SUMMARY
Many aspects of the porcine immune system remain poorly characterized, which poses a barrier to improving swine health and utilizing pigs as preclinical models. Here, we employ single-cell RNA sequencing (scRNA-seq) to create a cell atlas of the early-adolescent pig thymus. Our data show conserved features as well as species-specific differences in cell states and cell types compared with human thymocytes. We also describe several unconventional T cell types with gene expression profiles associated with innate effector functions. This includes a cell census of more than 11,000 differentiating invariant natural killer T (iNKT) cells, which reveals that the functional diversity of pig iNKT cells differs substantially from the iNKT0/1/2/17 subset differentiation paradigm established in mice. Our data characterize key differentiation events in porcine thymopoiesis and iNKT cell maturation and provide important insights into pig T cell development.
Graphical Abstract
In brief
Gu et al. use single-cell RNA sequencing to characterize thymopoiesis in pigs. This reveals subpopulations and regulatory networks relevant for understanding cellular immunity in swine. Their analysis includes a census of more than 11,000 thymic iNKT cells, which shows that pig iNKT cell differentiation differs from the iNKT0/1/2/17 paradigm established in mice.
INTRODUCTION
The thymus is responsible for development, selection, and maturation of T cells and therefore plays a critical role in adaptive immunity and central tolerance. Accordingly, thymic dysfunction is associated with a variety of diseases, such as cancer, autoimmune diseases, and infectious diseases (Miller, 2020). The thymus is a target for numerous physiological disorders, including malnutrition, autoimmunity and pathogen infection (Cheng and Anderson, 2018; Savino, 2006; Savino et al., 2007). Thus, a thorough understanding of thymopoiesis is essential for elucidating the mechanisms of cellular immunity and T cell-associated pathologies. Current knowledge about thymus function and cellular composition is based largely on rodent models.However, the dynamics of thymopoiesis and the resulting diversity of mature T cell subsets are unique to each species, which warrants studying the thymus in individual species.
Pigs (Sus scrofa) are an important agricultural species that have emerged as a valuable translational model to bridge the gap between rodent and non-human primate models (Bertho and Meurens, 2021; Dawson, 2011; Käser, 2021; Meurens et al., 2012; Sinkora and Butler, 2016). This is in part because pigs share many anatomical and physiological traits with humans. Genome sequence identity is approximately three times more similar between pigs and humans than between mice and humans (Humphray et al., 2007; Wernersson et al., 2005). In terms of the immune system, pigs express T cell subsets and that are phenotypically similar to humans, and both species share a high degree of nucleotide and protein sequence homology for immune molecules (Dawson, 2011; Käser, 2021; Starbæk et al., 2018). Nevertheless, the porcine immune system has several peculiarities that separate pigs from mice and humans. For instance, pigs have a high frequency of peripheral CD4+CD8a+ effector/memory T cells and γδ T cells (Pescovitz et al., 1990; Stepanova and Sinkora, 2013; Yang and Parkhouse, 1996; Zuckermann and Gaskins, 1996). The latter includes sublineages that are absent in mice and humans (Groh et al., 1989; Rakasz et al., 1997). The extent to which the pig and human immune systems overlap is not yet fully known. This gap can be addressed using next-generation sequencing platforms such as single-cell RNA sequencing (scRNA-seq), which provides unbiased transcriptional profiling of individual cells without marker-based sorting of unique cell subsets.
Here we used scRNA-seq to create a cell atlas of the prepubertal pig thymus. The primordial pig thymus appears by 22 days of gestation (DGs), completes development by 36 DGs, and undergoes a period of allometric growth from 36 DGs to the end of gestation (114 DGs) (Sinkora et al., 2005a). Additional growth occurs postnatally, when most cellular components, including lymphocytes and epithelio-reticular cells, trabeculae, and Hassal’s corpuscles, increase in size and number. Peak weight is reached during early adolescence, between 12 and 24 weeks of age, after which the thymus gradually decreases in size, especially after puberty (Igbokwe and Ezenwaka, 2017).
Our pig thymocyte dataset was combined with publicly available human thymic cell data to systematically compare thymopoiesis between pigs and humans. We identified common features shared between humans and swine as well as pig-specific gene signatures and cell states. Several of the differences were in innate-like T cell subsets that express memory markers and acquire functional competence in the thymus. However, the rarity of some innate T cell populations necessitates mapping their differentiation and subset composition using purified samples. As an illustration, our study included an analysis of purified semi-invariant natural killer T (iNKT) cells. iNKT cells recognize glycolipid antigens presented by the non-polymorphic CD1d molecule via a semi-invariant T cell receptor (TCR) (Godfrey et al., 2010,2015). Their ability to respond rapidly in the periphery is largely due to the presence of preexisting subsets that segregate in the thymus during their development (Stetson et al., 2003).
Our data provide a resource to better understand thymopoiesis and T cell-related diseases in swine. They also elucidate iNKT cell development in pigs. This is of interest for understanding the evolutionary conservation of the iNKT-CD1d system because pigs are distinct from ruminant artiodactyl species that express an alternative CD1D gene structure that cannot present the iNKT cell agonist α-galactosylceramide (Nguyen et al., 2013; Wang et al., 2012; Yang et al., 2019).
RESULTS
Cellular composition of the pig thymus
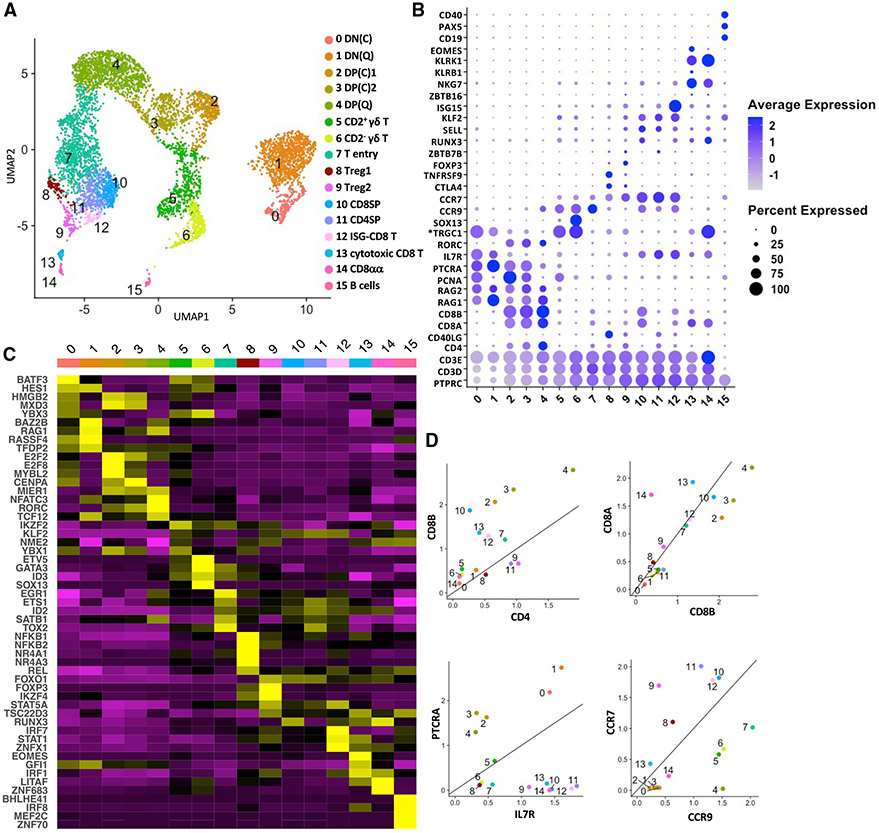
scRNA-seq was performed on the combined thymi of two 22-week-old mixed-breed pigs. We obtained 9,112 cells with 30,638 mean reads and 1,017 median genes per cell (Table S1). After removing cells with unusually low and high gene counts and high mitochondrial gene expression, the remaining cells were clustered using Seurat (v.3.2.2) (Stuart et al., 2019). Canonical cell cycle markers were then used to regress out cell cycle effects before the dimensional reduction step. We obtained 16 distinct clusters (clusters 0–15) (Figure 1A), which were annotated according to lineage marker genes that distinguish mouse and human thymocyte subsets (Figures 1B-1D and S1A; Table S2). Double-negative (DN) cells were separated into cycling clusters (DN(C); cluster 0) and quiescent clusters (DN(Q); cluster 1) according to expression of cell cycle and VDJ recombination genes (Figures 1B and S1B). Both populations expressed classical DN markers, including PTCRA and IL7R (Yui and Rothenberg, 2014) (Figures 1B and 1D). However, IL2RA (CD25) and CD44, which distinguish DN subsets in mice, were not upregulated until later in pig thymocyte development (Rothenberg et al., 2008; Yang et al., 2010; Yui and Rothenberg, 2014) (Figure S1B). Like humans, pig DN cells expressed *CD1A (Res et al., 1997) (Figure S1B). Double-positive (DP) thymocytes (clusters 2–4) consisted of one quiescent cluster (DP(Q)) and two rapidly cycling cell clusters (DP(C)1 and DP(C)2), which, respectively, upregulated G2M and a combination of G2M- and S-phase cell cycle genes (Figure S1B). Cluster 7 was designated as T entry cells because of their high CCR9 and low CCR7 expression (Hu et al., 2015; Uehara et al., 2006) (Figure 1D). Among post-committed thymocytes, we identified CD8 single-positive (CD8SP, cluster 10) and CD4 single-positive (CD4SP, cluster 11) cells, T regulatory (Treg) cells (clusters 8 and 9), γδ T cells (clusters 5 and 6), and three unconventional CD8+ T cell subsets (clusters 12–14). We also detected a minor cluster of B cells (cluster 15) that expressed high levels of SLA-DRA, SLA-DQB1, and CD40 (Figures 1A and 1B; Table S2). This is consistent with previous reports showing that pigs harbor a rare population of B cells that localize in the thymic medulla when mature (Sinkora et al., 2000; Sinkorova et al., 2019). A similar subset of B cells in mice contributes to negative selection (Perera and Huang, 2015).
Figure 1. Single-cell transcriptomics analysis of the cellular composition of the pig thymus.
(A) Uniform manifold approximation and projection (UMAP) visualization of pig thymus cell types, colored by cell clusters. Clusters were identified using the graph-based Louvain algorithm at a resolution of 0.5.
(B)Dot plot showing the Z-scored mean expression of marker genes that were used to designate cell types to cell clusters. The color intensity represents average expression of each marker gene in each cluster. The dot size indicates the proportion of cells expressing each marker gene. Genes with cluster-specific increases in expression are presented in Table S2.
(C) Heatmap showing row-scaled mean expression of the five highest differentially expressed transcription factors in each cluster.
(D) Scatterplots showing the ratio of various lineage marker genes for each thymocyte cluster (excluding B cells). Asterisks indicate non-annotated genes (described in Table S13).
Characterization of unconventional T cell populations
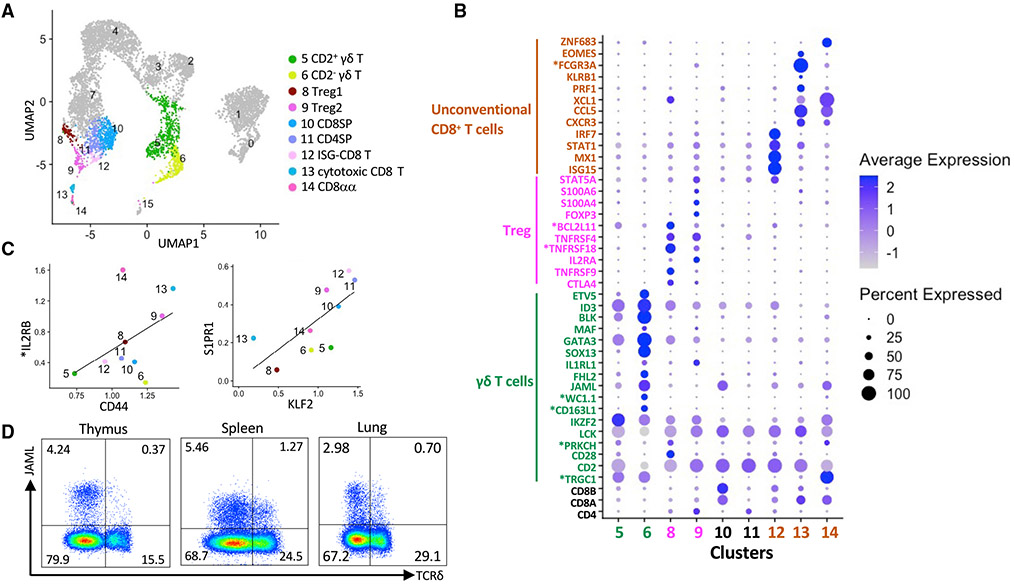
Next we assessed the gene profile of unconventional T cells (Figure 2A). Two clusters of Treg cells were identified, Treg1 and Treg2, which, respectively, correspond to CD25+Foxp3− Treg cell progenitors and CD25+Foxp3+ mature Treg cells in a previously described two-step model of murine Treg cell development (Burchill et al., 2008; Lio and Hsieh, 2008; Owen et al., 2019) (Figures 2B and S2A; Table S3). Step one is driven by strong TCR stimulation, which generates Treg cell progenitors expressing high-affinity CD25 and the TNF receptor (TNFR) superfamily members GITR, OX40, and TNFR2. Step two relies on cytokine signals that promote Treg cell maturation by phosphorylating Stat5 and upregulating Foxp3. Treg1 was enriched for *TNFRSF18 (GITR) and TNFRSF4 (OX40) in addition to other Treg cell signature genes, including CTLA4 and TNFRSF9 (CD137) (Vaeth et al., 2019; Wing et al., 2008) (Figure 2B). This subset also expressed several Nr4a nuclear receptor family members (NR4A1, NR4A2, and NR4A3) (Figure S2A) that are required for transducing high-affinity TCR signals into Foxp3 expression (Sekiya et al., 2013). Treg2 was enriched for FOXP3, IL2RA, and STAT5A (Figure 2B).
Figure 2. Characterization of unconventional T cells.
(A) UMAP visualization of post-committed thymocyte populations.
(B) Dot plot showing Z-scored mean expression of selected marker genes in clusters from (A).
(C) Scatterplots comparing the characteristics of mature T cells based on the ratio of genes associated with memory T cells (left panel) and thymic emigration (right panel).
(D) Representative flow cytometry plots showing JAML expression on thymic, splenic, and lung γδ T cells. Cells were gated on live CD3+ lymphocytes. Asterisks indicate non-annotated genes (described in Table S13).
Three unconventional CD8+ T cell subsets were identified that we designated interferon-stimulated gene (ISG)-CD8 T (cluster 12), cytotoxic CD8 T (cluster 13), and CD8αα cells (cluster 14). ISG-CD8 T cells were enriched for ISGs, including ISG15, MX1, STAT1, and IRF7, which mediate the antiviral activity of interferon (IFN)-α and type I IFN signaling (Perng and Lenschow, 2018) (Figure 2B). Our results are reminiscent of previous reports showing that type I IFN signaling is involved in inducing ISGs during the late stages of human and mouse CD4+ and CD8+ thymocyte development, even in the absence of infection (Colantonio et al., 2011; Xing et al., 2016). Cytotoxic CD8 T and CD8αα T cells expressed a high ratio of CD8A to CD8B (Figure 2B). Compared with conventional CD8SP cells, cytotoxic CD8 and CD8αα T cells were enriched for T cell memory markers (CD44, *IL2RB, CXCR3, and CCL5), NK cell receptors (NKG7, KLRK1, and *KLRD1), activation markers (SLA-DQB1 and SLA-DRA), and the chemokine XCL1, which is the ligand for the XCR1 receptor that is uniquely expressed by cross-presenting dendritic cells (Kroczek et al., 2018) (Figures 2B, 2C, S2B, and S2C; Table S3). Cytotoxic CD8 T cells expressed several additional NK cell signature genes, including PRF1 (perforin), KLRB1 (NK1.1), *FCGR3A (CD16), and EOMES (Figure 2B). These cells had barely detectable levels of KLF2 and S1PR1, which are required for thymic egress (Allende et al., 2004; Carlson et al., 2006) (Figure 2C). This may indicate that cytotoxic CD8 T cells are a thymus-resident population. CD8αα T cells expressed high levels of the transcription factor ZNF683 (Hobit) (Figure 2B), which regulates the transcriptional program of several tissue-resident T cell subsets, including iNKT cells and effector CD8+ T cells (Mackay et al., 2016; Verstichel et al., 2017). We noticed that CD8αα T cells strongly expressed TCR γ-chains (Figure 2B), which is consistent with previous reports showing that mouse and human CD8αα T cells have a mixed αβ and γδ T cell signature (Cheroutre et al., 2011; Dadi et al., 2016; Verstichel et al., 2017).
Like other Laurasiatheria, pigs express a high proportion of γδ T cells (Holderness et al., 2013). Our results agree with previous reports showing that porcine γδ T cells consist of two major subpopulations defined as WC1−GATA3loCD2+ (CD2+) and WC1+GATA3hiCD2− (CD2−) cells (Le Page et al., 2021; Rodríguez-Gómez et al., 2019; Stepanova and Sinkora, 2012) (Figures 2B, S2D, and S2E; Table S3). The CD2+ population preferentially accumulates in lymphoid organs, such as the spleen and lymph nodes, and can simultaneously secrete IFN-γ and tumor necrosis factor alpha (TNF-α) (Sedlak et al., 2014; Stepanova and Sinkora, 2012). The CD2− subset predominates in the blood and is capable of producing interleukin (IL)-17A (Sedlak et al., 2014; Stepanova and Sinkora, 2012). We observed that CD2+ γδ thymocytes (cluster 5) were enriched for genes associated with TCR signaling (CD28, *PRKCH, LCK, and IKZF2), which suggests that CD2+ γδ T cells are more reliant on TCR-mediated stimulation than CD2− γδ T cells. CD2− γδ T cells expressed several transcription factors that program the differentiation of IL17-producing γδ T cells (Tγδ17) in mice, including SOX13, GATA3, MAF, BLK, ETV5, and ID3 (Sagar et al., 2020; Spidale et al., 2018) (Figure 2B). However, we failed to detect RORC, which is required for Tγδ17 lineage commitment (Malhotra et al., 2013; Spidale et al., 2018), even though this transcription factor was abundantly expressed in DP thymocytes (Figure 1B). Like Tγδ17 cells, porcine CD2− γδ T cells are resident in a wide variety of tissues, including the thymus, lymph nodes, spleen, lungs, liver, and intestine (Saalmüller et al., 1990; Sedlak et al., 2014; Stepanova and Sinkora, 2012), where they appear to acquire specialized functions that modulate local immune responses. It is notable that CD2− γδ T cells strongly expressed JAML (Figures 2B and S2E), a costimulatory receptor expressed by mouse epithelial γδ T cells that promotes cellular proliferation and cytokine production by peripheral γδ and CD8+ T cells (McGraw et al., 2021; Verdino et al., 2010; Witherden et al., 2010). Unlike CD8+ T cells, surface expression of JAML molecules was mostly restricted to peripheral γδ T cells (Figures 2D and S2F), suggesting that JAML signaling is not essential for CD2− γδ T cell development.
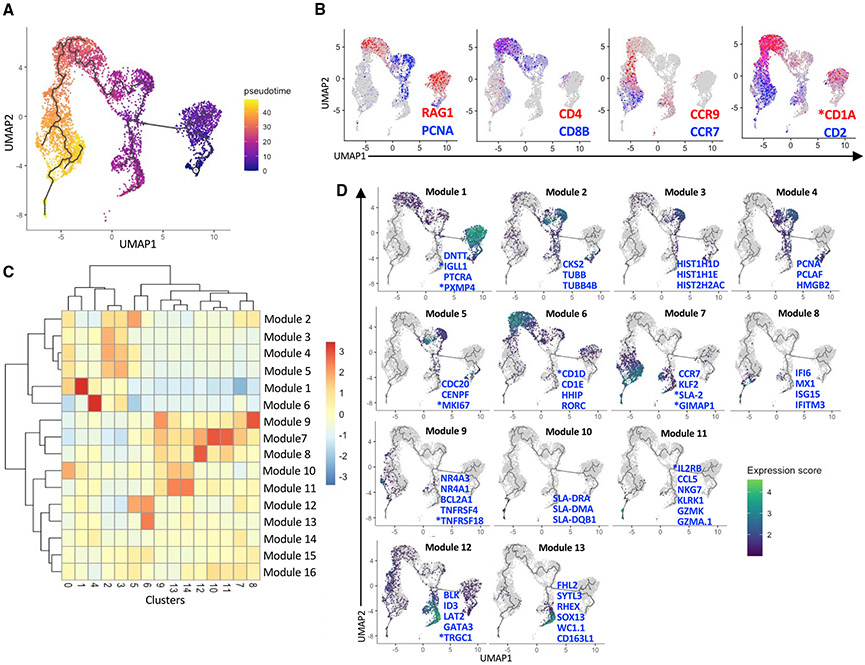
Pseudo-temporal analysis of thymocyte development
We ordered thymocytes along a differentiation trajectory using Monocle (v3) (Cao et al., 2019; Levine et al., 2015; Traag et al., 2019; Trapnell et al., 2014). Overall, the trajectory downstream of the DN(C) root cluster agreed with well-established αβ T cell development stages and stage-specific markers (Figures 3A and 3B). γδ T cells differentiated from DN thymocytes through a CD2+ precursor population enriched for cell cycling genes and *CD1A (Figures 3B and S1B) and then branched into two terminally differentiated subsets. Genes that varied with pseudotime clustered into 16 modules (Figures 3C and 3D; Table S4). Module 1 segregated with DN cells and included pre-T and pre-B cell receptors (PTCRA and *IGLL1), DNTT, which encodes terminal deoxynucleotidyl transferase, and *PXMP4, which purportedly modulates glycolipid availability for CD1d presentation (Fletcher et al., 2008). Modules 2–5 included histone and cell cycling genes that segregated with both DP(C) clusters. Module 6 segregated with DP(Q) cells and included two CD1 genes (*CD1D and CD1E); HHIP, which regulates thymic γδ T cell differentiation (Mengrelis et al., 2019); and RORC, a transcription factor that regulates DP cell survival (Kurebayashi et al., 2000; Yui and Rothenberg, 2014). Modules 7–11 contained genes that segregated with various post-committed populations. Module 7 was strongly upregulated in CD4SP, CD8SP, and γδ T cells and contained several thymic emigration genes, *SLA-2 (major histocompatibility complex [MHC] class I antigen 2), and *GIMAP1, a GTPase that is critical for mature T cell development and survival (Saunders et al., 2010). Module 8 included several ISGs and segregated with ISG-CD8 T cells and a small number of CD2+ γδ T cells. Module 9 varied with Treg cells (especially the Treg1 cluster) and included *TNFRSF18 and TNFRSF4, the antiapoptotic gene BCL2A1, as well as the Nr4a receptors NR4A1 (Nur77) and NR4A3 (Nor1), which are critical for Treg cell lineage commitment in mice (Owen et al., 2019; Sekiya et al., 2013; Tuzlak et al., 2017). Modules 10 and 11 varied with CD8αα and cytotoxic CD8 T cells. Module 10 was enriched for genes encoding MHC class II molecules, and module 11 contained genes encoding memory markers, NK receptors, and granzyme molecules. Module 12 was increased in all γδ T cells and included genes involved in the γδ TCR signaling cascade (*TRGC1, BLK, and LAT2) (Cibrian et al., 2020; Muro et al., 2019), GATA3, a transcription factor expressed by most porcine γδ T cells (Rodríguez-Gómez et al., 2019), and ID3, an E protein inhibitor that controls the survival and expansion of γδ thymocytes (Zhang et al., 2014). Module 13 segregated specifically with CD2− γδ T cells and included SYTL3, which regulates vesicular trafficking (Dong et al., 2021); *CD163L1 (also known as WC1) and its variant *WC1.1, which act as hybrid pattern recognition receptors and TCR coreceptors on bovine γδ T cells (Herzig et al., 2010; Hsu et al., 2015); FHL2, a transcriptional co-activator that regulates cell proliferation, survival, and motility (Hua et al., 2016); and RHEX, which controls erythroid cell expansion (Verma et al., 2014). The three remaining modules (14–16) exhibited subtle differences over pseudotime and did not segregate with specific cell clusters (Figure 3C).
Figure 3. Pseudotemporal analysis of pig thymocyte development.
(A) Pseudotime trajectory created by Monocle 3 using clusters 0–14 from Figure 1A.
(B) The same UMAP plot showing classical stage-specific markers of thymocyte development.
(C) Heatmap of 16 gene modules whose expression varied across pseudotime between clusters.
(D) UMAP plots showing the expression profiles of select genes from modules 1–13. See Table S4 for a complete list of module genes. Asterisks indicate non-annotated genes (described in Table S13)
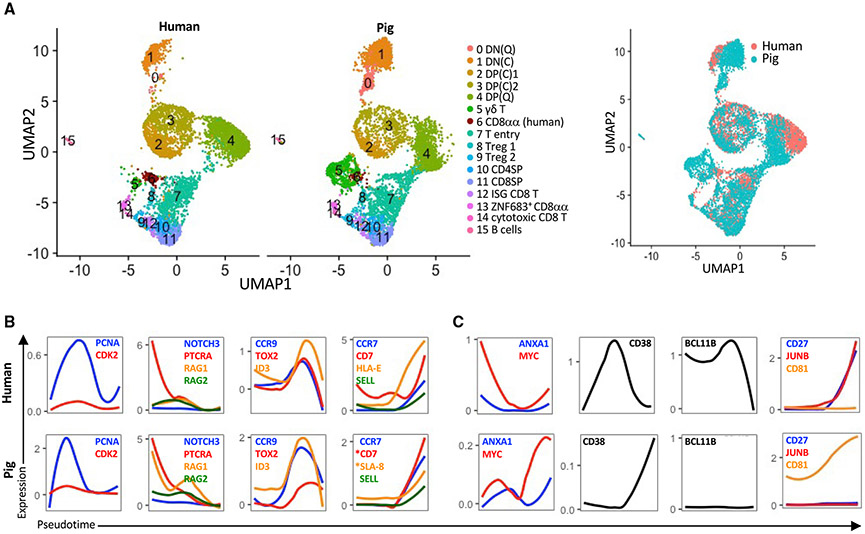
Porcine and human thymopoiesis are transcriptionally conserved
To compare the transcriptional landscape of pig and human thymocytes, we integrated our thymocyte dataset with published scRNA-seq data from CD34− thymic cells of a 19-month-old human (Le et al., 2020) (Figures S3A-S3C). We found a high degree of overlap for most clusters (Figure 4A). However, the pig DP(Q) cluster only partially overlapped with its human counterpart, in part because human DP(Q) cells expressed more CD3D, CD3G, *CD99, and MZB1 than pig DP(Q) cells (Table S5). The γδ T cell cluster was much larger in pigs. Pig and human γδ T cells were enriched for IKZF2, ID3, and CD44 (Table S6). However, SOX13 was not detected in human γδ T cells. Human thymocytes harbored two previously described subsets of CD8αα T cells, designated CD8αα+ T(I) (cluster 6) and CD8αα+ T(II) (cluster 13), which can be distinguished by their respective enrichment of GNG4 and ZNF683 (Park et al., 2020). CD8αα T(I) overlapped with a minor population of pig γδ T cells (cluster 6) that lacked CD8A. Both clusters expressed *PRKCH, PDCD1, and IKZF2 (Table S7), which facilitate high-affinity TCR interactions that agonist-selected T cells require for their development (Daley et al., 2013; Isakov and Altman, 2012; Pobezinsky et al., 2012). CD8αα T(II) overlapped with pig ZNF683+ CD8αα T cells (cluster 13), partly because of their common expression of γδ-TCR genes, NKG7, KLRB1, CXCR3, and CXCR6 (Table S8). However, there were also highly differentially expressed genes between them (Table S8), including the GIMAP family members GIMAP2 and *GIMAP6, which regulate thymocyte development (Filén and Lahesmaa, 2010). Pig cytotoxic CD8 T cells and the corresponding human cluster (cluster 14) expressed cytotoxic genes (NKG7, GZMK, and GZMH) (Table S9). However, only the human cluster expressed caspases involved in inflammation (CASP1 and *CASP4) (Galluzzi et al., 2016) and BST2, a type I IFN-inducible cellular protein (Kambara et al., 2015) (Table S9), indicating a possible difference in function.
Figure 4. Integrative analysis of human and pig thymocytes.
(A) UMAP showing an integrative analysis of human CD34− thymocytes and pig thymocytes using a canonical correlation analysis to identify shared genes between datasets.
(B and C) Transcription factor and lineage genes with conserved (B) and divergent (C) transcription profiles between pigs and humans. A public dataset containing human thymus samples was used (Le et al., 2020). Also see Figure S3. Asterisks indicate non-annotated genes (described in Table S13).
To compare temporal changes in gene expression between species, we performed a pseudotime analysis of the human dataset (Figure S3C). We found conserved profiles for several cell cycle (PCNA and CDK2), T lineage (PTCRA and NOTCH3), VDJ recombination (RAG1 and RAG2), DP-to-SP transition (ID3, TOX2, and CCR9), and terminal differentiation (CCR7, CD7, MHCI, and SELL) genes (Figure 4B). However, we also observed notable divergences (Figure 4C). MYC, a transcription factor required for thymocyte proliferation and differentiation (Broussard-Diehl et al., 1996), and ANXA1 (Annexin-A1), a phospholipid binding protein that plays a role in regulating the strength of TCR signaling during thymic selection (Paschalidis et al., 2010), were highly upregulated in human DN cells and became silenced with the transition to DP cells. In contrast, pigs upregulated these genes during the DP-to-SP transition. CD38, a cyclic ADP ribose hydrolase responsible for inducing apoptosis of DP thymocytes (Li et al., 2015), was highly expressed by human thymocytes at the DP stage. Conversely, this gene was undetected in pig thymocytes until the mature T cell stage, when it was expressed at low levels. The transcription factor BCL11B, which is a key regulator of thymocyte differentiation and survival (Hosokawa et al., 2020; Wakabayashi et al., 2003), was sustained at high levels until the commitment stage in humans but was barley detected in pig thymocytes. Expression of JUNB, a component of transcription factor AP-1, which is required for T cell differentiation (Katagiri et al., 2021), and CD27, which modulates T cell survival and memory formation (Hendriks et al., 2000), increased with commitment in the human thymus but were barely detected in pigs. Conversely, CD81, which modulates TCR signaling (Rocha-Perugini et al., 2013), was only upregulated in pig thymocytes. These data demonstrate a significant degree of conservation between pig and human thymocyte development as well as species-specific differences in several key regulators of thymopoiesis.
Cluster analysis of thymic iNKT cells
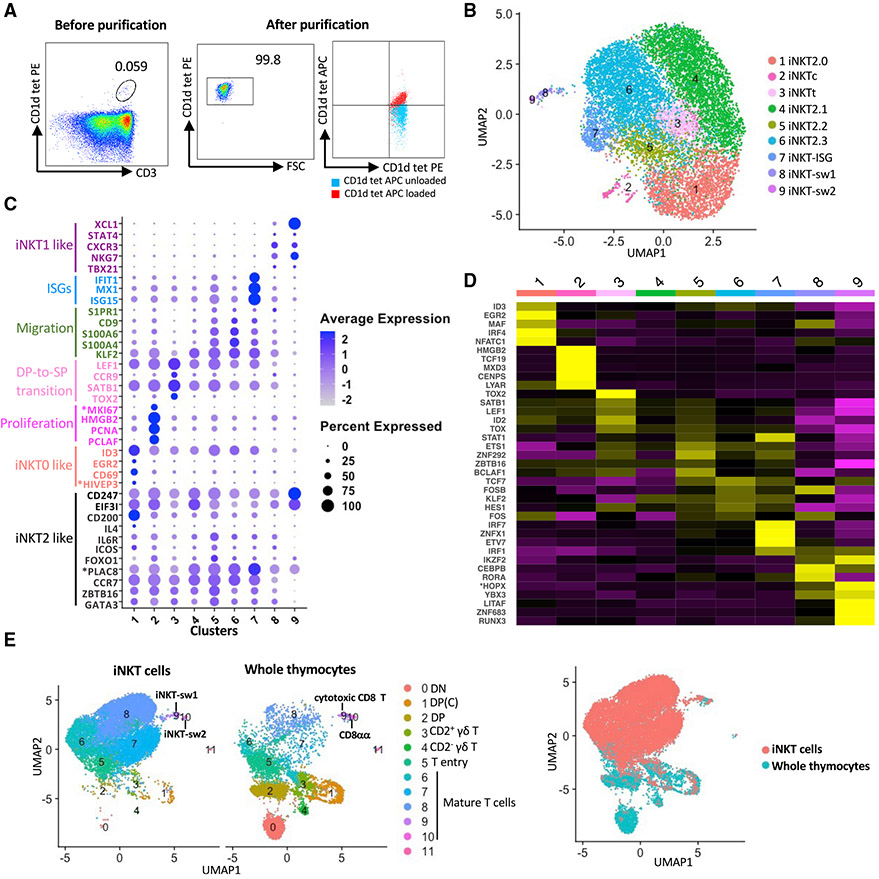
To perform a detailed assessment of iNKT cell differentiation, we interrogated the transcriptomes of more than 11,000 iNKT cells (Table S1) purified from the same thymi used to analyze porcine thymopoiesis. Thymocytes were stained with a phycoerythrin (PE)-conjugated mouse CD1d tetramer reagent that cross-reacts with the porcine-invariant TCR (Artiaga et al., 2014) (Figure 5A). CD1d tetramer+ cells were then enriched using magnetic bead separation and then sorted by fluorescence-activated cell sorting (FACS) to 99% purity (Figure 5A). Sorted cells were stained with PBS57-unloaded or -loaded allophycocyanin (APC)-conjugated CD1d tetramers to confirm the specificity of the CD1d tetramer-PBS57 antigen complex (Figure 5A).
Figure 5. scRNA-seq analysis of porcine thymic iNKT cells.
(A) Flow cytometry showing thymic phycoerythrin (PE)-conjugated mouse (m)CD1d tetramer+ cells before (left panel) and after (center panel) isolation with magnetic beads and FACS. Purity was confirmed by co-staining with allophycocyanin (APC)-conjugated PBS57-loaded or unloaded mCD1d tetramers (right panel).
(B) UMAP visualization of iNKT thymocyte clusters identified using the graph-based Louvain algorithm at a resolution of 0.5.
(C) Dot plot showing the Z-scored mean expression of selected genes encoding key transcription factors and thymocyte differentiation markers for each cluster.
(D) Heatmap showing row-scaled mean expression of the five highest differentially expressed transcription factors per cluster.
(E) Integrative analysis of iNKT cells and whole thymocytes (excluding B cells) from the same pigs. Asterisks indicate non-annotated genes (described in Table S13).
After completing read alignment and quality control steps, an unsupervised graph-based clustering analysis using the Louvain algorithm identified 9 clusters (Figures 5B-5D, S4A, and S4B; Table S10), which overlapped with emerging and mature αβ T cells in whole thymocytes (Figures 5E and S4C). Most clusters were immediately adjacent to each other, indicating that porcine iNKT thymocytes constitute a transcriptionally homogeneous population. A comparison of gene expression with past publications identified multiple genes associated with previously established mouse iNKT0, iNKT1, and iNKT2 subsets (Baranek et al., 2020; Harsha Krovi et al., 2020; Lee et al., 2016) (Figure 5C). Clusters 1–7 were closely grouped and collectively expressed iNKT2-associated genes, including ZBTB16 (PLZF), GATA3, CCR7, *PLAC8, FOXO1, ICOS, and IL6R (Baranek et al., 2020; Harsha Krovi et al., 2020). Cluster 1 (17.14%), designated iNKT2.0, upregulated EGR2, ID3, *HIVEP3, and CD69, which are expressed by mouse iNKT0 cells (Baranek et al., 2020; Harsha Krovi et al., 2020; Seiler et al., 2012). This cluster also upregulated the inhibitory receptor CD200 and the cytokine IL4, which are enriched in iNKT2 cells (Engel et al., 2016). Cluster 2 (1.72%), designated iNKTc, was enriched for cell cycle genes, including PCLAF, PCNA, HMGB2, and *MKI67. Cluster 3 (5.54%), designated iNKTt, upregulated genes associated with DP-to-SP thymocyte transition, including CCR9, LEF1, TOX2, and SATB1 (Aliahmad et al., 2012; Hu et al., 2015; Park et al., 2020; Uehara et al., 2006). Cluster 4 (31.82%), designated iNKT2.1, was similar to cluster 1 but lacked iNKT0-associated genes. Instead, cluster 4 upregulated EIF3I and CD247, which are additional iNKT2 genes (Baranek et al., 2020) (Figures 5C and S4B). Cluster 4 also expressed SOCS1, a critical regulator of iNKT cell differentiation (Hashimoto et al., 2011); HPGDS, a marker of a pathological Th2 cell subpopulation that potentiates allergic inflammation (Mitson-Salazar et al., 2016); and CPPED1, which likely regulates the phosphatidylinositol 3-kinase (PI3K)-protein kinase B (AKT) pathway (Haapalainen et al., 2021; Kim and Suresh, 2013) (Table S10). Cluster 5 (6.60%), designated iNKT2.2, and cluster 6 (31.60%), designated iNKT2.3, had relatively few unique differentially expressed genes (DEGs) compared with the other iNKT2-like clusters, except that both, along with cluster 7, were enriched for thymic emigration genes, including S1PR1, KLF2, CD9, S100A4, and S100A6 (Carlson et al., 2006; Reyes et al., 2018; Sreejit et al., 2020; Weinreich and Hogquist, 2008). Cluster 7 (4.47%) was unique for its strong enrichment of ISGs, including ISG15, MX1, and IFIT1, and was therefore designated iNKT-ISG. Cluster 8 (0.88%) and cluster 9 (0.23%) expressed genes associated with mouse iNKT1 cells, including TBX21, FCER1G, NKG7, CXCR3, STAT4, and XCL1 (Georgiev et al., 2016; Lee et al., 2016) (Figure 5C; Table S10). Cluster 8, designated iNKT-swine (sw)1, had several DEGs in common with the cytotoxic CD8 T cells identified in our whole-thymocyte dataset (Table S11). However, a comparative assessment of their transcriptomes identified that iNKT-sw1 cells were enriched for ZBTB16 and the TNF receptors *TNFRSF18 and TNFRSF4, whereas cytotoxic CD8 T cells were enriched for granzyme-related genes (GZMK, GZMA.1, and *GZMM) (Figure S4D). Cluster 9, designated iNKT-sw2, presented a transcriptional profile similar to CD8αα T cells, including expression of ZNF683 (Figure S4C; Table S11). However, CD8αα T cells were enriched for GATA3, whereas iNKT-sw2 cells expressed CD244 (SLAMF4) and EGR1 (Figure S4E), which are, respectively, associated with mouse iNKT1 cells (Lee et al., 2016) and the early stages of iNKT cell commitment (Seiler et al., 2012).
Pig iNKT cell clusters are transcriptionally distinct from iNKT cell subsets in mice
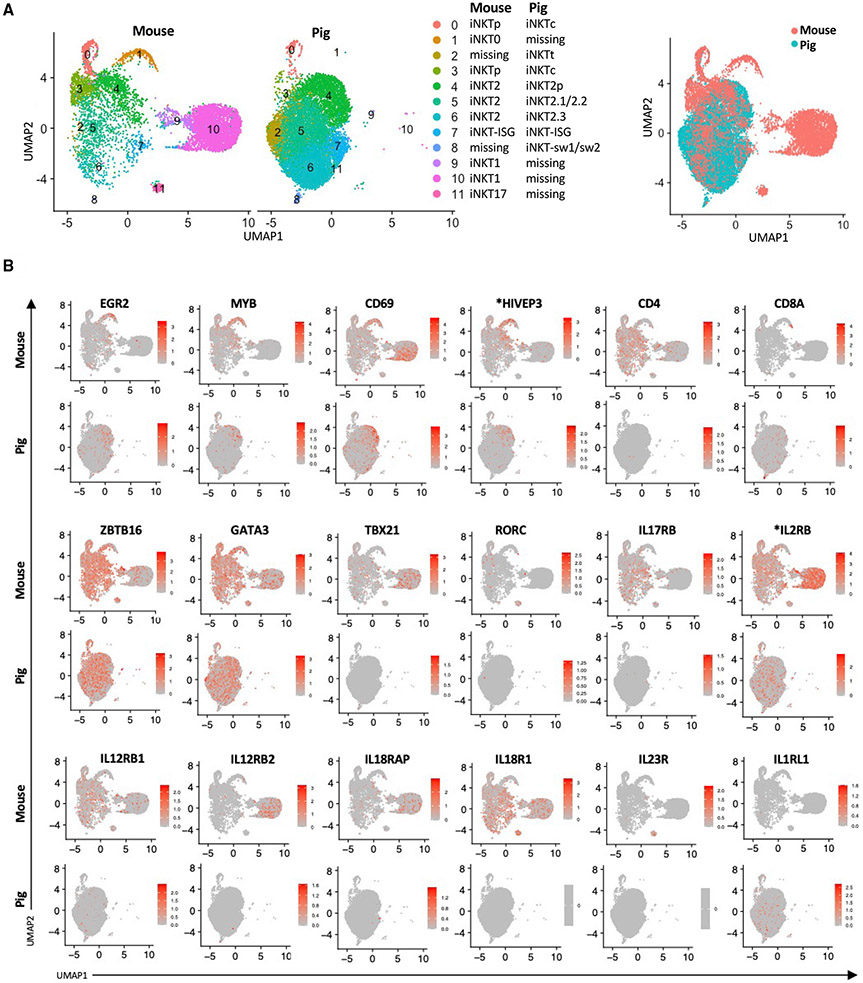
To compare the transcriptomes of porcine and murine iNKT cells, we integrated our data with a published scRNA-seq analysis of iNKT cells purified from the thymi of 8- to 9-week-old C57BL/6 mice (Harsha Krovi et al., 2020) (Figures 6A and S5A). The mouse dataset used CD1d tetramer+ cells sorted at a 50:50 ratio of CD44low to CD44high cells to enrich early precursors and developmental intermediates. We found that pigs lacked a population analogous to mouse iNKT0 cells (Figure 6A; cluster 1). However, several iNKT0 signature genes (EGR2, MYB, CD69, and *HIVEP3) were enriched in the pig iNKT2.0 cluster (Figure 6B). Cluster 2, which coincided with iNKTt cells, appeared to be unique to pigs. Clusters 0 and 3 in the mouse dataset were enriched for cell cycle genes as well as CCR7, CCR9, and IL13 (Figure S5A), which are expressed by a previously described multi-potent iNKT cell progenitor population (iNKTp) (Harsha Krovi et al., 2020). The corresponding pig clusters also upregulated cell cycle genes but lacked IL13 and did not differentially express CCR7 and CCR9 compared with the other pig iNKT cell clusters. Most pig iNKT cells appeared in clusters 4–6, which overlapped with three mouse iNKT2 clusters. However, the pig clusters did not express detectable CD4 or IL17RB, which are classic mouse iNKT2 markers (Baranek et al., 2020; Harsha Krovi et al., 2020; Tuttle et al., 2018) (Figure 6B). Cluster 7 coincided with pig iNKT-ISG cells and a corresponding population of mouse iNKT cells enriched for ISGs, including ISG15 and IFIT1 (Figures 6B and S5A). The mouse ISG cluster was similar to a previously identified iNKT cell subset displaying strong enrichment of type I IFN response genes (Baranek et al., 2020; Harsha Krovi et al., 2020). Strikingly, pig iNKT cells did not include clusters that overlapped with mouse iNKT1 (clusters 9 and 10) or iNKT17 (cluster 11) cells that are, respectively, enriched for TBX21 (T-bet) and RORC (RORγt) (Pellicci et al., 2020). However, several iNKT1 signature genes were enriched in pig cluster 8, which was a combination of iNKT-sw1 and iNKT-sw2 subsets. A comparison of DEGs between mouse iNKT1 cells and pig cluster 8 found several commonly expressed genes, including XCL1, CCL5, KLRK1, and NKG7 (Table S12). However, divergence was observed in genes associated with T cell activation (TRAT1, PDK1, and CD69) (de la Fuente et al., 2014; Kirchgessner et al., 2001; Lee et al., 2005) and lineage-determining transcription factors (IKZF2, JUNB, and KLF6) (Cao et al., 2010; Daley et al., 2013; Katagiri et al., 2021) (Figure S5B; Table S12). Because the transcription co-factor four and a half LIM domains 2 (FHL2) has been shown recently to control commitment to the iNKT1 lineage (Baranek et al., 2020), we examined FHL2 expression in pig iNKT cells. Consistent with our observation that swine lack a clear population of iNKT1 cells, we did not detect any subclusters enriched for FHL2 (Figure S5C). In contrast, FHL2 was upregulated in CD2− γδ T cells (Figure S2E), suggesting that this gene is involved in controlling commitment to this lineage.
Figure 6. Integrative analysis of pig and mouse thymic iNKT cells.
(A) UMAP plots showing the integrative data analyses of thymic iNKT cells from pig and mouse. A public dataset containing thymic iNKT cells isolated from 8- to 9-weeks old C57BL6/J mice was used (Harsha Krovi et al., 2020). Overlapping clusters are in the same figure legend row. Cell clusters that are absent in one species are annotated as “missing.”
(B) Expression of 18 genes typically used to distinguish the major subsets of mouse iNKT cells. Asterisks indicate non-annotated genes (described in Table S13).
We also compared the pig and mouse datasets for receptors of cytokines that are known to regulate iNKT cell activation and differentiation, such as IL-2, IL-12, IL-18, IL-23, and IL-33 (Bendelac et al., 2007; Ferhat et al., 2018). Mouse iNKT cells expressed genes encoding various IL-2, IL-12, and IL-18 receptor subunits (Figure 6B). Among these, *IL2RB, IL12RB2, and IL18RAP were enriched in iNKT1 cells, whereas IL23R was enriched in iNKT17 cells (Engel et al., 2016). IL-12 and IL-18 receptor subunits were barely detected in the pig dataset. However, pig iNKT cells did express *IL2RB as well as IL1RL1, which encodes the IL-33 receptor (Figure 6B). Our finding that pig iNKT cells express IL1RL1 agrees with a previous report that IL-33 strongly enhances in vitro expansion of pig iNKT cells when combined with the agonist α-galactosylceramide (Thierry et al., 2012).
These results demonstrate conserved features as well as species-specific differences in the transcriptional landscape underlying iNKT cell subsets in mice and pigs.
Pseudotime analysis of porcine iNKT cell differentiation
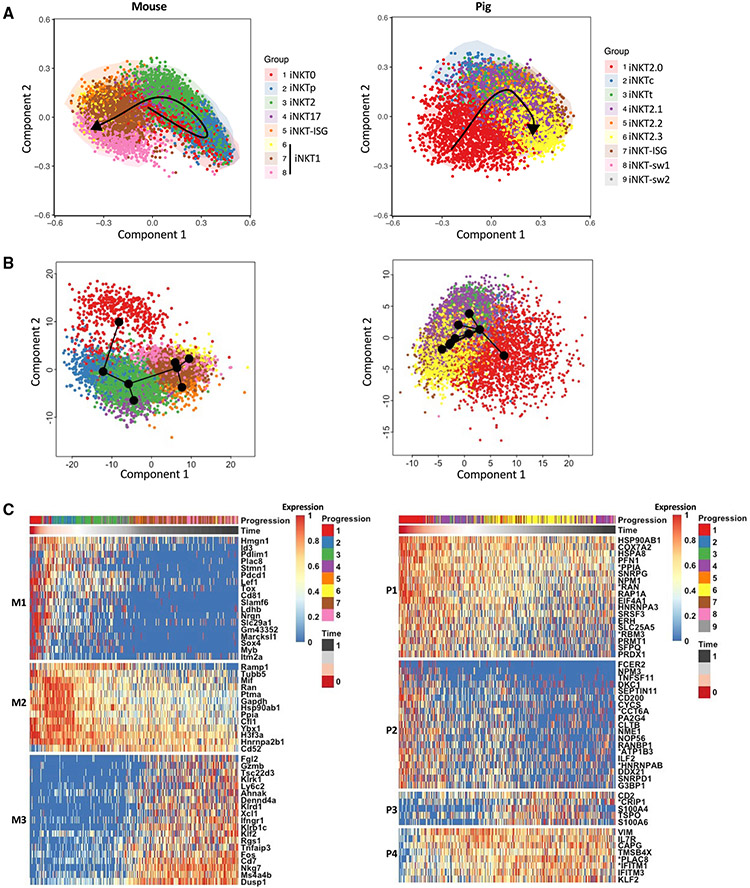
A pseudotime analysis was performed to compare the trajectories of pig and mouse iNKT cell differentiation. Because pig iNKT cells lacked a clear progenitor population that could be designated as a root cluster, we analyzed the datasets using the R packages SCORPIUS and Slingshot, which perform fully unsupervised pseudotime inferences of single-cell transcriptomics. However, SCORPIUS predicts developmental chronologies according to a linear dynamic process, whereas Slingshot uses a non-linear tree-shaped trajectory inference method (Cannoodt et al., 2016; Street et al., 2010). For mouse iNKT cells, both packages predicted a differentiation trajectory that closely agrees with the literature (Baranek et al., 2020; Harsha Krovi et al., 2020), where iNKT0 cells give rise to iNKT2 cells through iNKTp cells. Next, iNKT2 cells split into two branches leading to iNKT1 or iNKT17 cells (Figures 7A, 7B, and S6). Pig iNKT cells were predicted to follow a simpler pathway originating from iNKT2.0 cells and leading through overlapping clusters of the remaining iNKT cells (Figures 7A and 7B). Genes that varied according to pseudotime were clustered into three modules in mice (M1–M3) and four modules in pigs (P1–P4) (Figure 7C). The mouse modules segregated with genes upregulated by (1) iNKT0 and iNKTp cells (M1); (2) iNKT0, iNKTp, iNKT2, and iNKT17 cells (M2); and (3) iNKT1 cell subsets (M3). Pseudotime-dependent gene expression changes were less obvious in pig modules because pig iNKT cell development had fewer distinct phases. Nevertheless, we observed modules that segregated with genes that (1) gradually deceased over pseudotime (P1), (2) were enriched in iNKT2.0 cells (P2), (3) increased in iNKT2.3 cells and were important for thymic emigration (P3), and (4) were specifically silenced in iNKT2.0 cells (P4). Of the 50 DEGs that appeared in each of the pig and mouse modules, only four genes, *PLAC8, *RAN, KLF2, and HSP90AB1, were common to both datasets. These results indicate that the post-commitment differentiation of pig iNKT cells is shorter and transcriptionally less complex than in mice.
Figure 7. Unsupervised analysis of mouse and pig iNKT cell differentiation.
(A and B) Mouse and pig iNKT cells were ordered along their respective differentiation trajectories via unsupervised SCORPIUS (A) and Slingshot (B) analyses.
(C) The top 50 most important mouse (left panel) and pig (right panel) genes with respect to the inferred trajectory were, respectively, clustered into three (M1–M3) and four (P1–P4) gene modules by SCORPIUS. Normalized expression values are scaled from 0 to 1 using the scale_quantile function of SCORPIUS with default parameters.
DISCUSSION
We used scRNA-seq to characterize thymocyte maturation and iNKT cell differentiation in the thymi of 22-week-old pre-pubertal pigs. This age coincides with a period of maximum thymus size and output in swine.
Our map of αβ T cell development agrees with a previously proposed model of pig αβ T cell differentiation where CD1AhiCD4−CD8− precursor cells transform into a large CD3loCD4+CD8+ DP population that initially maintains pre-TCRα expression and eventually differentiates into CD4+CD8− and CD4−CD8+ SP T cells that express CD3 at high intensity (Sinkora and Butler, 2016). The high degree of overlap between our data and a published scRNA-seq dataset from CD34− thymic cells of a 19-month-old human (Le et al., 2020) indicates that the αβ T cell lineage is generally well conserved between pigs and humans compared with between mice and humans Figure S7(Canté-Barrett et al., 2017; Le et al., 2020; Park et al., 2020) (Figure S7). Nevertheless, we uncovered inter-species differences in key regulators of thymocyte differentiation, such as CD38, JUNB, BCL11B, CD27, and CD81, which has implications for translating some aspects of adaptive immunity and central tolerance from pigs to humans.
Our study analyzed the transcriptional profile of several unconventional T cell types, including Treg cell, γδ T cells, and memory-like CD8+ T cell subsets. Our results agree with previous reports showing that pig γδ T cells progress through a CD1Ahi γδ-TCR+ CD2+ intermediate stage that branches into mature CD2+ and CD2− subsets that downregulate CD1A (Sinkora and Butler, 2016; Sinkora et al., 2005b). Our findings suggest that the CD2− subset, which also appears in other high-frequency γδ T cell species, including sheep (Witherden et al., 1995), cattle (Mackay and Hein, 1989), and chickens (Vainio et al., 1991), are evolutionarily related to murine Tγδ17 cells. Among their similarities appears to be the ability of both subsets to develop TCR independently or with weak TCR signals (Haas et al., 2012; Parker and Ciofani, 2020). This contrasts with the CD2+ subset, which seems to require TCR stimulation for their induction, in a manner similar to IFN-γ-producing Tγδ1 cells, the other major lineage of mouse γδ T cells (Patil et al., 2015; Yang et al., 2021). We observed that CD2− γδ T cells were enriched for JAML, a costimulatory molecule expressed by monocytes, neutrophils, activated CD8+ T cells, and tissue-resident γδ T cells (Guo et al., 2009; Luissint et al., 2008; Verdino et al., 2010; Witherden et al., 2010). After binding to its cognate ligand, coxsackie and adenovirus receptor (CXADR), a cell adhesion molecule on non-hematopoietic cell types, JAML, stimulates a PI3K signaling cascade that induces T cell activation and proliferation (Ortiz-Zapater et al., 2017; Verdino and Wilson, 2011). This pathway is integral to epithelial γδ T cell biology and contributes to the antitumor effector responses of γδ T cell and CD8+ T cells (McGraw et al., 2021; Witherden et al., 2010). Our observation that JAML is intensely expressed by almost all pig CD2− γδ thymocytes (and CD8+ SP cells) suggests that the JAML-CXADR costimulatory axis plays an important role in these cells. We also observed that CD2− γδ T cells were the only thymocyte population enriched for FHL2, which is known to interact with several lymphocyte development factors, including PLZF, Id2, Id3, and Nur77 (Tran et al., 2016). This suggests that FHL2 is a key regulator of CD2− γδ T cell development and/or effector functions.
We identified two minor CD8+ T cell subsets (cytotoxic CD8 T and CD8αα cells) that resemble innate CD8+ T cell subpopulations resident in mouse and human thymi (Park et al., 2020; Verstichel et al., 2017; Yamagata et al., 2004). Both pig subsets have a memory T cell phenotype (Lee et al., 2011; White et al., 2017). Our pseudotime analysis predicted that the cytotoxic CD8 T cell subset may give rise to CD8αα T cells. Although genes associated with thymic emigration were barely detected in either subset, a significant population of CD8αα T cells are present in a recently published scRNA-seq dataset from unfractionated pig peripheral blood mononuclear cells (Herrera-Uribe et al., 2021). These cells expressed several genes in common with our thymic CD8αα T cells (ZNF683, CD44, *IL2RB, CCL5, NKG7, KLRK1, *KLRD1, and XCL1), supporting the intrathymic origin of this population. Although the human thymus clusters that overlapped with pig memory and CD8αα T cells expressed many of the same markers, species differences in several important lineage-defining genes point toward the acquisition of species-specific adaptations, perhaps for different pathogens.
Our analysis of thymic iNKT cells revealed substantial interspecies differences, which holds important implications for transposing paradigms of iNKT cell development from mice to pigs. Recent murine scRNA-seq studies have shown that post-committed iNKT0 cells differentiate through a cycling precursor population into a cluster of central and transitional iNKT2 cells, which give rise to terminally differentiated iNKT2 cells as well as iNKT1 and iNKT17 subsets (Baranek et al., 2020; Harsha Krovi et al., 2020). We observed unexpected homogeneity among porcine iNKT cells, where 97% of cells were enriched for mouse iNKT2 cell genes (Baranek et al., 2020; Engel et al., 2016; Georgiev et al., 2016; Harsha Krovi et al., 2020). Accordingly, pig iNKT cells followed a differentiation pathway that was considerably shorter and less distinctive than that of their murine counterparts. Two minor clusters (iNKT-sw1 and iNKT-sw2 cells) had unique transcriptional signatures characterized by loss of iNKT2 genes and gain of iNKT1 transcripts. These cells were most like cytotoxic CD8 T cells and CD8αα T cells identified in our pig thymocytes, sharing expression of *FCGR3A, ZNF683, NKG7, and MHC class II-encoding genes (SLA-DQB1, SLA-DMA, and SLA-DMB). Nevertheless, there were also highly differentially expressed genes, including increased ZBTB16, *TNFRSF18, TNFRSF4, CD244, and EGR1 transcripts in the iNKT cell clusters and increased GATA3 and granzyme related transcripts in the corresponding thymocyte clusters, indicating a significant difference in function. iNKT-sw1 and iNKT-sw2 cells downre-gulated tissue emigration genes and upregulated CXCR3 compared with the remaining iNKT cells, which suggests that both subsets are long-term thymus residents. Similar thymus-resident populations of innate T cells have been described before, including subsets of mucosa-associated invariant T (MAIT) cells, γδ T cells, and CD8αα T cells (Fan and Rudensky, 2016; Lee et al., 2020; Salou et al., 2019; Verstichel et al., 2017). Although the role of such cells remains unclear, it has been speculated that they modulate thymocyte differentiation to adapt to peripheral events, such as infection (Baranek et al., 2020; White et al., 2018).
One explanation for the surprisingly shortened differentiation pathway of pig iNKT thymocytes may be that pig iNKT cells emigrate from the thymus in a functionally immature state and undergo further differentiation in the periphery. This model has been proposed for human iNKT cell development because human iNKT thymocytes do not produce IFN-γ or IL-4 under steady-state conditions (Baev et al., 2004). Another possibility is that the iNKT subset diversity observed in mice is a peculiarity of this species, perhaps to compensate for innate T cell populations that are reduced or absent in mice compared with other species. For instance, mice lack group 1 CD1 (CD1a, CD1b, and CD1c)-restricted T cells, which are closely related to CD1d-restricted iNKT cells and relatively abundant in humans (Godfrey et al., 2015; Van Rhijn et al., 2015). Because all four types of CD1-restricted T cells recognize lipids presented by CD1 molecules on dendritic cells, it is possible that group 1-restricted T cells fulfill some of the roles performed by iNKT1 and iNKT17 cells in mice.
The substantial variability in iNKT cell differentiation between mice and pigs raises a cautionary note about interpreting the results of iNKT cell development studies in animal models, especially considering that a unifying model linking transcription factors and function in human and mouse iNKT cells has been difficult to establish. Given that innate-like T cells are emerging as key players in the immune system, insights gleaned from this study will prove valuable for evaluating innate-like T cell-associated diseases and immunotherapies in humans and live-stock, including those involving iNKT cells.
Limitations of the study
Although this study has provided a comprehensive transcriptional analysis of pig thymopoiesis, we only profiled pigs at a single age and did not include non-lymphoid cell populations. In the future, it will be important to expand our dataset to capture the full extent of thymus function and cellularity. Additionally, we cannot exclude that some of the interspecies differences we observed in thymopoiesis and iNKT cells were due to biological and technical effects, such as differences in physiological age, tissue preparation methods, and sequencing saturation; validation of our findings is required using additional datasets as they become available. Because of a lack of antibody reagents, our discovery that pig iNKT cell subsets differ substantially from mouse iNKT cell subsets cannot be confirmed at this time using flow cytometry of intracellular transcription factors; validation of our findings using this approach should be performed when these reagents become available.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, John P. Driver (driverjp@missouri.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The sequencing data have been deposited at GSE192520. Code used in this study are available on https://github.com/Driver-lab1/scRNAseq_pig_thymus. DOIs are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Live/Dead viability dye | Invitrogen | Cat# L34976 |
| Mouse anti-pig CD3ε antibody | BD | Clone BB23-8E6-8C8 |
| Anti-JAML antibody | Abcam | Clone EPR15289 |
| Alexa Fluor 488-conjugated anti-rabbit IgG secondary antibody | Abcam | Cat# ab150077; RRID: AB_2630356 |
| Mouse anti-pig TCRδ antibody | WSU mAb Center | Clone PGBL22A |
| Biological samples | ||
| Pig thymus | University of Florida’s Animal Sciences Department | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Mouse CD1d (mCD1d) tetramer | National Institutes of Health Tetramer Core Facility | N/A |
| Critical commercial assays | ||
| Chromium Next GEM Single Cell 3′ reagent kit | 10xGenomics | RRID: SCR_019145 |
| NovaSeq 6000 sequencer | Illumina | RRID: SCR_019152 |
| Deposited data | ||
| Raw data | This paper | GSE192520 |
| Human thymus dataset | Le et al. (2020) | GSE139042 |
| Mouse thymic iNKT cell dataset | Harsha Krovi et al. (2020) | GSE152786 |
| Software and algorithms | ||
| Cell Ranger v3.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| R (4.0.2) | CRAN | https://www.r-project.org/ |
| Seurat (v3.2.2) | Stuart et al. (2019) | https://satijalab.org/seurat/ |
| EnhancedVolcano | Blighe et al. (2018) | https://github.com/kevinblighe/EnhancedVolcano |
| Monocle 3 | Cao et al. (2019); Levine et al. (2015); Traag et al., 2019; Trapnell et al. (2014) | https://cole-trapnell-lab.github.io/monocle3/ |
| SCORPIUS (1.0.8) | Cannoodt et al. (2016) | https://github.com/rcannood/SCORPIUS |
| Slingshot (2.0.0) | Street et al. (2010) | https://bioconductor.org/packages/release/bioc/vignettes/slingshot/inst/doc/vignette.html |
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo |
| BioRender | https://biorender.com/ | RRID:SCR_018361 |
| Custom scripts | This paper | https://doi.org/10.5281/zenodo.6609557 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell isolation
Approximately 1 g of thymus tissue was collected from one male and one female mix-breed pig at 22 weeks of age that were slaughtered at the University of Florida’s Animal Sciences Department. Single cell suspensions were generated using glass homogenizers, after which mononuclear cells were isolated using Ficoll-Paque PREMIUM (GE Healthcare). Some cells were set aside for scRNA-seq of total thymocytes. The remaining cells were stained with Live/Dead Near-IR viability dye (Invitrogen) and a phycoerythrin (PE)-conjugated alpha-galactosylceramide (aGC) analog PBS57-loaded mouse CD1d (mCD1d) tetramer (National Institutes of Health Tetramer Core Facility). Cell suspensions were subsequently incubated with magnetically labeled anti-PE MicroBeads (Miltenyi Biotec), washed, and loaded onto an LS column (Miltenyi Biotec). After washing, the labeled cells were eluted and loaded onto an MS column for further enrichment. An equal number of cells from both pigs were combined and sorted for live mCD1d tetramer positive cells using a Sony SH800 Cell Sorter (Sony Biotechnology). Aliquots from the sorted cells were co-stained with allophycocyanin (APC)-conjugated PBS57-loaded or unloaded mCD1d tetramers to validate their specificity for the CD1d tetramer-PBS57 antigen complex by FACS. The thymocyte samples were stained with Live/Dead Near-IR viability dye and FACS sorted for live cells.
METHOD DETAILS
Single-cell RNA sequencing
Single-cell libraries were prepared using the 10x Chromium Next GEM Single Cell 3′ reagent kit (v3.1) according to the manufacturer’s instructions. Sequencing was performed on the S4 flow cell of the NovaSeq 6000 sequencer (Illumina) to obtain paired end reads.
Data processing and clustering analysis
Sequence reads were aligned to the pig reference genome Sscrofa 11.1 with gene transfer format file 11.1.98, following creation of barcode gene matrices using Cell Ranger v3.1 (10x Genomics). Then, clustering analyses were performed in R (4.0.2) using the Seurat package (v3.2.2) (Stuart et al., 2019). The data were pre-processed by removing genes expressed in <3 cells, excluding cells with aberrantly high or low gene counts and high mitochondrial gene expression, and regressing out cell cycle effects. After log-normalizing the data, the top 2,000 most variable genes in each dataset were identified using the FindVariableFeatures function. After scaling the data, we performed standard dimension reduction and clustering as follows: A principal component analysis (PCA) for linear dimension reduction was executed with the RunPCA function on the variable features; the most variable principal components were selected based on the elbow plot and then used to determine the k-nearest neighbors of each cell and construct a shared nearest neighbor graph using the FindNeighbors function; the FindCluster function using the Louvain algorithm was implemented to cluster cells; finally, a further non-linear dimensional reduction Uniform Manifold Approximation and Projection (UMAP) was performed to place similar cells together in low-dimensional space. The BuildClusterTree function was used to generate dendrograms with default arguments. Differentially expressed genes were identified within each cluster using the FindAllMarkers function with a minimum Log2 fold change threshold of + 0.25 using a Wilcoxon Rank-Sum test. The R package EnhancedVolcano (Blighe et al., 2018) was used to visualize results of differential expression analyses generated using the FindMarkers function (min.pct = 0.25, logfc.threshold = 0.25) in Seurat.
Trajectory analysis
Monocle 3 was used to perform a pseudotemporal analysis of thymocyte development (Cao et al., 2019; Levine et al., 2015; Traag et al., 2019; Trapnell et al., 2014). A subset of double-negative thymocytes was specified as the starting cluster. Cells were ordered onto a pseudotime trajectory according to their progress through their developmental program. The get_earliest_principal_node function was used to designate a node for which the highest fraction of closest cells belonged to the starting cluster as the root node. The learn_graph and order_cells functions were run to respectively learn the overall trajectory using the reversed graph embedding algorithm and to place each cell at its proper position through pseudotime. After constructing the trajectory, the graph_test function with the Moran I test was used to identify genes whose expression varied over pseudotime (q_value <0.05). Genes with similar expression patterns were grouped into modules using the find_gene_modules function. Next, the normalized expression levels and pseudotime values were extracted to generate individual gene dynamic expression profiles smoothed over pseudotime. To construct a pseudotime trajectory for iNKT cells, we used the R packages SCORPIUS (1.0.8) (Cannoodt et al., 2016) and Slingshot (v2.0.0) (Street et al., 2010) which both can reconstruct an ordering of cells without any prior knowledge of the dynamic process. For SCORPIUS, a classical Torgerson multi-dimensional scaling was applied to the normalized matrix and cluster data from Seurat using the reduce_dimensionality function. Next, cells were ordered according to the inferred timeline using the infer_trajectory function. To assess the importance of a gene with respect to the inferred trajectory, we ran the gene_importances function on the 2,000 most variable genes from Seurat. The top 50 most important genes were then segregated into coherent gene modules that were up- or down-regulated in different waves during iNKT cell development by running the extract_modules function. For Slingshot, the function slingshot was performed on clusters identified in Seurat, after which PCA reduction was used to determine dimensionality (reduced-Dims) and construct unbiased lineages.
Dataset integration
We used Seurat to integrate the thymocyte and iNKT cell data and to perform cross-species comparisons with published human thymocyte (GSE139042) (Le et al., 2020) and mouse thymic iNKT cell (GSE152786) (Harsha Krovi et al., 2020) datasets. The Ensembl genome browser (Ensembl Genes 105) was used to convert human (GRCh38) and mouse (GRCm39) gene names to the corresponding pig gene names prior to integration (https://www.ensembl.org/biomart/martview/). Only genes with one-to-one orthologs were included in the analyses. Low quality genes and cells were removed from each dataset as described above. Each dataset was independently normalized before identifying the most variable features. Afterward, we followed a standard integration workflow. Briefly, the SelectIntegrationFeatures function was applied to genes that were consistently variable across datasets. Next, the FindIntegrationAnchors function identified a set of anchors (pairs of cells from each dataset that are contained within each other’s neighborhoods) between datasets using the top 30 dimensions from the canonical correlation analysis to specify the neighbor search space. Next, an integrated dataset was created by running the IntegrateData function. Then, cell cycle effects were regressed out and the clustering analysis workflow was performed using RunPCA, FindNeighbours, FindClusters, and RunUMAP, as described above. The FindConservedMarkers function (min.pct = 0.1, only.pos = T) was used to identify DEGs that are conserved across datasets. Next, an analysis was performed to identify species-specific DEGs in select clusters. First, an additional column was added to the Seurat object listing each cluster according to its species origin. Next, the corresponding clusters were analyzed for DEGs using the FindMarkers function (min.pct = 0.25, logfc.threshold = 0.25), after which we removed genes that were differentially expressed due to dataset-specific effects and genes that were detected in only one species.
Flow cytometric analysis of JAML expression
Thymus, spleen, and lung samples were collected from 22-week-old mix-breed pigs. Tissues were dispersed into single cells as previously described (Artiaga et al., 2014), Fc receptor blocked with rat IgG, and stained with Alexa Fluor 647-conjugated mouse anti-porcine TCR δ chain antibody (PGBL22A, WSU mAb center), PE-Cy7-conjugated mouse anti-porcine CD3ε antibody (BB23-8E6-8C8, BD), and unconjugated rabbit anti-JAML antibody (EPR15289, Abcam). Next, the cells were incubated with an Alexa Fluor 488-conjugated anti-rabbit IgG secondary antibody (ab150077, Abcam). Viable stained cells were detected using an Attune NxT flow cytometer (Thermofisher, Grand Island, NY). A no primary antibody control was used to determine nonspecific binding of the secondary antibody (Figure S2F). Data were analyzed using FlowJo software v10 (BD, MA).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses in single cell analyses were performed using R (4.0.2) under the specific packages as described in the method details section. Briefly, for heatmap plots in Figures 1,5, S1, and S4, and volcano plots in Figures S2 and S5, the listed differentially expressed genes in each cluster were determined using the Wilcoxon Rank-Sum test. The module plot in Figure 3 were generated using the Moran I test. The genes in the heatmap plot in Figure 7 were selected using the Random Forest algorithm. For all tests, statistical significance was defined as p < 0.05.
Supplementary Material
Highlights.
A comprehensive atlas of thymocytes in the early-adolescent pig thymus
Detection of unconventional subsets and characterization of transcriptional heterogeneity
scRNA-seq on iNKT cells found more than 95% resemble murine iNKT2 and minor pig-specific subsets
Porcine iNKT cells lack clusters that overlap with mouse iNKT1 or iNKT17 subsets
ACKNOWLEDGMENTS
This work was supported by U.S. Department of Agriculture grant 2021-67015, National Institutes of Health grant HD092286 (to J.P.D.), as well as Merit (I01 BX001444) and Research Career Scientist (IK6 BX004595) awards from the VA (to S.J.). We thank the National Institutes of Health Tetramer Core Facility for provision of the CD1d tetramers under contract HHSN272201300006C. We also thank Dr. Rhonda Bacher for critical review of the manuscript. The graphical abstract was created using BioRender.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111050.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aliahmad P, Seksenyan A, and Kaye J (2012). The many roles of TOX in the immune system. Curr. Opin. Immunol 24, 173–177. 10.1016/j.coi.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende ML, Dreier JL, Mandala S, and Proia RL (2004). Expression of the sphingosine 1-phosphate receptor, S1P1, on T-cells controls thymic emigration. J. Biol. Chem 279, 15396–15401. 10.1074/jbc.m314291200. [DOI] [PubMed] [Google Scholar]
- Artiaga BL, Whitener RL, Staples CR, and Driver JP (2014). Adjuvant effects of therapeutic glycolipids administered to a cohort of NKT cell-diverse pigs. Vet. Immunol. Immunopathol 162, 1–13. 10.1016/j.vetimm.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Baev DV, Peng XH, Song L, Barnhart JR, Crooks GM, Weinberg KI, and Metelitsa LS (2004). Distinct homeostatic requirements of CD4+ and CD4-subsets of Vα24-invariant natural killer T cells in humans. Blood 104, 4150–4156. 10.1182/blood-2004-04-1629. [DOI] [PubMed] [Google Scholar]
- Baranek T, Lebrigand K, de Amat Herbozo C, Gonzalez L, Bogard G, Dietrich C, Magnone V, Boisseau C, Jouan Y, Trottein F, et al. (2020). High dimensional single-cell analysis reveals iNKT cell developmental trajectories and effector fate decision. Cell Rep. 32, 108116. 10.1016/j.celrep.2020.108116. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, and Teyton L (2007). The biology of NKT cells. Annu. Rev. Immunol 25, 297–336. 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Bertho N, and Meurens F (2021). The pig as a medical model for acquired respiratory diseases and dysfunctions: an immunological perspective. Mol. Immunol 135, 254–267. 10.1016/j.molimm.2021.03.014. [DOI] [PubMed] [Google Scholar]
- Blighe K, Rana S, and Lewis M (2018). EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. [Google Scholar]
- Broussard-Diehl C, Bauer SR, and Scheuermann RH (1996). A role for c-myc in the regulation of thymocyte differentiation and possibly positive selection. J. Immunol 156, 3141–3150. [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CWJ, Vegoe AL, Hsieh CS, Jenkins MK, and Farrar MA (2008). Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28, 112–121. 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannoodt R, Saelens W, Sichien D, Tavernier S, Janssens S, Guilliams M, Lambrecht B, Preter KD, and Saeys Y (2016). SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell developmentPriprint at. bioRxiv, 079509. 10.1101/079509. [DOI] [Google Scholar]
- Canté-Barrett K, Mendes RD, Li Y, Vroegindeweij E, Pike-Overzet K, Wabeke T, Langerak AW, Pieters R, Staal FJT, and Meijerink JPP (2017). Loss of CD44dim expression from early progenitor cells marks T-cell lineage commitment in the human thymus. Front. Immunol 8, 32. 10.3389/fimmu.2017.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Sun X, Icli B, Wara AK, and Feinberg MW (2010). Role of Krüppel-like factors in leukocyte development, function, and disease. Blood 116, 4404–414. 10.1182/blood-2010-05-285353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, and Jameson SC (2006). Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302. 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- Cheng M, and Anderson MS (2018). Thymic tolerance as a key brake on autoimmunity. Nat. Immunol 19, 659–664. 10.1038/s41590-018-0128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F, and Mucida D (2011). The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol 11, 445–456. 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibrian D, Castillo-González R, Fernández-Gallego N, de la Fuente H, Jorge I, Saiz ML, Punzón C, Ramírez-Huesca M, Vicente-Manzanares M, Fresno M, et al. (2020). Targeting L-type amino acid transporter 1 in innate and adaptive T cells efficiently controls skin inflammation. J. Allergy. Clin. Immunol 145, 199–214.e11. 10.1016/j.jaci.2019.09.025. [DOI] [PubMed] [Google Scholar]
- Colantonio AD, Epeldegui M, Jesiak M, Jachimowski L, Blom B, and Uittenbogaart CH (2011). IFN-α is constitutively expressed in the human thymus, but not in peripheral lymphoid organs. PLoS. One 6, e24252. 10.1371/journal.pone.0024252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadi S, Chhangawala S, Whitlock BM, Franklin RA, Luo CT, Oh SA, Toure A, Pritykin Y, Huse M, Leslie CS, and Li M (2016). Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell 164, 365–377. 10.1016/j.cell.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley SR, Hu DY, and Goodnow CC (2013). Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-κB. J. Exp. Med 210, 269–285. 10.1084/jem.20121458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson HD (2011). A comparative assessment of the pig , mouse and human genomes. Minipig Biomed. Res 1, 323–342. [Google Scholar]
- Dong X, Yang L, Liu K, Ji X, Tang C, Li W, Ma L, Mei Y, Peng T, Feng B, et al. (2021). Transcriptional networks identify synaptotagmin-like 3 as a regulator of cortical neuronal migration during early neurodevelopment. Cell Rep. 34, 108802. 10.1016/j.celrep.2021.108802. [DOI] [PubMed] [Google Scholar]
- Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, and Kronenberg M (2016). Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol 17, 728–739. 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, and Rudensky AY (2016). Hallmarks of tissue-resident lymphocytes. Cell 164, 1198–1211. 10.1016/j.cell.2016.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferhat MH, Robin A, Barbier L, Thierry A, Gombert JM, Barbarin A, and Herbelin A (2018). The impact of invariant NKT cells in sterile inflammation: the possible contribution of the alarmin/cytokine IL-33. Front. Immunol 9, 2308. 10.3389/fimmu.2018.02308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filén S, and Lahesmaa R (2010). GIMAP proteins in T-lymphocytes. J. Signal Transduct 2010, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JM, Jordan MA, Snelgrove SL, Slattery RM, Dufour FD, Kyparissoudis K, Besra GS, Godfrey DI, and Baxter AG (2008). Congenic analysis of the NKT cell control gene Nkt2 implicates the peroxisomal protein Pxmp4. J. Immunol 181, 3400–3412. 10.4049/jimmunol.181.5.3400. [DOI] [PubMed] [Google Scholar]
- de la Fuente H, Cruz-Adalia A, Martinez del Hoyo G, Cibrián-Vera D, Bonay P, Pérez-Hernández D, Vázquez J, Navarro P, Gutierrez-Gallego R, Ramirez-Huesca M, et al. (2014). The leukocyte activation receptor CD69 controls T cell differentiation through its interaction with galectin-1. Mol. Cell. Biol 34, 2479–2487. 10.1128/mcb.00348-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, López-Soto A, Kumar S, and Kroemer G (2016). Caspases connect cell-death signaling to organismal homeostasis. Immunity 44, 221–231. 10.1016/j.immuni.2016.01.020. [DOI] [PubMed] [Google Scholar]
- Georgiev H, Ravens I, Benarafa C, Förster R, and Bernhardt G (2016). Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets. Nat. Commun 7, 13116. 10.1038/ncomms13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, Stankovic S, and Baxter AG (2010). Raising the NKT cell family. Nat. Immunol 11, 197–206. 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Uldrich AP, Mccluskey J, Rossjohn J, and Moody DB (2015). The burgeoning family of unconventional T cells. Nat. Immunol 16, 1114–1123. 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- Groh V, Porcelli S, Fabbi M, Lanier LL, Picker LJ, Anderson T, Warnke RA, Bhan AK, Strominger JL, and Brenner MB (1989). Human lymphocytes bearing T cell receptor gamma/delta are phenotypically diverse and evenly distributed throughout the lymphoid system. J. Exp. Med 169, 1277–1294. 10.1084/jem.169.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL, Bai R, Chen CXJ, Liu DQ, Liu Y, Zhang CY, and Zen K (2009). Role of junctional adhesion molecule-like protein in mediating monocyte transendothelial migration. Arterioscler. Thromb. Vasc. Biol 29, 75–83. 10.1161/atvbaha.108.177717. [DOI] [PubMed] [Google Scholar]
- Haapalainen AM, Daddali R, Hallman M, and Rämet M (2021). Human CPPED1 belongs to calcineurin-like metallophosphoesterase superfamily and dephosphorylates PI3K-AKT pathway component PAK4. J. Cell. Mol. Med 25, 6304–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas JD, Ravens S, Düber S, Düber S, Sandrock I, Oberdörfer L, Oberdörfer L, Kashani E, Chennupati V, Föhse L, et al. (2012). Development of interleukin-17-producing γδ T cells is restricted to a functional embryonic wave. Immunity 37, 48–59. 10.1016/j.immuni.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Harsha Krovi S, Zhang J, Michaels-Foster MJ, Brunetti T, Loh L, Scott-Browne J, and Gapin L (2020). Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat. Commun 11, 6238. 10.1038/s41467-020-20073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Hiwatashi K, Ichiyama K, Morita R, Sekiya T, Kimura A, Sugiyama Y, Sibata T, Kuroda K, Takahashi R, and Yoshimura A (2011). SOCS1 regulates type I/type II NKT cell balance by regulating IFNγ signaling. Int. Immunol 23, 165–176. 10.1093/intimm/dxq469. [DOI] [PubMed] [Google Scholar]
- Hendriks J, Gravestein LA, Tesselaar K, Van Lier RAW, Schumacher TNM, and Borst J (2000). CD27 is required for generation and long-term maintenance of T cell immunity. Nat. Immunol 1, 433–440. 10.1038/80877. [DOI] [PubMed] [Google Scholar]
- Herrera-Uribe J, Wiarda JE, Sivasankaran SK, Daharsh L, Liu H, Byrne KA, Smith TPL, Lunney JK, Loving CL, and Tuggle CK (2021). Reference transcriptomes of porcine peripheral immune cells created through bulk and single-cell RNA sequencing. Front. Genet 12, 689406. 10.3389/fgene.2021.689406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig CT, Waters RW, Baldwin CL, and Telfer JC (2010). Evolution of the CD163 family and its relationship to the bovine gamma delta T cell co-receptor WC1. BMC. Evol. Biol 10, 181. 10.1186/1471-2148-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderness J, Hedges JF, Ramstead A, and Jutila MA (2013). Comparative biology of γδ T cell function in humans, mice, and domestic animals. Annu. Rev. Anim. Biosci 1, 99–124. 10.1146/annurev-animal-031412-103639. [DOI] [PubMed] [Google Scholar]
- Hosokawa H, Romero-Wolf M, Yang Q, Motomura Y, Levanon D, Groner Y, Moro K, Tanaka T, and Rothenberg EV (2020). Cell type-specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J. Exp. Med 217, e20190972. 10.1084/jem.20190972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H, Chen C, Nenninger A, Holz L, Baldwin CL, and Telfer JC (2015). WC1 is a hybrid γδ TCR coreceptor and pattern recognition receptor for pathogenic bacteria. J. Immunol 194, 2280–2288. 10.4049/jimmunol.1402021. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lancaster JN, and Ehrlich LIR (2015). The contribution of chemokines and migration to the induction of central tolerance in the thymus. Front. Immunol 6, 398. 10.3389/fimmu.2015.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua G, He C, Lv X, Fan L, Wang C, Remmenga SW, Rodabaugh KJ, Yang L, Lele SM, Yang P, et al. (2016). The four and a half LIM domains 2 (FHL2) regulates ovarian granulosa cell tumor progression via controlling AKT1 transcription. Cell Death Dis. 7, e2297. 10.1038/cddis.2016.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphray SJ, Scott CE, Clark R, Marron B, Bender C, Camm N, Davis J, Jenks A, Noon A, Patel M, et al. (2007). A high utility integrated map of the pig genome. Genome. Biol 8, R139. 10.1186/gb-2007-8-7-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbokwe CO, and Ezenwaka K (2017). Age-related morphological changes in the thymus of indigenous Large White pig cross during foetal and postnatal development. Anatomy 11, 12–20. 10.2399/ana.16.050. [DOI] [Google Scholar]
- Isakov N, and Altman A (2012). PKC-theta-mediated signal delivery from the TCR/CD28 surface receptors. Front. Immunol 3, 273. 10.3389/fimmu.2012.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Gunawardane L, Zebrowski E, Kostadinova L, Jobava R, Krokowski D, Hatzoglou M, Anthony DD, and Valadkhan S (2015). Regulation of interferon-stimulated gene BST2 by a lncRNA transcribed from a shared bidirectional promoter. Front. Immunol 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käser T (2021). Swine as biomedical animal model for T-cell research—success and potential for transmittable and non-transmittable human diseases. Mol. Immunol 135, 95–115. 10.1016/j.molimm.2021.04.004. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Kameda H, Nakano H, and Yamazaki S (2021). Regulation of T cell differentiation by the AP-1 transcription factor JunB. Immunol. Med 44, 197–203. 10.1080/25785826.2021.1872838. [DOI] [PubMed] [Google Scholar]
- Kim EH, and Suresh M (2013). Role of PI3K/Akt signaling in memory CD8 T cell differentiation. Front. Immunol 4, 20. 10.3389/fimmu.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgessner H, Dietrich J, Scherer J, Isomäki P, Korinek V, Hilgert I, Bruyns E, Leo A, Cope AP, and Schraven B (2001). The transmembrane adaptor protein TRIM regulates T cell receptor (TCR) expression and TCR-mediated signaling via an association with the TCR δ chain. J. Exp. Med 193, 1269–1284. 10.1084/jem.193.11.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczek AL, Hartung E, Gurka S, Becker M, Reeg N, Mages HW, Voigt S, Freund C, and Kroczek RA (2018). Structure-function relationship of XCL1 used for in vivo targeting of antigen into XCR1 + dendritic cells. Front. Immunol 9, 2806. 10.3389/fimmu.2018.02806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, and Jetten AM (2000). Retinoid-related orphan receptor γ (RORγ) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc. Natl. Acad. Sci. USA 97, 10132–10137. 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, Park JE, Ha VL, Luong A, Branciamore S, Rodin AS, Gogoshin G, Li F, Loh YHE, Camacho V, et al. (2020). Single-cell RNA-seq mapping of human thymopoiesis reveals lineage specification trajectories and a commitment spectrum in T cell development. Immunity 52, 1105–1118.e9, e9. 10.1016/j.immuni.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, D’Acquisto F, Hayden MS, Shim JH, and Ghosh S (2005). PDK1 nucleates T cell receptor-induced signaling complex for NF-κB activation. Science 308, 114–118. 10.1126/science.1107107. [DOI] [PubMed] [Google Scholar]
- Lee M, Lee E, Han SK, Choi YH, Kwon DI, Choi H, Lee K, Park ES, Rha MS, Joo DJ, et al. (2020). Single-cell RNA sequencing identifies shared differentiation paths of mouse thymic innate T cells. Nat. Commun 11, 4367. 10.1038/s41467-020-18155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Jameson SC, and Hogquist KA (2011). Alternative memory in the CD8 T cell lineage. Trends. Immunol 32, 50–56. 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Starrett GJ, Lee ST, Yang R, Henzler CM, Jameson SC, and Hogquist KA (2016). Lineage-specific effector signatures of invariant NKT cells are shared amongst γδ T, innate lymphoid, and Th cells. J. Immunol 197, 1460–1470. 10.4049/jimmunol.1600643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir EAD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197. 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Ji P, Lv Z, Wu Z, Shao X, Sun X, Song LG, Lei JX, Lv ZY, Wu ZD, et al. (2015). The expression of molecule CD28 and CD38 on CD4+/CD8+ T lymphocytes in thymus and spleen elicited by Schistosoma japonicum infection in mice model. Parasitol. Res 114, 3047–3058. 10.1007/s00436-015-4507-y. [DOI] [PubMed] [Google Scholar]
- Lio CWJ, and Hsieh CS (2008). A two-step process for thymic regulatory T cell development. Immunity 28, 100–111. 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luissint AC, Lutz PG, Calderwood DA, Couraud PO, and Bourdoulous S (2008). JAM-L-mediated leukocyte adhesion to endothelial cells is regulated in cis by α4β1 integrin activation. J. Cell. Biol 183, 1159–1173. 10.1083/jcb.200805061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Page L, Gillespie A, Schwartz JC, Prawits LM, Schlerka A, Farrell CP, Hammond JA, Baldwin CL, Telfer JC, and Hammer SE (2021). Subpopulations of swine γδ T cells defined by TCRg and WC1 gene expression. Dev. Comp. Immunol 125, 104214. 10.1016/j.dci.2021.104214. [DOI] [PubMed] [Google Scholar]
- Mackay CR, and Hein WR (1989). A large proportion of bovine T cells express the γδ T cell receptor and show a distinct tissue distribution and surface phenotype. Int. Immunol 1, 540–545. 10.1093/intimm/1.5.540. [DOI] [PubMed] [Google Scholar]
- Mackay LK, Minnich M, Kragten NAM, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. (2016). Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–63. 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- Malhotra N, Narayan K, Cho OH, Sylvia KE, Yin C, Melichar H, Rashighi M, Lefebvre V, Harris JE, Berg LJ, and Kang J (2013). A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity 38, 681–693. 10.1016/j.immuni.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw JM, Thelen F, Hampton EN, Bruno NE, Young TS, Havran WL, and Witherden DA (2021). JAML promotes CD8 and γδ T cell antitumor immunity and is a novel target for cancer immunotherapy. J. Exp. Med 218, e20202644. 10.1084/jem.20202644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengrelis K, Lau CI, Rowell J, Solanki A, Norris S, Ross S, Ono M, Outram S, and Crompton T (2019). Sonic hedgehog is a determinant of γδ T-cell differentiation in the thymus. Front. Immunol 10, 1629. 10.3389/fimmu.2019.01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurens F, Summerfield A, Nauwynck H, Saif L, and Gerdts V (2012). The pig: a model for human infectious diseases. Trends. Microbiol 20, 50–57. 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JFAP (2020). The function of the thymus and its impact on modern medicine. Science 369, eaba2429. 10.1126/science.aba2429. [DOI] [PubMed] [Google Scholar]
- Mitson-Salazar A, Yin Y, Wansley DL, Young M, Bolan H, Arceo S, Ho N, Koh C, Milner JD, Stone KD, et al. (2016). Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human TH2 cell subpopulation with enhanced function. J. Allergy. Clin. Immunol 137, 907–918.e9. 10.1016/j.jaci.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Muro R, Takayanagi H, and Nitta T (2019). T cell receptor signaling for γδT cell development. Inflamm. Regen 39, 6. 10.1186/s41232-019-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TKA, Koets AP, Vordermeier M, Jervis PJ, Cox LR, Graham SP, Santema WJ, Moody DB, Van calenbergh S, Zajonc DM, et al. (2013). The bovine CD1D gene has an unusual gene structure and is expressed but cannot present α-galactosylceramide with a C26 fatty acid. Int. Immunol 25, 91–98. 10.1093/intimm/dxs092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Zapater E, Santis G, and Parsons M (2017). CAR: a key regulator of adhesion and inflammation. Int. J. Biochem. Cell. Biol 89, 1–5. 10.1016/j.biocel.2017.05.025. [DOI] [PubMed] [Google Scholar]
- Owen DL, Sjaastad LE, and Farrar MA (2019). Regulatory T cell development in the thymus. J. Immunol 203, 2031–2041. 10.4049/jimmunol.1900662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Botting RA, Domínguez Conde C, Popescu DM, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, et al. (2020). A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224. 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker ME, and Ciofani M (2020). Regulation of γδ T cell effector diversification in the thymus. Front. Immunol 11, 42. 10.3389/fimmu.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschalidis N, Huggins A, Rowbotham NJ, Furmanski AL, Crompton T, Flower RJ, Perretti M, and D’Acquisto F (2010). Role of endogenous annexin-A1 in the regulation of thymocyte positive and negative selection. Cell Cycle 9, 785–794. 10.4161/cc.9.4.10673. [DOI] [PubMed] [Google Scholar]
- Patil RS, Bhat SA, Dar AA, and Chiplunkar SV (2015). The Jekyll and Hyde story of IL17-producing γδT cells. Front. Immunol 6, 37. 10.3389/fimmu.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci DG, Koay HF, and Berzins SP (2020). Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge. Nat. Rev. Immunol 20, 756–770. 10.1038/s41577-020-0345-y. [DOI] [PubMed] [Google Scholar]
- Perera J, and Huang H (2015). The development and function of thymic B cells. Cell. Mol. Life. Sci 72, 2657–2663. 10.1007/s00018-015-1895-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng YC, and Lenschow DJ (2018). ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol 16, 423–39. 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescovitz MD, Hsu S-M, Katz SI, Lunney JK, Shimada S, and Sachs DH (1990). Characterization of a porcine CD1-specific mAb that distinguishes CD4/CD8 double-positive thymic from peripheral T lymphocytes. Tissue. Antigens 35, 151–156. 10.1111/j.1399-0039.1990.tb01772.x. [DOI] [PubMed] [Google Scholar]
- Pobezinsky LA, Angelov GS, Tai X, Jeurling S, Van Laethem F, Feigenbaum L, Park JH, and Singer A (2012). Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat. Immunol 13, 569–578. 10.1038/ni.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakasz E, Hagen M, Sandor M, and Lynch RG (1997). Gamma delta T cells of the murine vagina: T cell response in vivo in the absence of the expression of CD2 and CD28 molecules. Int. Immunol 9, 161–167. 10.1093/intimm/9.1.161. [DOI] [PubMed] [Google Scholar]
- Res P, Blom B, Hori T, Weijer K, and Spits H (1997). Downregulation of CD1 marks acquisition of functional maturation of human thymocytes and defines a control point in late stages of human T cell development. J. Exp. Med 185, 141–152. 10.1084/jem.185.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes R, Cardeñes B, Machado-Pineda Y, and Cabañas C (2018). Tetraspanin CD9: a key regulator of cell adhesion in the immune system. Front. Immunol 9, 863. 10.3389/fimmu.2018.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Perugini V, Zamai M, González-Granado JM, Barreiro O, Tejera E, Yañez-Mó M, Caiolfa VR, and Sanchez-Madrid F (2013). CD81 controls sustained T cell activation signaling and defines the maturation stages of cognate immunological synapses. Mol. Cell. Biol 33, 3644–3658. 10.1128/mcb.00302-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Gómez IM, Talker SC, Käser T, Stadler M, Reiter L, Ladinig A, Milburn JV, Hammer SE, Mair KH, Saalmüller A, and Gerner W (2019). Expression of T-bet, eomesodermin, and GATA-3 correlates with distinct phenotypes and functional properties in porcine γδ T cells. Front. Immunol 10, 396. 10.3389/fimmu.2019.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, and Yui MA (2008). Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol 8, 9–21. 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalmüller A, Hirt W, and Reddehase MJ (1990). Porcine γ/δ Tlymphocyte subsets differing in their propensity to home to lymphoid tissue. Eur. J. Immunol 20, 2343–2346. 10.1002/eji.1830201026. [DOI] [PubMed] [Google Scholar]
- Sagar, Pokrovskii M, Herman JS, Naik S, Sock E, Zeis P, Lausch U, Wegner M, Tanriver Y, Littman DR, and Grun D (2020). Deciphering the regulatory landscape of fetal and adult γδ T-cell development at single-cell resolution. EMBO. J 39, e104159. 10.15252/embj.2019104159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salou M, Legoux F, Gilet J, Darbois A, Du Halgouet A, Alonso R, Richer W, Goubet AG, Daviaud C, Menger L, et al. (2019). A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J. Exp. Med 216, 133–151. 10.1084/jem.20181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Webb LMC, Janas ML, Hutchings A, Pascall J, Carter C, Pugh N, Morgan G, Turner M, and Butcher GW (2010). Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood 115, 3249–3257. 10.1182/blood-2009-08-237586. [DOI] [PubMed] [Google Scholar]
- Savino W (2006). The thymus is a common target organ in infectious diseases. PLoS Pathog. 2, e62. 10.1371/journal.ppat.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino W, Dardenne M, Velloso LA, and Dayse Silva-Barbosa S (2007). The thymus is a common target in malnutrition and infection. Br. J. Nutr 98, S11–S16. 10.1017/s0007114507832880. [DOI] [PubMed] [Google Scholar]
- Sedlak C, Patzl M, Saalmüller A, and Gerner W (2014). CD2 and CD8a define porcine γδ T cells with distinct cytokine production profiles. Dev. Comp. Immunol 45, 97–106. 10.1016/j.dci.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, Singh H, and Bendelac A (2012). Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol 13, 264–271. 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, Ichinose H, Metzger D, Chambon P, and Yoshimura A (2013). Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol 14, 230–237. 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- Sinkora M, and Butler JE (2016). Progress in the use of swine in developmental immunology of B and T lymphocytes. Dev. Comp. Immunol 58, 1–17. 10.1016/j.dci.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Sinkora M, Sinkora J, Reháková Z, Rehakova Z, and Butler JE (2000). Early ontogeny of thymocytes in pigs: sequential colonization of the thymus by T cell progenitors. J. Immunol 165, 1832–1839. 10.4049/jimmunol.165.4.1832. [DOI] [PubMed] [Google Scholar]
- Sinkora M, Butler JE, Holtmeier W, and Sinkorova J (2005a). Lymphocyte development in fetal piglets: facts and surprises. Vet. Immunol. Immunopathol 108, 177–184. 10.1016/j.vetimm.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Sinkora M, Sinkorova J, and Holtmeier W (2005b). Development of gammadelta thymocyte subsets during prenatal and postnatal ontogeny. Immunology 115, 544–555. 10.1111/j.1365-2567.2005.02194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkorova J, Stepanova K, Butler JE, and Sinkora M (2019). T cells in swine completely rearrange immunoglobulin heavy chain genes. Dev. Comp. Immunol 99, 103396. 10.1016/j.dci.2019.103396. [DOI] [PubMed] [Google Scholar]
- Spidale NA, Sylvia K, Narayan K, Miu B, Frascoli M, Melichar HJ, Zhihao W, Kisielow J, Palin A, Serwold T, et al. (2018). Interleukin-17-Pro-ducing γδ T cells originate from SOX13+ progenitors that are independent of γδTCR signaling. Immunity 49, 857–872.e5. 10.1016/j.immuni.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreejit G, Flynn MC, Patil M, Krishnamurthy P, Murphy AJ, and Nagareddy PR (2020). S100 family proteins in inflammation and beyond. Adv. Clin. Chem. 98, 173–231. 10.1016/bs.acc.2020.02.006. [DOI] [PubMed] [Google Scholar]
- Starbæk SMR, Brogaard L, Dawson HD, Smith AD, Heegaard PMH, Larsen LE, Jungersen G, and Skovgaard K (2018). Animal models for influenza A virus infection incorporating the involvement of innate host defenses: enhanced translational value of the porcine model. ILAR. J 59, 323–337. 10.1093/ilar/ily009. [DOI] [PubMed] [Google Scholar]
- Štěpánová K, and Sinkora M (2012). The expression of CD25, CD11b, SWC1, SWC7, MHC-II, and family of CD45 molecules can be used to characterize different stages of γδ T lymphocytes in pigs. Dev. Comp. Immunol 36, 728–740. 10.1016/j.dci.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Stepanova K, and Sinkora M (2013). Porcine γδ T lymphocytes can Be categorized into two functionally and developmentally distinct subsets according to expression of CD2 and level of TCR. J. Immunol 190, 2111–2120. 10.4049/jimmunol.1202890. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, and Locksley RM (2003). Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med 198, 1069–1076. 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, and Dudoit S (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC. Genomics 19, 477. 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A, Robin A, Giraud S, Minouflet S, Barra A, Bridoux F, Hauet T, Touchard G, Herbelin A, and Gombert JM (2012). Identification of invariant natural killer T cells in porcine peripheral blood. Vet. Immunol. Immunopathol 149, 272–279. 10.1016/j.vetimm.2012.06.023. [DOI] [PubMed] [Google Scholar]
- Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep 9, 5233. 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran MK, Kurakula K, Koenis DS, and de Vries CJM (2016). Protein-protein interactions of the LIM-only protein FHL2 and functional implication of the interactions relevant in cardiovascular disease. Biochim. Biophys. Acta - Mol. Cell Res. 1863, 219–228. 10.1016/j.bbamcr.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle KD, Krovi SH, Zhang J, Bedel R, Harmacek L, Peterson LK, Dragone LL, Lefferts A, Halluszczak C, Riemondy K, et al. (2018). TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat. Commun 9, 2650. 10.1038/s41467-018-05026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzlak S, Schenk RL, Vasanthakumar A, Preston SP, Haschka MD, Zotos D, Kallies A, Strasser A, Villunger A, and Herold MJ (2017). The BCL-2 pro-survival protein A1 is dispensable for T cell homeostasis on viral infection. Cell. Death. Differ 24, 523–533. 10.1038/cdd.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara S, Hayes SM, Li L, El-Khoury D, Canelles M, Fowlkes BJ, and Love PE (2006). Premature expression of chemokine receptor CCR9 impairs T cell development. J. Immunol 176, 75–84. 10.4049/jimmunol.176.1.75. [DOI] [PubMed] [Google Scholar]
- Vaeth M, Wang YH, Eckstein M, Yang J, Silverman GJ, Lacruz RS, Kannan K, and Feske S (2019). Tissue resident and follicular Treg cell differentiation is regulated by CRAC channels. Nat. Commun 10, 1183. 10.1038/s41467-019-08959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio O, Riwar B, Brown MH, and Lassila O (1991). Characterization of the putative avian CD2 homologue. J. Immunol 147, 1593–1599. [PubMed] [Google Scholar]
- Verdino P, and Wilson IA (2011). JAML and CAR: two more players in T-cell activation. Cell Cycle 10, 1341–1342. 10.4161/cc.10.9.15294. [DOI] [PubMed] [Google Scholar]
- Verdino P, Witherden DA, Havran WL, and Wilson IA (2010). The molecular interaction of CAR and JAML recruits the central cell signal transducer PI3K. Science 329, 1210–1214. 10.1126/science.1187996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Su S, McCrann DJ, Green JM, Leu K, Young PR, Schatz PJ, Silva JC, Stokes MP, and Wojchowski DM (2014). RHEX, a novel regulator of human erythroid progenitor cell expansion and erythroblast development. J. Exp. Med 211, 1715–1722. 10.1084/jem.20130624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rhijn I, Godfrey DI, Rossjohn J, and Moody DB (2015). Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol 15, 643–654. 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstichel G, Vermijlen D, Martens L, Goetgeluk G, Brouwer M, Thiault N, Van Caeneghem Y, De Munter S, Weening K, Bonte S, et al. (2017). The checkpoint for agonist selection precedes conventional selection in human thymus. Sci. Immunol 2, eaah4232. 10.1126/sciimmunol.aah4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, et al. (2003). Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat. Immunol 4, 533–539. 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- Wang J, Guillaume J, Pauwels N, van Calenbergh S, van Rhijn I, and Zajonc DM (2012). Crystal structures of bovine CD1d reveal altered aGalCer presentation and a restricted A’ pocket unable to bind long-chain glycolipids. PLoS One 7, e47989. 10.1371/journal.pone.0047989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich MA, and Hogquist KA (2008). Thymic emigration: when and how T cells leave home. J. Immunol 181, 2265–2270. 10.4049/jimmunol.181.4.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Schierup MH, Jørgensen FG, Gorodkin J, Panitz F, Stærfeldt HH, Christensen OF, Mailund T, Hornshøj H, Klein A, et al. (2005). Pigs in sequence space: a 0.66X coverage pig genome survey based on shotgun sequencing. BMC. Genomics 6, 70. 10.1186/1471-2164-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AJ, Lucas B, Jenkinson WE, and Anderson G (2018). Invariant NKT cells and control of the thymus medulla. J. Immunol 200, 3333–3339. 10.4049/jimmunol.1800120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JT, Cross EW, and Kedl RM (2017). Antigen-inexperienced memory CD8+ T cells: where they come from and why we need them. Nat. Rev. Immunol 17, 391–400. 10.1038/nri.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, and Sakaguchi S (2008). CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275. 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- Witherden DA, Abernethy NJ, Kimpton WG, and Cahill RNP (1995). Antigen-independent maturation of CD2, CD11a/CD18, CD44, and CD58 expression on thymic emigrants in fetal and postnatal sheep. Dev. Immunol 4, 199–209. 10.1155/1995/35075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherden DA, Verdino P, Rieder SE, Garijo O, Mills RE, Teyton L, Fischer WH, Wilson IA, and Havran WL (2010). The junctional adhesion molecule JAML is a costimulatory receptor for epithelial γδ T cell activation. Science 329, 1205–1210. 10.1126/science.1192698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Wang X, Jameson SC, and Hogquist KA (2016). Late stages of T cell maturation in the thymus involve NF-κB and tonic type i interferon signaling. Nat. Immunol 17, 565–573. 10.1038/ni.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T, Mathis D, and Benoist C (2004). Self-reactivity in thymic double-positive cells commits cells to a CD8αα lineage with characteristics of innate immune cells. Nat. Immunol 5, 597–605. 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- Yang H, and Parkhouse RME (1996). Phenotypic classification of porcine lymphocyte subpopulations in blood and lymphoid tissues. Immunology 89, 76–83. 10.1046/j.1365-2567.1996.d01-705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Kwon DI, Kim M, Im SH, and Lee YJ (2021). Commensal microbiome expands Tγδ17 cells in the lung and promotes particulate matter-induced acute neutrophilia. Front. Immunol 12, 645741. 10.3389/fimmu.2021.645741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Artiaga BL, Lomelino CL, Jayaprakash D, Sachidanandam R, Mckenna R, and Driver JP (2019). Next generation sequencing of the pig α β TCR repertoire identifies the porcine invariant NKT cell receptor. J. Immunol 202, 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jeremiah Bell J, and Bhandoola A (2010). T-cell lineage determination. Immunol. Rev 238, 12–22. 10.1111/j.1600-065x.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, and Rothenberg EV (2014). Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol 14, 529–545. 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Lin Y-Y, Dai M, and Zhuang Y (2014). Id3 and Id2 act as a dual safety mechanism in regulating the development and population sizeof innate-like γδ T cells. J. Immunol 192, 1055–1063. 10.4049/jimmunol.1302694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckermann FA, and Gaskins HR (1996). Distribution of porcine CD4/CD8 double-positive T lymphocytes in mucosa-associated lymphoid tissues. Immunology 87, 493–499. 10.1046/j.1365-2567.1996.494570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been deposited at GSE192520. Code used in this study are available on https://github.com/Driver-lab1/scRNAseq_pig_thymus. DOIs are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Live/Dead viability dye | Invitrogen | Cat# L34976 |
| Mouse anti-pig CD3ε antibody | BD | Clone BB23-8E6-8C8 |
| Anti-JAML antibody | Abcam | Clone EPR15289 |
| Alexa Fluor 488-conjugated anti-rabbit IgG secondary antibody | Abcam | Cat# ab150077; RRID: AB_2630356 |
| Mouse anti-pig TCRδ antibody | WSU mAb Center | Clone PGBL22A |
| Biological samples | ||
| Pig thymus | University of Florida’s Animal Sciences Department | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Mouse CD1d (mCD1d) tetramer | National Institutes of Health Tetramer Core Facility | N/A |
| Critical commercial assays | ||
| Chromium Next GEM Single Cell 3′ reagent kit | 10xGenomics | RRID: SCR_019145 |
| NovaSeq 6000 sequencer | Illumina | RRID: SCR_019152 |
| Deposited data | ||
| Raw data | This paper | GSE192520 |
| Human thymus dataset | Le et al. (2020) | GSE139042 |
| Mouse thymic iNKT cell dataset | Harsha Krovi et al. (2020) | GSE152786 |
| Software and algorithms | ||
| Cell Ranger v3.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| R (4.0.2) | CRAN | https://www.r-project.org/ |
| Seurat (v3.2.2) | Stuart et al. (2019) | https://satijalab.org/seurat/ |
| EnhancedVolcano | Blighe et al. (2018) | https://github.com/kevinblighe/EnhancedVolcano |
| Monocle 3 | Cao et al. (2019); Levine et al. (2015); Traag et al., 2019; Trapnell et al. (2014) | https://cole-trapnell-lab.github.io/monocle3/ |
| SCORPIUS (1.0.8) | Cannoodt et al. (2016) | https://github.com/rcannood/SCORPIUS |
| Slingshot (2.0.0) | Street et al. (2010) | https://bioconductor.org/packages/release/bioc/vignettes/slingshot/inst/doc/vignette.html |
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo |
| BioRender | https://biorender.com/ | RRID:SCR_018361 |
| Custom scripts | This paper | https://doi.org/10.5281/zenodo.6609557 |