenges posed by the specificity of these PBTK models, these type of detailed models are more likely to suffer from implementation and documentation errors [56]. McLanahan, El-Masri [57] observed that peer-reviewed publications alone are often insufficient to allow proper verification and reproducibility of PBTK models – for example, model code and parameters are often outdated, even when provided as supplemental material, and scripts for generating figures and other analyses often do not meet the threshold for inclusion in supplemental material. Reproducibility of PBTK models is highly desirable [58], but currently deficient due to the lack of adherence to published criteria or standards for evaluating these models [59–61]. For this reason, many tools and approaches have been developed to better document PBTK models [62–66], or to better identify the underlying chemical space that PBTK models have traditionally captured, and where there may be gaps [67]. Moreover, because of the uniqueness of these bespoke PBTK models, they require substantial chemical-specific TK data. Among non-pharmaceutical chemicals, comprehensive TK data only exist for relatively few, well-studied chemicals such as dioxin, lead, and trichloroethylene. The level of detailed data needed to build these models is not feasible in the context of HTTK as it can take years to develop for one of the thousands of chemicals potentially of interest.
A structurally-simpler approach to developing TK models is the empirical non-physiological compartmental approach that is not physiologically based, but is instead a best fit to in vivo measurements of the time course of body concentrations after single or repeated dosing of a chemical [13]. If concentrations appear to decrease with a single time constant, then an empirical model with a single compartment is developed; if concentrations appear to decrease with two time constants (for example, an initial rapid decrease and a second slower decrease), then an empirical model with two compartments is developed; and so forth. These empirical TK model structures are considered generic, since they can be parameterized to describe many different chemicals, and they do not attempt to describe chemical-specific ADME mechanisms [68]. However, parameterizing empirical models requires collecting species-specific in vivo measurements of concentration vs. dose and time, which is not feasible for large numbers of chemicals in an HTTK context. Moreover, because the parameters of empirical models are not physiologically based, empirical TK models do not allow the same degree of extrapolation (such as inter-species and route-to-route) as PBTK models, and therefore are less than ideal for many aspects of assessment for non-therapeutic chemicals where human studies are unlikely [39].
A third approach, “generic” PBTK models, uses a common structure across chemicals, bridging the gap between empirical and bespoke models. That is, the same physiological compartments and the same physiological processes are represented for all chemicals as opposed to customizing the physiological processes to those relevant for a specific chemical. Only the parameters representing chemical-body interaction differ between chemicals. These chemical-specific parameters may be derived from in vitro measurements [21,27–30], meaning that generic PBTK models are amenable to rapid parameterization for many chemicals, and therefore useful for HTTK applications. In fact, HTTK can be described as the combination of generic PBTK models with in vitro TK data. The consistent model structure helps improve reporting accuracy, fidelity of implementation, reproducibility, and statistical evaluation of generic PBTK models compared to bespoke PBTK models [62,69–73]. Several modeling software tools have been developed to facilitate the use of generic PBTK modeling [24,31,74–76]. Generic PBTK models vary with respect to the amount of chemical-specific data required, with more sophisticated models having “higher” data needs, such as enzyme-specific metabolism rates to address genetic polymorphisms, while those with “lower” data needs might only use a few parameters to characterize chemicals. An overview of some key generic PBTK modeling tools, including the httk package, are described in Table 1.
Table 1:
Overview of Generic PBTK Modeling Tools.
| SimCYP | ADMET Predictor / GastroPlus | PK-Sim | IndusChemFate | httk | |
|---|---|---|---|---|---|
| References | Jamei, Marciniak [19] | Lukacova, Woltosz [69] | Eissing, Kuepfer [70] | Jongeneelen and Berge [71] | Pearce, Setzer [27] |
| Availability | License, but inexpensive for research | License, but inexpensive for research | Free: http://www.open-systems-pharmacology.org/ | Free: http://cefic-lri.org/lri_toolbox/induschemfate/ | Free: https://CRAN.R-project.org/package=httk |
| Open Source | No | No | GitHub | No | CRAN and GitHub |
| Default PBTK Structure | Yes | Yes | Yes | Yes | Yes |
| Population Variability | Yes | Yes | Yes | No | Yes |
| Data Needs | High/Low | High/Low | High | High | Low |
| Typical Use Case | Drug Discovery | Drug Discovery | Drug Discovery | Environmental Assessment | Screening |
| Batch Mode | Yes | Yes | Yes | No | Yes |
| Graphical User Interface | Yes | Yes | Yes | Excel | No* |
| Built-in Chemical-Specific Library | Many Clinical Drugs | No | Many pharmaceutical- specific models available | 15 Environmental Compounds | 980 Pharmaceutical and ToxCast Compounds |
| Ionizable Compounds | Yes | Yes | Yes | No | Yes |
| Export Function | No | No | Matlab and R | No | SBML and Jarnac |
| R Integration | No | No | Yes (2017) | No | Yes |
| Reverse Dosimetry | Yes | Yes | Yes | No | Yes |
2. High-Throughput PBTK Modeling for IVIVE: The httk package
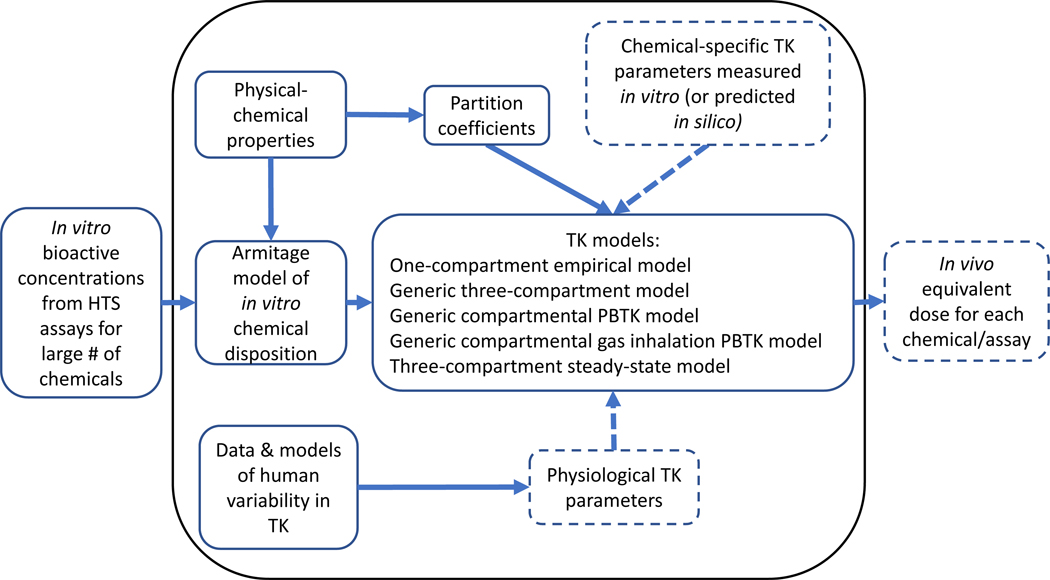
The httk package provides open source data and models with transparent documentation of the model design, the input parameters, and the differential equations used to calculate tissue concentrations. Since HTTK is the combination of in vitro data and generic TK modeling, the httk package [31] contains tables of chemical specific data (>1000 for humans and >200 for rats) as well as a suite of generic TK models that may be parameterized with those data. Released as an R package, the numerical modeling modules of httk are written in the C programming language for efficiency (that is, speed) of compiled code. The form of an R package was chosen to make httk open-source, transparent, freely available, and as platform-independent as possible. R and an integrated development environment (IDE) called RStudio are free software for statistical computing available for Windows, Mac, and Unix/Linux operating systems. Additional R packages are available to solve the required initial value problems (IVP) of ordinary differential equations (ODE). Using a command-line interface in R, the typical workflow of httk is shown in Figure 3. Rudimentary instructions for using httk is provided in Table 2 and further described in section 3.7.
Figure 3. Schematic of httk R package.
Dashed boxes/arrows represent parameters that can be probabilistic in a Monte Carlo simulation.
Table 2:
Getting Started with httk.
| Where Do I Get R? | R is freely available from the Comprehensive R Archive Network (CRAN): https://cloud.r-project.org/ Graphical user interface (GUI), RStudio, is freely available: https://rstudio.com/ |
| Getting Started with R Package httk | install.packages(httk) RStudio provides a menu “Install Packages” under “Tools” tab |
| Load the HTTK data, models, and functions | library(httk) |
| Check what version you are using | packageVersion(httk) |
| Getting help with R Package httk | help(httk) You can go straight to the index: help(package=httk) |
| List all CAS numbers for all chemicals with sufficient data to run httk | get_cheminfo() |
| List all information: | get_cheminfo(info=“all”) |
| Is a chemical with a specified CAS number available? | “80-05-7” %in% get_cheminfo() |
| All data on chemicals A, B, C | subset(get_cheminfo(info=“all”),Compound%in%c(“A”,”B”,”C”)) |
| Administrated equivalent dose (mg/kg BW/day) to produce 0.1 uM plasma concentration, 0.95 quantile, for a specified CAS number and species | calc_mc_oral_equiv(0.1,chem.cas=“34256–82-1”, species=“human”) |
| Calculate the mean, AUC, and peak concentrations for a simulated study (28-day daily dose, by default) for a specified CAS number and species | calc_tkstats(chem.cas=“34256–82-1”, species=“rat”) |
| Using the PBTK solver for a specified chem name | solve_pbtk(chem.name=“bisphenol a”, plots=TRUE) |
| List all vignettes for httk | vignette(package=httk) |
| Displays the vignette for a specified vignette | vignette(“Frank2018”) |
| Create data set, my_data, for all data on chemicals A, B, C, in R | my_data <- subset(get_cheminfo(info=“all”),Compound%in%c(“A”,”B”,”C”)) |
| Export data set, my_data, from R to csv file called my_data.csv in the current working directory | write.csv(my_data, file = “my_data.csv”) |
While httk is designed to be accessed through the R command line and scripts of commands, two graphical user interface (GUI) tools for httk are currently available with PLETHEM [77] and Web-ICE [78]. Additional custom httk web interfaces can be developed with R-Shiny (https://shiny.rstudio.com/). The httk R package can also be integrated with the Konstanz Information Miner (KNIME) [79]: “R nodes” for input/output calls and third party reporting (https://www.knime.com/nodeguide/scripting/r) allow the highly customizable KNIME user interface to integrate R with many other tools. Eventually, httk-specific web-service apps might facilitate broader usability.
3.1. Generic TK Models in httk
The current version of the httk package, version 2.0.3, has five options for TK models: a one-compartment empirical model [13], a generic three-compartment PBTK model [24], three-compartment steady-state model [18,21], a more elaborate generic PBTK model for intravenous and oral exposure [31], and a generic gas inhalation PBTK model [80]. The generic three-compartment PBTK model (contains a systemic blood compartment with separate tissue compartments for the liver and gut) is the condensed form of the generic compartmental PBTK model (contains separate tissue compartments for the gut, liver, lungs, arteries, veins, and kidneys) [31]. The three-compartment steady-state model is the simplest model, which describes the steady-state concentration in the liver of the three-compartment PBTK model without partitioning (contain only plasma without separate compartments for blood and tissue) [31]. The user selects which model to use, and can use the selected model to perform forward dosimetry (using functions such as solve_gas_pbtk()) or reverse dosimetry (using functions such as calc_mc_oral_equiv()).
For reverse dosimetry, as in HT risk screening, either the generic compartmental PBTK model for oral exposure or the generic compartmental gas inhalation PBTK model for inhalation exposure are typically recommended, depending upon the volatility of the compound and the scenario. For instance, the oral exposure model would be suitable for the calculation of AEDs for a chemical in drinking water. The one-compartment empirical model is included mainly for use when in vivo TK data are available for model evaluation: it allows comparison of model predictions with in vivo experiments [22,81], and uncertainty quantification for regulatory decision making [56]. The three-compartment steady-state model is applicable to the largest number of chemicals, specifically those which are missing information needed to predict tissue partitioning in the other models [31].
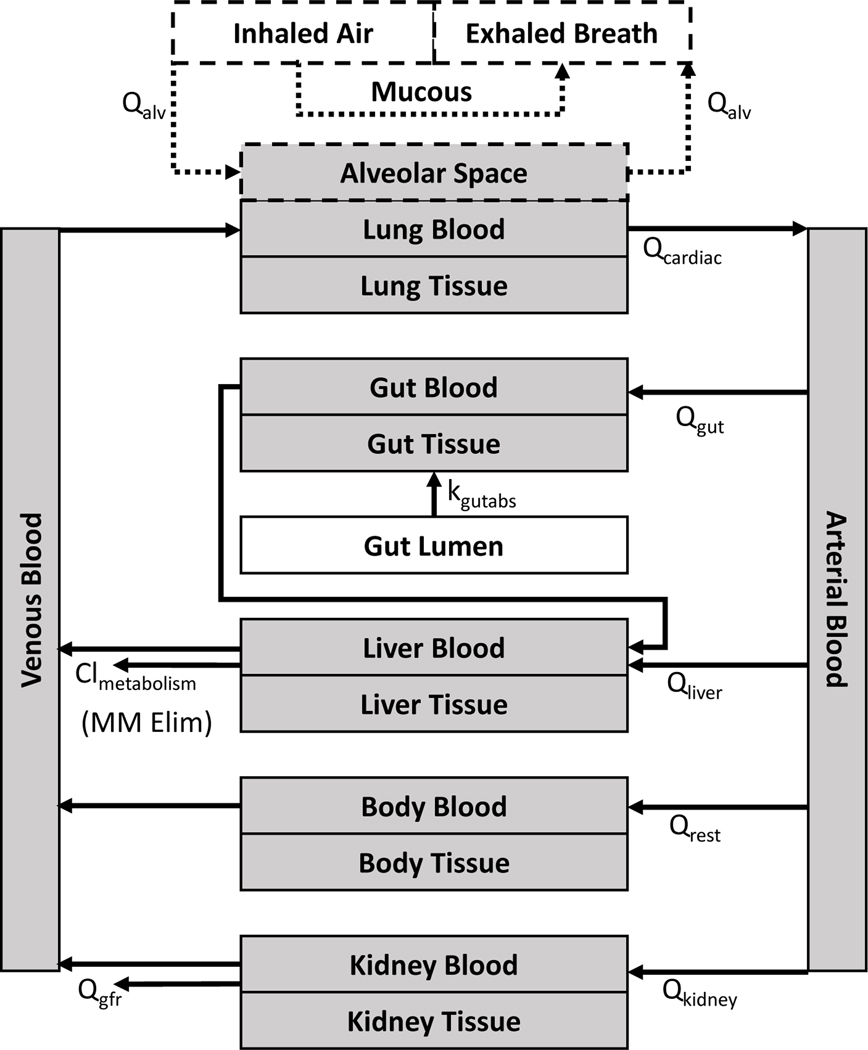
The structure of the httk generic compartmental PBTK models [31] — for example, the generic inhalation PBTK model in Figure 4 — includes compartments corresponding to blood and other tissues that are homogenous (well-mixed) and can be described by a volume and a single chemical concentration. Some tissues (for example arterial blood) are simple compartments, while others (for example kidney) are compound compartments consisting of separate blood and tissue sections. Some specific tissues (lung, kidney, gut, and liver) are modeled explicitly, while remaining tissues of body that are not from the organ of interest (for example fat, brain, bones) are lumped into the “Rest of Body” compartment for model simplifications. Following oral exposure, chemicals are absorbed from gut lumen, and chemicals are transformed through hepatic metabolism or excretion by passive renal clearance. The generic compartmental gas inhalation PBTK model [80] added a new component to allow modeling of inhalation exposures at a specified air concentration Figure 4. The structure of the inhalation model was developed from two previously published PBTK models from Jongeneelen and Berge [76] and Clewell III, Gentry [42], where chemicals are absorbed from an “alveolar space” compartment.
Figure 4. Example of a generic gas inhalation PBTK model. A single (generic) physiological structure is used for all appropriate chemicals.
Chemical-specific parameters can be predicted from a combination of in vitro measurements and QSARs. Qrest is defined as the difference between Qcardiac and the flow to the liver, kidney, and gut to preserve mass-balance.
Each TK model can yield either time-course concentrations in one or more compartments (including both peak and time-integrated “area under the curve”/AUC concentrations), or a steady-state concentration in one or more compartments, assuming repeated dosing at a constant level for a long duration. Typically, in the context of reverse dosimetry for HT risk screening, the steady-state concentration is used since analytical steady state solutions to the models can substantially shorten the computation time, and the resulting in vivo equivalent dose represents a long-term near-constant exposure. The httk package contains functions that automate the calculation of AEDs under the steady-state assumption. This allows the user to simply input the chemical and in vitro bioactive concentration, select the TK model, and then automatically obtain the in vivo equivalent dose which would produce a body concentration equal to the in vitro bioactive concentration. It relies on the linearity of the models to calculate a scaling factor to relate in vitro concentrations (μM) with AED. The scaling factor is the inverse of the steady-state plasma concentration (Css) predicted for a 1 mg/kg/day exposure dose rate (Figure 2):
where in vitro concentration [X] and Css must be in the same units. Note that it is typical for in vitro concentrations to be reported in units of μM and Css in units of mg/L, in which case one must be converted to the other using:
where MW is the molecular weight of the compound in units of g/mole.
If non-steady-state concentrations are needed (for example, to perform reverse dosimetry for an acute or subchronic exposure period), httk can automatically calculate peak concentration, average concentration, and AUC in a specified compartment with the functions calc_tkstats() and the Monte Carlo version calc_mc_tk(). As with Css, the user can again employ the linearity of the models in httk to determine AED by dividing the in vitro bioactive concentration with the relevant concentration summary statistic (peak, average, or AUC) for a dose of 1 mg/kg/day. The full time course of concentration in each compartment can be calculated using the function solve_model().
The generic TK models have parameters that can be divided into two main categories: chemical-specific parameters and physiological parameters. Chemical-specific parameters describe kinetics-related quantities that are different for different chemicals (for example, hepatic clearance rate, plasma protein binding, tissue partitioning, oral absorption). Physiological parameters describe kinetics-related quantities that depend on the anatomical and physiological characteristics of each person or animal and are not chemical-dependent (for example, body weight, organ and tissue volumes, blood flow rates, hepatocellularity). Below, we describe the details of these generic TK model parameters.
3.2. Chemical-specific Parameters: Experimental Data Needs/Inputs
Four chemical-specific parameters are used in httk:
intrinsic hepatic clearance (that is, disappearance of compound when incubated with primary hepatocytes)
fraction of compound unbound in plasma
Caco-2 membrane permeability, and
relative concentration in blood and plasma (that is, ratio of the blood concentration of a chemical to the plasma concentration).
Constant Rblood2plasma (ratio of the blood concentration of a chemical to the plasma concentration) is used throughout the body, predicted using hematocrit and the predicted partitioning between red blood cells and plasma when in vivo values are unavailable. These chemical-specific TK parameters for the httk models are based on in vitro measurements for a total of 1043 chemicals (see, for example, Rotroff, Wetmore [27], Wetmore, Wambaugh [29], Wetmore, Wambaugh [12], Honda, Pearce [20]). Of the four in vitro TK parameters mentioned above, the httk package primarily uses two: 1) intrinsic hepatic clearance [82] and 2) fraction unbound in plasma [83]. Currently only hepatic clearance estimates from primary hepatocyte suspensions [82] are used by httk, though this clearance might also be estimated by other means including microsomes and whole-body in vivo data. Protein binding in plasma, as characterized by the fraction unbound, is particularly important for predicting partitioning between tissues and renal excretion, and may impact the rate of metabolism [7,84] as only unbound chemical is able to freely partition and distribute into tissues. When data are available, the two remaining parameters, Caco2 permeability [85] (used to model oral absorption) and relative blood/plasma concentrations, can also be used by the httk package.
The httk package includes parameters (namely, intrinsic hepatic clearance and fraction unbound in plasma) from in vitro measurements with human-specific data for 1016 chemicals and rat-specific data for 212 chemicals. For other chemicals, user-provided model parameters can be entered into httk based on values obtained from other in vitro measurements, or from in silico models derived from applications such as OPEn structure-activity Relationship App (OPERA), Molecular Operating Environment (MOE) [86–89], or any cheminformatics tools that can predict molecular descriptors and derive QSAR (i.e. Schrödinger QikProp (New York, USA), Simulations Plus (Lancaster, USA) and Discovery Studio (Polouzane, France)).
Although physiological parameters (such as organ volumes and flows) are included for species beyond humans and rats, httk contains only very limited species-specific in vitro measured TK parameters for mouse and rabbit. While “chemical-specific” in vitro measured TK parameters might be considered more like inherent properties of a chemical than purely physiological parameters, properties like hepatic clearance do vary between species [29] and individuals [30] owing to inter-species and inter-individual differences in hepatic enzyme affinity, abundance, and polymorphism. When non-human parameter values are unavailable for a particular chemical, httk can automatically substitute human values (typically by setting the argument default.to.human = TRUE). Inter-individual variability in intrinsic hepatic clearance rate and fraction unbound in plasma is accounted for in httk, assuming a truncated normal distribution with 30% CV [47]. For hepatic clearance rate, a group of ultra-low metabolizers (with mean clearance ten times less than measured) are assumed to make up 5% of the population [47].
3.3. Chemical-Specific Parameters: Tissue:plasma partition coefficients
Since the chemical-specific tissue:plasma partition coefficients are more difficult to measure in vitro, httk contains a module to predict them based on physical-chemical properties and published tissue properties. It uses an empirically calibrated version of Schmitt’s method [90] that has been expanded with methods from Peyret, Poulin [91] using tissue data: cellular and water fractions of total volume, lipid and protein fractions of cellular volume, lipid fractions of the total lipid volume, the pH of each tissue, and the fractional volume of protein in plasma [31]. It calculates ionization with a default plasma pH of 7.4 [90], and the partition coefficient for the mass and volume of the body unaccounted for by the tissues included in Schmitt [90], which is determined with the averages of the fractional volumes and pH of these tissues, excluding red blood cells [31]. The model predictions are calibrated based on tissue-specific regressions of experimentally observed tissue partition coefficients on the predictions of the model (this can be turned off via “regression = FALSE”) [38].
3.4. Physiological parameters and Monte Carlo simulations to represent inter-individual variability
To support reverse dosimetry for a variety of species and inter-species extrapolation, httk contains default physiological parameter values for rat, rabbit, dog, mouse, and human. Here, physiological parameters represent values specific to the species of interest and independent of the chemical being simulated such as cardiac output, tissue volume, and respiratory rates. These default values are intended to represent an “average” or “standard” individual. However, default parameters may not be sufficient for the typical use case of prioritizing environmental chemicals based on potential human health risk in the population, because TK vary between individuals [47]. Each person’s body can process a chemical differently owing to inherent variability in organ size, blood flow rates, and enzymatic abundance. Therefore, different people with the same external exposure can have different internal concentrations. The TK models account for this inter-individual variability by varying both the physiological and chemical-specific parameters between individuals. The physiological parameters are varied to account for individual physiological differences, whereas chemical-specific parameters are varied to account for individual differences in the interaction between the body and the chemical.
The httk package includes the capability to simulate inter-individual TK variability using a Monte Carlo approach [92,93], which assumes distributions for the TK model parameters that represent population variability. The model is solved repeatedly, typically using an analytical steady-state solution, each time randomly drawing a set of parameter values from these distributions. The resulting set of model outputs approximates the distribution of AEDs among individuals. A lower percentile AED represents a more sensitive individual – a person that can reach the in vitro bioactive concentration with a lower exposure. A higher percentile AED represents a less-sensitive individual – a person that requires a higher exposure to reach the in vitro bioactive concentration.
The HTTK-Pop module of httk generates a “virtual population” of individual physiologies through Monte Carlo simulations that incorporate physiologies, incorporating observed correlations (for example, among age, sex, height, and weight) to produce a more realistic distribution of physiologies [47]. The httk package allows the user to specify attributes of the population to be simulated. For example, the user can specify a population for ages 6–11 to determine the range of AEDs for children, or a population of women ages 18–45 to determine the range of AEDs for reproductive-aged women. Since httk was developed primarily for risk prioritization in the US, it uses physiological data for the US population from the Centers for Disease Control and Prevention (CDC) the National Health and Nutrition Examination Survey (NHANES) database: demographics, body measurements, and certain direct measurements of physiology. For physiological parameters that are not measured by NHANES, such as tissue volumes and blood flows, httk uses allometric scaling or regression models from the literature to predict these parameters based on quantities that NHANES does measure (for example age, sex, body weight, height).
3.5. Uncertainty Propagation
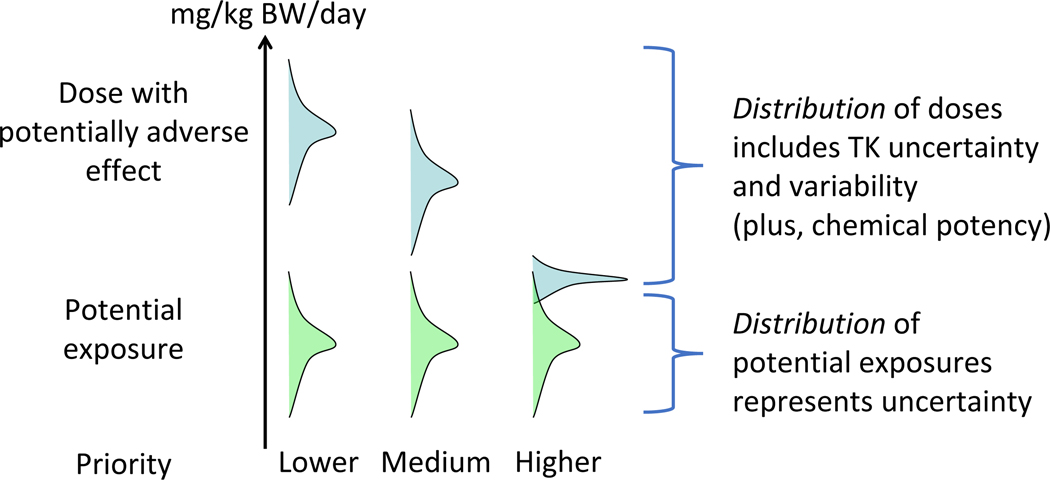
Taking advantage of much of the same Monte Carlo simulation approach that is used for population variability, propagation of uncertainty was recently included in httk. This allows for a quantitative assessment of the impact of experimental uncertainty in measuring the chemical-specific parameters [94], which is critical to assess the quality and accuracy of the data. Within httk, most parameters can be assigned distributions with means and standard deviations representing both variability and uncertainty where variability represents a range of observed parameter values while uncertainty represents a range of model-predicted values. Chemical-specific in vitro measurements can be assigned distributions reflecting both the constraints of the measurement process (for example, fraction unbound should be between zero and one) and any chemical specific uncertainty estimated reflecting the mass spectrometry signal-to-noise ratio [94]. Particularly of note, by assigning appropriate estimates of uncertainty, these methods allow the predictions of QSARs and experimental data to be used jointly. By using the Monte Carlo method to simulate population variability [47,93] and propagate uncertainty [95,96], an upper 95th percentile steady-state concentration (Css,95) can be calculated for individuals who have higher plasma concentrations from the same exposure in order to obtain a scaling factor for relating in vitro concentrations with AED (Figure 2) [94]. Moreover, this allows the high-throughput chemical risk prioritization to rapidly prioritize large numbers of chemicals by comparing distributions of dose with potentially adverse effect and potential exposure, which are estimated by accounting for both uncertainty and variability (Figure 5) [47].
Figure 5. High-throughput chemical risk prioritization to rapidly prioritize large numbers of chemicals.
Evaluation of potential risk by comparing distributions of dose with potentially adverse effect and potential exposure. For TK we can account for both uncertainty and variability. If the hazard and exposure distributions are far apart, as shown on the left, then potential risk is lower – it means exposure probably doesn’t reach a level where there would be an adverse effect. If the lower tail of the hazard distribution starts to overlap the upper tail of the exposure distribution, as shown in the middle, then potential risk is medium. And if the hazard and exposure distributions totally overlap, as shown on the right, then potential risk is higher – it means that exposure is more likely to reach a level where there might be an adverse effect.
3.6. Chemical-Specific Parameters: In Vitro Distribution
In the context of in vitro high-throughput toxicity testing, the bioactive concentration is typically reported as the nominal applied concentration [19], and reverse dosimetry is typically applied to estimate an AED for this nominal concentration. However, experimental evidence has indicated the importance of considering in vitro chemical disposition when performing IVIVE: a nominal applied concentration may partition into the different elements of an in vitro assay system differently depending on chemical properties, resulting in different free concentrations in culture media and cells that may affect in vitro bioactivity [97–100]. There are published models for predicting the freely dissolved cellular/tissue and membrane concentrations to characterize chemical distribution for in vitro assays [19,101]. For performing IVIVE with in vitro HTS assay data, httk contains a module that implements the Armitage model [19] of chemical disposition within an in vitro assay system. Although this is not a TK model, the Armitage model is important to correctly identify the in vitro concentration associated with bioactivity, and thereby derive the appropriate in vivo equivalent dose [20].
3.7. Getting Started with the R Package httk
The open-source nature of R and detailed documentation of its packages make httk readily accessible to the broader scientific community. This accessibility and the straightforward nature of R mean individuals with minimal coding experience should feel comfortable working with httk. RStudio is a particularly user-friendly platform for those with little to no coding background. Table 2 provides an overview of how to get started with httk. The provided tutorial is based on httk version 2.0.3, the most recent version as of this publication. Chemicals can be identified using name, CAS, or DTXSID (that is substance identifier for the Distributed Structure-Searchable Toxicity (DSSTox) database (https://comptox.epa.gov )). Available chemical-specific information includes logP, MW, pKa, intrinsic clearance, fraction unbound in plasma, and blood to plasma partitioning. Calculations can be performed to derive chemical properties, TK parameters, or IVIVE values. Functions are also available to perform forward dosimetry using the various models. As functions are typed at the RStudio command line, available arguments are displayed, with additional help available through the ‘?’ operator. Vignettes for the various available packages in httk are provided to give an overview of their respective capabilities. The aim of httk is to provide a readily accessible platform for working with HTTK models. Following the steps outlined in Table 2 should provide the user with a relatively solid introduction to using httk for their own work.
3. Evaluation of the HTTK Approach to IVIVE
Since HTTK is intended to support public health risk decision making, confidence in its predictions must be sufficiently established. Oreskes [102] wrote of models that “the goal of scientists working in a regulatory context should be not validation but evaluation, and where necessary, modification and even rejection. Evaluation implies an assessment in which both positive and negative results are possible, and where the grounds on which a model is declared good enough are clearly articulated”. By virtue of being applicable to many chemicals (therefore increasing the likelihood of evaluation data being available) and coupling to statistical software (as with the httk R package), it is possible to perform statistical evaluation to determine whether HTTK is “good enough”. Wambaugh, Wetmore [103] applied machine learning tools to develop a model for predicting the accuracy of HTTK methods for predicting Css, finding roughly half within a factor of 3 and most chemicals within a factor of ten. Thus a “domain of applicability” can be predicted, identifying for which chemicals HTTK may be suitable if an appropriate standard is identified.
The World Health Organization (WHO) indicates that PBTK models are “adequate” when predictions “are, on average, within a factor of 2 of the experimental data” [52]. However, they noted that both observations and model predictions are subject to uncertainty: “The experimental data, frequently obtained in a few experimental animals or human subjects, may constitute a biased sample...” Linakis, Sayre [80] have examined HTTK concentration versus time predictions using a generic PBTK model across roughly forty volatile, non-pharmaceutical chemicals and observed an overall (that is, all time points for all chemicals) root mean squared error (RMSE) of 1.11 (on a log10 scale, therefore a factor of 13x) and a coefficient of determination (R2) of 0.47. These results, while indicative of predictive ability, are nowhere near the WHO standard.
However, HTTK has been repeatedly shown to be close to the WHO standard for TK summary statistics (that is, dose metric predictions) such as peak concentration and time-integrated (“area under the curve” or AUC) concentration. Wang [25] found across 54 pharmaceutical clinical trials that the predicted AUC differed by 2.3x. Linakis, Sayre [80] found an RMSE = 0.46 or 2.9x for peak concentration and RMSE = 0.5 or 3.2x for AUC. Examining data for a mix of 45 chemicals of both pharmaceutical and non-pharmaceutical nature, Wambaugh, Hughes [22] found an RMSE of 2.2x for peak and 1.64x for AUC. The calibrated method for predicting tissue partitioning that is included in httk similarly predicted human volume of distribution with a RMSE of 0.48 (3x) [38].
The key predicted endpoint for the most common applications of HTTK is Css, which has been found by several investigations to be within a factor of three for some, but not all, chemicals [21,22,103,104]. This may be due to the use of in vitro measurements originally developed for pharmaceuticals, for which metabolism is driven by the liver, and therefore neglecting extra-hepatic metabolism that may be a larger factor for non-pharmaceuticals [22]. Error in prediction of TK may be further compounded by neglecting active transporters in the generic PBTK models [103].
The assumptions used for IVIVE with HTTK may vary between chemicals and applications. For example, it has long been recognized that the rate of metabolism for some compounds is “restricted” by the fraction unbound in plasma — that is, the off-binding rate of the chemical is slow relative to the rate of metabolism and therefore only the free fraction of chemical may be metabolized in any instant — while for other chemicals the metabolism is “unrestricted” [84]. In the absence of a model for predicting the off-binding rate for a chemical, one may choose to treat all chemicals as restricted [12,27,94,103], non-restricted [105], or try to make the determination using in vivo data [21]. The health protectiveness of the assumption varies on the application — restrictive clearance leads to higher estimated tissue concentrations which may be health-protective for human predictions but may underestimate toxic potency for animal experiments [6]. The httk package allows users to make choices as to how IVIVE is performed, but applies conservative default assumptions in the absence of user input (for example, clearance is assumed to be restricted unless the user specifies otherwise). Honda, Pearce [20] used httk to evaluate sixteen sets of assumptions for IVIVE, including restrictive vs. non-restrictive clearance. Regardless of the assumptions, Honda, Pearce [20] showed that applying a high-throughput PBTK model enhanced the apparent correlation between in vitro bioactive concentrations and toxic doses determined in vivo, compared with no TK at all.
Ominously, Oreskes [102] also wrote that “One may remove obvious errors in a model while more subtle ones remain.” It is hoped that modern, open-source, and modular tools may foster an environment in which subtle problems are more rapidly identified and remedied. Any sort of evaluation of HTTK requires data [6,7,56,106] and a recently-developed public database of TK concentration vs. time data now provides reference data for nearly 150 chemicals from more than 500 literature studies and provides a reporting standard and repository for more [81]. Empirical evaluation of HTTK across many chemicals and classes of chemicals allows quantification of the prediction uncertainty, potentially characterized by the RMSE or average fold error. In turn, regulators may decide whether the associated uncertainties are acceptable depending on context.
4. Conclusion
Generic, high-throughput PBTK models minimize the data requirements to generate chemical- and scenario-specific predictions. By sharing a common structure and software platform, these generic models help overcome reliability challenges with PBTK implementation and documentation [56,57] to allow greater reproducibility and potentially support regulatory decision making [57,58,62]. The httk R package is a key example of a suite of generic TK models and databases that allow users to rapidly predict ADME of chemicals within the body. Unlike many other similar tools, httk is open source software, and includes peer-reviewed chemical-specific data. The open accessibility of httk facilitates collaborations between scientists and enables continuous improvement of the tools, as researchers can identify and report potential issues within the httk package. The greatest strength of the httk tool over other similar tools is its ability to screen large numbers of chemicals at once due to a suite of generic TK models and databases. This capability makes it easier to identify outliers and other issues that would likely go unnoticed when screening a single chemical at a time. The httk tool makes important chemical prioritization information available for risk assessors and policymakers more rapidly than traditional models that focus on toxicity of single chemicals.
Inhalation is an important route of exposure, particularly for occupational settings. The latest version of the httk package includes an inhalation model [80]. Inhalation models are relatively new in the realm of HTS [107], since previous works mainly focused on the less volatile chemicals likely to enter the body orally (for example, Rotroff, Wetmore [27]). Inclusion of new routes of exposure and new classes of chemicals enhances the utility of HTTK.
There are some limitations with the httk package, many of which are addressed and supported by other available HTTK tools. One limitation is that httk focuses on generic, simple TK models, without a detailed description of ADME for a particular chemical and any physiological (or other) processes or compartments that may be especially relevant to that chemical. PK-Sim [75], for example, is a platform that readily supports development of chemical-specific models including potentially relevant physiology. In addition, metabolites are not currently considered in “httk,” as this package only reproduces metabolism as the elimination of the parent compound, whereas some proprietary software such as Simulations Plus [74] can account for the formation of metabolites. In httk, metabolism is based solely on whole hepatocyte clearance, instead of ascribing metabolism to specific enzymes, a strength of the proprietary software SimCYP. SimCYP has the additional advantage of being able to follow drug-chemical or chemical-chemical interactions [24]. Despite these limitations, httk provides an open-source, replicable interface for conducting high-throughput toxicological prioritization to assess risk potential across thousands of chemicals currently used throughout commerce.
5. Expert opinion
Using TK to understand the dose-response relationship is a critical part of assessing chemical risk posed to public health [108]. Chemicals still in need of triage, prioritization, and potentially full assessment number in the thousands [10]. HTTK can assist chemical risk assessors with its ability to support rapid chemical screening/prioritization, perform IVIVE, and perform forward TK modeling for a large number of chemicals with only limited available chemical-specific data. It is unlikely that HTTK will give better predictions than a bespoke model developed with detailed chemical-specific in vivo data that has been tailored to include all physiological processes identified as key to that chemical’s ADME. We expect a generic model to be less accurate with respect to reproducing in vivo ADME measurements, but more likely to be accurately reported, reproducible, and statistically evaluated. Therefore, generic models may, in some cases, be more suitable to decision making contexts. However, this hypothetical comparison may be irrelevant since it is unlikely that such detailed bespoke PBTK models will ever be developed for the vast majority of chemicals. For the thousands of chemicals for which detailed data and models will likely never be available, HTTK offers a rapid and scientifically defensible description of ADME.
The reproducibility of PBTK models is a necessity of public health risk assessment and a known weakness as documented in the scientific literature [57,60]. Model reporting/documentation criteria and templates are becoming available that may enhance reproducibility if a set of common standards are adopted by the community [62,66]. However, the longevity and portability of models become issues as languages change and, in some cases (notably the popular modeling language acslX), become discontinued [109]. Many existing chemical-specific models were written in acslX and these models now need to be translated if they are to continue to be used. To build detailed chemical-specific PBTK models, the free, open-source modeling software GNU MCSim might be used [110,111].
While many published PBTK models do not currently meet the Clark [62] criteria for sufficient documentation [57,61], generic models of HTTK offer a tantalizing alternative. The modular and open source design of tools like httk can enable the tools, models, and data to be used, evaluated, and continuously improved by a broad user community, including toxicological researchers, risk assessors, regulators, and discovery scientists. By trading specificity for reliability, the generic TK model design can increase model uncertainty. However, users can have greater confidence in the model structure (albeit simplified) and software implementation, since the generic model can be tested for hundreds to thousands of chemicals [56].
It should not be expected that the “one-size-fits-all” approach of HTTK will actually be appropriate for all chemicals; the goal is instead to develop something akin to “one-size-roughly-fits-many”. To do this, we must identify to which chemical classes HTTK should or should not be applied and with what confidence. Wambaugh, Wetmore [103] was an early attempt to use machine learning to anticipate what sorts of chemicals might be well predicted by HTTK, and which might be poorly predicted (for example, transporter substrates). However, thanks to significant advances in models and data since 2015, such approaches are already in need of an update.
Beyond the general suitability of HTTK to a chemical, work is needed to better anticipate and remedy chemical-specific difficulties with measurement of in vitro TK parameters. Some chemicals (for example, volatiles) are ill-suited to cell culture, while others (for example, curcumin-related compounds) prove difficult for mass spectrometry. Basic decision tree classifier methods can identify specific chemical features or molecular properties/descriptors that are associated with in vitro measurement difficulties. The development of alternative and improved in vitro assays to measure TK parameters, and tools to recognize when they should be applied, would enhance the applicability of HTTK across chemical classes. Similarly, substantial effort has gone into developing QSARs for in vitro TK parameters and a recent focus has been on broadening beyond pharmaceutical space to a wider variety of chemical classes [87,112–115]. As with all such tools, confidence must be quantified, and the domain of applicability established.
Development of the httk R package has followed the availability of chemical-specific data in the public domain. For example, the publicly available whole hepatocyte metabolism data is the result of multiple metabolizing enzymes, including “cytochrome P450 enzymes” or “CYPs”, which are known to vary in expression and functionality throughout the population. SimCYP and other tools oriented towards drugs have invested substantially in making use of less widely available data on enzyme-specific metabolism. While SimCYP includes a module on population variability in expression/function of specific enzymes, such a module has not been included in httk because of the lack of these data for non-pharmaceutical chemicals [47]. If efforts like that of Wetmore, Allen [30] for environmental chemicals could be expanded, then it would make sense to model enzyme-specific metabolism and ability to simulate population variability in enzyme abundance in httk. Other modules should be added as the need arises and the data to support them are developed.
Similarly, new high-throughput (generic) PBTK models should be added as both the need arises and the data to evaluate them are developed. One clear area of need is integration of early life-stage parameters for potentially sensitive life stages, including pregnancy and gestation and infancy. However, for any new model it will be necessary to identify and curate appropriate chemical datasets. Key needs include in silico QSAR models for (a) transplacental permeability (b) embryonic plasma binding (AFP or alpha-fetoprotein, an early life-stage-specific albumin ortholog) binding, and (c) milk:plasma partition coefficients for gestational pharmacokinetics/dosimetry estimation and post-gestational developmental models. Additionally, integrating gestational/embryonic tissue-specific expression profiles of phase I/II metabolizing enzymes will be vital.
Usability (that is, user interface) is a barrier to the adoption of any software tool, including those for TK [116]. Precalculated HTTK-based IVIVE predictions are available from on-line databases (such as https://comptox.epa.gov/dashboard [117]) in addition to graphical interfaces for PBTK models [77,78]. The goal of such tools is both to make the workings of the model more accessible as well as reduce software barriers (that is, lower need for third party software installation, command-line angst, drop down interaction as a web-service). Another enhancement to usability is the development of KNIME [79] workflows that take advantage of the R KNIME nodes, but enable a variety of manual structure (sketch) inputs, QSAR estimation and plugs to third party software such as MOE [89] or Schrodinger (https://www.schrodinger.com/drug-discovery), post simulation processing and reporting to third party visualization tools. Stewardship of HTTK tools is just as critical and will require the development of training material (including R “vignettes” and video examples) that provide a variety of user scenarios that are fully documented.
The longest-range goals of HTTK should include working toward “HTTD” (high-throughput toxicodynamics). Methods to link in vitro assays to in vivo health effects are needed. Currently, IVIVE of in vitro HTS data using HTTK involves the major assumptions that (1) a concentration that was bioactive in vitro will also be bioactive in the body and (2) even if the concentration is bioactive in the body, that activity in this one specific biological endpoint will have an actual effect on human health. Can those assumptions be refined, for instance by comparing in vitro predictions with the outcomes of legacy in vivo toxicity studies [20,29,118]? Better yet, can we integrate target specific information for receptors involved in signal transduction pathways and apical endpoints) — for instance adverse outcome pathway molecular initiating events [119] based on high-throughput receptor activity models [105]? HTTD, representing HTTK coupled to HTS, may eventually allow fully in vitro and computational prediction of public health effects of chemicals.
Article highlights:
In vitro high-throughput screening (HTS) to efficiently assess potential chemical risk to public health requires toxicokinetics for in vitro-in vivo extrapolation (IVIVE)
The high-throughput toxicokinetics (HTTK) method uses generic toxicokinetic (TK) models that can be parameterized for thousands of chemicals with in vitro TK data
The U.S. Environmental Protection Agency (U.S. EPA) provides HTTK methods through the publicly available software package called “httk”
The open accessibility of the httk package facilitates collaborations between scientists, and enables continuous improvement of the tools
The fundamental strength of the httk package is its ability to screen many chemicals for potential risk at once, using only limited data
Acknowledgements
The authors thank Drs. Elaina Kenyon and Todd Zurlinden for their helpful U.S. EPA internal reviews of the manuscript.
Annotated bibliography
- 1.Park YH, Lee K, Soltow QA, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012. May 16;295(1–3):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rager JE, Strynar MJ, Liang S, et al. Linking high resolution mass spectrometry data with exposure and toxicity forecasts to advance high-throughput environmental monitoring. Environment International. 2016;88:269–280. [DOI] [PubMed] [Google Scholar]
- 3.Egeghy PP, Judson R, Gangwal S, et al. The exposure data landscape for manufactured chemicals. Science of the Total Environment. 2012. Jan 1;414:159–66. [DOI] [PubMed] [Google Scholar]
- 4.Judson R, Richard A, Dix DJ, et al. The toxicity data landscape for environmental chemicals. Environmental Health Perspectives. 2009;117(5):685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breyer S. Breaking the Vicious Circle: Toward Effective Risk Regulation. Harvard University Press; 2009. [Google Scholar]
- 6. Bell SM, Chang X, Wambaugh JF, et al. In vitro to in vivo extrapolation for high throughput prioritization and decision making. Toxicology in Vitro. 2018. 2018/03/01/;47:213–227. This workshop report describes activities and resources that would promote inclusion of In vitro to in vivo extrapolation in regulatory decision-making.
- 7.Bessems JG, Loizou G, Krishnan K, et al. PBTK modelling platforms and parameter estimation tools to enable animal-free risk assessment: recommendations from a joint EPAA–EURL ECVAM ADME workshop. Regulatory Toxicology and Pharmacology. 2014;68(1):119–139. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt CW. TOX 21: new dimensions of toxicity testing. National Institute of Environmental Health Sciences; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dix DJ, Houck KA, Martin MT, et al. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicological Sciences. 2007;95(1):5–12. [DOI] [PubMed] [Google Scholar]
- 10.Kavlock RJ, Bahadori T, Barton-Maclaren TS, et al. Accelerating the Pace of Chemical Risk Assessment. Chemical Research in Toxicology. 2018. 2018/05/21;31(5):287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coecke S, Pelkonen O, Leite SB, et al. Toxicokinetics as a key to the integrated toxicity risk assessment based primarily on non-animal approaches. Toxicology in Vitro. 2013;27(5):1570–1577. [DOI] [PubMed] [Google Scholar]
- 12. Wetmore BA, Wambaugh JF, Allen B, et al. Incorporating High-Throughput Exposure Predictions With Dosimetry-Adjusted In Vitro Bioactivity to Inform Chemical Toxicity Testing. Toxicological Sciences. 2015. Nov;148(1):121–36. This study assesses toxicokinetics of ToxCast Phase II chemicals, which are incorporated into IVIVE to estimate oral equivalent doses. These oral equivalent doses are compared to exposure estimates from the US EPA’s ExpoCast program.
- 13.O’Flaherty EJ. Toxicants and drugs: kinetics and dynamics. John Wiley & Sons; 1981. [Google Scholar]
- 14. Tan Y-M, Liao KH, Clewell HJ. Reverse dosimetry: interpreting trihalomethanes biomonitoring data using physiologically based pharmacokinetic modeling. Journal of Exposure Science and Environmental Epidemiology. 2007;17(7):591–603. • Demonstrates the use of physiologically based pharmacokinetic (PBPK) modeling in a reverse dosimetry approach to assess a distribution of exposures that are consistent with biomonitoring data.
- 15.Sobus JR, DeWoskin RS, Tan YM, et al. Uses of NHANES Biomarker Data for Chemical Risk Assessment: Trends, Challenges, and Opportunities. Environmental Health Perspectives. 2015. Oct;123(10):919–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mage DT, Allen RH, Gondy G, et al. Estimating pesticide dose from urinary pesticide concentration data by creatinine correction in the Third National Health and Nutrition Examination Survey (NHANES-III). Journal of Exposure Science and Environmental Epidemiology. 2004;14(6):457. [DOI] [PubMed] [Google Scholar]
- 17.Lakind JS, Naiman DQ. Bisphenol A (BPA) daily intakes in the United States: estimates from the 2003–2004 NHANES urinary BPA data. Journal of Exposure Science and Environmental Epidemiology. 2008;18(6):608. [DOI] [PubMed] [Google Scholar]
- 18. Wetmore BA. Quantitative in vitro-to-in vivo extrapolation in a high-throughput environment. Toxicology. 2015;332:94–101. • This review covers the use of IVIVE from hepatic clearance and plasma protein binding assays to predict in vivo oral doses in a high throughput context that can inform risk assessment.
- 19. Armitage JM, Wania F, Arnot JA. Application of mass balance models and the chemical activity concept to facilitate the use of in vitro toxicity data for risk assessment. Environmental Science & Technology. 2014;48(16):9770–9779. • Given description of the in vitro testing conditions (such as well size and fraction of plasma protein in the media) the Armitage et al. model is extremely useful for predicting in vitro partitioning. Fischer et l. (2017) expanded the model to ionizable compounds.
- 20. Honda GS, Pearce RG, Pham LL, et al. Using the concordance of in vitro and in vivo data to evaluate extrapolation assumptions. PLOS ONE. 2019;14(5):e0217564. • This project was one of the first to statistically demonstrate the improvement in In Vitro-In Vivo extrapolation gained by using PBTK.
- 21. Wetmore BA, Wambaugh JF, Ferguson SS, et al. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicological Sciences. 2012. Jan;125(1):157–74. • Pharmacokinetics were measured for ToxCast Phase I chemicals and used in IVIVE to predict the human oral dose that could result in bioactivity. These estimates were then compared to human exposure estimates to assess risk.
- 22. Wambaugh JF, Hughes MF, Ring CL, et al. Evaluating in vitro-in vivo extrapolation of toxicokinetics. Toxicological Sciences. 2018;163(1):152–169. • For non-pharmaceutical compounds the correlation between in vitro-based TK predictions and in vivo observations have been weak – this study eliminated variability across in vivo studies as a factor and provided evidence for extra-hepatic metabolism being more important for non-pharmaceuticals.
- 23. Paul Friedman K, Gagne M, Loo LH, et al. Utility of In Vitro Bioactivity as a Lower Bound Estimate of In Vivo Adverse Effect Levels and in Risk-Based Prioritization. Toxicol Sci. 2020. Jan 1;173(1):202–225. • This case study demonstrated the suitability of IVIVE using ToxCast and R package httk for regulatory decision making as part of the Accelerating the Pace of Risk Assessment consortium (Kavlock et al, 2018).
- 24.Jamei M, Marciniak S, Feng K, et al. The Simcyp® population-based ADME simulator. Expert Opinion on Drug Metabolism & Toxicology. 2009;5(2):211–223. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y-H. Confidence Assessment of the Simcyp Time-Based Approach and a Static Mathematical Model in Predicting Clinical Drug-Drug Interactions for Mechanism-Based CYP3A Inhibitors. Drug Metabolism and Disposition. 2010 July 1, 2010;38(7):1094–1104. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Jin JY, Mukadam S, et al. Application of IVIVE and PBPK modeling in prospective prediction of clinical pharmacokinetics: strategy and approach during the drug discovery phase with four case studies. Biopharm Drug Dispos. 2012. Mar;33(2):85–98. [DOI] [PubMed] [Google Scholar]
- 27. Rotroff DM, Wetmore BA, Dix DJ, et al. Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicological Sciences. 2010;117(2):348–358. • This is the original bioactivity:exposure ratio IVIVIE framework for tiered toxicity testing in order to prioritize chemicals in a high-throughput fashion and reduce animal testing.
- 28.Tonnelier A, Coecke S, Zaldívar J-M. Screening of chemicals for human bioaccumulative potential with a physiologically based toxicokinetic model. Archives of Toxicology. 2012;86(3):393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wetmore BA, Wambaugh JF, Ferguson SS, et al. Relative impact of incorporating pharmacokinetics on predicting in vivo hazard and mode of action from high-throughput in vitro toxicity assays. Toxicological Sciences. 2013. Apr;132(2):327–46. • Study in which rat pharmacokinetics were measured and incorporated into IVIVE estimates for comparison with ToxCast activity values and prediction of in vivo effects.
- 30. Wetmore BA, Allen B, Clewell HJ, 3rd, et al. Incorporating population variability and susceptible subpopulations into dosimetry for high-throughput toxicity testing. Toxicological Sciences. 2014. Nov;142(1):210–24. • This study describes an approach to measure isozyme-specific clearance rates and incorporates this information into an IVIVE model to assess population and life stage pharmacokinetic variability, which could be used in high-throughput risk assessment.
- 31. Pearce RG, Setzer RW, Strope CL, et al. Httk: R package for high-throughput toxicokinetics. Journal of Statistical Software. 2017;79(1):1–26. • Describes the framework and organization of the “httk” R software package including four toxicokinetic models.
- 32. Health Canada. Science approach document - Bioactivity exposure ratio: Application in priority setting and risk assessment. 2021. • Health Canada report describes their determination that points of departure based on in vitro testing can serve as protective surrogates for traditional, animal testing-based hazard data, envisioning the use of the approach for future screening level assessments under the Canadian Environmental Protection Act. They advocate the use of HTTK methodology and the httk package.
- 33.Espié P, Tytgat D, Sargentini-Maier M-L, et al. Physiologically based pharmacokinetics (PBPK). Drug Metabolism Reviews. 2009;41(3):391–407. [DOI] [PubMed] [Google Scholar]
- 34.Grech A, Brochot C, Dorne J-L, et al. Toxicokinetic models and related tools in environmental risk assessment of chemicals. Science of the Total Environment. 2017;578:1–15. [DOI] [PubMed] [Google Scholar]
- 35. EPA US. Approaches for the application of physiologically based pharmacokinetic (PBPK) models and supporting data in risk assessment. National Center for Environmental Assessment, Washington, DC. 2006. • U.S. EPA report describing the evaluation and applications of physiologically based pharmacokinetic (PBPK) models in health risk assessment, by giving an overview of PBPK modeling, data needs, and evaluation of these models for risk assessment uses.
- 36.Andersen ME. Development of physiologically based pharmacokinetic and physiologically based pharmacodymamic models for applications in toxicology and risk assessment. Toxicology Letters. 1995;79(1):35–44. [DOI] [PubMed] [Google Scholar]
- 37.Campbell JL, Clewell RA, Gentry PR, et al. Physiologically based pharmacokinetic/toxicokinetic modeling. Computational Toxicology: Volume I. 2012:439–499. [Google Scholar]
- 38.Pearce RG, Setzer RW, Davis JL, et al. Evaluation and calibration of high-throughput predictions of chemical distribution to tissues. Journal of Pharmacokinetics and Pharmacodynamics. 2017. 2017/12/01;44(6):549–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barton HA, Chiu WA, Setzer RW, et al. Characterizing uncertainty and variability in physiologically based pharmacokinetic models: state of the science and needs for research and implementation. Toxicological Sciences. 2007;99(2):395–402. • This workshop report covers the problems and potential solutions with documenting, analyzing, and understanding the importance of and difference between uncertainty and variability in PBPK models for regulatory decision making.
- 40.Rostami-Hodjegan A. Physiologically based pharmacokinetics joined with in vitro–in vivo extrapolation of ADME: a marriage under the arch of systems pharmacology. Clinical Pharmacology & Therapeutics. 2012;92(1):50–61. [DOI] [PubMed] [Google Scholar]
- 41.Clewell III HJ, Andersen ME. Dose, species, and route extrapolation using physiologically based pharmacokinetic models. Drinking Water and Health, Volume 8: Pharmacokinetics in Risk Assessment. 1987;8:159. [Google Scholar]
- 42.Clewell III HJ, Gentry PR, Gearhart JM, et al. Development of a physiologically based pharmacokinetic model of isopropanol and its metabolite acetone. Toxicological Sciences. 2001;63(2):160–172. [DOI] [PubMed] [Google Scholar]
- 43.Sweeney LM, Kirman CR, Gannon SA, et al. Development of a physiologically based pharmacokinetic (PBPK) model for methyl iodide in rats, rabbits, and humans. Inhalation Toxicology. 2009;21(6):552–582. [DOI] [PubMed] [Google Scholar]
- 44.Jones HM, Barton HA, Lai Y, et al. Mechanistic pharmacokinetic modeling for the prediction of transporter-mediated disposition in humans from sandwich culture human hepatocyte data. Drug Metabolism and Disposition. 2012;40(5):1007–1017. [DOI] [PubMed] [Google Scholar]
- 45. Parham F, Kohn M, Matthews H, et al. Using structural information to create physiologically based pharmacokinetic models for all polychlorinated biphenyls: I. Tissue: blood partition coefficients. Toxicology and Applied Pharmacology. 1997;144(2):340–347. • This is one of the first examples of a semi-generic PBPK model for non-pharmaceutical chemicals – in this case, a generalized model for all PCB’s.
- 46.Clewell HJ, Gearhart JM, Gentry PR, et al. Evaluation of the uncertainty in an oral reference dose for methylmercury due to interindividual variability in pharmacokinetics. Risk Analysis. 1999;19(4):547–558. [DOI] [PubMed] [Google Scholar]
- 47. Ring CL, Pearce RG, Setzer RW, et al. Identifying populations sensitive to environmental chemicals by simulating toxicokinetic variability. Environment International. 2017. 2017/09/01/;106:105–118. • The “httk-pop” population simulator with a Monte Carlo approach using the National Health and Nutrition Examination Survey (NHANES) biometrics data to simulate toxicokinetic variability.
- 48.Rostami-Hodjegan A, Tucker GT. Simulation and prediction of in vivo drug metabolism in human populations from in vitro data. Nature Reviews Drug Discovery. 2007;6(2):140–148. [DOI] [PubMed] [Google Scholar]
- 49.Thomas RS, Bigelow PL, Keefe TJ, et al. Variability in biological exposure indices using physiologically based pharmacokinetic modeling and Monte Carlo simulation. American Industrial Hygiene Association Journal. 1996;57(1):23–32. [DOI] [PubMed] [Google Scholar]
- 50.Liao KH, Tan YM, Conolly RB, et al. Bayesian Estimation of Pharmacokinetic and Pharmacodynamic Parameters in a Mode-of-Action-Based Cancer Risk Assessment for Chloroform. Risk Analysis. 2007;27(6):1535–1551. [DOI] [PubMed] [Google Scholar]
- 51.Luecke RH, Wosilait WD, Pearce BA, et al. A physiologically based pharmacokinetic computer model for human pregnancy. Teratology. 1994;49(2):90–103. [DOI] [PubMed] [Google Scholar]
- 52. World Health Organization. Characterization and application of physiologically based pharmacokinetic models in risk assessment. World Health Organization, International Programme on Chemical Safety, Geneva, Switzerland. 2010. • This document articulated “factor of 2” criteria for judging accuracy of PBPK models that is often cited.
- 53.Corley RA, Bartels M, Carney E, et al. Development of a physiologically based pharmacokinetic model for ethylene glycol and its metabolite, glycolic acid, in rats and humans. Toxicological Sciences. 2005;85(1):476–490. [DOI] [PubMed] [Google Scholar]
- 54.Fisher JW, Twaddle NC, Vanlandingham M, et al. Pharmacokinetic modeling: prediction and evaluation of route dependent dosimetry of bisphenol A in monkeys with extrapolation to humans. Toxicol Appl Pharmacol. 2011. Nov 15;257(1):122–36. [DOI] [PubMed] [Google Scholar]
- 55.Docci L, Umehara K, Krähenbühl S, et al. Construction and Verification of Physiologically Based Pharmacokinetic Models for Four Drugs Majorly Cleared by Glucuronidation: Lorazepam, Oxazepam, Naloxone, and Zidovudine. Aaps j. 2020. Oct 8;22(6):128. [DOI] [PubMed] [Google Scholar]
- 56. Cohen Hubal EA, Wetmore B, Wambaugh J, et al. Advancing internal exposure and physiologically-based toxicokinetic modeling for 21st-century risk assessments. Journal of Exposure Science and Environmental Epidemiology. 2018. • This review covers toxicokinetic modeling tools used to assess internal exposures and inform risk assessment, including data and transparency needs.
- 57. McLanahan ED, El-Masri HA, Sweeney LM, et al. Physiologically based pharmacokinetic model use in risk assessment—why being published is not enough. Toxicological Sciences. 2012;126(1):5–15. • This report documents a 2011 SOT workshop in which the problems of PBPK documentation and reproducibility were discussed. When a semi-generic PBPK structure was discussed an audience member asked if saliva was included as a route of elimination.
- 58.Chiu WA, Barton HA, DeWoskin RS, et al. Evaluation of physiologically based pharmacokinetic models for use in risk assessment. Journal of Applied Toxicology. 2007;27(3):218–237. [DOI] [PubMed] [Google Scholar]
- 59.Tan YM, Worley RR, Leonard JA, et al. Challenges Associated With Applying Physiologically Based Pharmacokinetic Modeling for Public Health Decision-Making. Toxicol Sci. 2018. Apr 1;162(2):341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ioannidis JPA. Reproducible pharmacokinetics. J Pharmacokinet Pharmacodyn. 2019. Apr;46(2):111–116. [DOI] [PubMed] [Google Scholar]
- 61.Kirouac DC, Cicali B, Schmidt S. Reproducibility of Quantitative Systems Pharmacology Models: Current Challenges and Future Opportunities. CPT Pharmacometrics Syst Pharmacol. 2019. Apr;8(4):205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clark LH, Setzer RW, Barton HA. Framework for evaluation of physiologically-based pharmacokinetic models for use in safety or risk assessment. Risk Analysis. 2004;24(6):1697–1717. [DOI] [PubMed] [Google Scholar]
- 63.Loizou G, Spendiff M, Barton HA, et al. Development of good modelling practice for physiologically based pharmacokinetic models for use in risk assessment: the first steps. Regulatory Toxicology and Pharmacology. 2008;50(3):400–411. [DOI] [PubMed] [Google Scholar]
- 64.Madden JC, Pawar G, Cronin MTD, et al. In silico resources to assist in the development and evaluation of physiologically-based kinetic models. Computational Toxicology. 2019. 2019/08/01/;11:33–49. [Google Scholar]
- 65.Ruiz P, Ray M, Fisher J, et al. Development of a human physiologically based pharmacokinetic (PBPK) toolkit for environmental pollutants. International Journal of Molecular Sciences. 2011;12(11):7469–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan Y-M, Chan M, Chukwudebe A, et al. PBPK model reporting template for chemical risk assessment applications. Regulatory Toxicology and Pharmacology. 2020:104691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu J, Goldsmith M-R, Grulke CM, et al. Developing a physiologically-based pharmacokinetic model knowledgebase in support of provisional model construction. PLoS computational biology. 2016;12(2):e1004495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van der Molen G, Kooijman S, Slob W. A generic toxicokinetic model for persistent lipophilic compounds in humans: an application to TCDD. Fundamental and applied Toxicology. 1996;31(1):83–94. [DOI] [PubMed] [Google Scholar]
- 69.Sager JE, Yu J, Ragueneau-Majlessi I, et al. Physiologically Based Pharmacokinetic (PBPK) Modeling and Simulation Approaches: A Systematic Review of Published Models, Applications, and Model Verification. Drug Metab Dispos. 2015. Nov;43(11):1823–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller NA, Reddy MB, Heikkinen AT, et al. Physiologically Based Pharmacokinetic Modelling for First-In-Human Predictions: An Updated Model Building Strategy Illustrated with Challenging Industry Case Studies. Clinical Pharmacokinetics. 2019. 2019/06/01;58(6):727–746. [DOI] [PubMed] [Google Scholar]
- 71.Brightman FA, Leahy DE, Searle GE, et al. Application of a generic physiologically based pharmacokinetic model to the estimation of xenobiotic levels in human plasma. Drug Metabolism and Disposition. 2006;34(1):94–101. [DOI] [PubMed] [Google Scholar]
- 72.Cahill TM, Cousins I, Mackay D. Development and application of a generalized physiologically based pharmacokinetic model for multiple environmental contaminants. Environmental Toxicology and Chemistry: An International Journal. 2003;22(1):26–34. [PubMed] [Google Scholar]
- 73.Edginton AN, Schmitt W, Willmann S. Development and evaluation of a generic physiologically based pharmacokinetic model for children. Clinical pharmacokinetics. 2006;45(10):1013–1034. [DOI] [PubMed] [Google Scholar]
- 74. Lukacova V, Woltosz WS, Bolger MB. Prediction of modified release pharmacokinetics and pharmacodynamics from in vitro, immediate release, and intravenous data. The AAPS Journal. 2009;11(2):323–334. • Demonstrates the value of mechanistic simulations using the GastroPlus (Simulations Plus, Inc.) to describe the absorption, pharmacokinetics (PK) and pharmacodynamics (PD).
- 75. Eissing T, Kuepfer L, Becker C, et al. A computational systems biology software platform for multiscale modeling and simulation: integrating whole-body physiology, disease biology, and molecular reaction networks. Frontiers in physiology. 2011;2:4. Describes a modeling and simulation software platform PK-Sim which is capable of building and simulating models that integrate across biological scales: from the whole-body scale to the molecular level.
- 76. Jongeneelen FJ, Berge WFT. A generic, cross-chemical predictive PBTK model with multiple entry routes running as application in MS Excel; design of the model and comparison of predictions with experimental results. Annals of Occupational hygiene. 2011;55(8):841–864. • Describes development of a generic physiologically-based toxicokinetic (PBTK) model, IndusChemFate, which is suitable for ‘first tier assessments’ and for early explorations of the fate of chemicals and/or metabolites in the human body.
- 77.Pendse SN, Efremenko A, Hack CE, et al. Population Life-course exposure to health effects model (PLETHEM): An R package for PBPK modeling. Computational Toxicology. 2020. 2020/02/01/;13:100115. [Google Scholar]
- 78.Bell S, Abedini J, Ceger P, et al. An integrated chemical environment with tools for chemical safety testing. Toxicology in Vitro. 2020. 2020/09/01/;67:104916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berthold MR, Cebron N, Dill F, et al. KNIME-the Konstanz information miner: version 2.0 and beyond. AcM SIGKDD explorations Newsletter. 2009;11(1):26–31. [Google Scholar]
- 80. Linakis MW, Sayre RR, Pearce RG, et al. Development and Evaluation of a High Throughput Inhalation Model for Organic Chemicals Journal of Exposure Science & Environmental Epidemiology. 2020. • Generic gas inhalation physiologically based toxicokinetic model in the “httk” R software package, which was developed based on two previously published physiologically based models.
- 81. Sayre RR, Wambaugh JF, Grulke CM. Database of pharmacokinetic time-series data and parameters for 144 environmental chemicals. Sci Data. 2020. Apr 20;7(1):122. • This recent manuscript describes a public database schema for documenting TK concentration vs. time curves. In addition to providing standardized estimates of TK summary statistics such as half-life, these data also allow the evaluation of PBTK models. A template is provided for contributing to the database.
- 82.Shibata Y, Takahashi H, Chiba M, et al. Prediction of hepatic clearance and availability by cryopreserved human hepatocytes: an application of serum incubation method. Drug Metabolism and Disposition. 2002;30(8):892–896. [DOI] [PubMed] [Google Scholar]
- 83.Waters NJ, Jones R, Williams G, et al. Validation of a rapid equilibrium dialysis approach for the measurement of plasma protein binding. Journal of Pharmaceutical Sciences. 2008;97(10):4586–4595. [DOI] [PubMed] [Google Scholar]
- 84.Wilkinson GR, Shand DG. Commentary: a physiological approach to hepatic drug clearance. Clinical Pharmacology and Therapeutics. 1975;18(4):377–390. [DOI] [PubMed] [Google Scholar]
- 85.Hubatsch I, Ragnarsson EG, Artursson P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nature Protocols. 2007;2(9):2111. [DOI] [PubMed] [Google Scholar]
- 86. Mansouri K, Grulke CM, Judson RS, et al. OPERA models for predicting physicochemical properties and environmental fate endpoints. Journal of Cheminformatics. 2018;10(1):10. • The OPERA property models provide free and open source predictions of physico-chemical properties with well-defined domains of applicability. They are used by the R package “httk” for describing chemicals.
- 87.Ingle BL, Veber BC, Nichols JW, et al. Informing the Human Plasma Protein Binding of Environmental Chemicals by Machine Learning in the Pharmaceutical Space: Applicability Domain and Limits of Predictability. Journal of Chemical Information and Modeling. 2016;56(11):2243–2252. [DOI] [PubMed] [Google Scholar]
- 88.Yin Y, Chang D, Grulke C, et al. Essential Set of Molecular Descriptors for ADME Prediction in Drug and Environmental Chemical Space. Research. 2014. 08/22;1. [Google Scholar]
- 89.Vilar S, Cozza G, Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Current topics in medicinal chemistry. 2008;8(18):1555–1572. [DOI] [PubMed] [Google Scholar]
- 90.Schmitt W. General approach for the calculation of tissue to plasma partition coefficients. Toxicology in Vitro. 2008;22(2):457–467. [DOI] [PubMed] [Google Scholar]
- 91.Peyret T, Poulin P, Krishnan K. A unified algorithm for predicting partition coefficients for PBPK modeling of drugs and environmental chemicals. Toxicology and Applied Pharmacology. 2010;249(3):197–207. [DOI] [PubMed] [Google Scholar]
- 92. Metropolis N, Ulam S. The Monte Carlo method. J Am Stat Assoc. 1949. Sep;44(247):335–41. • Included for historical conext, this manuscript originally describes the Monte Carlo method as developed under the “Manhattan Project”.
- 93. Thomas RS, Lytle WE, Keefe TJ, et al. Incorporating Monte Carlo simulation into physiologically based pharmacokinetic models using advanced continuous simulation language (ACSL): a computational method. Toxicological Sciences. 1996;31(1):19–28. • As part of his Ph.D. work Russell Thomas first linked Monte Carlo simulation and PBPK.
- 94. Wambaugh JF, Wetmore BA, Ring CL, et al. Assessing Toxicokinetic Uncertainty and Variability in Risk Prioritization. Toxicol Sci. 2019. Dec 1;172(2):235–251. • This paper reported new toxicokinetic values for ToxCast chemicals, including a revised method, which resulted in a drop in measurement uncertainty. Using Monte Carlo simulations, measurement uncertainty and biological variability could be distinguished.
- 95.Hoffman FO, Hammonds JS. Propagation of uncertainty in risk assessments: the need to distinguish between uncertainty due to lack of knowledge and uncertainty due to variability. Risk analysis. 1994;14(5):707–712. [DOI] [PubMed] [Google Scholar]
- 96.Bois FY. Analysis of PBPK models for risk characterization. Annals of the New York Academy of Sciences. 1999;895(1):317–337. [DOI] [PubMed] [Google Scholar]
- 97.Kramer NI, Krismartina M, Rico-Rico A, et al. Quantifying processes determining the free concentration of phenanthrene in Basal cytotoxicity assays. Chem Res Toxicol. 2012. Feb 20;25(2):436–45. [DOI] [PubMed] [Google Scholar]
- 98.Henneberger L, Mühlenbrink M, Heinrich DJ, et al. Experimental Validation of Mass Balance Models for in Vitro Cell-Based Bioassays. Environmental Science & Technology. 2020. 2020/01/21;54(2):1120–1127. [DOI] [PubMed] [Google Scholar]
- 99.Groothuis FA, Heringa MB, Nicol B, et al. Dose metric considerations in in vitro assays to improve quantitative in vitro–in vivo dose extrapolations. Toxicology. 2015;332:30–40. [DOI] [PubMed] [Google Scholar]
- 100.Groothuis FA, Timmer N, Opsahl E, et al. Influence of in Vitro Assay Setup on the Apparent Cytotoxic Potency of Benzalkonium Chlorides. Chemical Research in Toxicology. 2019. 2019/06/17;32(6):1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fischer FC, Henneberger L, König M, et al. Modeling exposure in the Tox21 in vitro bioassays. Chemical Research in Toxicology. 2017;30(5):1197–1208. [DOI] [PubMed] [Google Scholar]
- 102.Evaluation Oreskes N. (not validation) of quantitative models. Environmental Health Perspectives. 1998;106(Suppl 6):1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wambaugh JF, Wetmore BA, Pearce R, et al. Toxicokinetic triage for environmental chemicals. Toxicological Sciences. 2015;147(1):55–67. • Literature review of HTTK models and generation of a HT PBTK concentration versus time model that identifies factors associated with predictive ability, allowing for categorization of HTTK predictive ability for chemicals.
- 104.Yoon M, Efremenko A, Blaauboer BJ, et al. Evaluation of simple in vitro to in vivo extrapolation approaches for environmental compounds. Toxicology in Vitro. 2014;28(2):164–170. [DOI] [PubMed] [Google Scholar]
- 105.Casey WM, Chang X, Allen DG, et al. Evaluation and Optimization of Pharmacokinetic Models for in vitro to in vivo Extrapolation of Estrogenic Activity for Environmental Chemicals. Environmental health perspectives. 2018;126(9):097001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kamiya Y, Otsuka S, Miura T, et al. Physiologically Based Pharmacokinetic Models Predicting Renal and Hepatic Concentrations of Industrial Chemicals after Virtual Oral Doses in Rats. Chemical Research in Toxicology. 2020. [DOI] [PubMed] [Google Scholar]
- 107.Zavala J, Freedman AN, Szilagyi JT, et al. New Approach Methods to Evaluate Health Risks of Air Pollutants: Critical Design Considerations for In Vitro Exposure Testing. International Journal of Environmental Research and Public Health. 2020;17(6):2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.National Research Council. Risk Assessment in the Federal Government: Managing the Process. Washington (DC): National Academies Press; 1983. [PubMed] [Google Scholar]
- 109.Hack C, Efremenko A, Pendse S, et al. Physiologically based pharmacokinetic modeling software. Physiologically Based Pharmacokinetic (PBPK) Modeling: Elsevier; 2020. p. 81–126. [Google Scholar]
- 110.Yoon M, Ring C, Van Landingham CB, et al. Assessing children’s exposure to manganese in drinking water using a PBPK model. Toxicol Appl Pharmacol. 2019. Oct 1;380:114695. [DOI] [PubMed] [Google Scholar]
- 111. Bois FY. GNU MCSim: Bayesian statistical inference for SBML-coded systems biology models. Bioinformatics. 2009. Jun 1;25(11):1453–4. • MCSim provides a free, open source language for describing a dynamic model (such as a PBPK model), converting that model to faster compiled code, and analyzing the compiled model with Markov Chain Monte Carlo methods for Bayesian inference. The models for R package “httk” are all developed first in MCSim.
- 112. Sipes NS, Wambaugh JF, Pearce R, et al. An Intuitive Approach for Predicting Potential Human Health Risk with the Tox21 10k Library. Environmental Science & Technology. 2017. 2017/09/19;51(18):10786–10796. • This publication used Simulations Plus to predict in vitro TK parameters for the entire Tox21 library. These predictions are available within httk via the command load_sipes2017().
- 113.Pradeep P, Patlewicz G, Pearce R, et al. Using chemical structure information to develop predictive models for in vitro toxicokinetic parameters to inform high-throughput risk-assessment. Computational Toxicology. 2020. 2020/11/01/;16:100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yun YE, Tornero-Velez R, Purucker ST, et al. Evaluation of Quantitative Structure Property Relationship Algorithms for Predicting Plasma Protein Binding in Humans. Computational Toxicology. 2020:100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kirman CR, Aylward LL, Wetmore BA, et al. Quantitative property–property relationship for screening-level prediction of intrinsic clearance: a tool for exposure modeling for high-throughput toxicity screening data. Applied In Vitro Toxicology. 2015;1(2):140–146. [Google Scholar]
- 116.Cohen Hubal EA, Richard A, Aylward L, et al. Advancing exposure characterization for chemical evaluation and risk assessment. Journal of Toxicology and Environmental Health, Part B. 2010;13(2–4):299–313. [DOI] [PubMed] [Google Scholar]
- 117.Williams AJ, Grulke CM, Edwards J, et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. Journal of Cheminformatics. 2017;9(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Paul Friedman K, Gagne M, Loo L-H, et al. Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Toxicological Sciences. 2020;173(1):202–225. • This case study demonstrated the suitability of IVIVE using ToxCast and R package httk for regulatory decision making as part of the Accelerating the Pace of Risk Assessment consortium (Kavlock et al, 2018).
- 119.Ankley GT, Bennett RS, Erickson RJ, et al. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environmental Toxicology and Chemistry. 2010;29(3):730–741. [DOI] [PubMed] [Google Scholar]