Abstract
The concept of high liquid repellency in multi-liquid-phase systems (e.g., aqueous droplets in an oil background) has been applied to areas of biomedical research to realize intrinsic advantages not available in single-liquid-phase systems. Such advantages have included minimizing analyte loss, facile manipulation of single cell samples, elimination of biofouling, and ease-of-use regarding loading and retrieving of the sample. In this manuscript, we present generalized design rules for predicting the wettability of solid-liquid-liquid systems (especially for discrimination between exclusive liquid repellency (ELR) and finite liquid repellency (FLR)) to extend the applications of ELR. We then apply ELR to two model systems with open microfluidic design in cell biology: 1) In situ underoil culture and combinatorial co-culture of mammalian cells in order to demonstrate directed single cell multi-encapsulation with minimal waste of sample as compared to stochastic cell seeding; 2) Isolation of a pure population of circulating tumor cells (CTCs), which is required for certain downstream analyses including sequencing and gene expression profiling.
Keywords: liquid repellency, underoil, cell culture, single cell, rare cell
Graphical Abstract

INTRODUCTION
Recently, phenotypic heterogeneity and the influence of rare cell populations are emerging themes in a number of different biomedical fields,[1,2] including cancer progression,[3,4] autoimmune disease,[5] and developmental biology.[6] Accompanying this emergence is a parallel need for new cell culture tools that are optimized to interrogate phenotype via ex vivo or in vitro culture of a single cell or a limited number of cells.[7–14] Specifically, these culture platforms would ideally possess a number of enabling characteristics, including: 1) amenable to handling precious samples (e.g., fine needle aspirates, liquid biopsies, bronchoalveolar lavages) and will retain cells with minimal loss; 2) minimize “observer effect” on the target population, as cells can be placed in the culture system with minimum shear stress and no contact with non-biological surfaces (while every in vitro culture system has artifacts, limiting exposure to exogenous components of the culture system will mitigate this effect); 3) scalable in terms of both target cell number and total cell number; 4) facilitate straightforward manipulation of specific target cells (e.g., combining specific cells for targeted multi-phenotype culture experiments, collection of selected cells-of-interest).
In response to this unique collection of requirements, cell culture platform developers have attempted to leverage the intrinsic advantages of microfluidics. While many creative solutions have been proposed in the literature, they can be largely classified by two dichotomous characteristics: 1) Closed vs. open microfluidics; 2) single-liquid-phase vs. multi-liquid-phase systems. Closed microfluidic devices offer precise control over liquid components and limit evaporation via physical enclosure.[15,16] However, closed systems typically rely on fluidic control components (pumps, tubing, valves) integrated into the platform, which introduce mechanisms of failure and increase costs. Additionally, closed systems limit accessibility to samples of interest and hinder cellular manipulation with external tools due to the physical microchannel barriers. In contrast, open microfluidic systems (e.g., traditional microtiter plate, microwells) offer direct access to individual cells, and enable high system simplicity from both fabrication and operation.[17–19] Medium loss via evaporation on open platforms is a significant issue especially when systems are scaled to single cell volumes (i.e., a few μL or less). While single-liquid-phase systems have proven immensely valuable, release and retrieval of adherent cells-of-interest is challenging and frequently involves cell loss (e.g., trypsin release of cells from a high density well plate).[20] Conversely, multi-liquid-phase platforms (e.g., droplet microfluidics) enable facile manipulation of specific cells with minimized sample loss and device fouling. Cells can be encapsulated and transported in micro emulsions in carrier oil and in no direct contact with solid.[21] Here we propose an open, multi-liquid-phase cell culture and screening system that leverages an underutilized combination of the aforementioned characteristics and enables novel applications in cell biology.
RESULTS AND DISCUSSION
Design rules, experimental realization and benefits of ELR for cellular studies
The underlying principles depicting the wetting behavior of multi-liquid-phase systems have been previously explored[22–26] and have been applied to several biomedical applications such as single cell isolation,[27,28] digital droplet polymerase chain reaction (PCR),[20,29,30] anti-thrombosis coating,[31] and tissue engineering.[32] While powerful and conceptually well understood by many, the principles remain known mainly to a “niche” within specific research communities and are complicated to reliably implement, limiting the use of this potentially powerful regime. Here, we outline a straightforward protocol for distinguishing two regimes of liquid repellency: ELR (the dispersed liquid phase is completely repelled and shows no adhesion on solid) and FLR (the dispersed liquid phase is non- or partially-repelled, and shows adhesion on solid). Further, we demonstrate the utility of these design rules through two applications for research in cell biology. First, in situ underoil cell culture and directed single cell multi-encapsulation of monocytes (THP-1) were achieved in ELR microdroplets. In addition, single cell subgrouping of a rare cell type (CTCs) was demonstrated, which could enhance the precision for downstream analysis (e.g., whole genome sequencing).
The repellency of a dispersed liquid droplet within a surrounding immiscible liquid against a solid is inherently determined by the interfacial energy between the two liquids as well as the interfacial energies between each liquid and the solid.[33] The principle can be silhouetted with thermodynamic modeling of a solid-liquid-liquid three phase system. Here we start by rewriting the subscript of each parameter in Young’s equation to address this situation: γS/Lcp = γS/Ldp + γLdp/Lcpcosθ, where θ is Young’s contact angle (CA), γS/Lcp, γS/Ldp, γLdp/Lcp are the interfacial energies of solid/liquid of continuous phase (S/Lcp), solid/liquid of dispersed phase (S/Ldp) and between the two liquids (Ldp/Lcp), respectively. Setting θ to 180o (e.g., ELR, or no contact between the dispersed liquid phase and the solid) yields γS/Ldp = γS/Lcp + γLdp/Lcp. If we next apply the thermodynamic boundary conditions of surface energy (γS/G, γLcp/G and γLdp/G > 0) and S/L and L/L interfacial energies (γS/Lcp < γS/G + γLcp/G, γS/Ldp < γS/G + γLdp/G, and γLdp/Lcp < γLdp/G + γLcp/G) (SI Figure 1), the relationship between γS/Ldp and γS/Lcp + γLdp/Lcp can be either “>”, “=”, or “<” (SI Figure 2).
To better visualize the balance between interfacial energies, we introduce a solid-water-oil (S/W/O) triangle and Venn diagram graphical representation to map the parameter space (Figure 1a). The three points of a S/W/O triangle correspond to the three phases of S, W, and O. Each side is formed by connecting any of two points with a solid line and represents the corresponding interfacial energy (e.g., S/W, S/O, and W/O). Thus, the relationship between γS/Ldp and γS/Lcp + γLdp/Lcp can be comparably expressed via “triangle inequality” (i.e., Is the sum of the lengths of any two sides of a triangle greater than the length of the third side?). In other words, ELR (CA = 180°) will occur if the sum of the two sides (γS/Lcp and γLdp/Lcp) of the S/W/O triangle is equal to or less than the third side (γS/Ldp), otherwise FLR (CA < 180°) will occur. To demonstrate how the S/W/O triangle is implemented, we chose three non-textured solid materials (O2 plasma-treated glass, polystyrene (PS), and (polydimethylsiloxane (PDMS)) and three W/O pairs (water/mineral oil, water/silicone oil, and water/Fluorinert FC-40) (SI Table 1, SI Table 2). The CAs of each test liquid on the three solid materials were measured in air to calculate the interfacial energies of S/W and S/O (SI Figure 3, SI Methods). On PDMS, ELR is only obtained against water when silicone oil is used as continuous phase. In contrast, ELR is only obtained against silicone oil and Fluorinert FC-40 on O2 plasma-treated glass when water is used as continuous phase (SI Figure 4). It is worth noting that on PDMS, if silicone oil is replaced with either mineral oil or Fluorinert FC-40, ELR is eliminated because the interfacial energy between water and oil is too large as per the S/W/O triangle (SI Figure 5, orange right column, SI Movie 1, SI Movie 2). An analogous effect can be observed on O2 plasma-treated glass (SI Figure 5, blue left column, SI Movie 3). Provided that a solid material shows no obvious preference between water and oil (e.g., PS), no matter which (water or oil) is set as continuous phase ELR will not be obtainable (SI Figure 5, orange and blue center column, SI Movie 4).
Figure 1.
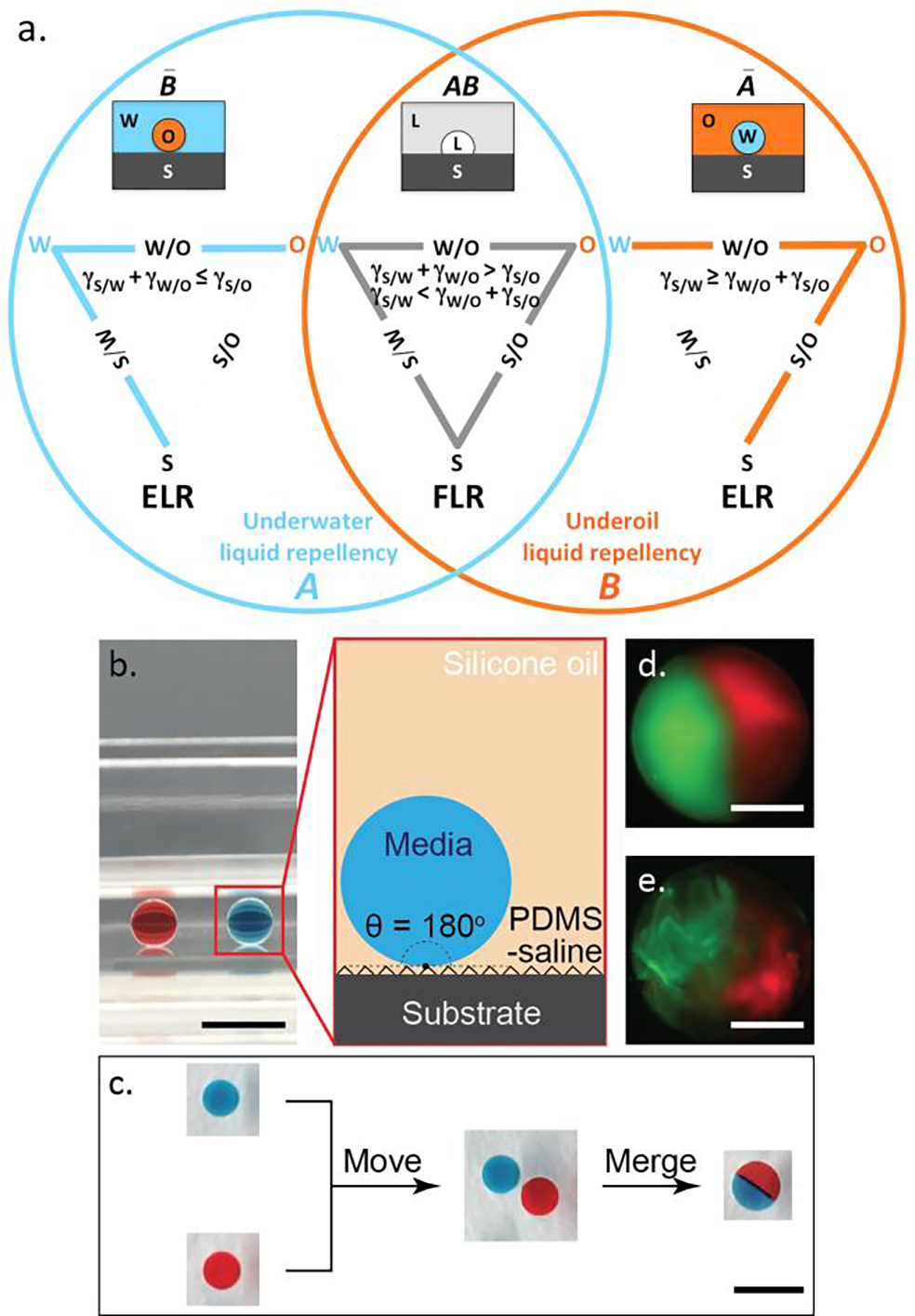
S/W/O triangle, Venn diagram of undermedia liquid repellency and water droplet manipulation in ELR systems. (a) The universal set to describe undermedia liquid repellency consists of two subsets, subset A for underwater liquid repellency and subset B for underoil liquid repellency. The intersection of A and B (AB) represents undermedia finite liquid repellency (i.e., FLR) with CA < 180°. The S/W, S/O and W/O interfaces are all present in a S/W/O triangle for FLR systems. The complement of AB (AB bar) is undermedia exclusive liquid repellency (i.e., ELR) with CA = 180°, which is also equal to the union of the complement of A (underoil exclusive water repellency, A bar) and the complement of B (underwater exclusive oil repellency, B bar). Either the S/O or the W/O interface disappears in a S/W/O triangle for ELR systems. (b) ELR water droplets (10 μL with red or blue dye, side view) sitting on PDMS-grafted glass, under silicone oil (5 cSt). (Inset) Schematic of ELR system consists of aqueous medium (dispersed phase), silicone oil (continuous phase), and PDMS-saline coated surface (solid phase). (c) Moving and merging of water droplets (top view) in the ELR system described in (b). The combined droplet showed a clear boundary of the two dyes after merging. Over time the dyes mix into each other via diffusion. Merging of two 0.5 μL fluorescently labeled collagen droplets before (d) and after (e) polymerization (37 °C/1 hour), respectively. The boundary of the two colored collagens is stable unless disturbed due to the high viscosity. Scale bars represent 5 mm in (b) and (c), 500 μm in (d) and (e).
ELR droplets of aqueous medium surrounded by a background of oil offer multiple advantages for cellular studies. First, all medium droplets regardless of size can rest in flow-free conditions and move freely on non-textured substrate without noticeable hysteresis and volume loss (Figure 1b and 1c). Next, no surfactant is required to maintain ELR and the addition of water-soluble molecules (including extracellular matrix (ECM) biomaterials such as collagen, SI Methods) in aqueous phase did not cause any obvious changes on droplet manipulation (Figure 1d and 1e). Thirdly, ELR mitigates cell and medium loss due to biofouling or evaporation. In general, the loss of material from the aqueous phase underoil can be attributed to either diffusion into the surrounding oil phase and/or adsorption onto a solid. ELR eliminates the second of these potential loss mechanisms by eliminating direct medium aqueous contact with any surface. This principle was demonstrated by observing fluorescently-labeled BSA in aqueous medium over time. In this experiment, all conditions of FLR show a time dependency of fouling, while ELR (PDMS-grafted glass/silicone oil) exhibits no fouling after 24 hours (SI Figure 6). The diffusion of molecules between aqueous phase and oil phase is determined by the partition coefficient of a given solute in the two phases. Less solubility and diffusion of polar compounds can be expected in oil phase compared with non-polar ones. A test done with Nile red (a hydrophobic dye) clearly showed that the extraction of non-polar molecules can be mostly inhibited if fluorinated oil used instead of silicone oil (SI Methods). The specific influence of depletion of non-polar molecules on cell culture will be for a further work. Based on the design rules, water drops show FLR under fluorinated oil on PDMS substrate. To achieve ELR with fluorinated oil in that situation, as reflected in the S/W/O triangle (SI Figure 4) a small amount of fluorosurfactant is needed to lower both S/O and W/O interfacial tensions in the meantime. In experiment, it turned out 0.2% of fluoro-PEG-ylated surfactant is enough for this purpose. Further testing demonstrated long-term stability (24 hours) of droplets in flow-free conditions, obviously reduced evaporation and medium loss of the dispersed phase all achieved in ELR systems (SI Figure 7, SI Methods).
In situ underoil cell culture in ELR droplets
Single cell isolation has been widely achieved on both closed and open microfluidic platforms.[13,27,28] Here, we aimed to further validate that single cells could be isolated and further cultured in situ underoil in ELR droplets. As a tutorial demo, THP-1 leukemia cancer cells were prepared in medium containing collagen I (~2 mg/mL, bovine,) at a concentration of 1 cell/μL and then distributed with 1 μL droplets, aiming for maximized single cell droplets (see protocol selection in SI Figure 8). Here, collagen I was used in order to increase the viscosity of medium and therefore keep cells from settling down during dispensing. Cells (with per droplet cell number in range of 1 to 5) were then cultured for four days underoil in ELR droplets without medium change (SI Methods), during which they were regularly imaged (Figure 2a to 2d). Proliferation efficiency (i.e., the fold change in cell number within a prescribed time period) was calculated for cells in each droplet over four days. Of the droplets interrogated, three out of the thirteen showed no proliferation, while the remaining ten proliferated with proliferation efficiencies ranging from 1.75 to 7 (Average = 3.05, SD = 1.98; Figure 2e). This variation in the proliferation efficiencies is typical as the heterogeneity in proliferation potential from cells derived from leukemia widely exists.[34] These findings provided a simple solution to isolate, culture (in situ), and recollect single cell derived colonies using an ELR system. It should be noted that the in situ underoil cell culture in ELR droplets can be extended to adherent cells. For example, a type of adherent cell from a prostate tumor cell line (C4-2) was tested (SI Methods). It is demonstrated that C4-2 can grow in ELR droplets in presence of ECM, showing normal cell viability within the culture time executed.
Figure 2.
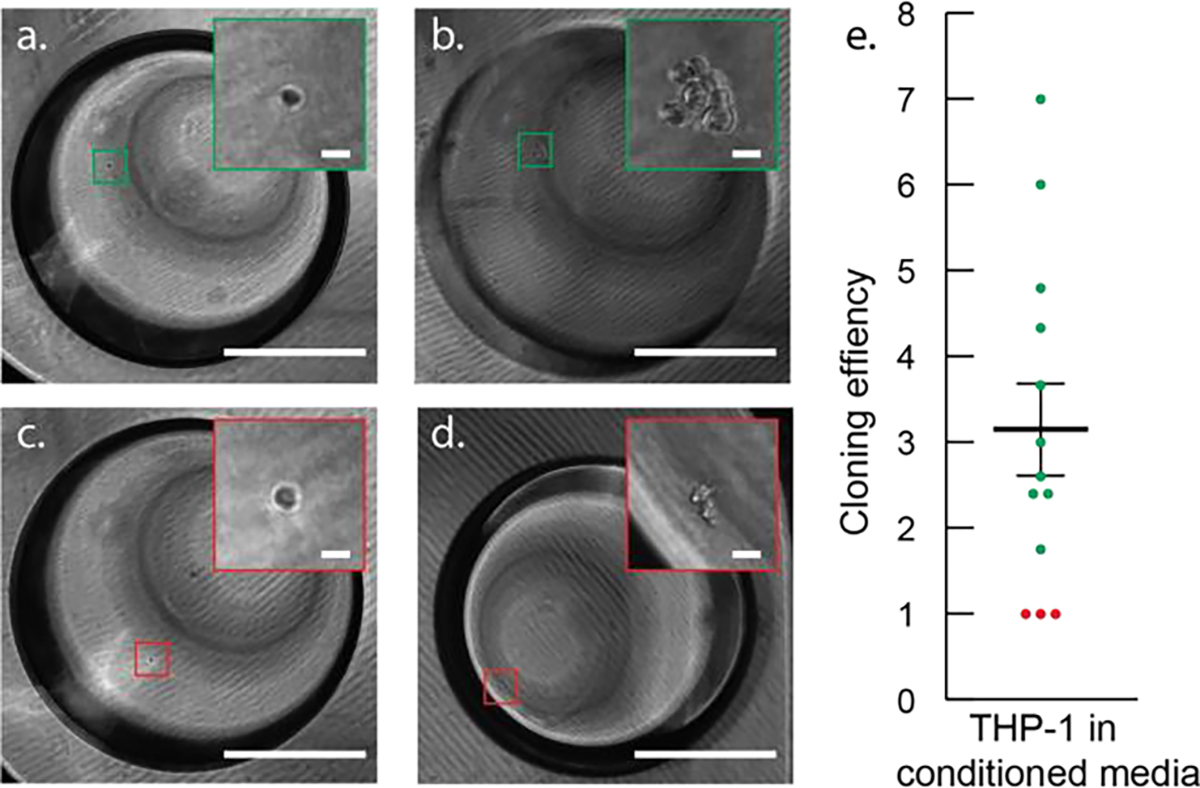
In situ underoil cell culture in ELR microdroplets. (a) and (c) A single THP-1 cell observed in a microdroplet (1 μL) 24 hours after seeding. (b) Proliferation of the single THP-1 cell in (a) resulted in a seven-cell cluster after four-day culture without medium change. (d) No proliferation was observed from the single THP-1 cell in (c) after four-day culture. (e) Proliferation efficiency of THP-1 from thirteen ELR microdroplets (green = proliferative, red = non proliferative) showing the heterogeneity in THP-1 proliferation dynamics. Scale bars represent 500 μm in (a), (b), (c), and (d) or 20 μm in all insets.
Single cell multi-encapsulation via directed merging of ELR droplets
Traditionally, single cell isolation is typically achieved by stochastically seeding cells into confined spaces (e.g., microdroplets, microwells). This stochasticity limits analysis efficiency by fundamentally limiting the number of “traps” (or droplets in the case of ELR) that contain a single cell. This limitation is compounded when using stochastic seeding to place multiple cell types within a single trap (i.e., while 36.8% of traps will contain a single cell type of interest (based on Poisson distribution), 36.8% × 36.8% = 13.5% of traps will contain exactly one of each cell type of interest when trying to co-culture two cellular phenotypes, and this declines to just 5.0% when three cell types are included) (SI Figure 9). In the event that one or more of these cell types is rare, this effect severely limits the information that can be obtained from the limited sample. Here, we demonstrate the ELR system enables us to overcome this fundamental limitation via a two-step process involving: 1) Stochastic seeding within ELR droplets of each cell type independently and imaging to identify ELR droplets containing single cells; 2) Directed combination of single cell ELR droplets to form target multi-culture combinations. While this method remains limited by the initial stochastic seeding (e.g., ELR droplets containing 2+ cells will be discarded), the compounded effects of stochastic seeding are eliminated (this principle is described in more detail in SI Methods). The experimental result is demonstrated in Figure 3, where ELR droplets containing different cell “phenotypes” (modeled here by different color staining) were generated and merged with high efficiency to produce a “tri-culture” system (SI Figure 10). Note that tradeoffs exist in step one between high efficiency stochastic seeding (maximize single cell droplets) and low loss stochastic seeding (minimize multi-cell droplets that must be discarded) (SI Figure 8). The suitable stochastic seeding method should be chosen based on the rarity of cell samples and required processing throughput.
Figure 3.
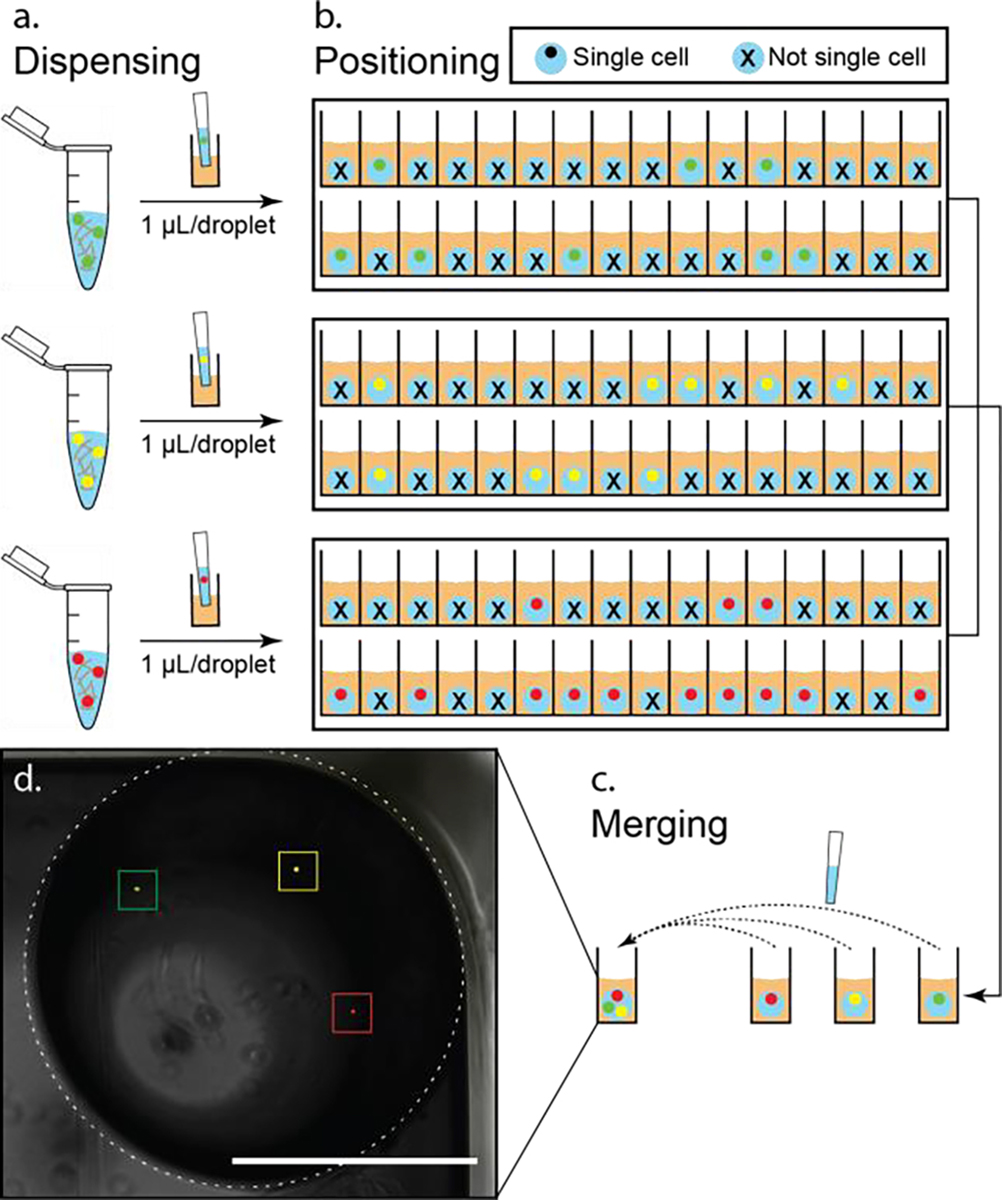
Tri-color single cell multi-encapsulation. (a) Generation of (single) cell containing droplets. Fluorescently labelled THP-1 cells (green, yellow, and red, stock concentration 1 cell/μL) were stabilized in collagen I (~2 mg/mL in growth medium) and then dispensed into 1 μL droplets by pipetting on a PDMS-grafted 384 well plate. (b) Using fluorescent microscope to identify and locate single cell droplets. Thirty-two droplets were generated for each condition. As illustrated in the schematic, eight (P1 = 8/32 = 25.0%), nine (P2 = 9/32 = 28.1%), and thirteen (P3 = 13/32 = 40.6%) single cell droplets were obtained (from top to bottom), respectively. (c) Recollection and merging of single cell droplets. With the obtained sample size of single cell droplets, the maximum rate of combination of single cell droplets (Pmin/n = P1/3 = 8.3%, n = 3 for tri-culture) was achieved (SI Figure 10). (d) Composite of fluorescent and phase micrographs of one merged droplet (denoted with dashed line circle) containing three different fluorescently labeled (left to right: green, yellow, and red) single cells. Scale bar represents 1 mm in (d).
Purification of CTCs by single cell encapsulation in ELR droplets
CTCs are a potential clinical marker for patients with metastatic disease and are typically found in very low concentrations of 1–10 CTCs per mL of whole blood. Our previous work has demonstrated isolation of hundreds of CTC candidate cells in 15 mL whole blood sampled from a patient-donor,[35] albeit with a typical purity of no more than 10%. This significant population of background cells adds difficulty and/or inconsistency in downstream readouts including gene expression analysis and next-generation sequencing (NGS). Here, the ELR system is used to further subgroup/purify rare cell samples by further allocating sorted populations into single cell ELR droplets. Specifically, we acquired a pre-isolated sample in total including 212 CTC candidate cells (including peripheral blood mononuclear cells (PBMCs) that remained after our sorting protocol stained against pan-Cytokeratin (pCK) and exclusion markers (CD11b/CD34/CD45) (see details of CTC VERSA isolation in our previous works and SI Methods).[35–37] The cell stock (0.5 cell/μL) was distributed into single droplets (1 μL) under oil (silicone oil) on a PDMS-grafted 384 well plate (see protocol selection in SI Figure 8). This distribution condition was chosen in order to minimize droplets containing multi cells (Figure 4a). Single cell droplets were then located and identified with fluorescent microscopic imaging (SI Figure 11). Further image analysis by gating high pCK signal (threshold > 1.8) and low exclusion signal (threshold < 2.3) concluded and recollected 12 CTCs (Figure 4b to 4d). This experiment demonstrated that isolated CTCs (and associated background cells) could be further subgrouped/purified by suspending all cells within an ELR system and then subsequently identifying and re-collecting only the confirmed CTCs, all without the risk of irreversible cell adhesion/loss within a device reliant on solid surfaces. Given the rarity of CTCs, this is a key advancement toward high fidelity selection and analysis of pure CTC populations.
Figure 4.
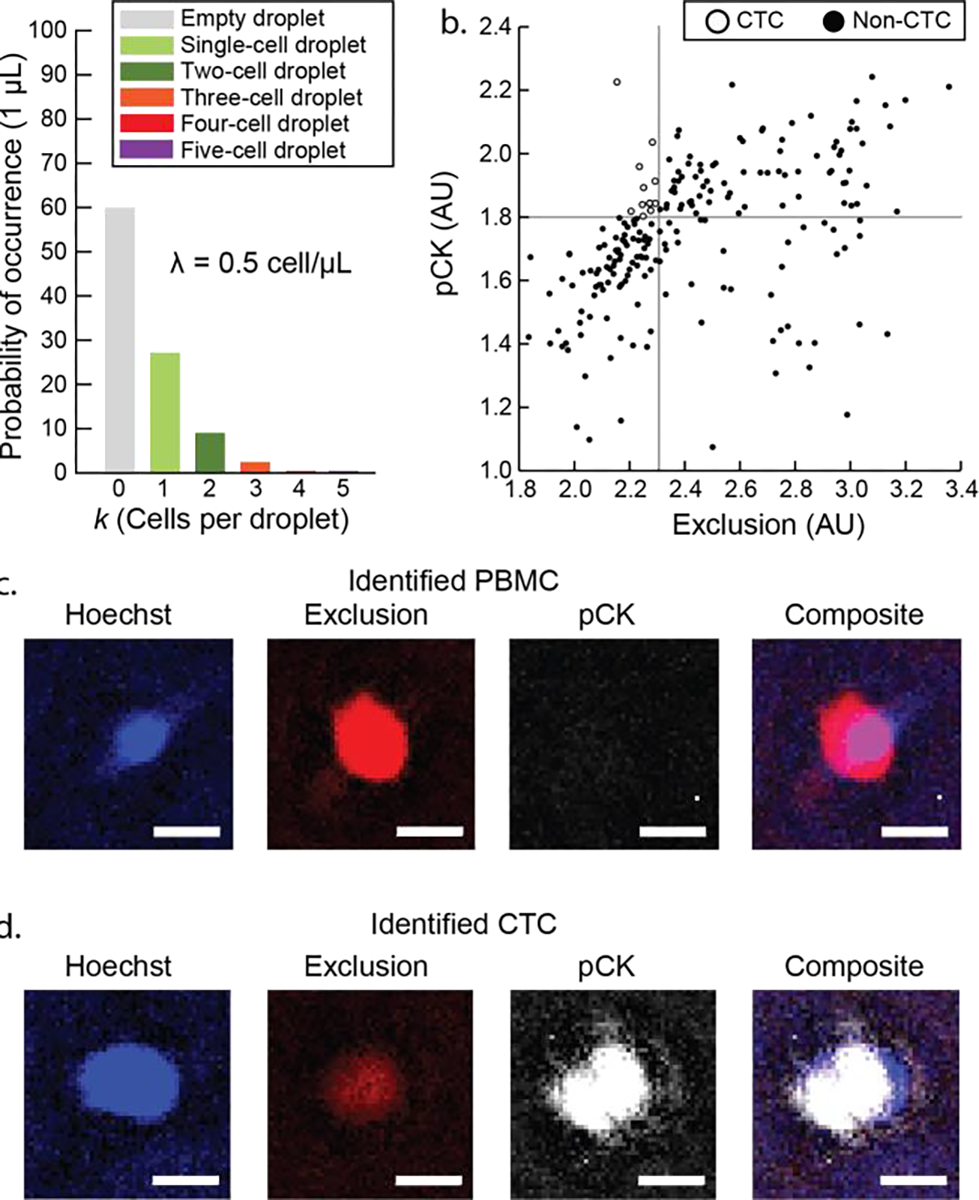
Single cell subgrouping for rare cell (CTCs) purification from patient sample. (a) Distribution of cell sample (212 cells after VERSA isolation) from patient sample (15 mL whole blood). The cell sample (stock concentration 0.5 cell/μL) were stabilized in collagen I (~2 mg/mL in PBS) and then dispensed into 1 μL droplets by pipetting on a PDMS-grafted 384 well plate. (b) Scatter plot of the 212 cells. CTCs (12 in total) were identified using presence of pan-cytokeratin (pCK) and the lack of exclusion markers (CD45, CD11b, CD34). (c) Fluorescent micrograph showing PBMCs identified from patient sample. PBMCs exhibit high intensity in exclusion channel. (d) Fluorescent micrograph showing potential CTCs identified from patient sample. Potential CTCs exhibit low intensity in exclusion channel but high intensity in pCK channel. Scale bars represent 10 μm in (c) and (d).
CONCLUSION
ELR, as defined by the design rules outlined in this manuscript, directly addresses a growing need in cell culture as themes of heterogeneity and single cell analysis emerge. The simplicity of both fabrication and operation offers a low acceptance barrier, making the proposed platform readily accessible to end users outside of engineering/surface science (e.g., clinics, biology research labs) and facilitating implementation in areas with limited infrastructure. ELR droplets can be generated, stored, and manipulated without contact with a solid in flow-free conditions. Additionally, this platform technology offers an ensemble of enabling attributes including low sample loss, open accessibility, high simplicity/reliability, and minimized evaporation. Further, ELR facilitates the ability to directly isolate and/or combine rare cell phenotypes in a minimal-loss process, enabling the interrogation of cell-cell interactions with high efficiency. This is demonstrated via applications involving monocyte culture and CTC purification. While these applications are important, we believe that they represent the “tip of the iceberg” regarding the potential of ELR and we hope that the design rules outlined in this manuscript will enable other researchers to adapt this technology to a wide range of functionality.
Supplementary Material
Funding Sources
This work is funded by National Science Foundation grant (EFRI-MKIS), Prostate Cancer Foundation Challenge Award, University of Wisconsin Carbone Cancer Center Cancer Center Support Grant P30 CA014520, NIH R01 EB010039 BRG, NIH R01 CA185251, NIH R01 CA186134, NIH R01 CA181648, EPA H-MAP 83573701, University of Wisconsin State Economic Engagement & Development (SEED) Research Program, Wisconsin Partnership Program Collaborative Health Sciences Grant, and The Creating Hope Fund at the UW Foundation.
ABBREVIATIONS
- ELR
exclusive liquid repellency
- FLR
finite liquid repellency
- CA
contact angle
- S
Solid
- L
Liquid
- G
Gas
- Lcp
liquid of continuous phase
- Ldp
liquid of dispersed phase
- PS
polystyrene
- PDMS
polydimethylsiloxane
- SD
standard deviation
- BSA
bovine serum albumin
- ECM
extracellular matrix
- CTCs
circulating tumor cells
- PBMCs
peripheral blood mononuclear cells
- pCK
pan-Cytokeratin
- CD
cluster of differentiation
- NGS
next-generation sequencing
- PCR
polymerase chain reaction
- VERSA
Vertical Exclusion-based Rare Sample Analysis
Footnotes
Conflict of Interest: The authors have potential conflicts of interest related to technologies presented here: J. Yu holds equity in Stacks to the Future, LLC. D. J. Beebe holds equity in Bellbrook Labs LLC, Tasso Inc., Stacks to the Future LLC, Lynx Biosciences LLC, Onexio Biosystems LLC and Salus Discovery LLC. S. Berry and J. Lang hold equity in Salus Discovery LLC.
REFERENCES
- (1).McGranahan N; Swanton C Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168 (4), 613–628. [DOI] [PubMed] [Google Scholar]
- (2).Cristofanilli M; Thomas Budd G; Ellis MJ; Stopeck A; Matera J; Craig Miller M; Reuben JM; Doyle GV; Jeffrey Allard W; Terstappen LWMM; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351 (8), 781–791. [DOI] [PubMed] [Google Scholar]
- (3).Pardal R; Clarke MF; Morrison SJ Applying the Principles of Stem-Cell Biology to Cancer. Nat. Rev. Cancer 2003, 3 (12), 895–902. [DOI] [PubMed] [Google Scholar]
- (4).Prasetyanti PR; Medema JP Intra-Tumor Heterogeneity from a Cancer Stem Cell Perspective. Mol. Cancer 2017, 16 (1), 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sakaguchi S; Ono M; Setoguchi R; Yagi H; Hori S; Fehervari Z; Shimizu J; Takahashi T; Nomura T Foxp3+ CD25+ CD4+ Natural Regulatory T Cells in Dominant Self-Tolerance and Autoimmune Disease. Immunol. Rev. 2006, 212, 8–27. [DOI] [PubMed] [Google Scholar]
- (6).Boiani M; Schöler HR Regulatory Networks in Embryo-Derived Pluripotent Stem Cells. Nat. Rev. Mol. Cell Biol. 2005, 6 (11), 872–884. [DOI] [PubMed] [Google Scholar]
- (7).Singhvi R; Kumar A; Lopez G; Stephanopoulos G; Wang D; Whitesides G; Ingber D Engineering Cell Shape and Function. Science 1994, 264 (5159), 696–698. [DOI] [PubMed] [Google Scholar]
- (8).Parker KK; Brock AL; Brangwynne C; Mannix RJ; Wang N; Ostuni E; Geisse NA; Adams JC; Whitesides GM; Ingber DE Directional Control of Lamellipodia Extension by Constraining Cell Shape and Orienting Cell Tractional Forces. FASEB J. 2002, 16 (10), 1195–1204. [DOI] [PubMed] [Google Scholar]
- (9).Sims CE; Allbritton NL Analysis of Single Mammalian Cells on-Chip. Lab Chip 2007, 7 (4), 423. [DOI] [PubMed] [Google Scholar]
- (10).Carlo DD; Di Carlo D; Wu LY; Lee LP Dynamic Single Cell Culture Array. Lab Chip 2006, 6 (11), 1445. [DOI] [PubMed] [Google Scholar]
- (11).Chen Y-C; Cheng Y-H; Kim HS; Ingram PN; Nor JE; Yoon E Paired Single Cell Co-Culture Microenvironments Isolated by Two-Phase Flow with Continuous Nutrient Renewal. Lab Chip 2014, 14 (16), 2941–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chua CW; Shibata M; Lei M; Toivanen R; Barlow LJ; Bergren SK; Badani KK; McKiernan JM; Benson MC; Hibshoosh H; et al. Single Luminal Epithelial Progenitors Can Generate Prostate Organoids in Culture. Nat. Cell Biol. 2014, 16 (10), 951–961, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gracz AD; Williamson IA; Roche KC; Johnston MJ; Wang F; Wang Y; Attayek PJ; Balowski J; Liu XF; Laurenza RJ; et al. A High-Throughput Platform for Stem Cell Niche Co-Cultures and Downstream Gene Expression Analysis. Nat. Cell Biol. 2015, 17 (3), 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Pushkarsky I; Tseng P; Black D; France B; Warfe L; Koziol-White CJ; Jester WF; Trinh RK; Lin J; Scumpia PO; et al. Elastomeric Sensor Surfaces for High-Throughput Single-Cell Force Cytometry. Nature Biomedical Engineering 2018, 2 (2), 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Mark D; Haeberle S; Roth G; von Stetten F; Zengerle R Microfluidic Lab-on-a-Chip Platforms: Requirements, Characteristics and Applications. Chem. Soc. Rev. 2010, 39 (3), 1153–1182. [DOI] [PubMed] [Google Scholar]
- (16).Liu Y; Lu H Microfluidics in Systems Biology—hype or Truly Useful? Curr. Opin. Biotechnol. 2016, 39, 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kaigala GV; Lovchik RD; Delamarche E Microfluidics in the “Open Space” for Performing Localized Chemistry on Biological Interfaces. Angew. Chem. Int. Ed. 2012, 51 (45), 11224–11240. [DOI] [PubMed] [Google Scholar]
- (18).Wang Y; Phillips C; Xu W; Pai J-H; Dhopeshwarkar R; Sims CE; Allbritton N Micromolded Arrays for Separation of Adherent Cells. Lab Chip 2010, 10 (21), 2917–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Casavant BP; Berthier E; Theberge AB; Berthier J; Montanez-Sauri SI; Bischel LL; Brakke K; Hedman CJ; Bushman W; Keller NP; et al. Suspended Microfluidics. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (25), 10111–10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Prakadan SM; Shalek AK; Weitz DA Scaling by Shrinking: Empowering Single-Cell “Omics” with Microfluidic Devices. Nat. Rev. Genet. 2017, 18 (6), 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Guo MT; Rotem A; Heyman JA; Weitz DA Droplet Microfluidics for High-Throughput Biological Assays. Lab Chip 2012, 12 (12), 2146. [DOI] [PubMed] [Google Scholar]
- (22).Wong T-S; Kang SH; Tang SKY; Smythe EJ; Hatton BD; Grinthal A; Aizenberg J Bioinspired Self-Repairing Slippery Surfaces with Pressure-Stable Omniphobicity. Nature 2011, 477 (7365), 443–447. [DOI] [PubMed] [Google Scholar]
- (23).Smith JD; David Smith J; Dhiman R; Anand S; Reza-Garduno E; Cohen RE; McKinley GH; Varanasi KK Droplet Mobility on Lubricant-Impregnated Surfaces. Soft Matter 2013, 9 (6), 1772–1780. [Google Scholar]
- (24).Zhang P; Wang S; Wang S; Jiang L Superwetting Surfaces under Different Media: Effects of Surface Topography on Wettability. Small 2015, 11 (16), 1939–1946. [DOI] [PubMed] [Google Scholar]
- (25).Biswas S; Pomeau Y; Chaudhury MK New Drop Fluidics Enabled by Magnetic-Field-Mediated Elastocapillary Transduction. Langmuir 2016, 32 (27), 6860–6870. [DOI] [PubMed] [Google Scholar]
- (26).Daniel D; Timonen JVI; Li R; Velling SJ; Aizenberg J Oleoplaning Droplets on Lubricated Surfaces. Nat. Phys. 2017, 13 (10), 1020–1025. [Google Scholar]
- (27).Gross A; Schoendube J; Zimmermann S; Steeb M; Zengerle R; Koltay P Technologies for Single-Cell Isolation. Int. J. Mol. Sci. 2015, 16 (12), 16897–16919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Eyer K; Doineau RCL; Castrillon CE; Briseño-Roa L; Menrath V; Mottet G; England P; Godina A; Brient-Litzler E; Nizak C; et al. Single-Cell Deep Phenotyping of IgG-Secreting Cells for High-Resolution Immune Monitoring. Nat. Biotechnol. 2017, 35 (10), 977–982. [DOI] [PubMed] [Google Scholar]
- (29).Zilionis R; Nainys J; Veres A; Savova V; Zemmour D; Klein AM; Mazutis L Single-Cell Barcoding and Sequencing Using Droplet Microfluidics. Nat. Protoc. 2017, 12 (1), 44–73. [DOI] [PubMed] [Google Scholar]
- (30).Lan F; Demaree B; Ahmed N; Abate AR Single-Cell Genome Sequencing at Ultra-High-Throughput with Microfluidic Droplet Barcoding. Nat. Biotechnol. 2017, 35 (7), 640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Leslie DC; Waterhouse A; Berthet JB; Valentin TM; Watters AL; Jain A; Kim P; Hatton BD; Nedder A; Donovan K; et al. A Bioinspired Omniphobic Surface Coating on Medical Devices Prevents Thrombosis and Biofouling. Nat. Biotechnol. 2014, 32 (11), 1134–1140. [DOI] [PubMed] [Google Scholar]
- (32).Juthani N; Howell C; Ledoux H; Sotiri I; Kelso S; Kovalenko Y; Tajik A; Vu TL; Lin JJ; Sutton A; et al. Infused Polymers for Cell Sheet Release. Sci. Rep. 2016, 6, 26109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chaudhury MK; Good RJ Retarded van Der Waals Force Theory of Stable Thick Films in Systems of Three Condensed Phases. J. Colloid Interface Sci. 1983, 94 (1), 292–294. [Google Scholar]
- (34).Chanput W; Mes JJ; Wichers HJ THP-1 Cell Line: An in Vitro Cell Model for Immune Modulation Approach. Int. Immunopharmacol. 2014, 23 (1), 37–45. [DOI] [PubMed] [Google Scholar]
- (35).Sperger JM; Strotman LN; Welsh A; Casavant BP; Chalmers Z; Horn S; Heninger E; Thiede SM; Tokar J; Gibbs BK; et al. Integrated Analysis of Multiple Biomarkers from Circulating Tumor Cells Enabled by Exclusion-Based Analyte Isolation. Clin. Cancer Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Strotman L; O’Connell R; Casavant BP; Berry SM; Sperger JM; Lang JM; Beebe DJ Selective Nucleic Acid Removal via Exclusion (SNARE): Capturing mRNA and DNA from a Single Sample. Anal. Chem. 2013, 85 (20), 9764–9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Schehr JL; Schultz ZD; Warrick JW; Guckenberger DJ; Pezzi HM; Sperger JM; Heninger E; Saeed A; Leal T; Mattox K; et al. High Specificity in Circulating Tumor Cell Identification Is Required for Accurate Evaluation of Programmed Death-Ligand 1. PLoS One 2016, 11 (7), e0159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


