Abstract
Conditional promoters allowing both induction and silencing of gene expression are indispensable for basic and applied research. The xylP promoter (pxylP) from Penicillium chrysogenum was demonstrated to function in various mold species including Aspergillus fumigatus. pxylP allows high induction by xylan or its degradation product xylose with low basal activity in the absence of an inducer. Here we structurally characterized and engineered pxylP in A. fumigatus to optimize its application. Mutational analysis demonstrated the importance of the putative TATA-box and a pyrimidine-rich region in the core promoter, both copies of a largely duplicated 91-bp sequence (91bpDS), as well as putative binding sites for the transcription factor XlnR and a GATA motif within the 91bpDS. In agreement, pxylP activity was found to depend on XlnR, while glucose repression appeared to be indirect. Truncation of the originally used 1643-bp promoter fragment to 725 bp largely preserved the promoter activity and the regulatory pattern. Integration of a third 91bpDS significantly increased promoter activity particularly under low inducer concentrations. Truncation of pxylP to 199 bp demonstrated that the upstream region including the 91bpDSs mediates not only inducer-dependent activation but also repression in the absence of inducer. Remarkably, the 1579-bp pxylP was found to act bi-bidirectionally with a similar regulatory pattern by driving expression of the upstream-located arabinofuranosidase gene. The latter opens the possibility of dual bidirectional use of pxylP. Comparison with a doxycycline-inducible TetOn system revealed a significantly higher dynamic range of pxylP. Taken together, this study identified functional elements of pxylP and opened new methodological opportunities for its application.
Keywords: Fungi, Aspergillus, xylP, Xylose, Promoter, XlnR
Abbreviations: AMM, Aspergillusminimal medium; 91bpDS, Duplicated 91bp sequence; Fru, Fructose; Glc, Glucose; pxylP, Xylanase promoter; Wt, Wild type; Xyl, Xylose
Highlights
-
•
Mutational analysis identified activating and repressing xylP promoter (pxylP) elements.
-
•
Reporter analysis revealed bidirectional activity of pxylP.
-
•
Promoter engineering improved inducer responsiveness of pxylP.
-
•
Comparison with the TetOn promoter indicated superior dynamic range of pxylP.
1. Introduction
Filamentous fungi are ubiquitously found in nature and are capable of adapting to diverse environments. As they are faced with numerous stressors, fungi represent an important source of natural products exhibiting a wide range of biological activities (Bills and Gloer 2016; Romsdahl and Wang 2019; Keller 2019). The majority of the microbial natural products currently reported in the Natural Products Atlas are derived from fungi (Sorokina et al., 2021). Several of these have been developed into applied drugs with high importance for human health including the first broad-spectrum antibiotic penicillin, hypolipidemic lovastatin and the immunosuppressants cyclosporine and mycophenolic acid (Bills and Gloer 2016; Skellam 2019; Keller 2019). Moreover, filamentous fungi such as Aspergillus niger serve as multipurpose cell factory for production of primary metabolites such as citric acid or heterologous proteins (Cairns et al., 2021). For both basic and applied research of these processes, conditional promoters that allow induction and alternatively silencing of gene expression are indispensable, e.g., to functionally characterize essential genes or to manipulate metabolism by modulating gene expression. The xylP promoter (pxylP) controlling expression of a xylanase from Penicillium chrysogenum allows high induction by xylan or its degradation product xylose with low basal activity in the absence of an inducer (Haas et al., 1993; Zadra et al., 2000). pxylP was demonstrated to permit conditional gene expression of diverse genes in various mold species including P. chrysogenum (Bugeja et al., 2010, 2013; Huber et al., 2019; Janus et al., 2009; Kopke et al., 2013; Pongsunk et al., 2005; Sigl et al. 2010, 2011), Penicillium marneffei (Bugeja et al., 2010, 2013; Pongsunk et al., 2005), Aspergillus nidulans (Monahan et al., 2006; Wong et al., 2007; Wong et al. 2008; Wong et al. 2009; Tribus et al., 2010; Ma et al., 2018; Pidroni et al., 2018; Wang et al., 2021; Li et al., 2021; Unkles et al., 2014), Aspergillus fumigatus (Yasmin et al., 2012; Gsaller et al., 2012; Fazius et al., 2012; Altwasser et al., 2015; Baldin et al. 2015, 2021; Vaknin et al., 2016; Misslinger et al., 2019; Bauer et al., 2019; López-Berges et al., 2021; Handelman et al., 2021; Fabri et al., 2021) and Sordaria macrospora (Kopke et al., 2010). A. fumigatus is a ubiquitous saprobic fungus but at the same time the most common mold pathogen of humans. This is one of the reasons why it has become an intensively studied model organism (Latgé and Chamilos 2019). Recently pxylP was demonstrated to even allow control of in vivo gene expression of A. fumigatus during murine infection (Bauer et al., 2019). In this invasive aspergillosis model the inducer xylose was supplemented in the drinking water of mice. Consequently, pxylP can serve as an alternative to TetOn promoter systems (Helmschrott et al., 2013).
The aim of this study was the characterization of functional elements of pxylP in A. fumigatus by truncations, deletions and mutagenesis to allow optimization of its application using genes encoding the yellow fluorescent protein mVenus (Kremers et al., 2006) and firefly luciferase (Galiger et al., 2013) as reporters for promoter activity. To exclude genomic position effects, we employed selection marker-free integration of all constructs in single copy at the fcyB locus (Birštonas et al., 2020).
2. Materials and methods
2.1. Fungal strains and growth conditions
All A. fumigatus strains in this study were generated in A. fumigatus AfS77, which is a derivative of the clinical isolate A. fumigatus ATCC 46645 (Hearn and Mackenzie 1980) lacking the akuA gene to impair non-homologous end joining (Krappmann et al., 2006; Carvalho et al., 2010). For spore production the strains were grown at 37 °C on Aspergillus complex medium (2% (w/v) glucose, 0.2% (w/v) peptone, 0.1% (w/v) yeast extract, 0.1% (w/v) casamino acids, salt solution and iron-free trace elements according to (Pontecorvo et al., 1953).
Plate growth assays were performed by point inoculating 1 x 103 conidia on solid Aspergillus minimal medium (AMM) according to (Pontecorvo et al., 1953). The used carbon source is described in the respective experiment. Xylan from oats spelts (SERVA) was used for the characterization of ΔxlnR mutants. If not described otherwise, 20 mM glutamine was used as nitrogen source. The plates were incubated for 48 h at 37 °C. All the strains used in this study are listed in Table 1.
Table 1.
Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| ATCC 46645 | Clinical strain isolated from a human infection (England) | Hearn and Mackenzie (1980) |
| AfS77 | ATCC46645, ΔakuA::loxP | (Hartmann et al., 2010; Krappmann et al., 2006) |
| ΔxlnR | AfS77; ΔxlnR::hph | This study |
| IG01V, IG02V, | Afs77, ΔfcyB∷pxylP versions driving expression of mVenus | This study |
| IG03V, IG04V, | ||
| IG05V, IG06V, | ||
| IG13V, IG15V, | ||
| IG16V, IG17V, | ||
| IG20V, IG21V, | ||
| IG22V, IG23V | ||
| IG01L, IG04L, | Afs77, ΔfcyB∷pxylP versions driving expression of luciferase | This study |
| IG06L, IG07L, | ||
| IG24L, IG25L | ||
| IG03L*, IG04L*, IG06L*, IG07L* IG24L* | Afs77, ΔxlnR::hph; ΔfcyB∷pxylP versions driving expression of luciferase | This study |
| TetOnoliC | Afs77, ΔfcyB∷tetOnolic version driving expression of luciferase | This study |
2.2. Generation of A. fumigatus mutant strains
Oligonucleotides used in this study to introduce the desired genetic manipulation are listed in Table S1. The plasmids containing the pxylP truncations and mutations in the reporter constructs were integrated into the fcyB locus of A. fumigatus, allowing selection for 5-flucytosine resistance without the need of another selection marker (Birštonas et al., 2020). To generate plasmid pIG01V, four DNA fragments were amplified with oligonucleotides shown in Table S1: (i) a plasmid backbone including fcyB flanking non-coding regions (NCR) amplified from template pfcyB (Birštonas et al., 2020), (ii) pxylP, amplified from template pMMHL15 (Misslinger et al., 2018), (iii) codon optimized yellow fluorescent protein derivative Venus encoding sequence (Gsaller et al., 2014) with mutations I152L and A206K to turn Venus into mVenus, and (iv) the trpC-terminator sequence, amplified from PgpdA_LacZ_AtTrpCTerm_pJET1.2 (Gressler et al., 2011). All fragments were subsequently assembled using NEBuilder® HiFi DNA Assembly (New England Biolabs). For pIG15V, pxylP was amplified from pIG01V and integrated in reverse-complementary direction in pIG01V backbone using NEBuilder. Plasmids pIG02V, pIG03V, pIG04V, pIG05V, pIG06V, pIG13V, pIG16V, pIG17V, pIG20V, pIG21V, pIG22V and pIG23V were generate from pIG01V via site directed mutagenesis using Q5® Site-Directed Mutagenesis Kit (New England Biolabs) and primers shown in Table S1. Mutations and deletions in the respective plasmids are shown in Supplementary Figure S1.
To generate the plasmid pIG01L with firefly luciferase as the reporter gene, four fragments were amplified using oligonucleotides shown in Table S1: (i) a plasmid backbone including the fcyB NCR, amplified from template pfcyB (Birštonas et al., 2020), (ii) pxylP fragments, amplified from template pIG01V, (iii) codon optimized Photinus pyralis luciferase gene (GenBank accession numberKC677695) (Galiger et al., 2013), and (iv) the trpC-terminator sequence amplified from pIG01V. All fragments were subsequently assembled using NEBuilder® HiFi DNA Assembly (New England Biolabs). Plasmids pIG03L, pIG04L, pIG06L, and pIG07L were generated from pIG01L via site directed mutagenesis using Q5® Site-Directed Mutagenesis Kit (New England Biolabs) and primers as shown in Table S1.
To generate pIG24L with both putative XlnR sites mutated, a fragment was generated by PCR using pIG17V as template. The fragment was assembled with the previously amplified plasmid backbone including the fcyB NCRs, luciferase and trpC terminator gene using NEBuilder® HiFi DNA Assembly (New England Biolabs).
To generate pIG25L containing a third 91bpDS (see below), a synthetic 107 bp DNA fragment with 70% similarity to the other two 91bpDS (generated by Integrated DNA Technologies, Inc, Iowa, USA) was assembled with the previously amplified plasmid backbone including fcyB sites, the pxylP fragment amplified from template pIG03V, and the previously amplified luciferase and trpC terminator using NEBuilder® HiFi DNA Assembly (New England Biolabs). In the third 91bpDS, non-conserved nucleotides between the two original 91bpDS were exchanged to other nucleotides (Supplementary Figure S1B) to avoid homologous recombination with the original 91bpDS due to sequence identity.
To generate the plasmid TetOnoliC with firefly luciferase as the reporter gene, four fragments were amplified using oligonucleotides shown in Table S1: (i) a plasmid backbone including the fcyB NCR, amplified from template pfcyB (Birštonas et al., 2020), (ii), the tetOnoliC fragment amplified from template pJW128 (Neubauer et al., 2015), (iii) codon optimized Photinus pyralis luciferase gene (GenBank accession numberKC677695) (Galiger et al., 2013), and (iv) the trpC-terminator sequence amplified from pIG01V. All fragments were subsequently assembled using NEBuilder® HiFi DNA Assembly (New England Biolabs).
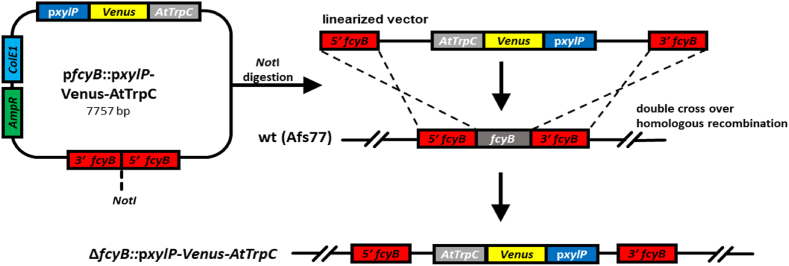
After NotI or PmeI (TetOnoliC) digestion-mediated linearization, all reporter constructs were integrated in single copy at the fcyB locus (Fig. 1), which allows selection for resistance to 5-flucytosine and evades the necessity for a heterologous selection marker (Birštonas et al., 2020).
Fig. 1.
Scheme of genomic insertion of the reporter gene cassettes in A. fumigatus by homologous recombination. The generated plasmids were linearized with NotI before transformation. The 3′- and 5′-fcyB NCR allowed an exchange of the original fcyB gene with the plasmid sequence. For simplification, the plasmid regions are not shown in the genomic integration.
The XlnR lacking mutant was generated by replacement of the xlnR coding region by the hygromycin resistance cassette (hph) via homologous recombination according to (Fraczek et al., 2013). Therefore, the 5′-NCR of xlnR, the hygromycin resistance cassette (hph) and the 3′-NCR of xlnR were amplified by PCR with oligonucleotides mentioned in Table S1, using genomic DNA as a template for the NCRs and the plasmid pMMHL69 for hph. Subsequently, the three fragments were then linked together via fusion PCR using the nested primers.
Plasmids and PCR products were purified (Monarch PCR and DNA Cleanup Kit, New England Biolabs) and used for transformation in A. fumigatus. The transformation of A. fumigatus AfS77 was performed according to (Tilburn et al., 1983). Selection of transformants was carried out on AMM plates with 0.2 mg/mL hygromycin B, or 10 μg/mL flucytosine (TCI©, Eschborn, Germany). Correct genetic manipulations were proven by Southern blot analysis (Supplementary Figures S2–S5) and growth assays.
2.3. In vivo determination of promoter activity
Promoter activities were assessed by measuring mVenus fluorescence intensity in 96-well microtiter plates (Nunc™). Each well contained 0.1 ml AMM inoculated with 104 spores. Plates were incubated for 18 h at 37 °C. Subsequently, absorbance and fluorescence signals were quantified using a CLARIOstar Plus® microplate reader (BMG LABTECH). Absorbance was measured at 280 nm, spiral well scan. For detection of mVenus fluorescence, 497-20 nm excitation and 540-20 nm emission was used. Each reporter strain was analyzed in biological triplicates followed by subtraction of background fluorescence recorded from untransformed recipient strain (wt).
For measuring the bioluminescence of luciferase reporter strains, LUMITRAC 96-well plates (Greiner Bio-ONE) were used. Spores were inoculated in AMM to obtain a final concentration of 1.5 × 104 spores in 0.1 ml. After incubation for 18 h at 37 °C, 20 μL of 0.6 mM D-luciferin (Synchem UG & Co.KG, Felsberg/Altenburg, Germany) in PBS was added. The bioluminescence was recorded at 580-80 nm using spiral well scan employing a CLARIOstar Plus® microplate reader (BMG LABTECH). Each reporter strain was analyzed in three biological triplicates followed by subtraction of background luminescence recorded from untransformed wt strain.
3. Results
3.1. In silico analysis reveals several putative regulatory sequences in pxylP
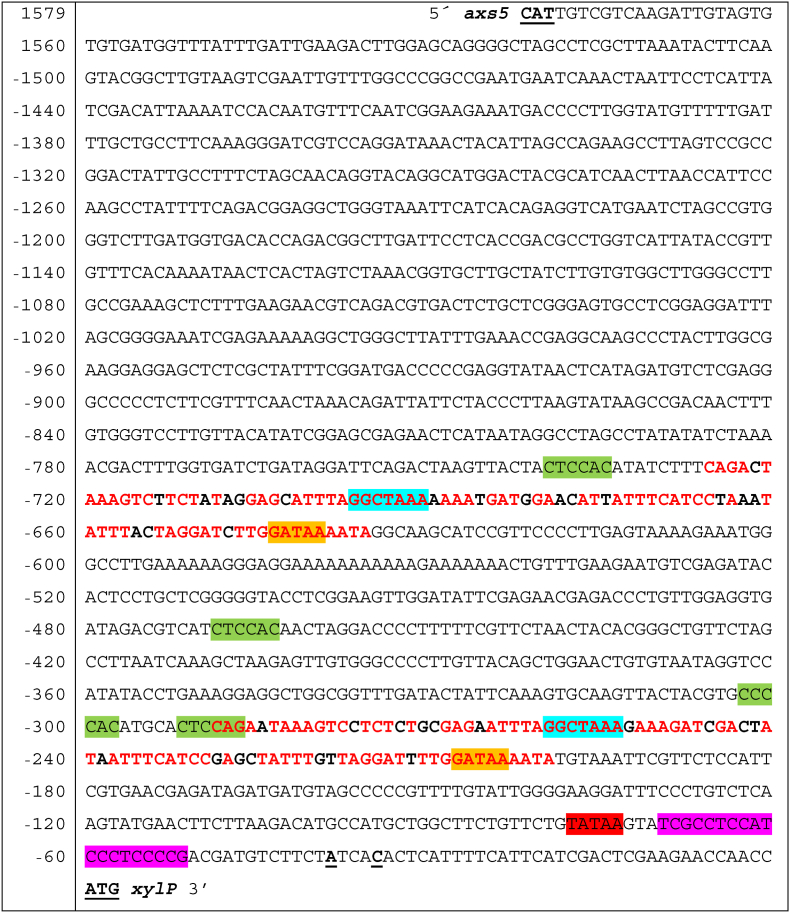
The pxylP nucleotide sequence is displayed in Fig. 2 (Zadra et al., 2000). A blastx search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) indicated that the 5′-upstream 64 bp of the originally used 1643-bp pxylP sequence encodes the N-terminus of the alpha-L-arabinofuranosidase Axs5 (Sakamoto et al., 2011), which suggests that this promoter functions bi-directionally. For the initial use of pxylP about 20 years ago, the translation start region 5′-caacATGa-3’ (translation start codon in capital letters) was mutated to 5′-caaccATGg-3’ (NcoI recognition site underlined) to allow digestion with the restriction enzyme NcoI because recombinant cloning strategies resided on restriction enzyme-mediated genetic engineering at that time. This pxylP variation was kept throughout the current study. As previously reported (Zadra et al., 2000) and shown in Fig. 2, pxylP contains a largely duplicated 91-bp sequence, here termed 91bpDS, displaying 80% sequence identity. The duplication of this sequence might indicate that it contains important regulatory sequences. Both 91bpDS copies contain a perfectly conserved putative XlnR binding motif and a perfectly conserved GATAA motif. The xylose-induced Gal4-type transcription factor XlnR has been shown to act as transcriptional activator of the xylanolytic system in A. niger, A. oryzae and A. nidulans (van Peij et al., 1998; Tamayo et al., 2008; de Souza et al., 2013; Kowalczyk et al., 2014). GATAA motifs are recognized by so called GATA-type transcription factors. Aspergillus species possess six GATA-type transcription factors termed AreA, AreB, SreA, LreA, LreB and NsdD. AreA and AreB mediate regulation of nitrogen and carbon metabolism (Chudzicka-Ormaniec et al., 2019; Haas et al., 1997), SreA controls iron acquisition (Oberegger et al., 2001; Schrettl et al., 2008), LreA and LreB allow light response (Purschwitz et al., 2008) and NsdD coordinates sexual and asexual development (Lee et al., 2014). Moreover, pxylP contains five putative CreA binding sites outside of the 91bpDS copies. The Cys2His2-type transcription factor CreA mediates carbon catabolite repression, which ensures that genes for the degradation of less preferred carbohydrates such as xylan are turned off in the presence of favorable carbon sources like glucose for economization (Kowalczyk et al., 2014). A putative TATA box is found 34 bp upstream of the most distal transcription start site identified (Haas et al., 1993); this motif might be recognized by the TATA-binding protein (TBP), which is the most conserved general transcription initiation factor (Kramm et al., 2019). Between the putative TATA box and the transcription start site, a pyrimidine-rich sequence is present. Transcriptome analysis in A. nidulans revealed an enrichment in pyrimidines immediately upstream of the first transcription start site (Haas et al., 1993; Sibthorp et al., 2013), which might be important to determine the transcription start site and the efficiency of transcription (Kinghorn and Turner 1992; Ballance 1986).
Fig. 2.
Nucleotide sequence of the bidirectional promoter driving xylP and axs5. The translation start sites of xylP and axs5 and the transcription start sites of xylP are in bold and underlined. The duplicated 91bpDS region is shown in bold letters with conserved nt in red. Putative functional sequences are shadded: 5′-TATAA-3′ in red, pyrimidine-rich sequence 5′-TCGCCTCCATCCCTCCCCG-3′ in pink, 5′-GATAA-3′ within 91bpDS in orange, putative XlnR binding sites (5′-GGCTAAA-3′) in blue, and CreA motifs (5′-SYGGRG-3′) in green. nt numbering refers to the xylP translation start codon. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Promoter studies employing mVenus as reporter enabled identification of functional sequences in pxylP
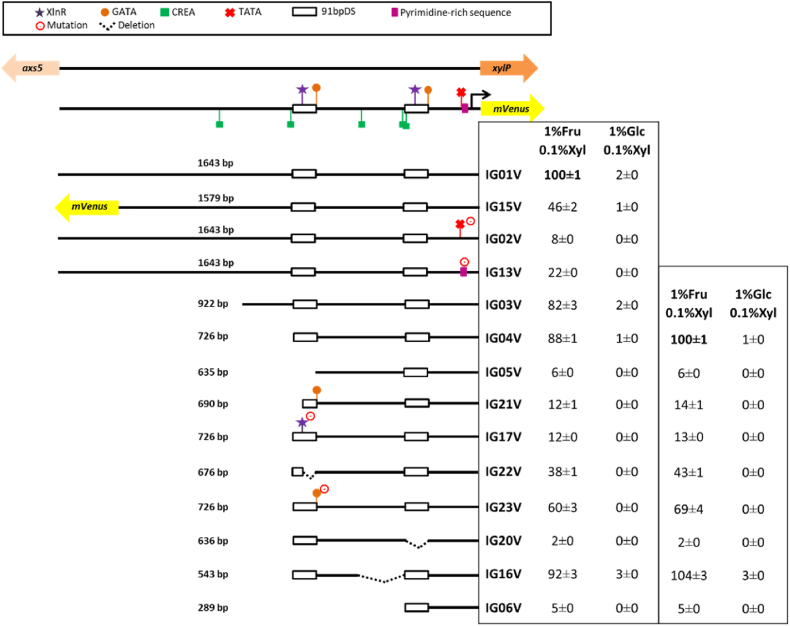
In order to identify functional elements in pxylP, the originally used promoter fragment and 13 versions with truncations, deletions or mutations were fused to the gene encoding yellow fluorescent protein mVenus (Kremers et al., 2006; Kodama and Hu 2010) as reporter for promoter activity (Fig. 3). To exclude influences from the integration locus in the A. fumigatus genome, all constructs were integrated in single copy at the fcyB locus (Fig. 1), which allows selection for resistance to 5-flucytosine and evades the necessity for a heterologous selection marker (Birštonas et al., 2020). For in vivo quantification of promoter activity, fungal strains were grown in minimal medium with different carbon sources in 96-well microtiter plates for 18 h at 37 °C (Table 2). As previously reported (Zadra et al., 2000), the originally used 1643-bp pxylP (IG01V) showed high activity with 1% xylose (1%Xyl) as carbon source, while no activity was detectable with 1% glucose (1%Glc) or 1% fructose (1%Fru) as sole carbon sources. Combination of 1%Glc with 0.1, 0.5 or 1%Xyl increased the promoter activity to 2, 21 and 33%, respectively. In 1%Fru combined with 0.1%Xyl (1%Fru/0.1%Xyl) pxylP activity reached 91% of that with 1%Xyl. Taken together, these data underline the repressive effect of glucose on pxylP activity in the presence of the inducer xylose and define fructose as a non-inducing and largely non-repressing carbon source. Therefore, in the following assays promoter activity of the pxylP versions was analyzed in 1%Fru/0.1%Xyl (non-repressing/inducing condition), 1%Glc/0.1%Xyl (repressing/inducing condition) and 1%Glc (repressing/non-inducing condition) to ensure similar availability of carbon source and inducer.
Fig. 3.
Truncation, deletion and mutation studies identified functional sequences in pxylP using mVenus as reporter for promoter activity. Promoter activity was measured as described in Material and Methods. Shown values are the mean ± STD of biological triplicates normalized to either IG01V (left) or IG04V (right) grown with 1%Fru/0.1%Xyl; original data are shown in Supplementary Table S2B. Strains were grown for 18 h at 37 °C. Values of growth with 1%Glc as carbon source are not shown as no promoter activity was detected under this condition for any of the promoter constructs.
Table 2.
Xylose-mediated induction of pxylP is repressed by glucose but not fructose. Promoter activity of IG01V was measured as described in Material and Methods. Shown values are the mean of biological triplicates normalized to 1%Xyl ± STD; original data are shown in Supplementary Table S2A.
| Carbon source |
|||||||
|---|---|---|---|---|---|---|---|
| 1%Glc | 1%Glc 0.1%Xyl |
1%Glc 0.5%Xyl |
1%Glc 1%Xyl |
1%Xyl | 1%Fru | 1%Fru 0.1%Xyl |
|
| Promoter activity [%] | 0 ± 0 | 2 ± 0 | 21 ± 1 | 33 ± 0 | 100 ± 9 | 0 ± 0 | 91 ± 2 |
To simplify comparison, the activity of all promoter versions was normalized to that of IG01V or IG04V, respectively (Fig. 3). The inverted 1579-bp pxylP construct driving expression of axs5 (IG15V) showed 46% of the activity of IG01V with 1%Fru/0.1%Xyl, which underlines that pxylP indeed functions bidirectionally and indicates that the arabinofuranosidase-encoding axs5 gene shows lower expression compared to the xylanase-encoding xylP gene. Mutation of the 5′-TATAAG-3′ sequence 34 bp upstream of the most distal transcription start site to 5′-GGATCC-3′ caused a dramatic drop of promoter activity to 8% under non-repressing/inducing conditions indicating that this sequence might indeed be the TATA box. Replacement of the pyrimidine-rich sequence 5′-TCGCCTCCATCCCTCCCCG-3′ downstream of the putative TATA box by the Tet operator sequence 5′-TCCCTATCAGTGATAGAGA-3’ (Helmschrott et al., 2013) in IG13V, which reduces the pyrimidine content in this region from 84% to 47%, decreased the promoter activity to 22%. Truncation of pxylP to 922 bp in IG03V or to 725 in IG04V, which eliminates one and two of the putative CreA sites, respectively, retained 82% and 88% of the promoter activity in 1%Fru/0.1%Xyl. These data indicate that the major regulatory elements are contained within the 725-bp pxylP fragment but that the upstream region contains further activating elements.
As all further promoter manipulations were conducted in IG04V, their promoter activity was normalized to that IG04V (Fig. 3). Truncation of the distal 91bpDS containing a putative XlnR recognition motif and a 5′-GATAA-3′motif in IG05V caused a dramatic decrease of activity to 6% in 1%Fru/0.1%Xyl, which emphasizes the importance of this duplicated region. Elimination of the putative XlnR motif in the distal 91bpDS by either truncation of pxylP to 690 bp (about the 5′-half of 91bpDS) in IG21V or replacement of the putative XlnR-recognition motif 5′-GGCTAAA-3′ by 5′-CATTAAA-3′ in IG17V decreased the promoter activity to 14% and 13%, respectively, compared to that of IG04V. These results strongly indicated the importance of XlnR for activation of pxylP. The higher activity of these promoter versions compared to lack of the entire distal 91bpDS in IG05V (14%/13% versus 6%) indicated additional regulatory elements in the distal 91bpDS element. In agreement, deletion of the 3′-half of the distal 91bpDS in IG22V, which conserves the XlnR binding motif, decreased the promoter activity to 43% compared to IG04V. Furthermore, mutation of the 5′-GGATAA-3′ sequence to 5′-GTCGAA-3′ in IG23V reduced the promoter activity to 69% compared to IG04V, which indicates a role of a GATA-factor in activation of pxylP. Comparison of promoter activity of IG22V and IG23V (43% versus 69% compared to IG04V) might indicate additional regulatory elements apart from the identified GATAA motif in the 3′-half of the distal 91bpDS. Deletion of the proximal 91bpDS in IG20V decreased the promoter activity to 2% compared to IG04V. Together with the decreased promoter activity caused by deletion of the distal 91bpDS in IG05V, these data indicate that both copies of 91bpDS are required for full activity of pxylP.
Notably, repression by glucose during growth with 1%Glc/0.1%Xyl was largely retained in all investigated promoter versions, even in IG16V, which lacks all predicted CreA motifs (Fig. 3). These results might indicate that pxylP is not subject to direct carbon catabolite repression. Moreover, neither of the analyzed pxylP versions displayed any detectable promoter activity under repressing/non-inducing conditions, i.e., with 1%Glc (data not shown).
3.3. Identification of A. fumigatus XlnR
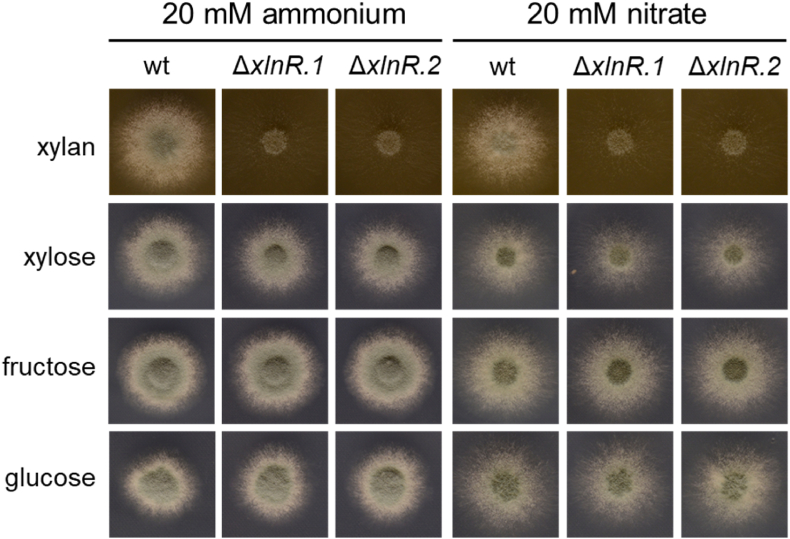
To further investigate the potential role of XlnR in pxylP regulation, we aimed to identify A. fumigatus XlnR. Therefore, a blastp search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with A. nidulans XlnR (Q5AVS0, Tamayo et al., 2008) was conducted, which identified Afu2g15620 as the most likely A. fumigatus homologue (73% identity over a length of 962 amino acids). Vice versa, a blastp search using Afu2g15620 indicated that A. nidulans XlnR, A. niger XlnR (A2R5W7; 77% identity over a length of 896 amino acids; van Peij et al., 1998) and Hypocrea jecurina (anamorph Trichoderma reesei) XynR (XP_006966092; 52% identity over a length of 962 amino acids; Rauscher et al., 2006) as the proteins with the highest similarity. These data strongly suggested that Afu2g15620 is indeed the A. fumigatus XlnR homologue and the encoded protein gene was therefore termed XlnR. To confirm its function, we generated a respective gene deletion mutant (ΔxlnR) in A. fumigatus Afs77 (termed wt here) by replacement of the XlnR coding sequence with the hph selection marker gene. In line with A. fumigatus XlnR functioning as xylanolytic regulator, two independently generated ΔxlnR mutants displayed negligible growth on xylan, slightly reduced growth on xylose but wt-like growth on glucose and fructose as carbon source (Fig. 4). This growth pattern matches that of A. nidulans ΔxlnR mutants (Tamayo et al., 2008). This mutant now allowed to directly test its role in regulation of pxylP (see below).
Fig. 4.
Deletion of the XlnR-encoding gene causes a strong growth defect of A. fumigatus on xylan, a slight growth defect on xylose but no growth defect on glucose or fructose as carbon source. 103A. fumigatus conidia were point-inoculated on solid AMM containing 1% of the indicated carbon source and either 20 mM ammonium or 20 mM nitrate as nitrogen source. The plates were incubated for 48 h at 37 °C.
3.4. Luciferase-mediated reporter assays elucidated the role of XlnR and in pxylP regulation
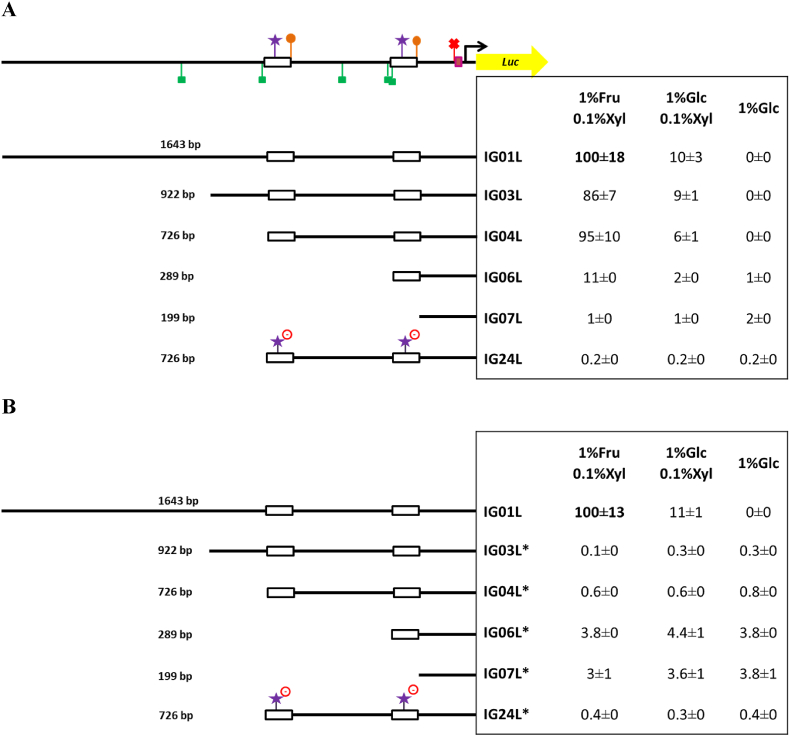
In vivo mVenus-mediated fluorescence measurements proved useful to analyze the pxylP activity under non-repressing/inducing conditions such as with 1%Fru/0.1%Xyl (Fig. 3). However, under conditions of low pxylP activity such as under repressing/inducing (1%Glc/0.1%Xyl) or repressing/non-inducing conditions (1%Glc), this methodology provided too low sensitivity, most likely due to the auto-fluorescence of hyphae. To increase sensitivity of the promoter analysis we tried luciferase instead of mVenus as reporter. Therefore, six pxylP versions including promoter truncations or mutations were fused to the codon optimized Photinus pyralis luciferase gene (GenBank accession numberKC677695) (Galiger et al., 2013) as reporter for promoter activity. All constructs were integrated in single copy at the fcyB locus as described above. For in vivo quantification of promoter activity, fungal strains were grown in minimal medium for 18 h at 37 °C and the promoter activity was determined as described in Materials and Methods.
To simplify comparison, the nomenclature of the promoter versions was kept the same with L (for luciferase) instead of V (for Venus) as suffix; the activity of all promoter versions was normalized to that of IG01L (Fig. 5A), which contains the original 1643-bp pxylP promotor and corresponds to the IG01V-Venus reporter construct. As found above using mVenus as reporter (Fig. 3), the truncated promoter versions IG03L and IG04L retained high promoter activity with 1%Fru/0.1%Xyl, displayed significant activity with 1%Glc/0.1%Xyl and lacked activity with 1%Glc (Fig. 5A). Interestingly, IG04L showed slightly higher activity than IG03L with 1%Fru/0.1%Xyl (Fig. 5A), which contrasts the Venus-reporter data (Fig. 3), and the activity with 1%Glc/0.1%Xyl was about 3-fold higher compared to the Venus-reporter experiments, which underlines the higher sensitivity of the luciferase-reporter system. Similar to the Venus-reporter experiments (IG06V, Fig. 3), truncation of the region upstream of the proximal 91bpDS in IG06L caused significant reduction of promoter activity but retained slight xylose-inducibility (Fig. 5A). Interestingly, this promoter version showed low promoter activity (1.0%) also with 1%Glc. Further truncation to 199 bp in IG07L, which eliminates both 91bpDS, resulted in low (1–2%) inducer-independent constitutive promoter activity (Fig. 5A). Taken together, the promoter activities mediated by IG06L and IG07L compared to IG04L, indicate that the promoter region between −726 bp (IG04L) and −199 bp (IG07L) contains not only all the major regulatory elements required for inducer-mediated activation of pxylP but also mediate repression in the absence of the inducer. Notably, mutation of both putative XlnR-recognition motifs (5′-GGCTAAA-3′ to 5′-CATTAAA-3′) in the 91bpDS of IG04L, leading to IG24L, abrogated xylose-induced promoter activity and caused weak constitutive promoter activity (0.2–0.4%) even with 1%Glc (Fig. 5A). These data indicate that these motifs, and consequently most likely XlnR, are essential not only for xylose-induced promoter activation but are also involved in full repression of pxylP under repressing/non-inducing conditions. The about 5.5-fold reduced promoter activity of IG06L in 1%Glc/0.1%Xyl compared to 1% Fru/0.1%Xyl (Fig. 5A) indicates that glucose repression is still functional in the absence of all CreA motifs and suggests that glucose repression of xylP is at least partially indirect.
Fig. 5.
Luciferase as reporter enabled to analyze effects of truncations and mutations on pxylP activity under conditions of low promoter activity in wt (A) and the ΔxlnR mutant strain (B). Promoter activity was measured as described in Material and Methods. Shown values are the mean ± STD of three biological replicates normalized to IG01L grown with 1%Fru/0.1%Xyl for 18 h at 37 °C. Before normalization, background wt values were subtracted. Promoter activity of promoter versions marked with * in B was determined in the ΔxlnR mutant strain. Original data are shown in Supplementary Table S3A.
To investigate the role of XlnR in regulation of pxylP, promoter activity of five pxylP versions was analyzed in the ΔxlnR strain. Lack of XlnR resulted in weak and largely constitutive promoter activity of all five versions, IG03, IG04, IG06, IG07 and IG24 (Fig. 5B). In other words, lack of XlnR eliminated xylose-induced promoter activity in IG03L, IG04L and IG06L, which proves that XlnR is indeed responsible for xylose-induced promoter activation. Compared to wt (IG03L, IG04L; Fig. 5A), lack of XlnR slightly increased (0.3%–0.8%) promoter activity of IG03L, IG04L also in 1%Glc (Fig. 5B), which indicates that XlnR is also involved in repression in the absence of inducer. Similar to analysis in the wt (Fig. 5A), IG06L and IG07L showed higher promoter activity than IG03L and IG04L in 1%Glc in ΔxlnR (Fig. 5B). These data indicate that the region upstream of the proximal 91bpDS is involved also in XlnR-independent repression of pxylP. Promotor version IG24L displayed similar low and largely constitutive promoter activity in both wt (Fig. 5A) and ΔxlnR (Fig. 5B) underlining that XlnR operates via the mutated XlnR consensus sequences that are mutated in these promoter versions.
Taken together, luciferase as reporter provided higher sensitivity of the promoter studies compared to mVenus. The luciferase-mediated promoter studies demonstrated that XlnR is responsible for xylose-mediated induction of xylP via the identified consensus binding motifs present in the two 91bpDS, that XlnR might also act negatively under non-inducing conditions, and that the region upstream of the proximal 91bpDS (−286 bp, IG06) is not only important for inducer-mediated activation but also for XlnR-independent repression in the absence of inducer.
Due to the identified importance of the two 91bpDS in transcriptional control of xylP, we investigated the effect of insertion of a third 91bpDS upstream of upstream of pIG03L, resulting in pIG25L (Supplementary Figure S1B). This genetic engineering increased promoter activity particularly during low inducing/repressing conditions, e.g., 5-fold with 1%Glc/0.05%Xyl and 2.8-fold with 1%Glc/0.1%Xyl (Table 3). Moreover, this manipulation increased maximal promoter activity in 1%Fru/0.1%Xyl about 1.3-fold without impacting repression in 1%Glc (Table 3).
Table 3.
Integration of a third 91bpDS in IG03L resulting in IG25L increases promoter activity during inducing/repressing and inducing non-repressing conditions. Promoter activity was measured as described in Material and Methods. The shown values are the mean ± STD of three biological replicates normalized to IG03L grown with 1%Fr/0.1%Xyl after growth for 18h at 37 °C. Original data are shown in Supplementary Table S4A.
| Carbon source |
|||||||
|---|---|---|---|---|---|---|---|
| 1%Glc | 1%Glc 0.01%Xyl |
1%Glc 0.02%Xyl |
1%Glc 0.03%Xyl |
1%Glc 0.05%Xyl |
1%Glc 0.1%Xyl |
1%Fru 0.1%Xyl |
|
| IG3L | 0 ± 0 | 0 ± 0 | 1 ± 1 | 1 ± 1 | 2 ± 2 | 11 ± 2 | 100 ± 10 |
| IG25L | 0 ± 0 | 0 ± 0 | 1.4 ± 1 | 3 ± 1 | 10 ± 1 | 31 ± 3 | 129 ± 9 |
3.5. Comparing the pxylP and tet-On promoter system
One of the most widely used conditional promoter is the tetracycline/doxycycline-induced TetOn system, here termed TetOnoliC (using the oliC minimal promoter for driving expression of the target gene) (Helmschrott et al., 2013). So far, comparison of xylP and TetOn systems suffered of their use for driving expression of different target genes at different genomic loci. Here we compared the pxylP promoter with TetOnoliC driving expression of the very same gene encoding luciferase integrated at the same genomic locus, the fcyB locus. Table 4 shows the mean of raw data ± STD of three biological replicates without normalization and without subtracting the wt background to better visualize the leakiness of the analyzed promoters. However, for calculating the fold-induction, the wt background was subtracted. pxylP showed about 2.6-fold higher basal promoter activity compared to wt background under repressing/non-inducing conditions (1%Glc) and about 2404-fold higher activity under non-repressing/inducing conditions (1%Fru/0.1%Xyl) compared to repressing/non-inducing conditions (1%Glc). TetOnoliC displayed a basal promoter activity that was 8-fold higher than the wt background and about 52-fold induction with 10 μg/ml and 121-fold induction with 20 μg/ml doxycycline (Dox). These data demonstrate that compared to pxylP, TetOnoliC displays an about 4.5-fold higher basal level and an about 4.4-fold lower maximal activity when comparing TetOnoliC with 20 μg/ml doxycycline and pxylP in 1%Fru/0.1%Xyl. Taken together pxylP showed a higher maximal promoter activity, lower leakiness and consequently higher dynamic range than TetOnoliC.
Table 4.
Comparison of pxylP and TetOnoliCpromoter activities. Promoter activity was measured as described in Material and Methods. Shown values are the mean ± STD of three biological replicates after growth for 18 h at 37 °C. Original data are shown in Supplementary Table S4B.
| Carbon source |
|||
|---|---|---|---|
| 1%Glc | 1%Glc 0.1%Xyl |
1%Fru 0.1%Xyl |
|
| IG01L | 46 ± 10 | 10550 ± 811 | 67358 ± 11051 |
|
wt |
18 ± 4 |
20 ± 5 |
29 ± 6 |
| 1%Glc |
1%Glc 10 μg/ml Dox |
1%Glc 20 μg/ml Dox |
|
| TetOnolic | 144 ± 7 | 6616 ± 458 | 15351 ± 1270 |
| wt | 18 ± 4 | 11 ± 1 | 12 ± 3 |
3.6. Discussion
As summarized in the Introduction, pxylP from Penicillium chrysogenum was shown to mediate conditional gene expression in various mold species. Despite its intensive use in different laboratories, the essential regulatory elements in pxylP remained uncharacterized so far. In this study, two different reporters were used for monitoring pxylP activity. mVenus as reporter (Fig. 3) enabled easy in vivo quantification of high promoter activities, while firefly luciferase provided less background and was therefore better suited for analysis of low promoter activities (Fig. 5).
Mutational analysis using mVenus as reporter (Fig. 3) demonstrated the importance of the putative TATA-box, a pyrimidine-rich region located between the putative TATA box and the transcription start sites, both copies of the 91bpDS, as well as putative binding sites for the xylanolytic transcription factor XlnR, and a GATAA motif within the distal 91bpDS element. To further analyze the impact of XlnR, we generated an A. fumigatus mutant lacking XlnR, which displayed significantly decreased growth on xylan, slightly decreased growth on xylose but wt-like growth on glucose or fructose as carbon source (Fig. 4). These data demonstrated conservation of the role of XlnR in activation of xylan degradation and xylose utilization in A. fumigatus as shown previously in other fungal species (Klaubauf et al., 2014). In agreement with XlnR-dependence of pxylP, XlnR inactivation as well as mutation of both putative XlnR binding motifs abrogated xylose-induction of pxylP (Fig. 5). Interestingly, lack of XlnR as well as mutation of both putative XlnR increased basal pxylP activity, which indicates that XlnR might also contribute to repression under repressing/non-inducing conditions (Fig. 5). Repressing activity of XlnR has been predicted previously in A. niger (Hasper et al., 2004; Stricker et al., 2008). Elimination of both 91bpDS copies, which each comprise a single XlnR binding motif (Fig. 5) or mutation of both XlnR binding motifs completely abrogated inducer response of pxylP (Fig. 3, Fig. 5). In contrast, elimination of only a single XlnR binding motif by individual deletion of the 91bpD copies significantly decreased but did not completely abrogate the inducer response (Fig. 3, Fig. 5). These data indicate that a single XlnR motif mediates weak xylose induction but that full pxylP activity requires synergism of two XlnR motifs. In this respect, it is noteworthy that the XlnR homologue of Hypocrea jecorina is predicted to activate as a dimer binding to two recognition motifs (Stricker et al., 2008). The crucial role of the GATAA motif in the distal 91bpDS element indicates regulation by a GATA-type transcription factor. Aspergillus species possess six functionally characterized GATA-type transcription factors termed AreA, AreB, SreA, LreA, LreB and NsdD; which are involved in control of utilization of nitrogen sources, carbon metabolism, iron acquisition, light response as well as sexual and asexual development (Jiang et al., 2021). It remains to be shown if the identified GATAA motif is indeed recognized by a GATA-type transcription factor. Interesting to note, NsdD was implicated in regulation of production of cellulases and xylanases in Penicillium oxalicum (He et al., 2018), while AreA was shown to be involved in regulation of cellulases in Trichoderma reesei (Qian et al., 2019).
Remarkably, the 1579-bp pxylP was found to act bi-bidirectionally with a similar regulatory pattern by driving expression of the upstream-located arabinofuranosidase-encoding axs5 gene (Sakamoto et al., 2011). In agreement with the presented reporter gene assays (Fig. 3), axs5 was previously found to be induced by xylose via semiquantitative RT-PCR analysis of transcript levels. Notably, pxylP displayed lower activity into the axs5 direction compared to the xylP direction. The co-regulation of xylP and axs5 is meaningful as cooperation of the encoded enzymes is required for degradation of xylan. The higher expression of xylP compared to axs5 might be physiologically relevant due to the higher abundance of xylose compared to arabinose in xylan (Rahman et al., 2003).
Truncation of pxylP to 199 bp demonstrated that the upstream region including the two 91bpDS copies mediates not only inducer-dependent activation but also XlnR-independent repression in the absence of inducer (Fig. 5).
The presence of glucose, but not of fructose was found to repress xylose-induction of pxylP activity. Due to the presence of five putative binding sites for CreA (Fig. 2), which mediates transcriptional downregulation of genes for the degradation of less-preferred sugars in the presence of favorable carbon sources such as glucose (Kowalczyk et al., 2014; Zadra et al., 2000) supposed that pxylP is subject to CreA-mediated carbon catabolite repression. Two lines of evidence, however, indicated that glucose mediated repression might not be CreA-dependent: (i) truncation of pxylP combined with deletion of all in silico predicted CreA motifs did not relieve glucose repression in 1%Glc/0.1%Xyl (Fig. 3, Fig. 5) and (ii) increasing the xylose concentration from 0.1% to 0.5 or 1% in the presence of 1%Glc increased pxylP activity approximately 8-fold and 12-fold, respectively (Table 2). Possibilities for glucose-repression independent of direct CreA regulation include (i) indirect CreA regulation via CreA transcriptional repression of the xylose-induced activator XlnR as indicated for the xylanase-encoding xlnA and xlnB genes in A. nidulans (Tamayo et al., 2008), (ii) inducer exclusion by CreA-mediated repression of xylose transporters as found for A. nidulans XtrD (Colabardini et al., 2014), (iii) inducer exclusion by competition of glucose and xylose for uptake by xylose transporters as all xylose transporters are competitively inhibited by glucose (Farwick et al., 2014) or (iv) a combination of the different possibilities.
Comparison with the widely used TetOnoliC promoter (Helmschrott et al., 2013) demonstrated that pxylP shows a higher maximal promoter activity, lower basal activity under repressing/non-inducing conditions and consequently a higher dynamic range than TetOnoliC.
Characterization of functional elements followed by engineering of pxylP also opened new methodological opportunities for its application: (i) truncation of the originally used 1643-bp promoter fragment to 725 bp (IG04Vand IG04L, Fig. 3, Fig. 5) and further deletional mutagenesis to 543 bp (IG16V, Fig. 3) largely preserved the regulatory pattern, which facilitates cloning procedures; (ii) the 1579-bp pxylP fragment was found to act bi-bidirectionally with a similar regulatory pattern (IG15V, Fig. 3), which opens the possibility of bidirectional use of pxylP for conditional co-expression of two genes; (iii) fusion of a third 91bpDS element to the 925-bp promoter fragment significantly increased promoter activity particularly during low inducer availability under repressed conditions (IG25L, Fig. 5); this changed regulatory pattern might be particularly useful for in vivo studies in murine infection models or in the presence of repressing glucose; (iv) truncation to 199 bp rendered pxylP insensitive to xylose induction and slightly increased the basal activity, which qualifies this promoter version as a minimal promoter for functional studies of promoter elements.
Taken together, this study revealed insights into regulation of the xylanolytic system in A. fumigatus, identified several functional elements of pxylP and opened new methodological opportunities for its application.
CRediT authorship contribution statement
Annie Yap: Investigation, Validation, Visualization, Methodology, Writing – original draft, Writing – review & editing. Irene Glarcher: Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. Matthias Misslinger: Investigation, Visualization, Writing – review & editing. Hubertus Haas: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Austrian Science Fund (FWF) doctoral program “host response in opportunistic infections (HOROS, W1253 to AY and HH). The funders had no role in study design, interpretation, decision to publish, in the writing of the manuscript, and in the decision to submit the manuscript for publication.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.mec.2022.e00214.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Data availability
No data was used for the research described in the article.
References
- Altwasser R., Baldin C., Weber J., Guthke R., Kniemeyer O., Brakhage A.A., et al. Network modeling reveals cross talk of MAP kinases during adaptation to caspofungin stress in Aspergillus fumigatus. PloS one. 2015;10(9) doi: 10.1371/journal.pone.0136932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldin C., Valiante V., Krüger T., Schafferer L., Haas H., Kniemeyer O., Brakhage A.A. Comparative proteomics of a tor inducible Aspergillus fumigatus mutant reveals involvement of the Tor kinase in iron regulation. Proteomics. 2015;15(13):2230–2243. doi: 10.1002/pmic.201400584. [DOI] [PubMed] [Google Scholar]
- Baldin C., Kühbacher A., Merschak P., Sastré-Velásquez L.E., Abt B., Dietl A.M., et al. Inducible selectable marker genes to improve Aspergillus fumigatus genetic manipulation. J. Fungi. 2021;7(7) doi: 10.3390/jof7070506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballance D.J. Sequences important for gene expression in filamentous fungi. Yeast. 1986;2(4):229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- Bauer I., Misslinger M., Shadkchan Y., Dietl A.M., Petzer V., Orasch T., et al. The lysine deacetylase RpdA is essential for virulence in Aspergillus fumigatus. Front. Microbiol. 2019;10:2773. doi: 10.3389/fmicb.2019.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bills G.F., Gloer J.B. Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 2016;4(6) doi: 10.1128/microbiolspec.FUNK-0009-2016. [DOI] [PubMed] [Google Scholar]
- Birštonas L., Dallemulle A., López-Berges M.S., Jacobsen I.D., Offterdinger M., Abt B., et al. Multiplex genetic engineering exploiting pyrimidine salvage pathway-based endogenous counterselectable markers. mBio. 2020;11(2) doi: 10.1128/mBio.00230-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugeja H.E., Hynes M.J., Andrianopoulos A. The RFX protein RfxA is an essential regulator of growth and morphogenesis in Penicillium marneffei. Eukaryot. Cell. 2010;9(4):578–591. doi: 10.1128/EC.00226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugeja H.E., Hynes M.J., Andrianopoulos A. HgrA is necessary and sufficient to drive hyphal growth in the dimorphic pathogen Penicillium marneffei. Mol. Microbiol. 2013;88(5):998–1014. doi: 10.1111/mmi.12239. [DOI] [PubMed] [Google Scholar]
- Cairns T.C., Barthel L., Meyer V. Something old, something new: challenges and developments in Aspergillus niger biotechnology. Essays Biochem. 2021;65(2):213–224. doi: 10.1042/EBC20200139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho N.D.S.P., Arentshorst M., Jin Kwon M.M., Vera R., Arthur F.J. Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl. Microbiol. Biotechnol. 2010;87(4):1463–1473. doi: 10.1007/s00253-010-2588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudzicka-Ormaniec P., Macios M., Koper M., Weedall G.D., Caddick M.X., Weglenski P., Dzikowska A. The role of the GATA transcription factor AreB in regulation of nitrogen and carbon metabolism in Aspergillus nidulans. FEMS Microbiol. Lett. 2019;366(6) doi: 10.1093/femsle/fnz066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colabardini A.C., Ries L.N.A., Brown N.A., Dos Reis T.F., Savoldi M., Goldman M.H.S., et al. Functional characterization of a xylose transporter in Aspergillus nidulans. InBiotechnol. Biofuels. 2014;7(1):46. doi: 10.1186/1754-6834-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza W.R., Maitan-Alfenas G.P., de Gouvêa P.F., Brown N.A., Savoldi M., Battaglia E., et al. The influence of Aspergillus niger transcription factors AraR and XlnR in the gene expression during growth in D-xylose, L-arabinose and steam-exploded sugarcane bagasse. Fungal Genet. Biol. 2013;60:29–45. doi: 10.1016/j.fgb.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Fabri J.H.T.M., Rocha M.C., Fernandes C.M., Persinoti G.F., Ries L.N.A., Da Cunha A.F., et al. The heat shock transcription factor HsfA is essential for thermotolerance and regulates cell wall integrity in Aspergillus fumigatus. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.656548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farwick A., Bruder S., Schadeweg V., Oreb M., Boles E. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. Proc. Natl. Acad. Sci. USA. 2014;111(14):5159–5164. doi: 10.1073/pnas.1323464111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazius F., Shelest E., Gebhardt P., Brock M. The fungal α-aminoadipate pathway for lysine biosynthesis requires two enzymes of the aconitase family for the isomerization of homocitrate to homoisocitrate. Mol. Microbiol. 2012;86(6):1508–1530. doi: 10.1111/mmi.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraczek M.G., Bromley M., Buied A., Moore C.B., Rajendran R., Rautemaa R., et al. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 2013;68(7):1486–1496. doi: 10.1093/jac/dkt075. [DOI] [PubMed] [Google Scholar]
- Galiger C., Brock M., Jouvion G., Savers A., Parlato M., Ibrahim-Granet O. Assessment of efficacy of antifungals against Aspergillus fumigatus: value of real-time bioluminescence imaging. Antimicrob. Agents Chemother. 2013;57(7):3046–3059. doi: 10.1128/AAC.01660-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressler M., Zaehle C., Scherlach K., Hertweck C., Brock M. Multifactorial induction of an orphan PKS-NRPS gene cluster in Aspergillus terreus. Chem. Biol. 2011;18(2):198–209. doi: 10.1016/j.chembiol.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Gsaller F., Eisendle M., Lechner B.E., Schrettl M., Lindner H., Müller Det al. The interplay between vacuolar and siderophore-mediated iron storage in Aspergillus fumigatus. Metallomics. 2012;4(12):1262. doi: 10.1039/c2mt20179h. [DOI] [PubMed] [Google Scholar]
- Gsaller F., Hortschansky P., Beattie S.R., Klammer V., Tuppatsch K., Lechner B.E., et al. The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J. 2014;33(19):2261–2276. doi: 10.15252/embj.201489468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H., Friedlin E., Stöffler G., Redl B. Cloning and structural organization of a xylanase-encoding gene from Penicillium chrysogenum. Gene. 1993;126(2):237–242. doi: 10.1016/0378-1119(93)90372-a. [DOI] [PubMed] [Google Scholar]
- Haas H., Angermayr K., Zadra I., Stöffler G. Overexpression of nreB, a new GATA factor-encoding gene of Penicillium chrysogenum, leads to repression of the nitrate assimilatory gene cluster. J. Biol. Chem. 1997;272(36):22576–22582. doi: 10.1074/jbc.272.36.22576. [DOI] [PubMed] [Google Scholar]
- Handelman M., Meir Z., Scott J., Shadkchan Y., Liu W., Ben-Ami R., et al. Point mutation or overexpression of Aspergillus fumigatus cyp51B, encoding lanosterol 14α-sterol demethylase, leads to triazole resistance. Antimicrob. Agents Chemother. 2021;65(10) doi: 10.1128/AAC.01252-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T., Dümig M., Jaber B.M., Szewczyk E., Olbermann P., Morschhäuser J., Krappmann S. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system. Appl. Environ. Microbiol. 2010;76(18):6313–6317. doi: 10.1128/AEM.00882-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasper A.A., Trindade L.M., van der Veen D., van Ooyen A.J.J., de Graaff L.H. Functional analysis of the transcriptional activator XlnR from Aspergillus niger. Microbiology (Reading, England) 2004;150(Pt 5):1367–1375. doi: 10.1099/mic.0.26557-0. [DOI] [PubMed] [Google Scholar]
- He Q.P., Zhao S., Wang J.X., Li C.X., Yan Y.Si, Wang L., et al. Transcription factor NsdD regulates the expression of genes involved in plant biomass-degrading enzymes, conidiation, and pigment biosynthesis in Penicillium oxalicum. Appl. Environ. Microbiol. 2018;84(18) doi: 10.1128/AEM.01039-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn V.M., Mackenzie D.W.R. Mycelial Antigens from Two Strains of Aspergillus fumigatus. An Analysis by Two-Dimensional Immunoelectrophoresis: myzeliale Antigene aus zwei Stämmen von Asperillus fumigatus: eke Analyse rnit der zweidimensionalen Immunelektrophorese. Mycoses. 1980;23(10):549–562. doi: 10.1111/j.1439-0507.1980.tb02557.x. [DOI] [PubMed] [Google Scholar]
- Helmschrott C., Sasse A., Samantaray S., Krappmann S., Wagener J. Upgrading fungal gene expression on demand: improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl. Environ. Microbiol. 2013;79(5):1751–1754. doi: 10.1128/AEM.03626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A., Lerchster H., Marx F. Nutrient excess triggers the expression of the Penicillium chrysogenum antifungal protein PAFB. Microorganisms. 2019;7(12) doi: 10.3390/microorganisms7120654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janus D., Hoff B., Kück U. Evidence for Dicer-dependent RNA interference in the industrial penicillin producer Penicillium chrysogenum. Microbiology. 2009;155(Pt 12):3946–3956. doi: 10.1099/mic.0.032763-0. [DOI] [PubMed] [Google Scholar]
- Jiang C., Lv G., Ge J., He B., Zhang Z., Hu Z., Zeng B. Genome-wide identification of the GATA transcription factor family and their expression patterns under temperature and salt stress in Aspergillus oryzae. Amb. Express. 2021;11(1):56. doi: 10.1186/s13568-021-01212-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N.P. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 2019;17(3):167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn J.R., Turner G. vol. 1. Blackie Academic & Professional; London: 1992. (Applied Molecular Genetics of Filamentous Fungi). [Google Scholar]
- Klaubauf S., Narang H.M., Post H., Zhou M., Brunner K., Mach-Aigner A.R., et al. Similar is not the same: differences in the function of the (hemi-)cellulolytic regulator XlnR (Xlr1/Xyr1) in filamentous fungi. Fungal Genet. Biol. 2014;72:73–81. doi: 10.1016/j.fgb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Kodama Y., Hu C.D. An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. Biotechniques. 2010;49(5):793–805. doi: 10.2144/000113519. [DOI] [PubMed] [Google Scholar]
- Kopke K., Hoff B., Kück U. Application of the Saccharomyces cerevisiae FLP/FRT recombination system in filamentous fungi for marker recycling and construction of knockout strains devoid of heterologous genes. Appl. Environ. Microbiol. 2010;76(14):4664–4674. doi: 10.1128/AEM.00670-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopke K., Hoff B., Bloemendal S., Katschorowski A., Kamerewerd J., Kück U. Members of the Penicillium chrysogenum velvet complex play functionally opposing roles in the regulation of penicillin biosynthesis and conidiation. Eukaryot. Cell. 2013;12(2):299–310. doi: 10.1128/EC.00272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk J.E., Benoit I., de Vries R.P. Regulation of plant biomass utilization in Aspergillus. Adv. Appl. Microbiol. 2014;88:31–56. doi: 10.1016/B978-0-12-800260-5.00002-4. [DOI] [PubMed] [Google Scholar]
- Kramm K., Engel C., Grohmann D. Transcription initiation factor TBP: old friend new questions. Biochem. Soc. Trans. 2019;47(1):411–423. doi: 10.1042/BST20180623. [DOI] [PubMed] [Google Scholar]
- Krappmann S., Sasse C., Braus G.H. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot. Cell. 2006;5(1):212–215. doi: 10.1128/EC.5.1.212-215.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers G.J., Goedhart J., van Munster E.B., Gadella T.W.J. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry. 2006;45(21):6570–6580. doi: 10.1021/bi0516273. [DOI] [PubMed] [Google Scholar]
- Latgé J.P., Chamilos G. Aspergillus fumigatus and aspergillosis in 2019. Clin. Microbiol. Rev. 2019;33(1) doi: 10.1128/CMR.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.K., Kwon N.J., Choi J.M., Lee I.S., Jung S., Yu J.H. NsdD is a key repressor of asexual development in Aspergillus nidulans. Genetics. 2014;197(1):159–173. doi: 10.1534/genetics.114.161430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Parsania C., Tan K., Todd R.B., Wong K.H. Co-option of an extracellular protease for transcriptional control of nutrient degradation in the fungus Aspergillus nidulans. Commun. Biol. 2021;4(1):1409. doi: 10.1038/s42003-021-02925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Berges M.S., Scheven M.T., Hortschansky P., Misslinger M., Baldin C., Gsaller F., et al. The bZIP transcription factor HapX is post-translationally regulated to control iron homeostasis in Aspergillus fumigatus. Int. J. Mol. Sci. 2021;22(14) doi: 10.3390/ijms22147739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Li W., Zhang P., Lyu H., Hu Y., Yin W.B. Rational design for heterologous production of aurovertin-type compounds in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2018;102(1):297–304. doi: 10.1007/s00253-017-8606-9. [DOI] [PubMed] [Google Scholar]
- Misslinger M., Lechner B.E., Bacher K., Haas H. Iron-sensing is governed by mitochondrial, not by cytosolic iron-sulfur cluster biogenesis in Aspergillus fumigatus. Metallomics. 2018;10(11):1687–1700. doi: 10.1039/c8mt00263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misslinger M., Scheven M.T., Hortschansky P., López-Berges M.S., Heiss K., Beckmann Nicola, et al. The monothiol glutaredoxin GrxD is essential for sensing iron starvation in Aspergillus fumigatus. PLoS Genet. 2019;15(9) doi: 10.1371/journal.pgen.1008379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan B.J., Askin M.C., Hynes M.J., Davis M.A. Differential expression of Aspergillus nidulans ammonium permease genes is regulated by GATA transcription factor AreA. Eukaryot. Cell. 2006;5(2):226–237. doi: 10.1128/EC.5.2.226-237.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer M., Zhu Z., Penka M., Helmschrott C., Wagener N., Wagener J. Mitochondrial dynamics in the pathogenic mold Aspergillus fumigatus: therapeutic and evolutionary implications. Mol. Microbiol. 2015;98(5):930–945. doi: 10.1111/mmi.13167. [DOI] [PubMed] [Google Scholar]
- Oberegger H., Schoeser M., Zadra I., Abt B., Haas H. SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 2001;41(5):1077–1089. doi: 10.1046/j.1365-2958.2001.02586.x. [DOI] [PubMed] [Google Scholar]
- Pidroni A., Faber B., Brosch G., Bauer I., Graessle S. A class 1 histone deacetylase as major regulator of secondary metabolite production in Aspergillus nidulans. Front. Microbiol. 2018;9:2212. doi: 10.3389/fmicb.2018.02212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongsunk S., Andrianopoulos A., Chaiyaroj S.C. Conditional lethal disruption of TATA-binding protein gene in Penicillium marneffei. Fungal Genet. Biol. 2005;42(11):893–903. doi: 10.1016/j.fgb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J.A., Hemmons L.M., Macdonald K.D., Bufton A.W.J. The genetics of Aspergillus nidulans. Adv. Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Purschwitz J., Müller S., Kastner C., Schöser M., Haas H., Espeso E.A., et al. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 2008;18(4):255–259. doi: 10.1016/j.cub.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Qian Y., Sun Yu, Zhong L., Sun N., Sheng Y., Qu Yi, Zhong Y. The GATA-type transcriptional factor Are1 modulates the expression of extracellular proteases and cellulases in Trichoderma reesei. Int. J. Mol. Sci. 2019;20(17) doi: 10.3390/ijms20174100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A.K.M.S., Sugitani N., Hatsu M., Takamizawa K. A role of xylanase, alpha-L-arabinofuranosidase, and xylosidase in xylan degradation. Can. J. Microbiol. 2003;49(1):58–64. doi: 10.1139/w02-114. [DOI] [PubMed] [Google Scholar]
- Rauscher R., Würleitner E., Wacenovsky C., Aro N., Stricker A.R., Zeilinger S., et al. Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot. Cell. 2006;5(3):447–456. doi: 10.1128/EC.5.3.447-456.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romsdahl J., Wang C.C.C. Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012-2018) MedChemComm. 2019;10(6):840–866. doi: 10.1039/c9md00054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Ogura A., Inui M., Tokuda S., Hosokawa S., Ihara H., Kasai N. Identification of a GH62 α-L-arabinofuranosidase specific for arabinoxylan produced by Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2011;90(1):137–146. doi: 10.1007/s00253-010-2988-2. [DOI] [PubMed] [Google Scholar]
- Schrettl M., Kim H.S., Eisendle M., Kragl C., Nierman W.C., Heinekamp T., et al. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 2008;70(1):27–43. doi: 10.1111/j.1365-2958.2008.06376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibthorp C., Wu H., Cowley G., Wong P.W.H., Palaima P., Morozov I.Y., et al. Transcriptome analysis of the filamentous fungus Aspergillus nidulans directed to the global identification of promoters. BMC Genom. 2013;14:847. doi: 10.1186/1471-2164-14-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigl C., Handler M., Sprenger G., Kürnsteiner H., Zadra I. A novel homologous dominant selection marker for genetic transformation of Penicillium chrysogenum: overexpression of squalene epoxidase-encoding ergA. J. Biotechnol. 2010;150(3):307–311. doi: 10.1016/j.jbiotec.2010.09.941. [DOI] [PubMed] [Google Scholar]
- Sigl C., Haas H., Specht T., Pfaller K., Kürnsteiner H., Zadra I. Among developmental regulators, StuA but not BrlA is essential for penicillin V production in Penicillium chrysogenum. Appl. Environ. Microbiol. 2011;77(3):972–982. doi: 10.1128/AEM.01557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skellam E. Strategies for engineering natural product biosynthesis in fungi. Trends Biotechnol. 2019;37(4):416–427. doi: 10.1016/j.tibtech.2018.09.003. [DOI] [PubMed] [Google Scholar]
- Sorokina Maria, Merseburger Peter, Rajan Kohulan, Mehmet Aziz Yirik, Steinbeck Christoph. COCONUT online: Collection of Open Natural Products database. J Cheminform. 2021;13(1/2) doi: 10.1186/s13321-020-00478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker A.R., Mach R.L., de Graaff L.H. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei) Appl. Microbiol. Biotechnol. 2008;78(2):211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- Tamayo E.N., Villanueva A., Hasper A.A., de Graaff L.H., Ramón D., Orejas M. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet. Biol. 2008;45(6):984–993. doi: 10.1016/j.fgb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Tilburn J., Scazzocchio C., Taylor G.G., Zabicky-Zissman J.H., Lockington R.A., Davies R.W. Transformation by integration in Aspergillus nidulans. Gene. 1983;26(2–3):205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- Tribus M., Bauer I., Galehr J., Rieser G., Trojer P., Brosch G., et al. A novel motif in fungal class 1 histone deacetylases is essential for growth and development of Aspergillus. Mol. Biol. Cell. 2010;21(2):345–353. doi: 10.1091/mbc.e09-08-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkles Shiela E, Valiante Vito, Mattern Derek J, Brakhage Axel A. Synthetic biology tools for bioprospecting of natural products in eukaryotes. Chem. Biol. 2014;21(4):502–508. doi: 10.1016/j.chembiol.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Vaknin Y., Hillmann F., Iannitti R., Ben B., Netali, Sandovsky-Losica H., Shadkchan Yona, et al. Identification and characterization of a novel Aspergillus fumigatus rhomboid family putative protease, RbdA, involved in hypoxia sensing and virulence. Infect. Immun. 2016;84(6):1866–1878. doi: 10.1128/IAI.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Peij N.N., Visser J., de Graaff L.H. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 1998;27(1):131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- Wang F., Sethiya P., Hu X., Guo S., Chen Y., Li Ang, et al. Transcription in fungal conidia before dormancy produces phenotypically variable conidia that maximize survival in different environments. Nat. Microbiol. 2021;6(8):1066–1081. doi: 10.1038/s41564-021-00922-y. [DOI] [PubMed] [Google Scholar]
- Wong K.H., Hynes M.J., Todd R.B., Davis M.A. Transcriptional control of nmrA by the bZIP transcription factor MeaB reveals a new level of nitrogen regulation in Aspergillus nidulans. Mol. Microbiol. 2007;66(2):534–551. doi: 10.1111/j.1365-2958.2007.05940.x. [DOI] [PubMed] [Google Scholar]
- Wong K.H., Todd R.B., Oakley B.R., Oakley C.E., Hynes M.J., Davis M.A. Sumoylation in Aspergillus nidulans: sumO inactivation, overexpression and live-cell imaging. Fungal Genet. Biol. 2008;45(5):728–737. doi: 10.1016/j.fgb.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.H., Hynes M.J., Todd R.B., Davis M.A. Deletion and overexpression of the Aspergillus nidulans GATA factor AreB reveals unexpected pleiotropy. Microbiology. 2009;155(Pt 12):3868–3880. doi: 10.1099/mic.0.031252-0. [DOI] [PubMed] [Google Scholar]
- Yasmin S., Alcazar-Fuoli L., Gründlinger M., Puempel T., Cairns T., Blatzer M., et al. Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proc. Natl. Acad. Sci. USA. 2012;109(8):E497–E504. doi: 10.1073/pnas.1106399108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadra I., Abt B., Parson W., Haas H. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl. Environ. Microbiol. 2000;66(11):4810–4816. doi: 10.1128/AEM.66.11.4810-4816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.