ABSTRACT
The critical roles of N6-methyladenosine (m6A) modification have been demonstrated by more and more evidence. However, the cross talk of m6A and long noncoding RNAs (lncRNAs) in non-small cell lung cancer (NSCLC) tumorigenesis is still unclear. Here, this work focused on the functions and molecular mechanism of m6A-modified lncRNA DLGAP1 antisense RNA 2 (DLGAP1-AS2) in NSCLC. Microarray analysis found that lncRNA DLGAP1-AS2 is upregulated in NSCLC cells. Clinical data showed that DLGAP1-AS2 high-expression was correlated with advanced pathological stage and poor prognosis. Functionally, DLGAP1-AS2 overexpression promoted the aerobic glycolysis and DLGAP1-AS2 knockdown suppressed the tumor growth of NSCLC cells. Mechanistically, m6A methyltransferase METTL3 enhanced the stability of DLGAP1-AS2 via m6A site binding. Moreover, DLGAP1-AS2 interacted with YTHDF1 to enhance the stability of c-Myc mRNA through DLGAP1-AS2/YTHDF1/m6A/c-Myc mRNA. In conclusion, our work indicates the functions of m6A-modified DLGAP1-AS2 in the NSCLC aerobic glycolysis, disclosing a potential m6A-dependent manner for NSCLC treatment.
KEYWORDS: N6-methyladenosine, non-small cell lung cancer, aerobic glycolysis, DLGAP1-AS2, YTHDF1
1. Introduction
Non-small cell lung cancer (NSCLC) is a worldwide malignant tumor with a significant high mortality and morbidity [1,2]. In spite of the achievement on the NSCLC therapy, including chemotherapy, radiotherapy and surgery, the overall survival rate or survival time of NSCLC sufferers remains unimproved [3]. The reason for the lethal trait of NSCLC is metastases and recurrence; however, the molecular mechanisms for NSCLC progression is still poorly understood [4]. Therefore, it is significantly critical to explore the underlying mechanism of NSCLC development, covering prognostic biomarkers, diagnostic and therapeutic targets.
Long noncoding RNAs (lncRNAs) are novel noncoding RNAs longer than 200 nucleotides length without protein-coding potential [5,6]. Over the years of research, growing number of lncRNAs are identified in the NSCLC occurrence [7,8]. For example, lncRNA DLX6-AS1 overexpresses in NSCLC and was inextricably correlates with NSCLC patients’ poor prognosis [9]. For another example, MAGI2-AS3 downregulates in NSCLC tissues and the overexpression of MAGI2-AS3 suppresses the NSCLC cell proliferation and invasion via miR-629-5p/TXNIP [10]. In summary, these findings suggest that lncRNA may function as an essential regulator in NSCLC progression.
N6-methyladenosine (m6A) is the most prevalent internal modification connected with eukaryotic mRNAs fate, including mRNA metabolism, splicing, export and stability [11–13]. Besides, in various tumor physiological events, m6A participates in these tumor progression-related progresses [14]. For example, IGF2BP2/IGF2BP3, HNRNPA2B1 and FTO significantly correlate with advanced-stage disease or clinical outcomes in NSCLC [15]. YTHDF2 expression upregulates in lung adenocarcinoma and YTHDF2 knockdown inhibits the proliferation but promotes the migration/invasion and epithelial-mesenchymal transition [16]. Thus, these evidences support that m6A modification significantly modulates the NSCLC carcinogenesis.
Herein, our work focused on the cross talk of m6A and lncRNAs in NSCLC tumorigenesis to uncover the functions and molecular mechanism of m6A-modified lncRNA DLGAP1 antisense RNA 2 (DLGAP1-AS2, Gene ID: 84,777, Chromosome 18, NC_000018.10). We found that lncRNA DLGAP1-AS2 was highly expressed in NSCLC, which was correlated with poor prognosis and advanced clinical stage. To investigate the function of DLGAP1-AS2 on NSCLC phenotype, cellular functional assays were performed using NSCLC cells. Moreover, m6A methyltransferase METTL3 enhanced the stability of DLGAP1-AS2 and then DLGAP1-AS2 interacted with YTHDF1 to enhance the stability of c-Myc mRNA in m6A-dependent manner. Overall, these findings provided a potential m6A-dependent therapeutic strategy for NSCLC treatment.
2. Materials and methods
2.1. Patient tissues specimens collection
A total of 50 NSCLC tissue samples in the cohort, which were formalin-fixed and paraffin-embedded, were recruited between January 2016 and December 2017 from patients who had underwent operation in Tianjin Medical University Cancer Institute and Hospital (Table 1). All samples were staged according to the criteria of the seventh edition of the AJCC Cancer Staging Manual [17]. This study was approved by the Tianjin Medical University Cancer Institute and Hospital Ethics Review Committee. Informed consent was written by each patient before the study.
Table 1.
DLGAP1-AS2 expression and NSCLC patients’ clinicopathological characteristics.
| 50 | DLGAP1-AS2 |
p-Value | |||
|---|---|---|---|---|---|
| Low | High | ||||
| Age | <55 | 29 | 17 | 12 | 0.152 |
| ≥55 | 21 | 8 | 13 | ||
| Gender | Male | 26 | 14 | 12 | 0.777 |
| Female | 24 | 11 | 13 | ||
| TNM stage | I/II | 22 | 7 | 15 | 0.022* |
| III/IV | 28 | 18 | 10 | ||
| Lymph node metastasis | Yes | 23 | 12 | 11 | 0.776 |
| No | 27 | 13 | 14 | ||
| Tumor differentiation | Well | 6 | 3 | 3 | 0.946 |
| Moderate | 13 | 7 | 6 | ||
| Poor | 31 | 15 | 16 | ||
| Distant metastasis | Yes | 17 | 7 | 10 | 0.370 |
| No | 33 | 18 | 15 | ||
Well, Well-differentiated adenocarcinoma; Moderate, moderately differentiated adenocarcinoma; Poor, poorly differentiated adenocarcinoma; TNM, tumor-node-metastasis. *p < 0.05 represents statistical differences.
2.2. Cell culture
NSCLC cell lines (H460, A549, H1299) and normal bronchial epithelial cells (NHBE) were purchased from the Cell Bank of the Chinese Academy of Sciences. The cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) and antibiotics at 37°C under 5% CO2.
2.3. Transfection vector construction
For the stable silencing of DLGAP1-AS2, lentivirus vector pLKD-U6-shRNA-EF1a-LUC-F2A-Puro-DLGAP1-AS2 (Obio Technology, Shanghai, China) or control vectors were packaged and transfected into A549 cells. Cells were grown in 1 μg/ml puromycin (Invitrogen) to select the stable transfected cells. Moreover, for the overexpression of DLGAP1-AS2, the cDNA of DLGAP1-AS2 was PCR-amplified ad then sub-cloned into pcDNA 3.1 vector (Invitrogen, Carlsbad, CA, USA). The efficiency of silencing or overexpression was subsequently quantified by qRT-PCR. The siRNAs and their controls (si-NC) were provided by GenePharma (Shanghai, China) and transfected using Lipofectamine 2000 (Invitrogen) when 70% confluent growth.
2.4. Quantitative real-time PCR (qRT-PCR)
Total RNA from NSCLC tissues or cell lines was extracted by TRIzol Reagent (Invitrogen, CA, USA). For the concentration and purity of RNA, RNA was detected with ultraviolet spectrophotometer using 260 and 280 nm. The real‐time PCR was performed using SYBR Premix Ex Taq (Takara) on the 7900 Real‐time PCR System. Reverse transcription was performed using PrimerScript RT Reagent Kit (TaKaRa, Kyoto, Japan). Fast SYBR Green Master Mix (Life Technologies) applied to perform qRT-PCR. The relative expression was calculated using the 2−ΔΔCT method. The β-actin acted as the internal mRNA control. The primer sequences are displayed in Table S1.
2.5. Western blot assay
The procedure details were conducted according to previous study. Proteins from transfected NSCLC cells were extracted using RIPA buffer (Beyotime, China) and authenticated using BCA Kit (Pierce, Rockford, IL, USA). Protein was resolved using the SDS-PAGE Electrophoresis System and then transferred to PVDF membranes (Millipore, USA). Specific primary antibodies (anti-METTL3, 1:1000, ab195352; anti-c-MYC, 1:1000, ab32072) were incubated at 4°C overnight. β-Actin was the internal reference.
2.6. Methylated RNA immunoprecipitation (MeRIP) sequencing and qPCR
All the specific manipulations were performed according to the protocol of Magna MeRIP™ m6A Kit (Merck Millipore). In brief, total RNAs were isolated from A549 cells and chemically fragmented into 100–300 nt. Fragmented fragments were incubated with m6A antibody for immunoprecipitation. The MeRIP was performed according to the manufacturer’s instructions. Samples were sequenced with the HiSeq PE150 platform. The enrichment of m6A containing mRNA was sent for quantitative RT-PCR. The RT-qPCR primers are listed in Table S1.
2.7. Quantitative analysis of glucose, lactate and ATP
The glucose uptake was quantificationally detected using Glucose Uptake Assay Kit (Fluorometric, Abcam, ab136956). The lactate production was quantificationally detected using Lactate Colorimetric/Fluorometric Assay Kit (BioVision, cat. K607–100). The ATP generation was quantificationally detected using ATP Assay Kit (Beyotime, S0026).
2.8. Measurement of extracellular acidification rate (ECAR)
ECAR was analyzed using Seahorse XFe96 analyzer (Seahorse Bioscience, Agilent) as previously described. Briefly, knockdown and overexpression of DLGAP1-AS2 transfected cells (1 × 104 cells/well) seeded into 96-well XF cell culture microplate. Medium (pH 7.4) was sequentially added with 10 mM glucose, 1 mM glutamine, 50 mM 2-DG and 1 µM oligomycin. ECAR was shown in mpH/min and measured using an XF96 analyzer.
2.9. Actinomycin D assay
A549 cells were seeded in 6-well plates (2 × 105 cells/well). Twenty-four hours later, cells were exposed to Actinomycin D (Act D, 2 μg/ml, Sigma) and harvested at an indicated time point. The RNA remaining level was analyzed using qRT-PCR and normalized to mock group (0 h).
2.10. RNA immunoprecipitation (RIP) assay
RIP assay was performed using EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) according to manufacturer’s protocol. Specific antibodies (anti-YTHDF1, Abcam) was conjugated with magnetic beads and then dissolved in RIP buffer. Approximate 90% confluence, cells were lysed using complete RIP lysis buffer (100 μl) containing protease inhibitor and RNase Inhibitor (Millipore). Mouse anti-IgG antibody (Cell Signaling Technology, USA) acted as negative control. Then, total RNA was retrieved and the relative expression was detected by qRT-PCR analysis.
2.11. Subcellular localization
Probes were obtained from Genepharma (Shanghai, China). Fluorescence in situ hybridization (FISH) was performed using fluorescent in situ hybridization kit according to the manufacturer’s protocols (Genepharma). Cy3-labeled probe for DLGAP1-AS2 and FAM-labeled probe for c-Myc, YTHDF1 were used for localization of DLGAP1-AS2, c-Myc and YTHDF1 in NSCLC cells. Nuclei was stained with 4,6-diamidino-2-phenylindole (DAPI), and images were obtained using confocal microscope (Olympus).
2.12. Animal models in vivo
Approximately 2 × 106 NSCLC cells (A549), transfected with sh-DLGAP1-AS2 or controls, were suspended in 100 μl PBS and then injected into flank of male BALB/c nude mice (5-wk old). In the following observation, tumor size was recorded by vernier caliper. At the indicated time, mice were anesthetized and sacrificed for tumors neoplasm acquisition. All animal assays were performed in the light of institutional Tianjin Medical University Cancer Institute and Hospital Animal Ethics Committee guidelines.
2.13. Dataset analysis
LncRNA and mRNA expression profile in lung cancer tissues and matched nontumor tissues was obtained from the NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/). Dataset GSE138172 consisted of five paired primary lung tissues (lung cancer tissues vs nontumor tissues) from non-small cell lung cancer patients. The GEO database subsequently analyzed by R (Version 3.4, http://www.bioconductor.org) with edgeR package. Fold change (FC) of gene expression was calculated with a cutoff of log2FC≥1, log2FC<1, and p value <0.05. Besides, the data and the corresponding clinical information of patients were collected from TCGA database (http://cancergenome.nih.gov/).
2.14. Statistical analysis
The data was expressed as mean ± S.D. (standard deviation). All results were conducted using SPSS Statistics software (Armonk, NY, USA) or GraphPad prism 7.0 (La Jolla, CA, USA). Difference within groups was calculated using one-way ANOVA test, non-parametric Mann–Whitney test and Student’s t-test. Overall survival rate was calculated using Kaplan–Meier analysis and log-rank test. *p ≤ 0.05 or **p ≤ 0.01 was considered statistically significant.
3. Results
3.1. DLGAP1-AS2 was an upregulated lncRNA in NSCLC
Using the microarray data (GSE138172), we found that numerous lncRNAs were dysregulated in the NSCLC tissue samples compared to normal tissue (Figure 1(a)). Moreover, a potential upregulated lncRNA (DLGAP1-AS2) might be correlated with the NSCLC carcinogenesis. In NSCLC cell lines, DLGAP1-AS2 was overexpressed as compared to the normal cell lines (Figure 1(b)). In NSCLC samples, DLGAP1-AS2 levels significantly overexpressed as compared to the normal adjacent tissues (Figure 1(c)). Kaplan–Meier analysis and log-rank test found that NSCLC tissue samples with high DLGAP1-AS2 levels displayed lower survival rate as compared to the control cohort (Figure 1(d)). Therefore, these data discovered an upregulated lncRNA DLGAP1-AS2 in NSCLC.
Figure 1.
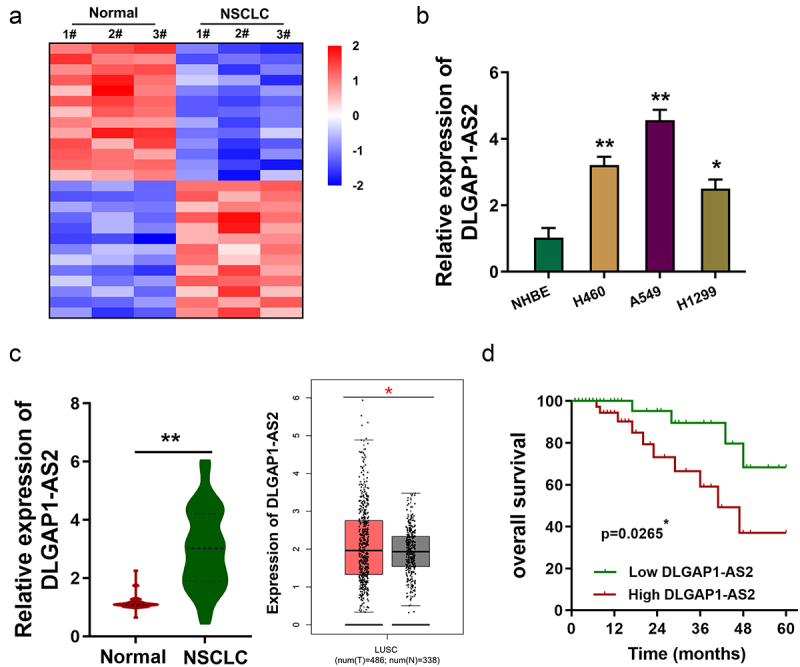
DLGAP1-AS2 was an upregulated lncRNA in NSCLC. (a) Based on the lncRNA microarray data (GSE138172), heatmap demonstrated the numerous deregulated lncRNAs in the NSCLC tissue as compared to normal tissue. (b) In NSCLC and normal cell lines, expression of DLGAP1-AS2 was detected using RT-qPCR. (c) In NSCLC samples, DLGAP1-AS2 level was detected using RT-qPCR as compared to the normal adjacent tissue. (d) Survival rate analysis showed the lower survival rate of NSCLC patients with high DLGAP1-AS2 levels, while the higher survival rate of NSCLC patients with low DLGAP1-AS2 levels. Data are expressed as mean ± SD. **p < 0.01; *p < 0.05.
3.2. MeRIP-Seq revealed the m6A modification of DLGAP1-AS2
MeRIP-Seq in NSCLC cells revealed the m6A peaks density, including 5’-UTR, coding region (CDS) and 3’-UTR (Figure 2(a,c)). The significant m6A motif for the m6A peaks was GGAC (Figure 2(b)). MeRIP-Seq demonstrated that there was a remarkable m6A modification site in the 3’-UTR of DLGAP1-AS2 (Figure 2(d)). In the 3’-UTR sequences of DLGAP1-AS2, there was a remarkable m6A site (AGAC) (Figure 2(e)). In the exploration which m6A enzyme catalyzes the m6A modification on DLGAP1-AS2, we found that m6A reader YTHDF1 was positively correlated to the level of DLGAP1-AS2 (figure 2(f)). Taken together, these MeRIP-Seq data indicated the m6A modification of DLGAP1-AS2.
Figure 2.
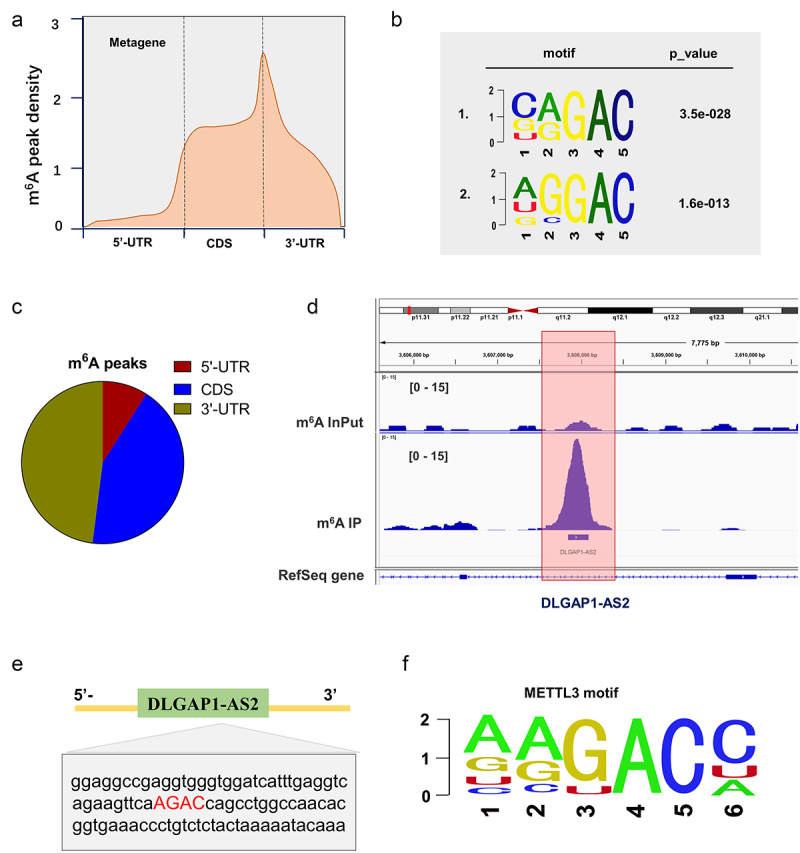
MeRIP-Seq revealed the m6A modification of DLGAP1-AS2. (a, c) MeRIP-Seq revealed the m6A peaks density in NSCLC cells, including 5’-UTR, coding region (CDS) and 3’-UTR. (b) The significant m6A motif for the m6A peaks was GGAC. (d) MeRIP-Seq showed that there was a remarkable m6A modification site in the 3’-UTR of DLGAP1-AS2. (e) Genomic analysis displayed the sequences of DLGAP1-AS2 3’-UTR containing m6A site (AGAC). (f) The motif of METTL3 targeting DLGAP1-AS2 m6A site.
3.3. METTL3 enhanced the stability of DLGAP1-AS2 in m6A-dependent manner
In the MeRIP-Seq, we noticed an m6A modification of DLGAP1-AS2; thus, DLGAP1-AS2 might be modulated in m6A-dependent manner. To investigate the possibility which m6A enzyme catalyzes the m6A modification on DLGAP1-AS2, we found that m6A methyltransferase METTL3 was positively correlated to the level of DLGAP1-AS2 (Figure 3(a)). In NSCLC cells, METTL3 overexpression was transfected (Figure 3(b)). After METTL3 overexpression, the m6A modification levels were found to be upregulated (Figure 3(c,d)). RNA stability analysis found that the METTL3 overexpression accelerated the RNA stability of DLGAP1-AS2 in H1299 cells and A549 cells (Figure 3(e,f)). In summary, these findings revealed that m6A-modified DLGAP1-AS2 demonstrated higher stability mediated by METTL3.
Figure 3.
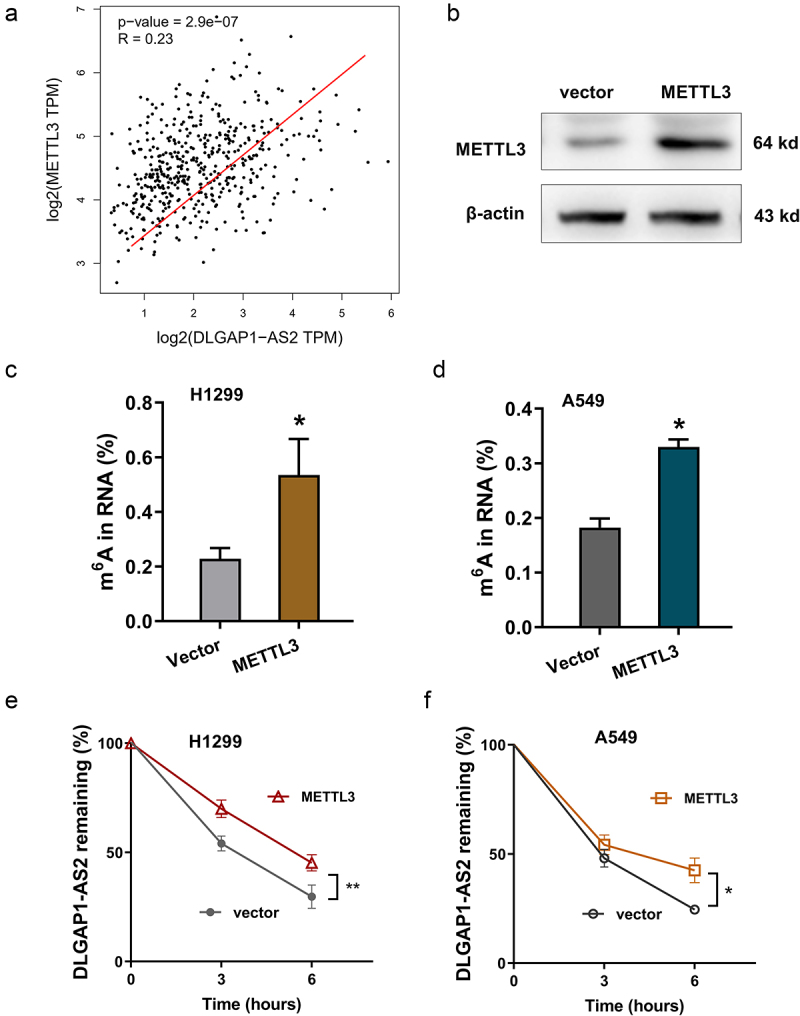
METTL3 enhanced the stability of DLGAP1-AS2 in m6A-dependent manner. (a) GEPIA dataset revealed the positive correlation within METTL3 and DLGAP1-AS2 in LUSC (Lung squamous cell carcinoma, http://gepia.cancer-pku.cn/). (b) The METTL3 protein was detected in NSCLC cells upon METTL3 overexpression transfection. (c, d) The m6A modification levels were detected using m6A RNA Methylation Quantification Kit (Colorimetric). (e, f) RNA stability analysis revealed the RNA remaining of DLGAP1-AS2 in H1299 cells and A549 cells treated by Act D (actinomycin D, 1 μg/m). LncRNA DLGAP1-AS2 was determined by qRT-PCR assay. *p < 0.05.
3.4. DLGAP1-AS2 promoted the aerobic glycolysis of NSCLC
In the functional experiments, enhanced expression or silencing of DLGAP1-AS2 was constructed in H1299 or A549 cells (Figure 4(a)). Subsequently, the biological functions of DLGAP1-AS2 on NSCLC aerobic glycolysis were investigated. Glucose uptake analysis illustrated that enforced DLGAP1-AS2 expression promoted the glucose consumption in H1299 cells, and knockdown of DLGAP1-AS2 repressed the glucose consumption in A549 cells (Figure 4(b)). Lactate production analysis unveiled that enforced DLGAP1-AS2 expression accelerated the lactate production in H1299 cells, and knockdown of DLGAP1-AS2 inhibited the lactate production in A549 cells (Figure 4(c)). ATP quantitative analysis illustrated that enforced DLGAP1-AS2 expression facilitated the ATP production in H1299 cells, and knockdown of DLGAP1-AS2 restrained the ATP production in A549 cells (Figure 4(d)). ECAR analysis for NSCLC glycolytic ability illustrated that enforced DLGAP1-AS2 expression promoted the extracellular acidification accumulation (Figure 4(e)), and knockdown of DLGAP1-AS2 repressed the acidification (Figure 4(f)). Moreover, the in vivo animal experiments indicated that knockdown of DLGAP1-AS2 inhibited the tumor neoplasm growth of NSCLC cells (Figure 4(g)). In conclusion, these findings demonstrated that DLGAP1-AS2 promoted the aerobic glycolysis of NSCLC
Figure 4.
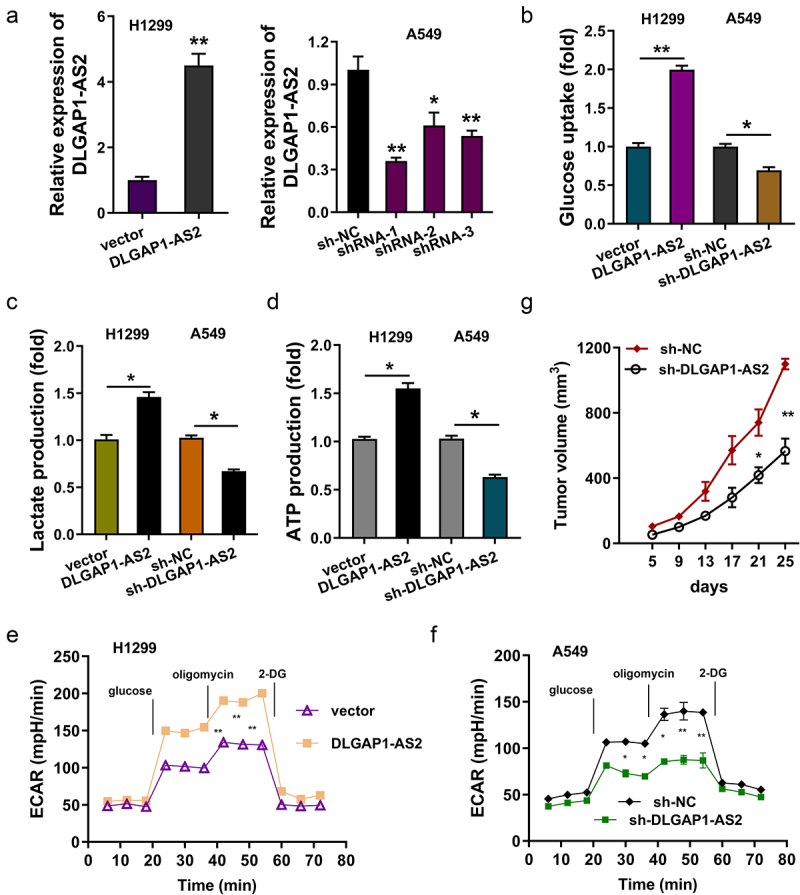
DLGAP1-AS2 promoted the aerobic glycolysis of NSCLC. (a) Detection of DLGAP1-AS2 levels by qRT-PCR in NSCLC cell lines (H1299, A549) respectively transfected with DLGAP1-AS2 overexpression plasmids or lentivirus shRNA. (b) Glucose uptake quantity was quantificationally detected using glucose uptake kit in H1299 and A549 cells. (c) Lactate production was quantificationally detected using lactate analysis kit. (d) ATP quantitative analysis was performed in H1299 or A549 cells. (e, f) The glycolysis levels of H1299 and A549 was detected using a seahorse analyzer. (g) In vivo animal experiments were performed to test the tumor neoplasm growth of NSCLC cells (A549) transfected with DLGAP1-AS2 silencing (sh-DLGAP1-AS2). The data are presented as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01.
3.5. DLGAP1-AS2 targeted c-Myc via YTHDF1-dependent manner
On the basis of finding that DLGAP1-AS2 regulated the aerobic glycolysis, we assumed that DLGAP1-AS2 might regulate certain key enzyme of glycolysis. First, the localization analysis using RNA-FISH found that the distribution of DLGAP1-AS2 was in cytoplasm, which was in accordance with the c-Myc and YTHDF1 (Figure 5(a)). The coincident cytoplasmic distribution of DLGAP1-AS2, YTHDF1 and c-Myc indicated the potential interaction within them. MeRIP-Seq presented that there was an m6A site in the c-Myc mRNA, suggesting a potential regulation for DLGAP1-AS2 via m6A-dependent manner (Figure 5(b)). In the GEPIA database, c-Myc was positively correlated with DLGAP1-AS2 and YTHDF1, indicating that c-Myc might act a target of DLGAP1-AS2 (Figure 5(c)). Besides, the c-Myc levels were highly expressed in the lung cancer (Lung squamous cell carcinoma) (Figure 5(d)). In summary, these data illustrated that c-Myc acted as a target of DLGAP1-AS2.
Figure 5.
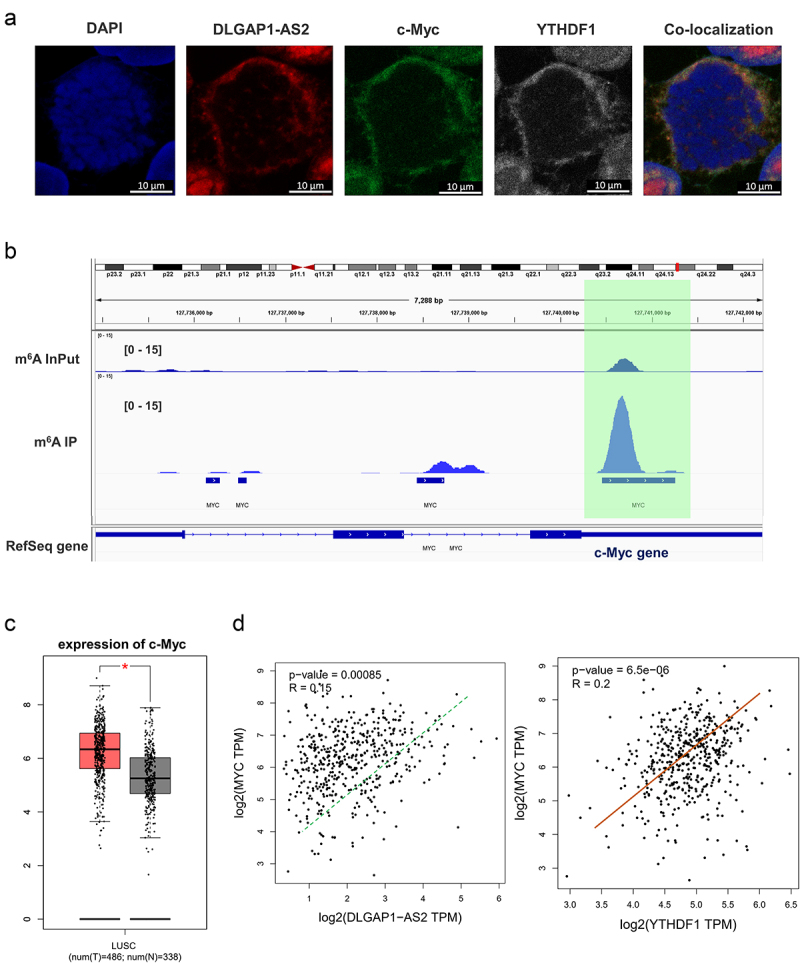
DLGAP1-AS2 targeted c-Myc via YTHDF1-dependent manner. (a) Co-localization analysis using fluorescence in situ hybridization (FISH) displayed the distribution of DLGAP1-AS2, YTHDF1 and c-Myc in A549 cells. (b) IGV viewer showed the m6A site in the c-Myc mRNA based on MeRIP-Seq data. (c) GEPIA database demonstrated the positive correlation within c-Myc and DLGAP1-AS2, or c-Myc with YTHDF1 (p < 0.001). (B) TCGA database illustrated the high-expression of c-Myc in LUSC (Lung squamous cell carcinoma, www.tcga.org/).
3.6. DLGAP1-AS2 promoted c-Myc mRNA stability through m6A-YTHDF1-dependent manner
The MeRIP-Seq demonstrated that there was an m6A modification site in the 3’-UTR of c-Myc mRNA, which was in accord with the CAGAC motif (Figure 6(a)). MeRIP-qPCR suggested us that the m6A enrichment on c-Myc mRNA was higher in the NSCLC cells (H1299, A549) (Figure 6(b)). Moreover, the RNA immunoprecipitation (RIP) unveiled that enhanced DLGAP1-AS2 expression promoted the c-Myc mRNA level precipitated by the anti-YTHDF1 antibody (Figure 6(b), left). Besides, knockdown of DLGAP1-AS2 inhibited the c-Myc mRNA level (Figure 6(b), right). RT-qPCR found that enhanced DLGAP1-AS2 expression amplified the c-Myc mRNA level (Figure 6(d), up). Moreover, the knockdown of DLGAP1-AS2 decreased the c-Myc mRNA level (Figure 6(d), down). RNA stability analysis found that DLGAP1-AS2 overexpression promoted the c-Myc mRNA remaining level, and the knockdown of DLGAP1-AS2 repressed the c-Myc mRNA remaining (Figure 6(e)). In summary, these findings illustrated that DLGAP1-AS2 promoted c-Myc mRNA stability through m6A-YTHDF1-dependent manner (Figure 7).
Figure 6.
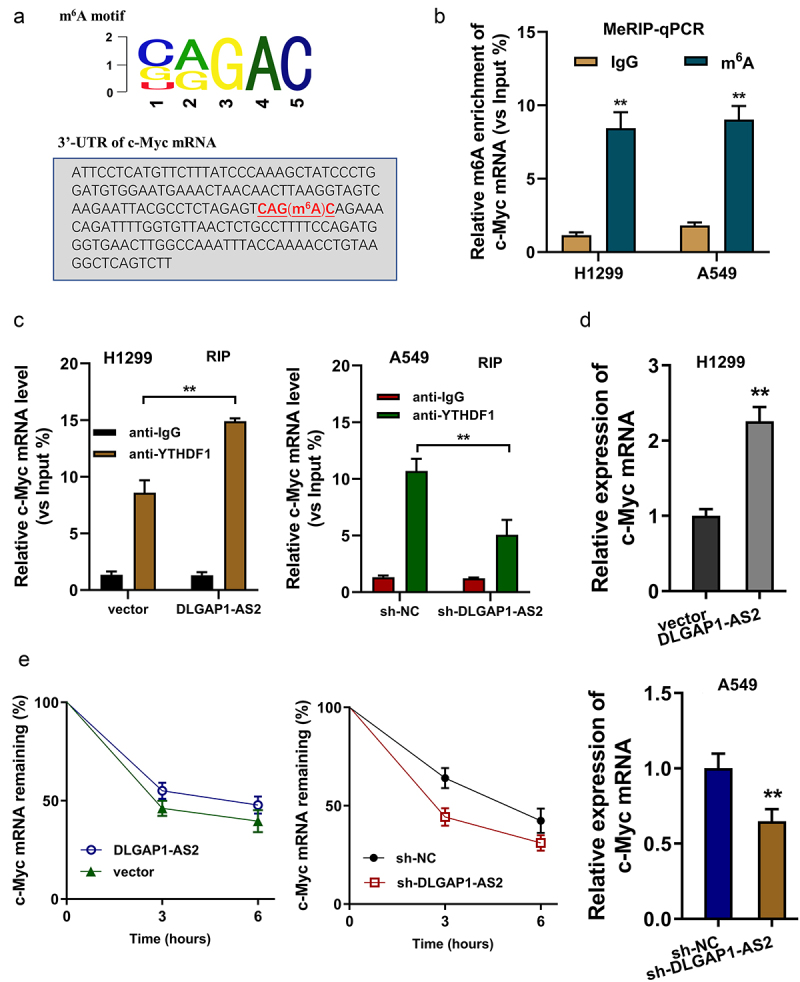
DLGAP1-AS2 promoted c-Myc mRNA stability through m6A-YTHDF1-dependent manner. (a) An m6A modification site in the 3’-UTR of c-Myc mRNA was discovered by MeRIP-Seq. The appropriate motif is CAGAC motif. (b) MeRIP-qPCR detected the m6A enrichment on c-Myc mRNA in NSCLC cells (H1299, A549) using m6A antibody and IgG control. (c) RNA immunoprecipitation (RIP) assays unveiled the c-Myc mRNA level precipitated by the anti-YTHDF1 antibody in NSCLC cells (H1299, A549) transfected with DLGAP1-AS2 overexpression (DLGAP1-AS2) and knockdown (sh-DLGAP1-AS2). (d) RT-qPCR detected the c-Myc mRNA level in NSCLC cells (H1299, A549) transfected with DLGAP1-AS2 overexpression and knockdown. (e) The levels of c-Myc expression in DLGAP1-AS2-overexpressing (DLGAP1-AS2), DLGAP1-AS2 knockdown (sh-DLGAP1-AS2) and their corresponding control were detected by qRT-PCR. NSCLC cells (H1299, A549) transfected NSCLC cells treated with actinomycin D (2 µg/mL). **p < 0.01 vs vector control.
Figure 7.
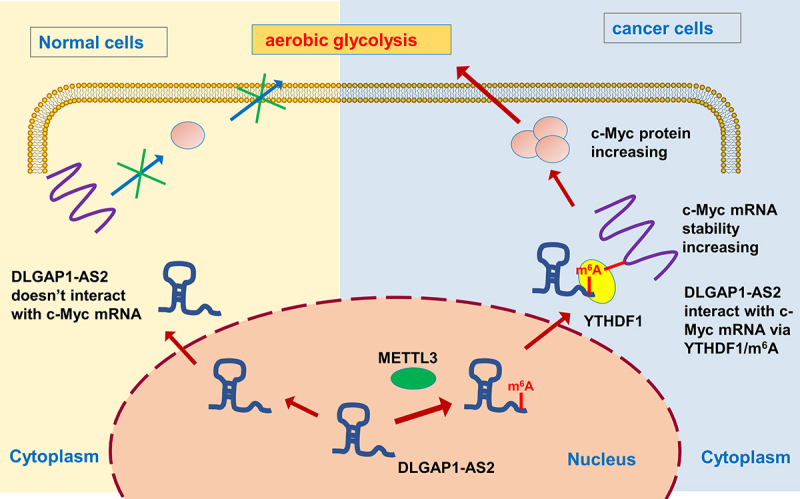
METTL3/DLGAP1-AS2/YTHDF1/m6A/c-Myc axis promotes the aerobic glycolysis of NSCLC.
4. Discussion
As one of the most prominent characteristics of malignant tumors, aerobic glycolysis (also known as Warburg effect) is a complex process affected by genetic and epigenetic modifications [18,19]. For the regulation of tumorigenesis, aerobic glycolysis exerts critical roles on the progress of tumor cell energy metabolism, providing the main energy supplement. In the current study, we demonstrated that m6A-modified DLGAP1-AS2 expression was increased in NSCLC tissues and cell lines. Furthermore, a high DLGAP1-AS2 expression implied a poor prognosis for NSCLC.
Here, we found that DLGAP1-AS2 expression was significantly upregulated in the NSCLC and moreover promoted the glucose uptake, lactate production and ATP generation. Our results reflect this possibility that DLGAP1-AS2 may act as an energy metabolism-related oncogene in NSCLC. Accumulating evidences have illustrated that lncRNA plays critical roles in cancer aerobic glycolysis. For instance, lnc-CYB561-5 is highly expressed in NSCLC and is associated with poor prognosis in lung adenocarcinoma. And, lnc-CYB561-5 knockdown inhibits cell migration, invasion and proliferation ability through interacting with Bsg/Hk2/Pfk1 [20]. These results consistently emphasized the regulation of lncRNAs for NSCLC aerobic glycolysis.
To further validate the biological function of lncRNA on NSCLC, we utilized the MeRIP-Seq and found that there was a potential m6A site in the 3’-UTR of DLGAP1-AS2, suggesting the m6A modification for DLGAP1-AS2. The specific m6A modification on DLGAP1-AS2 could significantly modulate its fate. In our research, we found that the m6A methyltransferase METTL3 could specifically bind with the m6A site on the DLGAP1-AS2. Besides, METTL3 could assist the stability increasing for DLGAP1-AS2. DLGAP1-AS2, on the other hand, connected with its downstream targets through m6A binding. Mechanistically, we found that m6A-modified DLGAP1-AS2 functions through a METTL3-involved m6A mechanism. DLGAP1-AS2 integrated with c-Myc mRNA via the m6A-mediated combination, constructing a DLGAP1-AS2/m6A/YTHDF1/c-Myc mRNA axis.
On this basis of m6A-DLGAP1-AS2, we realize the importance that m6A-modified lncRNA provides an important regulatory pattern for human cancer. For instance, lncRNA LINRIS is upregulated in colorectal cancer, and LINRIS knockdown attenuates the degradation of IGF2BP2 and especially MYC-mediated glycolysis [21]. For another example, m6A is highly enriched on the lncRNA THOR transcripts, including GAACA, GGACU and UGACU motifs. Besides, m6A-modified lncRNA THOR regulates the cancer cells’ proliferation via an m6A-reader-dependent manner [22]. In colorectal cancer, m6A-induced lncRNA RP11 triggers the progression and metastasis via post-translational Zeb1 upregulation through RP11/hnRNPA2B1/mRNA complex [23]. These results consistently demonstrated the m6A-lncRNA interaction regulation for human cancer.
In summary, our findings in this study revealed that DLGAP1-AS2 promoted the tumorigenesis of NSCLC through m6A/YTHDF1/c-Myc mRNA axis, thus triggering c-Myc mRNA stability and accelerating the aerobic glycolysis of NSCLC cells. The m6A-dependent RNA–protein interaction could maintain the oncogenic characteristic of lncRNA DLGAP1-AS2. Based on the findings, DLGAP1-AS2 may serve as a potential target in the NSCLC treatment.
Supplementary Material
Funding Statement
This study was supported by National Natural Science Foundation of China [82173038].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
No research data shared.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2105885
References
- [1].Machlowska J, Baj J, Sitarz M, et al. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;11(21):4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sexton RE, Al Hallak MN, Diab M, et al. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39:1179–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet. (London, England). 2020;396:635–648. [DOI] [PubMed] [Google Scholar]
- [4].Tang T, Zhang L, Li C, et al. Gastric and adrenal metastasis from breast cancer: case report and review of literature. Medicine (Baltimore). 2020;99:e18812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Han J, Kong H, Wang X, et al. Novel insights into the interaction between N6-methyladenosine methylation and noncoding RNAs in musculoskeletal disorders. Cell Prolif. 2022;e13294. DOI: 10.1111/cpr.13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wu X, Deng Z, Liao X, et al. Establishment of prognostic signatures of N6-Methyladenosine-related lncRNAs and their potential functions in hepatocellular carcinoma patients. Front Oncol. 2022;12:865917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xie P, Yan H, Gao Y, et al. Construction of m6A-Related lncRNA prognostic signature model and immunomodulatory effect in glioblastoma multiforme. Front Oncol. 2022;12:920926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang W, Zhang Q, Xie Z, et al. N (6) -methyladenosine-related long non-coding RNAs are identified as a potential prognostic biomarker for lung squamous cell carcinoma and validated by real-time PCR. Front Genet. 2022;13:839957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu C, Lin W, Fu F.. Long non-coding RNA DLX6-AS1 knockdown suppresses the tumorigenesis and progression of non-small cell lung cancer through microRNA-16-5p/BMI1 axis. Transl Cancer Res. 2021;10(8):3772–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gong J, Ma L, Peng C, et al. LncRNA MAGI2-AS3 acts as a tumor suppressor that attenuates non-small cell lung cancer progression by targeting the miR-629-5p/TXNIP axis. Ann Transl Med. 2021;9(24):1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ma Z, Ji J. N6-methyladenosine (m6A) RNA modification in cancer stem cells. Stem Cells. 2020;38(12):1511–1519. Dayton, Ohio. [DOI] [PubMed] [Google Scholar]
- [13].Zhu ZM, Huo FC, Pei DS. Function and evolution of RNA N6-methyladenosine modification. Int J Biol Sci. 2020;16:1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang YN, Yu CY, Jin HZ. RNA N(6)-Methyladenosine modifications and the immune response. J Immunol Res. 2020;2020:6327614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jin L, Chen C, Yao J, et al. The RNA N(6) -methyladenosine modulator HNRNPA2B1 is involved in the development of non-small cell lung cancer. Clin Exp Pharmacol Physiol. 2022;49:329–340. [DOI] [PubMed] [Google Scholar]
- [16].Zhao T, Wang M, Zhao X, et al. YTHDF2 inhibits the migration and invasion of lung adenocarcinoma by negatively regulating the FAM83D-TGFβ1-SMAD2/3 Pathway. Front Oncol. 2022;12:763341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Washington K. 7th edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17(12):3077–3079. [DOI] [PubMed] [Google Scholar]
- [18].Huang S, Guo Y, Li Z, et al. A systematic review of metabolomic profiling of gastric cancer and esophageal cancer. Cancer Biol Med. 2020;17(1):181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yuan LW, Yamashita H, Seto Y. Glucose metabolism in gastric cancer: the cutting-edge. World J Gastroenterol. 2016;22:2046–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li L, Li Z, Qu J, et al. Novel long non-coding RNA CYB561-5 promotes aerobic glycolysis and tumorigenesis by interacting with basigin in non-small cell lung cancer. J Cell Mol Med. 2022;26(5):1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Y, Lu JH, Wu QN, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu H, Xu Y, Yao B, et al. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 2020;11(8):613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wu Y, Yang X, Chen Z, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No research data shared.


