Significance
Hfq is a protein hexamer necessary for gene regulation by noncoding RNA in bacteria during infection or under stress. In the cell, Hfq must distinguish its RNA partners from many similar nucleic acids. Mass spectrometry dissociation patterns, together with molecular dynamics simulations, showed that flexible extensions of each Hfq subunit form a dense network that interconnects the entire hexamer. This network is disrupted by RNA binding, but the lost interactions are compensated by RNA-Hfq subunits. By measuring interactions that are too irregular to be counted by other methods, mass spectrometry shows how flexible protein extensions help chaperones like Hfq recognize their RNA partners in the messy interior of the cell.
Keywords: RNA chaperone, small RNA, intrinsically disordered protein, ion mobility mass spectrometry, surface-induced ion dissociation
Abstract
RNA-binding proteins contain intrinsically disordered regions whose functions in RNA recognition are poorly understood. The RNA chaperone Hfq is a homohexamer that contains six flexible C-terminal domains (CTDs). The effect of the CTDs on Hfq’s integrity and RNA binding has been challenging to study because of their sequence identity and inherent disorder. We used native mass spectrometry coupled with surface-induced dissociation and molecular dynamics simulations to disentangle the arrangement of the CTDs and their impact on the stability of Escherichia coli Hfq with and without RNA. The results show that the CTDs stabilize the Hfq hexamer through multiple interactions with the core and between CTDs. RNA binding perturbs this network of CTD interactions, destabilizing the Hfq ring. This destabilization is partially compensated by binding of RNAs that contact multiple surfaces of Hfq. By contrast, binding of short RNAs that only contact one or two subunits results in net destabilization of the complex. Together, the results show that a network of intrinsically disordered interactions integrate RNA contacts with the six subunits of Hfq. We propose that this CTD network raises the selectivity of RNA binding.
Many RNA-binding proteins (RBPs) contain intrinsically disordered regions (IDRs) (1) with overlapping functions that have been difficult to disentangle. For example, IDRs may augment specific RNA recognition, connect different RNA-binding modules, and enable the assembly of liquid condensates, while also serving as targets for posttranslational modification (2–4). The heterogeneous and dynamic structures of IDRs make their interactions especially challenging to quantify, and their functions in most RBPs remain poorly understood.
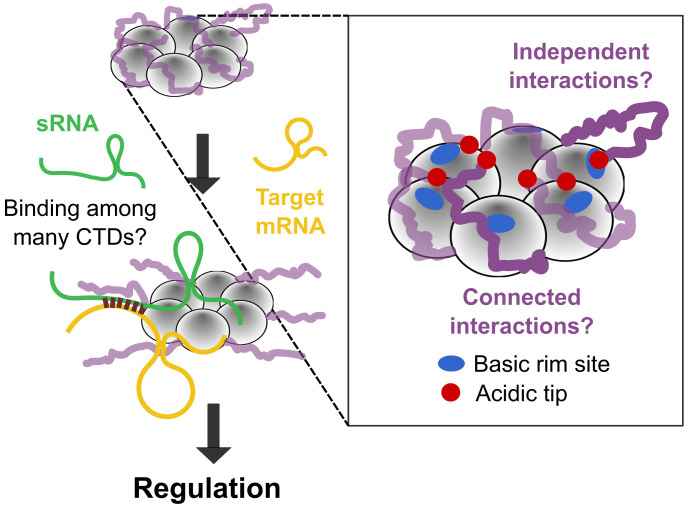
Hfq is a bacterial Sm protein that binds small noncoding RNA (sRNA) and chaperones sRNA regulation of complementary messenger RNAs (mRNAs) (5) (Fig. 1). Deletion of Hfq results in pleiotropic effects, including maladaptive responses to stress and decreased virulence (6). The well-folded core of the Hfq hexamer assembles into a symmetric ring that binds U- and A-rich sequence motifs in sRNA and mRNA substrates (7). Conserved arginine patches on the outer rim of the hexamer also bind RNA and are essential for its chaperone activity (8, 9).
Fig. 1.
Role of Hfq’s CTDs in sRNA regulation. Hfq chaperones the annealing of sRNAs with their target mRNAs, but it is not known how binding of RNAs occurs when the core of Hfq is occluded by many disordered CTDs. Although the acidic tips of the CTDs (red) can interact with basic patches on the rim (blue) (11), the organization and collective behavior of the CTDs is unknown.
Escherichia coli Hfq also has intrinsically disordered C-terminal domains (CTDs) that extend outward from the core of the hexamer (10). Each monomer containing 102 residues contributes a 37-amino-acid (aa) CTD, creating a crowded zone of disordered polypeptide around the protein. This ring-shaped organization, which is unlike disordered regions in other RBPs, raises the possibility that the Hfq CTDs act together rather than individually.
The CTD conformations of E. coli Hfq have never been fully resolved. Nevertheless, NMR chemical shift perturbations and molecular dynamics (MD) simulations determined that the CTDs interact with the rim of the hexamer (12). Additionally, unassigned electron density in a crystal structure of Hfq bound to RydC sRNA suggested that the CTDs make distributed contacts with the protein–RNA surface (13). These results aligned with the earlier observation that the CTDs (residues 65 to 102) stabilize the Hfq hexamer (14) and contribute to its function (15–20). We found that semiconserved acidic residues at the C terminus mimic nucleic acid, competing with RNA for binding to the rim (11, 21, 22). Competition with the CTDs can result in preferential dissociation of nonspecific RNA and retention of specific RNA ligands. More recently, it was shown that the bases and the tips of the CTDs interact synergistically with particular Hfq surfaces, leading to different effects depending on the RNA ligand (23).
Because of their intrinsic disorder and sixfold symmetry, how the CTDs organize around Hfq stabilizing the hexamer is still unknown. Additionally, it is not known if each CTD acts locally and independently, or if the six CTDs act together to accommodate or displace an incoming RNA (Fig. 1). Moreover, the energetic contributions of individual CTDs to RNA binding have been almost impossible to quantify.
We addressed these challenges by using native mass spectrometry (nMS) coupled with surface-induced dissociation (SID) (Fig. 2A and SI Appendix, Fig. S1A). In nMS, the protein complex is exchanged into a volatile electrolyte, allowing transfer of the intact native complex to the gas phase (24). After ionization, collision of the precursor ion with a surface (nMS-SID) dissociates the complex into product ions that provide information about the stabilities of the noncovalent interfaces within the complex and their molecular organization (25). This method has been used to characterize the stability, structure, and assembly pathways of many protein complexes, including RBPs and membrane proteins (26–28). Although nMS does not reveal atomic detail, it is uniquely capable of resolving mixtures of complexes by mass and shape. Yet, despite its promise for discovery, nMS-SID studies of large biomolecular complexes typically require customized instrumentation (SI Appendix, Fig. S1A).
Fig. 2.
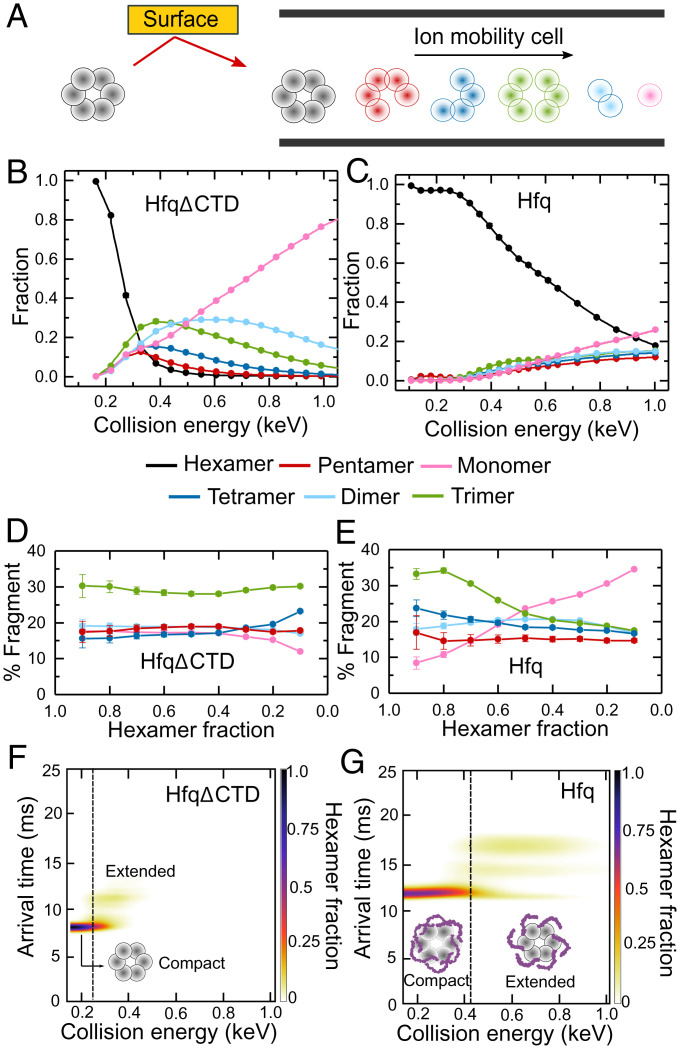
Disordered CTDs stabilize Hfq. (A) nMS-SID dissociates the Hfq hexamer precursor ions into oligomers that retain the connectivity of the native protein. Fragments are separated according to their arrival time after traversing an ion mobility cell (see also SI Appendix, Fig. S1 and Table S1). (B and C) Energy-resolved mass spectrum of (B) HfqΔCTD and (C) Hfq, showing the fraction of each fragment at different CE. The CE are corrected for the mass of the CTDs (SI Appendix, Eq. S1). Reported fractions are the sum of the intensities of each dissociation product normalized by the total intensity of all products. Symbols report the average of three replicates. Some SEs are smaller than the symbols. Solid lines represent a linear interpolation of the data. (D and E) Percentage of each oligomer (pentamer, tetramer, trimer, dimer, or monomer) in the dissociation products, as a function of the remaining hexamer fraction for (D) HfqΔCTD and (E) Hfq. Errors are the spread of the ERMS curves, normalized by the total dissociated fraction and converted to a percentage. Colored as in B and C. Solid lines are a visual guide. (F and G) Surface-induced unfolding (SIU) of (F) HfqΔCTD and (G) Hfq. Extended ions arrive later than compact ions. Color scale, fraction of hexamer; dashed vertical lines, CE at the transition from compact to extended protein, at which the hexamer fractions are ∼0.2 and ∼0.7, respectively.
Here, by using nMS-SID and all-atom MD simulations, we show that the six disordered CTDs of apo Hfq form extensive interactions that connect and stabilize the entire hexamer. When RNA binds any subunit of Hfq, these stabilizing interactions are disrupted throughout the hexamer. Taken together, our results show how disordered regions can integrate RNA–protein interactions across a multisubunit chaperone.
Results
Disordered CTDs Stabilize Hfq.
To better understand the interactions of the flexible CTDs, we first used nMS-SID on wild-type (WT) E. coli Hfq (102 aa per subunit) and a truncated Hfq lacking the CTDs (HfqΔCTD; 65 aa per subunit). Precursor ions (hexamer) and their product ions were resolved by m/z and by drift time (SI Appendix, Fig. S1 B and C). We analyzed the dissociation products (pentamer, tetramer, trimer, dimer, and monomer) obtained at increasing collision energies (CE) with the surface (Fig. 2A). The resulting energy-resolved mass spectra (ERMS) showed that the two proteins have different stabilities and fragmentation patterns (Fig. 2 B and C). HfqΔCTD reached 20% fragmentation (80% hexamer remaining) at ∼220 eV, compared to ∼390 eV for WT Hfq (Fig. 2 B and C, black lines). This large difference demonstrated that the WT protein is much more stable than the truncated version, in agreement with previous reports (14).
The fraction of HfqΔCTD hexamer sharply decreased with modest increases in collision energy, with 90% fragmentation of the hexamer at CE ∼350 eV. In contrast, dissociation of WT Hfq increased gradually over a wide range of CE, and ∼20% of the hexamer remained intact even at CE = 1,000 eV; Hfq fragmentation was less steep than for HfqΔCTD even when considering the broader collision energy range involved (SI Appendix, Fig. S2). This gradual response to higher CE suggested that the CTDs prevent dissociation of the complex across a range of energies. One explanation is that the CTDs form intersubunit interactions that reorganize upon activation. Additionally, the CTDs may be organized differently in each Hfq hexamer, causing dissociation over a continuum of CE.
Disordered CTDs Impact the Connectivity of Hfq.
The dissociation patterns for HfqΔCTD and Hfq generated fragments in different ratios, indicating a different degree of connectivity between subunits in the two proteins (Fig. 2 B–E). We compared the dissociation products of the two proteins at energies that deplete the hexamer equally. HfqΔCTD dissociated into twice as many trimers (∼30%) as other fragments (∼15 to 20%) regardless of the total amount of hexamer dissociated (Fig. 2D).
For WT Hfq, the distribution of dissociation products was markedly different from HfqΔCTD (Fig. 2E), indicating that the CTDs contribute to the subunit interfaces, as also reported earlier (14, 29). For example, the percentage of trimers decreased while the percentage of monomers increased as more hexamer was fragmented. This suggests that some hexamers dissociate at energies high enough to produce secondary fragmentation.
Inspection of ion mobility arrival times revealed that as the collision energy increases the initial WT hexamer converts into two complexes that migrate more slowly in the drift chamber, suggesting partial extension or restructuring of the protein hexamer (Fig. 2 F and G). We observed one additional conformation of HfqΔCTD upon activation (Fig. 2F), suggesting that one of the extended Hfq complexes comes from restructuring of the core beta sheet. The second extended form was only observed for WT Hfq and likely arises from extension of the CTDs (Fig. 2G). Altogether, the nMS results support a model in which the CTDs are bound to the core and each other, stabilizing the entire WT hexamer. At increasing CE, the CTDs disentangle, exposing the core and eliminating the stabilizing intersubunit connections. As a result, the fragile hexamer ruptures into smaller complexes.
MD Simulations Reveal a Network of CTD Interactions on Hfq.
To gain more insight into the organization of the disordered CTDs on Hfq, we performed multiple all-atom MD simulations on the WT protein. The simulations were started from 10 previous low-energy Rosetta models of the CTDs (models 1 to 10) (11) and an extended conformation started with four different initial velocities (models e1 to e4). Trajectories ranged from 110 ns to 1500 ns long, with total simulation time for all models = 8.9 µs (SI Appendix, Table S2). For all simulations, the radius of gyration (Rg) dropped after ∼100 ns and remained approximately constant thereafter (SI Appendix, Fig. S3). The obtained Rg agreed with the experimentally determined nMS collisional cross-section (SI Appendix, Table S1).
The simulated CTDs adopted a variety of conformations (Fig. 3A and SI Appendix, Fig. S4), in agreement with NMR experiments showing they are disordered (12, 30). These interactions continued to evolve after the first 100 ns, with some CTDs entering periods of mobility and others remaining less mobile (SI Appendix, Fig. S5). CTD movement was observed in all simulations, although no two CTDs adopted the same conformation (SI Appendix, Fig. S6). Thus, the diverse CTD structures confer heterogeneity to individual Hfq hexamers, as implied by the ERMS results (Fig. 2C).
Fig. 3.
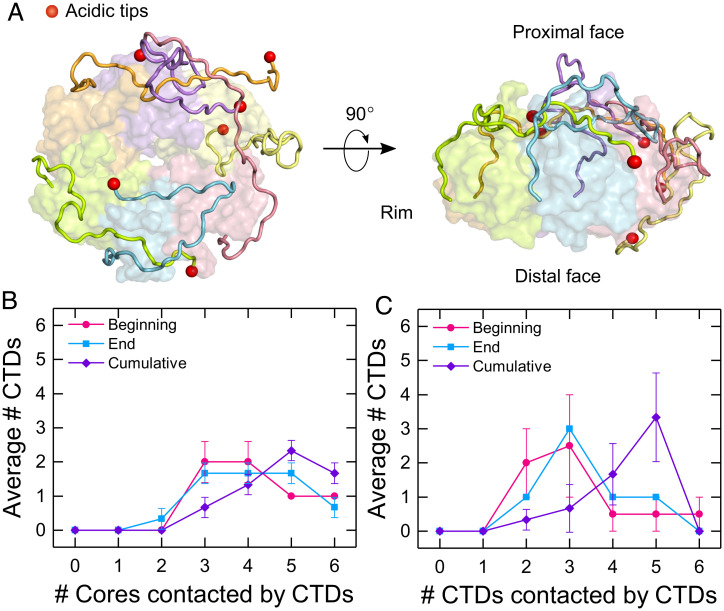
Disordered CTDs create a network of interactions with Hfq. (A) (Left) Top and (Right) side view of an Hfq model from one MD simulation (model 5, at 1,500 ns). Subunits are colored individually with each acidic C terminus shown as a red sphere. (B and C) Average number of CTDs interacting with zero, one, two, three, four, five, or six (B) cores or (C) CTDs. Interactions were summed over the first 100 to 200 ns (beginning), 1,400 to 1,500 ns (end), and for the entire simulation (cumulative) for models 1, 3, and 5. Symbols are the average and SEM (of the various simulations. Solid lines are a visual guide.
To evaluate the interchain interactions, we counted the number of subunit cores or the number of CTDs contacted by each CTD for all models (SI Appendix, Figs. S7 and S8). Most CTDs interacted with the core of the same subunit and adjacent subunits, but some CTDs engaged as many as five other subunits (SI Appendix, Figs. S7 and S8). These long-range intersubunit interactions were common (SI Appendix, Fig. S8). As illustrated for the three longest (1.5 μs) trajectories in Fig. 3 B and C, an average of two CTDs per hexamer interacted with three or four subunit cores over a 100-ns interval (Fig. 3B and SI Appendix, Figs. S7 and S8). This distribution was similar at the beginning and end of the simulations but shifted to larger values when averaged over the entire 1,400-ns simulation (Fig. 3B, purple), indicating that the CTDs sample different regions of Hfq over time, even though at each moment the total number of interactions is approximately the same. The CTDs also interacted among themselves, with a peak of three CTDs contacting five other CTDs when counted over the full 1,400-ns simulation (Fig. 3C, purple and SI Appendix, Figs. S7 and S8). Although the MD simulations cannot sample the full range of structures accessible to the Hfq CTDs, their disorder is apparent even on the 1-µs timescale and among 14 different starting points. The many interchain contacts in the simulations support the conclusion that the CTDs form a mobile network of interactions that connect the entire Hfq hexamer. The effect of this connectivity is illustrated in SI Appendix, Fig. S9 showing how changes in the interactions of one CTD affect the movements of other CTDs.
RNA Binding Stabilizes HfqΔCTD but Destabilizes WT Hfq.
Because nucleic acids compete with the CTDs for binding to Hfq’s rim (11), RNA binding has the potential to alter the organization of the CTDs around Hfq. As a result, the stability and fragmentation pattern of the Hfq•RNA complexes are expected to provide information on how the bound RNA perturbs the connections between Hfq subunits.
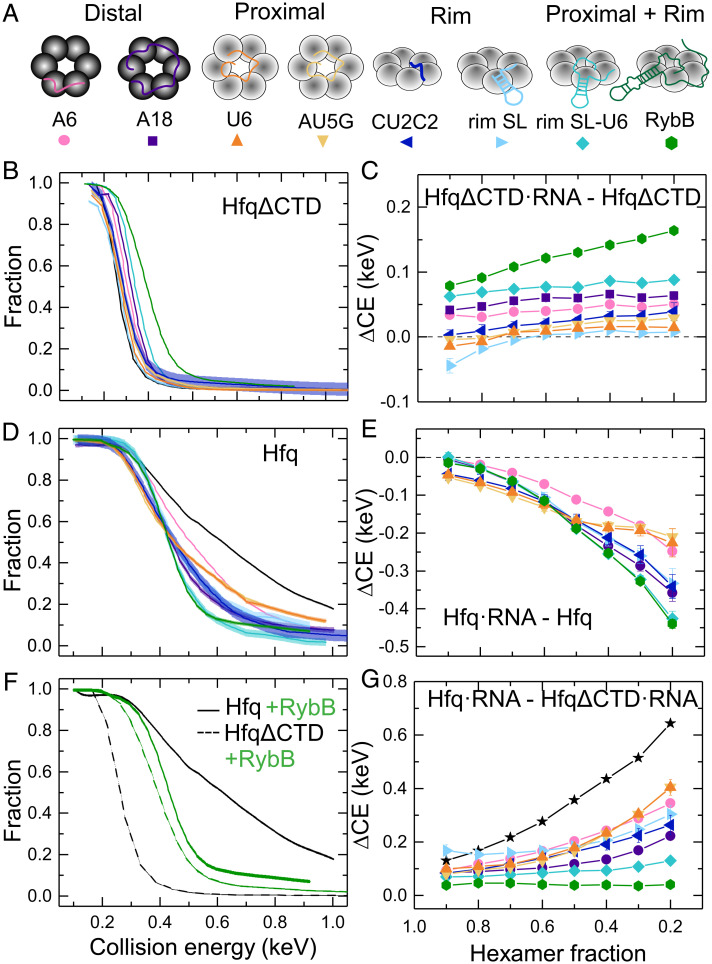
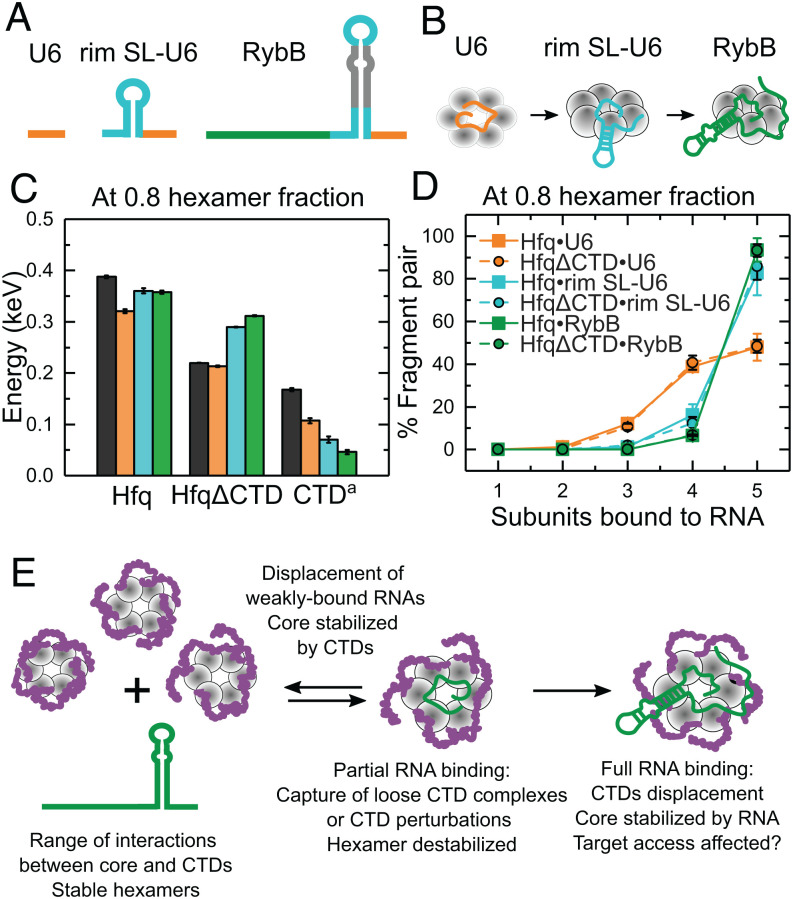
To determine whether RNA binding perturbs the structure of Hfq, we designed a series of short RNAs that mimic the Hfq binding motifs in natural sRNAs and mRNA targets of Hfq (Fig. 4A). The designed RNAs interact with different surfaces of the Hfq hexamer (SI Appendix, Fig. S10). rA6 and rA18 bind two or six subunits on the distal face of Hfq (31). rU6 and rAU5G mimic the sRNA 3′ end and contact five or six subunits around the proximal inner pore (32–34). rCU2C2 and rim-SL, which contains rCU2C2 plus a stable stem loop (SL), mimic RNA motifs that interact with the arginine patch on the rim (13, 21). Additionally, we studied two larger RNAs that interact with both the proximal face and rim: rim SL-U6, designed to mimic an entire sRNA 3′ Hfq binding region, and RybB, a 79-nt natural sRNA that contains both the 3′ Hfq binding region and a 5′ seed region responsible for targeting the mRNA (35, 36).
Fig. 4.
RNA binding stabilizes HfqΔCTD but destabilizes the WT protein. (A) RNAs used in this study (SI Appendix, Table S3) interact with different surfaces of Hfq. (B and D) Remaining fraction of hexamer or hexamer•RNA for (B) HfqΔCTD and (D) Hfq. Colors correspond to the RNAs in A. All CE are corrected for mass (SI Appendix, Eqs. S1–S3). Solid lines represent a linear interpolation of the data. The spread on the interpolated line represents the mean of the errors of individual data points. For clarity, data symbols and error bars are not shown. See SI Appendix, Figs. S11 and S12 for further data on all dissociation products. (F) ERMS of Hfq (solid lines) and HfqΔCTD (dashed lines) with (green) and without (black) bound RybB. (C, E, and G) Collision energy difference (ΔCE) between (C) HfqΔCTD•RNA–HfqΔCTD, (E) Hfq•RNA–Hfq, and (G) Hfq•RNA–HfqΔCTD•RNA as a function of hexameric precursor fraction. Colors and symbols as in A. Black line, star symbols depict Hfq–HfqΔCTD. Solid lines are a visual guide. Errors were calculated from the spread of the ERMS, as explained in SI Appendix, SI Materials and Methods.
We exploited nMS to monitor the dissociation of precursor ions corresponding to 1:1 RNA:Hfq complexes. The mass-corrected ERMS plots of HfqΔCTD revealed a greater dissociation energy when the protein was bound to RNA, indicating that in the absence of the CTDs the RNA stabilizes the Hfq hexamer (Fig. 4B and SI Appendix, Fig. S11). To determine how much stability HfqΔCTD gained from its interactions with the RNA, we calculated the difference in CE between HfqΔCTD•RNA and HfqΔCTD at each remaining fraction of hexamer precursor ion (Fig. 4C). We found that all RNAs stabilized HfqΔCTD over a range of fragmentation. Short RNAs contacting the proximal face (U6 and AU5G) and the rim (CU2C2 and rim-SL) provided minimal extra stability. The longest RNAs tested (RybB and rim SL-U6) provided the most extra stability, followed by RNAs that bind to the distal face (rA18 and rA6).
Unlike HfqΔCTD, RNA binding destabilized WT Hfq, relative to free Hfq. Not only did the RNA complexes fragment at lower CE than the unbound protein, but the shapes of the ERMS plots differed for the various RNAs (Fig. 4D and SI Appendix, Fig. S12), suggesting that the complexes have different structures. Surprisingly, the difference in CE between Hfq•RNA and Hfq showed that even short RNAs destabilized the complexes substantially in the presence of the CTDs (Fig. 4E).
RNA Binding Removes Stabilizing CTD Interactions.
We hypothesized that RNA binding destabilizes Hfq by perturbing the network of disordered CTDs. To determine how much the CTDs contribute to the stability of the complexes, we calculated the difference in collision energy needed to fragment free WT Hfq and HfqΔCTD with and without RNA (Fig. 4 F and G). In the absence of RNA, the CTDs significantly stabilized the Hfq hexamer (Fig. 4 F and G, black lines). In the presence of RybB sRNA, the complexes had similar stabilities that were intermediate between apo WT Hfq and apo HfqΔCTD (Fig. 4F, green lines). This result suggested that RNA binding opposes the effects of the CTDs while stabilizing the Hfq core.
For the free proteins, the observed stability gap widened as more hexamer was fragmented (Fig. 4G), suggesting a range of complexes with variable strengths of CTD interactions, as already deduced by comparison of the protein’s ERMS (Fig. 2 B and C). Upon RNA binding, the stability conferred by the CTDs became more uniform (Fig. 4G, colored lines). Thus, RNA binding results in complexes that dissociate more homogeneously.
All the RNAs tested interfered with stabilization by the CTDs. The effect was strongest for the longest RNAs (rim SL-U6 and RybB) that presumably displace more CTD contacts with the rim and proximal face of Hfq. However, even the smallest RNAs tested diminished the stabilizing effect of the CTDs (Fig. 4G). Interestingly, this loss was also substantial for RNAs binding the distal face, even though only the first few residues of the CTDs are close to this surface (10).
Next, we studied how RNA binding affected the dissociation pathways of Hfq and HfqΔCTD, by determining which subcomplexes retained the RNA after hexamer dissociation (SI Appendix, Fig. S13A). Comparison of the dissociation pathways revealed similar fragmentation of RNA complexes with Hfq and with HfqΔCTD (SI Appendix, Fig. S13 B and C). This result suggested that RNA binds the core of Hfq and that perturbation of the CTDs likely comes from their displacement from the core and not from direct interactions with the RNA. CTD displacement was greatest for the largest RNA (RybB sRNA), as the ERMS for Hfq and HfqΔCTD almost overlapped (Fig. 4F). The ERMS of shorter RNA complexes were closer to the ERMS of the free proteins, indicating that binding required less CTD displacement (Fig. 4 B and D).
Progressive RNA Binding Displaces the CTDs from Hfq.
The design of our RNAs allowed us to investigate the interplay between Hfq, the disordered CTDs, and RNA–protein interactions as an sRNA progressively binds to the protein (Fig. 5 A and B). For this, we compared the stabilities and dissociation pathways of WT Hfq and HfqΔCTD in the absence and presence of RNAs mimicking a stepwise binding process (Fig. 5B). On the one hand, we found that as the RNA interacts with more surfaces of Hfq, the RNA conferred stability to the protein core (Fig. 5C; HfqΔCTD). However, RNA binding was accompanied by a loss of favorable CTD interactions (Fig. 5C, CTD = Hfq•RNA-HfqΔCTD•RNA); see also Fig. 4G) that reduced stability overall (Fig. 5C, Hfq). Progressive RNA binding also shifted the dissociation products to increasingly larger fragments (Fig. 5D). This dissociation pattern was the same for Hfq and HfqΔCTD, indicating similar RNA interactions with the core in both proteins. Thus, stable interactions between RNA and the core are established as the CTDs are displaced.
Fig. 5.
RNA binding progressively displaces the Hfq CTDs. (A) RNAs mimicking segments of RybB sRNA that progressively interact with more Hfq surfaces: 3′ U tail (orange), terminal stem-loop (cyan), 5′ seed (dark green). (B) Cartoon of progressive RNA binding. (C) Stabilities of Hfq, HfqΔCTD (core) and conferred by the CTDs a(Hfq•RNA-HfqΔCTD•RNA) when bound to the RNAs shown in A. Colors correspond to the RNAs in B. Black bar, protein only. Stabilities and errors as in Fig. 4C (SI Appendix, SI Materials and Methods). (D) Percentage of fragment pairs after the collision of Hfq (solid lines, squares) or HfqΔCTD (dashed lines, filled circles) bound to the RNAs in A (see also SI Appendix, Fig. S13). Errors as in Figs. 2 D and E. (E) Assorted organization of the disordered CTDs results in heterogeneous hexamers. RNAs could bind hexamers with open surfaces or perturb a CTD. Local CTD disturbances destabilize the hexamer and are transmitted to the rest of the protein, promoting further CTD displacement and stable binding for favorable RNAs. Nonpreferred RNAs are displaced by competition with the CTDs (11), reestablishing hexamer stability.
Discussion
In this work, we employed nMS-SID to analyze Hfq complexes of defined stoichiometry, stability, and shape. The results demonstrate that the CTDs connect the Hfq subunits, stabilizing the hexamer (Fig. 2 C and G and SI Appendix, Fig. S13A). MD simulations using an improved force field for IDPs (37) suggest an explanation for this stabilization by revealing that each CTD can dynamically interact with several other CTDs and folded domains (Fig. 3 and SI Appendix, Figs. S4, S5, and S8). Although RNA binding disrupts the network of CTD interactions, RNA also stabilizes the folded core of Hfq. RNAs that form multiple favorable contacts with Hfq offset the loss of stabilization by the CTDs, while RNAs that do not form favorable contacts are displaced by the CTD interactions, explaining how the CTDs help Hfq discriminate among different RNAs (11, 22).
Based on our results, we propose that RNAs stably bind Hfq through a stepwise process (Fig. 5E). An RNA may first take advantage of configurations in which an Hfq region is exposed, or a CTD is loosely folded. The initial interaction with a segment of the RNA perturbs nearby CTDs. Because the CTDs form multiple contacts with the core and each other (Fig. 3 B and C and SI Appendix, Figs. S7 and S8), perturbations propagate to the rest of the hexamer (SI Appendix, Fig. S9), resulting in more CTD displacement and further RNA binding. If binding is favorable, the hexamer is stabilized. However, if binding is not favorable, the hexamer is destabilized, making it more favorable for the CTDs to regain their interactions with the rest of the protein. This search for stability could explain the removal of weakly bound RNAs when the CTDs are present (11).
The current results agree with previous binding experiments reporting a higher prevalence of nonspecific RNA binding to HfqΔCTD and lower populations of kinetically stable complexes (22). Additionally, even a flexible network of CTDs could facilitate sRNA competition and the search for mRNA targets, as binding of a second RNA will also perturb the CTDs, stimulating the disassembly of noncognate ternary complexes. Finally, the varied CTD conformations impart asymmetry to the Hfq homohexamer that may also contribute to the selection of RNA ligands.
Our nMS-SID results showed that the CTDs make the Hfq hexamer more resistant to dissociation by collision (Fig. 2C). This observation agrees with previous collision-induced dissociation MS experiments showing that E. coli Hfq is more stable than Hfq from Vibrio cholerae, which has a 22-residue CTD (38). Vincent et al. (38) attributed this stabilization to packing of the CTDs along the intersubunit interfaces. Based on the possibility that the CTDs unfold before the subunits dissociate (Fig. 2G and SI Appendix, Fig. S1) and MD results showing the CTD interact with multiple subunits (Fig. 3 B and C and SI Appendix, Figs. S4 and S5A), we propose that the Hfq hexamer is additionally stabilized by these intersubunit interactions that are facilitated by longer CTDs. Future experiments with longer or shorter CTDs will help elucidate how variable CTD contacts influence Hfq’s stability.
Network connectivity can explain why the binding of even short RNAs reduced the CTD’s stabilizing contribution (Figs. 4 D–G and 5C): Short RNAs such as rU6 still contact all six Hfq subunits and likely reduce the number of allowed CTD configurations, as suggested by the change in the shape of the ERMS plot (Fig. 4D). However, short RNAs may perturb fewer CTDs, or allow reestablishment of CTD contacts lost after RNA binding, explaining why they reduce the CTD contribution less than long RNAs. Since RNA-bound HfqΔCTD and Hfq fragmented similarly (Fig. 5D and SI Appendix, Fig. S13), stable RNA binding seems to involve contacts with the core and not the CTDs, as previously proposed (13, 30, 39). Finally, destabilization of Hfq but not HfqΔCTD when rA18 RNA is bound (Fig. 4C) supports a reported functional link between the distal RNA binding face and R66 at the start of the CTD (23).
Hfq is a model protein with disordered regions that act synergistically to communicate perturbations among its subunits. This feature is enabled by an architecture of identical monomers, each providing an identical disordered region. The single-stranded DNA binding (SSB) protein, a homotetramer involved in DNA repair, replication, and recombination (40), also contains disordered CTDs with parallels to Hfq: They impart stability to the SSB tetramer (41) but are displaced upon partial DNA engagement, modulating binding and SSB oligomerization (42). Importantly, partial deletion of SSB’s CTDs results in impaired activity, indicating a critical role for multiple CTD interactions in cellular function (43). Histone tails are also thought to fold heterogeneously around the DNA (44, 45) and to be disrupted by chaperone binding or posttranslational modification (46). It would be interesting to know if other RNA-binding proteins use interconnected IDPs to integrate the molecular interactions within RNA–protein complexes.
Materials and Methods
RNA Preparation.
RybB RNA was transcribed using phage T7 RNA polymerase followed by 8% polyacrylamide gel purification (8 M urea). The remaining short RNAs used in the study were purchased from IDT with high-performance liquid chromatography purification. See SI Appendix, Table S3 for RNA sequences.
Protein Expression and Purification.
HfqΔCTD and Hfq were purified as described before (21).
Sample Preparation for nMS.
Protein (HfqΔCTD and Hfq) and RNA (rA18, rim SL-U6 and RybB) samples were dialyzed overnight into 500 mM ammonium acetate, pH 6.8 (99.99%, MilliporeSigma), with eight buffer exchanges (3.5-kDa mass cutoff microdialysis; Pierce). This ionic strength prevented precipitation of protein–RNA complexes at the concentrations (10 µM hexamer) required for ion mobility MS. The remaining RNA samples were used as supplied by the manufacturer and did not require additional desalting. Protein–RNA complexes were prepared by mixing 1:1 RNA and HfqΔCTD or Hfq to a final concentration of 10 µM each; 400 mM ammonium acetate (final concentration) plus triethylammonium acetate (1 M, MilliporeSigma; 100 mM final concentration for charge reduction) were subsequently added to the samples.
MS.
All samples were introduced into the mass spectrometer using nano-electrospray emitters that were prepared in-house using a Sutter P-97 micropipette puller. All spectra in this work were acquired on a Waters Synapt G2 HDMS instrument (Waters Corporation) modified with an SID device between a shortened trap stacked ring ion guide and an ion mobility cell, as described previously (47). SID lenses can be tuned either to transmit ions for MS or to direct the ions onto the surface for collision. Typical voltage settings and instrument parameters used here for transmission mode and SID can be found in SI Appendix, Tables S4 and S5). ERMS were produced by acquiring data from tandem MS experiments with SID voltage potentials ranging from 15 and 140 V. Each experiment was repeated in technical triplicate. Additional information is provided in SI Appendix.
Analysis of MS data.
Ion mobility was used to separate product ions and selection rules for each SID product were made using Waters Corporation Driftscope 2.9 software. The intensity of subcomplexes were extracted from SID spectra with TWIMExtract v1.3 (48). CE were calculated as , where is the charge state of the precursor ions and is the SID voltage. ERMS were corrected by and (see also SI Appendix, Eqs. S1–S3). Additional information provided in SI Appendix.
MD Simulations.
All simulations were performed with the MD program OpenMM (49) and CHARMM36m force-field (50). Simulations were started from the Protein Data Bank ID 1HK9, which included residues 7 to 68 (29). The starting structures of the missing CTDs were obtained from 1) top 10 Rosetta models (15) (models 1 to 10) and 2) one structure in which CTDs are fully extended (11). Four simulations with extended CTDs were performed starting with different initial velocities. All protein structures were embedded in a water box and neutralized with 150 mM NaCl. Additional simulation and setup details are provided in SI Appendix. Following a 100-ns equilibration, the models were run for an additional 1,400 ns (models 1, 3, and 5), 400 ns (models 2 and 4), 100 ns (models 6 to 10), 1,000 ns (e1 and e4), or 10 ns (e2 and e3; SI Appendix, Table S2). To gain information about the short-term structures and their evolution, we analyzed contacts between CTDs and cores of various subunits during various time intervals of the simulation (SI Appendix, Figs. S7 and S8). To characterize the CTDs dynamics, we calculated root-mean-square deviation values for chosen individual CTD residues (SI Appendix, Figs. S5 and S6). Additional information is provided in SI Appendix.
Supplementary Material
Acknowledgments
The authors thank Benjamin Jones for his assistance with instrument maintenance and automated data acquisition and Andrew Santiago-Frangos for help with initial samples. This work was funded by grants from NIH (1P41GM128577-01 to V.H.W.; R35 GM136351-03 to S.A.W.) and the Johns Hopkins University Provost’s Office (PURA to H.L.). Part of this research was conducted using computational resources at the Maryland Advanced Research Computing Center.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208780119/-/DCSupplemental.
Data, Materials, and Software Availability
MD simulation trajectories and Energy Resolved mass spectra raw data have been deposited in the Johns Hopkins Data Archive Dataverse Network (https://doi.org/10.7281/T1/RTSGO0) (51).
References
- 1.Tompa P., Csermely P., The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 18, 1169–1175 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Ottoz D. S. M., Berchowitz L. E., The role of disorder in RNA binding affinity and specificity. Open Biol. 10, 200328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basu S., Bahadur R. P., A structural perspective of RNA recognition by intrinsically disordered proteins. Cell. Mol. Life Sci. 73, 4075–4084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp P. A., Chakraborty A. K., Henninger J. E., Young R. A., RNA in formation and regulation of transcriptional condensates. RNA 28, 52–57 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner E. G. H., Romby P., Small RNAs in Bacteria and Archaea: Who they are, what they do, and how they do it. Adv. Genet. 90, 133–208 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Sobrero P., Valverde C., The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 38, 276–299 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Santiago-Frangos A., Woodson S. A., Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 9, e1475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauer E., Schmidt S., Weichenrieder O., Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U.S.A. 109, 9396–9401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panja S., Schu D. J., Woodson S. A., Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 41, 7536–7546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beich-Frandsen M., Večerek B., Sjöblom B., Bläsi U., Djinović-Carugo K., Structural analysis of full-length Hfq from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 536–540 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santiago-Frangos A., Jeliazkov J. R., Gray J. J., Woodson S. A., Acidic C-terminal domains autoregulate the RNA chaperone Hfq. eLife 6, 1–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen B., et al. , Structural and dynamic properties of the C-terminal region of the Escherichia coli RNA chaperone Hfq: integrative experimental and computational studies. Phys. Chem. Chem. Phys. 19, 21152–21164 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Dimastrogiovanni D., et al. , Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. eLife 3, 1–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arluison V., et al. , The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 271, 1258–1265 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Večerek B., Rajkowitsch L., Sonnleitner E., Schroeder R., Bläsi U., The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 36, 133–143 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnleitner E., Moll I., Bläsi U., Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology (Reading) 148, 883–891 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Sonnleitner E., et al. , Functional effects of variants of the RNA chaperone Hfq. Biochem. Biophys. Res. Commun. 323, 1017–1023 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Olsen A. S., Møller-Jensen J., Brennan R. G., Valentin-Hansen P., C-terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J. Mol. Biol. 404, 173–182 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Updegrove T. B., Wartell R. M., The influence of Escherichia coli Hfq mutations on RNA binding and sRNA•mRNA duplex formation in rpoS riboregulation. Biochim. Biophys. Acta 1809, 532–540 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Salim N. N., Faner M. A., Philip J. A., Feig A. L., Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 40, 8021–8032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santiago-Frangos A., Kavita K., Schu D. J., Gottesman S., Woodson S. A., C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. U.S.A. 113, E6089–E6096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santiago-Frangos A., et al. , Caulobacter crescentus Hfq structure reveals a conserved mechanism of RNA annealing regulation. Proc. Natl. Acad. Sci. U.S.A. 116, 10978–10987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavita K., et al. , Multiple in vivo roles for the C-terminal domain of the RNA chaperone Hfq. Nucleic Acids Res. 50, 1718–1733 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leney A. C., Heck A. J. R., Native mass spectrometry: What is in the name? J. Am. Soc. Mass Spectrom. 28, 5–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvey S. R., et al. , Relative interfacial cleavage energetics of protein complexes revealed by surface collisions. Proc. Natl. Acad. Sci. U.S.A. 116, 8143–8148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiving A. Q., et al. , Surface-induced dissociation: An effective method for characterization of protein quaternary structure. Anal. Chem. 91, 190–209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M., Dagan S., Wysocki V. H., Protein subunits released by surface collisions of noncovalent complexes: Nativelike compact structures revealed by ion mobility mass spectrometry. Angew. Chem. Int. Ed. Engl. 51, 4336–4339 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Quintyn R. S., Yan J., Wysocki V. H., Surface-induced dissociation of homotetramers with D2 symmetry yields their assembly pathways and characterizes the effect of ligand binding. Chem. Biol. 22, 583–592 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Sauter C., Basquin J., Suck D., Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 31, 4091–4098 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beich-Frandsen M., et al. , Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 39, 4900–4915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Link T. M., Valentin-Hansen P., Brennan R. G., Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 106, 19292–19297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumacher M. A., Pearson R. F., Møller T., Valentin-Hansen P., Brennan R. G., Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 21, 3546–3556 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauer E., Weichenrieder O., Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. U.S.A. 108, 13065–13070 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W., et al. , Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 25, 2106–2117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wassarman K. M., Repoila F., Rosenow C., Storz G., Gottesman S., Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15, 1637–1651 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papenfort K., et al. , SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J., et al. , CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent H. A., et al. , Characterization of Vibrio cholerae Hfq provides novel insights into the role of the Hfq C-terminal region. J. Mol. Biol. 420, 56–69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson K. E., Orans J., Kovach A. R., Link T. M., Brennan R. G., Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 42, 2736–2749 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., Keck J. L., SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mason C. E., et al. , Escherichia coli single-stranded DNA-binding protein: nanoESI-MS studies of salt-modulated subunit exchange and DNA binding transactions. J. Am. Soc. Mass Spectrom. 24, 274–285 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Kozlov A. G., Cox M. M., Lohman T. M., Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 285, 17246–17252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antony E., et al. , Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 425, 4802–4819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison E. A., Bowerman S., Sylvers K. L., Wereszczynski J., Musselman C. A., The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. eLife 7, 1–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghoneim M., Fuchs H. A., Musselman C. A., Histone tail conformations: A fuzzy affair with DNA. Trends Biochem. Sci. 46, 564–578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Zhou M., Huang C., Wysocki V. H., Surface-induced dissociation of ion mobility-separated noncovalent complexes in a quadrupole/time-of-flight mass spectrometer. Anal. Chem. 84, 6016–6023 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Haynes S. E., et al. , Variable-velocity traveling-wave ion mobility separation enhancing peak capacity for data-independent acquisition proteomics. Anal. Chem. 89, 5669–5672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eastman P., et al. , OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLOS Comput. Biol. 13, e1005659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for computational chemistry softwares. J. Comput. Chem. 32, 174–182 (2012). [Google Scholar]
- 51.S. H. Sarni et al., Data from “Intrinsically disordered interaction network in an RNA chaperone revealed by native mass spectrometry.” Johns Hopkins Data Archive Dataverse Network. 10.7281/T1/RTSGO0. Deposited 31 October 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MD simulation trajectories and Energy Resolved mass spectra raw data have been deposited in the Johns Hopkins Data Archive Dataverse Network (https://doi.org/10.7281/T1/RTSGO0) (51).