ABSTRACT
Non-small cell lung cancer (NSCLC) is currently one of the malignant tumors with the highest incidence and mortality rate in China. Circular RNA hsa_circ_0000896 (circFARSA) has been reported as being an oncogene and a potential biomarker for NSCL. However, the functional role and action mechanism of circFARSA in NSCLC progression have not been fully elucidated. The present study demonstrated that circFRASA was upregulated in NSCLC tissues and cell lines, and its expression was positively correlated with poor prognosis of patients with NSCLC. Further experiments revealed that circFARSA knockdown inhibited cell proliferation, migration, and invasion in vitro experiments, but overexpression of circFARSA exhibited opposite results. Mechanistically, circFARSA facilitated the malignant phenotype of NSCLC cells by enhancing B7H3 expression through sponging miR-15a-5p. In vivo experiments, knockdown of circFARSA restricted tumor growth and metastasis. In conclusion, circFARSA served as a sponge of miR-15a-5p to promote tumorigenesis and development of NSCLC by upregulation of B7H3 expression, which provided evidence of circFARSA maybe act as a novel therapeutic target for NSCLC.
KEYWORDS: lung cancer, circFARSA, miR-15a-5p, B7H3, proliferation, metastasis
Introduction
Lung cancer is the leading cause of cancer-related deaths with the highest morbidity and mortality in the world [1]. Non-small cell lung cancer (NSCLC) is one of the most common types of lung cancer worldwide, accounting for about 80–85% [2]. Currently, diagnosis and therapeutic approaches for NSCLC are advanced, while the prognosis of advanced NSCLC is very poor with a low 5-year survival rate of approximately 15% [3]. Therefore, there is an urgent need to discover the molecular mechanism behind NSCLC progression in order to identify novel targets for the diagnosis and prognosis of NSCLC, which could serve as therapeutic targets.
Circular RNAs (circRNAs) are a novel class of non-coding RNAs with limited ability to encode proteins and usually form a closed circular structure covalently joined by the 3′ and 5′ ends [4]. Increasing evidence has demonstrated that circRNAs are tissue-specific and disease-specific, and they have been identified as candidate biomarkers for various malignant tumors [5]. CircRNA sequencing results confirmed that approximately thousands of differentially expressed circRNAs were identified in NSCLC tissues or cell lines [6,7]. In addition, previous studies have found that circRNAs may serve as an effective therapeutic target for anti-cancer drug development and personalized medicine in patients with cancer [8,9]. Yang et al. [10] showed that circPTK2 was a novel therapeutic target for metastatic colorectal cancer. Functionally, most of the circRNAs contain miRNA response elements, which can bind to miRNAs as competing endogenous RNA (ceRNA) to weaken their inhibitory effect on target genes. For example, circ-002178 acts as a ceRNA for miR-34 to facilitate the development of lung adenocarcinoma through enhancing PDL1/PD1 expression [11]. Another study has shown that circCdr1as served as a miR-1270 sponge to promote hepatocellular carcinoma progression [12]. Of note, a recent study demonstrated that circFARSA functioned as a potential diagnostic biomarker for NSCLC [13], but its molecular mechanism remains to be further explored.
In this study, we investigated the functional role and underlying mechanism of circFARSA in NSCLC progression. To begin with, the expression patterns of circFARSA, miR-15a-5p, and B7H3 were detected in NSCLC tissues and cell lines. In addition, we assessed the loss- and gain-of-function of circFARSA in the regulation of the malignant phenotype of NSCLC cells. Mechanistically, we verified that circFARSA acts as ceRNA to affect the proliferation, migration, and invasion of NSCLC cells via sponging miR-15a-5p and regulation of B7H3 expression. Therefore, this study will provide evidence to explain the regulatory role of circFARSA in NSCLC progression, and also supply an effective target for the early clinical diagnosis and treatment of NSCLC.
Materials and methods
Patients and specimens
The surgical specimens were obtained from September 2019 to October 2020 in the Department of Pulmonary and Critical Care Medicine of The First People’s Hospital of Yunnan Province. A total of 40 paired NSCLC tumor tissues and matched adjacent tissues were collected. All specimens were preserved in a − 70° refrigerator. Our study was approved by the Ethics committee of The First People’s Hospital of Yunnan Province (no. KHLL2021-038) and all patients signed an informed consent form.
Cell culture
Human bronchial epithelial cell line BEAS-2B, NSCLC cell lines (HCC827, NCI-H1299, NCI-H23, and H125), and 293 T cells were obtained from American Type Culture Collection (ATCC). Cells were inoculated in RPMI-1640 medium (Sangon Biotech, China) supplemented with 10% fetal bovine serum (FBS; Sangon Biotech, China) and 1% antibiotics mixture (Sangon Biotech, China). All cells were incubated in 5% CO2 at a 37° incubator.
Cell transfection
The small hairpin RNA (shRNA) against circFARSA (sh-FARSA), pLCDH-circFARSA vector for overexpression of circFARSA (OE-FARSA), and corresponding negative controls were synthesized by GeneChem (Shanghai, China). sh-FARSA or negative control vector (sh-NC) was established in NCI-H1299 cells, whereas OE-FARSA or negative control (pLCDH-circ-NC) was established in H125 cells. All plasmids were isolated using the cDNA Midiprep kit (Qiagen, Germany). For miR-15a-5p inhibitor (anti-miR-15a-5p), miR-15a-5p mimic (miR-15a-5p), B7H3 siRNA (si-B7H3), pcDNA3.1(+)-B7H3 (OE-B7H3), and negative controls were synthesized by GeneChem (Shanghai, China). Transfections were performed using Lipofectamine 3000 reagent (Invitrogen, USA) according to the manufacturer’s instructions. After 48 h, qRT-PCR and/or Western blot experiments were applied to verify the effect of transfection.
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA was extracted from NSCLC tissues and cells with TRIzol reagent (Invitrogen, USA). Then, cDNA was reversely synthesized using SuperScript cDNA Synthesis Kit (Invitrogen, USA). SYBR qRT-PCR kit (Tiangen, Beijing, China) and ABI 7500 System (ABI, USA) were applied to quantify the expression of circFARSA, miR-15a-5p and B7H3 followed to the manufacturer’s protocol. U6 was used for the normalization of the miRNA whereas GAPDH was used for the normalization of mRNA or circRNA. Relative expression analysis was carried out using the 2−ΔΔCt method. All primers used are listed in Table 1.
Table 1.
Sequences of primers used in qRT-PCR.
| Gene | Primer sequences |
|---|---|
| circFARSA | Forward: 5′-GCTCCTTCTGGAACTTTGAC-3′ |
| Reverse: 5′-TTGCTCACCCAGTAGGTCTT-3′ | |
| miR-15a-5p | Forward: 5′-GCCTAGCAGCACATAATGG-3′ |
| Reverse: 5′-GTGCAGGGTCCGAGGT-3′ | |
| B7H3 | Forward: 5′-CAAAGGATGCGATACACAGACCAC-3′ |
| Reverse: 5′-CAGCAGGCAGGATGACTTAGAGAA-3′ | |
| U6 | Forward: 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| GAPDH | Forward: 5′-TGCACCACCAACTGCTTAGC-3′ |
| Reverse: 5′-GGCATGGACTGTGGTCATGAG-3′ |
Western blot analysis
Protein was extracted from NSCLC tissues and cell lines with RIPA buffer containing protease inhibitors (Beyotime, China). Proteins were quantified using BCA Protein Assay Kit (Pierce, USA). Subsequently, the Western blot system was set up using a Biorad Bis-Tris Gel system according to the manufacturer’s instructions (Biorad, USA). Rabbit-anti-human PCNA antibody (Abcam, UK), B7H3 antibody (Abcam, UK), and β-actin (Abcam, UK) were prepared in 3% blocking buffer at a dilution of 1:1,000. The primary antibody was incubated with the membrane at 4° overnight followed by a brief wash and incubation with the secondary antibody (1:5,000; Abcam, UK) for 1 h at room temperature. Finally, peroxide and luminol solutions 40:1 were added to cover the blot surface for 5 min at room temperature and the membrane was placed in a developing cassette. The relative protein expression was determined using Image J software (National Institutes of Health, USA).
Cell counting kit-8 (CCK-8) assay
Transfected NSCLC cells were seeded in a 96-well plate with 2,000 cells per well in 2% FBS-containing RPMI-1640 and incubated at a 37° humidified incubator. 10 μL per well of CCK-8 reagent (Dojindo, Japan) was supplied 48 h post-transfection and cells were incubated for a further 1 h. OD450 nm readings were measured to represent the relative cell numbers of each well.
Transwell assay
Transwell migration and Matrigel invasion assays were performed using Transwell chambers according to the manufacturer’s instructions. In brief, a total of 1 × 104 cells/well in 100 μL RPMI-1640 were placed in the upper Transwell chamber (Corning Incorporated, USA), which was pre-coated with Matrigel (BD Biosciences, USA) for cell invasion assay, rather than cell migration assay. The bottom chambers were filled with RPMI-1640 medium containing 10% FBS. Once passed through the membranes, the cells were fixed and stained after 24 h. Cells in nine randomly selected fields were counted under an inverted microscope (Carl ZEISS, German) at 200 × magnification and an average value was used as the number of invaded or migrated cells.
Dual-luciferase reporter gene assay
To confirm that circFARSA and B7H3 were targets of miR-15a-5p, circFARSA-WT (containing the binding sites of miR-15a-5p at circFARSA), circFARSA-MUT (mutation of binding sites), B7H3-WT (containing the binding sites of miR-15a-5p at B7H3 3ʹUTR), and B7H3-MUT (mutation of binding sites) were cloned into Luciferase Reporter Vector (Invitrogen, USA). circFARSA-WT, circFARSA-MUT, B7H3-WT, or B7H3-MUT were co-transfected with miR-15a-5p mimic into 293 T cells using Lipofectamine 3000. After 48 h transfection, luciferase activity was measured by Dual-Glo Luciferase Assay System (Promega, USA) and normalized with Renilla luciferase.
Fluorescence in situ hybridization (FISH)
The FISH assay was performed to detect the colocalization between circFARSA and miR-15a-5p in NSCLC cells. In brief, NSCLC cells were digested and then cultured in RPMI-1640 medium, and then fixed with 4% paraformaldehyde for 30 min followed by PBS wash three times. Subsequently, the fixed cells were incubated with 0.5% TritonX-100 for 5 min. After that, the Hybridization buffer (Riobo biotech, China) containing Cy3-labeled circFARSA probe (Riobo biotech, China) and FITC-labeled miR-15a-5p probe (Riobo biotech, China) was co-cultured with cells at 37 ° overnight. Meanwhile, the DAPI was applied to stain the nucleus of NSCLC cells for 15 min. The confocal fluorescence microscope (Zeiss, Germany) was performed to observe and photograph the images.
RNA immunoprecipitation (RIP) assay
Cells were lysed by RIP lysis buffer (Millipore, USA) and then incubated with magnetic beads conjugated with IgG (1:100, Abcam, UK) or Ago2 (1:100, Abcam, UK) antibody. Then RNA was purified and investigated using the qRT-PCR assay.
Animal experiment
A total of 20 BALB/c nude mice (female, 5 weeks old) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). All mice were randomly divided into two groups (n = 10 mice per group). And then, NCI-H1299 cells (1 × 106) after transfection with circFARSA shRNA (sh-FARSA) or shRNA vector (sh-NC) were injected subcutaneously in nude mice to establish the NSCLC xenograft model. Mice were maintained with the standard diet. Tumor size was measured and tumor volume was calculated followed by the formula, tumor volume = 0.5× (length×width2). On day 28 post-injection, mice were executed and tumor tissue was isolated and weighted. For analysis of metastasis, nude mice were injected with NCI-H1299 cells with or without sh-FARSA using the tail vein containing 5 × 105 cells in 0.1 mL PBS. After 40 days, we killed the mice and counted the number of metastatic lung tumors. The lung tissues were dissected and fixed with 4% phosphate-buffered neutral formalin. Lung tissues were analyzed by hematoxylin and eosin (H&E) staining. This study was performed based on the guidance of the National Animal Care and Ethics Institution and approved by the Animal Research Committee of The First People’s Hospital of Yunnan Province (no. KHLL2021-KY104).
Statistical analysis
The mean ± SD represented data from three independent experiments. SPSS 20.0 (IBM Corp., USA) was applied for statistical analysis of all data. The student’s t-test was used for comparison between two groups, and one-way ANOVA and Tukey’s post-tests were used for multiple groups. Pearson correlation analysis was used to analyze the correlation between miR-15a-5p and circFARSA or B7H3. P < 0.05 were defined as statistically significant.
Results
CircFARSA expression was upregulated in NSCLC tissues and cell lines
The circBase showed that circFARSA (hsa_circ_0000896, genome position: chr19: 13,034,921–13,035,141) was derived from the exon 5–7 of the FARSA gene (Figure 1a). Subsequently, we analyzed the expression pattern of circFARSA in NSCLC using the GEPIA database. The results showed that circFARSA expression was highly expressed in tumor tissues compared with normal tissues (Figure 1b). Similarly, the qRT-PCR analysis revealed that circFARSA expression was upregulated in 40 paired NSCLC tumor tissues in contrast to corresponding adjacent normal tissues (Figure 1c). Moreover, we analyzed the association of circFARSA expression with clinicopathological characteristics in 40 patients with NSCLC. As expected, high circFARSA expression was positively associated with TNM stage, tumor size, and lymph node metastasis (Table 2), while its expression was not correlated with gender, age, and smoking history. Consistently, NSCLC patients with relatively high circFARSA expression exhibited a lower overall survival rate than those with low circFARSA expression via Kaplan-Meier curves analysis (Figure 1d). Subsequently, an increased level of circFARSA was detected in NSCLC cell lines (HCC827, NCI-H1299, NCI-H23, and H125) when compared to the human bronchial epithelial cell line (BEAS-2B) (Figure 1e). Furthermore, we found that circFARSA, but not linear FARSA, was resistant to the digestion of RNase R in both NSCLC cells (figure 1 f and g). Collectively, circFARSA may serve as an oncogene involved in the development and progression of NSCLC.
Figure 1.
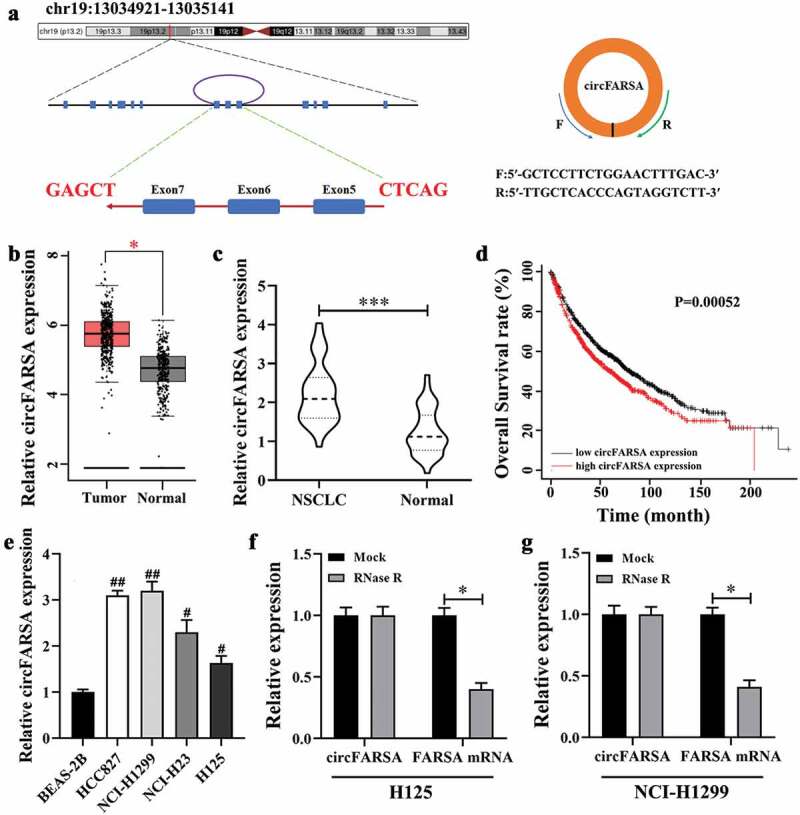
CircFARSA was highly expressed in NSCLC tissues and cell lines. (a), CircFARSA is derived from back-splicing of exons 5, 6, and 7 of FARSA gene. The back-splicing Orange arrow represents the special splicing junction of circFARSA. (b), CircFARSA expression in NSCLC tissues (n = 486) and normal tissues (n = 338) was analyzed from the Gene Expression Profiling Interactive Analysis (GEPIA) database. (c), The expression of circFARSA in NSCLC tumor tissues (n = 40) and matched adjacent normal tissues (n = 40) was examined using qRT-PCR. (d), Overall survival between NSCLC patients with high circFARSA expression and those with low expression was analyzed using Kaplan-Meier curves analysis. (e), CircFARSA levels in NSCLC cell lines and human bronchial epithelial cell line (BEAS-2B) were detected using qRT-PCR. (f and g), The RNase R assay was performed to determine the stability of circFARSA in NCI-H1299 and H125 cells. *P < 0.05, ***P < 0.001, Compared with the Normal group. #P < 0.05, ##P < 0.01, Compared with the BEAS-2B cells.
Table 2.
Association of circFARSA expression with clinicopathological characteristics in 40 patients with NSCLC [n (%)].
| Parameters | circFARSA expression |
P value | |
|---|---|---|---|
| Low (n = 22) | High (n = 18) | ||
| Gender | 0.771 | ||
| Male | 12(30.00) | 10(25.00) | |
| Female | 10(25.00) | 8(20.00) | |
| Age (years) | 0.565 | ||
| ≤60 | 9(22.50) | 9(22.50) | |
| >60 | 13(32.50) | 9(22.50) | |
| Smoking history | 0.842 | ||
| Smokers | 14(35.00) | 12(30.00) | |
| Never smokers | 8(20.00) | 6(15.00) | |
| TNM stage | 0.013 | ||
| I-II | 16(40.00) | 6(15.00) | |
| III-IV | 6(15.00) | 12(30.00) | |
| Tumor size (cm) | 0.014 | ||
| ≤5 | 17(42.50) | 7(17.50) | |
| >5 | 5(12.50) | 11(27.50) | |
| LNM | 0.024 | ||
| Yes | 8(20.00) | 13(32.50) | |
| No | 14(35.00) | 5(12.50) | |
Note: The median of relative circFARSA expression level is 2.22, so the number of high circFARSA expression is 18 (>2.22). TNM: Tumor, node, metastasis; LNM: lymph node metastasis.
Knockdown of circFARSA inhibited the proliferation, migration, and invasion abilities of NSCLC cells
To explore whether circFARSA affects the biological behavior of NSCLC cells, we transfected circFARSA shRNA (sh-FARSA) into NCI-H1299 or circFARSA overexpression vector (OE-FARSA) in H125 cells to alter circFARSA expression. The qRT-PCR analysis results showed that the expression of circFARSA was downregulated in NCI-H1299 cells after transfection with sh-FARSA, but its expression was enhanced in H125 cells after overexpression of circFARSA (Figure 2a). Moreover, knockdown of circFARSA in NCI-H1299 cells significantly inhibited cell proliferation ability using the CCK-8 assay (Figure 2b). In contrast, ectopic circFARSA expression markedly promoted cell proliferation compared with the negative control in H125 cells (Figure 2c) Furthermore, Transwell assay displayed that knockdown of circFARSA reduced the number of migration (Figure 2 d and e) and invasion (figure 2 f and g) of NCI-1299 cells, while circFARSA overexpression had the opposite effect (Figure 2 d–g). Taken together, knockdown of circFARSA restricted NCI-H1299 cell proliferation, migration, and invasion capabilities, while the elevation of circFARSA accelerated the malignant biological behavior of H125 cells.
Figure 2.
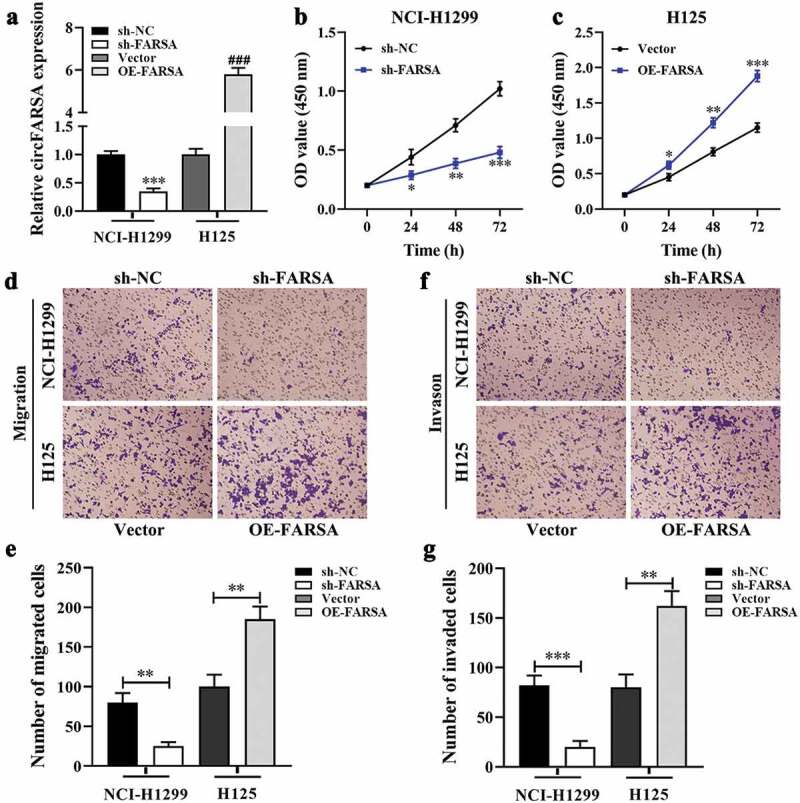
Effect of circFARSA on the proliferation, migration, and invasion of NSCLC cells. (a), qRT-PCR was performed to detect the expression of circFARSA in NSCLC cells after transfection with circFARSA shRNA, circFARSA overexpression, and corresponding negative control. (b and c), CCK-8 assay was used to evaluate cell proliferation in NCI-H1299 and H125 cells. (d-g), Transwell assay was employed to assess the migration (d and e) and invasion (f and g) capabilities of both NCI-1299 and H125 cells (magnification, ×200). *P < 0.05, **P < 0.01, ***P < 0.001.
Knockdown of circFARSA inhibited tumor growth and metastasis in vivo
Based on the inhibitory effect of circFARSA knockdown on the malignant biological behavior of NSCLC cells, we further verify the functional role of circFARSA-silenced in tumor growth using xenografts BALB/c nude mice. As expected, knockdown of circFARSA markedly inhibited tumor growth (Figure 3a) compared with the sh-NC group. H&E staining results showed that knockdown of circFARSA reduced the pathological mitotic figures and promoted nucleus disintegrated and disappeared (Figure 3b). Similarly, the tumor volume (Figure 3c) and tumor weight (Figure 3d) in the sh-FARSA group were much less than that in the sh-NC group. Moreover, we examined the expression of cell proliferation-related marker PCNA in tumor tissues using western blot. The results revealed that knockdown of circFARSA significantly reduced the protein level of PCNA compared with the sh-NC group (Figure 3e). Of note, H&E staining revealed that knockdown of circFARSA inhibited metastasis of NSCLC cells (figure 3f). Collectively, knockdown of circFARSA notably impeded tumor growth and metastasis.
Figure 3.
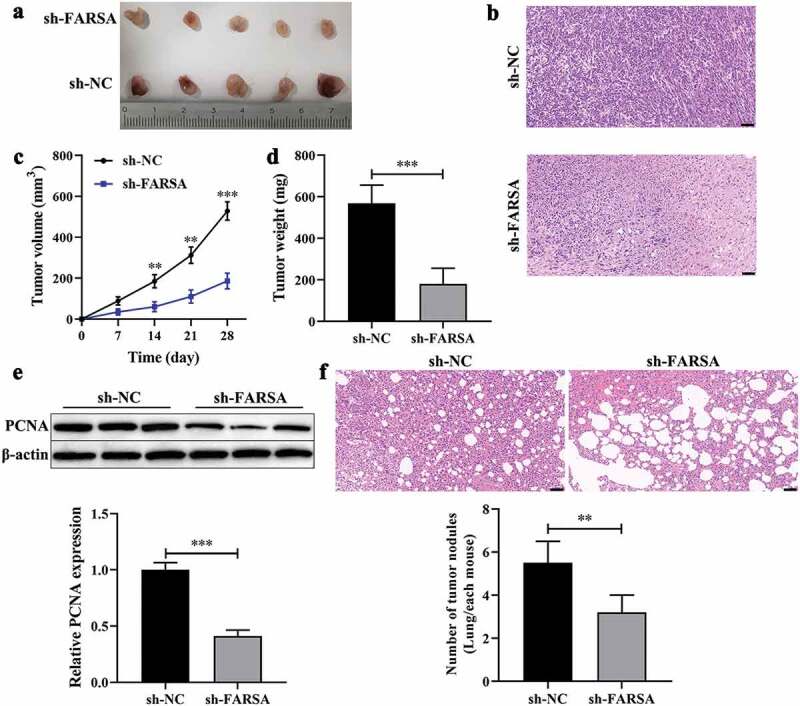
Knockdown of circFARSA inhibited tumor growth and metastasis in vivo. BALB/c nude mice have subcutaneously injected with NCI-1299 (1 × 106) cells with stably low-expressing circFARSA (sh-FARSA) or sh‑NC. (a), Tumors were dissected and imaged. (b), Histology examination of tumor slices by hematoxylin and eosin (H&E) staining, Scale bar = 50 μm. (c), Tumor volume was measured every seven days. (d), After 28 days of inoculation, the tumor weight of each group was examined. (e), Western blot was performed to detect the protein levels of PCNA in tumor tissues of xenograft BALB/c nude mice. (f), BALB/c nude mice were injected with NCI-H1299 cells with or without sh-FARSA using the tail vein containing 5 × 105 cells in 0.1 mL PBS. Lung tissues were analyzed by H&E staining, Scale bar = 50 μm. **P < 0.01, ***P < 0.001, Compared with the sh-NC group.
CircFARSA acted as a sponge of miR-15a-5p
Previous studies have shown that circRNA acts as a sponge of miRNA involve in the progression of cancer [14]. In this study, the online starBase3.0 (https://starbase.sysu.edu.cn/) database was performed to predict the potential target miRNA of circFARSA. Results showed that miR-15a-5p was selected as a candidate target miRNA of circFARSA (Figure 4a). Of note, qRT-PCR presented that miR-15a-5p expression was downregulated in NSCLC tumor tissues (Figure 4b) and both NSCLC cells (Figure 4c). Meanwhile, the expression of circFARSA and miR-15a-5p has a negative correlation in NSCLC tumor tissues (Figure 4d). Moreover, the dual-luciferase reporter gene assay showed that overexpression of miR-15a-5p notably reduced the luciferase activity of the luciferase reporters with circFARSA-WT in 293 T cells compared with the miR-NC group, but there was no significant change in the luciferase reporter with circFARSA-MUT (Figure 4e). RIP assay showed that circFARSA and miR-15a-5p were enriched in compounds precipitated by the Ago2 antibody, suggesting that there was an interaction between circFARSA and miR-15a-5p (figure 4 f and g). FISH identified that circFARSA and miR-15a-5p were co-located in the cytoplasm (Figure 4i). What’s more, the qRT-PCR analysis demonstrated that circFARSA knockdown significantly enhanced the expression of miR-15a-5p in NCI-H1299 cells, but the miR-15a-5p expression was decreased in H125 cells after transfection with circFARSA overexpression vector (Figure 4h). Taken together, circFARSA may act as a sponge for miR-15a-5p in NSCLC cells.
Figure 4.
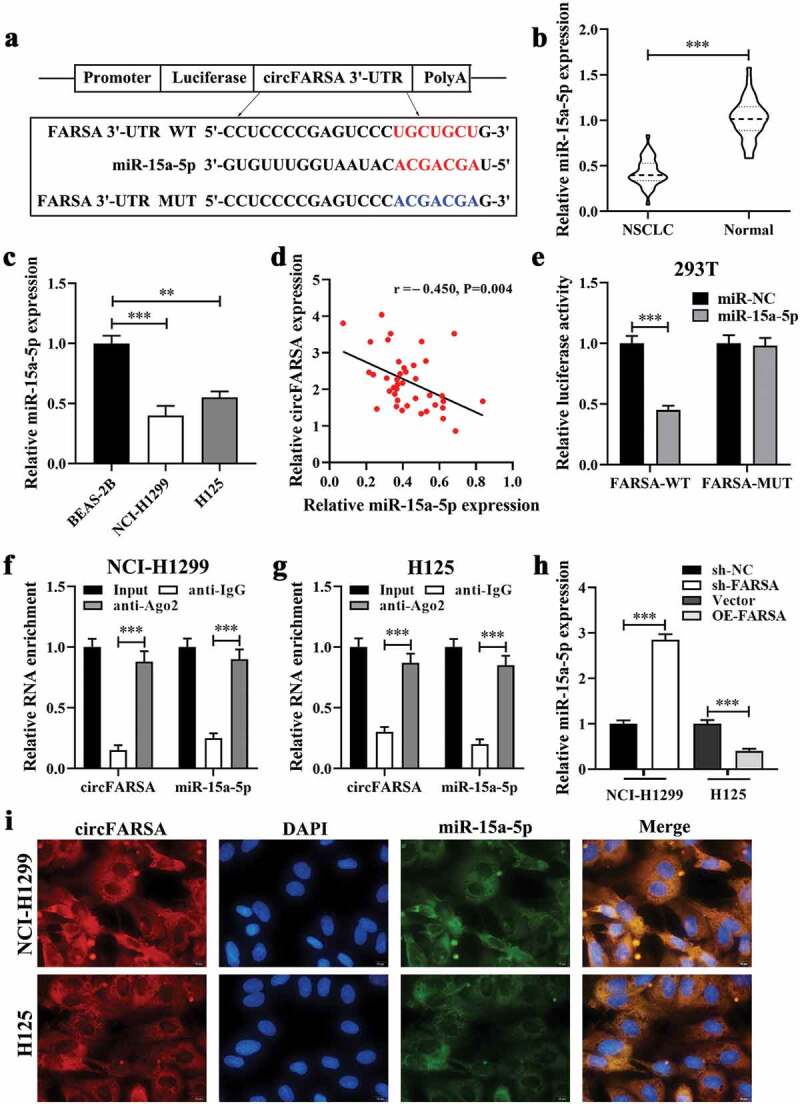
CircFARSA acted as a sponge of miR-15a-5p. (a), Wild-type (WT) and mutated-type (MUT) sequences of the putative binding sites between circFARSA and miR-15a-5p. (b), qRT-PCR was applied to detect the expression of miR-15a-5p in 40 paired NSCLC tumor tissues and matched adjacent tissues; (c), The expression of miR-15a-5p in BEAS-2B and both NSCLC cell lines was examined using qRT-PCR. (d), The correlation between the expression of circFARSA and miR-15a-5p in tumor tissues of 40 NSCLC patients was analyzed by Spearman’s correlation coefficient. (e-g), The interaction between circFARSA and miR-15a-5p was verified using dual-luciferase reporter gene assay (e) and RNA immunoprecipitation (f and g). (h), qRT-PCR was employed to examine the expression of miR-15a-5p in both NSCLC cells after transfection with circFARSA shRNA, circFARSA overexpression plasmid, and corresponding negative control. (i), The FISH assay was used to detect the colocalization of circFARSA and miR-15a-5p in both NSCLC cells. **P < 0.01, ***P < 0.001.
B7H3 was verified as a target for miR‑15a‑5p
In the online starBase3.0 database, B7H3 was selected and the binding sites between miR-15a-5p and B7H3 are shown in Figure 5a. Of note, B7H3 has been reported to play an important role in tumor progression as an oncogene [15]. Meanwhile, the GEPIA database analysis results showed that B7H3 mRNA expression in NSCLC tumor tissues was higher than that in the normal tissues (Figure 5b). Similarly, we found that the expression of B7H3 was upregulated in NSCLC tumor tissues (Figure 5c) and both NSCLC cell lines (Figure 5d). In addition, the expression of B7H3 was negatively correlated with miR-15a-5p expression in NSCLC (Figure 5e), but positively correlated with circFARSA expression (figure 5f). Kaplan-Meier curves analysis demonstrated that low B7H3 expression exhibited a higher overall survival rate than that of high B7H3 expression (Figure 5g). Next, we performed the luciferase reporter assay to verify the target association between B7H3 and miR-15a-5p. The results showed that luciferase activity of the 3ʹUTR region of B7H3 was significantly reduced when the 293 T cells were co-transfected with B7H3-WT and miR-15a-5p mimic. However, there was no significant difference in luciferase activity of B7H3-MUT between the miR-15a-5p mimic group (Figure 5h). Furthermore, western blot analysis results revealed that inhibition of miR-15a-5p significantly enhanced the protein level of B7H3 in NCI-H1299 cells, while co-transfection with circFARSA shRNA and miR-15a-5p inhibitor reduced B7H3 protein expression (Figure 5 i and j). In contrast, overexpression of miR-15a-5p notably decreased B7H3 protein expression in H125 cells, but overexpression of circFARSA alleviated the inhibitory effect of miR-15a-5p overexpression on the protein level of B7H3 (Figure 5 i and j). The above results showed that B7H3 was a direct target gene of miR-15a-5p and its expression can be regulated by circFARSA in NSCLC cells.
Figure 5.
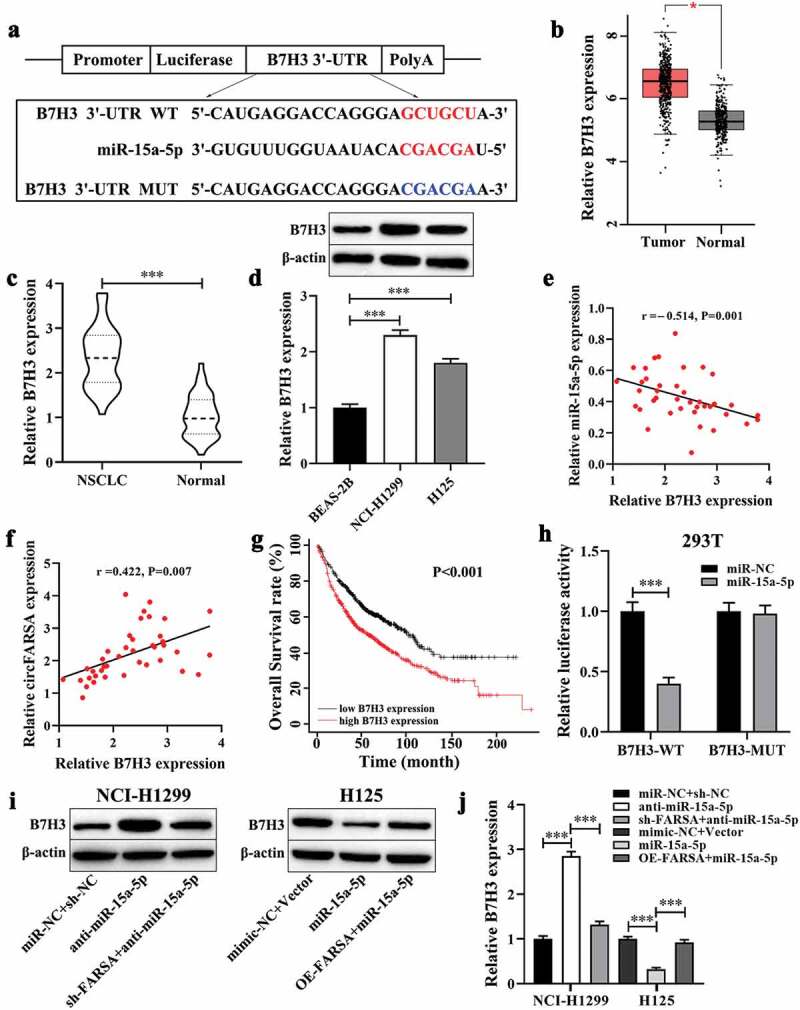
B7H3 was verified as a target for miR‑15a‑5p. (a), WT and MUT sequences of the putative binding sites between B7H3 and miR-15a-5p. (b), The mRNA levels of B7H3 in NSCLC tissues (n = 486) and normal tissues (n = 338) were analyzed from the GEPIA database. (c), qRT-PCR was performed to examine the mRNA expression of B7H3 in NSCLC tumor tissues (n = 40) and matched adjacent normal tissues (n = 40). (d), Western blot was used to measure the protein levels of B7H3 in NSCLC cell lines and BEAS-2B cells. Spearman’s correlation coefficient was applied to analyze the correlation between B7H3 and miR-15a-5p (e) or circFARSA (f) in 40 NSCLC tumor tissues. (g), Association between B7H3 expression and overall survival rate of patients with NSCLC was analyzed using Kaplan-Meier curves analysis. (h), Dual-luciferase reporter gene assay was employed to verify the target association between B7H3 and miR-15a-5p in 293 T cells. (i and j), The protein level of B7H3 in transfected NSCLC cells was determined using western blot. *P < 0.05, ***P < 0.001.
CircFARSA regulated NSCLC cell proliferation, migration, and invasion via miR-15a-5p/B7H3 axis
To further elucidate whether the effect of circFARSA on the malignant biological behavior of NSCLC cells was mediated via the miR-15a-5p/B7H3 axis. The CCK-8 and Transwell assays were performed to evaluate cell proliferation, migration, and invasion abilities in vitro. Both results showed the inhibitory effect of circFARSA knockdown on NCI-H1299 cell proliferation (Figure 6a), migration (Figure 6 c and d), and invasion (figure 6 f and g) was notably reversed by miR-15a-5p inhibitor or B7H3 overexpression (transfection with pcDNA3.1-B7H3). In addition, miR-15a-5p overexpression or B7H3 knockdown ameliorated the promotion effect of circFARSA overexpression on H125 cell proliferation (Figure 6b), migration (Figure 6 c and e), invasion (figure 6 f and h). Taken together, circFARSA functioned as a miR-15a-5p sponge to promote the malignant biological behavior of NSCLC cells through upregulating B7H3 expression.
Figure 6.
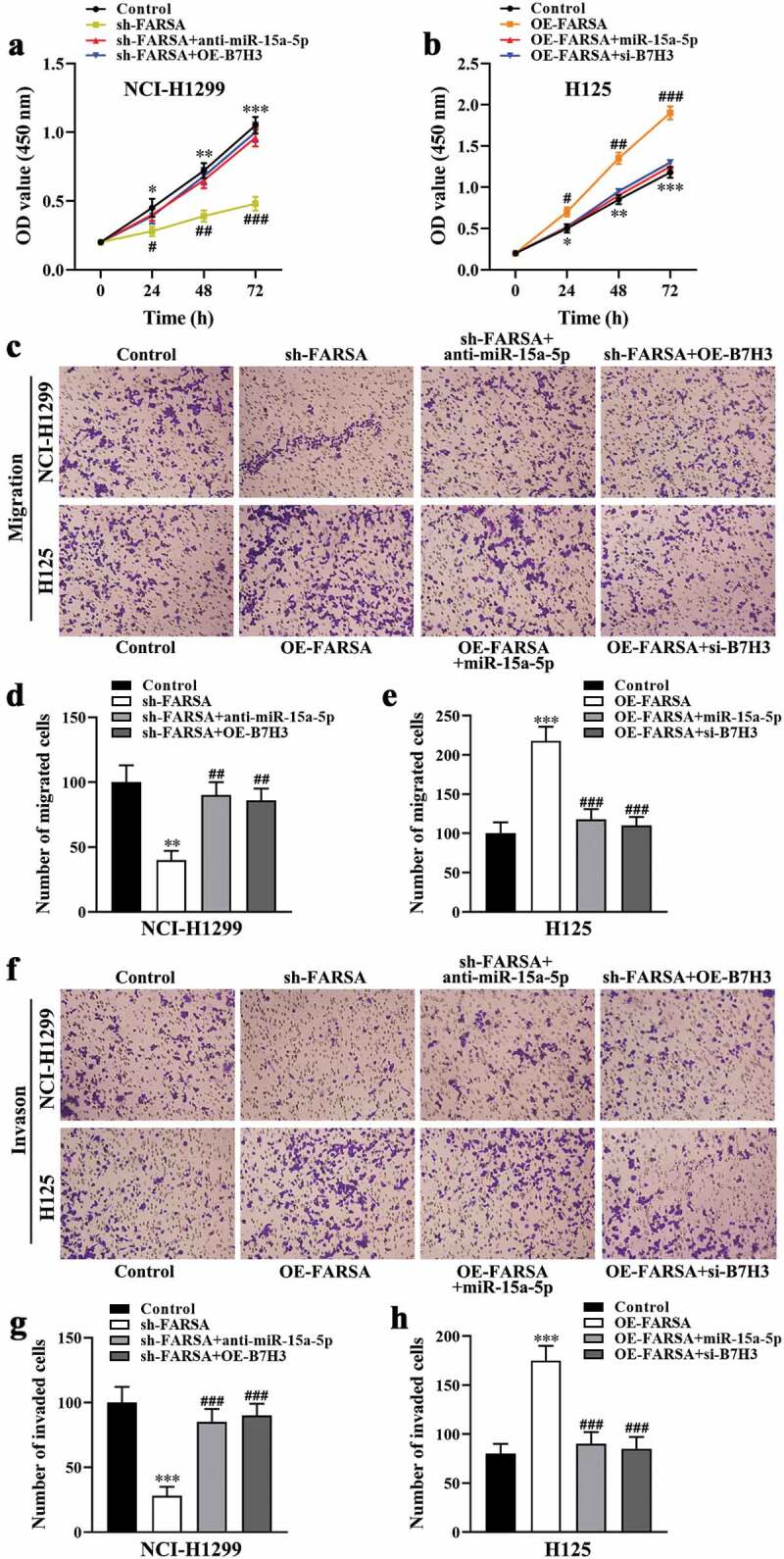
CircFARSA regulated NSCLC cell proliferation, migration, and invasion via miR-15a-5p/B7H3 axis. (a and b), CCK-8 assay was performed to examine cell proliferation in NCI-H1299 and H125 cells. (c-h), The cell migration (c-e) and invasion (f-h) ability in both NCI-H1299 and H125 cells were determined using Transwell assay (magnification, ×200). *P < 0.05, **P < 0.01, ***P < 0.001, Compared with the control group. #P < 0.05, ##P < 0.01, ###P < 0.001, Compared with the sh-FARSA or OE-FARSA group.
Discussion
Lung cancer progresses rapidly with a poor prognosis, which is one of the main culprits threatening the health of contemporary human beings [16]. Accumulating evidence has demonstrated that circRNAs exhibited an antitumor or cancerogenic role in the development and progression of many tumors [17]. Of note, previous studies have shown that abnormal circRNA expression was associated with the progression of NSCLC [18]. For example, circ_012515 expression was upregulated in NSCLC cells, and its expression was related to the TNM stage (III-IV) and lymph node metastasis in NSCLC patients, while overall survival and progression-free survival were significantly shorter in patients with high expression of circcirc_012515 [19]. Yao and colleagues found that circ_100876 may act as an effective target in carcinogenesis and progression of NSCLC due to its expression being a close association with the poor prognosis of patients with NSCLC [20]. In this study, our data showed that circFARSA expression was highly expressed in NSCLC cancer tissues and cell lines, and the poor prognosis of NSCLC patients with high circFARSA expression was evidenced by a shortened overall survival time. Moreover, knockdown of circFARSA hampered tumor growth and metastasis in vivo, as well as suppressed cell proliferation, migration, and invasion abilities in NCI-H129 cells in vitro. Similarly, circFARSA was reported to be overexpressed in tumor tissues of bladder cancer [21] and NSCLC [13], and it may be a useful predictive target for patients’ diagnosis and treatment in clinical. Collectively, the above results suggested that circFARSA served as an oncogene in NSCLC progression.
Increasing evidence has demonstrated that circRNAs act as an endogenous competitor of mRNA to regulate miRNAs for biological functions [14]. For instance, circPTPRA overexpression inhibited epithelial-mesenchymal transitioning and metastasis of NSCLC cells via sponging miR-96-5p [22]. Circ_0005585 binds to miR-15a as a ceRNA to facilitate proliferation and epithelial-mesenchymal transitioning of ovarian cancer cells by enhancing ESRP1 expression [23]. In the present study, we demonstrated that circFARSA contributed to the malignant phenotype of NSCLC cells by boosting B7H3 expression via sponging miR-15a-5p. Of note, previous studies have confirmed that miR-15a-5p served as a tumor suppressor in many tumors, such as hepatocellular carcinoma [24], multiple myeloma [25], and colorectal carcinoma [26]. Moreover, miR-15a-5p was involved in the tumorigenesis and progression of many tumors by regulating the malignant biological behavior of cancer cells, including cell proliferation, apoptosis, invasion, and angiogenesis [27]. A recent study showed that overexpression of miR-15a-5p could inhibit the proliferation, migration, and invasion of hepatocellular carcinoma cells by reducing PD1 in CD8+ T cells [28]. Similarly, Guo and colleagues revealed that miR-15a-5p plays an antitumor role in NSCLC progression by inhibiting A549 cell proliferation, migration, and invasion [29]. Another study demonstrated that overexpression of miR-15a-5p suppressed cell metastasis and lipid metabolism by reducing histone acetylation in lung cancer [30]. Besides, overexpression of miR-15a-5p contributed to enhancing the sensitivity of cancer cells to chemotherapeutic drugs [31,32]. Herein, our data revealed that overexpression of miR-15a-5p reversed the promotion effect of circFARSA on the malignant phenotype of NSCLC cells. These results indicated that miR-15a-5p functions as a cancer suppressor gene in NSCLC, which was a direct target gene of circFARSA.
The well-known oncogene B7H3 was reported as a promoter of metastasis and therapeutic target in human cancers [15]. For example, B7H3 can be used as a potential diagnostic biomarker for glioblastoma [33], as well as B7H3 overexpression facilitated the migration and invasion of tumor cells [34]. Zhong and colleagues demonstrated that upregulation of B7H3 promoted cell proliferation and invasion in glioma through activating the JAK2/STATS/Slug signaling pathway [35]. Moreover, several studies have shown that miRNA targets B7H3 to mediate tumor progression by regulating the malignant biological behavior of cancer cells, such as miR-506 [36], miR-199a [37], miR-187 [38]. In the present study, we verified that B7H3 was a direct target gene of miR-15a-5p, and its expression was positively associated with the poor prognosis of NSCLC patients. Meanwhile, knockdown of B7H3 significantly inhibited the proliferation, migration, and invasion of NSCLC cells induced by circFARSA overexpression.
In conclusion, this study confirmed that circFARSA exerted an oncogenic role in NSCLC by promoting tumor growth and metastasis, as well as facilitating the proliferation, migration, and invasion abilities of NSCLC cells. Mechanistically, circFARSA promoted the tumorigenesis and progression of NSCLC by sponging of miR-15a-5p and upregulation of B7H3 expression (Figure 7). These findings revealed the regulatory mechanisms by which circFARSA contributed to NSCLC progression, and circFARSA may serve as a novel therapeutic target for NSCLC.
Figure 7.
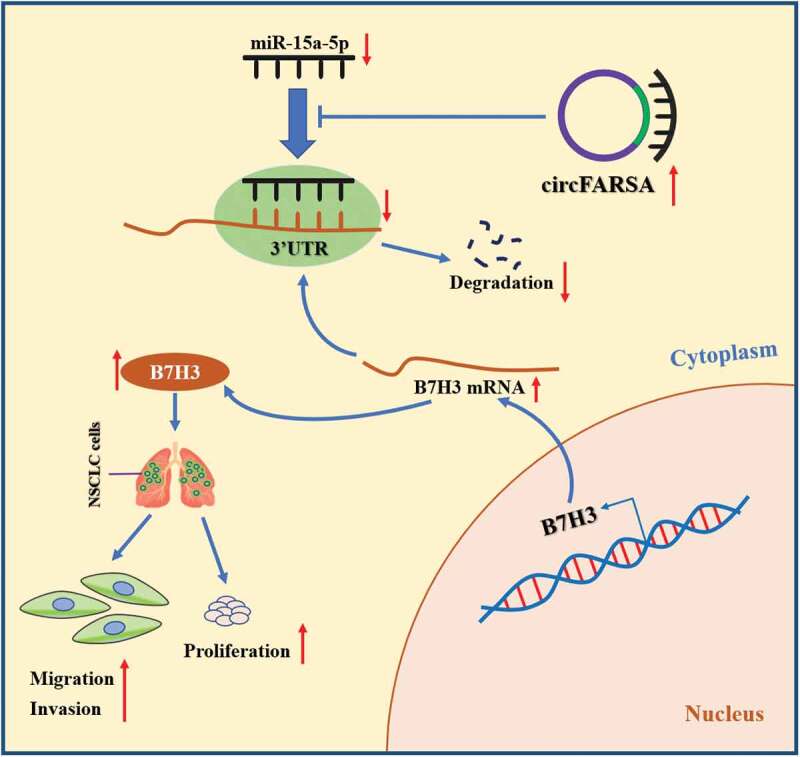
The schematic diagram of how circFARSA promotes the tumorigenesis and progression of NSCLC. circFARSA can competitively bind to miR-15a-5p and then up-regulate B7H3 expression, thereby stimulating the malignant phenotype of NSCLC cells.
Funding Statement
This work was partly supported by The National Natural Science Foundation of China (81660012, 81970053, 81570046, and 91739109), Yunnan Provincial Clinical Medical Center Open Project (2021LCZXXF-HX09), the Research Fund for the Doctoral Program of The First People’s Hospital of Yunnan Province(KHBS-2022-030), Joint Application Foundation Project of Kunming Medical University and Yunnan Provincial Science and Technology Department (202201AY070001-224), Yunnan Provincial Clinical Medical Research Center for Respiratory System Diseases-Clinical Translational Research on Respiratory Intractable Diseases (202102AA100057), Yunnan Provincial Clinical Medical Center for Respiratory System Diseases (ZX2019-01-03), and Academician Zhong Nanshan Workstation (2019IC032-1).
Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author(Yunhui Zhang) on a reasonable request. The data generated or used during the study are available online as follows:
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Siegel R, Naishadham D, Jemal A.. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. [DOI] [PubMed] [Google Scholar]
- [3].Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. [DOI] [PubMed] [Google Scholar]
- [4].Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jiang MM, Mai ZT, Wan SZ, et al. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144(4):667–674. [DOI] [PubMed] [Google Scholar]
- [7].Song L, Cui Z, Guo X. Comprehensive analysis of circular RNA expression profiles in cisplatin-resistant non-small cell lung cancer cell lines. Acta Biochim Biophys Sin (Shanghai). 2020;52(9):944–953. [DOI] [PubMed] [Google Scholar]
- [8].Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lei B, Tian Z, Fan W, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yang H, Li X, Meng Q, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang J, Zhao X, Wang Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11(19):8183–8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7(6):2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liang ZZ, Guo C, Zou MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 2020;20:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dong P, Xiong Y, Yue J, et al. B7H3 as a promoter of metastasis and promising therapeutic target. Front Oncol. 2018;8:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhu R, Liu Z, Jiao R, et al. Updates on the pathogenesis of advanced lung cancer-induced cachexia. Thorac Cancer. 2019;10(1):8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].de Fraipont F, Gazzeri S, Cho WC, et al. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet. 2019;10:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fu Y, Huang L, Tang H, et al. hsa_circRNA_012515 is highly expressed in NSCLC patients and affects its prognosis. Cancer Manag Res. 2020;12:1877–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pan J, Xie X, Li H, et al. Detection of serum long non-coding RNA UCA1 and circular RNAs for the diagnosis of bladder cancer and prediction of recurrence. Int J Clin Exp Pathol. 2019;12(8):2951–2958. [PMC free article] [PubMed] [Google Scholar]
- [22].Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Deng G, Zhou X, Chen L, et al. High expression of ESRP1 regulated by circ-0005585 promotes cell colonization in ovarian cancer. Cancer Cell Int. 2020;20:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Long J, Jiang C, Liu B, et al. MicroRNA-15a-5p suppresses cancer proliferation and division in human hepatocellular carcinoma by targeting BDNF. Tumour Biol. 2016;37(5):5821–5828. [DOI] [PubMed] [Google Scholar]
- [25].Li Z, Liu L, Du C, et al. Therapeutic effects of oligo-single-stranded DNA mimicking of hsa-miR-15a-5p on multiple myeloma. Cancer Gene Ther. 2020;27(12):869–877. [DOI] [PubMed] [Google Scholar]
- [26].Gopalan V, Ebrahimi F, Islam F, et al. Tumour suppressor properties of miR-15a and its regulatory effects on BCL2 and SOX2 proteins in colorectal carcinomas. Exp Cell Res. 2018;370(2):245–253. [DOI] [PubMed] [Google Scholar]
- [27].Liu T, Xu Z, Ou D, et al. The miR-15a/16 gene cluster in human cancer: a systematic review. J Cell Physiol. 2019;234(5):5496–5506. [DOI] [PubMed] [Google Scholar]
- [28].Zhang HY, Liang HX, Wu SH, et al. Overexpressed tumor suppressor exosomal miR-15a-5p in cancer cells inhibits PD1 expression in CD8+ T cells and suppresses the hepatocellular carcinoma progression. Front Oncol. 2021;11:622263. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [29].Guo K, Qi D, Huang B. LncRNA MEG8 promotes NSCLC progression by modulating the miR-15a-5p-miR-15b-5p/PSAT1 axis. Cancer Cell Int. 2021;21(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ni Y, Yang Y, Ran J, et al. miR-15a-5p inhibits metastasis and lipid metabolism by suppressing histone acetylation in lung cancer. Free Radic Biol Med. 2020;161:150–162. [DOI] [PubMed] [Google Scholar]
- [31].Vandewalle V, Essaghir A, Bollaert E, et al. miR-15a-5p and miR-21-5p contribute to chemoresistance in cytogenetically normal acute myeloid leukaemia by targeting PDCD4, ARL2 and BTG2. J Cell Mol Med. 2021;25(1):575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pang K, Song J, Bai Z, et al. miR-15a-5p targets PHLPP2 in gastric cancer cells to modulate platinum resistance and is a suitable serum biomarker for oxaliplatin resistance. Neoplasma. 2020;67(5):1114–1121. [DOI] [PubMed] [Google Scholar]
- [33].Zhang J, Wang J, Marzese DM, et al. B7H3 regulates differentiation and serves as a potential biomarker and theranostic target for human glioblastoma. Lab Invest. 2019;99(8):1117–1129. [DOI] [PubMed] [Google Scholar]
- [34].Chen YW, Tekle C, Fodstad O. The immunoregulatory protein human B7H3 is a tumor-associated antigen that regulates tumor cell migration and invasion. Curr Cancer Drug Targets. 2008;8(5):404–413. [DOI] [PubMed] [Google Scholar]
- [35].Zhong C, Tao B, Chen Y, et al. B7-H3 regulates glioma growth and cell invasion through a JAK2/STAT3/Slug-dependent signaling pathway. Onco Targets Ther. 2020;13:2215–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhu XW, Wang J, Zhu MX, et al. MicroRNA-506 inhibits the proliferation and invasion of mantle cell lymphoma cells by targeting B7H3. Biochem Biophys Res Commun. 2019;508(4):1067–1073. [DOI] [PubMed] [Google Scholar]
- [37].Yang X, Feng KX, Li H, et al. MicroRNA-199a inhibits cell proliferation, migration, and invasion and activates AKT/mTOR signaling pathway by targeting B7-H3 in cervical cancer. Technol Cancer Res Treat. 2020;19:1533033820942245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhao J, Lei T, Xu C, et al. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem Biophys Res Commun. 2013;438(2):439–444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author(Yunhui Zhang) on a reasonable request. The data generated or used during the study are available online as follows:


