Abstract
Typically, cancer is thought to arise due to DNA mutations, dysregulated transcription and/or aberrant signalling. Recently, it has become clear that dysregulated mRNA processing, mRNA export and translation also contribute to malignancy. RNA processing events result in major modifications to the physical nature of mRNAs such as the addition of the methyl-7-guanosine cap, the removal of introns and the addition of polyA tails. mRNA processing is a critical determinant for the protein-coding capacity of mRNAs since these physical changes impact the efficiency by which a given transcript can be exported to the cytoplasm and translated into protein. While many of these mRNA metabolism steps were considered constitutive housekeeping activities, they are now known to be highly regulated with combinatorial and multiplicative impacts i.e. one event will influence the capacity to undergo others. Furthermore, alternative splicing and/or cleavage and polyadenylation can produce transcripts with alternative messages and new functionalities. The coordinated processing of groups of functionally related RNAs can potently re-wire signalling pathways, modulate survival pathways and even re-structure the cell. As postulated by the RNA regulon model, combinatorial regulation of these groups is achieved by the presence of shared cis-acting elements (known as USER codes) which recruit machinery for processing, export or translation. In all, dysregulated RNA metabolism in cancer gives rise to an altered proteome that in turn elicits biological responses related to malignancy. Studies of these events in cancer revealed new mechanisms underpinning malignancies and unearthed novel therapeutic opportunities. In all, cancer cells coopt RNA processing, export and translation to support their oncogenic activity.
Keywords: cancer, capping, RNA export, RNA processing, splicing, translation
Introduction
Global analyses of cancer transcriptomes have yielded important insights into its pathogenesis. However, the transcriptome does not always predict the proteome [1]. This disconnect can arise because of the reprograming of RNA processing, export and/or translation which in turn enables the mRNA instructions sent from the DNA to be hijacked. Specifically, these events modify the physical nature of the transcripts and/or regulate the conversion of these messages into protein [2]. In cancer, mRNA metabolism can be reprogrammed to generate a proteome that drives malignancy even in the absence of altered mRNA levels or genetic mutation. In this review, the major mRNA processing steps, mRNA export and translation are described in relationship to their ability to subvert the mRNA message and thereby alter the proteome. As predicted by the RNA regulon model, the mRNA processing, export and translation of groups of mRNAs, often encoding factors in the same pathways, can be regulated simultaneously to elicit powerful biological responses [3,4]. These RNAs are selected based on cis-acting elements referred to as USER (untranslated sequence for regulation) codes which recruit the necessary machinery. Due to space limitations some important RNA modifications are not covered here but are reviewed elsewhere e.g. [5].
Starting the message: capping and re-capping RNAs
Typically, the first step in mRNA processing is referred to as ‘capping' and involves the addition of the methyl 7-guanosine (m7G) ‘cap' onto the 5′ end of coding and some non-coding RNAs [6–8]. While this step is usually considered to occur co-transcriptionally, re-capping after decapping can also occur in the cytoplasm [9–11]. Most subsequent mRNA processing steps rely on the cap to engage cap-binding proteins which permit recruitment of relevant processing machinery [2]. Two well-established cap-binding factors are the eukaryotic translation initiation factor eIF4E and the cap-binding complex (CBC) which is comprised of the cap-binding protein CBP20 (NCBP2) and its co-factor CBP80 (NCBP1) [2]. While it was once considered that CBC was restricted to the nucleus and eIF4E to the cytoplasm, it is now clear that these factors are found in both compartments where they act in a variety of cap-dependent RNA processing events [2]. In mammals, capping requires two enzymes RNGTT (RNA guanyltransferase and 5′ phosphatase) and RNMT (RNA guanine-7-methyltransferase) [8]. Indeed, these proteins are required for cell survival highlighting the importance of this modification [8]. RNGTT removes the 5′ phosphate on the 5′ end of the transcript followed by the addition of a guanosine forming a 5′–5′ pyrophosphate linkage. Then, RNMT methylates the cap guanylate at the 7-position; this activity is stimulated by its co-factor, RAM [8].
Intriguingly, capping of specific transcripts is modulated during development, differentiation and dysregulated in primary cancer specimens [7,12–14]. Interestingly, RNAs can undergo cycles of decapping and re-capping suggestive of cap homeostasis [10,11,15]. Newly developed methods revealed that capping levels for subsets of RNAs is much lower (∼30%) than the anticipated 100% [8,9,14]. The three best-defined factors known to regulate capping are Myc, RNMT and eIF4E, whereby each employs a distinct stratagem (Figure 1). Myc recruits RNGTT and RNMT to transcriptional sites of specific transcripts as well as increases levels of S-adenosyl hydrolase to neutralize inhibition of RNMT by the SAH by-product of methylation reactions [7]. RNMT overexpression only elevates capping of selected RNAs; to date this specificity is not yet understood [7]. Regulation of capping plays physiological roles. During differentiation of embryonic stem cells, RNMT and RAM elevation is required for the expression of pluripotency associated genes; then, ERK1/2 phosphorylates RAM to trigger its degradation which in turn represses these genes allowing differentiation [6]. eIF4E modulates capping likely by two mechanisms: (1) Increased nuclear export and translation efficiency of RNAs that encode the capping machinery thereby elevating levels of the resultant proteins and (2) Physical interaction with the RNMT protein [14,16]. eIF4E does not increase the RNA levels of RNMT or RNGTT. Similar to RNMT and MYC regulation of capping, the capping of selected (not all) transcripts is modulated by eIF4E. Many of eIF4E-capping target transcripts encode cancer related proteins such as Myc, MDM2, CTNNB1, CCND1 and interestingly RNMT [8,14]. A cap sensitivity element (CapSE) was identified in the 3′ UTR of model RNAs and this USER code engaged RNMT presumably to enhance eIF4E-dependent capping for some RNAs [14]. There are likely other such elements yet to be identified. The first study to show capping was dysregulated in any malignancy was in primary high-eIF4E AML specimens. Here, increased capping of specific transcripts was observed as was elevation of RNMT and RNGTT levels [14]. Collectively, these observations position capping as an important control point modulating the ultimate protein-coding capacity of mRNAs as well as impacting the biochemical activity of some non-coding RNAs [7,12–14]. In this way, these factors control the capability of transcripts to undergo other RNA processing steps as well as resultant protein-coding capacity.
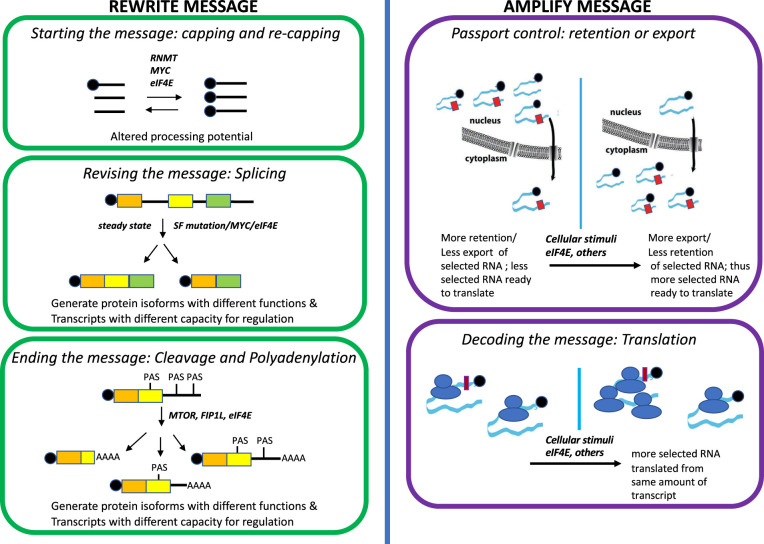
Figure 1. Major mRNA metabolism steps can be hijacked in cancer.
These are divided into two categories: those that can impact on the physical nature of the message and thus its content (left) and those that amplify of the message e.g. generate more protein per transcript (right). The m7G cap is represented as black balls on the 5′ end of the transcript, SF mutation indicated splice factor mutation and PAS, polyadenylation signal (see text). Capping section, black lines represent transcripts. Splicing and Cleavage and Polyadenylation section, coloured boxes represent exons. Right Side. Narrow rectangles in the retention/export and translation section represent USER codes in the 3′ or 5′ UTRs, respectively, that can sensitize RNAs to the levels of regulation shown. There are other processing steps than can be regulated and there are many other regulators of each of the steps shown, but for brevity only a few are highlighted.
Revising the message: RNA splicing
After transcripts are capped, many mammalian coding and some non-coding RNAs undergo splicing. mRNA splicing involves the removal of introns and joining of flanking exons in pre-mRNAs [17,18]. Splicing provides an important capacity to diversify the proteome and ncRNA isoforms [17,19,20]. The splicing machine, known as the spliceosome, is comprised of ∼150 proteins and five U-rich small nuclear RNAs (UsnRNAs) [17,18]. Approximately 80% of splicing occurs co-transcriptionally and the remaining ∼20% post-transcriptional splicing likely transpires in nuclear speckles [21,22]. Interestingly for a given RNA, some introns can be spliced co-transcriptionally and others post-transcriptionally [21]. Approximately 95% of multi-exonic transcripts undergo alternative splicing (AS) [18,19,23]. AS events include the skipping of exons, inclusion of mutually exclusive exons, use of alternative splice sites and intron retention [17,24]. All of these can alter the embedded coding information which can ultimately result in production of proteins with variant functions (Figure 1). Indeed, AS can generate proteins with opposing functions to their constitutive counterparts, alter localization of RNAs or protein products, and/or produce RNAs that are rapidly degraded resulting in reduced protein levels. For instance, the long isoform of MCL1 is anti-apoptotic and the short form is pro-apoptotic [24]. The use of alternative 3′ splice sites switch VEGF isoforms from angiogenic to anti-angiogenic [24].
Splicing is regulated during differentiation and development. Notably, the proteomic outputs of dysregulated splicing contribute to the hallmarks of cancer [24]. Mutation or dysregulation of splicing factors contributes to hematologic malignancies and solid tumours [19,25–27]. For example, AML is amongst the cancers with the largest number of AS events [25,26,28]. The most frequently mutated splicing factors in heme-malignancies are SF3B1, SRSF2, and U2AF1. These mutations are associated with progression from myelodysplastic syndromes to AML and are also prevalent in types of bladder cancer and glioma [19,25–28]. Interestingly, abnormal mRNA splicing is common in AML even in the absence of SF mutations suggesting other mechanisms also drive alternative splicing [29]. Some splicing factors are elevated rather than mutated in a diverse set of solid tumours including breast, colon, and lung [19]. Importantly, modulation of the core splicing machinery often elicits splicing alterations to specific pre-mRNAs, i.e. does not have global impacts [30]. Further, mutations within specific pre-mRNAs e.g. at splice sites, also contribute to AS and disease [24,25,28]. Recently it was found that eIF4E can impact splicing likely through its capacity to increase production of splice factors through elevation of their nuclear export of the corresponding mRNAs, and moreover, for some cases, via physical interaction with pre-mRNAs and spliceosome components [31]. eIF4E-dependent splicing impacts ∼1000 events in model U2Os cells and ∼4000 in AML primary specimens [31]. Many of these transcripts encode factors involved in malignancy.
Splicing dysregulation is the subject of therapeutic development in several malignancies. In this regard, there has been intense focus on the inhibition of SF3B1 [26,32] a core component of the spliceosome. Importantly, SF3B1 inhibitors such as Spliceostatin A or Pladienolide B only alter ∼10% of splicing events [25,26,32]. These inhibitors often target tumour cells with SF3B1 mutations more so than other tumour types, or normal cells, thereby providing a therapeutic window [25,33,34]. SF3B1 was targeted in early phase clinical trials in epithelial malignancies with E-7071 which was generally well tolerated but the trials were halted due to partial vision loss in two patients [35,36]. A next generation inhibitor (H3B-8800) is currently in trial for AML (ClinicalTrials.gov NCT02841540). A landmark use of splicing therapeutics is showcased in spinal muscular atrophy (SMA). Here, the therapeutic is an antisense-oligonucleotide (ASO) that targets splicing of SMN2 thereby reconstituting functional SMN protein activity in SMA patients [24,37]. This allows patients to regain mobility and the treatment received FDA approval. Investigations are underway into the utility of using a similar strategy in cancer cells vis-à-vis redirecting functionalities of proteins via antisense directed alteration of their splicing [24].
Ending the message: cleavage and polyadenylation
The formation of polyadenylated tails on 3′ end of mRNAs is another key step in RNA processing which also can impact the mRNA composition as well as its capacity to be exported from the nucleus and translated in the cytoplasm. This process, known as cleavage and polyadenylation (CPA) is comprised of two steps. First the transcript undergoes cleavage downstream from a polyadenylation signal (PAS) within the 3′ UTR of mRNAs followed by the addition of the polyA-tail via polyA polymerases [38,39]. Cleavage is considered a nuclear process; by contrast, polyA tail lengths can be modulated in the nucleus or cytoplasm. While the cleavage event is generally considered to be co-transcriptional, there are ample examples of post-transcriptional cleavage indicating that RNAs can be further modified after transcription. At least 70% of mammalian RNAs undergo alternative polyadenylation (APA) [40]. APA includes: (1) Altered PAS cleavage efficiency, (2) The cleavage of alternative PAS's within the 3′UTR, introns or exons and/or (3) Alterations in polyA-tail length [38–40] (Figure 1). APA can affect RNA stability, export, localization, translation efficiency, and sometimes protein product functionality by altering the coding sequence via production of C-terminal truncations [38–42].
In cancer, APA generally results in mRNAs with shorter 3′UTRs providing a mechanism to evade regulation by microRNAs or RNA binding proteins (RBPs) through the removal of key regulatory sequences [38–40,43]. These shorter UTRs are usually generated through selection of PAS's close to the stop codon [43]. However, shorter 3′UTRs are not always oncogenic and thus effects are context specific. Moreover, global shifts in 3′UTR length of the entire transcriptome are not observed during oncogenic transformation; but rather, the relative abundance of 3′UTR isoforms shift for specific mRNAs [42,44]. In colorectal cancers, the progression from adenomas to carcinomas is characterized by elevated expression of the APA machinery [38,45]. For instance, elevation of the cleavage co-factor Fip1L drives increased PAS cleavage efficiency of RNAs acting in self-renewal promoting this biological activity [38,46]. MTOR activation leads to shortening of 3′UTRs [42]. eIF4E overexpression alters production of CPA factors, physically interacts with some cleavage machinery [47] and impacts on the APA of many transcripts including in AML specimens (unpublished observations). These are just a few examples of the contribution of APA dysregulation to oncogenic capacity.
Passport control: retention or export?
The capacity of a transcript to be exported from the nucleus reflects the balance between retention and export. For instance, some mature mRNAs are retained in the nucleus for stockpiling so they are available for rapid export and translation once cellular conditions are suitable; or in some cases, retention is a means to auto-regulate factor levels e.g. the splicing factor SRSF1 [48]. Incompletely processed mRNAs can be retained as part of nuclear surveillance mechanisms to prevent aberrant protein production. Retention mechanisms are typically mRNA and cell context specific. For instance, some transcripts are retained at paraspeckles while partially spliced mRNAs can be held at nuclear speckles where they await post-transcriptional splicing. Retained mRNAs tend to be longer and can contain specific sequences linked to their retention [48].
Nuclear mRNA export controls the accessibility of mature transcripts to the translation machinery in the cytoplasm and thus contributes to increased protein levels without increasing the transcript levels or, a priori, translation efficiency [2,49–51]. RNAs are actively transported through the nuclear pore complex (NPC). mRNAs bind appropriate nuclear receptors and export factors which in turn associate with the nuclear basket of the NPC, transit through the central channel and exit via the cytoplasmic fibrils [49–51]. Bulk mRNA export employs the nuclear receptor NXF1 and its co-factor NXT1 (also known as TAP and p15, respectively). Typically, CBC-bound mRNAs bind the large export complex known as TREX and are then recruited by NXF1/NXT1 to the NPC for transit to the cytoplasm.
There are alternative routes through the NPC which can rely on mRNA containing specific USER codes [3], which permit engagement of selected export factors to facilitate their transit [49–51]. In this way, factors can promote the export of functionally related groups of mRNAs based on shared USER codes (Figure 1). For instance, both CRM1/XPO1 and NXF1/NXT1 act as nuclear receptors to chaperone mRNAs through the NPC [49–51]. In the case of eIF4E, it promotes the export of groups of cancer-causing RNAs via CRM1/XPO1 whereby these mRNAs typically have an eIF4E sensitivity element (4ESE) in their 3′UTR [52–55]. The 4ESE element is bound by the leucine rich pentatricopeptide repeat containing protein LRPPRC an assembly platform binding eIF4E, CRM1 and 4ESE RNA [52,55]. CRM1 can also host CBC-bound mRNAs in some instances [49,50]. Export of selected mRNAs can be enhanced under certain biological contexts. For instance, eIF4E elevation leads to increased export of mRNAs involved in oncogenic processes such as those that can alter the cell surface architecture supporting increased cell motility and invasion [56]. CRM1/XPO1 dysregulation is found in many cancers, which can in turn drive inappropriate RNA and protein export [57]. Repressing this export pathways is clinically useful. Indeed, the CRM1 inhibitor Selinxor is approved for treatment in relapsed multiple myeloma and in trial for other malignancies.
Decoding the message: translation
Once RNAs are released into the cytoplasm, many undergo translation while some transit to cytoplasmic bodies where they are sequestered for later translation or undergo degradation [58]. Clearly, the above processing steps will impact the composition of the protein generated as well as its translation efficiency. Increased translation efficiency is defined as more ribosomes loaded per transcript allowing more protein to be produced per RNA (Figure 1). Here, the focus is on translation initiation where eIF4E, in a complex known as eIF4F, recruits many capped mRNAs to the ribosome [58]. The eIF4F complex includes eIF4E, the eIF4A helicase which unwinds mRNAs, and eIF4G1 which acts as an assembly platform. eIF4F scans the 5′UTR of the target mRNA until it reaches a start codon. Under certain conditions, alternative start sites can be utilized to generate proteins with altered N-termini or sequence relative to their cognate counterparts. eIF4E overexpression is sufficient to increase translation efficiency of specific mRNAs typically with long, structured 5′ UTRs [58,59] (Figure 1). Other mRNAs such as ACTIN and GAPDH are not affected by eIF4E overexpression [58,59]. Importantly other translation initiation complexes recruit subsets of mRNAs to preferentially enhance their translation efficiency and thus their protein production [58]. For example, CBC in association with an eIF4G1 substitute, CTIF, play roles in translation of selected mRNAs in response to stresses and during the pioneer round of translation [2,58,60–62]. eIF3d recruits selected capped mRNAs using a distinct translation initiation complex including DAP5/eIF4G2 (rather than eIFG1) which impact ∼20–30% of the translatome of breast cancer cells [63]. Other specialized translation initiation complexes employ PARN or LARP to engage select mRNAs [64,65]. In all, translation responds to various environmental cues to modulate the production of specific factors without altering the entire proteome.
Controlling the message: therapeutic targeting
eIF4E has been a focal point for development of cancer therapeutics targeting RNA metabolism. Given its multiple cap-dependent functions, its targeting should have potent impacts (Figure 1). Supporting this direction, eIF4E is highly elevated in primary cancer specimens at the protein level and its activities in capping, splicing, export and/or translation are dysregulated in these specimens e.g. [14,31,66,67]. Strategies for its pharmacological inhibition can be divided into four categories: (1) m7G cap-competitors, (2) Antisense oligonucleotides (ASO) to reduce eIF4E levels, (3) MNK inhibitors to reduce eIF4E phosphorylation, and (4) Factors that interfere with the eIF4E–eIF4G translation initiation complex [68–72]. Cap-competitors or genetic reduction in eIF4E impacts on all its RNA processing activities making these strategies likely very potent. eIF4E phosphorylation inhibits RNA export [73] and may have impacts on translation although this is highly debated [74,75]. Phosphorylation may also modulate other eIF4E activities although this is yet to be determined. Thus, it is not yet known if inhibition of its phosphorylation will impact on multiple eIF4E activities. The last group including 4GI-1 may only impact eIF4E-dependent translation which may limit its clinical utility; however, its ability to inhibit other eIF4E activities has not yet been investigated and given the proximity of 4GI-1 to the cap-binding site [68] may well have inhibit other functions.
The first-in-class inhibitor administered to patients to target eIF4E was the cap-competitor ribavirin [76]. Ribavirin directly binds eIF4E and competes for cap-binding as seen by NMR, biophysical, biochemical and mass spectrometry methods [71,72,77,78]. RNAi-mediated reduction in eIF4E reduces ribavirin activity supporting that ribavirin acts through eIF4E [79]. Ribavirin impairs eIF4E-dependent splicing, RNA export and translation, and in animal models targets eIF4E activity, reduces tumours and metastases, and increases survival [31,56,66,79–84]. Its impacts on capping and APA are not yet known. In three clinical trials in poor prognosis AML patients, ribavirin targeted eIF4E which correlated with objective clinical responses including remissions ([76,85] and ClinicalTrials.govNCT02073838). These first studies serve as a proof-of-principle that eIF4E can be targeted in humans and that this targeting can lead to clinical benefit. Unfortunately, clinical trials employing ASOs were not successful as eIF4E levels were not decreased in patients [86]. Clinical studies using MNK inhibitors are ongoing in prostate cancer in combination with several other agents (ClinicalTrials.govNCT04261218). Whether the effects of MNK inhibitors are due to inhibition of eIF4E and/or reduction in phosphorylation of other MNK targets will be difficult to dissect clinically. To date, the last class of eIF4E inhibitor, 4GI-1 has not been tested in patients; however its use impaired proliferation of breast cancer stem cells [87] supporting this direction.
Selecting the message: RNA regulons and USER codes
A common theme emerging from the above discussion is that factors intricately involved in processing events can impact selected mRNA targets. Understanding the principles of selectivity in recruiting mRNAs is critical. How do certain factors select one set of mRNAs? How does one factor influence the same mRNA at one processing level but not at another? The specificity is best viewed through the lens of the mRNA regulon model [3,4]. In this model, mRNAs with specific USER codes recruit factors that act in a selected process. Thus, the correct combination of USER codes is required for combinatorial regulation of mRNAs at multiple levels (Figure 1). Further multiple USER codes can be needed. For example, eIF4E and CBC bind the m7G cap, a feature common to most mature mRNAs. Here, additional USER codes recruit relevant factors to customize processing, export and translation of the transcript. Indeed, mRNAs may switch between CBC and eIF4E cap-chaperones to modulate their processing and thus their ultimate product [2]. In this way, USER codes imbue selectivity of an mRNA to the given processing step under specific conditions. mRNAs with the same USER codes can be co-regulated enabling them to be similarly processed, exported and translated. Furthermore, co-regulation of the RNA binding proteins (RBPs) that bind USER codes is another potent way to co-regulate mRNAs in these groups [88]. These sorts of mechanisms also play important roles in genetic compensation observed in knockdown or knockout experiments [88]. In all, RNA regulons provide a powerful means by which to coordinate post-transcriptional processing in combinatorial and multiplicative ways which in turn potently drive important cellular processes in development, differentiation and malignancy [3,4].
One example of the biological impact of coordinated processing involves the capacity of eIF4E to alter the cell surface architecture which in turn is required for eIF4E-dependent cell motility. For example, the glycosaminoglycan hyaluronan (HA) and its receptor CD44 underpin tumour-stroma interactions and are important for association with the microenvironment and cell motility [89]. Interestingly, eIF4E elevates the RNA export of all the enzymes involved in the biosynthesis of HA as well as CD44 [56]. This enhanced RNA processing driven by eIF4E increased levels of both the HA sugar and the CD44 protein [56]. Notably, eIF4E does not alter the transcript levels of any of these factors. HA is usually extruded into the extracellular space where it is a major component of the microenvironment [89]. However in the high-eIF4E context, cells become encapsulated in coats of HA and CD44 on their cell surface [56]. Genetic or pharmacological targeting of HA or CD44 production in these high-eIF4E cells impairs eIF4E-mediated cell motility [56]. In all this provides an example of an eIF4E driven RNA regulon that underpins motility, an important feature of cancer cells.
Perspectives
Cancer cells can coopt RNA processing to reprogramme the proteome.
These changes can reprogramme entire biochemical networks, re-write protein functionalities, and can imbue cells with increased oncogenic capacity.
Diverted RNA processing can lead to wide-scale changes to the proteome. This information is not captured by traditional RNA-Seq methods which employ transcript levels as surrogates of protein levels.
Some factors such as eIF4E and CBC are positioned to combinatoraly re-write and amplify messages by influencing multiple processing events for groups of RNAs acting in the same biochemical pathways and thereby elicit potent biological outcomes.
Efforts to better understand the genetic flow of information through the RNA processing steps with a focus on identifying the USER codes to better predict functional outcomes from transcriptome analysis.
Targeting RNA processing can lead to clinical benefit and needs to be incorporated more broadly into therapeutic strategies.
Acknowledgements
Many thanks for critical reading of the manuscript by Drs Mehdi Ghram and Jean-Clement Mars.
Abbreviations
- APA
alternative polyadenylation
- CBC
cap-binding complex
- CPA
cleavage and polyadenylation
- HA
hyaluronan
- NPC
nuclear pore complex
- PAS
polyadenylation signal
- RBPs
RNA binding proteins
- SMA
spinal muscular atrophy
Competing Interests
The author declares that there are no competing interests associated with this manuscript.
Funding
KLBB is grateful for funding from the National Institutes for Health, Canadian Institute for Health Research, Leukemia and Lymphoma Society and holds a Canada Research Chair.
Author Contributions
Conception, writing and figure preparation were carried out by KLBB.
References
- 1.de Sousa Abreu, R., Penalva, L.O., Marcotte, E.M. and Vogel, C. (2009) Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mars, J.C., Ghram, M., Culjkovic-Kraljacic, B. and Borden, K.L.B. (2021) The cap-binding complex CBC and the eukaryotic translation factor eIF4E: co-conspirators in cap-dependent RNA maturation and translation. Cancers (Basel) 13, 6185 10.3390/cancers13246185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackinton, J.G. and Keene, J.D. (2014) Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin. Cell Dev. Biol. 34, 44–54 10.1016/j.semcdb.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene, J.D. and Tenenbaum, S.A. (2002) Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9, 1161–1167 10.1016/S1097-2765(02)00559-2 [DOI] [PubMed] [Google Scholar]
- 5.Zaccara, S., Ries, R.J. and Jaffrey, S.R. (2019) Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- 6.Grasso, L., Suska, O., Davidson, L., Gonatopoulos-Pournatzis, T., Williamson, R., Wasmus, L.et al. (2016) mRNA cap methylation in pluripotency and differentiation. Cell Rep. 16, 1352–1365 10.1016/j.celrep.2016.06.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galloway, A. and Cowling, V.H. (2019) mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 270–279 10.1016/j.bbagrm.2018.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borden, K., Culjkovic-Kraljacic, B. and Cowling, V.H. (2021) To cap it all off, again: dynamic capping and recapping of coding and non-coding RNAs to control transcript fate and biological activity. Cell Cycle 20, 1347–1360 10.1080/15384101.2021.1930929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenberg, D.R. and Maquat, L.E. (2009) Re-capping the message. Trends Biochem. Sci. 34, 435–442 10.1016/j.tibs.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee, C., Patil, D.P., Kennedy, B.A., Bakthavachalu, B., Bundschuh, R. and Schoenberg, D.R. (2012) Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep. 2, 674–684 10.1016/j.celrep.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trotman, J.B., Giltmier, A.J., Mukherjee, C. and Schoenberg, D.R. (2017) RNA guanine-7 methyltransferase catalyzes the methylation of cytoplasmically recapped RNAs. Nucleic Acids Res. 45, 10726–10739 10.1093/nar/gkx801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao, X., Xiang, S., Oh, C., Martin, C.E., Tong, L. and Kiledjian, M. (2010) Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467, 608–611 10.1038/nature09338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aregger, M., Kaskar, A., Varshney, D., Fernandez-Sanchez, M.E., Inesta-Vaquera, F.A., Weidlich, S.et al. (2016) CDK1-cyclin b1 activates RNMT, coordinating mRNA cap methylation with G1 phase transcription. Mol. Cell 61, 734–746 10.1016/j.molcel.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Culjkovic-Kraljacic, B., Skrabanek, L., Revuelta, M.V., Gasiorek, J., Cowling, V.H., Cerchietti, L.et al. (2020) The eukaryotic translation initiation factor eIF4E elevates steady-state m(7)G capping of coding and noncoding transcripts. Proc. Natl Acad. Sci. U.S.A. 117, 26773–26783 10.1073/pnas.2002360117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otsuka, Y., Kedersha, N.L. and Schoenberg, D.R. (2009) Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol. Cell. Biol. 29, 2155–2167 10.1128/MCB.01325-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborne, M.J., Volpon, L., Memarpoor-Yazdi, M., Pillay, S., Thambipillai, A., Czarnota, S.et al. (2022) Identification and characterization of the interaction between the methyl-7-guanosine cap maturation enzyme RNMT and the cap-binding protein eIF4E. J. Mol. Biol. 434, 167451 10.1016/j.jmb.2022.167451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahl, M.C., Will, C.L. and Luhrmann, R. (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 18.Wang, E. and Aifantis, I. (2020) RNA splicing and cancer. Trends Cancer 6, 631–644 10.1016/j.trecan.2020.04.011 [DOI] [PubMed] [Google Scholar]
- 19.Urbanski, L.M., Leclair, N. and Anczukow, O. (2018) Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 9, e1476 10.1002/wrna.1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan, M.R., Wellinger, R.J. and Laurent, B. (2021) Exploring the alternative splicing of long noncoding RNAs. Trends Genet. 37, 695–698 10.1016/j.tig.2021.03.010 [DOI] [PubMed] [Google Scholar]
- 21.Girard, C., Will, C.L., Peng, J., Makarov, E.M., Kastner, B., Lemm, I.et al. (2012) Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 3, 994 10.1038/ncomms1998 [DOI] [PubMed] [Google Scholar]
- 22.Vargas, D.Y., Shah, K., Batish, M., Levandoski, M., Sinha, S., Marras, S.A.et al. (2011) Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147, 1054–1065 10.1016/j.cell.2011.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan, Q., Shai, O., Lee, L.J., Frey, B.J. and Blencowe, B.J. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- 24.Bonnal, S.C., Lopez-Oreja, I. and Valcarcel, J. (2020) Roles and mechanisms of alternative splicing in cancer: implications for care. Nat. Rev. Clin. Oncol. 17, 457–474 10.1038/s41571-020-0350-x [DOI] [PubMed] [Google Scholar]
- 25.Saez, B., Walter, M.J. and Graubert, T.A. (2017) Splicing factor gene mutations in hematologic malignancies. Blood 129, 1260–1269 10.1182/blood-2016-10-692400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor, J. and Lee, S.C. (2019) Mutations in spliceosome genes and therapeutic opportunities in myeloid malignancies. Genes Chromosomes Cancer 58, 889–902 10.1002/gcc.22784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seiler, M., Peng, S., Agrawal, A.A., Palacino, J., Teng, T., Zhu, P.et al. (2018) Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296 10.1016/j.celrep.2018.01.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visconte, V., O Nakashima, M. and Rogers, H.J. (2019) Mutations in splicing factor genes in myeloid malignancies: significance and impact on clinical features. Cancers (Basel) 11, 1844 10.3390/cancers11121844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivera, O.D., Mallory, M.J., Quesnel-Vallieres, M., Chatrikhi, R., Schultz, D.C., Carroll, M.et al. (2021) Alternative splicing redefines landscape of commonly mutated genes in acute myeloid leukemia. Proc. Natl Acad. Sci. U.S.A. 118, e2014967118 10.1073/pnas.2014967118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papasaikas, P., Tejedor, J.R., Vigevani, L. and Valcarcel, J. (2015) Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell 57, 7–22 10.1016/j.molcel.2014.10.030 [DOI] [PubMed] [Google Scholar]
- 31.Ghram, M., Morris, G., Culjkovic-Kraljacic, B., Mars, J.C., Gendron, P., Skrabanek, L.et al. (2021) The eukaryotic translation initiation factor eIF4E reprogrammes the splicing machinery and drives alternative splicing. BioRxiv 10.1101/2021.12.07.471666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Effenberger, K.A., Urabe, V.K. and Jurica, M.S. (2017) Modulating splicing with small molecular inhibitors of the spliceosome. Wiley Interdiscip. Rev. RNA 8, 10.1002/wrna.1381 10.1002/wrna.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaida, D., Motoyoshi, H., Tashiro, E., Nojima, T., Hagiwara, M., Ishigami, K.et al. (2007) Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583 10.1038/nchembio.2007.18 [DOI] [PubMed] [Google Scholar]
- 34.Fan, L., Lagisetti, C., Edwards, C.C., Webb, T.R. and Potter, P.M. (2011) Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 6, 582–589 10.1021/cb100356k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eskens, F.A., Ramos, F.J., Burger, H., O'Brien, J.P., Piera, A., de Jonge, M.J.et al. (2013) Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin. Cancer Res. 19, 6296–6304 10.1158/1078-0432.CCR-13-0485 [DOI] [PubMed] [Google Scholar]
- 36.Hong, D.S., Kurzrock, R., Naing, A., Wheler, J.J., Falchook, G.S., Schiffman, J.S.et al. (2014) A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest. New Drugs 32, 436–444 10.1007/s10637-013-0046-5 [DOI] [PubMed] [Google Scholar]
- 37.Hua, Y., Vickers, T.A., Okunola, H.L., Bennett, C.F. and Krainer, A.R. (2008) Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 82, 834–848 10.1016/j.ajhg.2008.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curinha, A., Oliveira Braz, S., Pereira-Castro, I., Cruz, A. and Moreira, A. (2014) Implications of polyadenylation in health and disease. Nucleus 5, 508–519 10.4161/nucl.36360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gruber, A.R., Martin, G., Keller, W. and Zavolan, M. (2014) Means to an end: mechanisms of alternative polyadenylation of messenger RNA precursors. Wiley Interdiscip Rev. RNA 5, 183–196 10.1002/wrna.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian, B. and Manley, J.L. (2017) Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenal, M., Elkon, R., Loayza-Puch, F., van Haaften, G., Kuhn, U., Menzies, F.M.et al. (2012) The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553 10.1016/j.cell.2012.03.022 [DOI] [PubMed] [Google Scholar]
- 42.Chang, J.W., Zhang, W., Yeh, H.S., de Jong, E.P., Jun, S., Kim, K.H.et al. (2015) mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat. Commun. 6, 7218 10.1038/ncomms8218 [DOI] [PubMed] [Google Scholar]
- 43.Mayr, C. and Bartel, D.P. (2009) Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lianoglou, S., Garg, V., Yang, J.L., Leslie, C.S. and Mayr, C. (2013) Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396 10.1101/gad.229328.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris, A.R., Bos, A., Diosdado, B., Rooijers, K., Elkon, R., Bolijn, A.S.et al. (2012) Alternative cleavage and polyadenylation during colorectal cancer development. Clin. Cancer Res. 18, 5256–5266 10.1158/1078-0432.CCR-12-0543 [DOI] [PubMed] [Google Scholar]
- 46.Lackford, B., Yao, C., Charles, G.M., Weng, L., Zheng, X., Choi, E.A.et al. (2014) Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 33, 878–889 10.1002/embj.201386537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis, M.R., Delaleau, M. and Borden, K.L.B. (2019) Nuclear eIF4E stimulates 3′-end cleavage of target RNAs. Cell Rep. 27, 1397–1408 10.1016/j.celrep.2019.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wegener, M. and Muller-McNicoll, M. (2018) Nuclear retention of mRNAs: quality control, gene regulation and human disease. Semin. Cell Dev. Biol. 79, 131–142 10.1016/j.semcdb.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 49.Borden, K.L.B. (2020) The nuclear pore complex and mRNA export in cancer. Cancers (Basel) 13, 42 10.3390/cancers13010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Culjkovic-Kraljacic, B. and Borden, K.L. (2013) Aiding and abetting cancer: mRNA export and the nuclear pore. Trends in cell biology. 23, 328–335 10.1016/j.tcb.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Natalizio, B.J. and Wente, S.R. (2013) Postage for the messenger: designating routes for nuclear mRNA export. Trends Cell Biol. 23, 365–373 10.1016/j.tcb.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volpon, L., Culjkovic-Kraljacic, B., Sohn, H.S., Blanchet-Cohen, A., Osborne, M.J. and Borden, K.L.B. (2017) A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA 23, 927–937 10.1261/rna.060137.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Culjkovic, B., Topisirovic, I., Skrabanek, L., Ruiz-Gutierrez, M. and Borden, K.L. (2005) eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3′UTR. J. Cell Biol. 169, 245–256 10.1083/jcb.200501019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Culjkovic, B., Topisirovic, I., Skrabanek, L., Ruiz-Gutierrez, M. and Borden, K.L. (2006) eIF4E is a central node of an RNA regulon that governs cellular proliferation. J. Cell Biol. 175, 415–426 10.1083/jcb.200607020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Topisirovic, I., Siddiqui, N., Lapointe, V.L., Trost, M., Thibault, P., Bangeranye, C.et al. (2009) Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 28, 1087–1098 10.1038/emboj.2009.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zahreddine, H.A., Culjkovic-Kraljacic, B., Emond, A., Pettersson, F., Midura, R., Lauer, M.et al. (2017) The eukaryotic translation initiation factor eIF4E. eLife 6, e29830 10.7554/eLife.29830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun, Q., Chen, X., Zhou, Q., Burstein, E., Yang, S. and Jia, D. (2016) Inhibiting cancer cell hallmark features through nuclear export inhibition. Signal Transduct. Target. Ther. 1, 16010 10.1038/sigtrans.2016.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borden, K.L.B. and Volpon, L. (2020) The diversity, plasticity, and adaptability of cap-dependent translation initiation and the associated machinery. RNA Biol. 17, 1239–1251 10.1080/15476286.2020.1766179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rousseau, D., Kaspar, R., Rosenwald, I., Gehrke, L. and Sonenberg, N. (1996) Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl Acad. Sci. U.S.A. 93, 1065–1070 10.1073/pnas.93.3.1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garre, E., Romero-Santacreu, L., De Clercq, N., Blasco-Angulo, N., Sunnerhagen, P. and Alepuz, P. (2012) Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol. Biol. Cell 23, 137–150 10.1091/mbc.E11-05-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park, Y., Park, J., Hwang, J., Kim, L., Jeong, K., Song, H.K.et al. (2021) Translation mediated by the nuclear cap-binding complex is confined to the perinuclear region via a CTIF–DDX19B interaction. Nucleic Acids Res. 49, 8261–8276 10.1093/nar/gkab579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rambout, X. and Maquat, L.E. (2020) The nuclear cap-binding complex as choreographer of gene transcription and pre-mRNA processing. Genes Dev. 34, 1113–1127 10.1101/gad.339986.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de la Parra, C., Ernlund, A., Alard, A., Ruggles, K., Ueberheide, B. and Schneider, R.J. (2018) A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 9, 3068 10.1038/s41467-018-05539-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lahr, R.M., Fonseca, B.D., Ciotti, G.E., Al-Ashtal, H.A., Jia, J.J., Niklaus, M.R.et al. (2017) La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife 6, e24146 10.7554/eLife.24146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bukhari, S.I.A., Truesdell, S.S., Lee, S., Kollu, S., Classon, A., Boukhali, M.et al. (2016) A specialized mechanism of translation mediated by FXR1a-Associated microRNP in cellular quiescence. Mol. Cell 61, 760–773 10.1016/j.molcel.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Culjkovic-Kraljacic, B., Fernando, T.M., Marullo, R., Calvo-Vidal, N., Verma, A., Yang, S.et al. (2016) Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood 127, 858–868 10.1182/blood-2015-05-645069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Topisirovic, I., Guzman, M.L., McConnell, M.J., Licht, J.D., Culjkovic, B., Neering, S.J.et al. (2003) Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol. Cell. Biol. 23, 8992–9002 10.1128/MCB.23.24.8992-9002.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Papadopoulos, E., Jenni, S., Kabha, E., Takrouri, K.J., Yi, T., Salvi, N.et al. (2014) Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. Proc. Natl Acad. Sci. U.S.A. 111, E3187–E3195 10.1073/pnas.1410250111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joshi, S. and Platanias, L.C. (2015) Mnk kinases in cytokine signaling and regulation of cytokine responses. Biomol. Concepts 6, 85 10.1515/bmc-2011-2000 [DOI] [PubMed] [Google Scholar]
- 70.Graff, J.R., Konicek, B.W., Vincent, T.M., Lynch, R.L., Monteith, D., Weir, S.N.et al. (2007) Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 117, 2638–2648 10.1172/JCI32044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kentsis, A., Topisirovic, I., Culjkovic, B., Shao, L. and Borden, K.L. (2004) Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc. Natl Acad. Sci. U.S.A. 101, 18105–18110 10.1073/pnas.0406927102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kentsis, A., Volpon, L., Topisirovic, I., Soll, C.E., Culjkovic, B., Shao, L.et al. (2005) Further evidence that ribavirin interacts with eIF4E. RNA 11, 1762–1766 10.1261/rna.2238705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Topisirovic, I., Ruiz-Gutierrez, M. and Borden, K.L. (2004) Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 64, 8639–8642 10.1158/0008-5472.CAN-04-2677 [DOI] [PubMed] [Google Scholar]
- 74.McKendrick, L., Morley, S.J., Pain, V.M., Jagus, R. and Joshi, B. (2001) Phosphorylation of eukaryotic initiation factor 4E (eIF4E) at Ser209 is not required for protein synthesis in vitro and in vivo. Eur. J. Biochem. 268, 5375–5385 10.1046/j.0014-2956.2001.02478.x [DOI] [PubMed] [Google Scholar]
- 75.Morley, S.J. and Naegele, S. (2002) Phosphorylation of eukaryotic initiation factor (eIF) 4E is not required for de novo protein synthesis following recovery from hypertonic stress in human kidney cells. J. Biol. Chem. 277, 32855–9 10.1074/jbc.C200376200 [DOI] [PubMed] [Google Scholar]
- 76.Assouline, S., Culjkovic, B., Cocolakis, E., Rousseau, C., Beslu, N., Amri, A.et al. (2009) Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood 114, 257–260 10.1182/blood-2009-02-205153 [DOI] [PubMed] [Google Scholar]
- 77.Volpon, L., Osborne, M.J., Zahreddine, H., Romeo, A.A. and Borden, K.L. (2013) Conformational changes induced in the eukaryotic translation initiation factor eIF4E by a clinically relevant inhibitor, ribavirin triphosphate. Biochem. Biophys. Res. Commun. 434, 614–619 10.1016/j.bbrc.2013.03.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zahreddine, H.A., Culjkovic-Kraljacic, B., Assouline, S., Gendron, P., Romeo, A.A., Morris, S.J.et al. (2014) The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature 511, 90–93 10.1038/nature13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pettersson, F., Yau, C., Dobocan, M.C., Culjkovic-Kraljacic, B., Retrouvey, H., Puckett, R.et al. (2011) Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin. Cancer Res. 17, 2874–2884 10.1158/1078-0432.CCR-10-2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urtishak, K.A., Wang, L.S., Culjkovic-Kraljacic, B., Davenport, J.W., Porazzi, P., Vincent, T.L.et al. (2019) Targeting EIF4E signaling with ribavirin in infant acute lymphoblastic leukemia. Oncogene 38, 2241–2262 10.1038/s41388-018-0567-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettersson, F., del Rincon, S.V., Emond, A., Huor, B., Ngan, E., Ng, J.et al. (2015) Genetic and pharmacolgic inhibition of eIF4E reduces breast cancer cell migration, invasion and metastasis. Cancer Res. 75, 1102–1112 10.1158/0008-5472.CAN-14-1996 [DOI] [PubMed] [Google Scholar]
- 82.Bollmann, F., Fechir, K., Nowag, S., Koch, K., Art, J., Kleinert, H.et al. (2013) Human inducible nitric oxide synthase (iNOS) expression depends on chromosome region maintenance 1 (CRM1)- and eukaryotic translation initiation factor 4E (elF4E)-mediated nucleocytoplasmic mRNA transport. Nitric Oxide 30, 49–59 10.1016/j.niox.2013.02.083 [DOI] [PubMed] [Google Scholar]
- 83.Jin, J., Xiang, W., Wu, S., Wang, M., Xiao, M. and Deng, A. (2019) Targeting eIF4E signaling with ribavirin as a sensitizing strategy for ovarian cancer. Biochem. Biophys. Res. Commun. 510, 580–586 10.1016/j.bbrc.2019.01.117 [DOI] [PubMed] [Google Scholar]
- 84.Attar-Schneider, O., Drucker, L. and Gottfried, M. (2016) Migration and epithelial-to-mesenchymal transition of lung cancer can be targeted via translation initiation factors eIF4E and eIF4GI. Lab Invest. 96, 1004–1015 10.1038/labinvest.2016.77 [DOI] [PubMed] [Google Scholar]
- 85.Assouline, S., Culjkovic-Kraljacic, B., Bergeron, J., Caplan, S., Cocolakis, E., Lambert, C.et al. (2015) A phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia with elevated eIF4E. Haematologica 100, e7–e9 10.3324/haematol.2014.111245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hong, D.S., Kurzrock, R., Oh, Y., Wheler, J., Naing, A., Brail, L.et al. (2011) A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin. Cancer Res. 17, 6582–6591 10.1158/1078-0432.CCR-11-0430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yi, T., Kabha, E., Papadopoulos, E. and Wagner, G. (2014) 4EGI-1 targets breast cancer stem cells by selective inhibition of translation that persists in CSC maintenance, proliferation and metastasis. Oncotarget 5, 6028–6037 10.18632/oncotarget.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El-Brolosy, M.A. and Stainier, D.Y.R. (2017) Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 13, e1006780 10.1371/journal.pgen.1006780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Misra, S., Hascall, V.C., Markwald, R.R. and Ghatak, S. (2015) Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front Immunol. 6, 201 10.3389/fimmu.2015.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]