Abstract
To nominate novel disease genes for obesity and type 2 diabetes (T2D), we recently generated two mouse backcross populations of the T2D-susceptible New Zealand Obese (NZO/HI) mouse strain and two genetically different, lean and T2D-resistant strains, 129P2/OlaHsd and C3HeB/FeJ. Comparative linkage analysis of our two female backcross populations identified seven novel body fat-associated quantitative trait loci (QTL). Only the locus Nbw14 (NZO body weight on chromosome 14) showed linkage to obesity-related traits in both backcross populations, indicating that the causal gene variant is likely specific for the NZO strain as NZO allele carriers in both crosses displayed elevated body weight and fat mass. To identify candidate genes for Nbw14, we used a combined approach of gene expression and haplotype analysis to filter for NZO-specific gene variants in gonadal white adipose tissue, defined as the main QTL-target tissue. Only two genes, Arl11 and Sgcg, fulfilled our candidate criteria. In addition, expression QTL analysis revealed cis-signals for both genes within the Nbw14 locus. Moreover, retroviral overexpression of Sgcg in 3T3-L1 adipocytes resulted in increased insulin-stimulated glucose uptake. In humans, mRNA levels of SGCG correlated with body mass index and body fat mass exclusively in diabetic subjects, suggesting that SGCG may present a novel marker for metabolically unhealthy obesity. In conclusion, our comparative-cross analysis could substantially improve the mapping resolution of the obesity locus Nbw14. Future studies will throw light on the mechanism by which Sgcg may protect from the development of obesity.
Introduction
The pathogenesis of type 2 diabetes (T2D), which is characterized by chronically elevated blood glucose levels, is driven by genetic factors and their interaction with the environment (1,2). Approximately 90% of all patients with T2D are obese (3), underscoring obesity as the main risk factor in the disease development. Until today, the majority of genetic factors that predispose individuals to the development of obesity and T2D remain unknown (4,5). Genome-wide association studies (gWAS), in which clinical parameter are correlated with hundreds of thousands single nucleotide polymorphisms (SNPs), have nominated several hundred risk gene variants that are available in public databases (6). However, as the majority of the SNPs map into non-coding regions, functional evidence connecting genetic alterations with pathogenic processes of T2D is lacking for most of the genes. Therefore, mouse models that can be used for the engineering of gene mutations with well-established molecular genetic tools remain essential to study the molecular basis of T2D (7).
Numerous mouse strains are available that show a wide range of T2D-related phenotypes, similar as observed in the human population (7,8). By taking advantage of their divergent phenotypes, linkage studies on segregating outcross populations in combination with additional strategies, such as gene expression and bioinformatic analyses of the critical regions, have identified several novel T2D-modifier genes (9,10). However, linkage studies on a single population consisting of two inbred strains are limited; it will only survey loci that are genetically distinct between the two strains. Therefore, the comparative analysis of multiple inbred populations can substantially improve the mapping resolution of candidate disease genes (11–14).
The polygenetic New Zealand Obese (NZO) mouse strain has been frequently used as a model for spontaneous polygenetic obesity and T2D. Both genders develop impaired glucose tolerance and obesity in response to a high fat diet (15); however, subsequent T2D progressing into pancreatic islet failure is limited to the males, whereas the females benefit from the protective influence of the hormone oestrogen (16,17).
Several researchers (18–24), including ourselves (11–13,25,26), have used the NZO strain in genome-wide linkage studies attempting to uncover the gene variants that may drive the high susceptibility for obesity and T2D. All these studies were successful in identifying novel genomic regions, designated quantitative trait loci (QTL), associated with T2D-related phenotypes. However, due to huge nomination of candidate genes, the causal gene variants are still unknown for most of the identified QTL. Until today, only a few genes could have been functionally linked to the development of obesity and T2D in the obese NZO strain, including Ifgga2, Abcc8 and Pcpt (12,22,27).
To uncover novel gene variants that drive the prevalence for obesity and T2D in humans, we recently conducted crossbreeding experiments using the obese and T2D-susceptible NZO, and the two lean mouse strains, C3H/FeJ and 129P2/OlaHsd (11). Comparative linkage analysis of our two outcross populations and subsequent QTL mapping revealed two novel obesity-related gene variants that may influence adipose tissue function. For one of the two genes, sarcoglycan gamma (Sgcg), we provide functional evidence in glucose uptake in 3T3-L1 adipocytes.
Results
Diverging prevalence for obesity and insulin-resistance in the parental mouse strains
To validate the diverse metabolic features from our parental strains we monitored the development of body weight, body composition, blood glucose, and plasma insulin levels in female NZO, C3H and 129P2 mice. After weaning at 3 weeks of age, body weight was comparable between NZO and C3H, but significantly lower in 129P2 mice (NZO 13.4 ± 0.5 g, C3H 14.0 ± 0.4 g, 129P2 9.5 ± 0.2 g). As expected, during high-fat diet (HFD)-intervention NZO mice exhibited markedly higher body weight compared to the other two strains. At the age of 20 weeks, NZO mice gained 22 g and 42 g more than C3H and 129P2, respectively (NZO 66.4 ± 2 g, C3H 43.9 ± 0.5 g, 129P2 24.1 ± 0.8 g; Fig. 1A). These differences in body weight were mainly attributed to substantial differences in fat mass (week 15 of age: NZO 32.2 ± 1.6 g, C3H 13.9 ± 1 g, 129P2 5.76 ± 1.1 g; Fig. 1B), whereas lean mass was only slightly different between the strains (week 15 of age: NZO 25.7 ± 0.4 g, C3H 22.4 ± 0.7 g, 129P2 17.3 ± 0.7 g). Moreover, upon HFD NZO mice exhibited significantly higher blood glucose levels compared to the two lean strains. Maximal differences in glycaemia were observed at 18 week of age, when NZO mice exhibited 99 mg/dl and 123 mg/dl higher blood glucose levels compared to C3H and 129P2 mice, respectively (NZO 231 ± 22 mg/dl, C3H 132 ± 4 mg/dl, 129P2 108 ± 5 mg/dl; Fig. 1C). In addition, 6-hour fasting plasma insulin levels at the age of 22 weeks were 2-fold and 10-fold higher in NZO compared to C3H and 129P2 mice, respectively (NZO 9.9 ± 1.4 μg/L, C3H 5.1 ± 0.9 μg/L, 129P2 0.7 ± 0.3 μg/L; Fig. 1D).
Figure 1.
Metabolic characterization of females from parental mouse strains 129P2, C3H and NZO. Development of body weight (A) and blood glucose (C) was monitored at weeks 3, 6, 10, 15, 18 and 20 weeks of age, whereas fat mass (B) was measured at weeks 3, 6, 10 and 15. Six-hours-fasting plasma insulin levels (D) were measured at week 22 by ELISA. Dots represent single female animals (129P2: n = 11–12; C3H: n = 10–12; NZO: n = 12). Statistical differences between strains were calculated by 2-way (A-C) or one-way analysis of variance (D) followed by post hoc Bonferroni test, *P < 0.05, **P < 0.01, ***P < 0.001 by comparison to NZO unless otherwise stated.
Genome-wide linkage analysis on both N2 populations revealed seven novel obesity QTL
NZO females were bred either with C3H or 129P2 males to generate two F1 (F1(NZOxC3H) and F1(NZOx129P2)) generations. Subsequently, males from the F1 generations were bred with NZO females to generate two N2 populations. In total, 307 females from the N2(NZOx129P2) population and 310 females from the N2(NZOxC3H) population were metabolically phenotyped for T2D-associated traits. In this study, we present all significant obesity QTL, meaning loci that are linked to body weight and/or fat mass development, identified in our backcross females. In total, we identified seven novel obesity QTL (Fig. 2 and Table 1). Five QTL were identified exclusively in the N2(NZOxC3H) population (Nbw1p, Nbw4p, Nbw6, Nbw10 and Nbw13), whereas only one QTL on distal chromosome 1 (Nbw1d) was unique for the N2(NZOx129P2) cross. The QTL on proximal chromosome 14, designated Nbw14 (NZO body weight on chromosome 14) was the only QTL shared by both N2(NZOxC3H) and N2(NZOx129P2) crosses. Details on the seven different QTL, such as the confidence interval and the allelic effects are displayed in Table 1.
Figure 2.
Genome-wide linkage map of QTL for obesity-related traits in female mice derived from N2(NZOxC3H) and N2(NZOx129P2) outcross populations. Female NZO and male 129P2 or C3H mice, respectively, were used to generate the F1 generations, and male F1 offspring were subsequently backcrossed with NZO females to generate two backcross populations (N2(NZOxC3H) and N2(NZOx129P2)). The backcross populations were metabolically phenotyped, and genotyped for 105 N2(NZOxC3H) and 110 N2(NZOx129P2) informative SNP markers. Linkage analysis of the female backcross mice (n = 307–309) was conducted using the R-package as described in methods. Further information for each QTL is shown in Table 1. BW, body weight; FM, fat mass.
Table 1.
Summary of the major obesity QTL with significance for several weeks from female mice from both N2(NZOxC3H and NZOx129P2) populations
| Name | Chr | Traits | Peak-Pos (cM) | 95% CI (cM) | Closest SNP-marker (Mbp) | Max. LOD | Signif. weeks (max. effect) | Mean NZO/NZO | Mean NZO/C3H or NZO/129P2 | Cross |
|---|---|---|---|---|---|---|---|---|---|---|
| Nbw1p | 1 | BW | 9 | 0–20 | 46.3 | 4.0 | 10 | 45.2 g | 42.4 g | NZOxC3H |
| LM | 18 | 2–30 | 46.9 | 3.0 | 10 | 23.1 g | 22.3 g | NZOxC3H | ||
| FM | 8 | 0–22 | 27.3 | 3.6 | 6–10 (10) | 20.3 g | 18.3 g | NZOxC3H | ||
| Nbw1d | 1 | BW | 19 | 16–20 | 182.9 | 3.2 | 15–20 (20) | 53.0 g | 49.1 g | NZOX129P2 |
| BG | 19 | 13–20 | 182.9 | 3.5 | 20 | 147 mg/dl | 134 mg/dl | NZOX129P2 | ||
| Nbw4p | 4 | BW | 15 | 7–26 | 53 | 6.93 | 10–20 (19) | 62.1 g | 56 g | NZOxC3H |
| 4 | FM | 15 | 8–25 | 53 | 7.54 | 10–15 (15) | 28.7 g | 24.7 g | NZOxC3H | |
| Nbw6 | 6 | BW | 20 | 9–52 | 64.2 | 3.9 | 6–20 (6) | 31.7 g | 30 g | NZOxC3H |
| FM | 20 | 5–50 | 64.2 | 4.4 | 6–15 (6) | 10.5 g | 9.3 g | NZOxC3H | ||
| Nbw10 | 10 | BW | 7.2 | 5–11 | 89.1 | 9.3 | 6–20 (6) | 32.1 g | 29.5 g | NZOxC3H |
| FM | 7.2 | 3–10 | 89.1 | 6.3 | 6–20 (10) | 20.6 g | 17.8 g | NZOxC3H | ||
| Nbw13 | 13 | BW | 29.04 | 0–29 | 74.4 | 4.2 | 6–10 (6) | 31.8 g | 30 g | NZOxC3H |
| FM | 29 | 0–29 | 74.3 | 3.9 | 6–10 (6) | 10.4 g | 9.3 g | NZOxC3H | ||
| Nbw14 | 14 | BW | 23 | 17–30 | 63.6 | 5.2 | 6–20 (6) | 31.8 g | 29.8 g | NZOxC3H |
| FM | 23 | 17–32 | 63.5 | 6.3 | 6–15 (6) | 10.6 g | 9.2 g | NZOxC3H | ||
| BW | 32 | 25–40 | 65.4 | 4.4 | 15–20 (20) | 53.8 g | 49.2 g | NZOX129P2 | ||
| FM | 33 | 26–40 | 65.4 | 3.8 | 10–15 (15) | 22.7 g | 20.3 g | NZOX129P2 | ||
| BG | 29 | 22–37 | 65.4 | 3.1 | 20 | 148 mg/dl | 136 mg/dl | NZOX129P2 |
LOD scores, peak positions, and 95% confidence intervals (Bayesian method) were calculated by R/qtl software. NZOxC3H: n = 310, NZOx129P2: n = 307 females. BG, blood glucose; BW, body weight; Chr, chromosome; CI, confidence interval; FM, fat mass; LM, lean mass; LOD, logarithm of the odds; Pos, Position; Signif, significant.
Nbw14 represents the only obesity QTL that appeared in both crossbreedings
Out of our seven obesity QTL, only one of the loci showed overlap between the two crosses. Nbw14 revealed significant linkage with body weight (LOD 5.2 at 23 cM in N2(NZOxC3H) and LOD 4.4 at 32 cM in N2(NZOx129P2)) and fat mass (LOD 6.3 at 23 cM in N2(NZOxC3H) and LOD 3.7 at 33 cM in N2(NZOx129P2)) in both backcross populations. In addition, Nbw14 was further associated with blood glucose levels in N2(NZOx129P2) mice (LOD 3.1 at 29 cM, closest SNP marker at 65.4 Mb). In both crosses, the SNP marker associated with the highest LOD score for body weight and fat mass appeared almost at identical position (rs3702501 at 63.5 Mb in N2(NZOxC3H), Fig. 3A; rs3660830 at 65.4 Mb N2(NZOx129P2), Fig. 3B).
Figure 3.
Genetic linkage of body weight and fat mass to loci on Chr.14 in outcross populations of N2(NZOxC3H) and N2(NZOx129P2) female mice. Chr.14 revealed significant linkage with body weight and fat mass in both, N2(NZOxC3H) (A) and N2(NZOx129P2) (B) females. The SNP-markers that were used for genotyping with the corresponding Mb-position are plotted below. The SNP markers in closest proximity to the QTL peak position are marked in red. The genotypic effects are indicated with the arrows. LOD scores were calculated using R/qtl software as described in methods. LOD, logarithm of the odds; N, NZO; wk, week.
NZO-allele for Nbw14 is associated with increased body weight and fat mass in both crossbreedings
In both crossbreedings, NZO-allele carriers for Nbw14 gained more body weight compared to heterozygous allele carriers (N2(NZOxC3H): 62.4 g in Nbw14NZO/NZO compared to 58.9 g in Nbw14NZO/C3H at 20 weeks of age, N2(NZOx129P2: 53.8 g in Nbw14NZO/NZO compared to 49.2 g in Nbw14NZO/129P2 at 20 weeks of age; P < 0.001, Fig. 4A). These differences in body weight were attributed to significant differences in body fat (N2(NZOxC3H): 28.0 g in Nbw14NZO/NZO compared to 25.4 g in Nbw14NZO/C3H at 15 weeks of age, N2(NZOx129P2): 22.7 g in Nbw14NZO/NZO compared to 20.3 g in Nbw14NZO/129P2 at 15 weeks of age; P < 0.001, Fig. 4B). In contrast, the development of lean mass was similar between the different genotypes (Fig. 4C).
Figure 4.
Quantitative effect of Nbw14 in N2(NZOxC3H) and N2(NZOx129P2) mice. Mean values for body weight (± SEM) (A), fat mass (B) and lean mass (C) for homozygous NZO and heterozygous allele carriers for Nbw14 the N2(NZOxC3H) females. Mean values for body weight (± SEM) (D), fat mass (E) and lean mass (F) for homozygous NZO and heterozygous allele carriers for Nbw14 from N2(NZOx129P2) females. Nbw14NZO/NZO (N2(NZOxC3H): n = 181, Nbw14NZO/C3H (N2(NZOxC3H): n = 126, Nbw14NZO/NZO (N2(NZOx129P2): n = 133, Nbw14NZO/129P2 (N2(NZOx129P2): n = 170; N, NZO.
Combined haplotype- and gene expression analysis identifies two candidate genes for Nbw14
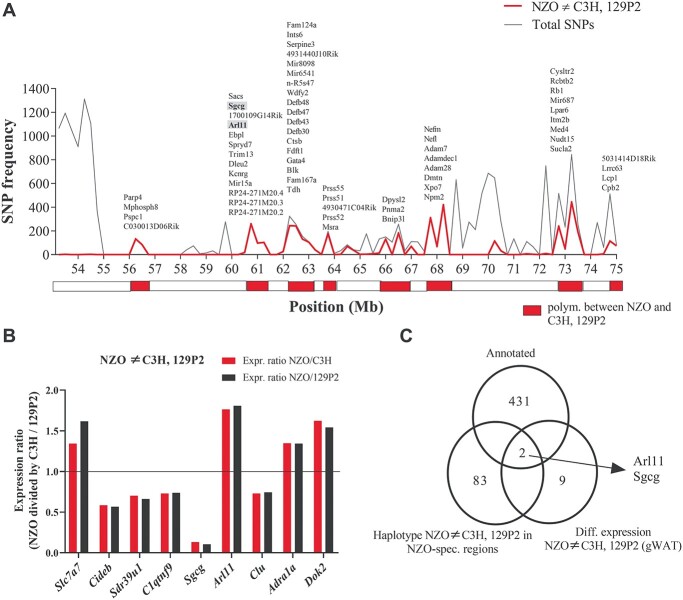
Based on the assumption that the causal gene variant for Nbw14 is different in NZO compared to C3H and 129P2 genomes, we used two major approaches to search for potential candidate genes. First, we used the SNP database from the Wellcome Trust Sanger Institute (28,29) to search for NZO polymorphisms in the defined critical region (53–75 Mb) that differed from C3H and 129P2. Since the genomic sequence for our C3H/FeJ substrain is not available in the database, the sequence from the closely related C3H/HeJ strain was used instead. As described before (11), for the dissection of the QTL peak into regions that are identical by decent (IBD) and polymorphic between NZO and the two lean strains, we determined the number of SNPs according to NZO ≠ C3H and 129P2 each 250 kbp. Regions exceeding a threshold of 100 SNPs/window were defined as polymorphic between NZO and C3H or 129P2, respectively (Fig. 5A). Out of 431 protein coding genes, only 83 were found to be located in regions defined as polymorphic between NZO and the other two strains. Supplementary Material, File S3 lists all annotated SNPs according to NZO ≠ C3H and 129P2 from all these 83 genes. We further analyzed these genes for coding nonsynonymous SNPs and found 42 protein polymorphisms where NZO differs from C3H and 129P2. According the ‘Sorting Tolerant From Intolerant’ (SIFT) algorithm (30), three variants in two genes (W277R in Pnma2, G285D and T330M in Rubcnl) are predicted to have deleterious impact on protein function (Supplementary Material, Table S2, Supplementary Material, File S2). However, none of these variants is unique for the NZO strain since these variants are shared by several lean mouse strains from the Sanger panel.
Figure 5.
Combined approach of haplotype- and gene expression analysis in gWAT of the parental strains for the identification of NZO-specific gene variants within Nbw14. (A) Haplotype analysis using Genome Reference Consortium Mouse Build 38 (GRC38) provided by the Wellcome Trust Sanger Institute. The red line represents the number of polymorphic SNPs according to NZO ≠ 129P2 and C3H, whereas the grey line shows the total number of SNPs (all SNPs annotated for C57B6/J reference genome with calls for C3H, 129 and NZO). Both lines overlap in the regions between 56.5–56.75 Mb, 60.25–61.5 Mb and 68.0–68.5 Mb. Genomic regions containing less than 100 polymorphic SNPs (NZO ≠ 129P2 and C3H) per window (250 kb) are represented with the white boxes. In contrast, regions with more than 100 polymorphic SNPs (NZO ≠ 129P2 and C3H) per window are highlighted by red boxes. Genes located in regions that were defined as polymorphic between NZO and the other strains are listed above. For a better overview, gene models are not included. (B) Significantly (P < 0.05) differential gene expression in NZO vs. C3H and 129P2 at the age of 6 weeks in gWAT detected by microarray analysis. Higher expression in NZO is indicated with an expression ratio (NZO/C3H and NZO/129P2) > 1, whereas downregulation in NZO is shown with an expression ratio (NZO/C3H and NZO/129P2) < 1. Differences between the strains were calculated by one-sided Wilcoxon signed rank test. (C) Venn diagram showing the total number of annotated genes (top), the number of genes revealed from the haplotype analysis (left), the number of genes revealed from the microarray analysis (right) and the overlap of genes (centre).
In addition, we implemented transcriptome data from gonadal white adipose tissue (gWAT) of our parental strains NZO, C3H and 129P2. In total, nine genes revealed to be either up- or downregulated in NZO gWAT compared to the other two strains. Whereas the expressions of Slc7a7, Arl11, Adra1a and Dok2 were upregulated, the five genes Cideb, Sdr39u1, C1qtnf9, Sgcg and Clu showed lower mRNA expression levels in NZO compared to the other two strains (Fig. 5B). However, only two (Arl11 and Sgcg) out of nine NZO-regulated genes were located in regions that were defined as highly polymorphic between NZO and the other strains. Thus, Arl11 and Sgcg, the only two genes that overlapped between our candidate gene approaches (Fig. 5C) were selected for further analysis as our main candidate genes for Nbw14.
Sgcg and Arl11 expressions are NZO-specifically regulated exclusively in adipose tissue
Using microarray data from several T2D-relevant tissues of our parental strains, we further compared the relative expression levels of Arl11 and Sgcg between white adipose tissue, liver, skeletal muscle, brown adipose tissue and pancreatic islets. In C3H and 129P2, expression levels for Sgcg could be exclusively observed in skeletal muscle, brown- and white adipose tissue, whereas liver and pancreatic islets did not show any Sgcg expression. In contrast, in NZO, Sgcg expression was limited to skeletal muscle (Fig. 6A). Arl11 revealed highest expression in gWAT tissue in all three strains. NZO-specific expression was observed exclusively in gWAT tissue, whereas no expression differences were detected in the other four tissues. In contrast to Sgcg, which was NZO specifically downregulated, Arl11 revealed to be NZO specifically upregulated in adipose tissue (Fig. 6C). NZO-specific gene expressions for Sgcg and Arl11 in gWAT were further validated by quantitative real-time polymerase chain reaction (qRT-PCR; Fig. 6B and D). Gene expression levels were markedly higher for Sgcg in the two lean strains (Ct values 23.0 in C3H, 23.2 in 129P2, 36.6 in NZO) compared to Arl11 in the obese strain (Ct values: 30.0 in NZO, 34.6 in C3H and 36.7 in 129P2). In addition, the differences in gene expression were bigger for Sgcg (ΔCt: 13.3 and 13.2) than for Arl11 (delta ΔCt: 6.7 and 4.6).
Figure 6.
Sgcg and Arl11 mRNA expressions across different tissues from the parental strains and validation in gWAT by qRT-PCR. All tissues were collected from male NZO, C3H and 129P2 mice at 6-weeks of age. The expressions of Sgcg and Arl11 in liver, skeletal muscle, gWAT, BAT and pancreatic islets was measured by microarray analysis and the bars show the relative fluorescence units (A and C). Differential expression in gWAT tissue was confirmed by qRT-PCR (B and D). Tbp was used as endogenous control. Data represent mean values ± SEM from 5 (microarray) or 7 (qRT-PCR) mice each strain, respectively. Differences between the strains were calculated by two-sided Wilcoxon test (microarray) or One-way analysis of variance followed by post hoc Bonferroni test (qRT-PCR), *P < 0.05, ***P < 0.001. gWAT, gonadal white adipose tissue; qPCR, qRT-PCR.
Sgcg and Arl11 expression QTL overlap with Nbw14
We further quantified the expression of Sgcg and Arl11 in gWAT of each male N2(NZOxC3H) mouse and mapped the mRNA levels to the genome. The linkage analysis revealed cis-expression QTL (eQTL) on chromosome 14 for the expression of both genes, Sgcg (LOD 41.1 at 20.5 cM, closest SNP marker at 63.54 Mb, Fig. 7A) and Arl11 (LOD 6.8 at 25 cM, closest SNP marker at 77.52 Mb, Fig. 7B). Both eQTL showed similar LOD score profiles as Nbw14 (body weight: LOD 5.2 at 23 cM; fat mass LOD 6.3 at 23 cM; Fig. 7C).
Figure 7.
Expression QTL analysis for Sgcg and Arl11 expressions in gWAT from all N2(NZOxC3H) mice. Genome-wide LOD scores distribution for the mRNA expressions of Sgcg (A) and Arl11 (B) in N2(NZOxC3H) males, Chr.14 is highlighted in grey and the significance threshold (P < 0.05) is indicated with the horizontal line. (C) Expression QTL curves and metabolic QTL curves (body weight and fat mass) on Chr.14 were superimposed. The SNP-markers that were used for genotyping with the corresponding Mb-positions are plotted below. The genotypic effects are indicated with the arrows. All LOD scores were calculated with R/qtl package as described in methods. LOD, logarithm of the odds; Expr, expression; N, NZO; wk, week.
Overexpression of Sgcg increases glucose uptake in 3T3-L1 adipocytes
For functional analysis, we focused on the gene Sgcg as most likely candidate for Nbw14. Undifferentiated 3T3-L1 cells were infected with either a retrovirus carrying cDNA from Sgcg (Sgcg OE) or the empty vector and differentiated into adipocytes. The success of the overexpression was confirmed by determining Sgcg mRNA levels in Sgcg OE vs control adipocytes using qRT-PCR. The Ct-value dropped from 34.5 in control- to 16.2 in Sgcg OE cells (Fig. 8A). 2-Deoxy-d-glucose uptake was measured in fully differentiated cells at basal state as well as after exposure to 100 nM insulin for 60 min. While basal 2-deoxy-d-glucose uptake was not different, insulin-stimulated 2-deoxy-d-glucose uptake was significantly increased by approx. 60% in Sgcg OE compared to control cells (Fig. 8B). However, lipid accumulation obtained by Oil Red O staining was not altered in Sgcg OE cells (Supplementary Material, File S2, Supplementary Material, Fig. S1). We further measured AKT and pAKT protein abundances in SGCG overexpressing cells, but could not detect any differences compared to control cells (Supplementary Material, File S2, Supplementary Material, Fig. S2).
Figure 8.
Measurement of 2-Deoxy-D-glucose uptake in Sgcg overexpressing 3T3-L1 vs. control adipocytes. Undifferentiated 3T3-L1 cells were infected with either a retrovirus carrying cDNA from Sgcg (Sgcg OE) or the empty pMSCV-puro vector as control and differentiated to adipocytes. The mRNA overexpression was confirmed by quantitative Realtime PCR (A). Hprt was used as endogenous control. 2-Deoxy-D-glucose uptake was determined in fully differentiated cells by measuring luminescence intensity at basal as well as after 60 min stimulation with 100 nM insulin (B). Bars represent mean values ± SEM from 3 (A) or 7 (B) different experiments. Statistical differences were calculated by two-way analysis of variance (B) followed by post hoc Bonferroni test, *P < 0.05.
SGCG expression correlates with body mass index and body fat in human fat tissue exclusively from T2D subjects
To study whether SGCG may be also relevant for human obesity and T2D, we analyzed its expression in human subcutaneous and visceral adipose tissues from both patients with T2D and healthy controls. SGCG mRNA levels significantly correlated with body mass index (BMI) in subcutaneous (r = 0.71, P = 0.002; Fig. 9A) as well as visceral (r = 0.699, P = 0.0025; Fig. 9B) adipose tissue exclusively from diabetic, but not from healthy subjects (subcutaneous fat: r = 0.12, P = 0.475; visceral fat: r = 0.229, P = 0.178). A similar pattern was observed for the correlation with body fat. Whereas SGCG expression significantly correlated with body fat in subcutaneous (r = 0.723, P = 0.0035; Fig. 9C) and visceral (r = 0.607, P = 0.0213; Fig. 9D) adipose tissue from diabetic patients, no correlation was observed in healthy subjects (subcutaneous fat: r = 0.094, P = 0.675; visceral fat: r = 0.299, P = 0.177).
Figure 9.
SGCG expression analysis in human adipose tissue. Correlation of SGCG mRNA levels with BMI in subcutaneous (A) and visceral adipose tissue (B) from healthy (blue circles) vs. patients with type 2 diabetes (T2D, red squares). Correlation of SGCG mRNA levels with body fat in subcutaneous (C) and visceral adipose tissue (D) from healthy controls (blue circles) vs. T2D (red squares) subjects. Correlations were calculated using Pearson correlation test. n = 16–36 for BMI and 14–22 for body fat.
Discussion
To identify novel obesity-modifier genes, we analyzed two backcross populations derived from diabetes-prone NZO mice and two lean, diabetes-protected strains, 129/P2 and C3H, respectively. Genome-wide linkage scans and transcriptome analysis of adipose tissue from mice identified Sgcg as a novel potential susceptibility gene for obesity. In human adipose tissue, SGCG expression significantly correlated with BMI and body fat exclusively in patients with T2D, suggesting that SGCG may represent a novel marker for diabetic obesity.
The NZO mouse strain represents a model for polygenetic obesity and T2D in humans. Both male and female mice develop obesity and severe insulin resistance. However, diabetes-related weight loss during the course of the disease may interfere with studies of obesity-related traits in male NZO mice (11,18). Nonetheless, female mice are protected from subsequent β-cell failure, which has been attributed to protective effects of oestrogen (16,17). In humans, the prevalence for T2D increases in women after menopause (31), also suggesting diabetes-protective effects of oestrogen.
Linkage analysis of female mice from two outcross populations, NZOxF1(129P2xNZO) and NZOxF1(C3HxNZO), revealed seven novel QTL for body weight and/or fat mass on six different chromosomes. Most of the QTL showed linkage exclusively in only one of the two backcross populations, suggesting that the underlying gene variants are derived from the respective lean strain, C3H or 129P2. In contrast, the locus Nbw14 showed linkage to obesity-related traits in both backcross populations, indicating that the causal gene variant is likely specific for the NZO strain as NZO allele carriers in both crosses displayed elevated body weight and fat mass. Based on this assumption, we applied two major criteria to filter for candidate genes that may drive the high prevalence for obesity in NZO: NZO-specific gene expression in gWAT and location in haplotype blocks that are highly polymorphic between NZO and the two lean strains.
Because underlying genes for a QTL are likely located in genomic regions where the parental strains have different haplotypes (32,33), out of 431 annotated genes we exclusively considered genes as potential candidates for Nbw14 that are residing in regions that are highly polymorphic between NZO and the other two strains. Our survey of the Sanger SNP database further revealed three missense mutations in the two genes Pnma2 and Rubcnl in NZO that are likely to affect protein function. However, we found that these variants were also shared by several lean mouse strains listed in the Sanger database, indicating that these protein polymorphisms are not associated with an obesity phenotype as seen in NZO. Moreover, both genes showed only marginal expression in adipose tissue, defined as our main QTL-target tissue, from our mouse strains and from humans as observed in GTEx transcriptome database (34). However, both genes should not be excluded as candidates for Nbw14, since other tissues than adipose tissue, in particular skeletal muscle or the central nervous system (CNS), may be responsible for the phenotype mediated by Nbw14. PNMA2 is highly expressed in the brain. The hypothalamus and other areas of the CNS have been established to play a key role in the regulation of energy homeostasis and food intake behaviour (35). In the future, it would be interesting to study a potential role of PNMA2 in relation to food intake in the CNS.
Our genome-wide transcriptome analysis in gWAT revealed nine genes that either were up- or down regulated in NZO, compared to C3H and 129/P2. Out of these nine candidates, only two genes Arl11 and Sgcg fulfilled our candidate gene criteria, thus representing the most likely candidates underlying the QTL.
Whereas Arl11 revealed to be upregulated in gWAT from NZO, Sgcg expression was exclusively detected in the two lean strains. Interestingly, both genes showed NZO-specific expression exclusively in adipose tissue, whereas in other T2D-relevant tissues, such as skeletal muscle or liver, the mRNA expression was not different between our parental strains. This observation strengthens our hypothesis that both genes are plausible candidates for Nbw14, which likely interferes with adipose tissue function in obese NZO mice. In addition, eQTL analysis in gWAT from our N2(NZOxC3H) population revealed cis-signals for both genes within the Nbw14 locus, thus providing evidence that both Arl11 and Sgcg harbour a genetic sequence variation between NZO and C3H, presumably within the genes, that may be causal for Nbw14. The eQTL approach was already successfully used in the past as a tool to exclude and nominate genes for physiological QTL (36–39).
Due to the results from the gene expression analysis in gWAT from our mouse strains, which revealed clearly higher mRNA levels and expression differences for Sgcg than for Arl11, we decided to focus on Sgcg in this study. Nevertheless, a potential role of Arl11 driving altered adipocyte function from NZO mice would need to be addressed in future studies. Arl11 is known to be expressed exclusively in murine and human macrophages where it regulates their activation in response to LPS stimulation (40), indicating a role in the inflammatory response. In context to Nbw14, Arl11 in pro-inflammatory adipose tissue macrophages from NZO mice may contribute to adipose tissue remodelling. It is broadly accepted that the amount of (pro-inflammatory) adipose tissue macrophages increases during obesity, leading to altered secretion profiles, surface marker expression and metabolic function of adipocytes, eventually contributing to the overall dysfunction of the adipose tissue (41). It remains to be investigated whether Arl11 may take a causal role in this scenario or whether increased mRNA expression in NZO gWAT rather occurs secondarily as a consequence of obesity-induced inflammation.
Sgcg belongs to the Sacroglycan (SG) gene family, each of them encoding for a transmembrane glycoprotein. Sgcg is a vital component of the SG complex, a subcomplex of the dystrophin–glycoprotein complex, which maintains the integrity of the sarcolemma by linking the cytoskeleton and the extracellular matrix. Research on the SG proteins has been mainly limited to skeletal muscle in relation to muscular dystrophies (42). Recently it was shown that the dystrophin–glycoprotein complex interacts with the insulin receptor and regulates insulin signalling in skeletal muscle (43), suggesting that mutations in this complex contribute to the development of insulin resistance and T2D. Moreover, the disease Duchenne muscular dystrophy, which is caused by dissociation of the dystrophin glycoprotein complex, is associated with reduced insulin sensitivity in humans (44). However, the molecular basis of this interaction and the role of the SG complex remains to be elucidated.
In the present study, we found that retroviral overexpression of Sgcg in 3T3-L1 mouse adipocytes results in substantially increased insulin-mediated glucose uptake without affecting basal glucose uptake, suggesting a role of Sgcg in insulin action and energy metabolism. However, measurement of cellular lipid levels by Oil Red O staining failed to reveal any differences compared to control cells. More detailed studies are required to clarify whether deficiency in Sgcg may affect fatty acid uptake or de novo lipogenesis. Moreover, additional gene variants, including Pnma2, Rubcnl and Arl11, may also contribute to the phenotype mediated by Nbw14. We did not observe any changes in phosphorylated AKT (pAKT) levels between SGCG overexpressing vs. control cells, indicating that SGCG affects glucose transport downstream of AKT signalling, possibly at the level of GLUT4 vesicle trafficking. While the role of the SG complex in adipocytes remains unclear, β-sarcoglycan null mice, which lack the SG complex in adipose tissue and skeletal muscle, develop glucose-intolerance and whole body insulin resistance specifically due to altered insulin-stimulated glucose uptake in skeletal muscle (45). Further studies of Sgcg function in adipocytes are required to investigate this mechanism.
Finally, our expression analysis in human adipose tissue as well as different gWAS indicates that SGCG is also relevant for the pathogenesis of obesity and T2D in humans. Interestingly, we found a significant correlation of SGCG expression with BMI and body fat mass in human subcutaneous as well as visceral adipose tissue exclusively in T2D subjects, whereas no correlation was found in the healthy control group. This observation shows that SGCG can be used as a novel genetic marker for metabolically unhealthy obesity in humans. In line with our mouse data, which argue for a beneficial role of Sgcg on glucose homeostasis, we speculate that elevated levels of SGCG in diabetic humans may be compensatory to insulin resistance.
Interestingly, a common SNP (rs679482) that disrupts a functional enhancer intronic to SGCG has been associated with weight loss in response to energy restriction (46). Another SNP (rs9552911) in the human SGCG gene has been associated with T2D in a Chinese (47) as well as in a South Asian population (48). These findings indicate that there may also be a causal relationship between SGCG function and T2D pathogenesis in humans.
In conclusion, we again provide evidence that the comparative analysis of multiple inbred populations generated with one common breeding can substantially improve the mapping resolution of disease genes. Our strategy allowed the identification of Sgcg as a novel potential obesity-modifier gene in adipose tissue, which was successfully validated by our in vitro studies. Moreover, our translational approach shows that Sgcg represents a novel genetic marker for metabolically unhealthy obesity. Future studies are necessary to elucidate the mechanism by which Sgcg increases insulin-sensitivity in adipocytes. Nevertheless, since there is evidence that QTL regions may include multiple genes that contribute to the phenotype (49), it may be possible that further gene variants, including Pnma2, Rubcnl and Arl11, that may also act in other tissues, exert effects on regulatory circuits in the Nbw14 locus. This may also include non-coding SNPs, indels, copy number polymorphisms, and yet unknown de novo mutations.
Materials and Methods
Animals and breeding strategy
All experiments were approved by the Ethics Committee (reference: 84–02.04.2013.A118) of the State Ministry of Agriculture, Nutrition and Forestry (State of North Rhine-Westphalia, Germany). Diabetes-prone NZO/Hl (NZO; German Diabetes Centre Duesseldorf) (50) and diabetes-resistant 129P2/OlaHsd (129P2; German Institute of Human Nutrition, Nuthetal, Germany) and C3HeB/FeJ (C3H; Helmholtz Center Munich, Germany) (51) mice were housed at three to six mice per cage (Macrolon type III) at a constant temperature of 22°C and a 12 h light–dark cycle (lights on at 6 a.m.). Animals had free access to food and water ad libitum. Female NZO and male 129P2 or C3H mice, respectively, were used to generate a F1 generation (NZOx129P2; NZOxC3H), and male F1 offspring was subsequently backcrossed with NZO females (N2: NZOxF1). For each backcross generation N2(NZOx F1(NZOxC3H)) and N2(NZOx F1(NZOx129P2)), approximately 300 females were generated, designated N2(NZOxC3H) and N2(NZOx129P2). After weaning at the age of 21 days, all experimental animals received a HFD containing 45 kcal% fat, 20 kcal% protein and 35 kcal% carbohydrates with 4.73 kcal/gm energy (D12451 Research Diets Inc., New Jersey, USA). At 21–22 weeks of age, mice were fasted for six hours before they were sacrificed by cardiac puncture under isoflurane anaesthesia. For the collection of gWAT for subsequent microarray analysis, NZO, C3H and 129P2 mice were sacrificed at 6 weeks of age.
Genotyping
Genomic DNA was isolated from mouse tail tips using the InViSorb Genomic DNA Kit II (Invitek, Berlin, Germany). The genotyping was performed by KASP (Kompetitive Allele Specific PCR) using appropriate SNP assays (LGC genomics, Teddington, UK). Informative SNP markers (105 for N2(NZOx129P2) and 110 for N2(NZOxC3H)) (Supplementary Material, File S1) polymorph between NZO and 129P2 or C3H, respectively, were selected in a distance of 20 Mbp for each chromosome.
Body weight and body composition
Body weight was determined at weeks 3, 6, 10, 15, 18 and 20 using an electronic scale. Body composition was measured at weeks 3, 6, 10 and 15 by noninvasive nuclear magnetic resonance spectroscopy (EchoMRI™-100 System, Echo Medical Systems, Houston, USA).
Blood glucose levels
Blood glucose was measured at weeks 3, 6, 10, 15, 18 and 20 in the morning between 8 and 10 am using a CONTOUR® XT glucometer (Bayer Consumer Care AG, Leverkusen, Germany).
RNA extraction and microarray analysis
Total RNA from gonadal adipose tissue (gWAT) (collected from 6-weeks old NZO, C3H and 129P2 mice, n = 5 per strain) and 3T3-L1 cells was isolated using the RNeasy mini kit (QIAGEN, Hilden, Germany) including DNAse digestion according to the manufacturer’s instructions. For microarray analysis of the gWAT, the quality of the isolated RNA was tested using an RNA 6000 nano kit (Agilent Technologies, Taufkirchen, Germany). Only samples with RIN values >8 were selected for subsequent microarray analysis. Genome wide expression analyses (n = 5 per genotype) were performed with 150 ng RNA using Affymetrix-Chip (GeneChip® Mouse Gene 1.0 ST Array) as previously described (11).
cDNA synthesis and qRT-PCR
cDNA was synthesized using GoScript™ Reverse Transcriptase Kit (Promega, Madison, USA) using 500 ng RNA. For qRT-PCR, the GoTaq® qPCR Master Mix (Promega, Madison, USA) on a QuantStudio 7 Flex PCR System (Applied Biosystems, Foster City, USA) was used. Tpb was used as an endogenous control for gWAT from NZO, C3H and 129P2 mice; Actb for gWAT from all N2 mice; and Hprt for 3T3-L1 cells. Gene expression was quantified using the 2−ΔΔCT method (52). For eQTL analysis, we used 2-ΔCT expression values for linkage scans with R/qtl package of R as described below.
Cell culture and differentiation of 3T3-L1 cells
3T3-L1 fibroblasts (Cl-173, ATCC, USA, negatively tested for mycoplasma contamination) were cultured in Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific) with 25 mM glucose, supplemented with 10% new-born calf serum and 1% penicillin/streptomycin. For differentiation into adipocytes, the fibroblast were grown to confluence and incubated with differentiation medium #1 (Dulbecco’s modified Eagle’s medium with 25 mM glucose, 10% fetal calf serum, 1% penicillin/streptomycin, 1 μg/ml insulin, 1 μM dexamethasone, 2 μM rosiglitazone and 0.5 μM IBMX. On day 7 of differentiation, differentiation medium #1 was replaced with differentiation medium #2 (medium #1 without dexamethasone, rosiglitazone and IBMX), and cells were cultured for additional 7 days until functional investigation.
Retroviral overexpression
ORF sequence of the Sgcg cDNA clone (Clone ID: OMu09100C, NM_011892.3, GenScipt) was subcloned into the retroviral vector pMSCV-puro (Addgene plasmid #68469) using the XhoI-EcoRI restriction site. Sgcg-pMSCV-puro and empty pMSCV-puro plasmids were transfected to Plat-E packaging cells (kindly provided from Prof. Dr Stork from the University of Duesseldorf, Germany) using Lipofectamin 2000 (Thermo Fisher Scientific). 48 hours post transfection, culture supernatants were centrifuged for 4 min at 300 x g, 6 μg/ml polybrene was added, and the viruses were added to 3T3-L1 fibroblasts overnight. Drug selection by 4 μg/ml puromycin was initiated 24 hours after infection.
Oil Red O staining
Retroviral transduced 3T3-L1 fibroblasts were seeded into collagen-coated 12-well plates (30 000 cells per well) and differentiated to adipocytes. Adipocytes were rinsed with PBS twice and fixed with 4% formalin in PBS for 60 min. The cells were washed with 60% isopropanol twice and dried before the addition of freshly diluted Oil Red O solution. Oil Red O was prepared by diluting a stock solution (0.5 g of Oil Red O; Sigma) in 100 mL of isopropanol with water (60:40 vol/vol), followed by filtration. Cells were stained for 20 min and then washed 4x with water before they were photographed. Finally, the Oil Red O dye was extracted by adding 100% isopropanol, the solution was transferred to 96-well plates and quantified at OD 500 nm.
Glucose uptake
Retroviral transduced 3T3-L1 fibroblasts were seeded into collagen-coated 96-well plates (7500 cells per well) and differentiated into adipocytes. One day prior the assay, cells were starved for 24 hours with DMEM without serum and antibiotics. Cells were either left untreated or stimulated with 100 nM insulin for 1 hour at 37°C. Uptake of 2-deoxy-D-glucose in the adipocytes was measured using glucose uptake-GloTM assay kit (Promega, Madison, WI, USA) and luminescence intensity (Relative light unit, RLU) was quantified according to the manufacturer’s instructions.
Expression analysis in humans
Paired samples of subcutaneous and omental visceral adipose tissue were obtained from 55 individuals (37 women, 18 men). The age ranged from 16 to 85 years and the BMI from 16 to 76 kg/m2. All adipose tissue samples were collected during laparoscopic abdominal surgery as described previously (53). Subsequently, adipose tissue biopsies were processed and gene expression was quantified as described previously (13). According to American Diabetes Association (ADA) criteria (54), the cohort was subdivided into patients with T2D and healthy normoglycemic controls.
Linkage analysis
Distributions of phenotypic data were tested for normality by the use of the D’Agostino-Pearson omnibus test (GraphPad Software Inc., La Jolla, CA, USA). While body weight and body composition data sets were normally distributed, blood glucose data sets were log2-transformed to achieve normal distribution. QTL analyses including the genetic map, genotyping errors and linkage between individual traits and genotypes were performed on N2(NZOx129P2) (307 females) and N2(NZOxC3H) (310 females) populations using the R/qtl 1.40–8 package (55) of R (version i386 3.3.2). Single-QTL genome scans were performed by interval mapping with the Expectation–maximization (EM) algorithm (56). The significance thresholds (P < 0.05) for linkage were estimated by 1000 permutations (57). For the eQTL analysis, mRNA expression levels (2-ΔCT) from the N2(NZOxC3H) population were used as quantitative traits and mapped to the genome in a non-parametric linkage analysis as described above.
Sequence and haplogroup analysis
Data for mouse SNPs and SNP-Gene assignments were from the Sanger Welcome Trust Institute Database (https://www.sanger.ac.uk/sanger/Mouse_SnpViewer). Coding nonsynonymous SNPs were analyzed for their potential impact on protein function using the ‘SIFT’ algorithm (http://sift.jcvi.org; (30). The chromosomal region was dissected into intervals of 250 kb to determine the frequency of polymorphic SNPs between the mouse strains as described previously (11). A window of 250 kb exceeding the threshold of 100 SNPs was defined as polymorphic according to the assumption NZO ≠ C3H, 129P2. For the determination of the total number, all SNPs annotated for the C57BL/6 J reference genome with calls for C3H/HeJ, 129P2/OlaHsd and NZO/HlLtJ were counted.
Statistical analysis
Data are presented as mean values ± SEM. Statistical significance was reported by two-tailed Student’s t-test or one/two-way analysis of variance followed by post hoc Bonferroni test as appropriate. The Pearson correlation test was used to determine the relationship between SGCG expression and metabolic parameters in human subjects. Differences were considered significant when P < 0.05. Statistical analysis was conducted by GraphPad Prism 8 (GraphPad Software Inc., La Jolla, CA, USA).
Data availability
Supplementary Material, File S1 contains R/qtl formatted mapping information, including mouse IDs, phenotype and gene expression data, and SNP marker identifiers, locations, and genotypes (A = NZO/NZO, H=NZO/C3H or NZO/129P2) from females of both N2(NZOxC3H and NZOx129P2) populations. Supplementary Material, File S2 contains further data supporting material. Supplementary Material, File S3 lists all annotated SNPs according to NZO ≠ C3H and 129P2 from all 83 genes located in regions that were defined as polymorphic between NZO and the other two strains. Microarray data are available under accession number GSE197101 (password: unqdeseynfcxnyf).
Supplementary Material
Acknowledgements
We gratefully thank Sylvia Jacob, Dagmar Grittner, Angelika Horrighs, Anette Kurowski, Peter Herdt and Cornelia Köllmer for technical assistance.
Conflict of Interest statement. The authors declare no conflict of interest.
Contributor Information
Tanja Kuhn, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Katharina Kaiser, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Sandra Lebek, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Delsi Altenhofen, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Birgit Knebel, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Ralf Herwig, Department of Computational Molecular Biology, Max Planck Institute for Molecular Genetics, Berlin D-14195, Germany.
Axel Rasche, Department of Computational Molecular Biology, Max Planck Institute for Molecular Genetics, Berlin D-14195, Germany.
Angela Pelligra, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Sarah Görigk, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Jenny Minh-An Khuong, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Heike Vogel, German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany; Department of Experimental Diabetology, German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal D-14558, Germany.
Annette Schürmann, German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany; Department of Experimental Diabetology, German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal D-14558, Germany.
Matthias Blüher, Helmholtz Institute for Metabolic, Obesity and Vascular Research (HI-MAG) of the Helmholtz Zentrum München at the University of Leipzig and University Hospital Leipzig, Leipzig D-04103, Germany.
Alexandra Chadt, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Hadi Al-Hasani, Medical Faculty, Institute for Clinical Biochemistry and Pathobiochemistry, German Diabetes Center (DDZ), Heinrich Heine University, Duesseldorf D-40225, Germany; German Center for Diabetes Research (DZD), Partner Düsseldorf / Partner Potsdam, Munich-Neuherberg, Germany.
Funding
German Ministry of Education and Research, the State of North-Rhine-Westphalia and the State of Brandenburg (82DZD00202 to H.A. and 82DZD00302 to A.S.); Deutsche Forschungsgemeinschaft (GRK2576); fellowships from the Deutscher Akademischer Austauschdienst (to D.A.); Deutsche Forschungsgemeinschaft (to T.K.).
References
- 1. Sanghera, D.K. and Blackett, P.R. (2012) Type 2 diabetes genetics: beyond GWAS. J. Diab. Metab., 03. 10.4172/2155-6156.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yazdi, F.T., Clee, S.M. and Meyre, D. (2015) Obesity genetics in mouse and human: back and forth, and back again. PeerJ, 3, e856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention (2020) National Diabetes Statistics Report website. https://www.cdc.gov/diabetes/data/statistics-report/index.html.
- 4. Fuchsberger, C., Flannick, J., Teslovich, T.M., Mahajan, A., Agarwala, V., Gaulton, K.J., Ma, C., Fontanillas, P., Moutsianas, L., McCarthy, D.J. et al. (2016) The genetic architecture of type 2 diabetes. Nature, 536, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mahajan, A., Taliun, D., Thurner, M., Robertson, N.R., Torres, J.M., Rayner, N.W., Payne, A.J., Steinthorsdottir, V., Scott, R.A., Grarup, N. et al. (2018) Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet., 50, 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buniello, A., MacArthur, J.A.L., Cerezo, M., Harris, L.W., Hayhurst, J., Malangone, C., McMahon, A., Morales, J., Mountjoy, E., Sollis, E. et al. (2019) The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res., 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kleinert, M., Clemmensen, C., Hofmann, S.M., Moore, M.C., Renner, S., Woods, S.C., Huypens, P., Beckers, J., de Angelis, M.H., Schurmann, A. et al. (2018) Animal models of obesity and diabetes mellitus. Nat. Rev. Endocrinol., 14, 140–162. [DOI] [PubMed] [Google Scholar]
- 8. Clee, S.M. and Attie, A.D. (2007) The genetic landscape of type 2 diabetes in mice. Endocr. Rev., 28, 48–83. [DOI] [PubMed] [Google Scholar]
- 9. Kebede, M.A. and Attie, A.D. (2014) Insights into obesity and diabetes at the intersection of mouse and human genetics. Trends Endocrinol. Metab., 25, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joost, H.G. and Schurmann, A. (2014) The genetic basis of obesity-associated type 2 diabetes (diabesity) in polygenic mouse models. Mamm. Genome, 25, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schallschmidt, T., Lebek, S., Altenhofen, D., Damen, M., Schulte, Y., Knebel, B., Herwig, R., Rasche, A., Stermann, T., Kamitz, A. et al. (2018) Two novel candidate genes for insulin secretion identified by comparative genomics of multiple backcross mouse populations. Genetics, 210, 1527–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwerbel, K., Kamitz, A., Krahmer, N., Hallahan, N., Jahnert, M., Gottmann, P., Lebek, S., Schallschmidt, T., Arends, D., Schumacher, F. et al. (2020) Immunity-related GTPase induces lipophagy to prevent excess hepatic lipid accumulation. J. Hepatol., 73, 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vogel, H., Kamitz, A., Hallahan, N., Lebek, S., Schallschmidt, T., Jonas, W., Jahnert, M., Gottmann, P., Zellner, L., Kanzleiter, T. et al. (2018) A collective diabetes cross in combination with a computational framework to dissect the genetics of human obesity and type 2 diabetes. Hum. Mol. Genet., 27, 3099–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keller, M.P., Rabaglia, M.E., Schueler, K.L., Stapleton, D.S., Gatti, D.M., Vincent, M., Mitok, K.A., Wang, Z., Ishimura, T., Simonett, S.P. et al. (2019) Gene loci associated with insulin secretion in islets from non-diabetic mice. J. Clin. Investig., 129, 4419–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kluge, R., Scherneck, S., Schurmann, A. and Joost, H.G. (2012) Pathophysiology and genetics of obesity and diabetes in the New Zealand obese mouse: a model of the human metabolic syndrome. Methods Mol. Biol., 933, 59–73. [DOI] [PubMed] [Google Scholar]
- 16. Lubura, M., Hesse, D., Kraemer, M., Hallahan, N., Schupp, M., von Loffelholz, C., Kriebel, J., Rudovich, N., Pfeiffer, A., John, C. et al. (2015) Diabetes prevalence in NZO females depends on estrogen action on liver fat content. Am. J. Physiol. Endocrinol. Metab., 309, E968–E980. [DOI] [PubMed] [Google Scholar]
- 17. Vogel, H., Mirhashemi, F., Liehl, B., Taugner, F., Kluth, O., Kluge, R., Joost, H.G. and Schurmann, A. (2013) Estrogen deficiency aggravates insulin resistance and induces beta-cell loss and diabetes in female New Zealand obese mice. Horm. Metab. Res., 45, 430–435. [DOI] [PubMed] [Google Scholar]
- 18. Kluge, R., Giesen, K., Bahrenberg, G., Plum, L., Ortlepp, J.R. and Joost, H.G. (2000) Quantitative trait loci for obesity and insulin resistance (Nob1, Nob2) and their interaction with the leptin receptor allele (LeprA720T/T1044I) in New Zealand obese mice. Diabetologia, 43, 1565–1572. [DOI] [PubMed] [Google Scholar]
- 19. Leiter, E.H., Reifsnyder, P.C., Flurkey, K., Partke, H.J., Junger, E. and Herberg, L. (1998) NIDDM genes in mice: deleterious synergism by both parental genomes contributes to diabetogenic thresholds. Diabetes, 47, 1287–1295. [DOI] [PubMed] [Google Scholar]
- 20. Vogel, H., Nestler, M., Ruschendorf, F., Block, M.D., Tischer, S., Kluge, R., Schurmann, A., Joost, H.G. and Scherneck, S. (2009) Characterization of Nob3, a major quantitative trait locus for obesity and hyperglycemia on mouse chromosome 1. Physiol. Genom., 38, 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reifsnyder, P.C., Churchill, G. and Leiter, E.H. (2000) Maternal environment and genotype interact to establish diabesity in mice. Genome Res., 10, 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andrikopoulos, S., Fam, B.C., Holdsworth, A., Visinoni, S., Ruan, Z., Stathopoulos, M., Thorburn, A.W., Joannides, C.N., Cancilla, M., Balmer, L. et al. (2016) Identification of ABCC8 as a contributory gene to impaired early-phase insulin secretion in NZO mice. J. Endocrinol., 228, 61–73. [DOI] [PubMed] [Google Scholar]
- 23. Scherneck, S., Nestler, M., Vogel, H., Bluher, M., Block, M.D., Berriel Diaz, M., Herzig, S., Schulz, N., Teichert, M., Tischer, S. et al. (2009) Positional cloning of zinc finger domain transcription factor Zfp69, a candidate gene for obesity-associated diabetes contributed by mouse locus Nidd/SJL. PLoS Genet., 5, e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Plum, L., Kluge, R., Giesen, K., Altmuller, J., Ortlepp, J.R. and Joost, H.G. (2000) Type 2 diabetes-like hyperglycemia in a backcross model of NZO and SJL mice: characterization of a susceptibility locus on chromosome 4 and its relation with obesity. Diabetes, 49, 1590–1596. [DOI] [PubMed] [Google Scholar]
- 25. Aga, H., Hallahan, N., Gottmann, P., Jaehnert, M., Osburg, S., Schulze, G., Kamitz, A., Arends, D., Brockmann, G., Schallschmidt, T. et al. (2020) Identification of novel potential type 2 diabetes genes mediating beta-cell loss and Hyperglycemia using positional cloning. Front. Genet., 11, 567191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chadt, A., Leicht, K., Deshmukh, A., Jiang, L.Q., Scherneck, S., Bernhardt, U., Dreja, T., Vogel, H., Schmolz, K., Kluge, R. et al. (2008) Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat. Genet., 40, 1354–1359. [DOI] [PubMed] [Google Scholar]
- 27. Pan, H.J., Agate, D.S., King, B.L., Wu, M.K., Roderick, S.L., Leiter, E.H. and Cohen, D.E. (2006) A polymorphism in New Zealand inbred mouse strains that inactivates phosphatidylcholine transfer protein. FEBS Lett., 580, 5953–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keane, T.M., Goodstadt, L., Danecek, P., White, M.A., Wong, K., Yalcin, B., Heger, A., Agam, A., Slater, G., Goodson, M. et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature, 477, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yalcin, B., Wong, K., Agam, A., Goodson, M., Keane, T.M., Gan, X., Nellaker, C., Goodstadt, L., Nicod, J., Bhomra, A. et al. (2011) Sequence-based characterization of structural variation in the mouse genome. Nature, 477, 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar, P., Henikoff, S. and Ng, P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 31. Szmuilowicz, E.D., Stuenkel, C.A. and Seely, E.W. (2009) Influence of menopause on diabetes and diabetes risk. Nat. Rev. Endocrinol., 5, 553–558. [DOI] [PubMed] [Google Scholar]
- 32. Park, Y.G., Clifford, R., Buetow, K.H. and Hunter, K.W. (2003) Multiple cross and inbred strain haplotype mapping of complex-trait candidate genes. Genome Res., 13, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manenti, G., Galbiati, F., Gianni-Barrera, R., Pettinicchio, A., Acevedo, A. and Dragani, T.A. (2004) Haplotype sharing suggests that a genomic segment containing six genes accounts for the pulmonary adenoma susceptibility 1 (Pas1) locus activity in mice. Oncogene, 23, 4495–4504. [DOI] [PubMed] [Google Scholar]
- 34. Consortium, G.T (2020) The GTEx consortium atlas of genetic regulatory effects across human tissues. Science, 369, 1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morton, G.J., Cummings, D.E., Baskin, D.G., Barsh, G.S. and Schwartz, M.W. (2006) Central nervous system control of food intake and body weight. Nature, 443, 289–295. [DOI] [PubMed] [Google Scholar]
- 36. Schadt, E.E., Monks, S.A., Drake, T.A., Lusis, A.J., Che, N., Colinayo, V., Ruff, T.G., Milligan, S.B., Lamb, J.R., Cavet, G. et al. (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature, 422, 297–302. [DOI] [PubMed] [Google Scholar]
- 37. Hubner, N., Wallace, C.A., Zimdahl, H., Petretto, E., Schulz, H., Maciver, F., Mueller, M., Hummel, O., Monti, J., Zidek, V. et al. (2005) Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet., 37, 243–253. [DOI] [PubMed] [Google Scholar]
- 38. Drake, T.A., Schadt, E.E., Davis, R.C. and Lusis, A.J. (2005) Integrating genetic and gene expression data to study the metabolic syndrome and diabetes in mice. Am. J. Ther., 12, 503–511. [DOI] [PubMed] [Google Scholar]
- 39. Bao, L., Peirce, J.L., Zhou, M., Li, H., Goldowitz, D., Williams, R.W., Lu, L. and Cui, Y. (2007) An integrative genomics strategy for systematic characterization of genetic loci modulating phenotypes. Hum. Mol. Genet., 16, 1381–1390. [DOI] [PubMed] [Google Scholar]
- 40. Arya, S.B., Kumar, G., Kaur, H., Kaur, A. and Tuli, A. (2018) ARL11 regulates lipopolysaccharide-stimulated macrophage activation by promoting mitogen-activated protein kinase (MAPK) signaling. J. Biol. Chem., 293, 9892–9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keuper, M. (2019) On the role of macrophages in the control of adipocyte energy metabolism. Endocr. Connect., 8, R105–R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tarakci, H. and Berger, J. (2016) The sarcoglycan complex in skeletal muscle. Front. Biosci. (Landmark Ed.), 21, 744–756. [DOI] [PubMed] [Google Scholar]
- 43. Eid Mutlak, Y., Aweida, D., Volodin, A., Ayalon, B., Dahan, N., Parnis, A. and Cohen, S. (2020) A signaling hub of insulin receptor, dystrophin glycoprotein complex and plakoglobin regulates muscle size. Nat. Commun., 11, 1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoffman, E.P., Brown, R.H., Jr. and Kunkel, L.M. (1987) Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell, 51, 919–928. [DOI] [PubMed] [Google Scholar]
- 45. Groh, S., Zong, H., Goddeeris, M.M., Lebakken, C.S., Venzke, D., Pessin, J.E. and Campbell, K.P. (2009) Sarcoglycan complex: implications for metabolic defects in muscular dystrophies. J. Biol. Chem., 284, 19178–19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikpay, M., Lau, P., Soubeyrand, S., Whytock, K.L., Beehler, K., Pileggi, C., Ghosh, S., Harper, M.E., Dent, R. and McPherson, R. (2020) SGCG rs679482 associates with weight loss success in response to an intensively supervised outpatient program. Diabetes, 69, 2017–2026. [DOI] [PubMed] [Google Scholar]
- 47. Chen, M., Zhang, X., Fang, Q., Wang, T., Li, T. and Qiao, H. (2017) Three single nucleotide polymorphisms associated with type 2 diabetes mellitus in a Chinese population. Exp. Ther. Med., 13, 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saxena, R., Saleheen, D., Been, L.F., Garavito, M.L., Braun, T., Bjonnes, A., Young, R., Ho, W.K., Rasheed, A., Frossard, P. et al. (2013) Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes, 62, 1746–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Buchner, D.A. and Nadeau, J.H. (2015) Contrasting genetic architectures in different mouse reference populations used for studying complex traits. Genome Res., 25, 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Herberg, L. and Coleman, D.L. (1977) Laboratory animals exhibiting obesity and diabetes syndromes. Metabolism, 26, 59–99. [DOI] [PubMed] [Google Scholar]
- 51. Gailus-Durner, V., Fuchs, H., Becker, L., Bolle, I., Brielmeier, M., Calzada-Wack, J., Elvert, R., Ehrhardt, N., Dalke, C., Franz, T.J. et al. (2005) Introducing the German mouse clinic: open access platform for standardized phenotyping. Nat. Methods, 2, 403–404. [DOI] [PubMed] [Google Scholar]
- 52. Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 53. Kloting, N., Fasshauer, M., Dietrich, A., Kovacs, P., Schon, M.R., Kern, M., Stumvoll, M. and Bluher, M. (2010) Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab., 299, E506–E515. [DOI] [PubMed] [Google Scholar]
- 54. American Diabetes Association (2014) Diagnosis and classification of diabetes mellitus. Diabetes Care, 37, S81–S90. [DOI] [PubMed] [Google Scholar]
- 55. Broman, K.W. and Sen, S. (2009) A Guide to QTL Mapping with R/qtl. Springer, New York. [Google Scholar]
- 56. Lander, E.S. and Botstein, D. (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics, 121, 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lander, E. and Kruglyak, L. (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet., 11, 241–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Material, File S1 contains R/qtl formatted mapping information, including mouse IDs, phenotype and gene expression data, and SNP marker identifiers, locations, and genotypes (A = NZO/NZO, H=NZO/C3H or NZO/129P2) from females of both N2(NZOxC3H and NZOx129P2) populations. Supplementary Material, File S2 contains further data supporting material. Supplementary Material, File S3 lists all annotated SNPs according to NZO ≠ C3H and 129P2 from all 83 genes located in regions that were defined as polymorphic between NZO and the other two strains. Microarray data are available under accession number GSE197101 (password: unqdeseynfcxnyf).