Significance
Meiosis is a special type of cell division occurring in germ cells to produce sexual gametes. Initially, germ cells contain two copies of each chromosome, one from the mother and one from the father, which are called homologs. During meiosis, cells divide twice to produce haploid gametes with only one copy of each chromosome. Here, we show that in Drosophila males, maternal and paternal chromosomes become associated during the four rounds of mitosis preceding the entry into meiosis. We further show that cytoplasmic forces are required to move chromosomes within the nucleus. These results reveal unsuspected similarities and differences with Drosophila female meiosis.
Keywords: spermatogenesis, sexual dimorphism, Dynein, germline, cell cycle
Abstract
In the early stages of meiosis, maternal and paternal chromosomes pair with their homologous partner and recombine to ensure exchange of genetic information and proper segregation. These events can vary drastically between species and between males and females of the same species. In Drosophila, in contrast to females, males do not form synaptonemal complexes (SCs), do not recombine, and have no crossing over; yet, males are able to segregate their chromosomes properly. Here, we investigated the early steps of homolog pairing in Drosophila males. We found that homolog centromeres are not paired in germline stem cells (GSCs) and become paired in the mitotic region before meiotic entry, similarly to females. Surprisingly, male germline cells express SC proteins, which localize to centromeres and promote pairing. We further found that the SUN/KASH (LINC) complex and microtubules are required for homolog pairing as in females. Chromosome movements in males, however, are much slower than in females and we demonstrate that this slow dynamic is compensated in males by having longer cell cycles. In agreement, slowing down cell cycles was sufficient to rescue pairing-defective mutants in female meiosis. Our results demonstrate that although meiosis differs significantly between males and females, sex-specific cell cycle kinetics integrate similar molecular mechanisms to achieve proper centromere pairing.
Meiosis is a two-step cell division process that generates haploid gametes from diploid parental germ cells (1, 2). During the first stages, homologous chromosomes face the daunting task of finding the correct homolog in the nuclear space in order to recombine and exchange genetic information. This requires chromosomes to move, assess their homology, align along their length, and pair. Pairing is then stabilized by the assembly of the synaptonemal complex (SC), which synapses both homologs together (3). Double-strand breaks (DSBs) are formed on parental chromosomes and some of these DSBs are repaired with the homologous chromosome leading to the formation of crossovers (COs) and the reciprocal exchange of parental genetic information. The whole process is highly conserved in all sexually reproducing organisms from yeasts to humans. However, the underlying mechanisms allowing meiosis are remarkably diverse from one species to the next (4). For example, although chromosome movements to promote pairing is a common theme in many species, these movements are actin dependent in Saccharomyces cerevisiae, while they depend on microtubules in Schizosaccharomyces pombe (5). Chromosomes move individually in Caenorhabditis elegans, whereas they follow global nuclei rotations in mice and in Drosophila. Chromosomes are anchored to the nuclear membrane by centromeres in flies, but by telomeres in mice (6, 7). Interestingly, this diversity of strategies between species also extends within the same species between male and female germ cells (8).
Indeed, beyond the obvious cellular dimorphism of a large female oocyte and small male sperm, there are many sex-specific differences in the meiotic process per se. Chromosomal axis and SC are known to have different lengths in males and females of mice, worms, and plants. Similarly, the number of COs and the response to DNA damage are sex specific in many species (8). Drosophila males are an interesting case, as in contrast to females, they do not form a synaptonemal complex nor make any crossovers (9). However, males segregate their homologous chromosomes just fine. How males are able to associate their homologs at meiotic entry without SC and recombination is not known.
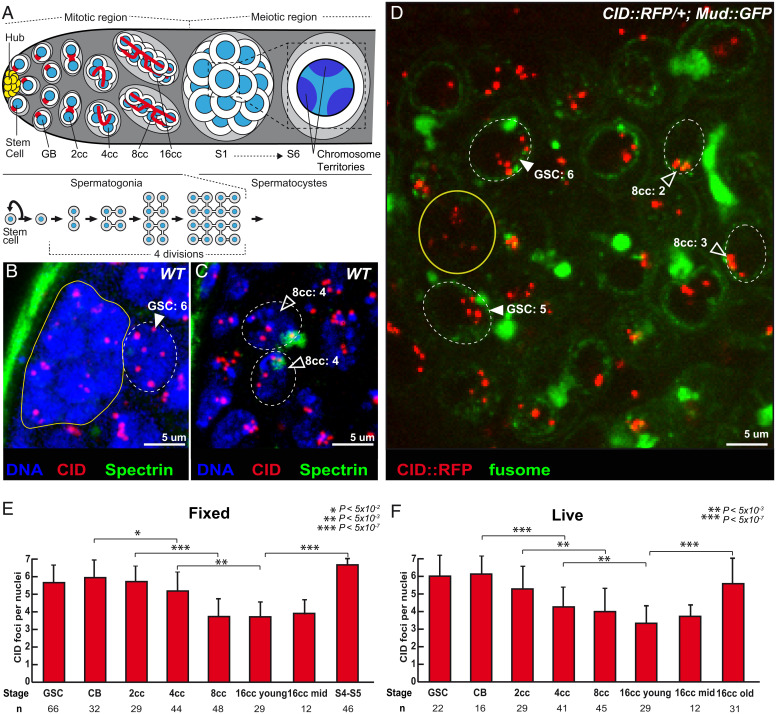
In male flies, meiosis starts at the anterior region of the testis (Fig. 1A). At the very tip, germline stem cells (GSCs) produce germ cells throughout adult life (10). GSCs divide asymmetrically to produce gonioblasts (GBs), which go through four rounds of mitosis with incomplete cytokinesis to produce germline cysts made of 16 spermatogonia. All 16 cells remain connected by ring canals and a germline-specific structure called the fusome, which is made of endoplasmic reticulum (ER)-derived vesicles. The branched shape of the fusome allows the identification of GSCs, GBs, and 2-, 4-, 8-, and 16-cell cysts (cc) in the mitotic region. These mitoses are followed by an extended G2 phase marked by a 25-fold increase in cell size. During this growth phase, homologous chromosomes are fully paired and then separate into chromosome territories (9). Pairing is then relaxed in these territories until chromosomes condense and form bivalents at metaphase I. Homologous chromosomes then segregate at anaphase I. At the spermatogonia–spermatocyte transition by the end of the mitotic region (Fig. 1A), homologous chromosomes are shown to be highly paired (11). It is, however, not known when and how they become paired. Here, we investigated whether the paired state is passively inherited from embryonic stages or whether there is de novo pairing in earlier stages of germ cell development in adults.
Fig. 1.
Unpaired centromeres in male germline stem cells become paired during cyst divisions. (A) Schematic overview of germ cell development in Drosophila testis. At the apical tip are the somatic hub cells (yellow), which serve as the niche for GSCs. GSCs divide asymmetrically giving rise to a GSC and a daughter cell, the GB, which undergoes four mitotic divisions by incomplete cytokinesis to produce interconnected 2-, 4-, 8-, and 16-cell spermatogonial cysts. Germ cells undergo an extended growth phase (S1 to S6) entering meiosis as primary spermatocytes. A zoom view of chromosome territories in one spermatocyte. The spectrosome (red circles) of GSCs and GB develops into a branched structure called the fusome (red) during each division. (B and C) Z-section projections of wild-type fixed testes stained for centromeres (CID, red), fusome (α-spectrin, green), and DNA (DAPI, blue). There are six centromeres in the GSC (B, arrowheads) but fewer in 8-cell cyst nuclei (C, open arrowheads), as quantified. (D) Z-section projection obtained by live imaging of a testis expressing the centromere marker CID::RFP (red) and the fusome marker MUD::GFP (green). Two GSCs (arrowheads) identified by their position next to the hub cells (yellow circle) and an 8-cell cyst (8cc), with cells linked by the fusome. Two nuclei of an 8-cell cyst (8cc, open arrowheads), with fusome GFP, are marked by dotted lines. (E and F) Graphs plot the number of CID foci for each developmental stage in the mitotic region of fixed (E) or live (F) wild-type testes. The number of analyzed cells is given below each stage. In B, C, and D, dotted lines surround germ cell nuclei. (Scale bars in B–D, 5 µm.) *P < 5 × 10−2, **P < 5 × 10−3, ***P < 5 × 10−7 (two-tailed Student’s t test).
We and others have shown in Drosophila females that although chromosomes start meiosis already paired in the oocyte, female GSCs have unpaired chromosomes that become paired de novo during the four mitotic divisions (12–14). We further have shown that homolog pairing requires extensive rotations of germ cell nuclei, and that these movements are driven by cytoplasmic microtubules (6). Microtubule forces are transmitted to the nuclear envelop and chromosomes by the SUN/KASH (LINC) complex encoded by the klaroid (koi) and klarsicht (klar) genes in Drosophila.
In this study, we investigated the pairing of homologous chromosomes in males before meiotic entry, i.e., in mitotic germ cells.
Results
Centromeres Are Unpaired in Male Germline Stem Cells and Become Progressively Associated in the Mitotic Region.
Germ cells were stained for α-spectrin, which marks the fusome and allows identification of GSCs and the different stages of cysts differentiation (Fig. 1A). To analyze chromosome organization, we labeled centromeres with an antibody against CID, the Drosophila homolog of Cenp-A. Drosophila diploid cells have eight chromosomes forming four pairs of homologs. If all homologs are paired, one should count four foci of CID, whereas if some chromosomes are unpaired, there should be more than four (15, 16). We found in GSCs and GBs an average of six dots of CID (Fig. 1 B and E), which indicated that some centromeres were unpaired at these stages. The number of CID dots went down to four, in 8- and 16-cell cysts, revealing that most chromosomes were associated at the end of the mitotic region (Fig. 1 C and E). In females, nonhomologous centromeres can cluster with those of homologous pairs to form one or two dots of CID (15, 16). In contrast, we found that in males, centromeres rarely clustered and remained in pairs (Fig. 1E). We further found that these pairs of centromeres and their corresponding telomeres were dispersed within the nucleus, indicating that chromosomes did not remain in the Rabl configuration between each division (Movie S1). Later during the growth phase in spermatocytes, when homologous chromosomes separate into different territories, the number of CID dots increased to six, as previously shown (Fig. 1E). We confirmed the results obtained on fixed testis by live imaging using a CID-RFP transgene to track centromeres and a fusome marker tagged with GFP (Fig. 1 D and F and Movies S2–S4).
We concluded that male centromeres are unpaired in GSCs and become de novo associated in pairs at the mitotic region similarly to females. However, they do not form stable nonhomologous clusters.
Centromeres of Chromosomes II and III Associate with Their Homologs in the Mitotic Region.
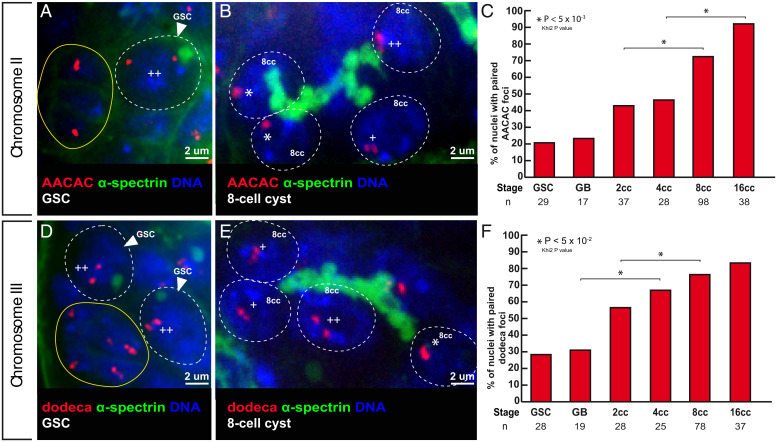
Next, we analyzed whether premeiotic pairing at centromeres occurred between homologous chromosomes by labeling pericentromeric repeated sequences specific for each chromosome. We performed fluorescent in situ hybridization (FISH) with the AACAC and dodeca probes, which identify pericentromeric regions of chromosomes II and III, respectively (Fig. 2 and SI Appendix, Fig. S1). We considered that chromosomes were paired when only one focus was visible or when two foci were visible separated by a distance of ≤0.70 μm (17, 18). With both probes, we found that less than 30% of chromosomes II and III were paired in GSCs, which went up to 90% (n = 38) and 80% (n = 37) for chromosomes II and III, respectively, in 16-cell cysts. We concluded that centromeres of chromosomes II and III become associated with their homologs in the mitotic region and at the meiotic entry.
Fig. 2.
Centromeres of chromosomes II and III become associated in the mitotic region. (A and B) Confocal Z-section projections of wild-type testes stained for chromosome II centromeres probe (AACAC, red), fusome (α-spectrin, green), and DNA (DAPI, blue). (A) GSC shows two foci separated by a distance >0.7 µm (++), thus unpaired; hub is marked in yellow. (B) The 8-cell cyst shows two nuclei with one focus (*), thus centromeres are paired and one nucleus with two foci (++) separated by a distance >0.7 µm and thus unpaired. (C) Graphs plot the percentage of paired chromosome II centromeres for each developmental stage in the mitotic region using the AACA probe. The number of cells analyzed is indicated under each stage.*P < 0.05 (khi2 test comparing 2cc with 8cc and 4cc with 16cc). (D and E) Confocal Z-section projections of wild-type testes stained for chromosome III centromere probe (dodeca, red), fusome (α-spectrin, green), and DNA (DAPI, blue). (D) Two GSCs with two foci separated by >0.7 mm (++), thus unpaired; hub is marked in yellow. (E) The 8-cell cyst shows a nucleus with one foci (*), thus paired; two nuclei with two foci (+) separated by a distance of ≤0.7 µm, thus paired; and one nucleus with two foci (++) separated by a distance of 0.7 µm, thus unpaired. (F) Graph plots percentage of paired chromosome III centromeres for each developmental stage in the mitotic region using the dodeca probe. The number of cells analyzed is indicated below each stage. In A, B, D, and E, dotted lines surround germ cell nuclei. (Scale bars, 2 µm.) *P < 0.005 (khi2 test comparing GB with 4cc and 2cc with 8cc).
Sex chromosomes X and Y share little homologous sequences outside the clusters of rDNA genes. The intergenic spacer (IGS) region located upstream of each rDNA repeat was shown to be necessary and sufficient for X and Y pairing (19). We used a probe labeling the IGS to investigate sex chromosome behavior. We found that it marked one unique but large subregion of the nucleus from GSCs to 16-cell cysts (SI Appendix, Fig. S1). This region was too broad to allow us to conclude on X/Y pairing (9). However, as in females, we counted between six and seven dots of centromeres in GSCs and not eight (Fig. 1E), indicating that some chromosomes were paired even in GSCs. In females, X chromosomes are already paired in GSCs (12, 13); it is thus possible, although it remains to be demonstrated, that in males the X and Y chromosomes are also paired in GSCs and cyst cells. Similarly, fourth chromosomes could also be paired in these cells.
Two Synaptonemal Complex Components Are Expressed in Male Germ Cells, Localize at Centromeres and Chromosome Arms, and Are Required for Efficient Pairing of Homologs.
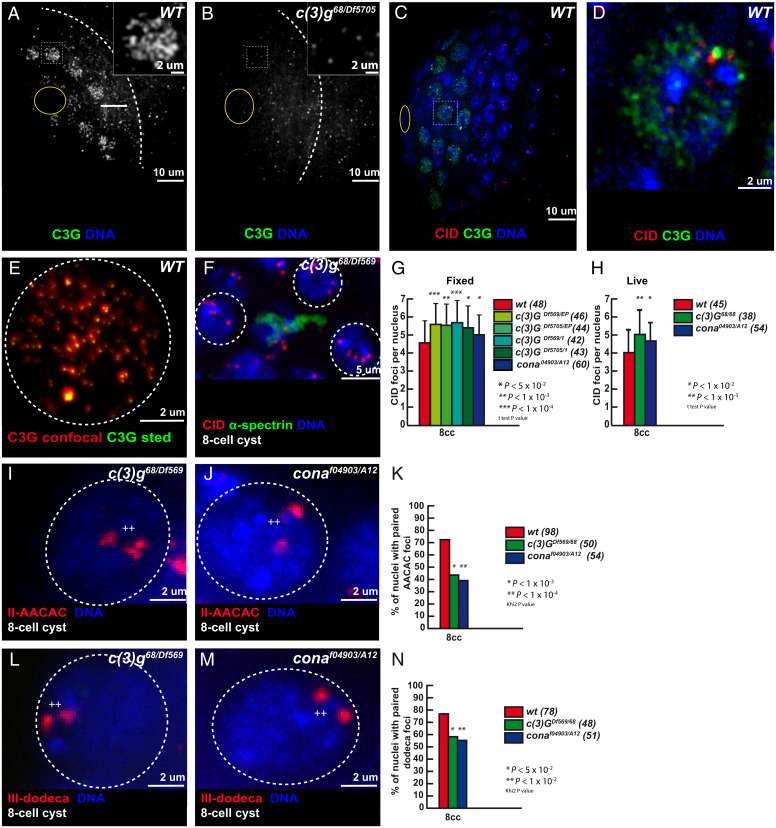
In Drosophila females, we showed that premeiotic pairing of autosome centromeres depends on components of the SC such as C(3)G and Corona (Cona), which localized at centromeres in the mitotic region (12). Interestingly, although no SC structure has ever been described in males, both the FlyAtlas2 and modENCODE datasets detected cona, and to a lesser extent c(3)G, expression in testis. We confirmed the expression of c(3)G and corona in testis by qRT-PCR (SI Appendix, Fig. S2A). We used antibodies against C(3)G and Cona to determine the localization of the endogenous proteins and found that both associated with chromosomes in the mitotic region (Fig. 3A and SI Appendix, Fig. S2 H–K). All signals disappeared completely in c(3)G mutant germ cells, demonstrating the antibody specificity (Fig. 3B). Superresolution microscopy revealed that C(3)G formed small foci thoughout the nucleus (Fig. 3E). The largest and brightest foci of C(3)G and Cona were associated with centromeres as in female germ cells (Fig. 3 C and D and SI Appendix, Fig. S2 D–K). We validated these findings with a tagged C(3)G-GFP and Corona-Venus (20) that colocalized with Cid-RFP by live imaging (SI Appendix, Fig. S2 B and C and Movies S5 and S6).
Fig. 3.
The synaptonemal complex component C(3)G is present in the mitotic region and promotes centromere association. (A and B) Confocal Z-section projections of wild-type (A) and c(3)G68/Df5705 (B) testes stained for C(3)G (green) and (DAPI, blue). Dotted white lines delimit the mitotic region; hub is marked by the yellow circle. (C and D) Confocal Z-section projections of wild-type testes stained for centromeres (CID, red), C(3)G (green), and DNA (blue); hub is marked by a yellow circle. Insets in A and B show magnified nuclei corresponding to outlined nuclei. Outlined nucleus is magnified in D. (Scale bars in A and B, 10 µm; Insets, 2 µm.) (E) Z-section projection acquired using Stimulated Emission Depletion Microscopy (STED) microscopy of a wild-type (WT) 2-cell cyst stained for C(3)G (red is confocal, green is STED). (Scale bar, 2 µm.) (F) Confocal Z-section projection of an eight-cell cyst from c(3)G68/Df(3R)BSC569 stained for centromere (CID, red), fusome (α-spectrin, green), and DNA (DAPI, blue). Three nuclei (dotted circles) show unpaired centromeres. (Scale bar, 5 µm.) (G and H) Graphs plot the number of CID foci of eight-cell cysts in fixed wild-type, Df(3R)BSC569/c(3)GEP, Df5705/c(3)GEP, Df(3R)BSC569/c(3)G1, Df5705/c(3)G1, and conaA12/cona04973 (G) and in live wild-type, c(3)G68/c(3)G68 and conaA12/cona04973 (H) testes. The number of analyzed cells is indicated next to each genotype. *P < 0.01, **P < 0.001, ***P < 0.0001, (two-tailed Student’s t test). (I and J) Confocal Z-section projections of c(3)G68/Df(3R)BSC569 (I) and conaA12/cona04973. (J) Eight-cell cyst nuclei stained for AACAC (red) and DNA. In both cases, nuclei (dotted circles) show two foci separated by >0.7 mm (++) indicating unpaired centromeres. (K) Graph plots the percentage of paired chromosomes III in eight-cell cysts using the AACAC probe in wild-type, c(3)G68/Df(3R)BSC569, and conaA12/cona04973 fixed testis. The number of analyzed cells is indicated next to each genotype. *P < 0.05, **P < 0.01 (khi2 test). (L and M) Confocal Z-section projections of c(3)G68/Df(3R)BSC569 (L) and conaA12/cona04973. (M) Eight-cell cyst nuclei stained for dodeca (red) and DNA. Both nuclei (dotted circles) show two foci separated by >0.7 µm (++), indicating unpaired centromeres. (Scale bars in I, J, L, and M, 2 µm.) (N) Graph plots the percentage of paired chromosome II in eight-cell cysts using the dodeca probe in wild-type, c(3)G68/Df(3R)BSC569, and conaA12/cona04973 fixed testes. The number of analyzed cells (n) is indicated next to each genotype. *P < 0.001, **P < 0.0001 (khi2 test). Chromosome II and III centromeres are considered paired when the distance between two foci is ≤0.7 µm.
To investigate c(3)G and cona function in male chromosome organization, we counted the number of CID foci in four different c(3)G mutant backgrounds and one cona mutant background at the eight-cell cyst stage. We found an average of 5.5 dots of CID, which was significantly higher than in wild-type germ cells (Fig. 3 F and G). We confirmed this difference by live imaging in c(3)G and in cona mutant cells (Fig. 3H). We next assessed the number of paired centromeres of chromosomes II and III by FISH. In c(3)G and cona mutant cells, less than 40% of chromosome II were paired and less than 50% for chromosome III compared to more than 70% in wild type eight-cell cysts (Fig. 3 I–N). We concluded that at least two SC components are expressed in the male mitotic region and are necessary for efficient pairing of homologous centromeres.
We next investigated whether these early pairing defects in c(3)G and cona mutant germ cells would affect the formation of chromosome territories later on during spermatogenesis. Firstly, by DNA staining, we did not observe abnormal chromosome territories in c(3)G mutants (SI Appendix, Fig. S3 A and B). Secondly, by FISH, we never observed the AACAC probe (chromosome II) and dodeca probe (chromosome III) signal both at the same time in the same territory either in wild-type (n = 34) or in c(3)G mutant conditions (n = 28) (SI Appendix, Fig. S3 C and D). We also measured the distance between chromosome II (AACAC) and chromosome III (dodeca) or the distance between the nearest AACAC and dodeca dots at S3 stage. We found that the distance between nonhomologous chromosome probes AACAC and dodeca was more than 10 times the distance between AACAC_AACAC or dodeca_dodeca both in wild-type and in c(3)G mutant conditions (SI Appendix, Fig. S3E). Finally, the distance between nonhomologous centromeres was not significantly different between wild type and c(3)G mutant conditions (SI Appendix, Fig. S3 C–E). These results indicated that the early pairing defects observed in c(3)G mutants do not affect the proper formation of chromosome territories.
Nevertheless, we tested whether these pairing defects induced chromosome segregation errors and/or reduced fertility. As previously observed by Gowen (21), we did not detect any nondisjunction of chromosome X or II in c(3)G and cona mutant males (SI Appendix, Table S1). Accordingly, analysis of c(3)G mutant spermatids did not reveal any cytological defects at the onion stage (SI Appendix, Fig. S4 A and B). We also tested for possible genetic interactions with genes known to be required for male chromosome segregation, such as mod(mdg4) (mnm), teflon, ord, and sunn. We did not find evidence of increased nondisjunction for both chromosomes X and II in single vs. double mutant combinations (SI Appendix, Table S2–S4). Furthermore, C(3)G localization at centromeres did not depend on sunn and ord (SI Appendix, Fig. S5). In addition, we looked for reduced fertility by using a sperm exhaustion assay (SEA) (Materials and Methods). We found that mutant males were as fertile as wild-type controls (SI Appendix, Fig. S4C) and also did not show increased cell death (SI Appendix, Fig. S4 D–F).
We concluded that C(3)G and Corona are required for premeiotic pairing, but not for chromosome segregation or fertility in males.
Dynamic Movements of Centromeres Depend on Microtubules and Dynein Motor.
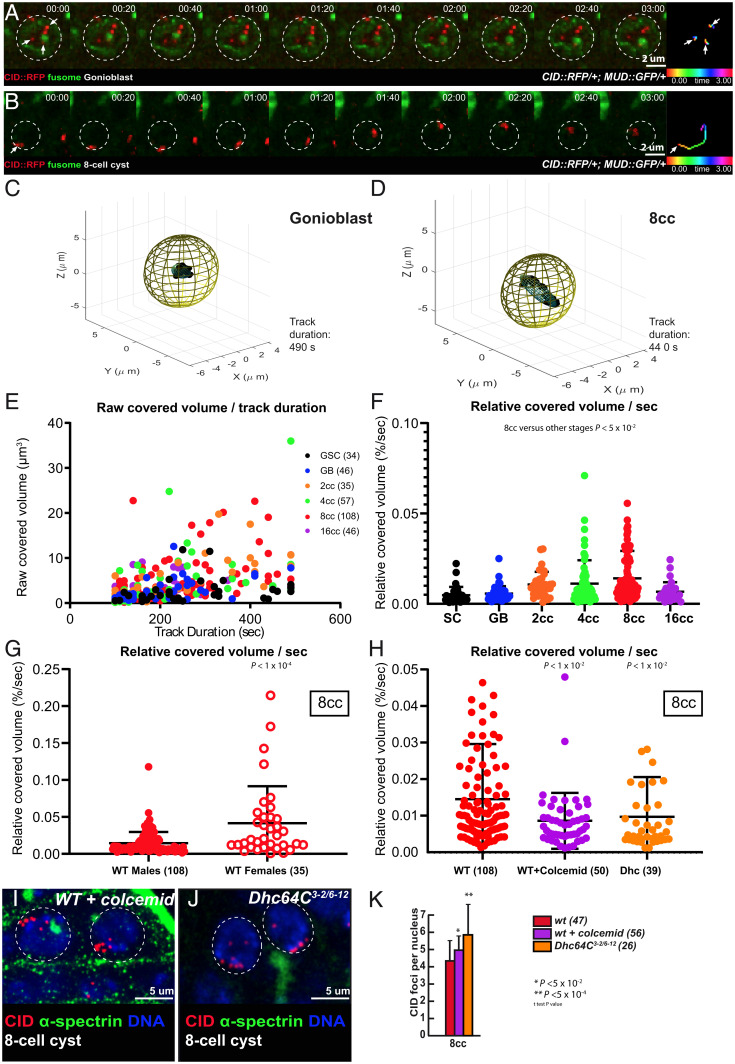
A common theme in meiosis is to set chromosomes in motion so that homologs have a chance to meet, assess their homology, and pair. In Drosophila females, we showed that extensive rotations of germ cell nuclei increased the efficiency of chromosome pairing (6). In males, we followed chromosome movements using a CID-RFP transgene, which marks the centromeres, and calculated the relative covered volume per second for each centromere by live imaging as previously done (Fig. 4 A and B) (6). We found that centromeres were dynamic and moved nonrandomly within the nuclear space. These movements were more prominent at the four- and eight-cell stages that coincided with the increase in chromosome pairing that we detected (Fig. 4 C–F). However, even at the eight-cell stage, the relative volumes covered by centromeres were markedly smaller than in females (Fig. 4G). For example, we never detected complete nuclear rotations of centromeres during the periods of recording. On average, we found that at the eight-cell stage the nuclear coverage in males was three times smaller than in females (Fig. 4G).
Fig. 4.
Centromere pairing of 8-cell cysts exhibit dynamic rotations mediated by microtubules and dynein. (A and B) Selected projections of nuclei from a GB (A) and an 8-cell cyst (B) over a 3-min time course (nuclear areas are indicated by dotted circles). Time-colored tracking of three CID–RFP dots (arrows) in A and one CID–RFP dot (arrowhead) in B are shown in the Rightmost panels. (C and D) Three-dimensional representations of the covered volume of centromeres in time from one representative track; an ellipsoid was arbitrarily centered, representing the nuclear volume in a GB (C) or an eight-cell cyst (D). (E) Raw covered volumes of each centromere track from GSC, GB, 2- to 16-cell cysts as a function of track duration. The number of tracks analyzed is given next to each cell type. (F) Relative covered volume (raw covered volume/nuclear volume) per second for each track at different cyst stages (mean ± SD, Mann–Whitney U test comparing the wild-type 8-cell cyst with other stages P < 0.05). (G) The relative covered volume (raw covered volume/nuclear volume) per second in 8-cell cyst nuclei of males is much less when compared to females. (mean ± SD, Mann–Whitney U test, P < 0.0001; the number of foci analyzed is indicated. (H) The relative covered volume (raw covered volume/nuclear volume) per second in 8-cell cyst nuclei of males treated with colcemid or in Dhc mutant is strongly reduced as compared to wild type. (mean ± SD, Mann–Whitney U test, P < 0.01; WT: n = 108 centromeric foci; WT + colcemid: n = 50 centromeric foci; Dhc: n = 39 centromeric foci) . (I and J) Confocal Z-section projections of testes from wild-type flies treated with colcemid (WT + colcemid) (I) and Dhc3-2/Dhc6-12 (J) stained for centromeres (CID, red), fusome (α-spectrin, green) and DNA (DAPI, blue). The 8-cell cyst nuclei are indicated by dotted circles. Note that in both conditions centromeres are unpaired. (Scale bars, 5 µm.) (K) Graph plots the number of CID foci in 8-cell cyst nuclei in WT, WT + colcemid, and in Dhc3-2/Dhc6-12 mutant. The number of analyzed cells is indicated next to each condition (two-tailed Student’s t test, *P < 0.05, **P < 0.0005).
We next tested whether these movements depended on the microtubule cytoskeleton and its associated motors. We fed male flies with the microtubule-polymerization inhibitor colcemid for 4 h and then dissected the testis for live imaging. We found that centromere movements were markedly reduced in the presence of colcemid (Fig. 4H and Movie S7). Likewise, in dynein Dhc64C mutant males, centromere dynamics were strongly reduced (Fig. 4H and Movie S8). Importantly, in both cases, centromere pairing was strongly affected at the eight-cell stage (Fig. 4 I–K and SI Appendix, Fig. S6 A, C, D, and F).
We concluded that centromeres are dynamic in males, albeit much less than in females. Nevertheless, as in females, these movements depend on microtubules and are required for efficient pairing of centromeres.
Drosophila LINC Complex Is Required for Efficient Centromere Pairing in Males.
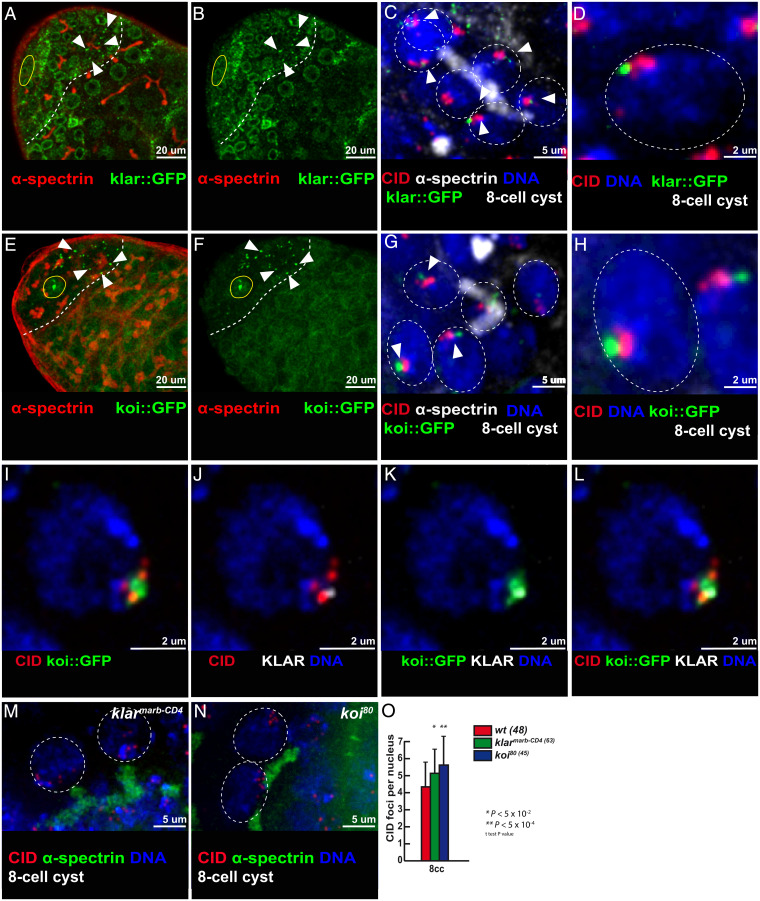
The LINC complex is formed by two transmembrane proteins, which bridge the inner and outer nuclear membranes with the cytoplasmic cytoskeleton. In Drosophila, there are two SUN-domain proteins, Klaroid (Koi) and Spag4, localizing at the inner nuclear membrane and two KASH-domain proteins, Klarsicht (Klar) and MSP-300, localizing at the outer membrane. We generated GFP knockin lines by CRISPR at the endogenous klaroid and klarsicht loci to investigate their localization. We also used an independent GFP-trap line inserted into the klaroid locus. All three lines showed a homogeneous localization at the nuclear envelop (NE) in GSCs, GBs, and two-cell cysts (Fig. 5 A, B, E, and F). However, both Klar and Koi also formed one or two dots at the NE of 4-, 8-, and early 16-cell cysts (Fig. 5 A, B, E, and F, arrowheads). Localization of Koi and Klar then became homogenous again at the NE in later stages. Interestingly, we noticed that Koi and Klar dots were often associated, colocalizing or in close proximity with centromeres at the NE. We observed this association mostly in 8-cell cysts, but also in some 4-cell and 16-cell cysts (Fig. 5 C, D, G, H, and I–L and SI Appendix, Fig. S7 A–F). We further noticed that when Klar and CID were in close proximity, short bundles of α-tubulin colocalized with Klar (SI Appendix, Fig. S7 G–L). This colocalization of Klar, Koi, centromeres and microtubules suggested that the LINC complex could be involved in centromere pairing in males.
Fig. 5.
Klarsicht and Klaroid localize near centromeres in the mitotic region and are required for 8-cell cyst chromosome pairing in males. (A and B) Confocal Z-section projections of klar::GFP (green) stained for fusome (α-spectrin, red). In A, B, E, and F, dotted lines delimit the mitotic region and the hub is marked by a yellow circle. Arrowheads point to klar::GFP dots at the NE. (Scale bars, 20 µm.) (C) Confocal Z-section projection of klar::GFP (green) stained for fusome (α-spectrin, white), centromeres (CID, red), and DNA (DAPI, blue). (D) Magnified view of a nucleus outlined in C showing CID–klar::GFP association. (E and F) Confocal Z-section projections of koi::GFP (green) stained for fusome (α-spectrin, red). Arrowheads point to Klar::GFP dots at the NE. (G) Confocal Z-section projections of koi::GFP (green) stained for fusome (α-spectrin, white), centromeres (CID, red), and DNA (DAPI, blue). (H) Magnified view of a nucleus outlined in G showing CID–koi::GFP association. (I–L) Confocal Z-section projections of a nucleus from an 8-cell cyst stained for centromere (CID, red), Klarsicht (KLAR, gray), koi::GFP (GFP, green), and DNA (DAPI, blue). (M and N) Confocal Z-section projections of klarmarb-CD4/klarmarb-CD4 (M) and koi80/koi80 (N) stained for centromeres (CID, red), fusome (α-spectrin, green), and DNA (DAPI, blue). (Scale bars, 2 µm in D, H–L, M, and N and 5 µm in C and G.) In C, D, G, H, M, and N, 8-cell cyst nuclei are indicated by a dotted circle. (O) Graph plots the number of CID foci in 8-cell cyst nuclei from wild-type, klarmarb-CD4/klarmarb-CD4, and koi80/koi80 mutants. The number of analyzed cells is indicated next to each genotype (two-tailed Student’s t test, *P < 0.05, **P < 0.0005).
We thus tested whether centromere movements depended on Koi and Klar. We measured centromere dynamic in the koi80/koi80 mutant as well as in sh-RNAs against klaroid and klarsicht using the nos::Gal4 driver. All of these conditions markedly reduced centromere movements at the eight-cell cyst stage compared to wild-type conditions (SI Appendix, Fig. S8 A and B). We next tested potential consequences on centromere pairing in these mutant backgrounds. We found an average of 5.7 dots of CID in koi mutants, and 5 dots in klar mutant germ cells, in contrast to 4 in wild-type control flies (Fig. 5 M–O). In addition, we found that pairing of centromeres II and III were down to 45% and 55%, respectively, in koi mutant cells (SI Appendix, Fig. S6 B, C, E, and F). However, chromosomes X and II segregated normally in koi mutant males (SI Appendix, Table S5). We did not investigate Spag4, as spag4 is not expressed in early male germ cells (22) and we did not test MSP-300 mutants, as MSP-300 is known to interact with the actin cytoskeleton (23, 24).
These results showed that the Drosophila LINC complex (Klar/Koi) is required for efficient pairing of chromosomes in males.
Mitotic Cell Cycle Durations in Males Are Much Longer Than in Females.
Our results revealed that chromosome pairing in males is premeiotic, depending on two SC components and on movements driven by the microtubule cytoskeleton and associated proteins, which is strikingly similar to females. Yet, centromere dynamics are much slower in males than in females. For comparison, the average covered volume per second for centromeres in wild-type males is less than in klaroid mutant females, which showed clear pairing defects (0.014%/s in wild-type males vs. 0.029%/s in koi females) (6). Thus, how could slow movements be sufficient for pairing in males but not in females, despite both using identical molecular machinery?
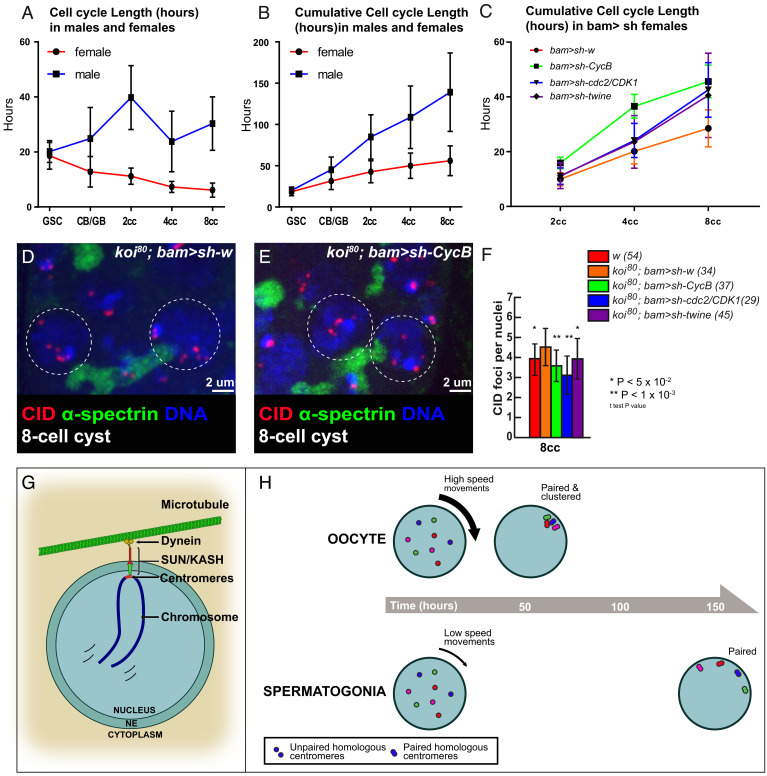
A first clue to this paradox came when we measured the relative volumes covered by centromeres in males. We noticed that the nuclear volumes of 2-, 4-, 8-, and 16-cell cysts were larger in males than in females (SI Appendix, Fig. S9A). It suggested to us that cells had more time to grow in males than in females during each interphase. To test this hypothesis, we determined the length of time spent at each stage of germ cell differentiation in wild-type males and females (SI Appendix, Experimental Procedures). We measured the frequency of mitosis at each stage by live imaging using a GFP marker for mitosis (survivin-GFP) (25). We recorded more than 254 h of 18 testis, and 235 h of 26 ovaries. We found that both female and male GSCs had a similar cell cycle duration of around 20 h, as previously published (26, 27). However, in female cysts, cell cycles became shorter at each division, reaching only 6 h in eight-cell cysts (Fig. 6A). In contrast, male cyst cell cycles were longer than in GSCs, with a maximum of 40 h for two-cell cysts (Fig. 6A). At the eight-cell cyst stage, we measured a 30-h duration in males, which is about five times longer than in females. By adding all the durations from GSCs to eight-cell cysts, we found that the length of time for a GSC to reach the eight-cell stage lasted 48 h in females and 148 h in males on average (Fig. 6B). The developmental window during which homolog chromosomes pair is thus about three times longer in males than in females. These observations support our hypothesis that slower chromosome movements in males could be compensated with an increase in the pairing period.
Fig. 6.
Slowing down the cell cycle rescues koi pairing defects in females. (A and B) Cell cycle length in males and females of developing cysts in the mitotic region represented as the number of hours spent per stage (A) or as the number of hours elapsed since GSC at each stage (B). (C) Cumulative cell cycle length hours in the mitotic region of females expressing a control bam > sh-w (red), bam > sh-CycB (green), bam > sh-cdc2/Cdk1 (blue), and bam > sh-twine (black). (D and E) Confocal Z-section projections of control koi80/koi80; bam/sh-w (D) and koi80/koi80; bam/sh-CycB (E) ovaries stained for centromeres (CID, red), fusome (α-spectrin, green), and DNA (DAPI, blue). Eight-cell cyst nuclei are indicated by a dotted circle in each image. (Scale bars, 2 µm.) (F) Graph plots the number of CID foci in eight-cell cyst nuclei in wild-type, koi80/koi80; bam > sh-w koi80/koi80; bam > sh-CycB, koi80/koi80; bam > sh-cdc2/CDK1, and koi80/koi80; bam > sh-twine. The number of analyzed cells is indicated next to each genotype (two-tailed Student’s t test, *P < 0.05, **P < 0.001). (G and H) Model comparing homolog chromosome pairing in Drosophila male and female.
Increasing Cell Cycle Length Is Sufficient to Rescue Pairing Defects in Females.
We wanted to test functionally the correlation between speed of chromosome pairing and cell cycle duration. We chose klaroid mutant females because centromeres are not attached to the NE, chromosome movements are inefficient, and homolog pairing is defective; however they have only mild SC defects and are fertile (6). To test whether increasing the cell cycle length would be sufficient to rescue pairing defects in koi females, we reduced the amount of Cyclin B, the limiting factor of the Cdk1/CycB complex, which promotes mitotic entry (28). We targeted cycB only during the pairing window of 2- to 8-cell cysts by using the bam promoter to express an shRNA targeting cycB (bam > sh-cycB). In this knockdown (KD) condition, 16-cell cysts could still form normally. Indeed, after recording 37 germarium for 257 h, we found that the duration of the pairing window had increased to 45 h in bam > sh-cycB females compared to 28 h in bam > sh-white control flies (Fig. 6C). These mutant conditions thus allowed us to almost double the length of the pairing period.
Next, we counted the number of CID dots in double mutant koi, bam > sh-cycB females, compared to koi, bam > sh-white control flies. In eight-cell cysts, the number of CID foci was significantly reduced to 3.6 in koi, bam > sh-cycB females, compared to 4.5 in koi, bam > sh-white control flies (Fig. 6 D–F).
To confirm that lengthening the cell cycle was sufficient to rescue pairing defects, we tested two additional cell cycle regulators to increase cell cycle duration: either knocking down the key kinase Cdk1/Cdc2 (bam > sh-cdc2) or by knocking down twine/Cdc25 (bam > sh-twe), a phosphatase, which activates CycB/Cdk1 activity (28). In cdc2 KD, we found that the duration of the pairing window increased to 42 h, and in twe KD to 41 h, which was about a 50% increase of the wild-type duration (Fig. 6C). We found a strong reduction in the number of CID foci in koi, bam > sh-cdc2 and a mild reduction in koi, bam > sh-twe females compared to koi, bam > sh-white control flies (Fig. 6F).
We concluded that increasing cell cycle length in three independent genetic backgrounds was sufficient to partially rescue pairing defects in klaroid mutant females.
Discussion
Drosophila males have always been considered outliers for meiosis as they do not form synaptonemal complexes nor do they need recombination between chromosomes to segregate homologs. Here, our results demonstrate that they share many unsuspected similarities with females. Firstly, timing of pairing is identical, there is de novo homolog pairing in the mitotic region from GSCs to 16-cell cysts. Secondly, underlying molecular mechanisms are similar, as males also depend on the microtubule cytoskeleton, SUN/KASH proteins and SC components for efficient pairing (Fig. 6G).
There are, however, interesting differences. In females, the early localization of SC components at centromeres may initiate the formation of the SC along chromosome arms and the complete synapsis of homologs (16). In the absence of C(3)G or Corona, there is no SC formation, an absence of crossing over, and high percentages of chromosome nondisjunction (21, 29, 30). Obviously in males, C(3)G and Corona at centromeres do not initiate the formation of the SC, and once homologs are paired, they are separated into different nuclear territories in spermatocytes. In these territories, homologs then become unpaired and centromeres dissociate. How these territories form remains unknown. The formation of these distinct territories for each chromosome pair may explain why we did not detect evidence of chromosome nondisjunction in males mutant for c(3)G or cona, despite early defects in centromere pairing. These are two different strategies to segregate homologs at metaphase I: In females, homologs are fully synapsed in the same nuclear space; whereas in males, homologs are unpaired but separated into different nuclear territories. Another interesting difference revealed by our study is the slower dynamics of male meiotic chromosomes as compared to females. Within the limitations of our live-imaging protocol, we never observed dramatic rotations of nuclei as we have described in females. Nevertheless, male centromeres follow the global movements of each nucleus. It remains thus possible that slow nuclear rotations occur, which could not be recorded within the time frame of our experiments. These coordinated movements are thus similar to what we described in Drosophila females and to chromosome movements in mouse spermatocytes (6, 31, 32). In contrast, in C. elegans, individual chromosomes move independently, although they also rely on the LINC complex and microtubules (33, 34).
Although, C(3)G and Corona do not initiate the formation of an SC in males, we found by superresolution microscopy that they form many foci associated with centromeres and along chromosome arms, specifically in the mitotic region. It is tempting to speculate that these foci could be sites of pairing between homologs, and that they could act as “button loci” as in somatic cells (35). Indeed, homolog chromosomes are also paired in Drosophila somatic cells (36). Recent studies have identified genomic regions, associated with topologically associated domains (TADs), which mediate somatic pairing of homologs. These button loci were identified by DNA FISH, Hi-C, and by biophysical modeling (35, 37, 38). Button loci are enriched with insulator or architectural proteins, such as CTCF, Cp190, or Mod(mdg4) (37). It would be interesting to test whether C(3)G and Corona associate with these proteins at button loci in germline cells. However, it remains a challenge to isolate enough premeiotic germline cells to perform co-Immunoprecipitation or Chromatin Immunoprecipitation sequencing experiments.
One major difference between male and female uncovered by our study is the difference in cell cycle durations (Fig. 6H). Previous studies had already reported that cell cycle regulation differs between males and females germ cells (39–41). However, by analyzing fixed samples, these studies could only estimate the relative ratios of cell cycle phases between GSCs and the different cyst stages. Here, using a live-imaging approach similar to Sheng and Matunis (26), we obtained more direct measurements of cell cycle length, not only of GSCs but also of all cyst stages. Our results are in very good agreement with those obtained with fixed samples. For example, we found a peak of duration at the two-cell stage in males, which lasted around 40 h, which is about twice as long as the four-cell stage that we measured at 20 h. This is very similar to the 2:1 ratio found with fixed samples (39). Overall, we found that the window when homologous chromosomes pair in males is three times longer than in females. These differences in timing and kinetics for the same process in males and females could well be widespread among species. For example, in C. elegans, prophase I of meiosis is completed in about 20 h in males, while in females it lasts around 60 h (8). This sexual dimorphism could explain the different sensitivity between males and females to the same mutation affecting meiosis. Indeed, mutations of checkpoint proteins or the presence of unrepaired DNA breaks could have stronger consequences in germ cells having a faster cell cycle than a slower one, which may be compensated by having longer cell cycles, males or females, depending on the species. It is also possible that cell cycle timing may impose different strategies for homolog pairing or synapsis between species. For example, yeast S. cerevisiae has to pair 16 pairs of chromosomes in a few hours and requires DSBs, compared to Drosophila males, which have only 4 pairs of chromosomes and about 100 h to achieve pairing and do not require DSBs. Our results show that time is an important dimension to understand the evolution of the different meiotic strategies between sexes and species.
Materials and Methods
Drosophila melanogaster.
Flies were maintained on standard medium in 25 °C incubators on a 12 h light/dark cycle. All koi rescue experiments were carried out at 29 °C. Wild-type controls alone and in combination with additional transgenes of fluorescently tagged proteins were in a w1118 background. The shRNA for white was used as control for knockdown experiments because white is not expressed during oogenesis and spermatogenesis.
Detailed Materials and Methods, including fly genetics, nondisjunction tests, RNA extraction, qRT-PCR, immunohistochemistry, colcemid treatments, image acquisition, data analysis, analysis of centromere trajectories, mean cyst number estimation, and cell cycle length estimation are listed in SI Appendix.
Supplementary Material
Acknowledgments
We thank Scott Hawley for the initial discussion on the expression of synaptonemal complex genes in males and Isabelle Bonnet for her help with centromere tracking. We are grateful to M. T. Fuller, S. Hawley, S. Roth, Y. Bellaiche, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank Hybridoma Center for antibodies and flies. We are grateful to the Orion Imaging facility at Centre interdisciplinaire de recherche en biologie (Collège de France). The J.-R.H. laboratory is supported by CNRS, INSERM, Collège de France, Fondation pour la Recherche Médicale (FRM) (Equipe FRM DEQ20160334884), Agence nationale de la recherche (ANR-13-BSV2-0007-02 PlasTiSiPi; ANR-15-CE13-0001-01, AbsCyStem) and the Bettencourt-Schueller Foundation (Fondation Schlumberger pour l'Education et la Recherche).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207660119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
References
- 1.Zickler D., Kleckner N., Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhalla N., Dernburg A. F., Prelude to a division. Annu. Rev. Cell Dev. Biol. 24, 397–424 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahoon C. K., Hawley R. S., Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 23, 369–377 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Gerton J. L., Hawley R. S., Homologous chromosome interactions in meiosis: Diversity amidst conservation. Nat. Rev. Genet. 6, 477–487 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Rubin T., Macaisne N., Huynh J. R., Mixing and matching chromosomes during female meiosis. Cells 9, 696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christophorou N., et al. , Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat. Cell Biol. 17, 1388–1400 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Ding X., et al. , SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12, 863–872 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Cahoon C. K., Libuda D. E., Leagues of their own: Sexually dimorphic features of meiotic prophase I. Chromosoma 128, 199–214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKee B. D., Yan R., Tsai J. H., Meiosis in male Drosophila. Spermatogenesis 2, 167–184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller M. T., “Spermatogenesis” in The Development of Drosophila, Bate M., Martinez-Arias A., Eds. (Cold Spring Harbor Laboratory Press, 1993), pp. 71–147. [Google Scholar]
- 11.Vazquez J., Belmont A. S., Sedat J. W., The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12, 1473–1483 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Christophorou N., Rubin T., Huynh J. R., Synaptonemal complex components promote centromere pairing in pre-meiotic germ cells. PLoS Genet. 9, e1004012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyce E. F., Apostolopoulos N., Beliveau B. J., Wu C. T., Germline progenitors escape the widespread phenomenon of homolog pairing during Drosophila development. PLoS Genet. 9, e1004013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin T., Christophorou N., Huynh J. R., How to pre-pair chromosomes for meiosis. Cell Cycle 15, 609–610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeo S., Lake C. M., Morais-de-Sá E., Sunkel C. E., Hawley R. S., Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr. Biol. 21, 1845–1851 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Tanneti N. S., Landy K., Joyce E. F., McKim K. S., A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr. Biol. 21, 1852–1857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blumenstiel J. P., Fu R., Theurkauf W. E., Hawley R. S., Components of the RNAi machinery that mediate long-distance chromosomal associations are dispensable for meiotic and early somatic homolog pairing in Drosophila melanogaster. Genetics 180, 1355–1365 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong W. J., McKim K. S., Hawley R. S., All paired up with no place to go: Pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet. 1, e67 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKee B. D., Karpen G. H., Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell 61, 61–72 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Page S. L., et al. , Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLoS Genet. 4, e1000194 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gowen J. W., Meiosis as a genetic character in Drosophila melanogaster. J. Exp. Zool. 65, 83–106 (1933). [Google Scholar]
- 22.Shi Z., et al. , Single-cyst transcriptome analysis of Drosophila male germline stem cell lineage. Development 147, dev184259 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volk T., A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development 116, 721–730 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Yu J., et al. , The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Dev. Biol. 289, 336–345 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Mathieu J., et al. , Aurora B and cyclin B have opposite effects on the timing of cytokinesis abscission in Drosophila germ cells and in vertebrate somatic cells. Dev. Cell 26, 250–265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheng X. R., Matunis E., Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 138, 3367–3376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris L. X., Spradling A. C., Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development 138, 2207–2215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindqvist A., Rodríguez-Bravo V., Medema R. H., The decision to enter mitosis: Feedback and redundancy in the mitotic entry network. J. Cell Biol. 185, 193–202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Page S. L., Hawley R. S., c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15, 3130–3143 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page S. L., Hawley R. S., Chromosome choreography: The meiotic ballet. Science 301, 785–789 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Lee C. Y., et al. , Mechanism and regulation of rapid telomere prophase movements in mouse meiotic chromosomes. Cell Rep. 11, 551–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibuya H., Morimoto A., Watanabe Y., The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet. 10, e1004821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiraoka Y., Dernburg A. F., The SUN rises on meiotic chromosome dynamics. Dev. Cell 17, 598–605 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Zetka M., Paouneskou D., Jantsch V., “The nuclear envelope, a meiotic jack-of-all-trades”. Curr. Opin. Cell Biol. 64, 34–42 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Viets K., et al. , Characterization of button loci that promote homologous chromosome pairing and cell-type-specific interchromosomal gene regulation. Dev. Cell 51, 341–356.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyce E. F., Erceg J., Wu C. T., Pairing and anti-pairing: A balancing act in the diploid genome. Curr. Opin. Genet. Dev. 37, 119–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowley M. J., et al. , Condensin II counteracts cohesin and RNA polymerase II in the establishment of 3D chromatin organization. Cell Rep. 26, 2890–2903.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Child M. B. VI, et al. , Live imaging and biophysical modeling support a button-based mechanism of somatic homolog pairing in Drosophila. eLife 10, e64412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gadre P., Chatterjee S., Varshney B., Ray K., Cyclin E and Cdk1 regulate the termination of germline transit-amplification process in Drosophila testis. Cell Cycle 19, 1786–1803 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinnant T. D., Alvarez A. A., Ables E. T., Temporal remodeling of the cell cycle accompanies differentiation in the Drosophila germline. Dev. Biol. 429, 118–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Insco M. L., Leon A., Tam C. H., McKearin D. M., Fuller M. T., Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc. Natl. Acad. Sci. U.S.A. 106, 22311–22316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.