Abstract
The genetic basis for the host adaptation of Salmonella serotypes is currently unknown. We have explored a new strategy to identify Salmonella enterica serotype Typhimurium (S. typhimurium) genes involved in host adaptation, by comparing the virulence of 260 randomly generated signature-tagged mutants during the oral infection of mice and calves. This screen identified four mutants, which were defective for colonization of only one of the two host species tested. One mutant, which only displayed a colonization defect during the infection of mice, was further characterized. During competitive infection experiments performed with the S. typhimurium wild type, the mutant was defective for colonization of murine Peyer's patches but colonized bovine Peyer's patches at the wild-type level. No difference in virulence between wild type and mutant was observed when calves were infected orally with 1010 CFU/animal. In contrast, the mutant possessed a sixfold increase in 50% lethal morbidity dose when mice were infected orally. The transposon in this mutant was inserted in a 2.9-kb pathogenicity islet, which is located between uvrB and yphK on the S. typhimurium chromosome. This pathogenicity islet contained a single gene, termed slrP, with homology to ipaH of Shigella flexneri and yopM of Yersinia pestis. These data show that comparative screening of signature-tagged mutants in two animal species can be used for scanning the S. typhimurium genome for genes involved in host adaptation.
Salmonella serotypes differ greatly with regard to host range and degree of host adaptation (8). Host-restricted serotypes have adapted to a small number of animal species where they are often associated with disease in all age groups. The prototypical host-restricted serotype is Salmonella enterica serotype Typhi (S. typhi), which causes disease only in humans and higher primates. However, for most other serotypes that display host specificity, their host range appears to be wider than simply a single vertebrate genus or related genera. For instance, S. enterica serotype Dublin (S. dublin), a bovine-adapted serotype, is associated with disease in calves and adult cattle but causes in addition a considerable number of disease incidents in sheep and pigs (54, 55). Broad-host-range serotypes, on the other hand, are able to infect a wide spectrum of animal species and are more frequently associated with disease in young animals than in adults. S. enterica serotype Typhimurium (S. typhimurium) is a typical broad-host-range pathogen which is among the serotypes most frequently associated with disease in a number of animal species, including humans, cattle, pigs, horses, poultry, rodents, and sheep (19, 53–55, 62). While epidemiological surveys have established the host range of Salmonella serotypes, little is known about the virulence factors responsible for this phenotype.
One approach to identify the virulence mechanisms involved in host adaptation is to compare serotypes, which differ with regard to host range during experimental infections of animals or in tissue culture models. This approach revealed that S. typhi is capable of invading M cells in murine Peyer's patches, but unlike S. typhimurium, it does not destroy these cells (43). Furthermore, S. typhi is unable to grow and survive in the hepatic and splenic tissue of mice, the major site of S. typhimurium multiplication in this host species (6). Differences between S. typhi and S. typhimurium are also apparent when entry, survival, or intracellular trafficking of these serotypes in murine macrophages in vitro is compared (1, 29, 43). However, which virulence factors may contribute to these host-restricted phenotypes remains an enigma.
To identify the genes required for host adaptation to mice, attempts were made to isolate fragments of S. typhimurium genomic DNA which, when introduced into a host-restricted serotype, would confer mouse virulence. One candidate for such a murine virulence factor is the spv operon, which is absent from S. typhi but present in all Salmonella serotypes capable of causing lethal infection in mice (47, 61). However, introduction of the spv operon into S. typhi does not confer mouse virulence to this host-restricted pathogen (47). Similarly, introduction of an S. typhimurium cosmid library into S. enterica serotype Gallinarum (S. gallinarum) does not convert this avian-adapted serotype into a mouse pathogen (42). A possible reason why host range factors have not been identified with this strategy is that adaptation to an animal species is a complex phenotype, involving a number of genes that may map to different locations on the chromosome. Thus, it may not be possible to transfer all missing host range factors by introducing a single cosmid from a gene bank. The finding that the genomes of different Salmonella serotypes differ substantially from each other seems to support this idea. For instance, based on subtractive hybridization analysis, it has been estimated that 20% of the S. typhimurium genome (approximately 900 kb) is not present in S. typhi (33).
These examples illustrate that the considerable amount of serotype-specific genetic material complicates the identification of an individual host range factor when two different Salmonella serotypes are compared. We have therefore chosen to explore an alternate strategy to identify host range factors. Instead of analyzing two serotypes with a single infection model, we compared the virulence of S. typhimurium mutants in two different host species. The S. typhimurium genome was sampled by generating a random bank of signature-tagged transposon mutants, each of which was tested for virulence in mice and cows. We reasoned that murine or bovine host range factors could be identified in this screen, because they are expected to be required for the colonization of only one of these animal species. This analysis provides a first insight into the genetic basis of the host adaptation of S. typhimurium.
MATERIALS AND METHODS
Construction and analysis of mutants.
A bank comprising 260 mutants of S. typhimurium IR715 (56), a nalidixic acid-resistant derivative of ATCC 14028, was generated by performing 260 individual conjugations with an Escherichia coli bank containing 10,000 uniquely tagged Tn5 derivatives carried on suicide plasmids (28). Auxotrophs were identified by streaking individual mutants on minimal medium and subsequently analyzing the mutants unable to grow by auxanography. All attenuated mutants except STN166 (rfaJ) were backcrossed into the wild type (IR715) with bacteriophage P22 and tested for growth defects by inoculating Luria-Bertani (LB) broth with mutant and wild-type bacteria at a 1:1 ratio. The final ratio of wild type to mutant was determined after an 18-h incubation at 37°C. Data were normalized by dividing the final ratio by the ratio present in the inoculum.
Cloning and sequence analysis.
Transposon-flanking DNA was cloned by inverse PCR as described previously (7) with RsaI for the digestion of chromosomal DNA and the primer pair SIGN-10 (5′-GCCGAACTTGTGTATAAGAGTCAG-3′) and SIGN-11 (5′-AAAGGTAGCGTTGCCAATG-3′). The DNA was cloned into cloning vector pCR II (Invitrogen), and plasmid DNA for sequencing was isolated from E. coli DH5α (22) with ion-exchange columns from Qiagen.
In addition, larger DNA fragments flanking the transposon insertions in STN35 and STN39 were cloned as described below. The transposon insertion of mutant STN39 was cloned by ligating PstI-restricted genomic DNA into PstI-restricted cloning vector pBluescript SK(+) (51) and selecting for the kanamycin resistance marker of the mini-Tn5 Km2. The resulting plasmid was termed pSTN39. In the case of STN35, the inverse PCR product was labeled with the NEN labeling and detection kit (nonradioactive) to clone the corresponding DNA region from a cosmid bank. The cosmid bank of S. typhimurium ATCC 14028 constructed in pLAFR2 and propagated in E. coli LE392 has been described previously (37). The bank was spread on LB plates containing tetracycline, and 450 colonies were picked and grown individually overnight. Cosmid DNA was prepared from 15 pools, each containing 30 overnight cultures. Each pool was digested with EcoRI, separated on an agarose gel, and hybridized with the STN35 probe. The DNA of two pools hybridized with the STN35 probe. The 60 strains representing these two pools were then grown individually, and cosmid DNA was isolated, digested with EcoRI, and separated on an agarose gel. Southern hybridization was performed to identify the cosmids hybridizing with the STN35 probe. These cosmids were termed pSTN350 and pSTN351.
Nucleotide sequences were analyzed with the MacVector 6.0.1 software package (Oxford Molecular Group) and the Identify program (12a). Sequence homology was determined with the BLAST2 search algorithm at the National Center for Biotechnology Information (2).
Hybridization analysis.
The Salmonella reference collection B (the SARB collection) and the Salmonella reference collection C (the SARC collection) have been described previously (15, 16). Isolation of chromosomal DNA, Southern transfer, hybridization, and detection were performed as described previously (4). A DNA fragment corresponding to bp 647 to 1553 of GenBank no. AF127079 was labeled to obtain an slrP-specific probe. Furthermore, a 300-bp DNA fragment encoding the 3′ end of sspH1 and a 1.7-kb DNA fragment consisting of the 3′ fragment of sspH2 were labeled as DNA probes. Hybridization was performed at 65°C in solutions without formamide. Two 15-min washes were performed under nonstringent conditions at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate.
Tissue culture.
HEp-2 cells were obtained from the American Type Culture Collection and maintained in Eagle's minimal essential medium with Earle's balanced salt solution (BioWhittaker), 1 mM l-glutamine and 10% fetal calf serum. Invasion was assayed as described previously (9). Briefly, 10 μl of a standing overnight culture was added to each well of a 24-well microtiter plate containing a monolayer of HEp-2 cells. Bacteria were allowed to invade for 1 h, and then the wells were rinsed five times with 1 ml of phosphate-buffered saline (PBS). Cells were overlaid with fresh medium containing 50 μg of gentamicin per ml to kill extracellular bacteria. After 90 min, cells were rinsed three times with 1 ml of PBS, lysed in 0.1% Triton X-100, and plated to determine the number of intracellular bacteria. Each experiment was performed twice using triplicate wells for each strain. The significance of observed differences was calculated with Student's t test.
Animal experiments.
Milk-fed male Holstein-Friesian calves, aged 3 to 4 weeks, were obtained from a commercial dairy calf-rearing operation. Prior to their use for experiments, calves were screened for elevated leukocyte counts, fever, and Salmonella infection. Salmonella strains were detected in fecal swabs by enrichment in tetrathionate broth (Difco) and plating on Brilliant Green agar (BBL). In a pilot experiment, groups of two calves were infected orally with six doses, which ranged from 104 to 1011 CFU/animal, to estimate the 50% lethal morbidity dose (LD50) of S. typhimurium IR715 (45). Lethal morbidity was recorded at day 10 postinfection.
The bank was divided into 10 input pools of 24 to 30 mutants, and each pool was used for the intragastric inoculation of pairs of 6-week-old BALB/c mice and the oral inoculation of pairs of 3- to 4-week old Holstein-Friesian calves at a dose of 109 CFU/animal. Animals were cared for according to AAALAC guidelines. Mice developed signs of terminal disease starting at day 5 postinfection. At this point, Peyer's patches and spleens were consistently colonized with high numbers of S. typhimurium. Tissues from calves were collected at 3 to 4 days postinfection, since signs of lethal morbidity (anorexia or inability to stand) developed at this time. Infection of animals and euthanasia were performed as described previously (36). Otherwise, the signature-tagged mutant (STM) screen was performed as described by Hensel et al. (28). If a mutant was not recovered from the same organ (either Peyer's patches or spleen) of either both mice or both calves, it was considered to have a colonization defect. These mutants were characterized further.
During competitive infection experiments, groups of four calves or four mice were inoculated at a total dose of approximately 109 CFU/animal with a 1:1 mixture of wild-type and mutant bacteria. Serial 10-fold dilutions of the inoculum were spread on LB plates to determine the exact challenge dose/animal. At 4 (calves) or 5 (mice) days postinfection, the animals were euthanized. Tissues were collected, homogenized in PBS, and plated in the presence of the appropriate antibiotics for the enumeration of mutant and wild-type bacteria to determine the output ratio. Data were normalized by dividing the output ratio (wild-type CFU to mutant CFU) by the input ratio (wild-type CFU to mutant CFU). All data were then converted logarithmically for statistical analysis. Student's t test was used to determine whether the wild type/mutant ratio recovered from infected organs was significantly different from the wild type/mutant ratio present in the challenge inoculum. Since calves develop a localized infection, the spleen was colonized in only approximately half of the calves and therefore statistical analysis was not performed on data obtained from the colonization of this organ.
During single-infection experiments performed with STN39 and IR715, groups of four calves were infected orally at a dose of 0.95 × 1010 CFU/animal (STN39) and 1.4 × 1010 CFU/animal (IR715). In addition, groups of four mice were infected orally with serial 10-fold dilutions of STN39 or IR715. Mortality in mice was recorded 20 days postinfection and the LD50 value was calculated as described previously (45).
Nucleotide sequence accession number.
The nucleotide sequences of bcfC and slrP are deposited in GenBank under the accession no. AF129435 and AF127079, respectively.
RESULTS
STM screen in mice and calves.
The STM screen allows for the direct comparison of a mixed pool of mutants used to infect an animal (input pool) with a pool of mutants recovered from host tissue (output pool) after the infection was allowed to progress for an appropriate time (28). Attenuated mutants are present in the input pool but absent from the output pool. Identification of individual mutants within a pool is possible because each contains a different signature-tagged transposon carrying a unique DNA sequence tag of 20 random nucleotides. Southern hybridization of a blot containing genomic DNA of all individual mutants of the input pool, performed with a DNA probe generated by the PCR amplification of tags from the output pool, identifies mutants that are absent from the output pool.
A bank of 260 STMs was generated and the randomness of the mutagenesis was tested by screening for auxotrophy. Four auxotrophs were identified in the mutant bank, each deficient for biosynthesis of a different amino acid (methionine, cysteine, tryptophan, and arginine), which suggested that the mutagenesis was random.
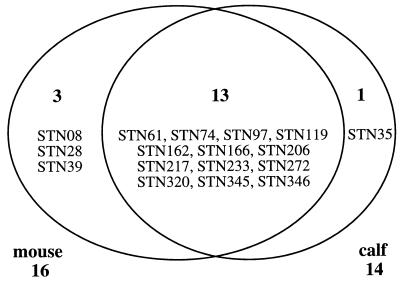
In a pilot experiment, the LD50 of S. typhimurium IR715 for calves was estimated to be 6 × 108 CFU/animal. No lethal morbidity was observed at doses of ≤1 × 108 CFU/animal. The bank of 260 mutants was divided into 10 pools. Each pool was used for the oral infection of a group of two mice and a group of two calves at a dose of 109 CFU/animal. Bacteria were recovered from the spleens and Peyer's patches of infected animals. Sixteen of the 260 mutants were not recovered from at least one of the output pools (spleen or Peyer's patches) of mice. Fourteen mutants were not recovered from at least one of the output pools (spleen or Peyer's patches) of infected calves. Comparison of the results from the STM screen in mice and calves revealed that 13 mutants were not recovered from the output pools of either animal (Fig. 1).
FIG. 1.
Results from the STM screen in mice and calves. A total of 260 mutants were screened for virulence in both mice and calves. Mutants not recovered from output pools of mice are shown in the left circle. Mutants not recovered from output pools of calves are shown in the right circle. The genotypes of these mutants are shown in Table 1.
In order to determine whether the reduced ability to compete with the wild type for organ colonization during the STM screen was caused by a general growth defect, all mutants missing from output pools were cocultured with the wild type (IR715) in LB broth. One mutant, STN74, was recovered in fourfold-lower numbers than the wild type after coculture (Table 1), suggesting that its failure to colonize organs may have resulted from a general growth defect. Cloning and sequence data supported this interpretation, since STN74 carried an insertion in ptsA, which encodes a putative enzyme I of the phosphoenolpyruvate-sugar phosphotransferase system and could be required for nutrient uptake (46).
TABLE 1.
Genotypes and characteristics of S. typhimurium signature-tagged transposon mutants
| Source and mutant | Sequence homology of transposon flanking DNA (organism) | Wild-type
CFU/mutant CFU determined after growth in:
|
||||
|---|---|---|---|---|---|---|
| LB broth | Murinea
|
Bovinea
|
||||
| Peyer's patchesb | Spleenb | Peyer's patchesb | Spleenc | |||
| Murine | ||||||
| STN08 | 0423b (E. coli) | 0.5 | 134.9* | 14.5* | 1.1 | 0.2 |
| STN28 | orf3 3′ to rcsF (E. coli) | 0.3 | 3.1 | 6.6* | 0.4 | 1.5 |
| STN39 | ipaH (S. flexneri) | 0.8 | 14.5* | 4.2 | 1.2 | 2.1 |
| Murine and bovine | ||||||
| STN61 | hilA (S. typhimurium) | 0.3 | 186.2* | 7.6* | 2,630.3* | 11,481.5 |
| STN74 | ptsA (E. coli) | 4.0 | 1.0 | 31.6* | 19.9* | 9.8 |
| STN97 | orgA (S. typhimurium) | 1.2 | 42.7* | 6.6* | NDd | ND |
| STN119 | spiB (S. typhimurium) | 1.0 | 14.8* | 301,995.7* | 67.6 | 32,359.4 |
| STN162 | prgH (S. typhimurium) | 1.4 | 69.2* | 46.8* | 363.1* | NRe |
| STN166 | rfaJ (E. coli) | 1.1 | 30.9* | 2,511.9* | 37.2* | 10.5 |
| STN206 | No homology | 1.0 | 0.6 | 31.6* | ND | ND |
| STN217 | spvB (S. typhimurium) | 0.7 | 17.4* | 426.6* | ND | ND |
| STN233 | ND | 0.1 | 3.2* | 12.9* | ND | ND |
| STN272 | spvR (S. typhimurium) | 1.3 | 0.04 | 13,489.6* | 4.0 | 5.5 |
| STN320 | yjeP (E. coli) | 0.1 | 12.9* | 239.9* | 13.8* | NR |
| STN345 | No homology | 0.7 | 8.1* | 10.0 | ND | ND |
| STN346 | selB (E. coli) | 0.4 | 5.0* | 3.0 | ND | ND |
| Bovine | ||||||
| STN35 | fimD-like (E. coli) | 0.5 | 6.5 | 26.3 | 3.4* | 1.5 |
Values are mean output ratios for groups of either four mice or four calves which were normalized to an input ratio of 1:1.
* denotes that the output ratio was significantly different (P < 0.05) from the input ratio.
Since bovine spleens were not consistently colonized, we did not obtain sufficient data points from this organ for statistical analysis with Student's t test.
ND, not determined.
NR, no bacteria recovered.
The STM screen provided qualitative data on the reduced recovery of individual mutants relative to the wild type but no information to quantify their colonization defect in an organ. Each mutant identified by the STM screen was therefore tested in a competitive infection assay to determine the magnitude of its colonization defect in Peyer's patches and spleens (Table 1). In these experiments, a mixed inoculum containing a 1:1 ratio of wild-type and mutant S. typhimurium was administered to groups of four mice. In addition, competitive infection experiments were performed for a subset of the mutants with groups of four calves. Statistical analysis was performed to determine whether the ratio of wild-type to mutant bacteria recovered from infected tissues differed significantly from the ratio of the inoculum. During mixed infection experiments, all 16 mutants which were not recovered from at least one output pool of mice in the STM screen were found to have a significant colonization defect for murine Peyer's patches and/or spleens (Fig. 1). Results from the STM screen in mice were thus in each case confirmed by detecting a statistically significant colonization defect during competitive infection experiments. Of these 16 mutants, 13 were either absent from at least one bovine output pool in the STM screen (Fig. 1) or exhibited a significant colonization defect detected during competitive infection experiments performed in calves (Table 1). Since the reduced ability of mutants STN97, STN206, STN217, STN233, STN345, and STN346 to colonize bovine spleen or Peyer's patches was not validated by performing competitive infection experiments, attenuation in calves should be considered putative for these strains. Three mutants, STN8, STN28, and STN39, were able to compete with the wild type for the colonization of bovine Peyer's patches and spleen (P > 0.05) but displayed a competitive colonization defect in murine Peyer's patches and/or spleen (P < 0.05) (Table 1). During the STM screen, one mutant (STN35) was not recovered from output pools of bovine Peyer's patches but was present in murine output pools (Fig. 1). During competitive infection experiments, this mutant (STN35) was recovered in reduced numbers from both murine and bovine tissues (Table 1). The colonization defect of STN35 for murine tissues was not statistically significant (P > 0.05), since the wild type/mutant ratios recovered from organs of different animals varied over a wide range, thereby resulting in a large standard deviation. In contrast, STN35 displayed a significant colonization defect for bovine Peyer's patches (P < 0.05).
Cloning and sequence analysis of transposon-flanking DNA.
Transposon-flanking DNA was cloned from 16 mutants (Table 1). Seven of the mutants defective for colonization of Peyer's patches and/or spleen of both host species carried insertions in genomic regions previously reported to be required for pathogenicity. These include Salmonella pathogenicity island 1, known as SPI-1 (mutants STN61, STN97, and STN162), SPI-2 (STN119), the rfa locus (STN166), and the spv operon (STN217 and STN272) (5, 17, 25, 30, 40, 44, 50). In STN35, the DNA flanking the transposon insertion site, which was cloned by inverse PCR, was used as a probe to clone the corresponding region from an S. typhimurium cosmid library. Sequence analysis revealed that the transposon was inserted in a 2,610-bp gene, termed bcfC. The deduced amino acid sequence of this gene was most similar to that of FimD (49% identity), the outer membrane usher of E. coli type 1 fimbriae, and that of SfaF (46% identity), the outer membrane usher of E. coli S fimbriae (31, 49).
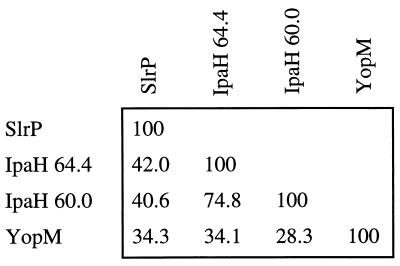
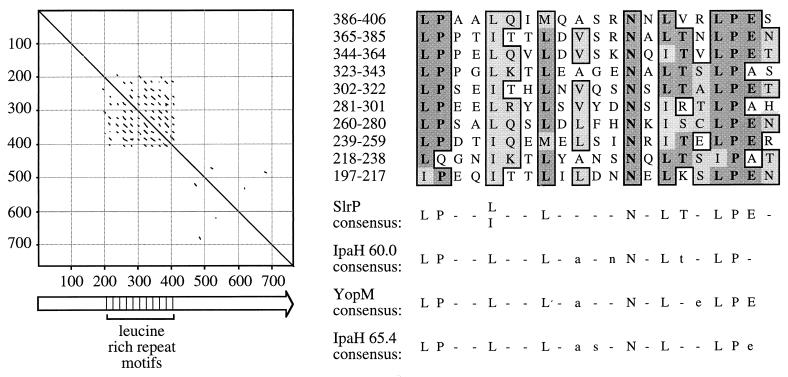
STN39 carried a transposon insertion 13 bp upstream of a 2,295-bp open reading frame with homology to yopM from Yersinia spp. and the ipaH genes from Shigella flexneri (Fig. 2) (13, 26, 35). The predicted protein sequence was inspected for protein motifs (3) with the Identify program. This analysis revealed the presence of 10 copies of a leucine-rich repeat signature, a protein motif involved in protein-protein interactions or the cellular adhesion of members of the leucine-rich glycoprotein (LRG) family (Fig. 3) (32). The S. typhimurium gene was thus termed slrP, for Salmonella leucine-rich repeat protein. The consensus sequence for the 21-amino-acid LRG-like repeat of SlrP was similar to that postulated for the 20-amino-acid LRG-like repeats found in IpaH 60.0, IpaH 65.4, and YopM (Fig. 3) (59).
FIG. 2.
Percent identity determined by pairwise alignment of amino acid sequences of SlrP, IpaH 60.0, IpaH 65.4, and YopM with the CLUSTAL program.
FIG. 3.
Pustell alignment of the SlrP amino acid sequence against itself (window size = 8; minimum % identity = 60%; hush value = 2) (left). Lines parallel to the diagonal identify direct amino acid repeats. The SlrP primary structure is shown as an arrow on the bottom (left). The positions of 10 copies of a 21-amino-acid repeat are indicated. A CLUSTAL alignment of the repeats is shown on the right (top). A comparison of the SlrP repeat consensus sequence with the consensus of 20-amino-acid repeats found in IpaH 60.0, IpaH 65.4, and YopM is shown on the right (bottom) (59). Capital letters in the consensus sequence indicate amino acids, which are conserved at this position in at least 50% of the repeats.
Characterization of STN39 for HEp-2 cell invasion and virulence in animal models.
Further analysis focused on STN39, since this mutant displayed a significant colonization defect in only one host species and carried a mutation in a DNA region with homology to virulence factors of enteric pathogens. To confirm that STN39 causes mortality at the wild-type level in cattle, groups of four calves were infected orally with either STN39 or IR715 (wild type) at a dose of approximately 1010 CFU/animal. All calves developed aqueous diarrhea with feces containing various combinations of blood, fibrin, and mucus. Terminal signs of illness, including anorexia and central nervous system depression, occurred between 1 and 3 days postinoculation. No evidence for attenuation was observed when the calves were infected at this dose, suggesting that the LD50 of STN39 is <3 × 109 CFU/animal. In contrast, an S. typhimurium prgH mutant (STN162) produces no mortality and no signs of disease in calves infected with 1010 CFU/animal (58). The possibility that the STN39 is only slightly (less than fivefold) attenuated was not ruled out, but no indications for reduced virulence were noted during competitive infection experiments (Table 1) or during single-infection studies in calves.
We next addressed the question whether STN39 is attenuated during the oral infection of mice. Groups of four mice were infected orally with serial 10-fold dilutions of STN39 or its isogenic parent (IR715). The LD50 determined for STN39 was 2.5 × 106 compared to an LD50 value of 4.4 × 105 determined for the wild type (IR715). Thus, STN39 is sixfold attenuated for mouse virulence after oral infection.
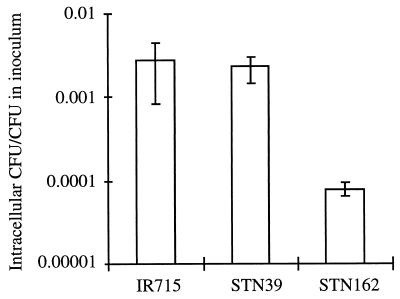
STN39 was recovered in 15-fold-lower numbers from murine Peyer's patches than its isogenic parent (P > 0.05) (Table 1) but did not display a colonization defect in spleens. The phenotype of STN39 was therefore similar to that reported for invA mutants, which exhibit reduced colonization of Peyer's patches but are able to colonize spleens when tested in competition with the wild type in mice (10, 20). Furthermore, the 6-fold increase in LD50 determined for STN39 during the infection of mice was similar to the 15-fold increase reported for an S. typhimurium ATCC 14028 derivative carrying a mutation in the invA gene (10, 20). Since mutations in SPI-1, including those in invA and prgH, result in reduced invasiveness for cultured epithelial cells (11, 21), we investigated whether STN39 is defective for entry into HEp-2 cells. While S. typhimurium STN162 (prgH) was defective for the invasion of HEp-2 cells (P < 0.05), there was no significant difference between the wild type (IR715) and STN39 in their ability to enter this epithelial cell line (P > 0.1) (Fig. 4).
FIG. 4.
Invasiveness of different S. typhimurium strains for HEp-2 cells. The number of gentamicin-protected bacteria recovered after the lysis of tissue culture cells is given as a fraction of the total number of bacteria added to each well at the beginning of the assay. Each bar represents the mean from six wells ± standard deviation.
The slrP gene is located on a pathogenicity islet at 18.4 centisomes.
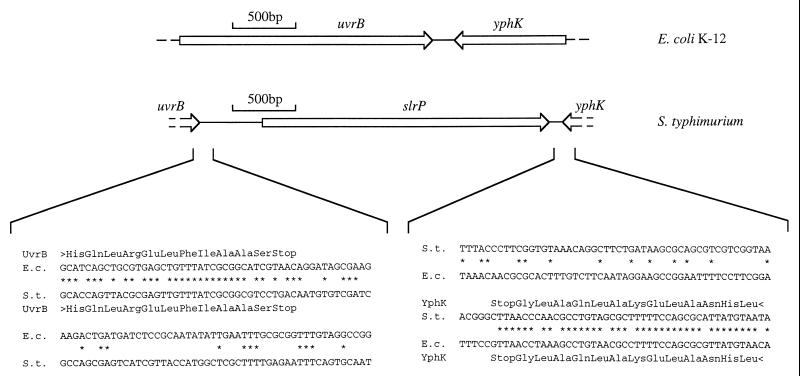
Further sequence analysis revealed that the slrP gene is located between uvrB and yphK, at 18.4 centisomes on the physical map of S. typhimurium (48). The slrP gene was located within a 2.9-kb DNA region with atypical G+C content. The G+C content of the S. typhimurium uvrB-yphK intergenic region averaged 45%, considerably lower than the overall G+C content of the S. typhimurium genome, which averages 52%. Previous studies have shown that only 4 of 87 regions sequenced from Salmonella serotypes have G+C contents of 45% or lower, and this has been taken as evidence for their acquisition by horizontal transfer (24, 52, 60). Consistent with its acquisition by horizontal transfer, the slrP gene was not present at the corresponding position in the E. coli K-12 genome. In E. coli K-12, uvrB and yphK are separated by only 152 bp (Fig. 5). Sequence homology between E. coli and S. typhimurium DNA ended immediately downstream of the stop codons of uvrB and yphK. If slrP was obtained by plasmid- or phage-mediated horizontal transfer, the mobile genetic element introducing this gene must have been lost subsequently by deletion, since no other genes were found in the uvrB-yphK intergenic region. This is consistent with the recent observation that E. coli, a close relative of S. typhimurium, acquires DNA horizontally at a rate of 16 kb/million years but subsequently loses the majority of this material by deletion (34). The DNA up- and downstream of slrP was therefore inspected for remnants of mobile genetic elements. No open reading frames were detected in the uvrB-slrP intergenic region. However, when translated, the DNA region located 250 to 400 bp upstream of slrP displayed homology (48% identity) to the N-terminal third of a transposase encoded by a gene (orf4) located on the enteroadherent factor plasmid of enteropathogenic E. coli (57). This DNA region may thus be the remnant of a mobile genetic element (possibly a plasmid), which may have introduced the slrP gene into the genome of an organism ancestral to S. typhimurium. Like other small DNA regions, which encode horizontally acquired virulence determinants (23), the uvrB-yphK intergenic region may thus be considered an S. typhimurium pathogenicity islet.
FIG. 5.
Comparison of the uvrB-yphK intergenic region of E. coli K-12 and S. typhimurium ATCC 14028 (top). Arrows indicate the positions of genes. Comparisons of nucleotide sequences from the left and right boundaries of the pathogenicity islet are shown on the bottom. E.c., E. coli; S.t., S. typhimurium.
Distribution of members of the leucine-rich repeat family among Salmonella serotypes.
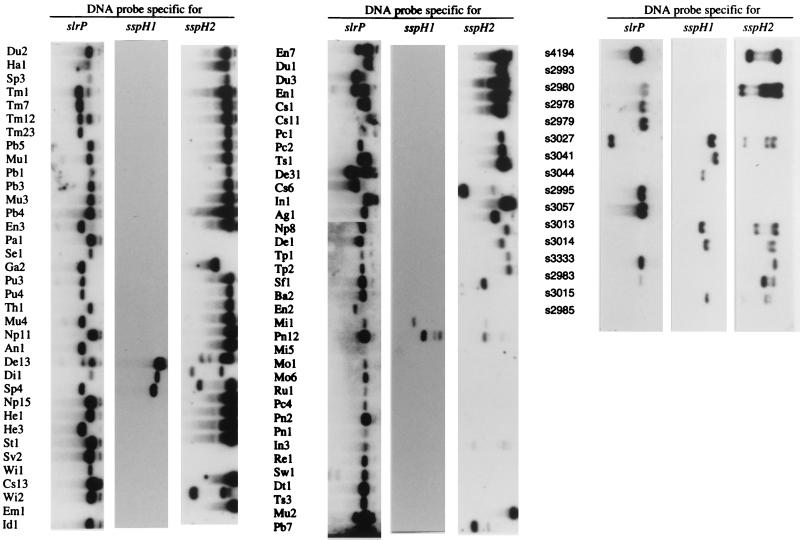
Recently, two genes with homology to slrP were identified in S. typhimurium and termed sspH1 and sspH2 (38). SspH1 and SspH2 are type III secreted proteins, which contain a leucine-rich repeat signature and share 42 and 41% amino acid identity with the deduced amino acid sequence of SlrP, respectively. To determine whether different combinations of members of the leucine-rich repeat family can be found among Salmonella serotypes, the distribution of slrP, sspH1, and sspH2 was determined among strains of two Salmonella reference collections. The SARC collection consists of 16 strains representing all phylogenetic lineages within the genus Salmonella, including Salmonella bongori and S. enterica subsp. I, II, IIIa, IIIb, IV, VI, and VII (16). The SARB collection consists of 72 strains representing 37 serotypes of S. enterica subsp. I (15). A number of different combinations of slrP, sspH1, and sspH2 were detected among Salmonella serotypes (Fig. 6). For instance, S. bongori serotypes hybridized only with the sspH1-specific DNA probe while S. enterica subspecies IIIb and VI and a number of subspecies I serotypes hybridized only with the slrP probe. Furthermore, one strain of S. enterica subsp. I (Em1) hybridized only with the sspH2-specific DNA probe and serotypes of S. enterica subsp. II did not hybridize with any DNA probe. Serotypes of S. enterica subsp. VII produced signals with the sspH1 and sspH2 probes but did not hybridize with the slrP probe. S. enterica subsp. IIIa and a number of S. enterica subsp. I serotypes produced signals with the slrP and sspH2 probes but did not hybridize with the sspH1 probe. Finally, all three probes hybridized with one serotype of S. enterica subsp. IV and five strains of S. enterica subsp. I (Sp4, De13, Di1, Mi1, and Pn12). This complex phylogenetic distribution of slrP, sspH1, and sspH2 suggests that extensive shuffling of these genes has occurred during the evolution of Salmonella serotypes.
FIG. 6.
Distribution of genes (slrP, sspH1, and sspH2) encoding leucine-rich repeat proteins among 16 SARC collection strains (right) and 72 SARB collection strains (left and center). The SARC collection contains serotypes of Salmonella bongori (s3041 and s3044) and of S. enterica subsp. I (s4194 and s3333), II (s2985 and s2993), IIIa (s2980 and s2983), IIIb (s2978 and s2979), IV (s3015 and s3027), VI (s2995 and s3057), and VII (s3013 and s3014). The SARB collection consists of 72 strains of S. enterica subsp. I. Ag, S. agona, An, S. anatum, Ba, S. brandenburg, Cs, S. choleraesuis, De, S. derby, Di, S. duisburg, Dt, S. decatur, Du, S. dublin, Em, S. emek, En, S. enteritidis, Ga, S. gallinarum, Ha, S. haifa, He, S. heidelberg, Id, S. indiana, In, S. infantis, Mi, S. miami, Mo, S. montevideo, Mu, S. muenchen, Np, S. newport, Pa, S. paratyphi A, Pb, S. paratyphi B, Pc, S. paratyphi C, Pn, S. panama, Pu, S. pullorum, Re, S. reading, Ru, S. rubislaw, Se, S. sendai, Sf, S. senftenberg, Sp, S. saintpaul, St, S. stanley, Sv, S. stanleyville, Sw, S. schwarzengrund, Th, S. thompson, Tm, S. typhimurium, Tp, S. typhi, Ts, S. typhisuis, Wi, S. wien.
DISCUSSION
The genus Salmonella contains a large number of serotypes, which are able to circulate in one or several species of domesticated animals. Since infections caused by Salmonella serotypes arise from the animal reservoir from which we draw our food supply, host adaptation to livestock or domestic fowl has a direct impact on human health. It is currently not known which virulence determinants are involved in the host adaptation of Salmonella serotypes. In this report, we show that S. typhimurium host range factors can be identified by comparative screening of STMs in two animal species, namely, mice and cows. Although the high cost of the calf model precludes its use for screening a large numbered bank, our sample of 10 pools of STMs provided a first sketch of the genetic makeup of a broad-host Salmonella serotype (Fig. 1).
We found that 13 S. typhimurium mutants (5% of mutants in the bank) were defective for colonization of both animal species (Fig. 1). Transposon-flanking DNA cloned from 12 of these mutants revealed that 7 insertions were located in known virulence determinants (Table 1). Four insertions were located in different genes located on either SPI-1 or SPI-2, two pathogenicity islands which together represent approximately 1.4% of the S. typhimurium genome (about 65 kb) (27, 39). This finding corresponds well with the prediction that a random bank of 260 transposon mutants is expected to contain, on average, 3.5 insertions in a 65-kb DNA region of the S. typhimurium genome. In addition to transposon insertions causing a colonization defect in both mice and calves, our screen identified 4 mutants (1.5% of mutants in the bank) which were missing from output pools of only one animal species (Fig. 1).
One mutant (STN39) which was present in output pools from calves but displayed a colonization defect in murine Peyer's patches was further characterized. The transposon insertion in STN39 was located in a 2.9-kb DNA region between uvrB and yphK on the S. typhimurium chromosome. Acquisition of this 2.9-kb DNA region by horizontal gene transfer was suggested by its absence from the uvrB-yphK intergenic region of E. coli (Fig. 5), its atypical G+C content (45%), and the remnant of a mobile genetic element identified by sequence homology. The uvrB-yphK intergenic region may thus be considered a pathogenicity islet, a term recently proposed for small DNA regions involved in virulence which have been obtained by horizontal gene transfer (23). Nucleotide sequence analysis revealed that the uvrB-yphK intergenic region contained a single open reading frame of 2,295 bp, which was termed slrP (Fig. 5).
Three lines of evidence supported the conclusion that STN39 displayed a virulence defect during interaction with the murine but not the bovine host. (i) During the STM screen, STN39 was missing from murine but not bovine output pools (Fig. 1). (ii) During competitive infection experiments, STN39 was recovered in significantly lower numbers than the wild type from murine but not bovine Peyer's patches (Table 1). (iii) STN39 was attenuated sixfold in mice but caused mortality at the wild-type level in calves infected with approximately 1010CFU/animal. The finding that STN39 was only modestly attenuated in mice may not be surprising, since our results suggest that a number of genes are required for host adaptation. Assuming that our bank represented a random sample of a genome containing about 4,400 genes, it can be extrapolated that S. typhimurium may encode as many as 50 virulence genes which are only required for the infection of a subset of the host species susceptible to this pathogen. Since each of these genes is expected to contribute to host adaptation, the inactivation of a single determinant may not result in marked attenuation.
Although there is currently no direct evidence for the expression of SlrP, the mutant phenotype of STN39 suggested a role for the slrP pathogenicity islet in host adaptation. The deduced amino acid sequence of slrP had homology with secreted targets of the type III export systems of S. flexneri (IpaH 60.0 and IpaH 65.4), Yersinia pestis (YopM), and S. typhimurium (SspH1 and SspH2) (13, 26, 35, 38). These type III secreted proteins have a leucine-rich repeat signature in common, a sequence motif involved in protein-protein interaction (32) (Fig. 3). Leucine-rich repeat domains have been implicated in the interaction between the products of plant disease resistance genes and those of avirulence (avr) genes of bacterial plant pathogens (12). The avr genes encode type III secreted proteins which are recognized by specific plant disease resistance proteins, thereby restricting the host range of the plant pathogen (14, 41). The avr gene compositions are highly variable, suggesting that alterations in the host range of plant pathogenic bacteria were mediated by horizontal gene transfer (18). Similarly, we found that the compositions of genes encoding leucine-rich repeat proteins vary greatly among Salmonella serotypes (Fig. 6). Whether the generation of new leucine-rich repeat gene combinations by horizontal gene transfer or deletion events was a mechanism to alter the host range of Salmonella serotypes remains to be determined.
We conclude that comparative screening of STMs in different animal species can be used to identify putative host range factors of S. typhimurium. Furthermore, this approach can easily be extended to include other host-pathogen combinations, thereby likely providing a more complete picture of host range factors present in S. typhimurium.
ACKNOWLEDGMENTS
The authors thank D. Holden for providing a plasmid bank of signature-tagged transposons and detailed protocols, C. Tanksley and T. Parsons for care of animals, R. Barthel and J.-A. Gutiérrez-Pabello for assistance with necropsies, J. R. Mock and P. C. Hong for assistance with competition assays, and R. A. Kingsley and M. Manson for critical comments on the manuscript.
Work in A.J.B.'s laboratory is supported by grant 9802610 from the U.S. Department of Agriculture, Public Health Service grants AI40124 and AI44170, and USDA Formula Animal Health Funding to T.A.F. and A.J.B. R.M.T. is supported by USDA/NRICGP fellowship 9702568.
REFERENCES
- 1.Alpuche-Aranda C M, Berthiaume E P, Mock B, Swanson J A, Miller S I. Spacious phagosome formation within mouse macrophages correlates with Salmonellaserotype pathogenicity and host susceptibility. Infect Immun. 1995;63:4456–4462. doi: 10.1128/iai.63.11.4456-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attwood T K, Beck M E, Bleasby A J, Degtyarenko K, Michie A D, Parry-Smith D J. Novel developments with the PRINTS protein fingerprint database. Nucleic Acids Res. 1997;25:212–217. doi: 10.1093/nar/25.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. J. New York, N.Y: Wiley & Sons; 1994. [Google Scholar]
- 5.Bajaj V, Hwang C, Lee C A. HilA is a novel OmpR/ToxR family member that activates the expression of Salmonella typhimuriuminvasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 6.Barrow P A, Huggins M B, Lovell M A. Host specificity of Salmonellainfection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect Immun. 1994;62:4602–4610. doi: 10.1128/iai.62.10.4602-4610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bäumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimuriumloci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bäumler A J, Tsolis R M, Ficht T A, Adams L G. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–4587. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäumler A J, Tsolis R M, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bäumler A J, Tsolis R M, Valentine P J, Ficht T A, Heffron F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimuriumto cause murine typhoid. Infect Immun. 1997;65:2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behlau I, Miller S J. A PhoP repressed gene promotes Salmonella typhimuriuminvasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Staskawicz B J. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 12a.Biochemistry Department, Stanford University 1 October 1999, revision date. EMOTIF Search. [Online.] http://dna.stanford.edu/identify. [18 October 1999, last date accessed.]
- 13.Boland A, Sory M P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocoliticais internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 14.Bonas U, Van den Ackerveken G. Gene-for-gene interactions: bacterial avirulence proteins specify plant disease resistance. Curr Opin Microbiol. 1999;2:94–98. doi: 10.1016/s1369-5274(99)80016-2. [DOI] [PubMed] [Google Scholar]
- 15.Boyd E F, Wang F-S, Beltran P, Plock S A, Nelson K, Selander R K. Salmonellareference collection B (SARB): strains of 37 serovars of subspecies I. J Gen Microbiol. 1993;139:1125–1132. doi: 10.1099/00221287-139-6-1125. [DOI] [PubMed] [Google Scholar]
- 16.Boyd E F, Wang F-S, Whittam T S, Selander R K. Molecular genetic relationships of the salmonellae. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carstenius P, Flock J I, Lindberg A. Nucleotide sequence of rfaI and rfaJ genes encoding lipopolysaccharide glycosyl transferases from Salmonella typhimurium. Nucleic Acids Res. 1990;18:6128. doi: 10.1093/nar/18.20.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collmer A. Determinants of pathogenicity and avirulence in plant pathogenic bacteria. Curr Opin Plant Biol. 1998;1:329–335. doi: 10.1016/1369-5266(88)80055-4. [DOI] [PubMed] [Google Scholar]
- 19.Edwards P R, Bruner D W. The occurrence and distribution of Salmonellatypes in the United States. J Infect Dis. 1943;72:58–67. doi: 10.1093/infdis/83.3.220. [DOI] [PubMed] [Google Scholar]
- 20.Galán J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimuriumto penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galán J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant S G N, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia colimethylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groisman E A, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 24.Groisman E A, Saier M H, Ochman H. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonellagenome. EMBO J. 1992;11:1309–1316. doi: 10.1002/j.1460-2075.1992.tb05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gulig P A, Caldwell A L, Chiodo V A. Identification, genetic analysis and DNA sequence of a 7.8-kb virulence region of the Salmonella typhimuriumvirulence plasmid. Mol Microbiol. 1992;6:1395–1411. doi: 10.1111/j.1365-2958.1992.tb00860.x. [DOI] [PubMed] [Google Scholar]
- 26.Hartman A B, Venkatesan M, Oaks E V, Buysse J M. Sequence and molecular characterization of a multicopy invasion plasmid antigen gene, ipaH, of Shigella flexneri. J Bacteriol. 1990;172:1905–1915. doi: 10.1128/jb.172.4.1905-1915.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hensel M, Shea J E, Bäumler A J, Gleeson C, Blattner F, Holden D W. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coliK-12. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 29.Ishibashi Y, Arai T. A possible mechanism for host-specific pathogenesis of Salmonellaserovars. Microb Pathog. 1996;21:435–446. doi: 10.1006/mpat.1996.0074. [DOI] [PubMed] [Google Scholar]
- 30.Jones B D, Falkow S. Identification and characterization of a Salmonella typhimuriumoxygen-regulated gene required for bacterial internalization. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klemm P, Christiansen G. The fimD gene required for cell surface localization of Escherichia colitype 1 fimbriae. Mol Gen Genet. 1990;220:334–338. doi: 10.1007/BF00260505. [DOI] [PubMed] [Google Scholar]
- 32.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 33.Lan R T, Reeves P R. Gene transfer is a major factor in bacterial evolution. Mol Biol Evol. 1996;13:47–55. doi: 10.1093/oxfordjournals.molbev.a025569. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence J G, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung K Y, Straley S C. The yopM gene of Yersinia pestisencodes a released protein having homology with the human platelet surface protein GPIb alpha. J Bacteriol. 1989;171:4623–4632. doi: 10.1128/jb.171.9.4623-4632.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Libby S J, Adams L G, Ficht T A, Allen C, Whitford H A, Buchmeier N A, Bossie S, Guiney D G. The spv genes of the Salmonella dublinvirulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect Immun. 1997;65:1786–1792. doi: 10.1128/iai.65.5.1786-1792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Libby S J, Goebel W, Ludwig A, Buchmeier N, Bowe F, Fang F C, Guiney D G, Songer J G, Heffron F. A cytolysin encoded by Salmonellais required for survival within macrophages. Proc Natl Acad Sci USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Bäumler, and S. I. Miller.Salmonella typhimurium leucine-rich repeat proteins are translocated by the SPI1 and SPI2 type III secretion systems. Mol. Microbiol., in press. [DOI] [PubMed]
- 39.Mills D M, Bajaj V, Lee C A. A 40kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coliK-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 40.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island for Salmonellasurvival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker J E, Coleman M J. Molecular intimacy between proteins specifying plant-pathogen recognition. Trends Biochem Sci. 1997;22:291–296. doi: 10.1016/s0968-0004(97)01089-x. [DOI] [PubMed] [Google Scholar]
- 42.Pascopella L, Falkow S, Small P L C. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Identification of a genetic determinant from Salmonella typhimurium that confers upon Salmonella gallinarum enhanced survival in the mouse, abstr. B-109; p. 173. [Google Scholar]
- 43.Pascopella L, Raupach B, Ghori N, Monack D, Falkow S, Small P L. Host restriction phenotypes of Salmonella typhi and Salmonella gallinarum. Infect Immun. 1995;63:4329–4335. doi: 10.1128/iai.63.11.4329-4335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimuriuminvasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 45.Reed L J, Muench H. A simple method for estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 46.Reizer J, Reizer A, Saier M H., Jr Novel phosphotransferase system genes revealed by bacterial genome analysis—a gene cluster encoding a unique enzyme I and the proteins of a fructose-like permease system. Microbiology. 1995;141:961–971. doi: 10.1099/13500872-141-4-961. [DOI] [PubMed] [Google Scholar]
- 47.Roudier C, Krause M, Fierer J, Guiney D G. Correlation between the presence of sequences homologous to the vir region of Salmonella dublin plasmid pSDL2 and the virulence of twenty-two Salmonellaserotypes in mice. Infect Immun. 1990;58:1180–1185. doi: 10.1128/iai.58.5.1180-1185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanderson K E, Hessel A, Rudd K E. Genetic map of Salmonella typhimurium, edition VIII. Microbiol Rev. 1995;59:241–303. doi: 10.1128/mr.59.2.241-303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmoll T, Morschhauser J, Ott M, Ludwig B, van Die I, Hacker J. Complete genetic organization and functional aspects of the Escherichia coli S fimbrial adhesion determinant: nucleotide sequence of the genes sfa B, C, D, E, F. Microb Pathog. 1990;9:331–343. doi: 10.1016/0882-4010(90)90067-z. [DOI] [PubMed] [Google Scholar]
- 50.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Short J M, Fernandez J M, Sorge J A, Huse W D. λZAP: a bacteriophage expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon M, Zieg J, Silverman M, Mandel G, Doolittle R. Phase variation: evolution of a controlling element. Science. 1980;209:1370–1374. doi: 10.1126/science.6251543. [DOI] [PubMed] [Google Scholar]
- 53.Sojka W J, Field H I. Salmonellosis in England and Wales 1958–1967. Vet Bull. 1970;40:515–531. [Google Scholar]
- 54.Sojka W J, Wray C, Hudson E B, Benson J A. Incidence of salmonella infection in animals in England and Wales, 1968–73. Vet Rec. 1975;96:280–284. [Google Scholar]
- 55.Sojka W J, Wray C, Shreeve J E, Bell J C. The incidence of salmonella infection in sheep in England and Wales, 1975 to 1981. Br Vet J. 1983;139:386–392. doi: 10.1016/s0007-1935(17)30383-4. [DOI] [PubMed] [Google Scholar]
- 56.Stojiljkovic I, Bäumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutHgene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tobe T, Schoolnik G K, Sohel I, Bustamante V H, Puente J L. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 58.Tsolis R M, Ficht T A, Adams L G, Bäumler A J. Contribution of Salmonella typhimuriumvirulence factors to diarrheal disease in calves. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venkatesan M M, Buysse J M, Hartman A B. Sequence variation in two ipaH genes of Shigella flexneri5 and homology to the LRG-like family of proteins. Mol Microbiol. 1991;5:2435–2445. doi: 10.1111/j.1365-2958.1991.tb02089.x. [DOI] [PubMed] [Google Scholar]
- 60.Verma N, Reeves P. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J Bacteriol. 1989;171:5694–5701. doi: 10.1128/jb.171.10.5694-5701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woodward M J, McLaren I, Wray C. Distribution of virulence plasmids within salmonellae. J Gen Microbiol. 1989;135:503–511. doi: 10.1099/00221287-135-3-503. [DOI] [PubMed] [Google Scholar]
- 62.Wray C, Sojka W J, Bell J C. Salmonellainfection in horses in England and Wales, 1973 to 1979. Vet Rec. 1981;109:398–401. doi: 10.1136/vr.109.18.398. [DOI] [PubMed] [Google Scholar]