Significance
Cytochrome c (cyt c) is an abundant heme protein that, in its native conformation, plays essential roles in mitochondrial bioenergetics and apoptotic signaling in mammalian cells. The dynamic properties of cyt c confers substantial plasticity to produce alternative conformations in vitro, but their detection in cellula remains essentially unexplored. We have restored by de novo sequencing a unique monoclonal antibody (R1D3) recognizing conformationally altered cyt c proteoforms. R1D3 application to different cell models showed that alternative cyt c conformers are translocated from mitochondria to the nuclei without triggering apoptosis. R1D3 represents a unique immunochemical tool for mitochondrial and cell biology, encompassing knowledge of protein dynamics and plasticity with the identification of noncanonical structures and functions of cyt c in vivo.
Keywords: cytochrome c, monoclonal antibody, alternative conformation, redox signaling, oxidation
Abstract
Cytochrome c (cyt c) can undergo reversible conformational changes under biologically relevant conditions. Revealing these alternative cyt c conformers at the cell and tissue level is challenging. A monoclonal antibody (mAb) identifying a key conformational change in cyt c was previously reported, but the hybridoma was rendered nonviable. To resurrect the mAb in a recombinant form, the amino-acid sequences of the heavy and light chains were determined by peptide mapping–mass spectrometry–bioinformatic analysis and used to construct plasmids encoding the full-length chains. The recombinant mAb (R1D3) was shown to perform similarly to the original mAb in antigen-binding assays. The mAb bound to a variety of oxidatively modified cyt c species (e.g., nitrated at Tyr74 or oxidized at Met80), which lose the sixth heme ligation (Fe-Met80); it did not bind to several cyt c phospho- and acetyl-mimetics. Peptide competition assays together with molecular dynamic studies support that R1D3 binds a neoepitope within the loop 40–57. R1D3 was employed to identify alternative conformations of cyt c in cells under oxidant- or senescence-induced challenge as confirmed by immunocytochemistry and immunoaffinity studies. Alternative conformers translocated to the nuclei without causing apoptosis, an observation that was further confirmed after pinocytic loading of oxidatively modified cyt c to B16-F1 cells. Thus, alternative cyt c conformers, known to gain peroxidatic function, may represent redox messengers at the cell nuclei. The availability and properties of R1D3 open avenues of interrogation regarding the presence and biological functions of alternative conformations of cyt c in mammalian cells and tissues.
Cytochrome c (cyt c) is an abundant, small (12 kDa) heme protein, evolutionarily conserved in mammals. It is a nuclear-encoded protein located in the mitochondrial intermembrane space and functions as a single-electron carrier between complex III and complex IV of the electron transfer chain (1). The translocation of cyt c to the cytoplasm is provoked by different death stimuli and leads to the assembly of the apoptosome, with the final activation of caspase 3 resulting in the execution of intrinsic apoptosis (2). The activity of cyt c can be modulated by reversible posttranslational modifications (PTMs), such as phosphorylation at serine, threonine, and tyrosine residues or lysine acetylation, which impact different biochemical activities, such as the inhibition of caspase 3 and cyt c oxidase activities (3–15).
The native structure of cyt c involves the covalently linked heme moiety to the peptide chain by thioether bonds with cysteine residues (C14 and C17). Additionally, His18 and Met80 (M80) occupy the fifth and sixth coordination sites of the heme iron, respectively. In the native configuration, the heme possesses an absorption band centered at 695 nm (16). Under relevant cellular conditions, such as interactions with different phospholipids (cardiolipin) (17–23), electric fields (24, 25), or amino acid oxidative PTMs (tyrosine nitration and methionine oxidation) (26–29), cyt c can give rise to nonnative, alternative protein conformations (24, 25). All these alternative conformations share a gain-of-peroxidase activity and the disruption of the heme ligand at the sixth axial M80-Fe position, causing a loss of the absorption at 695 nm (17–19, 23, 27, 30, 31). These changes are associated with modifications of the heme coordination that favor the release of cyt c from the mitochondria into the cytosol. Spectroscopic analysis of peroxynitrite*-mediated nitration of cyt c at the solvent-exposed Tyr74 (Y74) revealed that nitration causes a conformational transition at physiological pH (26, 28, 30, 32). Indeed, in NO2-Tyr74 cyt c, the conformational change known as “alkaline transition” (33–35) shifts its pKa from 9.2 to 7.2 (26). This transition results in the disruption of the M80-Fe bond and the replacement of M80 with K73 or K79, yielding the alternative low spin conformation (26). Using site-specific Tyr to Phe mutants of the four conserved tyrosine residues of cyt c, it was proposed that the transition is generated by a destabilizing steric effect of the nitro group in a mobile loop of cyt c transmitted to the iron center via the nearby Y67. The key role of Y67 in promoting this transition through interactions with the M80 was confirmed using the Y67F mutant, which reveals how the nitration of distant tyrosine residues triggers profound structural changes in the heme microenvironment, impacting protein function (32). Importantly, Y67 donates a hydrogen bond to the Fe–S bond to further tune electron transfer reactions. Early observations by Margoliash and colleagues suggested that conserved residue Y67 survived mutational replacement during evolution to preserve cyt c structural flexibility (36). Other oxidative modifications, such as methionine sulfoxidation mediated by hydrogen peroxide (H2O2), also provoke the loss of the M80–Fe bond, leading to increased peroxidase activity (37). In vitro, cyt c oxidation by H2O2 begins with oxidation of Y67 followed by irreversible displacement of the M80–Fe bond and side-chain carbonylation of K72 and K73 (38). Recently, mass spectrometry (MS) analysis revealed that the initial modification of cyt c upon H2O2 treatment also involves a major pathway of radical transfer from the heme region to the protein surface, involving M80-Y67-Y74 (39).
At the cellular level, revealing the existence and functions of alternative cyt c proteoforms is challenging and can benefit from immunochemical approaches. A monoclonal antibody that recognizes a conformational change in mouse cyt c of apoptotic and necrotic T hybridoma cells (monoclonal antibody [mAb] 1D3) was previously discovered and characterized (40). The conformational change in cyt c is an early event in apoptosis, which can be identified in preapoptotic cells that are negative for other indicators of apoptosis (40). Using this mAb, a decade ago, we reported that, upon peroxynitrite treatment of HeLa cells, cyt c translocates to the nucleus in an alternative conformation probably involving tyrosine nitration (41). Notably, this result was obtained under low, nonlethal, peroxynitrite concentrations, indicating that cyt c translocation from mitochondria to other cell compartments could occur in nonapoptotic cells (41). Moreover, the subcellular distribution of the cyt c mutant M80A (losing the M80–Fe ligation) was evaluated in HeLa cells and shown to have a nuclear and cytoplasmic distribution without induction of apoptosis. Cyt c nitration at Y67 and 97 was detected in the cytosol of control cells by MS (42). Indeed, neither the M80A cyt c mutant (41) nor nitrated cyt c (43) were able to substantially activate caspase 3 as native cyt c, suggesting a novel function for this proteoform.
The translocation of cyt c to the nucleus was also reported after the induction of DNA damage and after treatment with proapoptotic agents, although there is no information regarding cyt c conformation (44–47). The functions of accumulated cyt c in the nucleus following apoptotic stimuli are related to nuclear pyknosis, DNA fragmentation, and chromatin remodeling (44, 45). Moreover, cyt c translocates early to the nucleus in response to DNA-damaging agents, but not upon stimulation of the TRAIL-activated extrinsic death pathway (46). It was previously shown that cyt c interacts with several histone chaperones, as is the case for the oncoprotein SET/TAF-Iβ. This interaction impairs the histone chaperone activity and prevents the formation of the core histone complex (46–48). These results suggest a wider role of cyt c in DNA damage response independent caspase activation. Nuclear cyt c was also detected in the brains of rats with lithium-pilocarpine–induced seizures even before signs of irreversible neuronal death, suggesting its participation in nuclear pyknosis and DNA fragmentation (45). Recently, cyt c–dependent activation of the heme-regulated inhibitor kinase was observed in drug-tolerant cells as a consequence of sublethal mitochondrial outer membrane permeabilization, leading to cyt c release into the cytosol (49). This sublethal cyt c release drives cell reprogramming in the cells that survive the initiation of apoptosis, uncovering new functions of cyt c after a sublethal insult (49).
Only two studies have identified the presence of these nonnative cyt c proteoforms at the cellular level using the mAb 1D3 antibody (40, 41). Some of the cellular conditions that favor the appearance of these cyt c conformations include heat shock, presence of dexamethasone or staurosporine, γ-irradiation, and oxidant treatment (peroxynitrite and H2O2). Detection of cyt c with the mAb 1D3 was observed in early apoptotic and necrotic cells (40), indicating the presence of these alternative conformations in stressed cells. Importantly, nonnative cyt c was also detected in cells not committed to death (41). The extramitochondrial appearance of nonnative cyt c species, which may not be related to the apoptotic pathway, opens the way forward for identifying conditions that promote these conformational changes as well as novel cellular functions for this well-studied heme protein.
Unfortunately, the mAb 1D3 producing hybridoma was lost in a defective liquid-nitrogen tank, and several attempts to reproduce it employing conventional hybridoma technology were unproductive. Knowing the importance of this antibody as a unique tool for the detection of alternative conformations of cyt c, we aimed to “resurrect” it using the available remaining amounts of mAb 1D3. For this purpose, the entire amino-acid sequences of the antibody light (L) and heavy (H) chains were determined by MS and used for the recombinant expression of 1D3 in eukaryotic cells. The resurrected antibody obtained was then biochemically characterized in vitro against different nonnative cyt c species, extending the proteoforms that the antibody recognizes, and applied to different cellular models where the nonnative conformation was identified. The data presented herein restore and boost the potential uses in cells of what we consider a key immunochemical tool for cyt c, mitochondrial, and cell biology research and underscore how the dynamic properties of cyt c confer substantial plasticity to produce alternative conformations and functions in vivo.
Results
De Novo Sequencing and Recombinant Expression of the mAb 1D3.
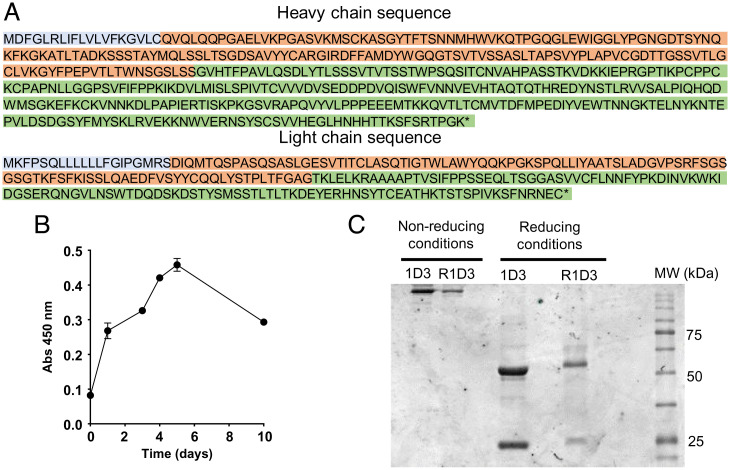
The complete amino-acid sequences of the H and L chains of purified mAb 1D3 were obtained by MS-based peptide sequencing coupled with bioinformatics analysis strategies. This iteration determined the entire H and L chain sequences (Fig. 1 and SI Appendix, Fig. S1). Sequence homology to publicly available antibodies was then used to resolve amino acids with similar masses (e.g., Ile and Leu, Lys and Gln), with ambiguities identified for subsequent empirical testing. These analyses led to several potential versions of the H chain, which were expressed and functionally tested to identify authentic 1D3 antibody (SI Appendix, Fig. S2). The antibody production in transfected HEK293 cells (evaluated in culture supernatant) increased up to day 6 of culture, and cells maintain productivity over 10 d (Fig. 1B). The purified recombinant mAb antibody (mAb R1D3) showed the expected molecular weight size for both the H (50 kDa) and the L (25 kDa) chains (Fig. 1C and SI Appendix, Fig. S3).
Fig. 1.
Generation of the R1D3 antibody based on mAb 1D3. (A) Predicted polypeptide sequences for mAb 1D3 H and L chains. Colors indicate leader (light blue), variable (red), and constant regions (green). * denotes the terminal carboxy group. (B) Cell production of R1D3 antibody. Time course of the production of soluble R1D3 in transfected HEK293 cells (1/50 dilution of the culture media) analyzed by indirect ELISA with adsorbed native cyt c (0.5 µM). Plates were developed and absorbance at 450 nm was recorded. (C) SDS–PAGE analysis of mAb R1D3. SDS–PAGE (10%) of purified mAb 1D3 (lane 1 and 3, 20 µg) and R1D3 (lane 2 and 4, 5 µg) expressed in HEK293 cells under nonreducing and reducing conditions (dithiothreitol, 1 mM) following Coomassie blue stain.
Immunochemical characterization of 1D3 and R1D3 antibodies.
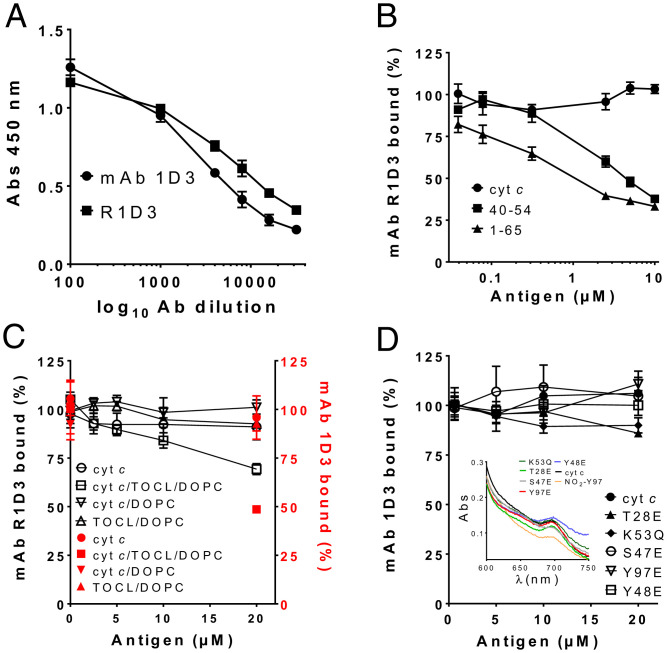
Although mAb 1D3 does not bind to native cyt c in solution, it recognizes a conformation of the protein obtained when some molecules adsorb to microtiter wells in enzyme-linked immunosorbent assay (ELISA) plates (40). The binding capacity of the mAb R1D3 antibody was then evaluated by indirect ELISA, confirming that the recombinant antibody titrates similarly to mAb 1D3 (Fig. 2A). The antibody concentration that produces a 50% decrease of the initial signal at 450 nm was calculated and corresponded to ∼0.18 ng/µL for both antibodies. Next, using this antibody concentration, competitive ELISA were performed in the presence of soluble native cyt c. Results were comparable to those previously reported for mAb 1D3 (Fig. 2B). Thus, the 1–65 CNBr-cleaved peptide was more effective in binding 1D3 than the 40–54 synthetic peptide and native cyt c (Fig. 2B) (40). These results support that the binding region of R1D3 to adsorbed cyt c is the same as for the original mAb 1D3 antibody. This region comprises the loop containing the residue Pro-44 (40). We further characterized the ability of mAb 1D3 and R1D3 to bind cyt c in complex with cardiolipin. For this purpose, di-oleoyl phosphatidyl choline (DOPC) and DOPC-containing cardiolipin liposomes (tetra-oleoyl cardiolipin [TOCL]/DOPC) were generated containing or not native cyt c. As shown in Fig. 2C, the cyt c-cardiolipin–containing liposomes inhibit the binding of mAb 1D3 and R1D3 to the adsorbed cyt c on the ELISA plate, whereas no inhibition was observed for the cyt c/DOPC liposome and for the liposomes alone. Finally, several cyt c mutants that mimic PTMs (phosphorylation and acetylation) at different residues were tested (3, 8, 10–12, 15). As shown in Fig. 2D, none of the cyt c phospho- and acetyl-mimetics used herein were able to compete with the mAb 1D3 for the binding, suggesting that these mimetics do not change the cyt c structure to an alternative conformation (7, 11). This was further confirmed spectrophotometrically, showing that all the cyt c mimetics have absorbance at 695 nm, indicating the presence of the M80–Fe bond (Fig. 2 D, Inset). As it has been reported that one cyt c phosphomimetic may undergo conformational changes [e.g., Y48pCFM (5)], there is room for future characterization of the immunoreactivity of R1D3 with biologically relevant phospho-cyt c proteoforms (3, 9, 11, 13, 50–52).
Fig. 2.
Immunochemical characterization of mAb 1D3 and R1D3 binding. (A) Binding analysis of mAb 1D3 and R1D3 antibodies. Binding of mAb 1D3 (circles) and R1D3 (squares) to adsorbed cyt c (0.5 µM) using different antibody dilutions (1:500–1:30,000 in PBS-Tween) was assayed by indirect ELISA. Plates were developed with OPD and absorbance at 450 nm recorded. (B) Binding of R1D3 (0.18 ng/µL) was evaluated in the presence of soluble native cyt c (circles), 40–54 peptide (squares), and 1–65 cyt c peptide (triangles) (0–10 µM) by competitive ELISA. Results are expressed as % of bound antibody. (C) The binding of mAb 1D3 (black squares) and R1D3 (red circles) (0.18 ng/µL) to phosphatidylcholine liposomes (DOPC) or phosphatidylcholine and cardiolipin liposomes (TOCL/DOPC) with or without native cyt c (200 µM) was analyzed by competitive ELISA as for B. (D) Competitive ELISA was performed as in B in the presence or absence of soluble recombinant native cyt c and the mutants: Y48E, T28E, Y97E, K53Q, and S47E (0–20 µM). Plates were developed with OPD and absorbance recorded at 450 nm. Results are expressed as % of bound antibody. Inset: spectroscopic analysis of cyt c and the different cyt c mimetics showing the peak of absorbance at 695 nm, indicating the presence of M80–Fe bond. NO2Y97 is included as an oxidatively modified cyt c with disrupted heme-M80 ligation (28).
Binding of R1D3 to nonnative, alternative conformations of cyt c.
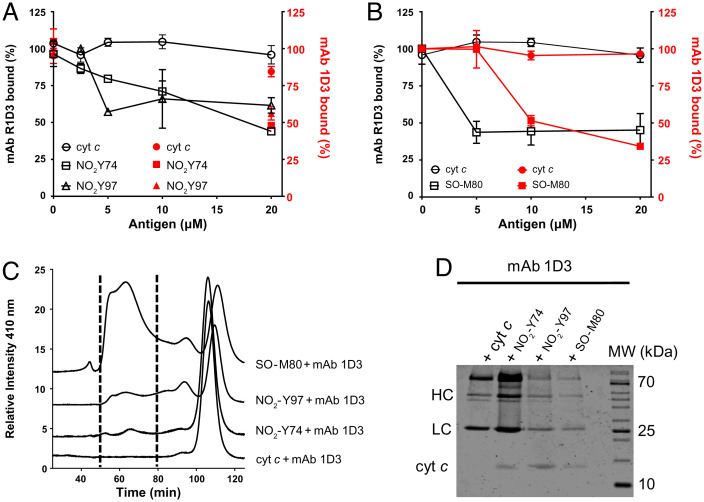
The previously characterized mAb 1D3 was shown to bind to nonnative conformations of cyt c, including those adopted when adsorbed to microtiter wells, when the protein is associated to negatively charged synthetic phospholipid vesicles, and to the alkaline conformation obtained by cross-linking it to ovalbumin at alkaline pH (40). The antibody fails to detect these nonnative cyt c conformations in western blot analysis, indicating the conformational nature dependence of the neoepitope generated upon the structural change (40). Since the disruption of the M80–Fe bond is observed during the alkaline transition of cyt c, when bound to phospholipids (20–23), and also when cyt c is treated with different oxidants (peroxynitrite, HOCl, and H2O2) (26, 27), we aimed to measure the ability of mAb 1D3 and R1D3 to recognize site-specific mononitrated (NO2Y74 and NO2Y97) and methionine-sulfoxidized (SO-M80) cyt c proteoforms that also lack the Fe–M80 ligation. The gain-of-peroxidase activity was also checked for these conformations, and all of them were shown to be increased (SI Appendix, Fig. S4), as previously reported (28, 37). Purified NO2Y74 and NO2Y97 as well as SO-M80 cyt c species were used in competitive ELISA (Fig. 3). Both antibodies efficiently recognized NO2Y74 and NO2Y97 (Fig. 3A) as well as SO-M80 cyt c (Fig. 3B). These results show the ability of mAb 1D3/R1D3 to bind to these oxidatively modified cyt c species. To further confirm the binding, native and oxidatively modified cyt c (NO2Y74, NO2Y97, and SO-M80) were allowed to bind to mAb 1D3 for 16 h in solution (phosphate buffer 50 mM, pH 7.4) followed by size exclusion chromatography and the elution of the cyt c-mAb 1D3 complex monitored at 410 and 280 nm (Fig. 3C and SI Appendix, Fig. S5, respectively). Higher-molecular-weight complexes that absorb at 410 nm were detected for the oxidatively modified cyt c proteins, indicating the binding of the antibody to these oxidized proteoforms, whereas no immune complex was observed with native cyt c (Fig. 3C). The eluted peaks were collected, concentrated, and analyzed by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS–PAGE) (Fig. 3D). Cyt c protein was detected for NO2Y74, NO2Y97, and SO-M80 cyt c and was absent for the native cyt c mixture, confirming the binding of mAb 1D3 to these alternative conformations. Antibody–cyt c complexes were also observed following absorbance at 280 nm (SI Appendix, Fig. S5).
Fig. 3.
Binding of mAb 1D3/R1D3 antibodies to alternative conformations of oxidized cyt c species. (A) Competitive ELISA was performed adsorbing native cyt c (0.5 µM) to the assay plate and the mAb R1D3 (black symbols) or the mAb R1D3 (red symbols) (0.18 ng/µL) allowed to bind in the presence or absence of soluble native cyt c, NO2Y74, and NO2Y97 cyt c (0–20 µM). (B) Binding of mAb R1D3 (black lines) and R1D3 (red lines) to native and SO-M80 cyt c (0–20 µM). Plates were developed with OPD and absorbance recorded at 450 nm. Results are expressed as % of bound antibody. (C) Following incubation of mAb 1D3 with native cyt c, NO2Y74, NO2Y97, and SO-M80 cyt c, the immune complexes were separated by size exclusion chromatography and monitored at 410 nm. Fractions containing the complexes (between dotted lines, 50–80 min) were collected. (D) SDS–PAGE (15%) and Coomassie Blue staining of the proteins eluted during the size exclusion chromatography (fractions between 50 and 80 min). MW: molecular weight marker. Arrows indicate H (HC) and L chains (LC) of mAb 1D3 and cyt c.
Structural and dynamic variations of cyt c upon changes to heme coordination.
Several modifications and/or interactions reported (previously and herein) mediate cyt c transition to alternative conformations, which then can be recognized by the mAb R1D3. However, structural details regarding these alternative proteoforms are scarce. Nevertheless, these conformations share in common that M80 is displaced from the sixth heme-coordination position, where Lys, His, water molecules, or other small ligands occupy the coordination site (17, 19, 26, 37, 53). For instance, the alkaline transition of cyt c involves several steps, but the crucial events are the partial unfolding of the loop 40–57 that leads to deprotonation of an unidentified internal group, followed by the Met → Lys ligand exchange reaction itself. The last step has been identified as rate limiting and requires deprotonation of the ligating surface lysine residue (pKaLys = 11.4) and rearrangement of the loops 70–85 and 23–26 (26).
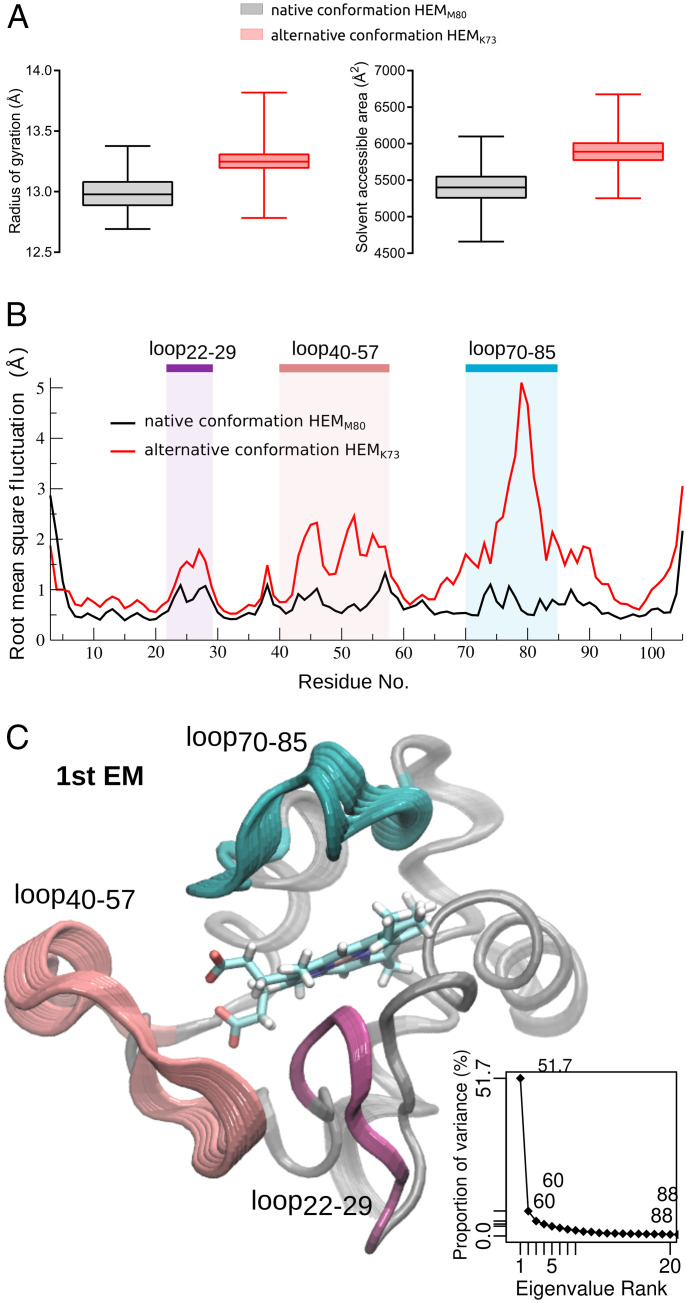
In order to understand the structural and dynamic basis that drives mAb R1D3 recognition of these cyt c alternative conformations, molecular dynamics (MD) simulations and exhaustive analysis of the dynamic properties of native and alternative cyt c conformations were performed. As an alternative conformation model, we selected the previously characterized NO2Y74, in which K73 occupies the axial coordination position (26). The comparison of the dynamic behavior of native and alternative conformations is presented in Fig. 4 and SI Appendix, Fig. S6. Global dynamic properties of both proteoforms are similar, but subtle changes can be observed. The alternative conformation presents a higher degree of gyration than the native, which is accompanied by an increase in its solvent accessible area (Fig. 4A). These changes are due to significant gains of flexibility when going from native to this alternative conformation (Fig. 4B). These regions comprise three loops (22–29, 40–57, and 70–85), which undergo breathing-like dynamics, exposing several residues within these regions to the solvent (Fig. 4 B and C and SI Appendix, Fig. S7).
Fig. 4.
Comparative dynamic behavior of cyt c in native and alternative conformations. (A) Protein radius of gyration (Å) (Left) and total solvent accessible surface area (Å2) (Right), calculated from the MD simulations of native conformation HEMM80 (black) and alternative conformation HEMK73 (red). (B) Protein backbone rms fluctuation (Å) in a pair residue basis shows the regions that present differential flexibility, which are highlighted. (C) Essential dynamics analysis. Cartoon representation of first essential mode (1st EM). Regions are colored as in B. Inset: accumulated proportion of variance explained by essential mode rank.
Detection of alternative conformations of cyt c in living cells.
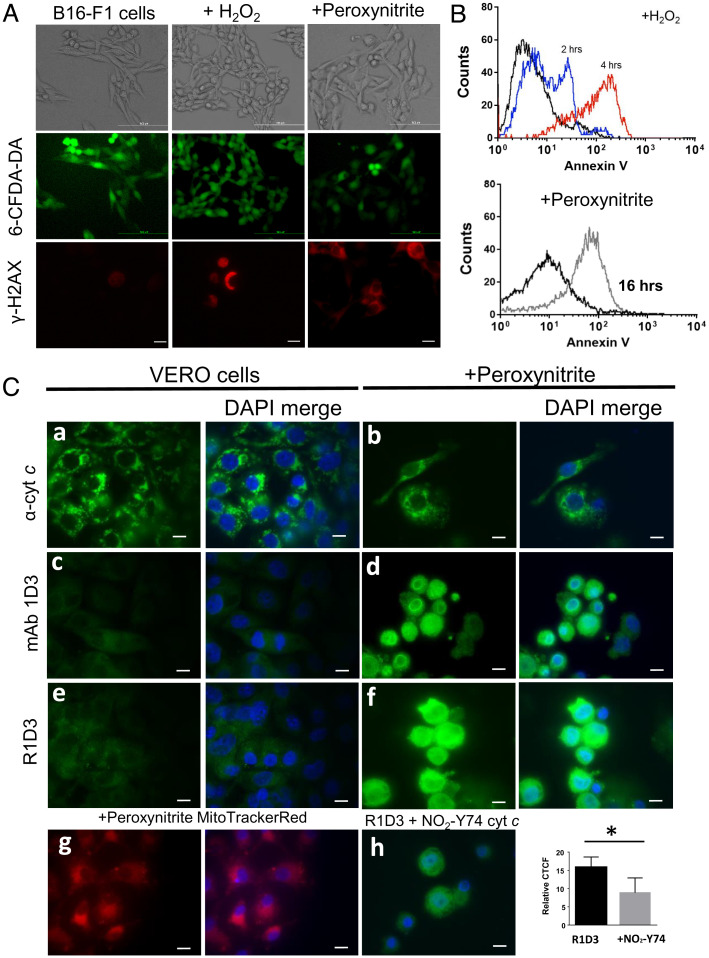
The detection and identification of alternative conformations of cyt c in cellular systems are challenging due to the different conformations that this protein may adopt (such as those seen by tyrosine nitration (54), methionine oxidation (42), and cardiolipin and phospholipid binding (17, 53)). The mAb 1D3 was previously shown to recognize cyt c translocated to the nucleus following peroxynitrite treatment of HeLa cells (41). Herein, the ability of R1D3 to detect this nuclear localization was tested in different cell types following oxidant treatment with peroxynitrite, H2O2, or in a previously described model of cell senescence (55). Both oxidants led to the generation of DNA damage evidenced by phosphorylation of the histone H2AX (Fig. 5A) and commitment to cell death via apoptosis showed by the externalization of phosphatidyl serine to the outer leaflet of the plasma membrane (Fig. 5B), while plasma membrane integrity was preserved (Fig. 5A).
Fig. 5.
Immunocytochemical detection of an alternative conformation of cyt c after peroxynitrite treatment. (A) Plasma membrane integrity and DNA damage. After oxidant treatment (H2O2 or peroxynitrite), B16-F1 living cells were stained with 6CF-DA for 30 min and analyzed by confocal microscopy. Green stain indicates the integrity of the plasma membrane. After oxidant treatment, B16-F1 cells were fixed and probed with antiphosphorylated H2AX antibody followed by antimouse-Cy3 conjugate antibody (red) by confocal microscopy. (B) Phosphatidyl serine exposure was observed using Annexin V-FITC and analyzed by flow cytometry after 2 h (blue histogram) or 4 h (red histogram) for H2O2 (2 mM) (Upper) or peroxynitrite (300 µM) treatment (gray histogram) (Lower). (C) Immunocytochemical detection of an alternative conformation of cyt c. Control conditions and cells treated with peroxynitrite (300 µM in PBS) were incubated for 16 h at 37 °C and 5% CO2. Fixed cells (control or oxidant treated) were probed with antinative cyt c (6H2.B4, a and b), mAb 1D3 (c and d), or mAb R1D3 (e and f) followed by antimouse FITC-labeled conjugated antibody (green) and DAPI (DNA, blue) and then analyzed by confocal microscopy. Mitochondrial distribution of peroxynitrite-treated cells was evaluated with Mito Tracker Red (g). Inhibition of the mAb R1D3 binding was performed by coincubation of the antibody with soluble NO2Y74 cyt c (50 µM, h). Relative CTCF of at least 50 cells in the presence or absence of soluble NO2Y74 were analyzed. * denotes statistical difference P < 0.05. Horizontal scale bars indicate 10 µm.
Then, using peroxynitrite, we evaluated cyt c translocation from mitochondria to the cell nuclei in different cell types. First, using Vero cells after oxidant treatment, cyt c localization was evaluated immunocytochemically using anti-cyt c (native) or mAb 1D3 or R1D3 antibody (Fig. 5C). For both antibodies, a clear nuclear immunostaining was observed in the oxidant-treated cells (Fig. 5 C, d and f, respectively) that was absent in the control conditions (Fig. 5 C, c and e, respectively). As observed, the pattern of the native cyt c immunostaining employing a mAb specific for the native conformation (Fig. 5 C, a and b) as well as the mitochondrial cell distribution (Fig. 5 C, g) were clearly different from that detected with the mAb 1D3/R1D3 antibodies, confirming the binding to an alternative conformation of cyt c in this cellular model. In order to confirm the binding of R1D3 to an alternative conformation of cyt c in this subcellular localization, immunostaining was performed in the presence of soluble NO2Y74 cyt c (50 µM). As shown in Fig. 5 C, h, addition of soluble NO2Y74 cyt c resulted in less-intense staining, confirming recognition of R1D3 of a nitrated/oxidized cyt c in the nucleus, as was previously suggested (41).
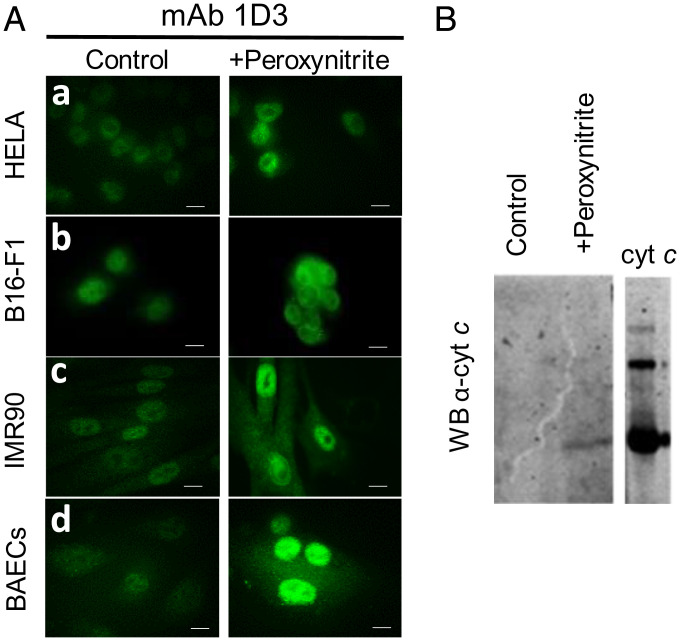
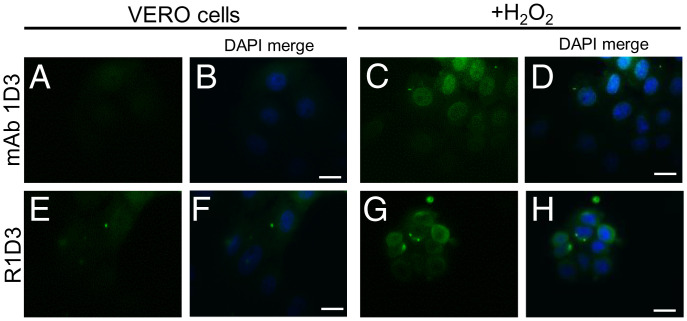
Other cell types were examined in order to evaluate whether the observed cyt c translocation upon peroxynitrite treatment was a general phenomenon. Bovine aortic endothelial cells (BAECs), cells from cancerous cervical tumor (HeLa), human fibroblast (IMR-90), and the mouse-derived melanoma cell line (B16-F1) were exposed to peroxynitrite as above, and the binding of mAb 1D3 was evaluated by confocal microscopy. As shown in Fig. 6A, after peroxynitrite addition, nuclear binding of mAb 1D3 was evident for all the cellular models tested. To further explore that the antibody was recognizing an alternative conformation of cyt c upon oxidant treatment, immunoaffinity purification of cell lysates from control and oxidant-treated cells was performed using affinity chromatography with coupled mAb 1D3. Cell extracts were loaded onto a mAb 1D3 affinity column, and bound proteins were eluted and subjected to western blot analysis using anti-cyt c mAb (7H8.2C12) (Fig. 6B). Cyt c was only detected in the oxidant-treated cells, supporting that the mAb 1D3 recognized an alternative conformation of cyt c. In order to expand the results obtained with peroxynitrite to other relevant biological oxidants, cells were exposed to H2O2 (0.05%, 4 h at 37 °C) and probed with the mAb 1D3 and R1D3 antibodies. As shown in Fig. 7, H2O2 treatment also induced the nuclear localization of cyt c. This result suggests that, following oxidant treatment, either with peroxynitrite or H2O2, some subpopulation of cyt c molecules translocates to the nucleus in an alternative conformation that is recognized by mAb 1D3 and R1D3 antibodies.
Fig. 6.
Detection of an alternative conformation of cyt c following peroxynitrite treatment in different cell models using mAb 1D3. (A) Control (PBS, Left) and peroxynitrite treated (300 µM in PBS, Right) were incubated overnight at 37 °C in DMEM. Fixed HeLa cells (a), melanoma murine cells (B16-F1, b), human fibroblast (IMR90, c), and BAECs (D) were incubated with mAb 1D3 antibody (1/50 dilution) followed by antimouse FITC-labeled conjugated antibody (green). Horizontal scale bars indicate 10 µm. (B) Purification of cyt c in nonnative conformation from oxidant-treated cells. BAECs were treated or not with peroxynitrite as in A and cell extracts affinity purified using mAb 1D3 linked to agarose. Bound proteins were eluted, run on 15% SDS–PAGE, and transferred to nitrocellulose membranes. Transferred proteins were probed with anti-cyt c mAb (7H8.2C12, Sigma) and developed with antimouse-IREDye-800–conjugated antibody (Odyssey). Commercially obtained cyt c was used as standard.
Fig. 7.
mAb 1D3 detection of an alternative conformation of cyt c following H2O2 treatment of Vero cells. Cells were treated with H2O2 (0.05% in PBS) and incubated for 2 h in DMEM media without FBS and fixed. Cells were then probed with mAb 1D3 and R1D3 followed by antimouse-FITC (green) conjugate antibody and analyzed by confocal microscopy. Horizontal scale bars indicate 10 µm.
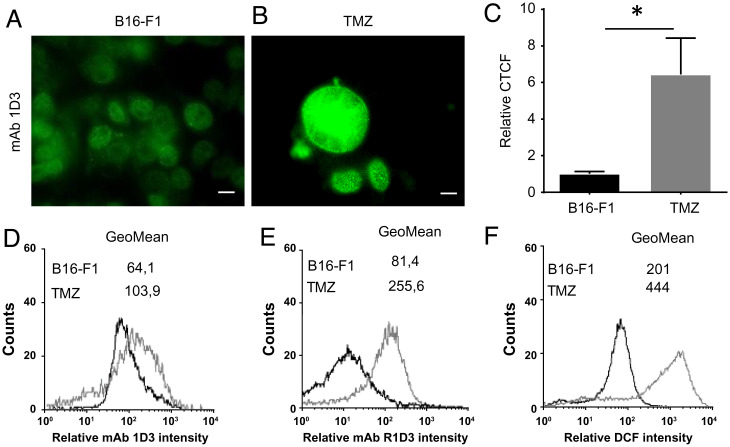
Finally, we used a pre-established model of therapy-induced cell senescence by temozolomide (TMZ) treatment of melanoma cells in which a persistent DNA damage response is observed (55). Following TMZ treatment, an increase in the cyt c detected at the cell nuclei with respect to control cells was observed by immunocytochemistry (Fig. 8 A–C) and by flow cytometry using mAb 1D3/R1D3 (Fig. 8 D and E). Interestingly, in this model, an increase (twofold) in di-chloro-fluorescein diacetate oxidation, indicative of enhanced cellular oxidation events (56), is also observed that may be connected with the generation of this alternative conformation of cyt c (Fig. 8F).
Fig. 8.
Detection of an alternative conformation of cyt c in a senescent cell model. (A and B) mAb 1D3 staining. B16-F1 (control, DMSO-treated) (A) or TMZ-treated cells (B) were probed with the mAb 1D3 following antimouse-FITC–conjugated antibody (green) and analyzed by confocal microscopy. Horizontal scale bars indicate 10 µm. (C) CTCF is shown as fold change in fluorescence levels compared with control. Differences shown with an asterisk (*) were statistically significant, with P < 0.05. (D) Flow cytometry analysis of the mAb 1D3 bound to control (black histogram) or senescent cells (gray histogram). (E) Flow cytometry analysis of the R1D3 bound to control (black histogram) or senescent cells (gray histogram). (F) Flow cytometry analysis of reactive species generated in control (B16-F1, black histogram) or senescent cells (gray histogram) detected by the increase in DCF.
Pinocytic Loading and Immunolocalization of Alternative cyt c Conformers in Cells.
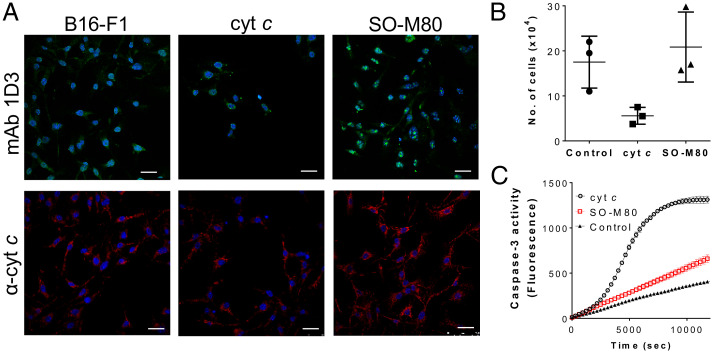
Finally, to unambiguously establish that oxidatively modified cyt c translocates to the cell nuclei, we performed pinocytic loading (57) of B16F1 cells with native and purified SO-M80 cyt c (Fig. 9). A decrease in the cell No. in respect to control conditions was observed for native cyt c loading following 6 h after pinocytosis (Fig. 9 A and B), indicating cell death by apoptosis as previously described (57). On the other hand, there was no change in the No. of cells loaded with SO-M80 cyt c, showing a clear nuclear localization (Fig. 9 A and B). Importantly, SO-M80 cyt c is not effective in activating caspase-3 when compared with native cyt c (Fig. 9C). This failure in the activation of the apoptosome was also previously observed for NO2-Y74 and NO2-Y97 (43) and M80A cyt c (41). Overall, this result shows the ability of alternative conformations of cyt c to translocate to the cell nuclei without activating the apoptotic death pathway.
Fig. 9.
Pinocytic loading of B16-F1 cells with SO-M80 cyt c does not activate caspases. (A) Cells were loaded with nothing (control) or native or SO-M80 cyt c (20 µM) via pinocytosis with overnight incubation. After loading, cells were fixed and probed with mAb 1D3 (green), anti-cyt c (6H2.B4, red), and nuclear staining (DAPI) and analyzed by confocal microscopy. Horizontal scale bars indicate 25 µm. (B) Cell viability was measured by trypan blue assay. No differences in cell population were observed between control and cell pinocytically loaded with SO-M80 cyt c. (C) Purified cyt c or SO-M80 cyt c (20 µg) were added to cytosolic extracts of Jurkat cells (2 mg/mL) in the presence of dATP and ATP. Caspase-3 activity was measured using the fluorogenic substrate DEVD-AFC at λex = 400 and λem = 505 nm.
Discussion
In the present work, we regenerated and performed biochemical and cellular characterizations of a previously lost mAb that recognizes nonnative, alternative conformations of cyt c, named the R1D3 (Figs. 1 and 2 and SI Appendix, Figs. S1–S3). The source hybridoma producing the original antibody mAb 1D3, reported in only two published studies (40, 41), was accidentally lost. This loss hindered all prospects for the detection of physiologically relevant conformations of cyt c that are not specifically recognized by commercial anti-cyt c antibodies. Now, this tool will be available to test in different cellular models that require tracking of cyt c translocation to the cell nuclei elicited by different stimuli (44–46). The binding properties of the recombinant mAb R1D3 produced in this study behave identically to the original mAb 1D3 antibody in indirect (adsorbed cyt c) and competitive ELISA using the cyt c short and long peptides containing Pro44 (Fig. 2 A and B, respectively).
The change of cyt c to an alternative conformation involves, in all cases, the disruption of the M80 bond and can be achieved by different conditions, such as interaction with negative-charged species (like cardiolipin) and different oxidative modifications (tyrosine nitration and methionine oxidation). All these changes lead to an increase in the peroxidase activity of cyt c. In this study, we show the ability of mAb 1D3 to recognize cyt c in cardiolipin-containing liposomes (Fig. 2C). Phosphorylation of cyt c at multiple Ser, Thr, and Tyr sites has been previously shown to occur in different tissues as well as acetylation. These PTMs are involved in the regulation of cell respiration and other mitochondrial processes (3, 4, 8–15). Using cyt c mutants that mimic different phosphorylation and acetylation sites, it was shown that the mAb 1D3/R1D3 fails to recognize them in agreement with the spectroscopic studies that indicate the presence of the M80–Fe bond in all these cyt c mimetics (Fig. 2D). This lack of recognition of the cyt c phospho- and acetyl-mimetics (used in this study) by the mAb 1D3/R1D3 is important, since these reversible PTMs account for the most-frequent cyt c modifications in cells and tissues (58). The results obtained herein strongly suggest that the antibody recognizes only cyt c proteoforms with disrupted M80–Fe ligation.
Herein, it was demonstrated that oxidatively modified cyt c either by nitration at Y74 and Y97 or the oxidized cyt c at M80 (SO-M80) are recognized by mAb 1D3/R1D3, extending the proteoforms that this antibody can recognize (Fig. 3). These oxidatively modified cyt c species lead to the disruption of the M80–Fe bond, as observed by the loss in absorbance at 695 nm and the gain in peroxidase activity (SI Appendix, Fig. S4) (17–19, 23, 27, 30, 31). Further oxidative modifications in cyt c may also occur under excess H2O2, such as lysine carbonylation following the oxidation of other residues (M67/80; Y72/73) (59, 60).
Earlier, it was revealed that the antibody recognizes a conformational epitope that is lost upon cyt c denaturation, and thus, it can only be used in solution [ELISA and immunocytochemical assays using carefully controlled fixation procedures (40)]. In this study, we established that the mAb 1D3 can be also used in immunoprecipitation studies, as observed by the binding of this antibody to an alternative conformation of cyt c in solution (Fig. 3) and by the recovery of cyt c from cell extracts after oxidant treatment by immunoaffinity chromatography (see above Fig. 6). In this sense, R1D3 offers the possibility of identification of other PTMs that may trigger conformational changes by accurate MS measurement and mapping of immunoprecipitated proteins from affected cells.
The disruption of the M80–Fe ligation in cyt c leads, in all cases, to the opening of the heme crevice, causing a gain-of-peroxidase activity of these nonnative conformations with several structural changes that ultimately destabilize the loop 40–57 (24, 40) (SI Appendix, Fig. S7). Our simulation results also suggest that changes in the sixth-coordination position at the axial cavity have repercussions on protein dynamics that can be traced beyond this region, resulting in significant exposure of the 22–29, 40–57, and 70–80 loops (Fig. 4 and SI Appendix, Fig. S7). Taking these data together with the previously reported evidence suggesting that mAb 1D3 recognizes cyt c regions containing P44 that appear after protein conformational changes, we hypothesize that these variations together with flexibility gains in other loops may represent the molecular basis of the mAb 1D3 recognition of alternative cyt c conformations. Further structural studies, including crystallization of the mAb 1D3 bound to cyt c, are ongoing in order to evaluate these hypotheses.
The translocation of cyt c to cellular nucleus has been reported in several cell models (44–46, 48), although the native or the alternative conformation of this cyt c was not determined. Herein, and using different stimuli (oxidants and/or drug treatment), we search for the nuclear cyt c translocation from mitochondria in an alternative conformation. Peroxynitrite and/or H2O2 challenge led to DNA damage and the commitment to cell death by apoptosis (Fig. 5 A and B), conditions that were previously observed to provoke the translocation of cyt c from mitochondria to the nucleus (Fig. 5C). Immunostaining was effectively blocked by the coincubation of mAb 1D3 with the NO2-Y74 cyt c in solution, clearly indicating the presence of an alternative conformation of cyt c at the nuclear level following oxidant treatment (Fig. 5 C, h). Moreover, for all the cell lines analyzed, a positive nuclear cyt c staining was observed after oxidant treatment, indicating that this may represent a common cell response (Figs. 6 and 7). Importantly, and using an affinity column with bound mAb 1D3, cyt c was recovered only from the peroxynitrite-treated cells (Fig. 6B). Then, we used a previously characterized therapy-induced senescent melanoma cell model (B16-F1 cells) in which an increase in mitochondrial mass, length, and energy metabolism was observed after TMZ treatment (55). This model induces a persistent DNA damage response, ATM activation, and increase in γ-H2AX nuclear foci in treated cells, factors that have been shown to be associated with cyt c translocation to the nucleus (44–46). Cyt c translocation was observed in this model as well as a rise in oxidant production (Fig. 8).
Finally, and in order to unequivocally show the localization of alternative conformations of cyt c at the cell nuclei, we conducted pinocytic loading of native and/or SO-M80 cyt c to B16-F1 cells as previously performed (57). Native cyt c loading induces caspase 3 activation and the commitment of cells to die by apoptosis (Fig. 9), as was previously shown (57). A strong nuclear localization was observed for SO-M80 cyt c (Fig. 9A), and importantly, at the cytosol, this proteoform did not activate caspases (Fig. 9C), similarly to what was previously shown for M80A (41), NO2-Y74, and NO2-Y97 cyt c (43). The biological role of alternative cyt c conformations present at the cell nuclei is still difficult to establish but may involve the regulation of gene expression and the preconditioning of cells to deal with further oxidative stress, as previously suggested (41). It is also interesting to consider that the peroxidatic activity of alternative cyt c conformers at the nuclei may catalyze the oxidation of nucleotides and promote DNA damage, including DNA strand breaks (61). Activation of the DNA damage response was evidenced in B16-F1 cells along with cyt c translocation to the nuclei (Figs. 5, 6, and 8), suggesting that these alternative conformations could play a role in mitochondrial retrograde signaling, communicating mitochondrial status to the nuclei, and leading to changes in gene expression.
The research efforts communicated herein will now permit the time- and space-resolved interrogation of the formation, lifetime, and biological functions of alternative conformers of cyt c under physiological or pathophysiological conditions. R1D3 represents a unique tool encompassing knowledge of protein dynamics and plasticity with the identification of noncanonical structures and functions of cyt c in vivo.
Materials and Methods
Equine cyt c was purchased from Sigma (C7752). cyt c was repurified to 100% homogeneity through high-performance liquid chromatography (HPLC) separation as previously reported (28). Peroxynitrite solutions were prepared from acidified H2O2 and sodium nitrite as described (62) and stored at −80 °C. H2O2 contamination was eliminated by the addition of manganese dioxide. Peroxynitrite concentration was determined measuring the absorbance at 302 nm (ε302 = 1.70 mM−1 cm−1) (62). The concentration of nitrite present as contaminant was measured with the Griess reagent after decay of peroxynitrite to nitrate in monobasic sodium phosphate solution and was always less than 20% of peroxynitrite. H2O2 solutions were prepared in Nanopure water, and the concentration was determined from the absorbance at 240 nm (ε = 43.6 M−1.cm−1) (63). o-phenylenediamine (OPD) and diethylenetriamine penta-acetic acid (DTPA) were purchased from Sigma (P9029).
De Novo Sequencing and Expression of Recombinant 1D3 Antibody (R1D3).
To achieve maximum coverage for antibody sequencing, 5 μg of the antibody original 1D3 was digested in parallel with five different proteases: trypsin, elastase, chymotrypsin, subtylysin, and Asp-N. For each digestion mixture, peptides were loaded onto a nanoflow C18 HPLC column, and peptides were resolved using an aqueous to organic gradient over the course of 90 min. As they eluted from the column, peptides were directly ionized on a Thermo Fisher Orbitrap Velos mass spectrometer. In a data-dependent manner, both high-resolution full mass measurements and multiple different tandem mass fragmentation (MS/MS) modalities were collected to give the greatest likelihood of correct sequence interpretation. These include standard collision-induced dissociation, higher-energy dissociation, and electron transfer dissociation. After acquisition, data were transferred to Abterra Bioscience for analysis using their proprietary Valens platform. Briefly, an analysis of bottom-up mass spectra generated by the Vanderbilt University proteomics facility using multiple enzymes was conducted. The framework sequence was identified by performing a database search of the spectra against the germline immunoglobulin gene sequences. Next, de novo peptide sequencing was used to assemble spectra from the hypervariable, complementarity-determining regions. The analysis determined the entire L and the H chain sequences. The gene fragments representing the predicted H and L chain variable domains of mAb 1D3 were synthesized using gBLOCK technology (IDT Inc., https://www.idtdna.com/). The mAb 1D3 H and L chain sequences were cloned into separate expression vectors for cotransfection production studies. For transfection of the pcDNA 3.1 1D3 H chain and pcDNA 3.1 1D3 L chain plasmids, cells were grown to 70% confluence in Dulbecco’s modified Eagle’s medium (DMEM) supplement with 5% fetal bovine serum (FBS) (Gibco) (five flasks). HEK293 cells were transfected by adding H chain (36 µg) and L chain (36 µg) plasmids in the presence of polyethylenimine (PEI) “Max” (1 mg/mL) per bottle. The PEI–DNA mixture was prepared and added to the cells on the same day for 30 min at room temperature (RT). After 30 min of incubation, fresh media were added. Cells were cultured for 10 d, and the medium was collected daily. The medium was centrifuged at 800 × g for 10 min at RT and kept sterile at 4 °C until used. The recombinant mAb 1D3 (R1D3) expression levels with these vectors were tested in HEK293 and CHO cell lines. mAb R1D3 was recovered daily (for 10 d) from the culture media of transfected HEK293 cells, and antibody production was verified by indirect ELISA (see below) using adsorbed native cyt c. The antibody concentration was measured by SDS–PAGE (10%) followed by Coomassie staining using bovine serum albumin (BSA) as standard (2–30 µg) under reducing (beta-mercaptoethanol, 0.2 M) and nonreducing conditions. Protein bands were scanned and quantified using ImageJ, and antibody concentration was estimated using the slope obtained from the BSA standard calibration curve (SI Appendix, Fig. S3), giving a global production of 80–100 µg of pure mAb R1D3 obtained from 200 mL of a 10-d HEK293 supernatant culture (five cell-confluent 175 cm2 culture flasks).
Binding Analysis of mAb 1D3 and R1D3 by Indirect or Competitive ELISA.
In this study, mAb 1D3 and R1D3 binding to cyt c species was characterized in parallel by indirect and competitive ELISA, as previously described (40). Native cyt c (0.5 µM) was adsorbed to Nunc Maxisorp microtiter plates (Gibco, Coon Rapids, MN) in phosphate-buffered saline (PBS) (pH 7.4) for 1 h at 37 °C. Plates were then blocked (1 h, 3% wt/vol milk in PBS) and washed in PBS containing Tween-20 (0.05% vol/vol). One hundred microliters of the mAb 1D3 and/or R1D3 antibodies (1:500–1:30,000 dilution in PBS-Tween from stock solution of 1.44 mg/mL) were incubated for 1 h at 37 °C followed by 1 h with antimouse-horseradish peroxidase (100 µL, 1/8,000). Plates were developed in citrate/phosphate buffer (0.05 M, pH 5) containing OPD (0.4 mg/mL), and absorbance was read at 450 nm in a microplate reader. The mAb 1D3/R1D3 concentrations that caused a 50% decrease in the absorbance at 450 nm was used for competitive ELISA (1:8,000 dilution ∼0.18 ng/µL). For competitive ELISA, cyt c was adsorbed to microtiter plates, and various soluble antigens (2.5–20 µM in PBS) were examined for their ability to block the binding of mAb 1D3/R1D3 antibody (∼0.18 ng/µL) to adsorbed cyt c.
Purification of the Nitrated and Sulfoxidized cyt c Species.
Nitration of cyt c was performed by reaction with peroxynitrite, with minor modification as described (28). Peroxynitrite was added to cyt c (67 mM) in potassium phosphate (200 mM, pH 7), NaHCO3 (25 mM), and DTPA (100 µM) at 25 °C as continuous flux (0.13 mM min−1) using a motor-driven syringe (SAGE Instruments, Boston, MA), under vigorous vortexing. The reaction was stopped by passing the mixture through a desalting column (HiTrap, 5 mL, Amersham Biosciences) equilibrated with potassium phosphate (10 mM, pH 7.4). For the purification of mononitrated species, the reaction mixture was passed through a cation exchange sulfopropyl-TSK preparative column (21.5 mm × 15.0 cm; Tosoh Biosep) at a flow rate of 3 mL/min. The column was equilibrated with ammonium acetate buffer (5 mM, pH 9.0), kept for 5 min in this buffer, and then eluted using a linear gradient of ammonium acetate as follows: 5 mM–150 mM from 5 to 30 min, 150–400 mM from 30 to 75 min, and 500 mM from 76 to 90 min. Sulfoxidized cyt c (SO-M80) was obtained by the reaction with chloramine-T, as was previously described with minor modifications (37). Equal volumes of cyt c (1 mM) were mixed with chloramine-T (5 mM) and the pH adjusted to 8.4. After 3 h at RT, the reaction was stopped by passing the mixture through a desalting column (HiTrap, 5 mL, Amersham Biosciences) equilibrated with potassium phosphate (10 mM, pH 7.4). For the purification of sulfoxidized cyt c (SO-M80), the same purification methods were used as for the nitrated mix (28, 37). Following purification of the oxidatively modified cyt c, the species were used (0–20 μM) in competitive ELISA in order to assess the antibody binding.
Liposome Preparation.
Small, unilamellar liposomes were prepared using a tip sonicator (Vibracel, Sonic Materials, Danbury CT, USA) as described on the website https://avantilipids.com. Liposomes were made from TOCL combined with DOPC at a 50:50 ratio in 20 mM sodium phosphate containing DTPA (100 μM) (pH 7.4) by sonicating 10 times at 4 °C for 30 s with 1-min intervals. Samples were centrifuged after sonication in order to eliminate any titanium particles. Native cyt c (50 μM) was mixed with liposomes (400 μM) in a 1:20 ratio in phosphate buffer (20 mM, pH 7.4) and incubated for 30 min at room temperature. With this procedure, all cyt c interacts with liposomes and no free cyt c is detected (37, 53).
Recombinant Bacterial Expression of Mutant cyt c Mimicking PTMs.
The protocol for the recombinant expression and purification of phosphomimetic recombinants (Y48E, T28E, Y97E, and S47E) and acetyl-mimetic recombinant (K53Q) was performed as previously described (8, 9, 11, 13, 51). For all mutants and for the NO2Y97, the spectrum around 695 nm was measured to confirm the presence of the Fe–M80 bond (Fig. 2 D, Inset).
Purification of mAb 1D3-cyt c Complex.
mAb 1D3 (0.8 mg) and native, NO2Y74, NO2Y97, or SO-M80 cyt c (0.07 mg) were incubated overnight at 4 °C with orbital shaking. Proteins were each loaded on a Sephacryl S-200 column (GE Healthcare, 60 cm × 16 mm) and eluted using phosphate buffer (50 mM, pH 7.4) containing NaCl (150 mM) at a flow rate of 0.8 mL/min. Absorbance at 410 and 280 nm was recorded. Fractions absorbing at 410 nm were collected (55–80 min) for the different samples, concentrated, and analyzed by SDS–PAGE 15% with Coomassie Brilliant Blue-R250 staining.
MD Simulations.
We performed MD simulations of the native conformation cyt c (HEMM80), in which M80 occupies the sixth iron-coordination position and an alternative conformation cyt c (HEMK73), with Lys73 replacing M80 in the metal coordination. For the native conformation, the equine cyt c crystal structure (Protein Data Bank [PDB]: 1hrc) (16) was used as the starting model, and the alternative conformation initial structure was taken from Capdevila and coworkers (26), which was created from a previously solved crystal structure of an alkaline conformation (PDB: 1lms) (64). These two systems were subjected to the same MD protocol. Briefly, systems were solvated using a default method, with an octahedral box of 12 Å in radius with transferable intermolecular potential 3P water molecules (65). Protein residue parameters correspond to the parm14SB Assisted Model Building with Energy Refinement (AMBER) force field (66), and heme C parameters correspond to the previously developed ones (26). All simulations were performed using periodic boundary conditions with a 10-Å cutoff and particle mesh Ewald summation method for treating the electrostatic interactions. The hydrogen bond lengths were kept at their equilibrium distance by using the SHAKE algorithm, while temperature and pressure were kept constant with a Langevin thermostat and barostat, respectively, as implemented in the AMBER program (67). For both cases, the system was optimized in 1,000 steps (10 with steep gradient and the rest with conjugate gradient). Then, it was slowly heated from 0 K to 300 K for 20 ps at constant pressure, with a Berendsen thermostat, and pressure was equilibrated at 1 bar for 5 ps. After these two steps, a 10-ns MD long simulation at constant temperature (300 K) and constant volume was performed. Afterward, three independent 1-μs-long independent production MD replicas at constant pressure were performed. Analysis was performed over these production trajectories, using the cpptraj (68) module of the AMBER suite and Bio3D (69). All MD visualizations and molecular drawings were performed with Visual Molecular Dynamics 1.9.1 (70).
Cell Culture and Treatment.
The mouse melanoma B16-F1 cell line (American Type Culture Collection [ATCC] CRL-6323), IMR-90 (ATCC CCL-186), BAECs (Genlantis, San Diego, CA), Vero (ATCC CCL-81), and HEK-293 (ATCC CRL-1573) were grown at 37 °C and 5% CO2 in complete DMEM (Sigma), supplemented with 10% or 5% FBS (Gibco), L-glutamine (0.2 mM), penicillin (10 U/mL), and streptomycin (10 µg/mL). The cell types used in this study were grown to 70% confluence in 35-mm glass-bottom dishes. For experiments involving peroxynitrite, cells (1 × 108) were washed with PBS, trypsinized, centrifuged for 5 min at 800 × g, and resuspended in 200 µL of PBS. Peroxynitrite (or PBS, control condition) was added to cell suspension at a final concentration of 300 µM by a single bolus addition and incubated for 10 min with gentle swirling. Following incubation, 10 mL of complete DMEM, supplemented with 10% of FBS, penicillin (50 U/mL), and streptomycin 50 (mg/mL), was added to the cell suspension, which was plated and incubated overnight (∼16 h) at 37 °C with 5% CO2. For experiments involving H2O2, cells were washed three times with PBS, and H2O2 was added (final concentration 0.05%, 4 mM) in DMEM without FBS. Following H2O2 treatment (4 h), cells were washed, fresh complete media were added, and the cells were incubated for 6 h at 37 °C in 5% CO2. For the senescence cell model, the mouse melanoma B16-F1 cell line (CRL-6323, ATCC) was cultured in complete DMEM containing glucose (4.5 g/L) at 37 °C and 5% CO2. B16-F1 cells were treated with TMZ (Sigma) (200 µM) for 5 h, and this procedure was performed twice with a 24-h interval, as previously described (55). Treatment with the vehicle, dimethyl sulfoxide (DMSO), was used as a control in all experiments. Measurements were made 4 d after the second exposure to the drug unless otherwise specified.
Confocal Microscopy.
After cell treatment, cells were fixed in two steps with freshly prepared paraformaldehyde (PFA) (2% vol/vol) in DMEM for 30 min at RT and a second step with PFA (4% vol/vol) in PBS (0.1 M pH 7.4) for 60 min at 4 °C. Fixed cells were permeabilized with Triton X-100 (0.5% vol/vol, 10 min). Blockage was carried out with PBS containing FBS (3% vol/vol) followed by a 60-min incubation at RT with the anti-cyt c primary antibody (Sigma 6H2.B4, 1:100 dilution), mAb 1D3, R1D3 (1:50 dilution), or anti-phospho-histone γ-H2AX (Ser139) clone JBW301 antibody (05-636, Merck Millipore, MA). Fluorescein isothiocyanate (FITC)- and/or Cy3-conjugated rabbit antimouse was added as the secondary antibody for 60 min in the dark. Finally, cells were incubated with DAPI (1:1,000 dilution, Sigma) to stain DNA at RT for 10 min. Cold PBS washes were carried out between all steps. The coverslips were mounted on glass slides with Prolong Gold Antifade and analyzed by confocal microscopy. Confocal imaging was performed with a Leica DMIRE2 inverted microscope and Leica True Confocal Scanner TCS SP5. Fluorescence intensity was analyzed using the corrected total cell fluorescence (CTCF) parameter of ImageJ software. CTCF was calculated as CTCF = integrated density − (area of selected cell × mean fluorescence of background readings). mAb 1DR and R1D3 immunostaining of B16-F1 was also evaluated by flow cytometry (FACS-Calibur). Cells were collected in culture media and labeled with mAb 1D3/R1D3 as for microscopy. At least 10,000 events were acquired in each condition.
Cell Viability and Oxidant Production.
Following oxidant treatment (peroxynitrite or H2O2), B16-F1 cells were trypsinized, collected in culture medium, and stained with annexin-V-Alexa Fluor 488 (Invitrogen) for the determination of phosphatidylserine externalization by flow cytometry (FACS-Calibur, Becton Dickinson) according to manufacturer instructions. Plasma membrane integrity was evaluated by incubating plated cells with 6-chloro fluorescein di acetate (25 µM, 30 min at 37 °C in the dark). Esterases from viable cells allowed the desesterification of the probe, yielding green fluorescence that was analyzed by confocal microscopy. Oxidant production was evaluated in TMZ-treated cells following the oxidation of di-chloro-fluorescein-DA (H2DCF-DA) (Molecular Probes). Briefly, cells were incubated in PBS for 30 min at 37 °C with H2DCF-DA (30 µM) in the dark. After incubation, cells were washed and immediately analyzed by flow cytometry (FACS-Calibur).
Purification of cyt c in Alternative Conformations from Oxidant-Treated Cells.
BAECs were grown to 70% confluence in 100-mm bottom dishes. Cells (1 × 108) were washed with PBS, trypsinized, and treated with peroxynitrite as described above. After a 16-h incubation period, cells were trypsinized as above and subjected to three cycles of freeze/thaw in liquid N2. Samples were centrifuged at 1,000 × g for 10 min at 4 °C, and the supernatant was used to purify the nonnative conformations of cyt c using a mAb 1D3-affinity column CNBr-activated Sepharose (performed following GE HealthCare protocol using 1.2 mg of mAb 1D3 with 2 g of activated resin). The samples were incubated with the resin in batch overnight at 4 °C. Unbound proteins were eluted with Tris⋅HCl buffer (20 mM, pH 7.5). Bound proteins were eluted with 10 volumes of glycine (100 mM, pH 2.5) and with 10 volumes ethanolamine (1 M, pH 11). Eluted proteins were dialyzed with phosphate buffer (10 mM, pH 7.4), lyophilized, and resuspended in water (500 µL). Samples obtained were run on 15% SDS–PAGE and transferred to nitrocellulose membranes. Transferred proteins were probed with anti-cyt c antibody (Sigma, 7H8.2C12) and developed with antimouse immunoglobulin G-IREDye-800 conjugate (Odyssey).
Pinocytosis of Native and SO-M80 cyt c.
Pinocytic loading of native and SO-M80 cyt c into B16-F1 cells was performed similarly to the method described by Gilmore et al. (57). Briefly, 0.5 × 106 cells were trypsinized, collected in culture medium, centrifuged (500 × g, 5 min), and resuspended in 83 µL of hyperosmotic buffer (10 mM Hepes, pH 7.4, 10% (wt/vol) PEG1000, 0.5 M sucrose) with or without 20 µM of native or purified SO-M80 cyt c. After a 10-min incubation at 37 °C, the cells were plated onto 35-mm glass-bottom slides with complete culture medium and incubated overnight at 37 °C, 5% CO2. Cells were processed for confocal microscopy as above. Cell viability was measured by the trypan blue exclusion assay. Results are expressed as No. of viable cell population in each condition.
Preparation of Cell-Free Extracts.
Cell-free extracts were generated from Jurkat T lymphocyte cells (ATCC TIB-152) as previously described (71). Cells (5 × 106/mL) were pelleted and washed twice with ice-cold PBS. The cell pellet was suspended in five volumes of ice-cold buffer (Hepes-KOH, pH 7.5 [20 mM], KCl [10 mM], MgCl2 [1.5 mM], EDTA [1 mM], ethylene glycol tetra-acetic acid [1 mM], and dithiothreitol [1 mM]) supplemented with protease inhibitors (protease inhibitor mixture P8340, Sigma). After sitting on ice for 15 min, the cells were disrupted with 10 strokes at 500 rpm with a Teflon pestle. Nuclei and undisrupted cells were removed by centrifugation at 1,000 × g for 10 min at 4 °C. The supernatant was then centrifuged at 100,000 × g for 1 h at 4 °C. The resulting supernatant was carefully removed and stored at −80 °C until use.
Caspase 3 Activity Assay.
Caspase 3 activity was evaluated following the cleavage of the fluorogenic substrate DEVD-AFC using 200 µL final volume of cell-free extract (2–4 mg/mL) containing adenosine triphosphate (ATP) (2 mM), dATP (2 mM), AcDEVD-AFC (0.2 mM) and different concentrations of native and SO-M80 cyt c. The fluorescence of the cleavage product (AFC) was monitored at λem = 505 nm and λex = 400 nm and the fluorescence registered using a Varioskan Flash Multimode Reader (Thermo). In the control condition, the cell extract was omitted.
Statistics.
Statistical difference was determined using a two-tailed t test for paired samples.
Supplementary Material
Acknowledgments
This work was supported by grants of Universidad de la República (CSIC_2018 and EI_2020) and Agencia Nacional de Investigación e Innovación (FCE_1_2014_1_104233) to R.R., Universidad de la República (CSIC I+D 2020) and Agencia Nacional de Investigación e Innovación (FCE_1_2017_1_136021) to C.Q., NIH (R01 GM116807) to M.H., and NIH (SBIR 2R44GM103346) to N.C. The work performed in the Vanderbilt Antibody Core received support from the Vanderbilt Institute of Chemical Biology and the Vanderbilt Ingram Cancer Center Support grant (NIH grant P30 CA68485). Additional funding was obtained from Programa de Desarrollo de Ciencias Básicas (Uruguay). F.T. was partially supported by a fellowship from Universidad de la República, Uruguay. J.M. was partially supported by a fellowship from Agencia Nacional de Investigación e Innovación.
Footnotes
Reviewers: M.D.l.R., Universidad de Sevilla; and M.M., Yale University.
Competing interest statement: N.C. declares to have a competing interest as an employee and shareholder of Abterra Biosciences.
*Peroxynitrite is a biological oxidant formed as the product of the reaction of superoxide radical with nitric oxide. The term peroxynitrite refers to the sum of peroxynitrite anion (ONOO-) and peroxynitrous acid (ONOOH, pKa = 6.8).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2213432119/-/DCSupplemental.
Data, Materials, and Software Availability
The plasmids 1D3LC and 1D3HC encoding the light and heavy chains, respectively, have been deposited at Addgene (https://www.addgene.org/).
LC/MS data have been deposited in ProteomeXchange (PXD036958) (72).
References
- 1.Heineman W. R., Norris B. J., Goelz J. F., Measurement of enzyme E’values by optically transparent thin layer electrochemical cells. Anal. Chem. 47, 79–84 (1975). [DOI] [PubMed] [Google Scholar]
- 2.Hüttemann M., et al. , The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: From respiration to apoptosis. Mitochondrion 11, 369–381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H., Lee I., Salomon A. R., Yu K., Hüttemann M., Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim. Biophys. Acta 1777, 1066–1071 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee I., et al. , New prospects for an old enzyme: Mammalian cytochrome c is tyrosine-phosphorylated in vivo. Biochemistry 45, 9121–9128 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Guerra-Castellano A., et al. , Mimicking tyrosine phosphorylation in human cytochrome c by the evolved tRNA synthetase technique. Chemistry 21, 15004–15012 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Pérez-Mejías G., Guerra-Castellano A., Díaz-Quintana A., De la Rosa M. A., Díaz-Moreno I., Cytochrome c: Surfing off of the mitochondrial membrane on the tops of complexes III and IV. Comput. Struct. Biotechnol. J. 17, 654–660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerra-Castellano A., et al. , Oxidative stress is tightly regulated by cytochrome c phosphorylation and respirasome factors in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 115, 7955–7960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pecina P., et al. , Phosphomimetic substitution of cytochrome C tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Biochemistry 49, 6705–6714 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Bazylianska V., et al. , Lysine 53 acetylation of cytochrome c in prostate cancer: Warburg metabolism and evasion of apoptosis. Cells 10, 802 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Márquez I., et al. , Structural and functional insights into lysine acetylation of cytochrome c using mimetic point mutants. FEBS Open Bio 11, 3304–3323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalpage H. A., et al. , Serine-47 phosphorylation of cytochrome c in the mammalian brain regulates cytochrome c oxidase and caspase-3 activity. FASEB J. 33, 13503–13514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerra-Castellano A., Díaz-Moreno I., Velázquez-Campoy A., De la Rosa M. A., Díaz-Quintana A., Structural and functional characterization of phosphomimetic mutants of cytochrome c at threonine 28 and serine 47. Biochim. Biophys. Acta 1857, 387–395 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Mahapatra G., et al. , Phosphorylation of cytochrome c threonine 28 regulates electron transport chain activity in kidney: Implications for AMP kinase. J. Biol. Chem. 292, 64–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan J., et al. , Regulation of respiration and apoptosis by cytochrome c threonine 58 phosphorylation. Sci. Rep. 9, 15815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Beltrán B., et al. , Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48. Proc. Natl. Acad. Sci. U.S.A. 114, E3041–E3050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushnell G. W., Louie G. V., Brayer G. D., High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol. 214, 585–595 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Hanske J., et al. , Conformational properties of cardiolipin-bound cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 109, 125–130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong Y., Muenzner J., Grimm S. K., Pletneva E. V., Origin of the conformational heterogeneity of cardiolipin-bound cytochrome C. J. Am. Chem. Soc. 134, 18713–18723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagan V. E., et al. , Cytochrome c/cardiolipin relations in mitochondria: A kiss of death. Free Radic. Biol. Med. 46, 1439–1453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rytömaa M., Kinnunen P. K., Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J. Biol. Chem. 269, 1770–1774 (1994). [PubMed] [Google Scholar]
- 21.Pinheiro T. J. T., Elöve G. A., Watts A., Roder H., Structural and kinetic description of cytochrome c unfolding induced by the interaction with lipid vesicles. Biochemistry 36, 13122–13132 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Tuominen E. K. J., Wallace C. J. A., Kinnunen P. K. J., Phospholipid-cytochrome c interaction: Evidence for the extended lipid anchorage. J. Biol. Chem. 277, 8822–8826 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Kagan V. E., et al. , Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 1, 223–232 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Hannibal L., et al. , Alternative conformations of cytochrome c: Structure, function, and detection. Biochemistry 55, 407–428 (2016). Correction in: Biochemistry 55, 2062 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Alvarez-Paggi D., et al. , Multifunctional cytochrome c: Learning new tricks from an old dog. Chem. Rev. 117, 13382–13460 (2017). Correction in: Chem. Rev. 117, 14014 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Capdevila D. A., et al. , Coupling of tyrosine deprotonation and axial ligand exchange in nitrocytochrome c. Chem. Commun. (Camb.) 50, 2592–2594 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Capdevila D. A., et al. , Active site structure and peroxidase activity of oxidatively modified cytochrome c species in complexes with cardiolipin. Biochemistry 54, 7491–7504 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Batthyány C., et al. , Time course and site(s) of cytochrome c tyrosine nitration by peroxynitrite. Biochemistry 44, 8038–8046 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Díaz-Moreno I., García-Heredia J. M., Díaz-Quintana A., Teixeira M., De la Rosa M. A., Nitration of tyrosines 46 and 48 induces the specific degradation of cytochrome c upon change of the heme iron state to high-spin. Biochim. Biophys. Acta 1807, 1616–1623 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Cassina A. M., et al. , Cytochrome c nitration by peroxynitrite. J. Biol. Chem. 275, 21409–21415 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Kagan V. E., et al. , Oxidative lipidomics of apoptosis: Redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free Radic. Biol. Med. 37, 1963–1985 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Abriata L. A., et al. , Nitration of solvent-exposed tyrosine 74 on cytochrome c triggers heme iron-methionine 80 bond disruption. Nuclear magnetic resonance and optical spectroscopy studies. J. Biol. Chem. 284, 17–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaurasia G., et al. , UniHI: An entry gate to the human protein interactome. Nucleic Acids Res. 35, D590–D594 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenwood C., Palmer G., Evidence for the existence of two functionally distinct forms cytochrome c manomer at alkaline pH. J. Biol. Chem. 240, 3660–3663 (1965). [PubMed] [Google Scholar]
- 35.Scott R. A., Mauk A. G., Cytochrome c: A Multidisciplinary Approach (University Science Books, 1996). [Google Scholar]
- 36.Luntz T. L., Schejter A., Garber E. A., Margoliash E., Structural significance of an internal water molecule studied by site-directed mutagenesis of tyrosine-67 in rat cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 86, 3524–3528 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capdevila D. A., et al. , Specific methionine oxidation of cytochrome c in complexes with zwitterionic lipids by hydrogen peroxide: Potential implications for apoptosis. Chem. Sci. (Camb.) 6, 705–713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin V., Shaw G. S., Konermann L., Cytochrome c as a peroxidase: Activation of the precatalytic native state by H2O2-induced covalent modifications. J. Am. Chem. Soc. 139, 15701–15709 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Tomášková N., et al. , Early modification of cytochrome c by hydrogen peroxide triggers its fast degradation. Int. J. Biol. Macromol. 174, 413–423 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Jemmerson R., et al. , A conformational change in cytochrome c of apoptotic and necrotic cells is detected by monoclonal antibody binding and mimicked by association of the native antigen with synthetic phospholipid vesicles. Biochemistry 38, 3599–3609 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Godoy L. C., et al. , Disruption of the M80-Fe ligation stimulates the translocation of cytochrome c to the cytoplasm and nucleus in nonapoptotic cells. Proc. Natl. Acad. Sci. U.S.A. 106, 2653–2658 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oursler M. J., Bradley E. W., Elfering S. L., Giulivi C., Native, not nitrated, cytochrome c and mitochondria-derived hydrogen peroxide drive osteoclast apoptosis. Am. J. Physiol. Cell Physiol. 288, C156–C168 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Souza J. M., Castro L., Cassina A. M., Batthyány C., Radi R., Nitrocytochrome c: Synthesis, purification, and functional studies. Methods Enzymol. 441, 197–215 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Nur-E-Kamal A., et al. , Nuclear translocation of cytochrome c during apoptosis. J. Biol. Chem. 279, 24911–24914 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Zhao S., Aviles E. R. Jr., Fujikawa D. G., Nuclear translocation of mitochondrial cytochrome c, lysosomal cathepsins B and D, and three other death-promoting proteins within the first 60 minutes of generalized seizures. J. Neurosci. Res. 88, 1727–1737 (2010). [DOI] [PubMed] [Google Scholar]
- 46.González-Arzola K., et al. , Structural basis for inhibition of the histone chaperone activity of SET/TAF-Iβ by cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 112, 9908–9913 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.González-Arzola K., et al. , Mitochondrial cytochrome c shot towards histone chaperone condensates in the nucleus. FEBS Open Bio 11, 2418–2440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Díaz-Moreno I., Velázquez-Cruz A., Curran-French S., Díaz-Quintana A., De la Rosa M. A., Nuclear cytochrome c - a mitochondrial visitor regulating damaged chromatin dynamics. FEBS Lett. 592, 172–178 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Kalkavan H., et al. , Sublethal cytochrome c release generates drug-tolerant persister cells. Cell 185, 3356–3374.e22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanderson T. H., et al. , Cytochrome C is tyrosine 97 phosphorylated by neuroprotective insulin treatment. PLoS One 8, e78627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hüttemann M., Lee I., Grossman L. I., Doan J. W., Sanderson T. H., Phosphorylation of mammalian cytochrome c and cytochrome c oxidase in the regulation of cell destiny: Respiration, apoptosis, and human disease. Adv. Exp. Med. Biol. 748, 237–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arunachalam M., et al. , Current perspectives of healthy mitochondrial function for healthy neurons. Curr. Drug Targets 22, 1688–1703 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Demicheli V., et al. , Cardiolipin interactions with cytochrome c increase tyrosine nitration yields and site-specificity. Arch. Biochem. Biophys. 703, 108824 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Alonso D., et al. , Coexistence of translocated cytochrome c and nitrated protein in neurons of the rat cerebral cortex after oxygen and glucose deprivation. Neuroscience 111, 47–56 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Martínez J., et al. , Mitofusins modulate the increase in mitochondrial length, bioenergetics and secretory phenotype in therapy-induced senescent melanoma cells. Biochem. J. 476, 2463–2486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence A., Jones C. M., Wardman P., Burkitt M. J., Evidence for the role of a peroxidase compound I-type intermediate in the oxidation of glutathione, NADH, ascorbate, and dichlorofluorescin by cytochrome c/H2O2. Implications for oxidative stress during apoptosis. J. Biol. Chem. 278, 29410–29419 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Gilmore K. J., Quinn H. E., Wilson M. R., Pinocytic loading of cytochrome c into intact cells specifically induces caspase-dependent permeabilization of mitochondria: Evidence for a cytochrome c feedback loop. Cell Death Differ. 8, 631–639 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Kalpage H. A., et al. , Tissue-specific regulation of cytochrome c by post-translational modifications: Respiration, the mitochondrial membrane potential, ROS, and apoptosis. FASEB J. 33, 1540–1553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barayeu U., et al. , Cytochrome c autocatalyzed carbonylation in the presence of hydrogen peroxide and cardiolipins. J. Biol. Chem. 294, 1816–1830 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guerra-Castellano A., et al. , Post-translational modifications of cytochrome c in cell life and disease. Int. J. Mol. Sci. 21, 8483 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adam W., Kurz A., Saha-Möller C. R., Peroxidase-catalyzed oxidative damage of DNA and 2′-deoxyguanosine by model compounds of lipid hydroperoxides: Involvement of peroxyl radicals. Chem. Res. Toxicol. 13, 1199–1207 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Saha A., Goldstein S., Cabelli D., Czapski G., Determination of optimal conditions for synthesis of peroxynitrite by mixing acidified hydrogen peroxide with nitrite. Free Radic. Biol. Med. 24, 653–659 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Claiborne A. L., Catalase Activity (CRC Handbook of Methods for Oxygen Radical Research, 1985), vol. 1. [Google Scholar]
- 64.Assfalg M., et al. , Structural model for an alkaline form of ferricytochrome C. J. Am. Chem. Soc. 125, 2913–2922 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 66.Maier J. A., et al. , ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Case D. A., et al. , Amber 2018 (University of California, San Francisco, 2018), pp. 1–923. [Google Scholar]
- 68.Roe D. R., Cheatham T. E. III, PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Grant B. J., Skjaerven L., Yao X. Q., The Bio3D packages for structural bioinformatics. Protein Sci. 30, 20–30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996). [DOI] [PubMed] [Google Scholar]
- 71.Slee E. A., et al. , Ordering the cytochrome c-initiated caspase cascade: Hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144, 281–292 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.W. H. McDonald et al., Sequencing of monoclonal antibody 1D3 (mAb1D3). ProteomeXchange: Accession code: PXD036958. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD036958. Accessed 24 September 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plasmids 1D3LC and 1D3HC encoding the light and heavy chains, respectively, have been deposited at Addgene (https://www.addgene.org/).
LC/MS data have been deposited in ProteomeXchange (PXD036958) (72).