Abstract
Coiled-coil-helix-coiled-coil-helix domain containing 2 (CHCHD2) is a mitochondrial protein that plays important roles in cristae structure, oxidative phosphorylation and apoptosis. Multiple mutations in CHCHD2 have been associated with Lewy body disorders (LBDs), such as Parkinson’s disease (PD) and dementia with Lewy bodies, with the CHCHD2-T61I mutation being the most widely studied. However, at present, only CHCHD2 knockout or CHCHD2/CHCHD10 double knockout mouse models have been investigated. They do not recapitulate the pathology seen in patients with CHCHD2 mutations. We generated the first transgenic mouse model expressing the human PD-linked CHCHD2-T61I mutation driven by the mPrP promoter. We show that CHCHD2-T61I Tg mice exhibit perinuclear mitochondrial aggregates, neuroinflammation, and have impaired long-term synaptic plasticity associated with synaptic dysfunction. Dopaminergic neurodegeneration, a hallmark of PD, is also observed along with α-synuclein pathology. Significant motor dysfunction is seen with no changes in learning and memory at 1 year of age. A minor proportion of the CHCHD2-T61I Tg mice (~10%) show a severe motor phenotype consistent with human Pisa Syndrome, an atypical PD phenotype. Unbiased proteomics analysis reveals surprising increases in many insoluble proteins predominantly originating from mitochondria and perturbing multiple canonical biological pathways as assessed by ingenuity pathway analysis, including neurodegenerative disease-associated proteins such as tau, cofilin, SOD1 and DJ-1. Overall, CHCHD2-T61I Tg mice exhibit pathological and motor changes associated with LBDs, indicating that this model successfully captures phenotypes seen in human LBD patients with CHCHD2 mutations and demonstrates changes in neurodegenerative disease-associated proteins, which delineates relevant pathological pathways for further investigation.
Introduction
Lewy body disorders (LBDs) include Parkinson’s disease (PD), dementia with Lewy bodies (DLBs) and multiple system atrophy. LBDs are characterized by the presence of intracellular aggregates of α-synuclein and ubiquitin proteins, known as Lewy bodies (LBs) (1,2). Mitochondrial dysfunction is a hallmark of neurodegenerative disease, including LBDs (3–5). 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), a mitochondrial complex I inhibitor, has been shown to induce parkinsonian-like motor dysfunction associated with dopaminergic (DA) neuronal loss (6–8). Furthermore, familial mutations in Parkin and PINK1, two important proteins involved in mitophagy, are also associated with PD and can contribute to the PD phenotype (9–13). These findings strongly suggest that mitochondrial dysfunction and LBDs are intertwined.
Coiled-coil helix coiled-coil helix domain containing 2 (CHCHD2/MNRR1) is a mitochondrial protein localized to the intermembrane space (IMS). CHCHD2 plays a multitude of roles in the mitochondria, including regulating electron flow in the electron transport chain (ETC) (14) and acting as an anti-apoptotic protein (15–17). Under hypoxic stress, CHCHD2 can also translocate to the nucleus and act as a transcription factor for a complex IV subunit and itself (18,19). In 2015, two mutations of CHCHD2 (T61I and R145Q) were identified in autosomal dominant PD patients with a familial pedigree (20). Interestingly, autopsy data from a female patient with the CHCHD2-T61I mutation showed the presence of LBs, neurofibrillary tau tangles, and amyloid-β plaques (21). Furthermore, this patient exhibited loss of midbrain DA neurons and insoluble accumulation of CHCHD2 and CHCHD10, a homologous sister protein to CHCHD2 (21).
Animal models have been used to study the functional roles of CHCHD2, and the consequences of mutant CHCHD2 expression. Har-1 is orthologous to CHCHD2 and CHCHD10 in Caenorhabditis elegans, and knockout of har-1 induces reactive oxygen species (ROS) production, shortened lifespan and an abnormal curling phenotype (22). WT, but not FTD-ALS associated mutant CHCHD10, can rescue these phenotypes (22). In Drosophila, CG5010 is an orthologous protein of both CHCHD2 and CHCHD10. Knockout of CG5010 decreases survival, increases toxic ROS and induces motor dysfunction and DA neuron loss (15,23). WT, but not mutant CHCHD2, can rescue these phenotypes (15), suggesting a loss-of-function mutation. Transgenic flies expressing human WT and mutant CHCHD2 also exhibit decreased survival and motor dysfunction (24), suggesting that excessive CHCHD2 may be detrimental or that Drosophila may not be the ideal model organism to study the consequences of mutant CHCHD2.
Thus far, whole-body knockout of CHCHD2, or double knockouts of both CHCHD2 and CHCHD10 (DKO) mice have been reported and characterized (25,26). Two-year-old CHCHD2 knockout mice exhibit motor dysfunction, loss of DA neurons in the substantia nigra and the formation of p62/SQSTM1 inclusions (25). While DKO mice show disorganized mitochondrial cristae in the heart due to OPA1 cleavage by OMA1 (26), no CNS pathology has been reported in this mouse model.
To date, there have not been any transgenic or knock-in mouse models of mutant CHCHD2 reported. Here, we generated the first transgenic mouse model neuronally expressing human CHCHD2-T61I and characterized behavioral phenotype and pathologic features. Our data suggest that the CHCHD2-T61I mutation contributes to LB pathologies commonly observed in PD and other LBDs while perturbing multiple biological pathways originating from mitochondria in vivo, thereby representing a useful mammalian model that recapitulates CHCHD2-T61I-mediated pathologies.
Results
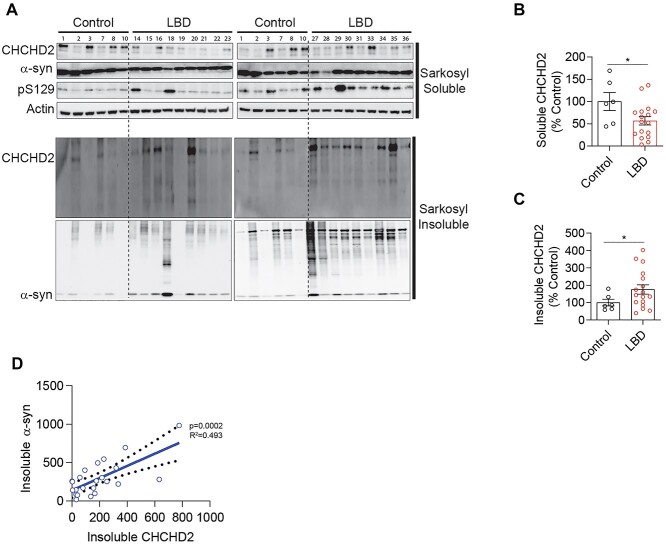
Insoluble CHCHD2 positively correlates with insoluble α-synuclein in LBD patient brain
Recent studies have shown that CHCHD2 mRNA and soluble CHCHD2 protein levels are significantly reduced in the postmortem substantia nigra of PD patients and A53T α-synuclein mice (27). To determine whether decreased CHCHD2 expression correlates with α-synuclein pathology in LBD patients, we investigated changes in sarkosyl soluble and insoluble levels of CHCHD2 protein in the frontal cortex of sporadic LBD patients compared with control subjects. Sarkosyl fractionation and western blotting data revealed significantly decreased soluble CHCHD2 levels (Fig. 1A and B) and increased insoluble CHCHD2 levels (Fig. 1A and C) in the frontal cortex of LBD patients compared with controls (Supplementary Material, Table S1). Interestingly, we found that sarkosyl insoluble α-synuclein significantly correlates with insoluble CHCHD2 levels (Fig. 1D) with a coefficient of determination (R2 = 0.493) by linear regression analysis, indicating a pathological association between CHCHD2 and α-synuclein in the frontal cortex of LBD patients.
Figure 1.
Insoluble CHCHD2 is increased and correlates with insoluble α-synuclein levels in the brains of LBD patients. (A) Sarkosyl soluble and insoluble representative immunoblots for CHCHD2, α-synuclein and S129 phospho-α-synuclein from the frontal cortex of control subjects and LBD patients. (B) Quantitative analysis of sarkosyl soluble and (C) insoluble CHCHD2 levels. Error bars represent mean ± SEM; n = 6 control, n = 17 LBD patients; t-test; *P < 0.05. (D) Correlation of insoluble CHCHD2 and α-synuclein levels. n = 6 control, n = 17 LBD patients.
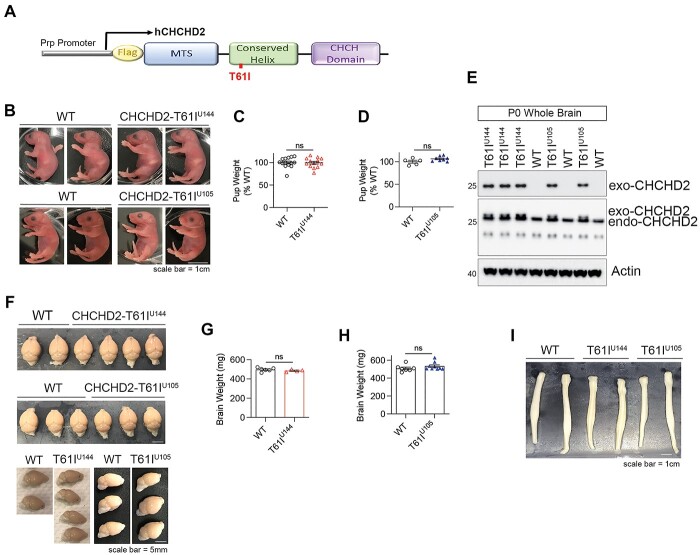
CHCHD2-T61I transgenic mice are viable and lack gross abnormalities
To express the transgene neuronally in mice, transgene expression of human CHCHD2-T61I was controlled by the murine prion protein promoter (mPrp) (Fig. 2A). Founders were identified after initial genotyping (Supplementary Material, Fig. S1A and B) with progenies generated from four founders with the highest copy numbers of the transgene. Two lines (U144 and U105) were established from the two founders that showed successful neuronal expression, as evaluated by Western blotting (Supplementary Material, Fig. S2B). Sequencing of DNA extracted from CHCHD2-T61I Tg mice lines U144 and U105 show the correct c.182C > T mutation, which corresponds to the p.T61I mutation (Supplementary Material, Fig. S1C and D). CHCHD2-T61I Tg pups from lines U144 and U105 are phenotypically normal compared with WT littermates (Fig. 2B), and feed well as indicated by the presence of the milk spot and birth weight (Fig. 2B–D). Whole brain lysates from P0 pups indicate transgene expression as detected by M2 (flag), and CHCHD2 immunoblots (Fig. 2E). Furthermore, neither significant differences between the WT and CHCHD2-T61I Tg lines in brain weights (Fig. 2F–H) nor anatomical morphologies of the brain and spinal cord (Fig. 2F and I) were noted at 1 year of age. These data indicate that CHCHD2-T61I mice are viable, fertile and do not exhibit gross morphological abnormalities.
Figure 2.
CHCHD2-T61I Tg mice are viable, fertile and lack gross abnormalities of the brain and spinal cord morphology. (A) Schematic representation of the human CHCHD2-T61I-Flag transgene driven by the mouse Prp promoter. (B) Representative images of postnatal (P0) CHCHD2-T61I Tg line U144 and U105 mice and WT littermates. P0 T61I Tg line U144 and U105 mice are phenotypically normal and feed well, as indicated by the presence of the milk spot (Scale bar: 1 cm). (C) Quantitative analysis of birth weights of CHCHD2-T61I Tg line U144 pups and WT littermates. Error bars represent mean ± SEM; n = 14 WT, n = 13 T61I U144; n.s., not significant. (D) Quantitative analysis of birth weights of CHCHD2-T61I Tg line U105 pups and WT littermates. Error bars represent mean ± SEM; n = 5 WT, n = 8 T61I U105; n.s., not significant. Both strains do not differ in birth weight compared with nontransgenic WT littermates. (E) Representative immunoblots for M2, CHCHD2 and loading control actin from whole brains extracted from P0 CHCHD2-T61I Tg mice. Flag-CHCHD2-T61I transgenic expression is detected by M2 and CHCHD2 antibodies. (F) Representative images of whole perfused brains from 1-year-old CHCHD2-T61I Tg line U144, line U105 and WT littermates (Scale bar: 5 mm). (G) Quantitative analysis of whole brain weights from 1-year-old CHCHD2-T61I Tg line U144 and WT mice. Error bars represent mean ± SEM; n = 7 WT, n = 4 T61I U144; n.s., not significant. (H) Quantitative analysis of whole brain weights from 1-year-old CHCHD2-T61I Tg line U105 and WT mice. Error bars represent mean ± SEM; n = 7 WT, n = 9 T61I U144; n.s., not significant. (I) Representative images of whole spinal cords from 1-year-old CHCHD2-T61I Tg line U144, line U105 and WT littermates. CHCHD2-T61I Tg mice do not show any abnormalities of spinal cord morphology compared with nontransgenic littermates (Scale bar: 1 cm).
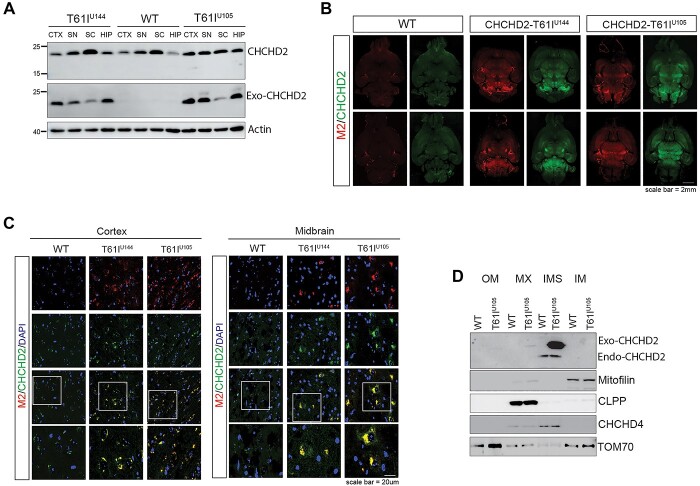
The hCHCHD2-T61I transgene is successfully expressed, and hCHCHD2-T61I protein is localized to mitochondria in vivo
Next, we performed western blotting to monitor the expression of the CHCHD2-T61I transgene from different brain regions in 1-year-old CHCHD2-T61I Tg mice. We found that 1-year-old CHCHD2-T61I Tg mice show transgene expression in the cortex (CTX), midbrain (SN), spinal cord (SC) and hippocampus (HIP) (Fig. 3A). To confirm these data in a different way, we performed immunohistochemistry and found that whole brain sections from 1-year-old CHCHD2-T61I Tg mice exhibit increased CHCHD2 and M2 (Flag) expression compared with WT littermates (Fig. 3B). Indeed, secondary antibody only staining shows no immunoreactivities (Supplementary Material, Fig. S3A). Gene expression analysis showed 2–2.5× increase in CHCHD2 expression compared with endogenous mouse CHCHD2 expression in both Tg lines at 1 year of age (Supplementary Material, Fig. S2A). These results indicate successful transgene expression in multiple brain regions. The 60× images of both cortical and midbrain regions revealed clear CHCHD2 and M2 colocalization, confirming transgene expression (Fig. 3C). CHCHD2 is normally imported to the mitochondrial IMS (28), and the T61I mutation does not interfere with proper mitochondrial IMS targeting (29). However, loss of mitochondrial localization of CHCHD2 was observed in the brain of a PD patient harboring the CHCHD2-T61I mutation (21), which was measured by CHCHD2’s co-localization with ATP5a. Therefore, we tested whether human CHCHD2-T61I successfully localizes to the mitochondrial IMS in vivo. We performed mitochondrial subfractionation from whole brains of CHCHD2-T61I Tg mice and found that endogenous CHCHD2 and exogenous human CHCHD2-T61I proteins reside in the IMS of mitochondria (Fig. 3D). We also confirmed successful subfractionation of the IMS by looking at CHCHD4/MIA40, which is another known IMS protein (Fig. 3D). Taken together, these results indicate successful transgene expression and correct mitochondrial targeting, thus allowing for further study of the consequences of such mitochondrial CHCHD2-T61I expression in the brain.
Figure 3.
CHCHD2-T61I is expressed in different regions of the brain and localizes to the mitochondrial IMS. (A) Representative immunoblots of RIPA-soluble lysate from the cortex (CTX), midbrain (SN), spinal cord (SC) and hippocampus (HIP) from 1-year-old CHCHD2-T61I Tg mice and nontransgenic WT littermates. Exogenous CHCHD2 expression is clearly seen in all regions, with the highest expression seen in the cortex, midbrain and hippocampus. (B) Representative images of whole brain horizontal slices of 1-year-old CHCHD2-T61I Tg line U144, line U105 and WT littermates immunostained for M2 (flag) and CHCHD2 (Scale bar: 2 mm). (C) Representative images of cortex and midbrain regions immunostained for M2 (flag) and CHCHD2 from 1-year-old CHCHD2-T61I Tg line U144, line U105 and WT littermates (Scale bar: 20 μm). (D) Representative immunoblots of mitochondrial subfractionation from whole brains of 3- to 4-month-old CHCHD2-T61I Tg line U105 mice and WT littermates (n = 4 mice/genotype). The exogenous CHCHD2-T61I and endogenous CHCHD2 target correctly to the mitochondrial IMS in vivo.
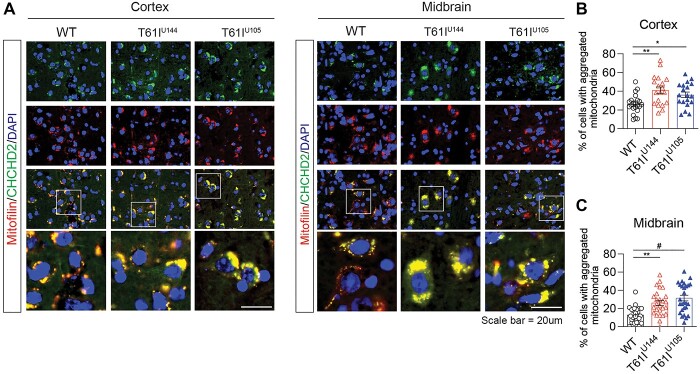
CHCHD2-T61I Tg mice exhibit perinuclear aggregation of mitochondria in the brain
While CHCHD2 is a mitochondrial IMS protein that plays key roles in oxidative phosphorylation and apoptosis (16,19,28,30), CHCHD2 also plays important roles in mitochondrial cristae maintenance (15,25). In Drosophila models, loss of CHCHD2 leads to abnormal cristae structure in indirect flight muscles, which can be rescued by dCHCHD2 or hCHCHD2 WT expression, but not PD-linked mutations T61I or R145Q (15). Furthermore, 2-year-old CHCHD2 knockout mice also exhibit fragmented mitochondria in DA neurons, suggesting that CHCHD2 regulates mitochondrial morphology (25). Therefore, we tested whether CHCHD2-T61I Tg mice have altered mitochondrial morphology in vivo. Interestingly, we found that CHCHD2-T61I Tg mice exhibit perinuclear aggregation of mitochondria in both the cortex and midbrain at 1 year of age, as the number of cells with aggregated mitochondria in the cortex (Fig. 4A and B) and midbrain (Fig. 4A and C) was significantly increased compared with nontransgenic littermates. As expected, we confirmed that secondary antibody only staining shows no immunoreactivities (Supplementary Material, Fig. S3B).
Figure 4.
Expression of CHCHD2-T61I induces the perinuclear aggregation of mitochondria in vivo. (A) Representative images from cortical and midbrain tissues immunostained with CHCHD2 and mitofilin. CHCHD2-T61I Tg mice exhibit the perinuclear aggregation of mitochondria (Scale bar: 20 μm) in the cortex and midbrain. (B and C) Quantitative analysis of cells with aggregated mitochondria in 1-year-old CHCHD2-T61I Tg line U144, line U105 and WT littermates. CHCHD2-T61I Tg show significantly increased mitochondrial aggregation in the cortex and midbrain. Error bars represent mean ± SEM; n = 4 mice/genotype; one-way ANOVA with Tukey post hoc test; *P < 0.05, **P < 0.005, #P < 0.0001.
CHCHD2-T61I expression induces gliosis and synaptic dysfunction in vivo
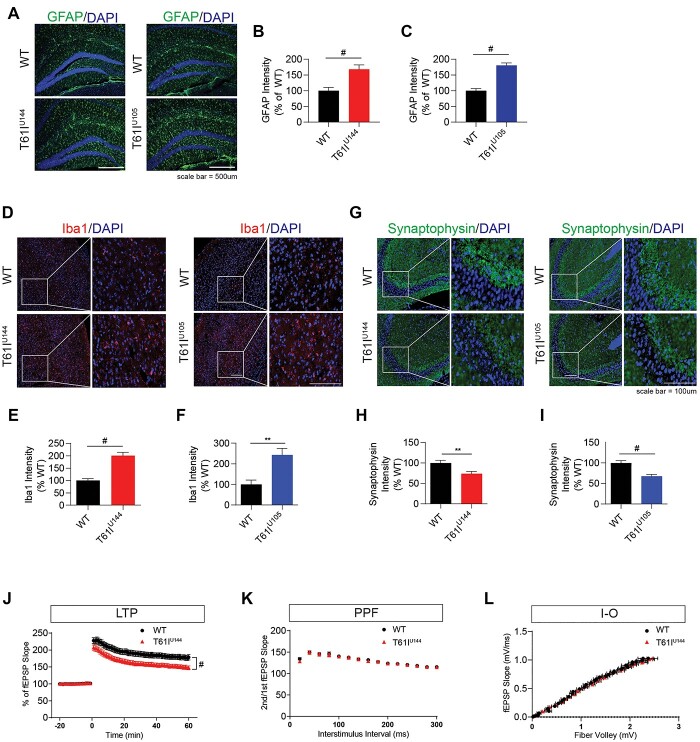
Microgliosis is a hallmark in neurodegenerative disease, including LBDs (31–33). CHCHD2 is also expressed in >90% of astrocytes and in 10–30% of microglia (27). Therefore, we performed immunohistochemistry to measure astrogliosis and microgliosis in 1-year-old CHCHD2-T61I Tg mice. We found that hippocampal immunostaining for GFAP, a marker for activated astrocytes, is significantly increased in 1-year-old CHCHD2-T61I Tg mice line U144 and U105 compared with nontransgenic littermates (Fig. 5A–C). Cortical immunostaining for Iba1, a marker for activated microglia, is also significantly increased in 1-year-old CHCHD2-T61I Tg mice line U144 and U105 compared with their WT littermates (Fig. 5D–F). These data suggest that CHCHD2-T61I expression induces reactive hippocampal and cortical gliosis, a hallmark of LBDs and other neurodegenerative diseases.
Figure 5.
CHCHD2-T61I Tg mice exhibit gliosis and synaptic dysfunction in vivo. (A) Representative images of 1-year-old CHCHD2-T61I Tg line U144, line U105 and WT hippocampus immunostained for GFAP and DAPI. (Scale bar: 500 μm.) (B) Quantitative analysis of hippocampal GFAP immunofluorescence intensity. One-year-old CHCHD2-T61I Tg line U144 mice exhibit increased hippocampal GFAP compared with nontransgenic littermates. Error bars represent mean ± SEM; n = 5 WT, n = 6 U144; t-test; #P < 0.0001. (C) Quantitative analysis of hippocampal GFAP immunofluorescence intensity. One-year-old CHCHD2-T61I Tg line U105 mice exhibit increased hippocampal GFAP compared with WT littermates. Error bars represent mean ± SEM; n = 4 WT, n = 5 U105; t-test; #P < 0.0001. (D) Representative images of 1-year-old CHCHD2-T61I Tg line U144, line U105 and WT cortex immunostained for Iba1 and DAPI. (Scale bar: 100 μm.) (E) Quantitative analysis of cortical Iba1 immunofluorescence intensity. One-year-old CHCHD2-T61I Tg line U144 mice exhibit increased cortical microgliosis compared with WT littermates. Error bars represent mean ± SEM; n = 5 WT, n = 6 U144; t-test; #P < 0.0001. (F) Quantitative analysis of cortical Iba1 immunofluorescence intensity. One-year-old CHCHD2-T61I Tg line U105 mice exhibit increased cortical microgliosis compared with WT littermates. Error bars represent mean ± SEM; n = 4 WT, n = 5 U144; t-test; **P < 0.005. (G) Representative images of 1-year-old CHCHD2-T61I Tg line U144, line U105 and WT hippocampus immunostained for synaptophysin and DAPI. (Scale bar: 100 μm.) (H) Quantitative analysis of synaptophysin intensity in the hippocampal CA3 region in 1-year-old CHCHD2-T61I Tg line U144 mice compared with WT littermates. CHCHD2-T61I Tg line U144 mice exhibit decreased synaptophysin immunoreactivity. Error bars represent mean ± SEM; n = 6/genotype; t-test **P < 0.005. (I) Quantitative analysis of synaptophysin intensity in the hippocampal CA3 region in 1-year-old CHCHD2-T61I Tg line U105 mice compared with nontransgenic WT littermates. CHCHD2-T61I Tg line U105 mice exhibit decreased synaptophysin immunoreactivity. Error bars represent mean ± SEM; n = 6/genotype; t-test; #P < 0.0001. Quantification of electrophysiological recordings. Ten-month-old CHCHD2-T61I Tg line U144 mice show significantly reduced hippocampal long-term potentiation induced by theta-burst stimulation (J) with no change in PPF (K) or input–output (L). Error bars represent mean ± SEM; n = 4/genotype; WT: 30 slices, T61I-Tg line U144: 32 slices, two-way ANOVA with Dunnett’s post hoc test; #P < 0.0001.
Synaptophysin is a presynaptic protein, and reduction of synaptophysin in the hippocampus is associated with cognitive decline (34). Therefore, we next tested whether CHCHD2-T61I expression alters synaptophysin immunoreactivity in vivo. Indeed, we found that 1-year-old CHCHD2-T61I Tg line U144 and U105 mice exhibit significantly reduced synaptophysin immunoreactivity in the CA3 region of the hippocampus, indicating that the CHCHD2-T61I mutation results in the loss of synaptic integrity (Fig. 5G–I). We also confirmed that the secondary antibody only staining image does not show immunoreactivity (Supplementary Material, Fig. S3C). To measure functional changes in synaptic plasticity imposed by the CHCHD2-T61I mutation, we carried out electrophysiological studies from ex vivo brain slices as we previously showed (35). Long-term potentiation (LTP) induced by theta-burst stimulation showed that 10-month-old CHCHD2-T61I Tg line U144 mice exhibit a significant reduction in hippocampal LTP compared with WT littermates (Fig. 5J). We also measured paired-pulse facilitation (PPF), a form of short-term synaptic plasticity and input–output (I-O), a form of basal synaptic plasticity, in CHCHD2-T61I Tg line U144 mice and WT littermates. We confirmed that there were no significant differences at all interstimulus intervals in PPF experiments (Fig. 5K). Likewise, no changes in basal synaptic efficacy were detected in I-O analysis of CHCHD2-T61I Tg and littermate WT mice (Fig. 5L). These results suggest that CHCHD2-T61I expression promotes gliosis and long-term synaptic plasticity deficits in vivo.
CHCHD2-T61I Tg mice exhibit dopaminergic neurodegeneration and α-synuclein mislocalization and accumulation in vivo
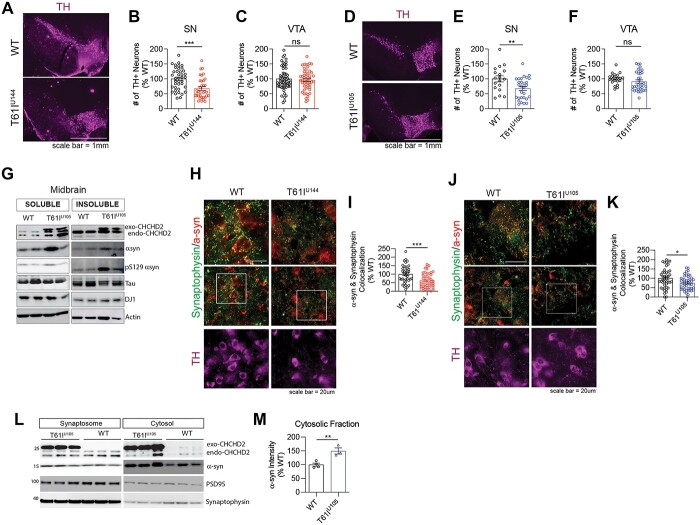
DA neurodegeneration is a pathological hallmark of PD, and the loss of dopamine and DA projections into the striatum is thought to contribute to motor dysfunction (6–8,36). Given that CHCHD2-T61I is associated with late-onset autosomal dominant PD (20) and CHCHD2-T61I enhances the toxicity of α-synuclein in Drosophila dopaminergic neurons (21), we tested whether CHCHD2-T61I Tg mice exhibit DA neuronal loss in vivo. Therefore, we immunostained midbrain slices from 1-year-old CHCHD2-T61I Tg mice line U144 and U105 and their WT littermates for tyrosine hydroxylase (TH) to visualize DA neuron cell bodies (Fig. 6A and D). Interestingly, we found a significant reduction in the number of TH+ cell bodies in the substantia nigra (SN) in CHCHD2-T61I Tg line U144 (Fig. 6B) and U105 (Fig. 6E), with no changes in the ventral tegmental area (VTA) (Fig. 6C and F), suggesting that mutant CHCHD2-induced DA neurodegeneration is specific to the SN. This result is consistent with that reported in 2-year-old CHCHD2 knockout mice, where DA neurodegeneration was explicitly seen in the SNcc (25). Given that DA neurons project forward into the striatum, we visualized these projections by immunostaining striatum tissue for TH from 1-year-old CHCHD2-T61I Tg mice line U144 and U105. We found that CHCHD2-T61I expression significantly reduced striatal TH intensity compared with WT littermates (Supplementary Material, Fig. S4B and C), further confirming the loss of DA neurons as a consequence of neuronal CHCHD2-T61I expression.
Figure 6.
CHCHD2-T61I Tg mice exhibit dopaminergic neuronal loss and α-synuclein mislocalization. (A) Representative images of CHCHD2-T61I Tg line U144 midbrain immunostained for tyrosine hydroxylase to visualize dopaminergic neurons (Scale bar: 1 mm). (B) Quantification of TH+ cells in the substantia nigra (SN). One-year-old CHCHD2-T61I Tg line U144 mice exhibit DA neurodegeneration in the SN. Error bars represent mean ± SEM; n = 10 WT, n = 7 U144; t-test; ***P < 0.0005. (C) Quantification of TH+ cells in the VTA. One-year-old CHCHD2-T61I Tg line U144 mice do not exhibit DA neurodegeneration in the VTA. Error bars represent mean ± SEM; n = 10 WT, n = 7 U144; t-test; n.s., not significant. (D) Representative images of CHCHD2-T61I Tg line U105 midbrain immunostained for tyrosine hydroxylase to visualize DA neurons. (E) Quantification of TH+ cells in the SN. One-year-old CHCHD2-T61I Tg line U105 mice exhibit DA neurodegeneration in the SN. Error bars represent mean ± SEM; n = 5/genotype; t-test, **P < 0.005. (F) Quantification of TH+ cells in the VTA. One-year-old CHCHD2-T61I Tg line U105 mice do not exhibit DA neurodegeneration in the VTA. Error bars represent mean ± SEM; n = 5 WT, n = 5 U105; t-test; n.s., not significant. (G) Representative immunoblots of RIPA soluble and insoluble lysates from 1-year-old CHCHD2-T61I Tg line U105 midbrain. CHCHD2-T61I Tg line U105 mice exhibit insoluble accumulation of α-synuclein and S129 phospho-α-synuclein in the midbrain. (H) Representative images of midbrain tissue from 1-year-old CHCHD2-T61I Tg line U144 mice immunostained for α-synuclein, synaptophysin and tyrosine hydroxylase (Scale bar: 20 μm). (I) Quantification of colocalization between α-synuclein and synaptophysin. Error bars represent mean ± SEM; n = 4/genotype; t-test, ***P < 0.0005. (J) Representative images of midbrain tissue from 1-year-old CHCHD2-T61I Tg line U105 mice immunostained for α-synuclein, synaptophysin and tyrosine hydroxylase (Scale bar: 20 μm). (K) Quantification of colocalization between α-synuclein and synaptophysin. Error bars represent mean ± SEM; n = 4/genotype; t-test, *P < 0.05. (L) Representative immunoblots of RIPA-soluble synaptosomal and cytosolic lysates from brains of 1-year-old CHCHD2-T61I Tg line U105 mice. (M) Quantification of cytosolic α-synuclein. Error bars represent mean ± SEM; n = 4 WT, 3 U105; t-test, **P < 0.005.
α-synuclein is an abundant presynaptic protein linked genetically and pathologically to PD and other LBDs (1,2,37–39). Given that the pathological hallmark of PD includes dopaminergic neuronal loss and aberrant accumulation of α-synuclein, we tested the hypothesis that CHCHD2-T61I Tg mice exhibit α-synuclein aggregation. Western blotting of RIPA soluble and insoluble lysates from the midbrain of 1-year-old CHCHD2-T61I Tg line U105 mice revealed insoluble accumulation of α-synuclein and S129 phospho-α-synuclein compared with WT littermates (Fig. 6G), which are known to contribute to DA neuron loss. Under physiological conditions, α-synuclein localizes to presynaptic terminals and assists with SNARE-dependent vesicle fusion (40–43), which is essential for proper neuronal signaling. However, under pathological conditions, α-synuclein is mislocalized from distal presynaptic terminals, becomes aggregated in proximal neuronal compartments (44,45) and forms LBs. Therefore, we performed immunohistochemical analysis to assess colocalization between α-synuclein and the presynaptic marker synaptophysin in brain tissue. In the midbrains of one-year-old CHCHD2-T61I Tg mice line U144, we observed a significant reduction in colocalization between α-synuclein and synaptophysin compared with WT littermates (Fig. 6H and I). We also confirmed the same finding in the CHCHD2-T61I Tg mice line U105 (Fig. 6J and K). To assess this finding in a different way, we separated synaptosome and cytosol fractions from 1-year-old CHCHD2-T61I Tg line U105 mice and WT littermates. As expected, we found a significant increase in α-synuclein in the cytosolic fraction of CHCHD2-T61I Tg line U105 mice compared with WT littermates (Fig. 6L and M), indicating that the CHCHD2-T61I mutation leads to α-synuclein mislocalization.
CHCHD2-T61I Tg mice show motor deficits without cognitive impairments at 1 year of age
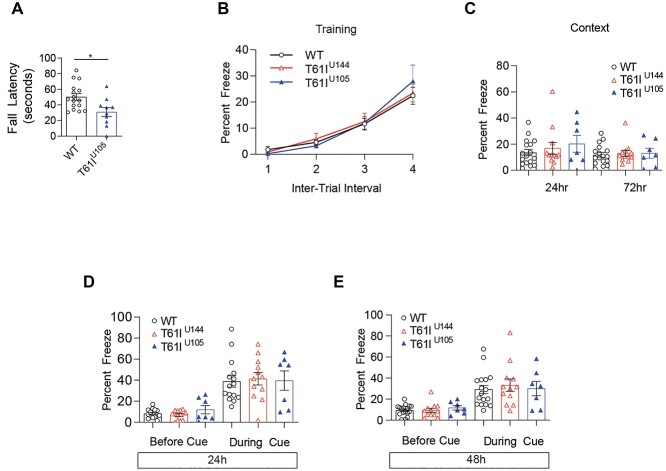
The CHCHD2-T61I mutation is primarily associated with late-onset autosomal dominant PD, and patients harboring this mutation exhibit motor dysfunction (20,21). Previous studies have shown that the CHCHD2-T61I mutation leads to locomotor impairments in Drosophila (24) and that CHCHD2-T61I and α-synuclein synergistically exacerbate motor deficits in Drosophila (21). However, these motor deficits were assessed in Drosophila models, and no studies have characterized CHCHD2-T61I-induced motor deficits in mammalian models. We performed rotarod testing with 1-year-old CHCHD2-T61I Tg line U105 mice and WT littermates. Rotarod testing data indicate a significant decrease in fall latency in CHCHD2-T61I Tg line U105 mice compared with WT littermates (Fig. 7A), suggesting motor dysfunction at 1 year of age. Next, we assessed hind limb clasping, where the hind limbs of the mouse come together and the hind paws curl. This phenotype is indicative of midbrain lesioning and is seen in other PD mouse models (46,47). Hind limb clasping was scored on a scale of 0–3, with 0 indicating a lack of clasping, 1 indicating partial clasping, 2 indicating full clasping and 3 indicating severe clasping. Indeed, we found that CHCHD2-T61I Tg line U144 mice exhibit a significantly increased hind limb clasping score compared with WT littermates (Supplementary Material, Fig. S5A and B). These data further suggest that CHCHD2-T61I Tg mice exhibit motor impairments associated with PD.
Figure 7.
CHCHD2-T61I Tg mice exhibit motor dysfunction without cognitive impairments at 1 year of age. (A) Rotarod testing in 1-year-old CHCHD2-T61I Tg mice compared with WT littermates. CHCHD2-T61I Tg mice line U105 exhibit motor dysfunction as indicated by a significant decrease in fall latency (n = 16 WT, n = 10 U105; one-way ANOVA; *P < 0.05). (B) Fear conditioning training indicates strong association between the conditioned stimulus (tone) and unconditional stimulus (shock) due to an increase in freezing over time. CHCHD2-T61I Tg mice do not differ in learning ability at 1 year of age compared with nontransgenic WT littermates. (C) One-year-old CHCHD2-T61I Tg mice do not exhibit dysfunction in hippocampal-driven or (D and E) amygdala-driven memory at 1 year of age (n = 15 WT, n = 12 T61I U144, n = 7 T61I U105).
Next, we assessed whether the CHCHD2-T61I mutation leads to associative fear learning and memory impairments. Fear conditioning is a well-known method of determining changes in contextual (hippocampal) and cued (amygdala-driven) memory in rodent models (48,49). First, we trained the animals to associate a conditional stimulus (tone) with an unconditional stimulus (mild foot shock). Later, the mice were tested to determine if they remember the association. Contextual memory, associated with hippocampal learning, was tested by returning the animal to the environment they were trained in. Fear is measured as percent freeze, as freezing indicates the animals remember the training period. Cued memory, associated with amygdala, was then tested by placing the animal into a new environment, replaying the conditional stimulus and measuring freezing. Interestingly, we found that 1-year-old CHCHD2-T61I Tg line U144 and U105 mice do not show differences in learning ability during the testing period compared with WT littermates (Fig. 7B). Furthermore, contextual testing at 24 and 72 h after training showed no statistically significant differences between CHCHD2-T61I Tg mice and WT littermates (Fig. 7C), indicating the preservation of hippocampal memory. We also confirmed that cued testing 24 and 72 h after training was not significantly altered between WT and CHCHD2-T61I Tg mice (Fig. 7D and E). Collectively, these data indicate that while 1-year-old CHCHD2-T61I Tg mice exhibit motor deficits, they do not show any deficiencies in either hippocampal or amygdala-driven memory at this age.
Major clinical symptoms of PD and atypical parkinsonism patients is abnormal postures, including lateral flexion. Previous studies have reported a very high frequency (80~95%) of lateral flexion in PD (50,51). Pisa syndrome is characterized by a lateral deviation of the spine with a corresponding tendency to lean to one side and has an estimated prevalence of 7.4–10.3% in PD (52–54). However, the pathophysiology of Pisa syndrome in PD remains to be elucidated. Recent studies have shown that PD patients with Pisa syndrome show significantly worse performances in certain cognitive tests, including visuo-spatial tests (55). Interestingly, we found that ~10% of CHCHD2-T61I Tg mice sporadically develop a severe head tilt at 1 year of age (Video 1), similar to that observed in human patients with Pisa syndrome. These mice were unable to complete rotarod testing and spun when held up by their tails (Video 2). Obvious hind limb clasping was also observed in these mice, indicative of DA neurodegeneration. We next performed immunohistochemistry to assess α-synuclein accumulation in CHCHD2-T61I Tg line U105 mice with Pisa syndrome. Indeed, we found the aggregation and loss of synaptic localization of α-synuclein (Supplementary Material, Fig. S6A), as indicated by a significant reduction in α-synuclein puncta number in the midbrain (Supplementary Material, Fig. S6B). Furthermore, Western blotting data revealed the insoluble accumulation of S129 phospho-α-synuclein in the midbrains of CHCHD2-T61I Tg line U105 mice with Pisa syndrome (Supplementary Material, Fig. S6C). These results suggest that the severe head tilt and motor phenotype observed in these mice are associated with the aggregation and phosphorylation of α-synuclein in the midbrain, which is consistent with LBD pathology seen in human patients. Collectively, these results suggest that CHCHD2-T61I Tg mice recapitulate the motor phenotypes and pathologic features of human LBD patients with CHCHD2 mutations.
The CHCHD2-T61I mutation induces insoluble protein accumulation in vivo
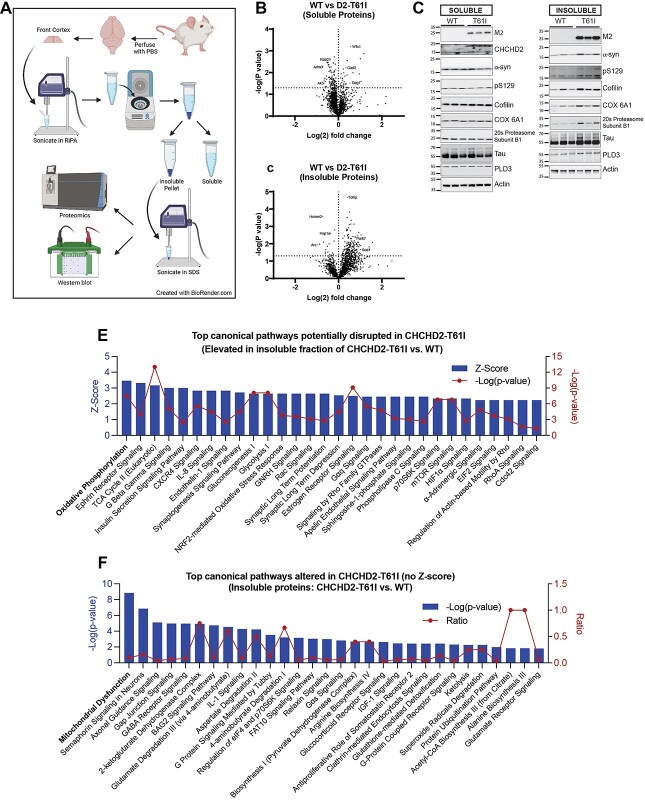
As the autopsied CHCHD2-T61I PD patient exhibited pathological phenotypes beyond α-synuclein accumulation, including neurofibrillary tangles, we next conducted an unbiased mass spectrometry-based proteomics assessment with subsequent ingenuity pathway analysis (IPA) to identify molecular pathways altered in CHCHD2-T61I Tg brains versus WT littermate brains. For this purpose, we extracted both RIPA-soluble and RIPA-insoluble fractions from 10-month-old CHCHD2-T61I line U144 mice and WT littermates to determine changes in physiological (soluble) and potentially pathological (insoluble) proteins by LC–MS/MS (Fig. 8A). Of the 4477 proteins detected in the soluble fraction, 158 proteins were significantly altered in CHCHD2-T61I brains compared with WT littermates, of which most proteins (115/158) were significantly reduced in CHCHD2-T61I Tg brains (Supplementary Material, Table S2). Surprisingly, of the 2301 proteins detected in the insoluble fraction, 273 insoluble proteins were significantly altered in CHCHD2-T61I brains compared with WT littermates, with most of them (245/273) being significantly increased in CHCHD2-T61I brains (Supplementary Material, Table S3). Volcano plot analysis of fold changes versus −Log (P-value) demonstrates a clear decrease in soluble proteins and an increase in insoluble proteins in CHCHD2-T61I brains (Fig. 8B and C), indicating that the CHCHD2-T61I mutation drives the insolubility of many proteins. Remarkably, of the 245 insoluble proteins significantly increased in CHCHD2-T61I brains, the majority (141) are known to localize to mitochondria, indicating that the CHCHD2-T61I mutation disproportionately and negatively impacts the solubility of mitochondrial proteins (Table 1), which is consistent with the localization of CHCHD2 to the IMS of mitochondria (Fig. 3D). Furthermore, 97 of the insoluble proteins significantly increased in CHCHD2-T61I brains were also associated with neurological or neurodegenerative diseases (Table 1). Notably, mitochondrial and neurodegenerative disease-associated proteins, including DJ-1, cofilin-1, SOD1, apoE and tau, were significantly increased in the RIPA-insoluble fraction as a consequence of CHCHD2-T61I expression (Supplementary Material, Table S3). We chose a subset of insoluble proteins detected by mass spectrometry and confirmed that they are indeed elevated in the insoluble fraction of CHCHD2-T61I Tg brains compared with WT littermates by western blotting, including cofilin, COX 6A1, 20S proteasome subunit β1, PLD3, tau and α-synuclein (Fig. 8D).
Figure 8.
Accumulation of insoluble proteins in CHCHD2-T61I Tg brains. (A) Schematic diagram of tissue lysate preparation for downstream proteomic and western blotting analysis (created with BioRender.com). Volcano plots showing Log (2) fold change of LFQ intensities and corresponding −Log (P-value) of proteins in 10-month-old CHCHD2-T61I brains compared with WT littermate brains from the RIPA-soluble fraction (B) and RIPA-insoluble fraction (C). The cut-off for significance (dotted horizontal line) is obtained by Welch’s t-test. Plots indicate reduced numbers of soluble proteins and increased numbers of insoluble proteins in CHCHD2-T61I brains. (D) Representative western blots from RIPA-soluble and RIPA-insoluble fractions of 10-month-old CHCHD2-T61I and WT littermate brains for a subset of proteins detected by mass spectrometry. Blots confirm increases in specific insoluble proteins in CHCHD2-T61I brains. (E) Graph represents IPA of top canonical pathways potentially disrupted in the insoluble fraction of CHCHD2-T61I brains compared with WT littermates plotted by Z-score and corresponding −Log (P-value). (F) Graph represents IPA of top canonical pathways (without Z-scores) altered in the insoluble fraction of CHCHD2-T61I brains compared with WT littermates plotted by −Log (P-value) and ratio of proteins mapped to the corresponding canonical pathway.
Table 1.
Insoluble proteins accumulated in CHCHD2-T61I Tg cortex
| T61I versus WT (protein categories) | |
|---|---|
| Mitochondrial | 141 up, 7 down |
| Ribosome/RNA-binding | 37 up, 4 down |
| ER/membrane | 104 up, 13 down |
| Neurological/neurodegenerative disease | 97 up, 13 down |
| Protein degradation | 17 up, 3 down |
| Chaperone/chaperone binding | 10 up, 2 down |
| Kinases/phosphatases | 19 up, 1 down |
| Myelin proteins | 3 up, 0 down |
| GTPases and related proteins | 35 up, 12 down |
| Cytoskeletal | 38 up, 13 down |
| DNA/histone binding | 7 up, 7 down |
| Glucose metabolism | 8 up, 0 down |
A complete list of genes for Table 1 can be found in Supplementary Material, Tables S6 and S7.
We next conducted IPA to determine the canonical biological pathways impacted by the CHCHD2-T61I mutation initially by pathways showing positive Z-scores (increased insolubility) and statistical significance [−Log(P-value)]. In the context of insoluble proteins, higher Z-scores likely reflect increased perturbation of the corresponding pathways. Accordingly, among the 111 canonical pathways showing significant positive Z-scores in CHCHD2-T61I versus WT (Supplementary Material, Table S4) in the insoluble fraction, ‘oxidative phosphorylation’ showed the highest Z-score, likely representing the highest pathway perturbation, followed by ‘Ephrin receptor signaling’ and ‘TCA cycle II’ (Fig. 8E). Statistical significance in −Log (P-values) are plotted in red and overlaid on Z-score blue bars, with highest values representing highest statistical significance (Fig. 8E). Other notable canonical pathways of potential perturbation in the insoluble fraction included ‘Nrf2-mediated oxidative stress response’ and ‘synaptic long-term potentiation’ (Fig. 8E), the latter which we experimentally confirmed in the CHCHD2-T61I Tg mice (Fig. 5G). In addition, many other canonical pathways were shown to be potentially disrupted in CHCHD2-T61I brains by increased protein insolubility (Fig. 8E). In contrast to the large numbers of canonical pathways demonstrating significant positive Z-scores, only a few pathways showed negative Z-scores in the insoluble fraction of CHCHD2-T61I brains (Supplementary Material, Fig. S7A), consistent with the observed predominant increase in protein insolubility. Of the top canonical pathways altered in the insoluble fraction of CHCHD2-T61I Tg brains that could not be assigned Z-scores by IPA (no specific directionality), ‘mitochondrial dysfunction’ showed the highest statistical significance (−Log[P-value]) (Fig. 8F). The ratios of proteins that map to the corresponding pathway are plotted in red (Fig. 8F). Notably, ‘GABA receptor signaling’ and ‘Glutamate degradation III (via 4-aminobutyrate)’ canonical pathways were among the most significantly altered in both the insoluble (Fig. 8F) and soluble fractions (Supplementary Material, Fig. S7B), indicating generalized imbalance in these pathways in CHCHD2-T61I Tg brains. In addition, ‘gluconeogenesis’ and ‘glycolysis’ pathways were also altered in both insoluble (Fig. 8E) and soluble (Supplementary Material, Fig. S7B) fractions of CHCHD2-T61I versus WT brains, suggesting that these metabolic pathways are also perturbed downstream of ‘mitochondrial dysfunction’ and disruptions in ‘oxidative phosphorylation’. These results taken together demonstrate that the CHCHD2-T61I mutation impacts not only α-synuclein pathogenesis but also a multitude of other pathways originating from the insoluble accumulation of mitochondrial proteins and deregulating multiple signaling and metabolic pathways. Tables containing a list of all canonical pathways altered in CHCHD2-T61I Tg mice compared with WT littermates can be found in Supplementary Material, Tables S4 and S5.
Discussion
Mutations in CHCHD2 have been shown to contribute to LBDs (20,21), while the precise mechanistic underpinnings remain unknown. CHCHD2 knockout or CHCHD2/CHCHD10 double knockout mouse models fail to recapitulate the pathology seen in human patients with CHCHD2 mutations (25,26). While Drosophila models have been used to study the role of CHCHD2 and its mutations in PD (15,21,24), no transgenic or knock-in models of CHCHD2 mutations have been reported in the mammalian setting.
In this study, we generated the first transgenic mouse model neuronally expressing PD-linked CHCHD2-T61I driven by the mouse Prp promoter. CHCHD2-T61I Tg mice are viable and successfully express the transgene. Furthermore, they exhibit mitochondrial aggregation in the midbrain, increased gliosis and reduced synaptic integrity in vivo. They also exhibit impaired LTP, with no changes in PPF and I-O, indicating that the CHCHD2-T61I mutation impairs long-term synaptic plasticity. Furthermore, mislocalization and accumulation of α-synuclein from the synapse is observed in CHCHD2-T61I Tg brains. Unbiased proteomic analysis indicates robust insoluble protein accumulation, particularly in mitochondrial proteins, indicating proteostasis imbalance originating from the mitochondria. IPA reveals that the CHCHD2-T61I mutation significantly alters multiple canonical pathways, including oxidative phosphorylation, mitochondrial dysfunction, multiple metabolic pathways and proteins associated with neurodegenerative disease. Behavioral tests show hind limb clasping and motor dysfunction, without cognitive impairments, at 1 year of age in CHCHD2-T61I Tg mice, suggesting a phenotype similar to those seen in human patients with movement disorders. Interestingly, a minor proportion of the CHCHD2-T61I Tg mice (~10%) develop a much more severe motor phenotype resembling human Pisa syndrome, which was never observed in WT mice. Collectively, these results indicate that neuronal CHCHD2-T61I expression induces pathological and motor changes consistent with LBDs.
Previously reported CHCHD2 mouse models include a CHCHD2/CHCHD10 double knockout (DKO) (26), and a recently reported CHCHD2 knockout model (25). DKO mice exhibit disrupted mitochondrial cristae and cardiomyopathy (26). Two-year-old CHCHD2 knockout mice present motor dysfunction, loss of midbrain DA neurons and accumulation of p62/SQSTM1 inclusions (25). CHCHD2-T61I Tg mice exhibit similar changes at 1 year of age, indicating that the T61I mutation induces a phenotype consistent with at least partial loss-of-function. It has been suggested that due to sequence similarities between CHCHD2 and CHCHD10, they are able to compensate for one another if one is lost. However, this compensatory effect declines with age, as indicated by the loss-of-function phenotype seen in 2-year-old CHCHD2 knockout mice (25). CHCHD2-T61I expression induces motor dysfunction and DA neurodegeneration at 1 year of age, indicating a more rapid phenotypic progression than CHCHD2 knockout mice, likely through gain-of-function effects. As CHCHD2-T61I per se is known to readily misfold and become insoluble (29), misfolded CHCHD2-T61I is likely to exert gain-of-function activities. Indeed, our unbiased proteomics observations underscore such mode of action, as CHCHD2-T61I Tg mice exhibit accumulation of many proteins in the insoluble fraction, suggesting that misfolded CHCHD2-T61I impairs proteostasis machinery. Such impairment likely originates from the mitochondria, as mitochondrial proteins are highly overrepresented (~57%) among the elevated insoluble proteins in CHCHD2-T61I Tg mice. Indeed, oxidative phosphorylation and mitochondrial dysfunction pathways are the most significantly altered among the IPA canonical pathways in the insoluble fraction of CHCHD2-T61I Tg brains. Such insoluble phenotype is consistent with previous studies showing functional deficits in mitochondrial respiration and electron flow by CHCHD2-T61I expression (15,29). Secondary insoluble alterations particularly in metabolic (i.e. TCA cycle, gluconeogenesis, glycolysis), anti-oxidant (i.e. Nrf2) and glutamate/GABA signaling pathways suggest broader pathophysiological consequences downstream of mitochondrial dysfunction that likely impact the CHCHD2-T61I phenotype. Our observation that insoluble CHCHD2 positively correlates with insoluble α-synuclein in human LBD brains suggests that spontaneous misfolding of WT CHCHD2 in the disease setting may also drive the accumulation of insoluble α-synuclein and multiple mitochondrial proteins that may impact disease progression in LBDs.
Autopsy data from a PD patient with the T61I mutation demonstrate loss of midbrain DA neurons and widespread LB, neurofibrillary tau tangle and amyloid-β pathology (21). One-year-old CHCHD2-T61I Tg mice exhibit DA neuron loss and increased insoluble α-synuclein, S129 phospho-α-synuclein, tau, cofilin and many mitochondrial proteins, indicating that these mice capture the pathology observed in human patients. Interestingly, recent observations show that α-synuclein exacerbates tau pathology (56), while tau mediates postsynaptic dysfunction associated with A53T α-synuclein expression (57). Additionally, cofilin associates with α-synuclein and promotes its aggregation and transmission in vivo (58). Furthermore, pull down of S129 phospho-α-synuclein from LBD patients reveals that tau, DJ-1, and many other mitochondrial proteins are significantly enriched compared with control patients (59), which is consistent with our proteomics data from CHCHD2-T61I Tg brain. Therefore, further study of CHCHD2-T61I Tg mice may help to elucidate the molecular mechanisms underlying mutant CHCHD2-induced pathology, leading to improve in vivo understanding of LBDs. Although we observe insoluble accumulation of neurodegenerative disease-associated proteins and impaired LTP at 10 months of age, no changes in learning and memory are detected at 1 year of age as measured by fear conditioning testing. Nonetheless, it is likely that CHCHD2-T61I Tg mice will develop learning and memory dysfunction at more advanced ages, similar to that seen in PD dementia or DLB. Therefore, further studies in 18–24-month-old CHCHD2-T61I Tg mice may reveal more pronounced motor and cognitive decline.
Materials and Methods
Mouse models
Wild-type (WT) C57BL6 and hemizygous transgenic mice expressing CHCHD2-T61I were bred in the C57BL6 background. Mice were kept at standard vivarium conditions with food and water supplied ad libitum with a 12-h light/dark cycle.
Transgenic mice were generated as previously described (26). To generate transgenic mice expressing human CHCHD2-T61I, a transgenic expression construct containing the mouse Prp promoter (MoPrP.Xho) was obtained from Dr David Borchelt (University of Florida, FL, USA). This construct has previously been used to create other mouse models of neurodegeneration, such as the α-synuclein A53T (60), PS19 tauP301S (61) and APP/PS1 (62) transgenic models. Flag-CHCHD2-T61I was subcloned into the XhoI site of the MoPrP.Xho vector and we sequenced the region for validity. A valid transgenic plasmid was sent for embryo injection to the University of Utah Gene Targeting Core (Salt Lake City, Utah, USA). The transgenic construct was microinjected into at least 200 fertilized C57BL6 embryos and implanted into pseudo-pregnant mice. Genomic DNA isolated from tail snips from over 100 offspring was used for genotyping by PCR with the following primer sequences for human CHCHD2: forward primer, 5′-ATG AGA GCT GCA CCC AGG CCA-3′ and reverse primer, 5′-CTG CTG CTG CTG TGC TGG CT-3′. Positive founders were bred with WT C57BL6 mice for a minimum of three generations prior to characterization. To confirm the presence of the intended T61I mutation, the CHCHD2 transgene was amplified by PCR from genomic DNA, and the open reading frame was sequenced. To quantify expression of the CHCHD2-T61I transcript compared with endogenous expression of CHCHD2, total RNA isolated from both C57BL6 and CHCHD2-T61I mice were subjected to qRT-PCR using primers common to both mouse and human CHCHD2: forward primer, 5′-ATG CCG CGT GGA AGC CGA AG-3′ and reverse primer, 5′-CCT GAG GCT CCT GGT AAG TG-3′.
Behavior testing
Hindlimb clasping
Hindlimb clasping severity was quantified as previously described (63). Briefly, mice were held by the tail and suspended for 10 s while clasping behavior was observed. Clasping severity was scored on a scale of 0–3. A score of 0 is assigned when both hindlimbs are splayed outwards from the abdomen. A score of 1 is assigned when one hindlimb is retracted toward the abdomen. A score of 2 is assigned when both hindlimbs are partially retracted toward the abdomen. Finally, a score of 3 is assigned when both hindlimbs are completely retracted, touching the abdomen.
Rotarod Mice underwent Rotarod testing at 12 months of age to assess motor performance. White noise was played continuously to lessen the impact of environmental noise. Each mouse was allowed a training period prior to the start of the test. During this training period, the mouse was placed onto the stationary rod of the Panlab Harvard rotarod apparatus. When the mouse fell, it was placed back on the stationary rod until it was stable. The training period continued by rotating the rod at a constant speed (4 r.p.m.) to allow the mice to acclimate to walking on the rotarod.
When the mice were comfortable with the constant-velocity rotations, an accelerated rotation began, signaling the start of the assessment. The assessment began with rotations paced at 4 r.p.m. and accelerated to 40 r.p.m. gradually over 300 s. A timer was started at the beginning of each trial and stopped when the mouse fell off of the rod and landed on the platform below. A total of four trials were completed and the time elapsed from each trial was averaged for each mouse.
Fear conditioning Fear conditioning was used to measure learning and memory in the forms of contextual and cued responses. White noise was played continuously to lessen the impact of environmental noise. During training, mice were individually placed into a Coulbourn HABITEST mouse test cage. After 175 s, a monotone sound was played for 30 s followed by a 2-s mild foot shock (0.5 mA) administered by a Coulbourn precision animal shocker. This was repeated for a total of three times. Freezing times during training were used to assess learning for the following trials. The following trials were conducted 24 and 72 h after the initial training to test both contextual and cued responses. Contextual response testing consisted of re-introducing mice into the test cage (Coulbourn, Whitehall, PA, USA) and recording freezing behavior for 180 s. Cued response testing involved altering the environment of the chamber by adding a different platform, siding and scented odor. Mice were then introduced to the altered chamber, and after 180 s, the same monotone sound played for 180 s. Freezing behavior was recorded with FreezeFrame 5 software (ActiMetrics) with a threshold setting of 1.0 and bout setting of 0.25 s.
Tissue harvesting
Extracted samples from various regions of the brain were harvested in RIPA lysis buffer (50 mm Tris–HCl pH 7.4, 150 mm NaCl, 2 mm ethlenediaminetetra acetic acid, 1% NP-40, 0.1% sodium dodecyl sulfate) with protease and phosphatase inhibitors added (GeneDEPOT, P3200-005 and P3100-010).
Tissues were sonicated in RIPA buffer with a QSonica Q125 sonicator at 20% amplitude with a pulse lasting 2 s then resting for 1 s before pulsing again. This was done until the sample was fully homogenized. Sonicated samples were then centrifuged at 4°C and 15 000 r.p.m. (Eppendorf 5424R) for 15 min to separate the RIPA-soluble and RIPA-insoluble fractions. The supernatant was decanted into a new microcentrifuge tube; this is the RIPA-soluble fraction. The pellet was washed with 100 μL of RIPA and inhibitor solution and centrifuged at 4°C and 15 000 r.p.m. (Eppendorf 5424R) for 5 min. The RIPA was decanted and discarded.
The pellet was then sonicated in sample buffer part 1 (10% sodium dodecyl sulfate, 250 mm Tris–HCl pH 6.8) in a room-temperature water bath sonicator (QSonica, Q800R3) in 3-min intervals at 50% amplitude with a pulse lasting 2 s then resting for 1 s before pulsing again. Sonication was repeated until the sample was fully homogenized; this is the RIPA-insoluble sample.
A BCA kit (BCA protein assay, Pierce, 23 225) and a Biotek microplate reader were used to determine protein concentration. Soluble and insoluble samples were equalized to the same concentration; 4× sample buffer (Alfa Aesar) with Dithiothreitol (DTT) was added to each soluble sample for a final concentration of 1×. Sample buffer part 2 (250 mm Tris–HCl pH 6.8, 0.1% bromophenol blue, 40% glycerol) and DTT were added to each insoluble sample in 1:1, v:v ratio. Soluble samples were stored at −20°C and insoluble samples were stored at room temperature for further analysis.
Synaptosomes were isolated from fresh tissue using the Syn-PER reagent and manufacturer’s protocol (ThermoFisher, 87 793). Isolated synaptosomes were sonicated in RIPA buffer plus inhibitors to harvest synaptic proteins.
Sarkosyl extraction of human cortex
Protein extraction from human cortex was performed as previously described (64,65). Briefly, brain tissue was sonicated in cold extraction buffer (25 mm Tris–HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA) with inhibitors, centrifuged at 80 000×g for 15 min at 4°C and the supernatants collected; this is the soluble fraction. A68 buffer (10 mm Tris–HCl, pH 7.4, 0.8 M NaCl, 10% Sucrose, 1 mm EGTA) with inhibitors was then added to the pellet, sonicated and centrifuged at 400×g for 20 min at 4°C. The supernatant was moved to a new tube, a 1% Sarkosyl solution was added, and incubated at room temperature for 1.5 h. Tubes were then centrifuged at 80 000×g for 30 min at room temperature. The supernatant was discarded, and the pellets were resuspended in 50 mm Tris–HCl pH 7.4; this is the insoluble fraction.
Mitochondrial subfractionation
Mitochondrial subfractionation was performed as previously described (66). Briefly, mitochondria were isolated from fresh tissue using the ThermoFisher mitochondrial isolation kit for tissue (89801). Isolated mitochondria were washed and then resuspended in swelling buffer (10 mm KH2PO4, pH 7.4) for 20 min at 4°C. An equal volume of shrinking buffer (10 mm HEPES, 10 mm MgCl2, 32% sucrose, 30% glycerol, pH 7.4) was added and resuspended for 15 min followed by centrifugation at 10 000×g for 15 min at 4°C. The pellet (mitoplast) contains the inner membrane (IM) and matrix, while the supernatant contains the outer membrane (OM) and IMS fractions. The supernatant was ultracentrifuged at 100 000×g for 40 min at 4°C to separate the OM and IMS fractions. The mitoplast was washed twice with washing buffer (250 mm sucrose, 1 mm EGTA, 10 mm HEPES, pH 7.4), resuspended in swelling buffer, sonicated and subjected to centrifugation at 12 000×g for 15 min at 4°C. The supernatant was then ultracentrifuged at 100 000×g for 40 min at 4°C to separate the IM and matrix fractions. All fractions were harvested with RIPA buffer with inhibitors, plus 4× sample buffer (Alfa Aesar) with Dithiothreitol (DTT) prior to western blotting.
Western blotting
The equalized soluble and insoluble tissue samples were boiled at 100°C for 10 min, then loaded into acrylamide gels and subjected to SDS-PAGE electrophoresis. The volume of sample loaded into the gel was based upon concentration of the sample that was determined by the BCA kit and Biotek microplate reader. The proteins were transferred from the acrylamide gel to 0.45 μm nitrocellulose membranes (GE Healthcare, 10600002) in a transfer tank (BIO-RAD, 1704070) by applying a 300 mA current to the gel for 100 min.
Membranes were blocked with 5% skim milk in TBS-T for 1 h at room temperature prior to primary antibody application. Primary antibodies in TBS-T buffer and 0.01% sodium azide were applied overnight at 4°C with continuous shaking. HRP secondary antibodies (1:1000, v:v) diluted in blocking solution were applied overnight at 4°C or for 4 h at room temperature with continuous shaking. Blots were developed with ECL western blot reagents (Pierce, 34 578) on a Fuji LAS-4000 imager (LAS-4000, Pittsburgh, PA, USA). Image quantification was performed using ImageJ Software (National Institutes of Health, Bethesda, MD, USA). All measurements were normalized to their respective actin levels.
Immunohistochemistry
Mice were perfused with 1× PBS and brain tissue samples were collected. The extracted brains were post-fixed in 4% paraformaldehyde at 4°C for 72 h. The 4% paraformaldehyde solution was then replaced with 30% sucrose for cryoprotection. The tissue samples were sectioned (25 μm) coronally or horizontally (American Optical Microtome, AO-860) and prepared for staining. Tissue samples were washed with 0.2% Triton in TBS, boiled in citrate buffer for 5 min for epitope retrieval, then blocked in a 3% NGS and 0.2% Triton in PBS solution for 1 h at room temperature. Tissues were then incubated with primary antibody in blocking solution at 4°C overnight with continuous shaking. The tissues were washed with PBS, and secondary Alexa fluor antibodies (1:1000, v:v) in blocking solution were added for 45 min at room temperature with continuous shaking. Tissues were washed with PBS for the final time, mounted on slides (Fisher Scientific, 12-544-7) and fully dried. Mounting solution (Invitrogen, 00-4958-02) was applied along with a glass coverslip (Fisher Scientific, 125485P) and left to dry for at least 12 h. Slides were imaged with the Keyence BZ-800 microscope (Keyence, Osaka, Japan) or ZEISS LSM880 confocal microscope (ZEISS, Jena, Germany). Signal intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Electrophysiology
Hippocampal brain slice preparation This acute hippocampal slice preparation was performed as previously described (67). Briefly, 10-month-old mice (C57BL6 or CHCHD2 T61I) were first decapitated. The brain was removed from the skull and put into ice-cold and oxygenated cutting solution (110 mm sucrose, 60 mm NaCl, 3 mm KCl, 28 mm NaHCO3, 1.25 mm NaH2PO4, 5 mm glucose, 0.6 mm ascorbate, 7 mm MgCl2 and 0.5 mm CaCl2). A Leica VT1200 was used to generate 400 mm parasagittal sections in cutting solution.
The hippocampus slice was dissected and transferred to room temperature cutting solution diluted 1:1 with artificial cerebrospinal fluid (ACSF) (125 mm NaCl, 2.5 mm KCl, 26 mm NaHCO3, 1.25 mm NaH2PO4, 25 mm glucose, 1 mm MgCl2 and 2 mm CaCl2), for 10 min. Then, slices were incubated in ACSF with constant 95% O2/5% CO2 perfusion for 30 min before being loaded into the brain slice recording chamber (Scientific Systems Design Inc.).
Ex vivo electrophysiology recording The long-term potentiation (LTP) recording was previously described (67). Slices were loaded into the recording chamber after a 30 min recovery. The recording chamber was held at 30 ± 0.5°C with an ACSF flow rate of 1 mL/min. Field excitatory postsynaptic potential (fEPSP) was recorded from stratum radiatum in the hippocampal CA1 area via a glass electrode (1–4 MΩ) loaded with ACSF. A stimulating electrode made by the formvar-coated nichrome wire (A-M Systems) was positioned on the Schaffer collaterals arising from the CA3 region, which was used to deliver biphasic stimulus pulses (1–18 V, 100 μs duration). The electric stimulation, controlled by pClamp 11 software (Molecular Devices), was delivered via the Digidata 1550B interface (Molecular Devices) and a stimulus isolator (model 2200; A-M Systems). Signals were amplified using a differential amplifier (model 1800; A-M Systems), filtered at 1 kHz and digitized at 10 kHz. The input–output relationship was determined by stimulating slices from 0 to 18 V at 0.5 V increments. A stimulus intensity, which would evoke ∼50% of the maximum fEPSP response, was determined from the input–output. This stimulus intensity was applied to the electrophysiological recording. PPF, which is a short-term plasticity, was induced with two sequential pulses and the interval was increased incrementally every 20 ms from 20 to 300 ms. A 20 min baseline fEPSP was recorded at 0.05 Hz as a control before the LTP evocation. LTP was induced by a theta burst stimulation (TBS), which consisted of five trains of four pulses at 200 Hz separated by 200 ms, repeated six times with an inter-train interval of 10 s. A 60-min fEPSP was recorded at 0.05 Hz following the TBS.
Statistical analysis Data in bar graphs are shown as the mean ± SEM. The evoked fEPSP measured by the initial slope after the TBS was normalized to the averaged baseline slope of each hippocampal brain slice. For the PPF, the second fEPSP slope was normalized to the first fEPSP slope. For the input–output experiment, the amplitude of fiber volley (FV) and the fEPSP slope were measured. FV½, which would be the fiber volley that evokes a ½ maximal fEPSP response, is a value that will normalize the fiber volley. The maximal fEPSP response will be used to normalize the fEPSP slope. A sigmoidal function (FV versus fEPSP) for each slice and a predicted value from this curve will be generated.
Proteomics
Equal amounts of protein (100 μg) were processed for LC–MS/MS using s-traps (Protifi) (68,69). Briefly, proteins are reduced with dithiothreitol (DTT), alkylated with iodoacetamide (IAA), acidified using phosphoric acid, and combined with s-trap loading buffer (90% MeOH, 100 mm TEAB). Proteins were loaded onto s-traps, washed with s-trap loading buffer, and finally digested with Trypsin/Lys-C (1:100, w:w; enzyme:protein) overnight at 37°C. Peptides were eluted and dried with a vacuum concentrator. Peptides were resuspended in H2O/1% acetonitrile/0.1% formic acid for LC–MS/MS analysis.
Peptides were separated using a 75 μm × 50 cm C18 reversed-phase-HPLC column (Thermo Scientific) on an Ultimate 3000 UHPLC (Thermo Scientific) using a 120 min gradient (2–32% ACN with 0.1% formic acid) and analyzed on a hybrid quadrupole-Orbitrap instrument (Q Exactive Plus, Thermo Fisher Scientific). Full MS survey scans were acquired at 70 000 resolution. The top 10 most abundant ions were selected for MS/MS analysis.
Raw data files are processed in MaxQuant (v 1.6.14.0, www.maxquant.org) and searched against the current Uniprot Mus musculus protein sequences database with the sequence for human CHCHD2 added. Search parameters included constant modification of cysteine by carbamidomethylation and the variable modifications, methionine oxidation and protein N-term acetylation. Proteins were identified using the filtering criteria of 1% protein and peptide false discovery rate. Protein intensity values were normalized using the MaxQuant LFQ function (70).
LFQ Intensity values were Log2-transformed for the creation of volcano plots. If a protein was detected in all three replicates in one experimental group and not detected in any replicates in the other experimental group, we assigned an LFQ value of 1 to represent below the threshold of detection. Statistically significant changes in protein abundance are determined using Welch’s t-test P-values and z-scores.
Statistical analysis
All graphs were created and analyzed with GraphPad Prism software (GraphPad Software, San Diego, CA, USA) using student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey post hoc test, two-way ANOVA followed by Dunnett’s, Sidak post hoc test, or Fisher’s exact test. Differences were deemed significant when P < 0.05. Quantified results are expressed as mean ± S.E.M.
Antibodies and reagents
Anti-CHCHD2 (Cat#: ab220688), anti-CHCHD10 (Cat#: ab121196), and anti-synaptophysin (Cat#: ab8049, ab32127) primary antibodies were obtained from Abcam (Cambridge, UK). Anti-Beta Actin (Cat#:66009-1-Ig), anti-COX6A1 (Cat#: 11460-1-AP) and anti-PLD3 (Cat#: 17327-1-AP) primary antibodies were obtained from Proteintech Group (Rosemont, IL, USA). Anti-mitofilin (D-3) (Cat#: sc-390 707), anti-Tau (A-10) (Cat#: sc-390 476), anti-20s Proteasome Beta 1 (D-9) (Cat#: sc-374 405), anti-CHCHD4 (C-12) (Cat#: sc-365 137) and anti-Tom70 (A-8) (Cat#: sc-390 545) were obtained from Santa Cruz (Dallas, TX, USA). Anti-Alpha Synuclein (Cat#: 2642S), anti-Phospho-alpha-Synuclein (Ser129) (D1R1R) (Cat#: 23706S), anti-Cofilin (D3F9) (Cat#: 5175S), anti-PSD95 (Cat#: 3409S) and anti-DJ-1 (D29E5) (Cat#: 5933S) were purchased from Cell Signaling (Danvers, MA, USA). Anti-GFAP (2.2B10) (Cat#: 13-0300) and anti-Tyrosine Hydroxylase (Cat#: PA1-4679, Cat#: MA1-24654) were obtained from Invitrogen (Camarillo, CA, USA). Anti-Flag M2 (Cat#: F1804) was obtained from Sigma-Aldrich (St. Louis, MO, USA). Anti-Iba1 (Cat#: 09-19741) was purchased from Fujifilm Wako Chemicals (Richmond, VA, USA). Anti-CLPP (Cat#: NBP1-89557) was obtained from Novus Biologicals (Littleton, CO, USA). All antibodies are listed in Table 2.
Table 2.
Antibody information
| Target | Host | Supplier | Catalog number | WB dilution | IHC dilution |
|---|---|---|---|---|---|
| CHCHD2 | Rabbit Polyclonal | Abcam | ab220688 | 1:500 | 1:100 |
| CHCHD10 | Rabbit Polyclonal | Abcam | ab121196 | 1:1000 | – |
| Synaptophysin | Rabbit Monoclonal | Abcam | ab32127 | – | 1:500 |
| Synaptophysin | Mouse Monoclonal | Abcam | ab8049 | 1:1000 | 1:200 |
| β-Actin | Mouse Monoclonal | Proteintech | 66 009-1-Ig | 1:5000 | – |
| COX6A1 | Rabbit Polyclonal | Proteintech | 11 460-1-AP | 1:1000 | – |
| PLD3 | Rabbit Polyclonal | Proteintech | 17 327-1-AP | 1:1000 | – |
| Mitofilin | Mouse Monoclonal | Santa Cruz | sc-390707 | – | 1:200 |
| Tau | Mouse Monoclonal | Santa Cruz | sc-390476 | 1:1000 | – |
| 20s Protβ1 | Mouse Monoclonal | Santa Cruz | sc-374405 | 1:1000 | – |
| CHCHD4 | Mouse Monoclonal | Santa Cruz | sc-365137 | 1:1000 | – |
| Tom70 | Mouse Monoclonal | Santa Cruz | sc-390545 | 1:1000 | – |
| Alpha Synuclein | Rabbit Polyclonal | Cell Signaling | 2642S | 1:1000 | 1:200 |
| Phospho α-syn | Rabbit Monoclonal | Cell Signaling | 23706S | 1:1000 | 1:200 |
| Cofilin | Rabbit Monoclonal | Cell Signaling | 5175S | 1:1000 | – |
| PSD95 | Rabbit Monoclonal | Cell Signaling | 3409S | 1:1000 | – |
| DJ-1 | Rabbit Monoclonal | Cell Signaling | 5933S | 1:1000 | – |
| GFAP | Rat Monoclonal | Invitrogen | 13-0300 | – | 1:500 |
| TH | Sheep Polyclonal | Invitrogen | PA1-4679 | – | 1:200 |
| TH | Mouse Monoclonal | Invitrogen | MA1-24654 | – | 1:200 |
| M2 (Flag) | Mouse Monoclonal | Sigma-Aldrich | F1804 | 1:1000 | 1:200 |
| Iba1 | Rabbit Polyclonal | Fujifilm | 09-19 741 | – | 1:500 |
| CLPP | Rabbit Polyclonal | Novus Biologicals | NBP1-89557 | 1:1000 | – |
Supplementary Material
Acknowledgements
We thank the USF proteomics core facility for performing mass spectrometry studies, Dr David Borchelt at the University of Florida for providing the transgenic expression construct containing the mouse Prp promoter (MoPrP.Xho) and the University of Utah transgenic core facility for generating the CHCHD2-T61I Tg mice. We also thank Drs Allan Levey and Marla Gearing for providing us with patients’ brain samples. Patient brain samples were obtained from Emory ADRC center (P50 AG025688).
Conflict of Interest statement. The authors declare that they have no conflict of interest.
Contributor Information
Teresa R Kee, Department of Pathology, CWRU School of Medicine, Cleveland, OH 44106, USA; Department of Molecular of Medicine, USF Health College of Medicine, Tampa, FL 33613, USA.
Jessica L Wehinger, Department of Molecular of Medicine, USF Health College of Medicine, Tampa, FL 33613, USA.
Pamela Espinoza Gonzalez, Department of Molecular of Medicine, USF Health College of Medicine, Tampa, FL 33613, USA.
Eric Nguyen, Department of Molecular of Medicine, USF Health College of Medicine, Tampa, FL 33613, USA.
Kyle C McGill Percy, Department of Pathology, CWRU School of Medicine, Cleveland, OH 44106, USA.
Sophia A Khan, Department of Pathology, CWRU School of Medicine, Cleveland, OH 44106, USA.
Dale Chaput, Department of Cell Biology, Microbiology, and Molecular Biology, University of South Florida, Tampa, FL 33620, USA.
Xinming Wang, Department of Pathology, CWRU School of Medicine, Cleveland, OH 44106, USA.
Tian Liu, Department of Pathology, CWRU School of Medicine, Cleveland, OH 44106, USA.
David E Kang, Department of Pathology, CWRU School of Medicine, Cleveland, OH 44106, USA; Louis Stokes Cleveland VA Medical Center, Cleveland, OH 44106, USA.
Jung-A A Woo, Department of Pathology, CWRU School of Medicine, Cleveland, OH 44106, USA.
Funding
National Institutes of Health (NIH) (R01AG059721 to J.-A.A.W., R01AG067741 to D.E.K. and J.-A.A.W., R01NS122350 to D.E.K., RF1AG053060 to D.E.K.); Veterans Affairs (BX004680 to D.E.K.).
Authors’ Contributions
T.R.K.: data curation, validation, investigation, software, formal analysis, methodology, visualization and writing—original draft, review and editing.
J.L.W.: data curation, validation, investigation, methodology and writing—review and editing.
P.E.G.: data curation, validation, investigation, methodology and writing—review and editing.
E.N.: data curation, validation, investigation, methodology and writing—review and editing.
K.C.M.P.: data curation, validation, investigation, methodology and writing—review and editing.
S.A.K.: data curation, validation, investigation, methodology and writing—review and editing.
D.C.: investigation, methodology, software and writing—review and editing.
X.W.: data curation, validation, investigation, methodology and writing—review and editing.
T.L.: data curation, validation, visualization and methodology—review and editing.
D.E.K.: conceptualization, resources, funding acquisition, project administration, formal analysis, supervision and writing—original draft, review and editing.
J.-A.A.W.: conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, project administration and writing—original draft, review and editing.
References
- 1. Jellinger, K.A. (2018) Dementia with Lewy bodies and Parkinson's disease-dementia: current concepts and controversies. J. Neural Transm. (Vienna), 125, 615–650. [DOI] [PubMed] [Google Scholar]
- 2. Spillantini, M.G., Schmidt, M.L., Lee, V.M., Trojanowski, J.Q., Jakes, R. and Goedert, M. (1997) Alpha-synuclein in Lewy bodies. Nature, 388, 839–840. [DOI] [PubMed] [Google Scholar]
- 3. Beal, M.F. (1998) Mitochondrial dysfunction in neurodegenerative diseases. Biochim. Biophys. Acta, 1366, 211–223. [DOI] [PubMed] [Google Scholar]
- 4. Johri, A. and Beal, M.F. (2012) Mitochondrial dysfunction in neurodegenerative diseases. J. Pharmacol. Exp. Ther., 342, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin, M.T. and Beal, M.F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature, 443, 787–795. [DOI] [PubMed] [Google Scholar]
- 6. Kopin, I.J. and Markey, S.P. (1988) MPTP toxicity: implications for research in Parkinson's disease. Annu. Rev. Neurosci., 11, 81–96. [DOI] [PubMed] [Google Scholar]
- 7. Langston, J.W. (2017) The MPTP story. J. Parkinsons Dis., 7, S11–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mustapha, M., and Taib, C. N. M. (2021) MPTP-induced mouse model of Parkinson's disease: a promising direction of therapeutic strategies. Bosn. J. Basic Med. Sci, 21, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bentivoglio, A.R., Cortelli, P., Valente, E.M., Ialongo, T., Ferraris, A., Elia, A., Montagna, P. and Albanese, A. (2001) Phenotypic characterisation of autosomal recessive PARK6-linked parkinsonism in three unrelated Italian families. Mov. Disord., 16, 999–1006. [DOI] [PubMed] [Google Scholar]
- 10. Hattori, N., Matsumine, H., Asakawa, S., Kitada, T., Yoshino, H., Elibol, B., Brookes, A.J., Yamamura, Y., Kobayashi, T., Wang, M. et al. (1998) Point mutations (Thr240Arg and Gln311Stop) [correction of Thr240Arg and Ala311Stop] in the Parkin gene. Biochem. Biophys. Res. Commun., 249, 754–758. [DOI] [PubMed] [Google Scholar]
- 11. Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y. and Shimizu, N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature, 392, 605–608. [DOI] [PubMed] [Google Scholar]
- 12. Matsumine, H., Saito, M., Shimoda-Matsubayashi, S., Tanaka, H., Ishikawa, A., Nakagawa-Hattori, Y., Yokochi, M., Kobayashi, T., Igarashi, S., Takano, H. et al. (1997) Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2-27. Am. J. Hum. Genet., 60, 588–596. [PMC free article] [PubMed] [Google Scholar]
- 13. Valente, E.M., Bentivoglio, A.R., Dixon, P.H., Ferraris, A., Ialongo, T., Frontali, M., Albanese, A. and Wood, N.W. (2001) Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am. J. Hum. Genet., 68, 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palikaras, K., Lionaki, E. and Tavernarakis, N. (2015) Balancing mitochondrial biogenesis and mitophagy to maintain energy metabolism homeostasis. Cell Death Differ., 22, 1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meng, H., Yamashita, C., Shiba-Fukushima, K., Inoshita, T., Funayama, M., Sato, S., Hatta, T., Natsume, T., Umitsu, M., Takagi, J. et al. (2017) Loss of Parkinson's disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat. Commun., 8, 15500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu, Y., Clegg, H.V., Leslie, P.L., Di, J., Tollini, L.A., He, Y., Kim, T.H., Jin, A., Graves, L.M., Zheng, J. et al. (2015) CHCHD2 inhibits apoptosis by interacting with Bcl-x L to regulate Bax activation. Cell Death Differ., 22, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kee, T.R., Espinoza Gonzalez, P., Wehinger, J.L., Bukhari, M.Z., Ermekbaeva, A., Sista, A., Kotsiviras, P., Liu, T., Kang, D.E. and Woo, J.A. (2021) Mitochondrial CHCHD2: disease-associated mutations, physiological functions, and current animal models. Front. Aging Neurosci., 13, 660843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ajroud-Driss, S., Fecto, F., Ajroud, K., Lalani, I., Calvo, S.E., Mootha, V.K., Deng, H.X., Siddique, N., Tahmoush, A.J., Heiman-Patterson, T.D. et al. (2015) Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics, 16, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aras, S., Pak, O., Sommer, N., Finley, R., Jr., Huttemann, M., Weissmann, N. and Grossman, L.I. (2013) Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res., 41, 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Funayama, M., Ohe, K., Amo, T., Furuya, N., Yamaguchi, J., Saiki, S., Li, Y., Ogaki, K., Ando, M., Yoshino, H. et al. (2015) CHCHD2 mutations in autosomal dominant late-onset Parkinson's disease: a genome-wide linkage and sequencing study. Lancet Neurol., 14, 274–282. [DOI] [PubMed] [Google Scholar]
- 21. Ikeda, A., Nishioka, K., Meng, H., Takanashi, M., Hasegawa, I., Inoshita, T., Shiba-Fukushima, K., Li, Y., Yoshino, H., Mori, A. et al. (2019) Mutations in CHCHD2 cause alpha-synuclein aggregation. Hum. Mol. Genet., 28, 3895–3911. [DOI] [PubMed] [Google Scholar]
- 22. Woo, J.-A., Liu, T., Trotter, C., Fang, C.C., De Narvaez, E., LePochat, P., Maslar, D., Bukhari, A., Zhao, X. and Deonarine, A. (2017) Loss of function CHCHD10 mutations in cytoplasmic TDP-43 accumulation and synaptic integrity. Nat. Commun., 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Imai, Y., Inoshita, T., Meng, H., Shiba-Fukushima, K., Hara, K.Y., Sawamura, N. and Hattori, N. (2019) Light-driven activation of mitochondrial proton-motive force improves motor behaviors in a Drosophila model of Parkinson's disease. Commun. Biol., 2, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tio, M., Wen, R., Lim, Y.L., Zukifli, Z.H.B., Xie, S., Ho, P., Zhou, Z., Koh, T.W., Zhao, Y. and Tan, E.K. (2017) Varied pathological and therapeutic response effects associated with CHCHD2 mutant and risk variants. Hum. Mutat., 38, 978–987. [DOI] [PubMed] [Google Scholar]
- 25. Sato, S., Noda, S., Torii, S., Amo, T., Ikeda, A., Funayama, M., Yamaguchi, J., Fukuda, T., Kondo, H., Tada, N. et al. (2021) Homeostatic p62 levels and inclusion body formation in CHCHD2 knockout mice. Hum. Mol. Genet., 30, 443–453. [DOI] [PubMed] [Google Scholar]
- 26. Liu, Y.T., Huang, X., Nguyen, D., Shammas, M., Wu, B., Dombi, E., Springer, D.A., Poulton, J., Sekine, S. and Narendra, D.P. (2020) Loss of CHCHD2 and CHCHD10 activates OMA1 peptidase to disrupt mitochondrial cristae phenocopying patient mutations. Hum. Mol. Genet., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu, X., Wang, Q., Yang, Y., Stewart, T., Shi, M., Soltys, D., Liu, G., Thorland, E., Cilento, E.M., Hou, Y. et al. (2021) Reduced erythrocytic CHCHD2 mRNA is associated with brain pathology of Parkinson's disease. Acta Neuropathol. Commun., 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aras, S., Bai, M., Lee, I., Springett, R., Huttemann, M. and Grossman, L.I. (2015) MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion, 20, 43–51. [DOI] [PubMed] [Google Scholar]
- 29. Cornelissen, T., Spinazzi, M., Martin, S., Imberechts, D., Vangheluwe, P., Bird, M., De Strooper, B. and Vandenberghe, W. (2020) CHCHD2 harboring Parkinson's disease-linked T61I mutation precipitates inside mitochondria and induces precipitation of wild-type CHCHD2. Hum. Mol. Genet., 29, 1096–1106. [DOI] [PubMed] [Google Scholar]
- 30. Aras, S., Arrabi, H., Purandare, N., Huttemann, M., Kamholz, J., Zuchner, S. and Grossman, L.I. (2017) Abl2 kinase phosphorylates Bi-organellar regulator MNRR1 in mitochondria, stimulating respiration. Biochim. Biophys. Acta Mol. Cell Res., 1864, 440–448. [DOI] [PubMed] [Google Scholar]
- 31. Mrak, R.E. and Griffin, W.S. (2005) Glia and their cytokines in progression of neurodegeneration. Neurobiol. Aging, 26, 349–354. [DOI] [PubMed] [Google Scholar]
- 32. Czlonkowska, A. and Kurkowska-Jastrzebska, I. (2011) Inflammation and gliosis in neurological diseases--clinical implications. J. Neuroimmunol., 231, 78–85. [DOI] [PubMed] [Google Scholar]
- 33. Vila, M., Jackson-Lewis, V., Guegan, C., Wu, D.C., Teismann, P., Choi, D.K., Tieu, K. and Przedborski, S. (2001) The role of glial cells in Parkinson's disease. Curr. Opin. Neurol., 14, 483–489. [DOI] [PubMed] [Google Scholar]
- 34. Sze, C.I., Troncoso, J.C., Kawas, C., Mouton, P., Price, D.L. and Martin, L.J. (1997) Loss of the presynaptic vesicle protein synaptophysin in hippocampus correlates with cognitive decline in Alzheimer disease. J. Neuropathol. Exp. Neurol., 56, 933–944. [DOI] [PubMed] [Google Scholar]
- 35. Woo, J.A., Yan, Y., Kee, T.R., Cazzaro, S., McGill Percy, K.C., Wang, X., Liu, T., Liggett, S.B. and Kang, D.E. (2022) β-arrestin1 promotes tauopathy by transducing GPCR signaling, disrupting microtubules and autophagy. Life Sci. Alliance, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kopin, I.J. (1992) Features of the dopaminergic neurotoxin MPTP. Ann. N. Y. Acad. Sci., 648, 96–104. [DOI] [PubMed] [Google Scholar]
- 37. Singleton, A.B., Farrer, M., Johnson, J., Singleton, A., Hague, S., Kachergus, J., Hulihan, M., Peuralinna, T., Dutra, A., Nussbaum, R. et al. (2003) alpha-Synuclein locus triplication causes Parkinson's disease. Science, 302, 841. [DOI] [PubMed] [Google Scholar]
- 38. Baba, M., Nakajo, S., Tu, P.H., Tomita, T., Nakaya, K., Lee, V.M., Trojanowski, J.Q. and Iwatsubo, T. (1998) Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am. J. Pathol., 152, 879–884. [PMC free article] [PubMed] [Google Scholar]
- 39. Goedert, M. (2001) Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci., 2, 492–501. [DOI] [PubMed] [Google Scholar]
- 40. George, J.M., Jin, H., Woods, W.S. and Clayton, D.F. (1995) Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron, 15, 361–372. [DOI] [PubMed] [Google Scholar]
- 41. Kahle, P.J., Neumann, M., Ozmen, L., Muller, V., Jacobsen, H., Schindzielorz, A., Okochi, M., Leimer, U., van Der Putten, H., Probst, A. et al. (2000) Subcellular localization of wild-type and Parkinson's disease-associated mutant alpha -synuclein in human and transgenic mouse brain. J. Neurosci., 20, 6365–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maroteaux, L., Campanelli, J.T. and Scheller, R.H. (1988) Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J. Neurosci., 8, 2804–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burre, J., Sharma, M., Tsetsenis, T., Buchman, V., Etherton, M.R. and Sudhof, T.C. (2010) Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science, 329, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saha, A.R., Hill, J., Utton, M.A., Asuni, A.A., Ackerley, S., Grierson, A.J., Miller, C.C., Davies, A.M., Buchman, V.L., Anderton, B.H. et al. (2004) Parkinson's disease alpha-synuclein mutations exhibit defective axonal transport in cultured neurons. J. Cell Sci., 117, 1017–1024. [DOI] [PubMed] [Google Scholar]
- 45. Jensen, P.H., Nielsen, M.S., Jakes, R., Dotti, C.G. and Goedert, M. (1998) Binding of alpha-synuclein to brain vesicles is abolished by familial Parkinson's disease mutation. J. Biol. Chem., 273, 26292–26294. [DOI] [PubMed] [Google Scholar]
- 46. Lieu, C.A., Chinta, S.J., Rane, A. and Andersen, J.K. (2013) Age-related behavioral phenotype of an astrocytic monoamine oxidase-B transgenic mouse model of Parkinson's disease. PLoS One, 8, e54200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Filali, M. and Lalonde, R. (2016) Neurobehavioral anomalies in the Pitx3/ak murine model of Parkinson's disease and MPTP. Behav. Genet., 46, 228–241. [DOI] [PubMed] [Google Scholar]
- 48. Phillips, R.G. and LeDoux, J.E. (1992) Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav. Neurosci., 106, 274–285. [DOI] [PubMed] [Google Scholar]
- 49. Maren, S. (2001) Neurobiology of Pavlovian fear conditioning. Annu. Rev. Neurosci., 24, 897–931. [DOI] [PubMed] [Google Scholar]
- 50. Marsden, C.D. and Duvoisin, R. (1980) Scoliosis and Parkinson's disease. Arch. Neurol., 37, 253–254. [DOI] [PubMed] [Google Scholar]
- 51. Duvoisin, R.C. and Marsden, C.D. (1975) Note on the scoliosis of Parkinsonism. J. Neurol. Neurosurg. Psychiatry, 38, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tinazzi, M., Geroin, C., Gandolfi, M., Smania, N., Tamburin, S., Morgante, F. and Fasano, A. (2016) Pisa syndrome in Parkinson's disease: an integrated approach from pathophysiology to management. Mov. Disord., 31, 1785–1795. [DOI] [PubMed] [Google Scholar]
- 53. Kashihara, K. and Imamura, T. (2012) Clinical correlates of anterior and lateral flexion of the thoracolumbar spine and dropped head in patients with Parkinson's disease. Parkinsonism Relat. Disord., 18, 290–293. [DOI] [PubMed] [Google Scholar]
- 54. Doherty, K.M., van de Warrenburg, B.P., Peralta, M.C., Silveira-Moriyama, L., Azulay, J.P., Gershanik, O.S. and Bloem, B.R. (2011) Postural deformities in Parkinson's disease. Lancet Neurol., 10, 538–549. [DOI] [PubMed] [Google Scholar]
- 55. Artusi, C.A., Montanaro, E., Tuttobene, S., Romagnolo, A., Zibetti, M. and Lopiano, L. (2019) Pisa syndrome in Parkinson's disease is associated with specific cognitive alterations. Front. Neurol., 10, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bassil, F., Meymand, E.S., Brown, H.J., Xu, H., Cox, T.O., Pattabhiraman, S., Maghames, C.M., Wu, Q., Zhang, B., Trojanowski, J.Q. et al. (2021) α-Synuclein modulates tau spreading in mouse brains. J. Exp. Med., 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Teravskis, P.J., Covelo, A., Miller, E.C., Singh, B., Martell-Martinez, H.A., Benneyworth, M.A., Gallardo, C., Oxnard, B.R., Araque, A., Lee, M.K. et al. (2018) A53T mutant alpha-synuclein induces Tau-dependent postsynaptic impairment independently of neurodegenerative changes. J. Neurosci., 38, 9754–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yan, M., Xiong, M., Dai, L., Zhang, X., Zha, Y., Deng, X., Yu, Z. and Zhang, Z. (2022) Cofilin 1 promotes the pathogenicity and transmission of pathological alpha-synuclein in mouse models of Parkinson's disease. NPJ Parkinsons Dis., 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Killinger, B.A., Marshall, L.L., Chatterjee, D., Chu, Y., Bras, J., Guerreiro, R. and Kordower, J.H. (2022) In situ proximity labeling identifies Lewy pathology molecular interactions in the human brain. Proc. Natl. Acad. Sci. U. S. A., 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee, M.K., Stirling, W., Xu, Y., Xu, X., Qui, D., Mandir, A.S., Dawson, T.M., Copeland, N.G., Jenkins, N.A. and Price, D.L. (2002) Human alpha-synuclein-harboring familial Parkinson's disease-linked Ala-53 --> Thr mutation causes neurodegenerative disease with alpha-synuclein aggregation in transgenic mice. Proc. Natl. Acad. Sci. U. S. A., 99, 8968–8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoshiyama, Y., Higuchi, M., Zhang, B., Huang, S.M., Iwata, N., Saido, T.C., Maeda, J., Suhara, T., Trojanowski, J.Q. and Lee, V.M. (2007) Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron, 53, 337–351. [DOI] [PubMed] [Google Scholar]
- 62. Jankowsky, J.L., Fadale, D.J., Anderson, J., Xu, G.M., Gonzales, V., Jenkins, N.A., Copeland, N.G., Lee, M.K., Younkin, L.H., Wagner, S.L. et al. (2004) Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet., 13, 159–170. [DOI] [PubMed] [Google Scholar]
- 63. Guyenet, S.J., Furrer, S.A., Damian, V.M., Baughan, T.D., La Spada, A.R. and Garden, G.A. (2010) A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J. Vis. Exp., 39, e1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Woo, J.A., Liu, T., Fang, C.C., Castano, M.A., Kee, T., Yrigoin, K., Yan, Y., Cazzaro, S., Matlack, J., Wang, X. et al. (2020) beta-Arrestin2 oligomer.s impair the clearance of pathological tau and increase tau aggregates. Proc. Natl. Acad. Sci. U. S. A., 117, 5006–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Woo, J.A., Liu, T., Fang, C.C., Cazzaro, S., Kee, T., LePochat, P., Yrigoin, K., Penn, C., Zhao, X., Wang, X. et al. (2019) Activated cofilin exacerbates tau pathology by impairing tau-mediated microtubule dynamics. Commun. Biol., 2, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Park, S., Choi, S.G., Yoo, S.M., Son, J.H. and Jung, Y.K. (2014) Choline dehydrogenase interacts with SQSTM1/p62 to recruit LC3 and stimulate mitophagy. Autophagy, 10, 1906–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Woo, J.A., Zhao, X., Khan, H., Penn, C., Wang, X., Joly-Amado, A., Weeber, E., Morgan, D. and Kang, D.E. (2015) Slingshot-Cofilin activation mediates mitochondrial and synaptic dysfunction via Abeta ligation to beta1-integrin conformers. Cell Death Differ., 22, 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zougman, A., Selby, P.J. and Banks, R.E. (2014) Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics, 14, 1006–1000. [DOI] [PubMed] [Google Scholar]
- 69. HaileMariam, M., Eguez, R.V., Singh, H., Bekele, S., Ameni, G., Pieper, R. and Yu, Y. (2018) S-Trap, an ultrafast sample-preparation approach for shotgun proteomics. J. Proteome Res., 17, 2917–2924. [DOI] [PubMed] [Google Scholar]
- 70. Cox, J., Hein, M.Y., Luber, C.A., Paron, I., Nagaraj, N. and Mann, M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics, 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.