Abstract
The mechanistic understanding of how DNA double-strand breaks (DSB) are repaired is rapidly advancing in part due to the advent of inducible site-specific break model systems as well as the employment of next-generation sequencing (NGS) technologies to sequence repair junctions at high depth. Unfortunately, the sheer volume of data produced by these methods makes it difficult to analyze the structure of repair junctions manually or with other general-purpose software. Here, we describe methods to produce amplicon libraries of DSB repair junctions for sequencing, to map the sequencing reads, and then to use a robust, custom python script, Hi-FiBR, to analyze the sequence structure of mapped reads. The Hi-FiBR analysis processes large data sets quickly and provides information such as number and type of repair events, size of deletion, size of insertion and inserted sequence, microhomology usage, and whether mismatches are due to sequencing error or biological effect. The analysis also corrects for common alignment errors generated by sequencing read mapping tools, allowing high-throughput analysis of DSB break repair fidelity to be accurately conducted regardless of which suite of NGS analysis software is available.
1. INTRODUCTION
DNA double-strand breaks (DSBs) are an extremely deleterious form of DNA damage that, if not repaired accurately, can result in cell death (Bennett, Lewis, Baldwin, & Resnick, 1993; Jeggo, 1990; Resnick & Martin, 1976) or carcinogenic mutations (Aparicio, Baer, & Gautier, 2014; Hromas, Williamson, Lee, & Nickoloff, 2016). Historically, DSBs have been thought to be repaired via homologous recombination (HR) or classical nonhomologous end joining (C-NHEJ) (Ciccia & Elledge, 2010). HR is recognized as error-free since it uses homologous template DNA to direct the repair process (Nickoloff, 2017). In contrast, C-NHEJ involves local processing of the DNA sequence surrounding the break until the DNA ends are compatible for direct ligation (Lieber, 2010). As a consequence, C-NHEJ is frequently error-prone, resulting in insertions, deletions, and base substitutions, and yet it is the primary repair pathway for induced DSBs in mammalian cells (Jeggo, 1990). Recent studies have begun to delineate the mechanism of a third pathway for DSB repair, called alternative end joining (alt-EJ) (Decottignies, 2013) (also called microhomology-mediated end joining (MMEJ) (Lee & Lee, 2007; Moore & Haber, 1996; Sharma et al., 2015) or theta-mediated end joining (TMEJ) (Beagan et al., 2017; Chan, Yu, & McVey, 2010; Kent, Chandramouly, McDevitt, Ozdemir, & Pomerantz, 2015; Mateos-Gomez et al., 2015; Wyatt et al., 2016; Yousefzadeh et al., 2014)). This pathway is believed to be a backup pathway for breaks that are unrepairable by HR or C-NHEJ (Nussenzweig & Nussenzweig, 2007; Sfeir & Symington, 2015). As this pathway appears to be used following the initiation of end resection (Seol, Shim, & Lee, 2017), alt-EJ is even more mutagenic than C-NHEJ, often causing hypermutation in flanking DNA (Sinha et al., 2017) and large deletions mediated by microhomology (Soni, Siemann, Pantelias, & Iliakis, 2015; van Schendel, van Heteren, Welten, & Tijsterman, 2016).
The choice between which repair mechanism to use, as well as the degree of mutagenicity, appears to be affected by a number of factors, including the presence of microhomologies in the DNA sequence. To further understand what factors affect the pathway used for DSB repair, and the types of mutations they cause, model cell systems have been developed that enable delineation of repair events at specific nuclease target sites (Chu, Wu, Xu, & Her, 2013; Gunn, Bennardo, Cheng, & Stark, 2011; Holmes & Haber, 1999; Liang, Han, Romanienko, &Jasin, 1998; Lin, Lukacsovich, & Waldman, 1999; Moynahan & Jasin, 1997; Nick McElhinny et al., 2005; Roth & Wilson, 1986; Rouet, Smih, & Jasin, 1994; Rudin & Haber, 1988; Xu, Wu, & Her, 2015), such as those generated by CRISPR-Cas9 technology (Vriend, Jasin, & Krawczyk, 2014; Vriend et al., 2016; Wyatt et al., 2016), or at ends of transfected linear DNA (Waters et al., 2014). Traditionally, these break sites have been induced, given time for repair, and then up to 100 individual isolates are chosen and sequenced. With the advent of next-generation sequencing (NGS) technology (Goodwin, McPherson, & McCombie, 2016), it is possible to sequence tens of thousands of events, from hundreds of samples, each with high fidelity and low cost. This shift in technology has led to a standard of high volume sequencing that accurately reveals the frequency of a broad spectrum of repair events including any specific mutations that occur (Huefner, Mizuno, Weil, Korf, & Britt, 2011; IJspeert et al., 2016; Liang, Sunder, Nallasivam, & Wilson, 2016; Mateos-Gomez et al., 2015; Soong et al., 2015; Waters et al., 2014; Wyatt et al., 2016), and even enabled single nucleotide resolution assessment of randomly occurring translocations (Chiarle et al., 2011). Because of the large number of events that are accessed, these methods are capable of detecting subtle differences in DSB repair junctions among model systems and how they are influenced by sequences around the break sites (Khodaverdian et al., 2017).
However, these large data sets demand robust methods for alignment of the sequences and analysis of the break junctions. The sequencing and alignment methods used to produce and analyze these data sets must be highly accurate to avoid misinterpretation of some repair events. For instance, sequencing errors near a break junction can cause misalignment of a repair product. Likewise, in downstream analysis sequencing error might be incorrectly called a base substitution, insertion, or deletion event.
In this chapter, we discuss methods for robustly sequencing and analyzing DSB repair junctions. We describe how to construct high-throughput sequencing libraries of DNA amplicons containing break-induced recombination events, how to align NGS sequence reads to a reference sequence, and ultimately how to analyze the alignments for base substitutions, insertions, and deletions at the repair junction (Fig. 1). The downstream analyses are largely performed by our custom script (written in Python3; https://github.com/Alexander-Brown13/Hi-FiBR), called Hi-FiBR (high-throughput fidelity of break repair) analysis. We describe how this tool works and compare alignment and sequencing strategies to determine how to maximize the accuracy and utility of the Hi-FiBR analysis.
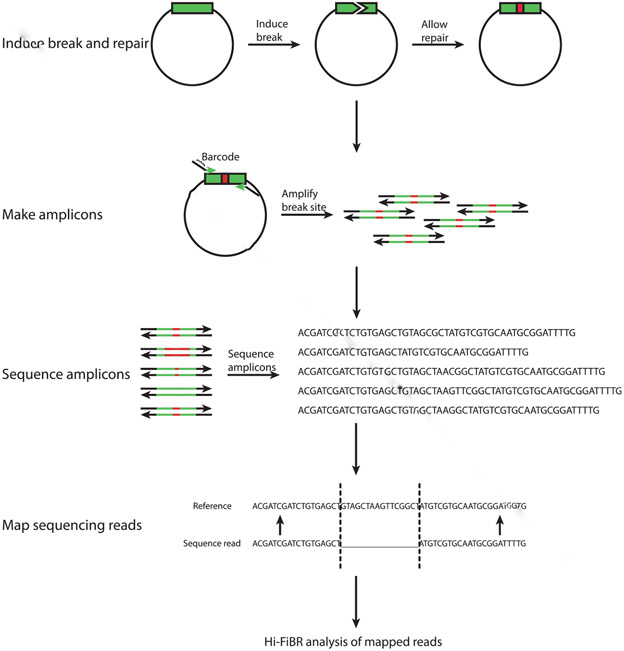
Fig. 1.
Steps in building and sequencing an amplicon library of DSB repair events. First, linear or circular DNA with an inducible break site must undergo a break and be given time for repair. The sequence surrounding the break site (shown in green) may experience an error such as an insertion, deletion, or base substitution (shown in red). To capture the frequency and variety of these errors the sequence surrounding the break site is PCR amplified, optionally with primers that contain barcodes. The amplicon products are then sequenced, and the sequenced reads are mapped to the reference sequence. Finally, the mapped reads are analyzed by the Hi-FiBR analysis.
2. CREATING COMPLEX LIBRARIES OF DSB REPAIR JUNCTIONS
High-throughput evaluation of the sequence of DSB repair junctions requires either the high efficiency induction of a site-specific DSB within chromosomal DNA, or the transfection of linearized episomal DNA in a large population of cells whose repair machinery can then rejoin the broken DNA ends (Soong et al., 2015; Waters et al., 2014; Wyatt et al., 2016). Nearly synchronous and uniform creation and repair of the break in the cell population is critical to obtain samples with a high degree of complexity in the repair products. Ultimately, this complexity is essential to effectively and robustly determine the roles of specific repair pathways or proteins in rejoining a DSB.
The first step in generating complex libraries of repair junction amplicons is to either isolate genomic DNA from a population of cells having undergone repair of a site-specific break or to harvest bulk circularized episomal DNA (i.e., linear DNA that has been joined into a circle) from transfected or injected cells. For mammalian cells containing site-specific DSB repair events, a typical genomic DNA isolation kit, such as a Qiagen DNeasy Blood and Tissue kit, is sufficient.
Once isolated, the repair junctions must be PCR amplified using unique sequence surrounding the break site. The position of the primers plays a crucial role in dictating the type of repair events that can be observed. More distantly spaced primers allow larger deletion events to be recovered while also providing additional sequences that may be used in microhomology-mediated events. The maximum length of the amplicon, however, is determined by the type of high-throughput sequencing technology that is to be employed. For example, paired-end sequencing of amplicons on an Illumina HiSeq platform would limit analysis to amplicons in the 250–300 bp product size (150 bp of sequence is acquired from each end), while MiSeq sequencing would maximally allow analysis of 500 bp fragments. Other sequencing platforms like Ion Proton or PacBio can accommodate even larger amplicons (up to several kilobases for PacBio); however, systematic sequencing error common to these platforms is problematic for analysis (see Section 5.2).
To produce amplicons we recommend following a protocol using a high-fidelity DNA polymerase, as standard DNA Taq polymerase may introduce errors. The following PCR protocol uses NEB’s high-fidelity DNA polymerase. The reagents and their corresponding final concentrations are as follows: 1× Phusion HF Buffer, 200 μM dNTPs, 0.5 μM forward primer, 0.5 μM reverse primer, 1.0 U/50 μL PCR Phusion DNA polymerase, and ~250 ng template genomic DNA. For repair junctions induced by Cas9 cleavage of human genomic DNA, 250 ng of input DNA would correspond to ~25,000–50,000 independent repair events that could be assessed. The PCR mix should then be cycled in a thermocycler using the following steps: initial denaturation at 98°C for 30 s, 22–25 cycles of amplification which are 98°C for 5–10 s, 45–72°C for 10–30 s, and 72°C for 15–30 s per kb, the final extension at 72°C for 5–10 min, and an optional hold step at 4–10°C.
Following the PCR amplification, the amplicon must be purified to add adapters for NGS. The following Qiagen PCR-cleanup kit protocol works well for this purpose:
Mix PCR with 5 reaction volumes of Buffer PB,
Place spin column in collection tube, apply sample, centrifuge for 1 min, discard flow-through, and place column back into collection tube,
Add 750 μL Buffer PE, let stand for 2–5 min, centrifuge for 1 min, discard flow-through, place column back into collection tube, and centrifuge for 1 min to remove residual wash buffer,
Place column into clean 1.5-mL microcentrifuge tube,
Add 30 μL Buffer EB or water to center of column membrane, let stand for 1–4 min, and centrifuge for 1 min.
Once a pure amplicon is obtained, one can proceed with a NGS library preparation protocol appropriate for the type of sequencing that will be conducted. These preparations involve addition of adapter sequences used as the template sequence to prime sequencing. Adapter sequences can either be appended to PCR primers, or ligated to the amplicon ends. If the adapters are to be added by ligation, amplicon products first need to be 5′ phosphorylated by a 15-min incubation with 10 U T4 Polynucleotide kinase (New England Biosciences) and 10 mM ATP at 37°C. Then a 3′ terminal A nucleotide is added by addition of 15 U Klenow exo− polymerase (New England Biosciences), 0.2 mM dATP, and an additional incubation at 30 min. For Illumina libraries, adapters (for sequences, see https://support.illumina.com/downloads/illumina-customer-sequence-letter.html) are annealed and ligated to amplicon DNA with a 10:1 ratio of linker to amplicon, 10 mM ATP, 2000 U T4 DNA ligase (New England Biosciences), and 30% PEG. About 1–2 μg of DNA is typically used for a library preparation, but this equates to far more molecules of PCR product than the original ~20,000 molecules of repair events placed in the original PCR. Consequently, sequencing of a library from a single sample will likely result in a massive oversampling of the repair events in the sample. For instance, an Illumina HiSeq 2500 is capable of generating ~150 million reads per lane; thus, sequencing of a single sample/library per lane would be expected to sample the same repair event 10,000 times. To maximize cost effectiveness, amplicons from different treatment conditions can be pooled together prior to the library preparation. To differentiate between amplicons from different treatments, barcoded primers need to be used during the PCR step. Barcodes can be designed in multiple ways and can be assigned to both the forward and reverse primer, which increases the permutations available for barcoding each amplicon. After sequencing, the barcodes are used to parse the sequence reads into separate samples, and then the reads are examined for specific repair events.
To remove possible PCR products of incorrect size and unligated adapters from the NGS library preparation, it is recommended to do a gel extraction or PEG purification. The following abridged gel extraction protocol uses Qiagen’s QIAquick gel extraction kit:
Excise the DNA fragment,
Weigh the gel and add 3 volumes Buffer QG to 1 volume gel,
Incubate at 50°C for 10 min,
Add 1 gel volume isopropanol to sample and mix,
Place spin column in collection tube, apply sample, centrifuge for 1 min, discard flow-through, and place column back into collection tube,
Add 500 μL Buffer QG to column, centrifuge for 1 min, discard flow-through, and place column back into collection tube,
Add 750 μL Buffer PE, let stand for 2–5 min, centrifuge for 1 min, discard flow-through, place column back into collection tube, and centrifuge for 1 min to remove residual wash buffer,
Place column into clean 1.5-mL microcentrifuge tube,
Add 30 μL Buffer EB or water to center of column membrane, let stand for 1–4 min, and centrifuge for 1 min.
Alternatively, small molecular weight contaminants can be separated efficiently from an otherwise clean amplicon by PEG purification. An abridged PEG purification protocol is as follows:
Add an equal volume PEG-8000/2.5 M NaCl to the PCR product and mix by brief gentle vortex (the final percentage of PEG is based on the PCR fragment size: 10% for >100 bp, 8.3% for >200 bp, 6.7% for >650 bp, and 5.6% for >800 bp),
Incubate PCR + PEG mix at room temperature for 5 min,
Centrifuge mix at ~15,000 × g for 20 min (at room temperature),
Remove and discard the supernatant,
Add the same volume of cold 80% ethanol (as PEG solution),
Centrifuge 5 min at max RCF,
Remove and discard supernatant,
Centrifuge tubes for 10 s at max RCF,
Remove and discard residual ethanol,
Place tubes open in a 37°C incubator for 10 min to remove ethanol,
Add an appropriate volume of molecular grade water, heat for 5 min at 37°C, and then vortex briefly to suspend DNA.
We recommend following this protocol with an additional QIAquick PCR-cleanup column purification (outlined above) to remove contaminating enzymes and trace levels of adapter that may still be present in the sample.
3. ALIGNING SEQUENCE READS
3.1. Merging Paired Reads
If a sequencing technology is used that generates pair-end reads (i.e., HiSeq or MiSeq), the reads will need to be merged. This can be done using a variety of tools, including CLC Genomics Workbench or BBMerge. BBMerge is an open access tool, written in Java, and can be installed on any operating system that runs Java, but is also a plug-in for Geneious. Here, we describe its usage in Geneious 10.2.2, but if used as a stand-alone tool, refer to the BBMap guide on GitHub (https://github.com/BioInfoTools/BBMap) for installation and execution.
To run BBMerge in Geneious, first select the unmerged, paired reads file. Then select “Sequence” in the top bar, and choose “Merge Paired Reads.” A prompt will appear asking for Merge Rate and Maximum memory to use. Normal merge rate and 500 MB of memory is sufficient. Select “OK.” The tool will produce two output files with the same name as the input file labeled either “Merged” or “Unmerged” as a prefix.
3.2. Trimming and Padding fastq Files
Once the paired-end sequences have been merged, the resulting fastq reads need to be cropped so that they all start and end at the same 5′ and 3′ sequences. This provides common reference index points that are critical for downstream automated determination of the repair junction structure. We have designed a “Trim_and_Pad” script that searches for user-defined sequences at the 5′ and 3′ ends of the reads and trims the fastq reads prior to the 5′ matching sequence and after the 3′ matching sequence. The 5′ sequence is looked for starting at the 5′ end of the read and progresses to the 3′ end, while the 3′ sequence starts at the 3′ end and passes to the 5′ end. This progression ensures that short sequences are not found prematurely in the rest of the sequence. Also, if the 3′ and 5′ sequences are both found, but in the incorrect relative orientation (i.e., the 3′ sequence is 5′ of the 5′ sequence), then it is not considered a match and the read is ignored. Reads containing both the 5′ and 3′ sequences in correct orientation are trimmed and printed to an output file, “file_Match,” in fastq format, where “file” is the original file name. In addition to the sequence of the read being trimmed, the corresponding quality scores will be trimmed as well; most aligners will not accept fastq files if the read sequence length is different than the quality score length. Alternatively, reads lacking either the 5′ or the 3′ sequence are printed to a different file, “file_NoMatch.” Nonmatching reads are primarily due to PCR contaminants from off-target amplification and low level sequencing error that modifies the corresponding sequence in a specific read. This step of read processing typically removes ~3% of the total reads in a sequenced library.
The user must decide what an appropriate length of sequence to search for is; if the sequence is too short, it will likely appear by random chance, but the longer the sequence is, the greater the chance of a sequencing error occurring in the search sequence, which will result in the removal of a larger number of reads. We suggest scaling the length of the searching sequence by the read length, using longer sequences for longer reads.
Some repair amplicon libraries may contain reads with large deletions, or mismatches near the ends of the sequence. Many aligners will have difficulty properly aligning the ends of these sequences which is required to generate the appropriate position indexes. This problem can be rectified using the padding functionality, which adds user-defined sequence to the ends of the read that will help anchor these sequences to the appropriate index. When the script prompts for padding sequence, the user can define nothing and the script will continue without adding sequence, but if padding is included it will add it to the corresponding end of the sequence and add the appropriate number of quality scores “G” (a perfect score). Once again, the user should make sure the reference sequence has the same sequence added to it. If the padding is being used to replace what is already present in the reference, the aligner should work correctly. If it is instead being used to act as an anchor and includes a highly repetitive sequence such as “AAAAAAAAAA,” then that sequence must also be added to the reference.
The Trim_and_Pad script runs on all the files in a directory, and will output two files for each file in the directory with the same name as the file being processed, but with the extensions “Match.fastq” and “NoMatch.fastq,” as described earlier. When the script finishes running, compare the sizes of the two files to each other. If the size of the Match file is significantly larger, then the sequencing data were probably very clean and the file is ready for aligning to the reference. If the sizes are close or the NoMatch is significantly larger, the sequencing data may have high frequency of sequencing error or the library may be significantly contaminated with off-target PCR product.
3.3. Choosing an Aligner
After processing the raw fastq reads, they are now ready to be aligned to a reference sequence to determine the sequence structure of each repair junction. Because the Hi-FiBR analysis operates on SAM files (Li et al., 2009), which is a standard high-throughput sequencing read alignment format, nearly any commercial or open source alignment software can be used to align your sequencing reads to a reference sequence. Using an aligner with appropriate parameters, however, can have a dramatic impact on the accuracy of the data analysis. In the following sections we provide tutorials for a few of the more widely used aligning softwares: commercial products Geneious 10.2.2 (BioMatters) and CLC Genomics Workbench 7.5 (Qiagen) and open source aligners Bowtie2 (Langmead & Salzberg, 2012) and Burrows–Wheeler Aligner (BWA) (Li & Durbin, 2009).
3.3.1. Mapping With Geneious 10
The trimmed fastq files can be batch imported into Geneious by selecting them all and dragging them into a new, empty folder within the Geneious database. The program will try to recognize the sequencing that was done based on the quality scores from the file, and the user must confirm which they are. The reference sequence should also be imported as a fasta file (reference file). If all the fastq files can use the same reference sequence for mapping, they can be batch mapped by selecting all of them, then choosing “Align/Assemble,” followed by “Map to Reference.”
A window will open allowing several parameters to be changed. Start by unchecking all the boxes. Check “Do not trim” (under Trim Before Mapping) and “Save contigs” (under Results). Under Data (at the top) choose the appropriate reference sequence. Under Method, Geneious should be the default Mapper, but to obtain the most accurate alignments, the Sensitivity should be changed to “Highest Sensitivity/Slow” and Fine Tuning should be set to “None (fast/read mapping).” Assembly name can be chosen by the user, but a convenient format is to use the exact phrase “{Reads Name} assembled to {Reference Name}” which will autopopulate the {Name} and produce a file clearly stating which reads were assembled to which reference file. Select “OK” when all the parameters have been set to launch the mapping. The high sensitivity setting may trigger a warning such as “Sensitivity is too high,” which can be ignored.
Once all the files have been mapped, they can be individually or batch exported as SAM files. They must be exported with the following settings for the Hi-FiBR breakpoint analysis to work properly. Choose the files and select export, then make sure to change the file format to SAM, not just the extension. When you choose export, there may be a warning “Potential Data Loss,” which can be ignored by choosing “Proceed.” A window will appear called “SAM sequences/alignments Export.” Select “More Options.” Then uncheck all the boxes except “Replace IUPAC ambiguities with N.” Be sure not to export “padded” CIGAR strings as this option will include information comparing each read to each other (as opposed to just a pairwise comparison of each read to the reference) into the format of the CIGAR string (Compact Idiosyncratic Gapped Alignment Report: an alphanumeric description of the alignment of each read to the reference). The CIGAR string is the primary field used in Hi-FiBR analysis to determine the sequence structure of repair junctions. Therefore, failure to properly format it will ultimately cause the breakpoint analysis to fail. Select OK and the files will begin exporting. Once the exporting is finished, the breakpoint analysis can be done.
3.3.2. Mapping With CLC Genomics Workbench 7.5
The fastq files containing merged and trimmed sequencing reads of DSB repair junctions can be batch imported into CLC by selecting “Import,” followed by the appropriate sequencing platform (in this example, Illumina), then finding the location of the files and selecting all of them. Before choosing “Next,” make sure there are no options selected under “General options” or “Illumina options”; however, Remove failed reads may optionally be chosen. Make sure the “Quality scores” are correct, which should be 1.8 and later. After choosing “Next” another screen will prompt the user for “Result handling,” and Save should be chosen. The next window will show folders in CLC that are mapped, and one must be chosen for the data to be saved to, then select “Finish.”
Alternatively, the directory containing the data can be mapped to CLC by choosing “Add File Location” near the top of the Navigation Area, and finding the directory and adding it. The reference file can be similarly imported, instead as a fasta file, and preferably in a separate folder for batching purposes.
Once the data have been imported, choose “Toolbox,” “NGS Core Tools,” and “Map Reads to Reference.” Choose the Batch option near the bottom of the new window and select the folder containing the fastq files by moving them into “Selected elements” with the blue arrow. The next window allows you to choose or exclude particular files in the folder to analyze; if all files in the folder are to be mapped, simply proceed to the next screen. In the next screen, choose the reference file and “No masking.” Following is a screen with alignment parameters which are important for determining how well the aligner opens gaps. This can be left with the default settings which use linear gap penalties (2, 3, 3, 0.5, 0.8), no global alignment, and “Ignore” nonspecific match handling. Newer versions of CLC Genomics Workbench include an option to utilize affine gap penalties designed to favor generation of long, continuous gaps. Affine gap penalties tend to produce more accurate alignments as long as the insertion and deletion extension costs are set to 1. In the final screen choose “Create stand-alone read mappings,” and Save; you may also choose to “Create report” for additional details. After choosing Finish the program will run and map the reads to the reference one at a time.
As with mapping in Geneious, read alignments generated in CLC need to be exported as SAM files. First select Export near the top of the screen, then choose SAM and Open. As before, choose the Batch option and move the folder into Selected elements. In the next window, do not use a compression, and we recommend making a Custom file name with the exact phrase “{1}.{2}” which will produce a SAM file with the original file name and the SAM extension. The final window requests the location for where the files will be exported. Once the exporting is finished, the breakpoint analysis can be done.
3.3.3. Mapping With Bowtie2
Bowtie2 (Langmead & Salzberg, 2012) runs in Linux or Mac OS from the Command Line, and thus requires no data importing. However, Bowtie2 does require an index for the reference genome to run properly. This is easily done through the command line by entering the directory in which the reference file is located and running “bowtie2-build reference.fa reference” where “reference.fa” is the name of the file and “reference” is the output prefix used for the index files. Once finished, Bowtie2 will have generated a number of index files with the given prefix and a suffix ending with “bt2” (bowtie2). When launching the mapping portion of the program, it is easiest to do in the directory containing these indices, so keep them together and in a directory in which you want to run the program.
Once the index files are finished, the trimmed fastq files can be mapped. Enter the directory where the bt2 index files are, and run the command “bowtie2 -x reference -U read.fastq -S output.sam” where “reference” is the prefix for the bt2 index files, “read.fastq” is the fastq file to be mapped, and “output.sam” is the name of the SAM file where the results of the analysis will be written. It is highly recommended to give the output SAM the same name as the input file. Also be sure to include the extensions in the input/output names, but not the reference prefix. The program runs fast and automatically creates SAM files, so there is no need to export them. However, it is difficult to batch run on multiple fastq files; multiple files can be listed, but they export to the same output SAM file. It may be simpler to use other software when a large number of fastq files are being analyzed. Regardless, once the program is finished, it will produce percentages of reads that aligned 0, 1, and >1 times. The vast majority of reads should map exactly once.
3.3.4. Mapping With BWA
BWA (Li & Durbin, 2009) is run almost exactly the same as Bowtie2. Like Bowtie2, BWA runs in Linux or Mac OS from the Command Line. BWA also requires an index of the reference genome, and it is generated by entering the directory with the reference file, through the command line, and running “bwa index reference.fa” where “reference.fa” is the name of the reference file. The output index files will share the same prefix as the reference file name and should be kept in the same directory to conveniently run the mapping function.
Once the index files are complete enter the directory that contains the BWA index files and run the command “bwa mem reference.fa read.fastq > output.sam” where “reference.fa” is the reference file, “read.fastq” is the fastq file to be mapped, and “output.sam” is the output SAM file. Include the appropriate extensions in the file names and, preferably, give the SAM file the same name as the input files. The program runs quickly, but it suffers from the same difficulty as Bowtie2 with an inability to batch run multiple fastq files. Consequently, it may be simpler to use other software when many fastq files are being analyzed. After the mapping is finished, the breakpoint analysis can be done on the SAM files.
4. BREAKPOINT ANALYSIS
4.1. Running Hi-FiBR
Hi-FiBR requires six inputs from the user: (1) the directory of SAM files, (2) a reference sequence file in fasta format, (3) the number of nucleotides from the start of the reference sequence to the position in the reference sequence where the DSB was generated (left breakpoint), (4) the number of nucleotides from the DSB site in the reference to the end of the reference sequence (right breakpoint), and (5) and (6) the number of nucleotides left and right of the break site in which base substitutions will be looked for. For convenience, once the script starts running it will ask for these inputs. The file and directory inputs need to be in the same directory that the script is in. Once all the inputs are provided, the script will run from start to finish on all files in the directory. This allows a large number of samples that utilize the same reference sequence to be analyzed all at once.
All the output files are generated in the same directory as the input directory in order to keep the files grouped. Please note that if the analysis needs to be rerun on the same directory of files, the output files from the initial analysis will also be considered as new inputs, so be sure to separate these files from the old inputs prior to reinitiating a second analysis on the same directory.
4.2. How Hi-FiBR Works
Hi-FiBR utilizes the CIGAR string for each read within the SAM files alignments to determine the nature of the repair that has occurred. A CIGAR string is, as its name (Compact Idiosyncratic Gapped Alignment Report) suggests, a report for the gapped alignment performed on the read to the reference sequence. In the string, “M” represents matching nucleotides, “D” indicates deleted nucleotides, and “I” designates nucleotides present in the read, but not the reference, i.e., insertions. The numbers indicate how many nucleotides are attributed to M, I, or D. For example, in our data set of repair events from a 197-bp amplicon, a “perfect” repair event would be indicated with a CIGAR string of 197M, as the reference is 197 nucleotides long and no processing of the break occurred.
In the Hi-FiBR analysis, first, the CIGAR string is examined for the character “S,” which is used in programs such as CLC to denote soft clips, “*” which is used in some programs like Bowtie2 to signify reads that did not map properly, and “H” which is used in BWA to denote hard clips. As reads containing any of these characters are unlikely to result from appropriate amplification of repair junctions, they are removed from further analysis. Next, the CIGAR is broken down into its respective components (e.g., 20M5D25M→20M, 5D, 25M) and the matching lengths are used to determine the region of each read where processing of the DSB occurred (called the processing window). This window can result from relatively simple processing of the break, such as loss of nucleotides surrounding the breakpoint (a deletion), or insertion of nucleotides at the break site. However, more complex events also occur, involving both resection and DNA synthesis from the break site. Errors during the resynthesis of these ends can result in small insertions, deletions, or base substitutions in regions flanking break sites. Mathematically, the processing window of these complex events is determined by calculating the distance between the breakpoint and the distal most error (i.e., nonreference base) on both sides of the repair junction. Immediately after determining the processing window, the class of repair event is determined as “exact,” “insertion,” “deletion,” or “complex.”
If the amplification, sequencing, and alignment algorithms were all perfectly accurate, then the number of times each specific event (or read sequence) is observed in the sequencing library could be counted at this time. However, each of these steps produces error that can cause mis-classification of a repair event. For example, PCR amplification and high-throughput sequencing technologies produce errors in the reported sequence of individual reads that are intrinsic to the preparation and sequencing of the DNA and are not related to the actual repair event. Likewise, small insertions during the repair event (both templated and non-templated) are frequently misaligned by most alignment algorithms. Such reads are frequently miscategorized. To overcome these issues, Hi-FiBR utilizes a local, realignment of the sequence near the DSB repair junction to ensure accurate calling of the repair processing window. This process specifically checks that the ends of inserted sequences are not from the reference sequence and looks for base substitutions within a user-defined distance on each side of the repair junction. If any of these events occur, the size and location of the processing window is adjusted (as shown in Fig. 2).
Fig. 2.
Determining the processing window of a DSB repair event. Standard alignment tools will often treat base substitutions as sequencing error, which causes them to be called matches to the reference sequence. The Hi-FiBR analysis can recognize mismatches in a user-defined window around the break site (here shown as 4 bp on either side) and adjust the matching sequences. This also changes what the program considers as the processing window for repair event classification.
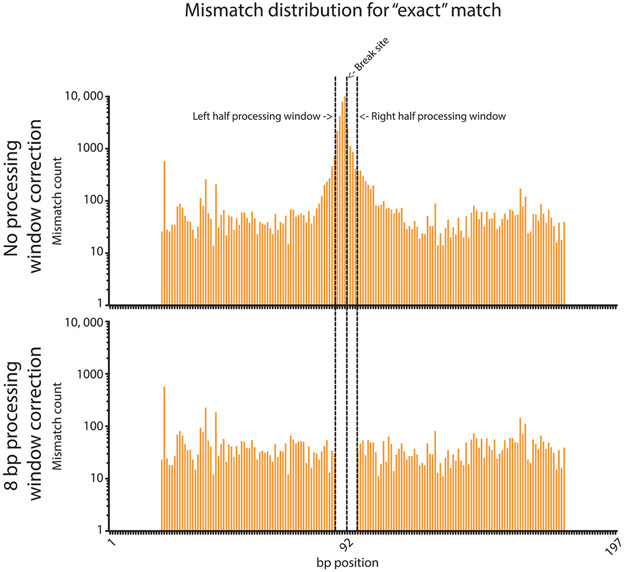
Once adjusted, all remaining base substitutions are likely due to sequencing error. This is apparent as sequencing error should be evenly distributed across the amplicon, while sequence changes stemming from biological processing of the repair junction should be enriched near the breakpoint of the reference. Indeed, in MiSeq-sequenced amplicons, small insertions, small deletions, and base substitutions all cluster near repair breakpoints (Fig. 3).
Fig. 3.
Distribution of base substitutions in Illumina sequencing DSB repair amplicons. The distribution of mismatched bp is given for reads that were considered perfect repair events by Geneious, before and after the 8 bp search for base substitutions correction. The majority of mismatches occur homogenously across the read, as expected of sequencing error, with counts of ~10–100. However, the mismatches peak around the break site with a maximum of 11,982 immediately at the break site (~120–1200-fold higher than the surrounding sequence). The 8 bp processing window correction recategorizes these counts and leaves the rest that are likely sequencing error. No mismatches occur at the ends of the reads due to the “Trim_and_Pad” script used to filter reads.
However, after excluding base pair substitutions within eight nucleotides surrounding a repair junction, all remaining base substitutions are evenly distributed across the amplicon. This is consistent with the major source of Illumina sequencing error being random base substitutions. Hi-FiBR subsequently removes any base pair substitutions outside of the defined processing window by constructing a hybrid sequence for each read (called a “reconstructed read”), consisting of actual read sequence in the processing window surrounded by the corresponding reference sequence for the flanking regions (as shown in Fig. 4). The only variation in this reconstructed read should now be due to processing of the DSB during the repair event.
Fig. 4.
Removing sequencing error by generating reconstructed reads. Sequencing error in the flanks of reads around the break site causes the same type of repaired read to appear as a different repair event. To correct for this discrepancy the reads are reconstructed in the regions of matching sequence to become the same as the reference sequence, thus creating a common, reconstructed read.
The number of nucleotides processed on each side of the breakpoint, deletion lengths, insertion lengths, insertion sequences, and reconstructed read sequences is added as additional columns for each read in the original SAM file and written as a new SAM with the same name plus the extension “_Extended.sam.” Furthermore, a new data file containing the list of repair events observed is generated with the same name as the original SAM file plus the extension “_Final.sam.” The “Final” output file contains a summary of all the analyses Hi-FiBR conducts. It is a tab-delimited table containing 22 columns, where each column contains unique information describing the repair event (see Table 1 for a detailed description of each column’s contents). This file is generated by listing each reconstructed read observed in the data set once. The script then proceeds to count how many times each reconstructed read occurs in the full data set. It is also important to know the percent of mismatch between the reconstructed read and the original reads, as it may help identify systemic mapping errors or biological events that are being masked by the reconstruction. A small, nonzero value is always expected as sequencing error will occur at a low frequency, and thus, some portion of reads will always contain a mismatch. As the value increases, this suggests that there is some other issue, such as PCR cross-contamination, or real biological events being called sequencing error by the mapping tool. To help determine the cause, the script compares the original reads and the reconstructed read and outputs a read error distribution file which contains counts of the mismatches at each individual bp position among raw reads categorized as the same repair event (i.e., they produce the same reconstructed read). If the errors occur in a homogeneous distribution, it is likely just sequencing error. If there is a peak near the break site, then there is a biological event being ignored. In the event that the same reconstructed read sequence can be generated by multiple alignments of the read sequence, all possible alignments are included in _Final.sam file; however, they will contain the same count for the number of times that event was observed due to them having the same reconstructed read sequence. Hi-FiBR marks these events in column 21 of the _Final.sam file (which indicates the number of times a sequence occurs in the _Final.sam file) so that the user can manually determine which alignment is most likely.
Table 1.
Descriptions Are Given for the Contents of Each Column in the “Final” Output File
| Column | Description |
|---|---|
| 1 | Name of reference sequence to which read was mapped |
| 2 | Original CIGAR string |
| 3 | Length of read |
| 4 | Original CIGAR string; later may contain a reconstructed CIGAR string |
| 5 | Corrected length of sequence matching the left flank of the reference |
| 6 | Corrected length of sequence matching the right flank of the reference |
| 7 | Distance between the break and the end of the left matching sequence |
| 8 | Distance between the break and the end of the right matching sequence |
| 9 | Number of nucleotides deleted on the left side of the break |
| 10 | Number of nucleotides deleted on the right side of the break |
| 11 | Total number of deleted nucleotides |
| 12 | Start of inserted sequence |
| 13 | End of inserted sequence |
| 14 | Length of inserted sequence |
| 15 | Sequence of inserted nucleotides |
| 16 | Repair event class |
| 17 | Reconstructed read sequence |
| 18 | Number of times reconstructed read occurred in read population |
| 19 | Potential microhomology that mediated deletion |
| 20 | Length of microhomology |
| 21 | Number of times reconstructed read is observed in this file (in a row) |
| 22 | Percentage of reads that contain mismatches to reconstructed read |
The output does not follow a conventional format, and so the descriptions should be carefully examined.
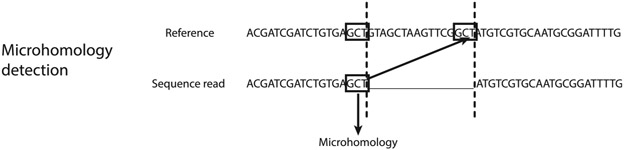
For repair events that are simple deletions, Hi-FiBR also determines whether microhomology mediated the event. This is done by comparing the matching indices to the position of the breakpoint and then moving along the working sequence and reference sequence in single base iterations to search for homology (see Fig. 5). Specifically, the sequence on the left side of the repair junction is compared to the rightmost portion of the deleted sequence. Similarly, the sequence to the right of repair junction is compared to the leftmost portion of the deleted sequence. If microhomologies are found on each half, they are added together to make the complete sequence and the homology length is calculated.
Fig. 5.
Determination of microhomology usage. For reads classified as deletions, sequence microhomologies may mediate the repair event. These microhomologies are detected by examining the sequence to the left of the deletion in the read, and the sequence inside the deletion on the right side, as shown here. The same process is repeated for the sequence to the right of the deletion in the read (not shown here). The microhomologies detected are then combined into a single sequence.
Finally, each line in the newly written file is reread by the script and the insert sequences are exported into fasta files based on their class as “complex” or “insertion,” and all the working (reconstructed) sequences are exported into a single fasta file.
5. RESULTS OF ANALYSIS AND DISCUSSION
5.1. Effects of Using Different Aligners for Hi-FiBR Analysis
Many alignment tools besides those described here are used by the scientific community to analyze sequencing data. However, scoring penalties associated with features such as gap creation, extension, and mismatching nucleotides are not universal, neither are the methods for alignment (local vs global, gapped vs ungapped, etc.), and, ultimately, lead to differences in mapping. Here, we compared four of the most commonly used aligners to see if differences in their algorithms made a significant impact on the number and diversity of events they called, as denoted by the CIGAR strings. Additionally, to test the versatility of each aligner, we used a data set that had a highly varied array of sequences (suggesting many different repair event types) (Fig. 6). This helps show how individual alignment tools handle events with a high degree of complexity, and how efficaciously Hi-FiBR corrects errors that the aligners make when aligning complex reads.
Fig. 6.
Overview of the data set’s complexity. The data set used here mostly consisted of deletion events, followed by insertion events, complex events, and finally exact matches (i.e., error-free repair). A cursory examination shows that in each event there is a diverse number of repair events (based on size of insertion or deletion) and the sizes tend to follow a normal distribution from smallest size to largest (although the scale here is logarithmic and, consequently, is not immediately obvious as normal).
The data set we used was first trimmed, then mapped using each alignment tool, individually, and the SAM files from each mapper were analyzed using the Hi-FiBR analysis script with a 0- and an 8-bp window around the break site, creating eight permutations. The count and percentage of reads remaining after each step are listed in Table 2, and show that most of the reads are kept throughout the entire analysis. Importantly, the change in window size around the break site does not remove reads, but reclassifies them. It is worth noting, though, that the final outputs for Geneious and BWA retain the highest number of reads at ~97% and ~96.7%, respectively, and CLC and Bowtie2 retain the least at ~93% and ~94%, respectively. For CLC, this is because ~4% of the reads are mapped using methods such as soft clipping that the script does not process and removes from the analysis. For Bowtie2, this is because ~3% of the reads do not map, and never make it to the analysis (even if the reads are left in the SAM file, the Hi-FiBR analysis will skip them as they are denoted with a “*” symbol). Based on the percent of reads retained, all aligners appear to be highly similar; however, Geneious would maintain the highest number of reads to use in conjunction with the Hi-FiBR analysis.
Table 2.
The Number of Sequencing Reads Retained Following Each Processing Step in the Analysis
| Processing Step | Counts | Percent Retained |
|---|---|---|
| fastq | 1134957 | 100 |
| Trimmed | 1101132 | 97.01971088 |
| Geneious | 1100535 | 96.96710977 |
| Hi-FiBR Geneious (No Processing Window Correction) | 1100535 | 96.96710977 |
| Hi-FiBR Geneious (8 bp Processing Window Correction) | 1100535 | 96.96710977 |
| CLC | 1099030 | 96.83450562 |
| Hi-FiBR CLC (No Processing Window Correction) | 1054561 | 92.91638362 |
| Hi-FiBR CLC (8 bp Processing Window Correction) | 1054561 | 92.91638362 |
| Bowtie2 | 1070588 | 94.3285076 |
| Hi-FiBR Bowtie2 (No Processing Window Correction) | 1070588 | 94.3285076 |
| Hi-FiBR Bowtie2 (8 bp Processing Window Correction) | 1070588 | 94.3285076 |
| BWA | 1100458 | 96.96032537 |
| Hi-FiBR BWA (No Processing Window Correction) | 1097364 | 96.68771592 |
| Hi-FiBR BWA (8 bp Processing Window Correction) | 1097364 | 96.68771592 |
About 3% of reads are lost to the trimming step, and the remaining lost are due to the alignment tools. The order of most reads retained from highest to lowest is Geneious (~97%), BWA (~96.7%), Bowtie2 (~94%), and CLC (~93%). No difference occurs from the size of the processing window adjustment.
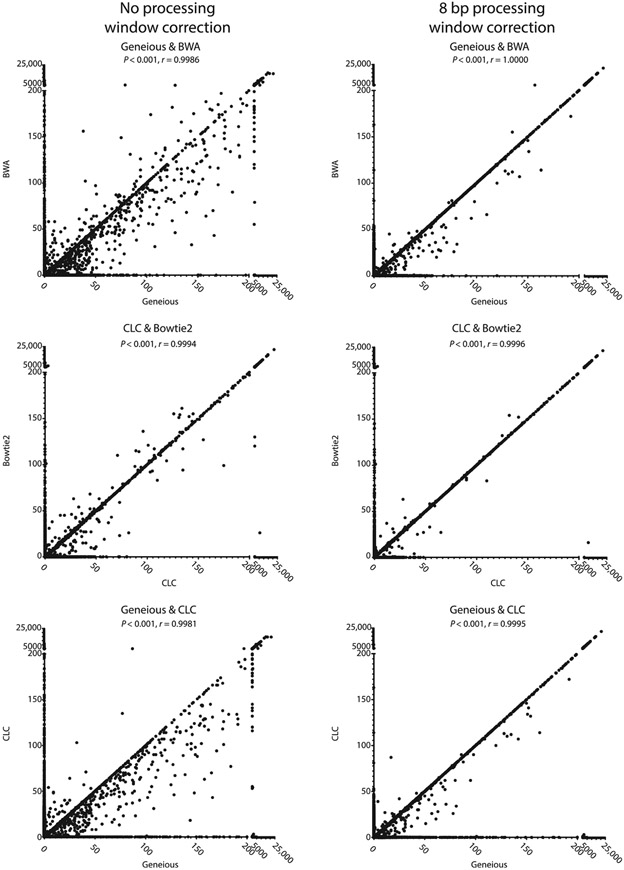
Pearson correlation coefficients were calculated between the final reconstructed reads for each of the alignment tools to determine how similarly they processed the data. This showed high correlation between all mappers (see Fig. 7). Interestingly, when ignoring base substitutions near the break site, a small percentage of reads show vastly different counts among the mappers, with Geneious tending to produce higher counts per read than the other alignment tools. Many of these reads can be traced back to reads that were lost during the mapping and processing of CLC and Bowtie2 (as noted by the 0 counts with these aligners). Most of the remaining differentially counted reads appear to be caused by mismatches near the break site that influence how each mapper aligns the read. Once the processing windows of each repair event were enlarged to include base substitutions occurring within 8 bp of the break site, nearly all reads are counted equally, regardless of the aligner used, and the Pearson correlation coefficients increased (see Table 3).
Fig. 7.
Comparison of NGS alignment softwares for analyzing DSB repair junctions. Example Pearson correlations are given for Geneious, CLC, Bowtie2, and BWA. After processing window correction the aligning tools are more similar. However, there are still many reads that Geneious calls that tools, like CLC, do not catch, as shown by the many 0 reads. This is unsurprising, as CLC retained fewer reads than Geneious.
Table 3.
Pearson Correlation Coefficients for Pairwise Comparison of NGS Alignment Softwares
| Aligner Pairs | No Processing Window Correction |
8 bp Processing Window Correction |
|---|---|---|
| Geneious & CLC | 0.9981 | 0.9995 |
| Geneious & Bowtie2 | 0.9985 | 0.9999 |
| Geneious & BWA | 0.9986 | 1 |
| CLC & Bowtie2 | 0.9994 | 0.9996 |
| CLC & BWA | 0.9994 | 0.9995 |
| Bowtie2 & BWA | 0.9994 | 0.9999 |
After processing window adjustment, all aligners become more similar to each other. However, Geneious and BWA remain the most similar.
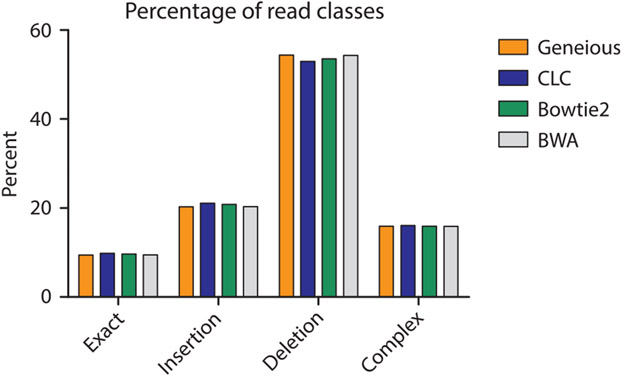
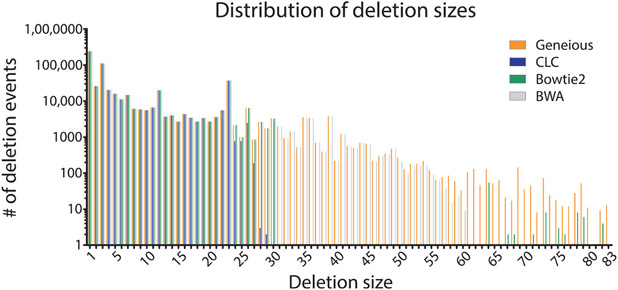
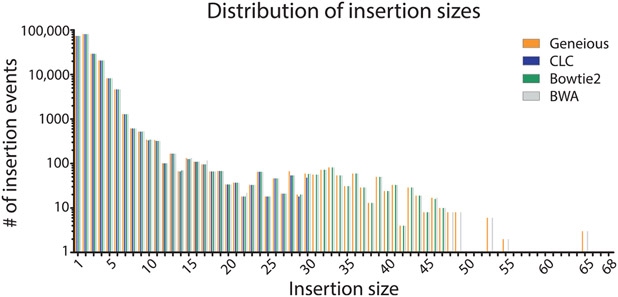
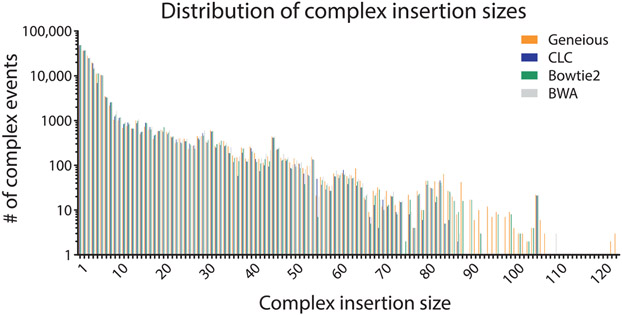
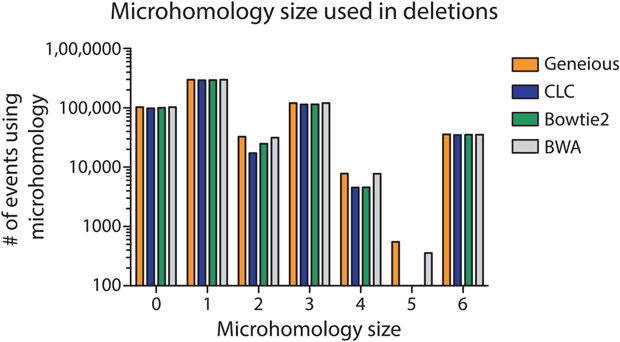
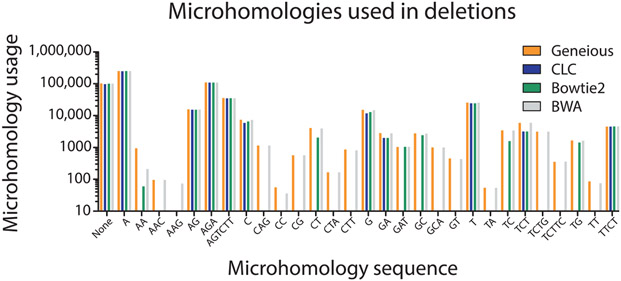
To determine if specific aligners, particularly CLC and Bowtie2 which mapped the lowest number of reads, had difficulty mapping specific types of repair events, we compared (using a nonparametric one-way ANOVA; i.e., Kruskal–Wallis test) the mappers ability to count repair events parsed by class (P = 0.9778, Fig. 8), the size of read deletions (P < 0.0001, Fig. 9), the size of read insertions (P = 0.0098, Fig. 10), the size of read complex insertions (P = 0.4905, Fig. 11), the size of microhomologies used in repair (P = 0.9207, Fig. 12), and the types of microhomologies used in repair (P = 0.0108, Fig. 13).
Fig. 8.
Distribution of repair classes identified by Hi-FiBR analysis of an example data set. The different aligners have the same distribution of repair class types (P = 0.9778). For our data set, deletion events occurred the most frequently, followed by insertions, complex events, and exact matches (i.e., error-free repair).
Fig. 9.
Distribution of observed deletion events based on deletion size. The different aligners produced different distributions of deletion sizes (P < 0.0001). Geneious and BWA called larger deletion sizes. However, they are a small percentage of the total deletions.
Fig. 10.
Distribution of observed insertion events based on insertion size. The different aligners produce statistically different distributions of insertion sizes (P = 0.0098). Manual observation shows that Geneious, BWA, and Bowtie2 called larger insertion sizes, but only a small percent of the reads are larger.
Fig. 11.
Distribution of observed complex repair events based on insertion size. The complex insertion sizes are the same for all alignment tools (P = 0.4905), unlike the deletion and insertion sizes.
Fig. 12.
Distribution of microhomology size observed in deletion events. The size of the microhomologies that mediate deletion events was the same for all aligners (P = 0.9207). The sizes tended to be small (0–3 bp), but this would likely shift depending on the sequence context around the break site.
Fig. 13.
Microhomology sequence usage. The sequence of the microhomologies used differs between all alignment tools (P = 0.0108). Since Geneious and BWA detect events with larger deletion sizes, the difference in microhomologies are likely linked to the larger deletions.
This analysis indicated that the aligners differed primarily based upon the size of deletion and insertion events as well as the specific microhomology sequences utilized during repair. Post-ANOVA Dunn’s multiple comparison tests showed that Geneious and BWA called events with larger deletion sizes more efficiently than CLC and Bowtie2 (P < 0.05, see Table 4). The ability of Geneious and BWA to accurately map these larger deletion events also accounts for the difference in the specific microhomology sequences observed by the different mapping methods, as these additional reads increased the number of sequences that can potentially be used to find microhomology. The distribution of insertion sizes between mappers was also statistically different (see Table 5). Manual observation of the insertion graph indicates Geneious, Bowtie2, and BWA call events with larger insertion sizes than CLC; however, these events generally constitute a small fraction of the total number of repair events that occur. Cumulatively, this may suggest that Geneious and BWA are better at recognizing biological events in the flanks of DNA sequences around the break site.
Table 4.
The Deletion Size Dunn’s Multiple Comparison P values Are Listed Here Between the Different Aligners
| Aligner Pairs | P < 0.05 |
|---|---|
| Geneious & CLC | Yes |
| Geneious & Bowtie2 | Yes |
| Geneious & BWA | No |
| CLC & Bowtie2 | No |
| CLC & BWA | Yes |
| Bowtie2 & BWA | No |
Table 5.
The Insertion Size Dunn’s Multiple Comparison P values Are Listed Here Between the Different Aligners
| Aligner Pairs | P < 0.05 |
|---|---|
| Geneious & Bowtie2 | No |
| Geneious & BWA | No |
| CLC & Bowtie2 | No |
| CLC & BWA | Yes |
| Bowtie2 & BWA | No |
5.2. Using Different Sequencing Platforms With Hi-FiBR
The data set used here was generated on an Illumina sequencing platform; however, because Hi-FiBR operates on a standard SAM file format, other sequencing methods could theoretically be used for sequencing repair junction amplicons. PacBio (Eid et al., 2009) and Ion Proton sequencing both have potential benefits that may make them particularly appealing for such analyses. They both produce longer reads, with PacBio easily able to accommodate multiple kb amplicons, which would allow even longer deletion events to be assessed. Moreover, these platforms cost less per SMRT cell or semiconductor, operate on scales that produce less reads per run (50,000 and 80,000,000 reads, respectively), and run faster than Illumina platforms (providing results sooner). Consequently, these sequencing methods would allow smaller scale experiments to be easily and cheaply conducted. This is opposed to experiments utilizing Illumina sequencing, where many libraries need to be accumulated and sequenced on a single lane to gain the greatest cost effectiveness.
As mentioned previously however, both Ion Proton and PacBio can experience systematic sequencing error that significantly inhibits sequence analysis of repair junctions. In particular, both platforms can produce frequent deletions or insertions in homopolymer runs of greater than three nucleotides in length. All NGS alignment tools identify these sequencing errors within the CIGAR string of the SAM file. This will ultimately cause Hi-FiBR to propagate the error and inaccurately report the sequence between the error and the break site as part of the repair processing window. If the sequencing reads can be corrected to account for these errors, namely, in the CIGAR string, Hi-FiBR will function normally. In the future, it may be desirable to design additional functions that examine a population of sequencing reads for highly recurrent errors, and that cleanse these sequences prior to mapping and sequence analysis.
6. SUMMARY AND CONCLUSION
The large data sets generated by NGS-based analysis of DSB repair events require robust alignment and downstream analyses. We report here an automated method for quantifying the relative frequency of repair events from amplicons sequenced with NGS technologies. The analysis is accurate and highly efficient. The 615 MB data set analyzed in this chapter was processed by the Hi-FiBR analysis on average in 8 min 13 s (for five runs), on a 3.50-GHz processor with 32 GB RAM available (although the RAM usage only peaked at ~0.7 GB, not including other background processes). Thus, it should easily run on most computers with Python3 installed.
Our study suggests that aligners map reads differently in regard to mismatches near breaks sites, but the Hi-FiBR analysis effectively corrects for this alignment error. These differences are normally small and the analysis for the majority of events strongly correlates between mappers even without Hi-FiBR’s alignment correction. However, in data sets where base substitution near break sites is high, Hi-FiBR can identify events that would otherwise be improperly classified.
Despite the fact that Hi-FiBR can produce universally robust results regardless of the alignment software used to map the NGS reads to the reference sequence, Geneious’s and BWA’s alignment algorithms inherently allow them to call larger deletion/insertion events than CLC and Bowtie2. Most of these events are lost from the analysis due to failed or improper mapping of the specific reads, which may result from penalties associated with gap creation and extension. We therefore recommend using Geneious or BWA in association with Hi-FiBR analysis to preserve as many repair events as possible. However, it is likely that use of a version of CLC Genomics Workbench that allows affine gap penalties or adjusting the alignment parameters in CLC or Bowtie2 would increase the mappability of reads with larger deletion and insertions.
We believe that expanded utilization of NGS sequencing of DSB repair events will greatly enhance our understanding of the mechanisms employed in cells to repair these extremely deleterious lesions. Moreover, as inappropriate repair of DSBs commonly underlies potentially carcinogenic genome rearrangements (Arlt, Casper, & Glover, 2003; Byrne et al., 2014; Huebner & Croce, 2001), developing highly detailed signatures of error-prone DSB repair processes will greatly facilitate our ability to determine what enzymatic processes contribute to cancer genetic heterogeneity. NGS sequencing of DSB repair junctions provides an easy means to produce these signatures. Already, it is clear that the translocation junctions in many different cancer types bear resemblance to sequences used by alt-EJ (Chiarle et al., 2011; Hromas et al., 2016; Nussenzweig & Nussenzweig, 2007; Soni et al., 2015). In particular, BRCA1- and BRCA2-deficient breast cancers maintain a mutation signature similar to the spectra of sequences produced by pol theta-mediated repair of DSBs (Wyatt et al., 2016). It will be important to further outline what initiates the usage of these pathways and the genomic environments in which it is most mutagenic. This will be accomplished through the high data production of NGS technologies and the accurate analyses of these data, as shown here by the Hi-FiBR analysis.
ACKNOWLEDGMENTS
We would like to thank Drs. Varandt Khodaverian, Mitch McVey, and David Weinstock, as well as Jacob Layer for their input on this analysis pipeline as it was being developed. This research was supported by grants from the National Institute of Environmental Health Sciences (NIEHS) (R00ES022633 to S.A.R.), the National Cancer Institute (NCI) (R01ES002614 to S.A.R.), and the Department of Defense Congressionally Directed Medical Research Programs (BC141727 to S.A.R.).
ABBREVIATIONS
- BWA
Burrows–Wheeler Aligner
- CIGAR
Compact Idiosyncratic Gapped Alignment Report
- C-NHEJ
classical nonhomologous end joining
- DSB
double-strand break
- Hi-FiBR
high-throughput fidelity of break repair
- MMEJ
microhomology-mediated end joining
- NGS
next-generation sequencing
REFERENCES
- Aparicio T, Baer R, & Gautier J (2014). DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst), 19, 169–175. 10.1016/j.dnarep.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Casper AM, & Glover TW (2003). Common fragile sites. Cytogenetic and Genome Research, 100(1–4), 92–100. 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- Beagan K, Armstrong RL, Witsell A, Roy U, Renedo N, Baker AE, et al. (2017). Drosophila DNA polymerase theta utilizes both helicase-like and polymerase domains during microhomology-mediated end joining and interstrand crosslink repair. PLoS Genetics, 13(5), e1006813. 10.1371/journal.pgen.1006813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CB, Lewis AL, Baldwin KK, & Resnick MA (1993). Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proceedings of the National Academy of Sciences of the United States of America, 90(12), 5613–5617. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8516308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M, Wray J, Reinert B, Wu Y, Nickoloff J, Lee SH, et al. (2014). Mechanisms of oncogenic chromosomal translocations. Annals of the New York Academy of Sciences, 1310, 89–97. 10.1111/nyas.12370. [DOI] [PubMed] [Google Scholar]
- Chan SH, Yu AM, & McVey M (2010). Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genetics, 6(7), e1001005. 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, et al. (2011). Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell, 147(1), 107–119. 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu YL, Wu X, Xu Y, & Her C (2013). MutS homologue hMSH4: Interaction with eIF3f and a role in NHEJ-mediated DSB repair. Molecular Cancer, 12(51). 10.1186/1476-4598-12-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, & Elledge SJ (2010). The DNA damage response: Making it safe to play with knives. Molecular Cell, 40(2), 179–204. 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A (2013). Alternative end-joining mechanisms: A historical perspective. Frontiers in Genetics, 4(48). 10.3389/fgene.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science, 323(5910), 133–138. 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, & McCombie WR (2016). Coming of age: Ten years of next-generation sequencing technologies. Nature Reviews. Genetics, 17(6), 333–351. 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn A, Bennardo N, Cheng A, & Stark JM (2011). Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. The Journal of Biological Chemistry, 286(49), 42470–42482. 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A, & Haber JE (1999). Physical monitoring of HO-induced homologous recombination. Methods in Molecular Biology, 113, 403–415. 10.1385/1-59259-675-4:403. [DOI] [PubMed] [Google Scholar]
- Hromas R, Williamson E, Lee SH, &Nickoloff J (2016). Preventing the chromosomal translocations that cause cancer. Transactions of the American Clinical and Climatological Association, 127, 176–195. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/28066052. [PMC free article] [PubMed] [Google Scholar]
- Huebner K, & Croce CM (2001). FRA3B and other common fragile sites: The weakest links. Nature Reviews. Cancer, 1(3), 214–221. 10.1038/35106058. [DOI] [PubMed] [Google Scholar]
- Huefner ND, Mizuno Y, Weil CF, Korf I, & Britt AB (2011). Breadth by depth: Expanding our understanding of the repair of transposon-induced DNA double strand breaks via deep-sequencing. DNA Repair (Amst), 10(10), 1023–1033. 10.1016/j.dnarep.2011.07.011. [DOI] [PubMed] [Google Scholar]
- IJspeert H, Rozmus J, Schwarz K, Warren RL, van Zessen D, Holt RA, et al. (2016). XLF deficiency results in reduced N-nucleotide addition during V(D)J recombination. Blood, 128(5), 650–659. 10.1182/blood-2016-02-701029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA (1990). Studies on mammalian mutants defective in rejoining double-strand breaks in DNA. Mutation Research, 239(1), 1–16. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/2195330. [DOI] [PubMed] [Google Scholar]
- Kent T, Chandramouly G, McDevitt SM, Ozdemir AY,& Pomerantz RT (2015). Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nature Structural & Molecular Biology, 22(3), 230–237. 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodaverdian VY, Hanscom T, Yu AM, Yu TL, Mak V, Brown AJ, et al. (2017). Secondary structure forming sequences drive SD-MMEJ repair of DNA double-strand breaks. Nucleic Acids Research, 45(22), 12848–12861. 10.1093/nar/gkx1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, & Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, & Lee SE (2007). Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics, 176(4), 2003–2014. 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14), 1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko PJ, &Jasin M (1998). Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America, 95(9), 5172–5177. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9560248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Sunder S, Nallasivam S, & Wilson TE (2016). Overhang polarity of chromosomal double-strand breaks impacts kinetics and fidelity of yeast non-homologous end joining. Nucleic Acids Research, 44(6), 2769–2781. 10.1093/nar/gkw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR (2010). The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual Review of Biochemistry, 79, 181–211. 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Lukacsovich T, & Waldman AS (1999). Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes. Molecular and Cellular Biology, 19(12), 8353–8360. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10567560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, & Sfeir A (2015). Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature, 518(7538), 254–257. 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, & Haber JE (1996). Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair ofdouble-strand breaks in Saccharomyces cerevisiae. Molecular and Cellular Biology, 16(5), 2164–2173. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8628283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, & Jasin M (1997). Loss of heterozygosity induced by a chromosomal double-strand break. Proceedings of the National Academy of Sciences of the United States of America, 94(17), 8988–8993. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9256422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, et al. (2005). A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Molecular Cell, 19(3), 357–366. 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Nickoloff JA (2017). Paths from DNA damage and signaling to genome rearrangements via homologous recombination. Mutation Research, 806, 64–74. 10.1016/j.mrfmmm.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A, & Nussenzweig MC (2007). A backup DNA repair pathway moves to the forefront. Cell, 131(2), 223–225. 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Resnick MA, &Martin P (1976). The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Molecular & General Genetics, 143(2), 119–129. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/765749. [DOI] [PubMed] [Google Scholar]
- Roth DB, & Wilson JH (1986). Nonhomologous recombination in mammalian cells: Role for short sequence homologies in the joining reaction. 6, 4295–4304. http://www.ncbi.nlm.nih.gov/pubmed/3025650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, & Jasin M (1994). Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Molecular and Cellular Biology, 14(12), 8096–8106. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7969147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N, & Haber JE (1988). Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Molecular and Cellular Biology, 8(9), 3918–3928. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/3065627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Shim EY, & Lee SE (2017). Microhomology-mediated end joining: Good, bad and ugly. In Mutation Research, pii: S0027-5107(17)30041-6. 10.1016/j.mrfmmm.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, & Symington LS (2015). Microhomology-mediated end joining: A back-up survival mechanism or dedicated pathway? Trends in Biochemical Sciences, 40(11), 701–714. 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Javadekar SM, Pandey M, Srivastava M, Kumari R, & Raghavan SC (2015). Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death & Disease, 6, e1697. 10.1038/cddis.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Li F, Villarreal D, Shim JH, Yoon S, Myung K, et al. (2017). Microhomology-mediated end joining induces hypermutagenesis at breakpoint junctions. PLoS Genetics, 13(4), e1006714. 10.1371/journal.pgen.1006714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni A, Siemann M, Pantelias GE, & Iliakis G (2015). Marked contribution of alternative end-joining to chromosome-translocation-formation by stochastically induced DNA double-strand-breaks in G2-phase human cells. Mutation Research, Genetic Toxicology and Environmental Mutagenesis, 793, 2–8. 10.1016/j.mrgentox.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Soong CP, Breuer GA, Hannon RA, Kim SD, Salem AF, Wang G, et al. (2015). Development of a novel method to create double-strand break repair fingerprints using next-generation sequencing. DNA Repair (Amst), 26, 44–53. 10.1016/j.dnarep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- van Schendel R, van Heteren J, Welten R, & Tijsterman M (2016). Genomic scars generated by polymerase theta reveal the versatile mechanism of alternative end-joining. PLoS Genetics, 12(10), e1006368. 10.1371/journal.pgen.1006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend LE, Jasin M, & Krawczyk PM (2014). Assaying break and nick-induced homologous recombination in mammalian cells using the DR-GFP reporter and Cas9 nucleases. Methods in Enzymology, 546, 175–191. 10.1016/B978-0-12-801185-0.00009-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend LE, Prakash R, Chen CC, Vanoli F, Cavallo F, Zhang Y, et al. (2016). Distinct genetic control of homologous recombination repair of Cas9-induced double-strand breaks, nicks and paired nicks. Nucleic Acids Research, 44(11), 5204–5217. 10.1093/nar/gkw179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CA, Strande NT, Pryor JM, Strom CN, Mieczkowski P, Burkhalter MD, et al. (2014). The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nature Communications, 5, 4286. 10.1038/ncomms5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt DW, Feng W, Conlin MP, Yousefzadeh MJ, Roberts SA, Mieczkowski P, et al. (2016). Essential roles for polymerase theta-mediated end joining in the repair of chromosome breaks. Molecular Cell, 63(4), 662–673. 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu X, & Her C (2015). hMSH5 facilitates the repair of camptothecin-induced double-strand breaks through an interaction with FANCJ. The Journal of Biological Chemistry, 290(30), 18545–18558. 10.1074/jbc.M115.642884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefzadeh MJ, Wyatt DW, Takata K, Mu Y, Hensley SC, Tomida J, et al. (2014). Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genetics, 10(10), e1004654. 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]