Abstract
The broad objective of the research presented here is to develop a noncatalytic plasmid maintenance system for the stabilization of multicopy expression plasmids encoding foreign antigens in a Salmonella typhi live-vector vaccine strain such as CVD 908-htrA. We have enhanced the maintenance of expression plasmids at two independent levels. First, we removed dependence upon balanced-lethal maintenance systems that involve catalytic enzymes expressed from multicopy plasmids; we accomplished this through incorporation into expression plasmids of a postsegregational killing system based on the noncatalytic hok-sok plasmid addiction system from the antibiotic resistance factor pR1. We also included at least one naturally occurring plasmid partition function in our expression plasmids, which eliminates random segregation of these plasmids, thereby enhancing their inheritance and stability; to accomplish this, we incorporated either the par locus from pSC101, the parA locus from pR1, or both. We monitored the stability of optimized expression plasmids within CVD 908-htrA by quantitating expression of a variant of green fluorescent protein (GFPuv) by using flow cytometry. In this report, we demonstrate the utility of this novel plasmid maintenance system in enhancing the stability of our expression plasmids and go on to show that as the copy number of stabilized plasmids increases, the toxicity of GFPuv synthesis also increases. The implications of these observations for the rational design of immunogenic and protective bacterial live vector vaccines are discussed.
Bacterial live-vector vaccines represent a vaccine development strategy that offers exceptional flexibility. In this approach, genes that encode protective antigens of unrelated bacterial, viral, or parasitic pathogens are expressed in a live vector that carries the foreign antigens to the immune system, thereby eliciting an appropriate immune response. The attenuated Salmonella typhi vaccine strain CVD 908-htrA is a particularly attractive live vector in that it is well tolerated and elicits a broad immune response to S. typhi antigens, which includes intestinal soluble immunoglobulin A antibodies, serum immunoglobulin G antibodies, and cellular immune responses (47, 48). In addition, genetic methods have been developed to express foreign antigens within attenuated S. typhi vaccine strains, and a murine intranasal model has been developed as a practical animal model for examining the immunogenicity and protective efficacy of a wide variety of heterologous antigens within S. typhi-based live-vector vaccines, prior to initiating clinical trials (3, 16).
The efficacy of any bacterial live-vector vaccine rests with its ability to present sufficient foreign antigen to the human immune system to initiate the desired protective immune response. Controlled expression of heterologous antigens from multicopy expression plasmids represents one obvious solution for synthesis of high levels of antigen within live vectors. However, these plasmids may become unstable in vivo, resulting in the loss of foreign genes and a decrease in the intended immune response.
One method of enhancing the inheritance of expression plasmids by live vectors involves construction of a “balanced-lethal” system for plasmids expressing heterologous antigens (34). In a plasmid-based balanced-lethal system, plasmids replicating in the cytoplasm of the bacterium express a critical protein required by the bacterium to grow and replicate; loss of such plasmids removes the ability of the bacterium to express the critical protein and results in cell death. This phenomenon of plasmid loss during bacterial replication, which results in the death of any plasmidless bacterium, is also referred to as “postsegregational killing.” Such a system has been successfully employed in Salmonella typhimurium and is based on expression of the asd gene encoding aspartate β-semialdehyde dehydrogenase (Asd) (15, 34). Asd is a critical enzyme involved in the synthesis of structural components essential for the formation of the cell wall in gram-negative bacteria. Therefore, loss of plasmids encoding such a critical enzyme would be lethal for any bacterium incapable of synthesizing Asd from the chromosome. Although the asd balanced-lethal system has been successfully employed in attenuated S. typhimurium-based live-vector strains for immunization of mice with a variety of prokaryotic and eukaryotic antigens (12, 28, 45), use of this method for stabilizing plasmids within attenuated S. typhi vaccine strains has, to date, been unsuccessful (47).
Here, we present the design and initial testing of a novel set of isogenic multicopy expression plasmids into which we have incorporated a noncatalytic postsegregational killing function, coupled with both active (14, 27) and passive plasmid partition functions (1, 31, 54), to provide a plasmid maintenance system designed to optimize expression of heterologous antigens within CVD 908-htrA for delivery to the human immune system. Since this method of improving plasmid maintenance involves no chromosomal mutagenesis of the live vector strain, in principle, such stabilized plasmids can be introduced into any live vector strain to improve the immunogenicity of heterologous antigens expressed, without additional genetic manipulations.
The approach is based on the use of the naturally occurring hok-sok postsegregational killing system residing on the R factor pR1 (19, 20). The hok-sok system is a two-component toxin-antitoxin system in which hok encodes a lethal pore-forming Hok protein. Synthesis of Hok is blocked by hybridization of a small antisense sok mRNA to hok mRNA, preventing translation and synthesis of Hok. However, sok mRNA is highly susceptible to degradation by nucleases, and its protective intracellular concentration must be maintained by constitutive transcription from resident plasmids carrying hok-sok. Therefore, bacteria that spontaneously lose such plasmids are postsegregationally killed because existing levels of the protective sok mRNA rapidly drop and levels of the more stable toxin-encoding hok mRNA quickly lead to Hok synthesis and cell death.
Inheritance of these expression plasmids has been enhanced through insertion of at least one partition function. A passive par locus from pSC101 (4, 31, 32, 52) was tested, in which no de novo partitioning proteins were encoded. In addition, the parA centromere-like active partitioning system from pR1 (14, 27) was investigated, since the combination of hok-sok and parA naturally occurs within pR1 and was not expected to present any compatibility complications. Plasmid maintenance was monitored by using flow cytometry to measure synthesis of a variant green fluorescent protein (GFPuv) test antigen within plasmid-bearing CVD 908-htrA. We show that although individual maintenance functions contribute to various degrees to the observed stability of expression plasmids, the highest levels of sustained synthesis of the test heterologous antigen GFPuv were detected from expression plasmids carrying the full complement of maintenance functions.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All plasmid constructions were recovered in Escherichia coli DH5α (Gibco BRL). Construction of the hok-sok gene cassette used pR1 template DNA isolated from E. coli J53(pR1), a generous gift from James B. Kaper. The live vector S. typhi CVD 908-htrA is an auxotrophic derivative of the wild-type strain Ty2 with deletions in aroC, aroD, and htrA (48). All strains used in this work were grown in media supplemented with 2,3-dihydroxybenzoic acid (DHB) as previously described (16, 26). When grown on solid medium, plasmid-bearing strains of CVD 908-htrA were streaked from frozen (−70°C) master stocks onto 2× Luria-Bertani (LB) agar containing 20 g of Bacto Tryptone, 10 g of Bacto yeast extract, and 50 mM NaCl (2× LB agar) plus carbenicillin at a concentration of 50 μg/ml. Plates were incubated at 30°C for 24 to 36 h to obtain isolated colonies ∼2 mm in diameter; strains were incubated at 30°C to minimize the toxicity of GFPuv expression in CVD 908-htrA.
For induction experiments involving comparison of the osmotically induced promoters PompC1 and PompC3, strains were streaked from frozen master stocks onto 2× LB agar supplemented with DHB and carbenicillin and incubated for 36 h at 30°C. Induction conditions were adapted from those of Puente et al. (40) and Tartera and Metcalf (49); isolated colonies were pooled into 300 μl of nutrient broth supplemented with DHB and carbenicillin, from which 25 μl was inoculated into 25 ml of supplemented nutrient broth, with and without 150 mM NaCl, and incubated at 37°C at 250 rpm in an orbital shaker (Forma Scientific) for 24 h. Bacteria were then pelleted, resuspended in 1 ml of phosphate-buffered saline (PBS [pH 7.4]), and then diluted 1:1,000 into PBS for analysis by flow cytometry as described below.
For plasmid retention studies, all strains were streaked from frozen master stocks onto 2× LB agar supplemented with DHB and carbenicillin and incubated for 36 h at 30°C. Isolated colonies were pooled into 300 μl of 1× LB broth supplemented only with DHB, from which 25 μl was inoculated into 25 ml of 1× LB broth containing DHB and either 50, 150, or 300 mM NaCl; cultures were incubated at 37°C at 250 rpm for 24 h. Bacteria were then pelleted, resuspended in 1 ml of PBS (pH 7.4), and diluted 1:1,000 into PBS for analysis by flow cytometry.
Molecular genetic techniques.
Standard techniques were used for the construction of the plasmids represented here (42). Unless otherwise noted, native Taq DNA polymerase (Gibco BRL) was used in PCRs. S. typhi strains were prepared for electroporation of recombinant plasmids after being harvested from Miller's LB broth (Gibco BRL) supplemented with DHB; after pelleting of bacteria, the cells were washed three times with one culture volume of sterile distilled water and resuspended in sterile distilled water to a final volume of 1/100 of the original culture volume. Electroporation of strains was performed in a Gene Pulser apparatus (Bio-Rad) set at 2.5 kV, 200 Ω, and 25 μF. Following electroporation, bacteria were repaired with SOC medium (Biofluids) and incubation at 37°C and 250 rpm for 45 min; bacteria were then plated on 1× LB medium containing DHB plus 50 μg of carbenicillin per ml, and incubated at 30°C for 24 h. Isolated colonies were then swabbed onto supplemented 2× LB medium and incubated at 30°C for 16 h. Frozen master stocks were prepared by harvesting bacteria into SOC medium without further supplementation and freezing at −70°C.
Construction of expression plasmids. (i) Construction of pJN1 and pJN2.
All primers used in this work are listed in Table 1; essential plasmids created from these primers are listed in Table 2 to illustrate the flow of logic in designing the final isogenic expression plasmids. The expression plasmids constructed for these studies are composed of three basic cassettes encoding (i) expression of a heterologous antigen, (ii) a plasmid origin of replication, and (iii) selection and maintenance functions. To accomplish this, a basic replicon was constructed in which these cassettes were separated by unique restriction sites. pTETnir15 (Table 2) (38) was reengineered such that the oriE1 origin of replication and bla gene were separated by a unique SpeI site. Toward this end, an oriE1 cassette was synthesized by PCR with Vent polymerase with primers 1 and 2 and pCVD315 (17) as the template. The resulting 735-bp fragment carries engineered SpeI and BglII sites 5′ proximal to the promoter controlling transcription of RNA II, and an engineered AvrII site 675 bases from these sites. A separate PCR was carried out by using primers 3 and 4 to create a 1,234-bp bla cassette containing an engineered XbaI site 5′ proximal to the original EcoRI site. The products from these two PCRs were gel purified and used in an overlapping PCR with primers 1 and 4 to yield a final 1,916-bp oriE1-bla fragment which was self-ligated to create pJN1. The Pnir15-toxC fragment from pTETnir15 was excised as an EcoRI (partial digestion)-AvaI fragment, in which the AvaI terminus was polished, and inserted into the multiple cloning region from pSL1180 (9) cleaved with EcoRI and StuI; this cassette was then reexcised as an EcoRI (partial digestion)-AvrII fragment and inserted into pJN1 cleaved with EcoRI-AvrII, creating pJN2 (Table 2).
TABLE 1.
Primers used in construction of the plasmid cassettes
| Primer no. | Sequencea | Cassette created | GenBank accession no. | Template | Source or reference |
|---|---|---|---|---|---|
| 1 | 5′-GCAGGAAAGAACATGTGAGCCTAGGGCCAGCAAAAGGCCAGGAAC-3′ | oriE1 | J01749 | pCVD315 | 17 |
| 2 | 5′-CATGACCAAAATCCCTTAACTAGTGTTTTAGATCTACTGAGCGTCAGACCCCG-3′ | oriE1 | J01749 | pCVD315 | |
| 3 | 5′-CGGGGTCTGACGCTCAGTAGATCTAAAACACTAGTTAAGGGATTTTGGTCATG-3′ | bla | J01749 | pCVD315 | |
| 4 | 5′-GCTGTCAAACATGAGAATTCTAGAAGACGAAAGGGCCTCGTGATACGCC-3′ | bla | J01749 | pCVD315 | |
| 5 | 5′-ACAGCCTGCAGACAGATCTTGACAGCTGGATCGCACTCTGGTATAATTGGGAAGCCCTGCAAAG-3′ | aphA-2 | V00618 | pIB279 | 5 |
| 6 | 5′-CGAAGCCCAACCTTTCATAGAAGCTAGCGGTGGATCCGAAATCTCGTGATGGCAGGTTG-3′ | aphA-2 | V00618 | pIB279 | |
| 7 | 5′-AACAAGCGTTATAGGAATTCTGTGGTAGCA-3′ | PompC | K00541 | E. coli DH5α | Gibco BRL |
| 8 | 5′-ACTTTCATGTTATTAAAGATCTGTTATATG-3′ | PompC | K00541 | E. coli DH5α | |
| 9 | 5′-AGATCTTAATCATCCACAGGAGGCTTTCTGATGAGTAAAGGAGAAGAACTTTTCACTGG-3′ | gfpuv | U62636 | pGFPuv | 13 |
| 10 | 5′-GCTAGCTCATTATTTGTAGAGCTCATCCATGC-3′ | gfpuv | U62636 | pGFPuv | |
| 11 | 5′-AGATCTGAATTCTAGATCATGTTTGACAGCTTATCATCGATAAGCTTTAATGCG-3′ | tetA | J01749 | pBR322 | 2 |
| 12 | 5′-AGATCTTATCAGGTCGAGGTGGCCCGGCTCCATGCACCGCGACGCAACGCG-3′ | tetA | J01749 | pBR322 | |
| 13 | 5′-CGCGAATTCTCGAGACAAACTCCGGGAGGCAGCGTGATGCGGCAACAATCACACGGATTTC-3′ | hok-sok-tetA | X05813 | pR1 | 19, 24 |
| 14 | 5′-ATGAGCGCATTGTTAGATTTCATTTTTTTTTCCTCCTTATTTTCTAGACAACATCAGCAAGGAGAAAGG-3′ | hok-sok-tetA | J01749, X05813 | pR1 | |
| 15 | 5′-CCTTTCTCCTTGCTGATGTTGTCTAGAAAATAAGGAGGAAAAAAAAATGAAATCTAACAATGCGCTCAT-3′ | hok-sok-tetA | X05813, J01749 | pBR322 | 2 |
| 16 | 5′-GCTACATTTGAAGAGATAAATTGCACTGGATCCTAGAAATATTTTATCTGATTAATAAGATGATC-3′ | ori15A | X06403 | pACYC184 | 43 |
| 17 | 5′-CGGAGATTTCCTGGAAGATGCCTAGGAGATACTTAACAGGGAAGTGAGAG-3′ | ori15A | X06403 | pACYC184 | |
| 18 | 5′-GTCTGCCGGATTGCTTATCCTGGCGGATCCGGTTGACAGTAAGACGGGTAAGCCTGTTGAT-3′ | par | X01654 | pSC101 | 33 |
| 19 | 5′-AGGCTTAAGTAGCACCCTCGCAAGATCTGGCAAATCGCTGAATATTCCTTTTGTCTCCGAC-3′ | par | X01654 | pSC101 | |
| 20 | 5′-GAGGGCGCCCCAGCTGGCAATTCTAGACTCGAGCACTTTTGTTACCCGCCAAACAAAACCCAAAAACAAC-3′ | aphA-2–parA | V00618, X04268 | pR1 | 19, 24 |
| 21 | 5′-AGAAGAAAAATCGAATTCCAGCATGAAGAGTTTCAGAAAATGACAGAGCGTGAGCAAGTGC-3′ | aphA-2–parA | X04268 | pR1 | |
| 22 | 5′-GTTGTTTTTGGGTTTTGTTTGGCGGGTAACAAAAGTGCTCGAGTCTAGAATTGCCAGCTGGGGCGCCCTC-3′ | aphA-2–parA | X04268, V00618 | pIB279 | 5 |
| 23 | 5′-CGAAGCCCAACCTTTCATAGAAACTAGTGGTGGAATCGAAATCTCGTGATGGCAGGTTG-3′ | aphA-2–parA | V00618 | pIB279 |
Relevant restriction sites are designated in boldface, underlined, and referred to in the text; ribosome binding sites and start codons are designated in italics.
TABLE 2.
Selected plasmids used in this work
| Plasmid | Size (kb) | Relevant genotype | Source or reference |
|---|---|---|---|
| pTETnir15 | 3.7 | oriE1 toxC bla | 38 |
| pJN1 | 1.9 | oriE1 bla | This work |
| pJN2 | 3.4 | oriE1 toxC bla | This work |
| pGFPuv | 3.3 | pUC19ori gfpuv bla | Clontech |
| pGFPompC | 3.5 | oriE1 gfpuv bla | This work |
| pNRB1 | 3.5 | oriE1 gfpuv tetA | This work |
| pGEN2 | 4.2 | oriE1 gfpuv tetA hok-sok | This work |
| pGEN3 | 4.1 | ori15A gfpuv tetA hok-sok | This work |
| pJN5 | 3.1 | oriE1 gfpuv bla | This work |
| pJN6 | 3.7 | oriE1 gfpuv bla hok-sok | This work |
| pJN7 | 4.1 | oriE1 gfpuv bla hok-sok par | This work |
| pJN8 | 5.4 | oriE1 gfpuv bla hok-sok parA | This work |
| pGEN51 | 3.6 | oriE1 gfpuv bla | This work |
| pGEN71 | 4.2 | oriE1 gfpuv bla hok-sok | This work |
| pGEN84 | 4.5 | oriE1 gfpuv bla hok-sok par | This work |
| pGEN183 | 5.9 | oriE1 gfpuv bla hok-sok parA | This work |
| pGEN211 | 6.2 | oriE1 gfpuv bla hok-sok par parA | This work |
| pGEN91 | 3.5 | ori15A gfpuv bla | This work |
| pGEN111 | 4.1 | ori15A gfpuv bla hok-sok | This work |
| pGEN121 | 4.5 | ori15A gfpuv bla hok-sok par | This work |
| pGEN193 | 5.8 | ori15A gfpuv bla hok-sok parA | This work |
| pGEN222 | 6.2 | ori15A gfpuv bla hok-sok par parA | This work |
(ii) Construction of pGFPompC.
To facilitate screening of a functional osmotically regulated PompC allele from E. coli, an aphA-2 cassette was constructed, encoding resistance to the aminoglycosides neomycin and kanamycin (44). A PCR was carried out with primers 5 and 6 with the template pIB279 (5) to generate a 1,044-bp product, from which a promoterless 903-bp aphA-2 BglII-NheI fragment was cleaved for replacement of a BglII-NheI toxC cassette encoding fragment C of tetanus toxin in pTETnir15. The anaerobically regulated Pnir15 promoter was replaced with a 459-bp EcoRI-BglII PompC allele constructed with primers 7 and 8 with chromosomal template DNA from E. coli DH5α to create pKompC. After confirming osmotic induction of PompC by examining the increase in resistance to kanamycin with increasing osmolarity, the aphA-2 cassette was then replaced with a gfpuv gene encoding a prokaryotic codon-optimized GFPuv allele (Clontech) (13). The gfpuv gene was recovered by PCR with primers 9 and 10 with the template pGFPuv to generate a 751-bp BglII-NheI fragment, which was inserted into pKompC, to generate pGFPompC. Colonies were screened for functional GFPuv, and the brightest colonies were then examined for induction of fluorescence with increasing concentrations of NaCl. A PompC1-gfpuv cassette (see Results below) was cleaved from pGFPompC1 as an EcoRI-NheI fragment and inserted into a derivative of pJN2 cleaved with EcoRI-NheI to create pJJ4.
(iii) Construction of pNRB1, pGEN2, and pGEN3.
Since it was intended that copy number not be influenced by transcription originating from promoters outside the origin of replication, it was necessary to ensure that all replication cassettes were flanked at both ends by transcription terminators. Because the origin and antigen cassettes of pJN2 are separated by the trpA terminator, it was only necessary to insert one additional terminator between the origin and bla cassettes.
To facilitate construction of additional plasmids later on, a tetA-T1T2 cassette was created. pYA292 (15) was first cleaved with HindIII and BglII, and the T1T2 terminator fragment was polished and inserted into the SmaI site of the pBluescript II KS (Stratagene) multiple cloning region; when the proper orientation was identified, this cassette was reexcised as a BamHI-PstI fragment and inserted into pIB307 (5) cleaved with BamHI-PstI, creating pJG14. It was later determined by sequence analysis that the cassette had undergone a deletion of approximately 100 bp, removing half of the T2 terminator.
Using pBR322 as a template, primers 11 and 12 were used to synthesize a 1,291-bp tetA BglII fragment. This tetA BglII fragment was then inserted into the BamHI site of pJG14, such that transcription of the tetA gene is terminated at the T1T2 terminator, creating pJG14tetA. Finally, this tetA-T1T2 cassette was cleaved from pJG14tetA as an EcoRI-PstI fragment in which the PstI site was removed by polishing; the resulting fragment was inserted into pJJ4 cleaved with SpeI, polished, and recleaved with EcoRI to replace the bla cassette and create pNRB1.
The noncatalytic postsegregational killing function to be incorporated into the plasmid maintenance systems of the expression plasmids described here was the hok-sok locus, from the multiple drug resistance R factor pR1. Initial attempts at recovering the hok-sok locus after PCR were unsuccessful. It was therefore necessary to use overlapping PCR to generate a cassette in which hok-sok was transcriptionally fused to a promoterless tetA gene such that transcription originating from the hok promoter would continue into tetA and result in a transcript encoding both Hok and resistance to tetracycline. pR1 plasmid DNA was purified from E. coli J53(pR1) in which pR1 encodes resistance to both carbenicillin and chloramphenicol. A 640-bp hok-sok fragment was synthesized by using primers 13 and 14; a promoterless 1,245-bp tetA fragment was recovered in a separate PCR by using primers 15 and 12 with pNRB1 as the template. The products from these two PCRs were then used in an overlapping PCR with primers 12 and 13 to yield the final 1,816-bp hok-sok-tetA fragment. This fragment was inserted as an EcoRI-SphI fragment into pNRB1 cleaved with EcoRI-SphI, regenerating the tetA gene and creating pGEN1.
Two isogenic plasmids were then constructed, differing only in copy number, from which all further expression plasmids would be derived. The BglII-AvrII origin of replication cassette of pGEN1 was replaced by a BglII-AvrII oriE1 cassette from pJN2 to generate pGEN2. In addition, an ori15A replication cassette was synthesized by PCR by using primers 16 and 17 with pACYC184 template to generate a 629-bp BamHI-AvrII fragment, which was inserted into pGEN2 cleaved with BglII-AvrII to create pGEN3.
(iv) Construction of pJN5, pGEN51, and pGEN91.
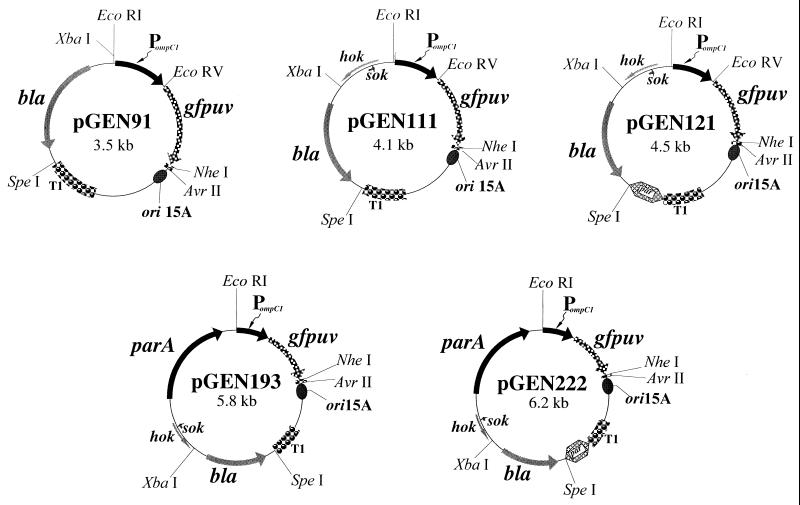
The principle set of isogenic expression plasmids, to which individual elements of a plasmid maintenance system were sequentially added, was composed of pGEN51 (containing oriE1) and pGEN91 (containing ori15A). The basic replicon from which these two plasmids were constructed was pJN5, which was assembled by cleaving the PompC-gfpuv cartridge as an EcoRI-NheI fragment from pGFPompC to replace the Pnir15-toxC cassette of pJN2. Construction of pGEN51 was then accomplished by removal of the replication cassette from pGEN2 as a BamHI fragment and replacement of the origin of replication within pJN5 digested with BglII and BamHI, thereby regenerating the gfpuv gene. Construction of pGEN91 was accomplished in an identical manner by excision of the origin cassette from pGEN3 as a BamHI fragment and insertion into pJN5 (Fig. 1 and Table 2).
FIG. 1.
Genetic maps of a representative set of isogenic expression plasmids, carrying an ori15A origin of replication, which differ only in the introduction of increasingly complex combinations of maintenance functions. The plasmids pGEN111, pGEN121, pGEN193, and pGEN222 contain maintenance functions encoded by the hok-sok, hok-sok plus par, hok-sok plus parA, and hok-sok plus par plus parA loci, respectively. Other expression plasmids were constructed by replacing each ori15A origin of replication cassette (copy number, ∼15) with an oriE1 origin (copy number, ∼60); a list of the plasmids used in this work is given in Table 2. The key restriction sites shown represent unique sites in the expression plasmids. PompC1, modified osmotically controlled ompC promoter from E. coli; gfpuv, gene encoding the prokaryotic codon-optimized GFPuv; T1, transcriptional terminator from the rrnB rRNA operon of E. coli; par, passive partitioning system from pSC101; bla, β-lactamase gene conferring resistance to carbenicillin; hok-sok, postsegregational killing locus from the multiple antibiotic resistance R plasmid pR1; parA, active partitioning system from pR1.
(v) Construction of pJN6, pGEN71, and pGEN111.
The hok-sok locus was then excised as an XbaI-SalI fragment from pGEN2 and inserted into pJN5 cleaved with XbaI and SalI, again regenerating the gfpuv gene to create pJN6. Construction of pGEN71 and pGEN111 was then carried out exactly as described for pGEN51 and pGEN91 by insertion into pJN6 of origin cassettes as BamHI fragments from pGEN2 and pGEN3, respectively (Fig. 1 and Table 2).
(vi) Construction of pJN7, pGEN84, and pGEN121.
Construction of oriE1 and ori15A expression plasmids containing a plasmid maintenance system, composed of both a postsegregational killing system and at least one partition function, was first attempted by using the par function from pSC101. A 377-bp BamHI-BglII fragment was synthesized with primers 18 and 19 with pSC101 template DNA; this fragment was inserted into pJN6 cleaved with BglII to create pJN7. As in the constructions above, origin cassettes from pGEN2 and pGEN3 were then excised as BamHI fragments and inserted into pJN7 digested with BglII and BamHI to create pGEN84 and pGEN121.
(vii) Construction of pJN8, pGEN183, pGEN193, pGEN211, and pGEN222.
The final expression plasmids were constructed by introduction of the parA active partitioning locus from pR1. As with hok-sok, initial attempts at recovering the parA locus after PCR were unsuccessful. It was necessary to use overlapping PCR to generate an aphA-2–parA cassette, in which aphA-2 and parA were divergently transcribed and separated by XbaI and XhoI sites, to enable subcloning of the parA locus. A 1,737-bp parA fragment was synthesized by using primers 20 and 21 with pR1 template; a 1,076-bp aphA-2 fragment was recovered in a separate PCR by using primers 22 and 23 with pIB279 as the template. The products from these two PCRs were then used in an overlapping PCR with primers 21 and 23 to yield the final 2,743-bp aphA-2–parA fragment. This fragment was inserted as a 2,703-bp EcoRI-SpeI fragment into pJN6. The parA cassette was then reexcised as an XhoI fragment and inserted again into pJN6 cleaved with XhoI, regenerating the gfpuv gene and creating pJN8.
Plasmids carrying a plasmid maintenance system, composed of the postsegregational killing hok-sok function and parA, were constructed by excision of oriE1 and ori15A BamHI-SpeI cassettes from pGEN51 and pGEN91, respectively, and insertion into pJN8 cleaved with BamHI and SpeI, regenerating gfpuv and creating pGEN183 and pGEN193, respectively. Plasmids containing the full complement of hok-sok, par, and parA maintenance functions were constructed by insertion of par-containing origin cassettes as BamHI-SpeI cassettes from pGEN84 and pGEN121 into pJN8 cleaved with BamHI and SpeI, again regenerating gfpuv to create pGEN211 and pGEN222, respectively.
Flow cytometry.
Quantitation of GFPuv and plasmid maintenance were analyzed by measuring the fluorescence of plasmid-bearing live vectors by using an Epics Elite ESP flow cytometer/cell sorter system (Coulter) with the argon laser exciting bacteria at 488 nm and emissions detected at 525 nm. Twenty-five-milliliter 1× LB cultures grown as described above were pelleted, and bacteria were resuspended in 1 ml of PBS. Cells were then diluted 1:1,000 in PBS prior to determination of viable counts and flow analysis. Forward versus side light scatter, measured with logarithmic amplifiers, was used to gate on bacteria. A minimum of 30,000 events were acquired from each sample at a collection rate of approximately 3,500 events per second. Mean fluorescence intensity for a given bacterial population was determined by using the Epics Elite Software Analysis Package. The fluorescence for plasmidless S. typhi CVD 908-htrA and E. coli DH5α strains was quantitated to establish that autofluorescence from either host strain was negligible and that fluorescence measured for plasmid-bearing strains was directly related to synthesis of GFPuv.
RESULTS
Rationale for construction of the expression plasmids.
Although balanced-lethal plasmid stabilization systems based on expression of Asd have been created to maintain plasmids within Salmonella, a potential limitation of the asd system is its reliance on synthesis of an enzyme with catalytic activity. Since complementation with only a single copy of asd is sufficient to remove auxotrophy (15), it is not clear why all copies of a multicopy expression plasmid should remain stable and maintain maximum gene dosage, especially if they encode an especially problematic antigen which inhibits growth of the bacterium. Another potential limitation of the asd system is that it does not enhance the inheritance of resident plasmids, which continue to segregate randomly with or without the presence of the asd system. Therefore, if resident expression plasmids carrying asd genes are inherently unstable for some undetermined reason, they will be lost regardless of the requirement of the bacterium for Asd. Here, we present the design and initial testing of a set of isogenic multicopy expression plasmids into which we have incorporated a noncatalytic postsegregational killing function, coupled with both active (14, 27) and passive plasmid partition functions (1, 31, 54), to provide a plasmid maintenance system designed to optimize expression of heterologous antigens within CVD 908-htrA for delivery to the human immune system.
Two series of isogenic expression plasmids were constructed for use in E. coli and Salmonella, with expected copy numbers of ∼60 copies per chromosomal equivalent (from pAT153, carrying a derivative of oriE1 [11, 38, 50]) or ∼15 copies per equivalent (derived from pACYC184 and carrying ori15A [10, 25]). Each plasmid comprises three basic cassettes encoding the origin of replication; a plasmid selection cassette encoding β-lactamase, which confers resistance to carbenicillin; and a heterologous antigen expression cassette. A representative set of expression plasmids containing an ori15A origin of replication is shown in Fig. 1. The heterologous antigen cassette of the basic expression vector pGEN91 is composed of individual cassettes encoding an inducible promoter to control transcription of the heterologous antigen cassette, which for the work presented here encodes the test antigen GFPuv. Components of a plasmid maintenance system were then systematically inserted into pGEN91 to assess any individual or synergistic influence of these functions on plasmid stability in the presence and absence of selection. A complete plasmid maintenance system will be defined here as being composed of a postsegregational killing function and both a passive and an active plasmid-partitioning function. For the expression plasmids reported here, the postsegregational killing function is represented by the hok-sok locus; this locus was inserted into pGEN91 to create pGEN111 and ensure that flanking transcription from surrounding loci, such as the antigen and selection cassettes, was divergent and would not significantly disturb the wild-type transcription levels which control the lethality of this locus. We also examined the effects of both passive and active partitioning loci on expression plasmid stability and synthesis of GFPuv. We inserted the par passive partition locus between the origin of replication and selection cassettes (Fig. 1, pGEN121). Interestingly, it was noted that the orientation of the par locus enhanced synthesis of GFPuv on solid medium when inserted in the natural orientation found within ori101 of pSC101, and this orientation was adopted for all of the expression plasmids. The active partitioning locus chosen for this work was the parA locus from the same pR1 resistance plasmid from which hok-sok was adapted; it was expected that the compatibility of these two loci within pR1 would be maintained within our expression plasmids. Again, to preserve natural transcription levels and regulation within this locus, the cassette was positioned within an area of the expression plasmids such that flanking transcription progressed away from parA (Fig. 1, pGEN193 and pGEN222).
Osmotic control of PompC.
It was intended that any promoter controlling transcription of a heterologous gene be responsive to an environmental signal of biological relevance. For the expression plasmids described here, an ompC promoter cassette (PompC) from E. coli was used, which is induced by increases in osmolarity. Construction of this cassette was based on the sequence of PompC published by Norioka et al. (37) and was engineered to control expression of a test antigen cassette containing the gfpuv allele encoding GFPuv; this PompC-gfpuv cassette was inserted into a derivative of pBR322 to create pGFPompC. During the visual screening of E. coli DH5α(pGFPompC) colonies subilluminated with UV light, one very brightly fluorescing colony and another representative fluorescent colony were chosen for further study, designated clones 1 and 3, respectively. Upon purification of the plasmids involved, it was determined that clone 1 contained a plasmid that no longer carried a BglII site separating PompC and gfpuv, while clone 3 carried the expected BglII site. We examined the induction of GFPuv expression when clones 1 and 3 were grown on nutrient agar in the presence or absence of NaCl and determined by visual inspection that clone 3 displayed very little fluorescence when grown in the absence of NaCl but fluoresced brightly when plated on medium containing 300 mM NaCl (data not shown). Clone 1, however, had a higher background level of fluorescence when uninduced, but fluoresced intensely when induced with 300 mM NaCl. To rule out mutations within the gfpuv gene which might affect fluorescence, we replaced PompC from clone 1 with PompC from clone 3 and confirmed the expected decrease in fluorescence as judged by subillumination (data not shown). We therefore concluded that differences in observed fluorescence were controlled by two genetically distinct versions of the PompC promoter, which we designate as PompC1 (higher transcription levels with less osmotic control) and PompC3 (moderate transcription levels but more responsive to osmolarity); we have designated the plasmids containing these expression cassettes as pGFPompC1 and pGFPompC3, respectively.
To quantify the differences between induced and uninduced expression of gfpuv controlled by PompC1 and PompC3, GFPuv synthesis was monitored within both E. coli DH5α and S. typhi CVD 908-htrA by flow cytometry. This powerful technique has the unique advantage of allowing rapid measurement of GFPuv expression within large numbers of individual bacteria, as well as accurately determining the intensity of fluorescence due to GFPuv synthesis within each bacterium analyzed. As summarized in Table 3, the basal level of expression for the PompC1-gfpuv cassette is 2.5-fold higher than that for the PompC3-gfpuv cassette, when expressed in DH5α, and 2.1-fold higher when expressed within CVD 908-htrA; however, the basal level of fluorescence detected for synthesis of GFPuv never exceeded a mean fluorescent intensity of 5.4, regardless of host background. If we define the induction ratio as the ratio of mean fluorescent intensity measured after induction divided by the basal level of mean fluorescent intensity, it was observed that when induced with 150 mM NaCl, PompC1 and PompC3 displayed within DH5α induction ratios of 1.7 and 2.4, respectively. Surprisingly, the induction ratio for PompC1 when measured in CVD 908-htrA was 4.4, and PompC1 produced a maximum mean fluorescence intensity of 23.4 for these experiments. Although the induction ratio for PompC3 within CVD 908-htrA was 6.7, the mean fluorescence intensity of 17.1 was lower than that measured for PompC1.
TABLE 3.
Comparison of induction of PompC1 and PompC3, controlling expression of GFPuv, within the host strains E. coli DH5α and CVD 908-htrA
| Strain | Induction result with:
|
Induction ratioc | |||
|---|---|---|---|---|---|
| Low
osmolarity
|
150 mM NaCl
|
||||
| OD600a | Mean fluorescence intensityb | OD600 | Mean fluorescence intensityb | ||
| DH5α | 0.61 | 0.3 | 0.95 | 0.3 | NAd |
| DH5α(pGFPompC1) | 0.56 | 4.5 | 0.72 | 7.7 | 1.7 |
| DH5α(pGFPompC3) | 0.58 | 1.8 | 0.73 | 4.2 | 2.4 |
| CVD 908-htrA | 0.58 | 0.3 | 0.65 | 0.3 | NA |
| CVD 908-htrA(pGFPompC1) | 0.60 | 5.4 | 0.54 | 23.4 | 4.4 |
| CVD 908-htrA(pGFPompC3) | 0.54 | 2.6 | 0.53 | 17.1 | 6.7 |
OD600, optical density at 600 nm.
All standard error values are less than 0.1.
Defined as the ratio of mean fluorescent intensity measured after induction with 150 mM NaCl divided by the basal level of mean fluorescent intensity measured at low osmolarity.
NA, not applicable.
Since PompC3 was noted to possess the intended 3′-terminal BglII site, which was not detected for PompC1, we determined the nucleotide sequence for PompC1 to perhaps detect point mutations which might explain the strength of PompC1. The only differences identified were located at the 3′ terminus of the cassette. The intended sequence within this region was 5′-…catataacAGATCTtaatcatccacAGGAGGatatctgATG-3′. (From left to right, uppercase denotes the BglII site, ribosome binding site, and GFPuv start codon, respectively.) The actual sequence proved to be 5′-…catataacAGATCGATCTtaaAcatccacAGGAGGAtAtctgATG-3. (Inserted or changed bases are denoted by underlined bold uppercase letters.) These changes detected within the ompC1 promoter sequence are apparently responsible for increasing the observed strength of PompC1 by an unknown mechanism, since neither the basic ompC promoter sequence nor the optimized ribosome binding site has been spontaneously altered.
These data clearly show that when driving expression of gfpuv within the live-vector strain CVD 908-htrA, PompC1 and PompC3 are inducible with increasing osmolarity, although the basal level of transcription is still noteworthy in both cases. It appears that PompC1 is the strongest of the two osmotically responsive ompC promoters; PompC1 was therefore chosen for synthesis of the widest possible range of heterologous test antigen and the effects of such synthesis on plasmid stability.
Stability of expression plasmids in the absence of selection.
Since the broad objective of the research presented here is to develop a noncatalytic plasmid maintenance system to enhance the stability of multicopy expression plasmids encoding foreign antigens within CVD 908-htrA, we initiated experiments to monitor plasmid stability by quantitating expression of GFPuv by flow cytometry when strains were passaged in the absence of antibiotic selection. These experiments were designed to address three fundamental questions. (i) What is the effect of copy number on the stability of plasmids expressing GFPuv? (ii) What is the effect of the induction level of PompC1 on the stability of plasmids encoding a heterologous antigen such as GFPuv? (iii) How do the hok-sok, par, and parA maintenance functions affect plasmid retention, both as individual components and synergistically?
Initial flow cytometry experiments were carried out in which CVD 908-htrA carried isogenic replicons with various maintenance functions and either the oriE1 or ori15A origin of replication. (Figure 1 depicts the isogenic series of ori15A replicons.) It was quickly determined that replicons carrying the higher-copy-number oriE1 origins with maintenance functions were very unstable, even when strains were grown in the presence of antibiotic selection. Flow cytometry results indicated that even when cultured in the presence of carbenicillin, the percentage of the bacterial populations no longer expressing detectable GFPuv ranged from approximately 50% for constructs carrying either hok-sok or hok-sok plus par to 62% for constructs with hok-sok plus par plus parA. Since replicons carrying an oriE1 origin clearly did not allow for optimal synthesis of the heterologous GFPuv test antigen within the majority of a growing population of live-vector bacteria, this series of expression plasmids was not examined further.
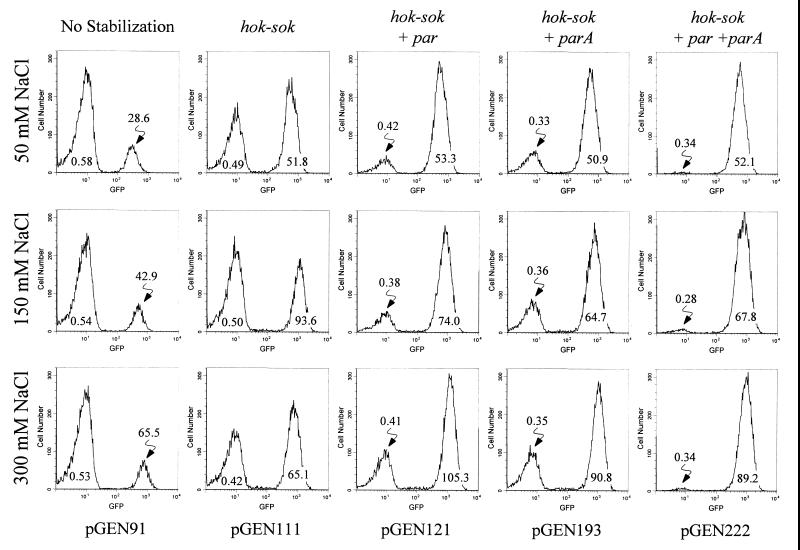
Maintenance of expression plasmids containing an ori15A origin of replication was then examined. Results for CVD 908-htrA harboring a particular expression plasmid and passaged for 24 h in the absence of selection are listed in Table 4; histograms representing these data are shown in Fig. 2. In general, as osmolarity increased and induction of PompC1 rose, the percentage of the live-vector population expressing GFPuv decreased; nevertheless, the mean level of fluorescence intensity increased as expected. For example, in the presence of 50 mM NaCl, 80.5% of a population of CVD 908-htrA(pGEN121) expressed GFPuv with a mean fluorescence intensity of 53.3; as the concentration of NaCl increased to 300 mM, only 56.7% of the population expressed GFPuv, but the mean fluorescence intensity jumped to 105.3. However, it is notable that for strains carrying pGEN222 with a complete plasmid maintenance system (i.e., hok-sok plus par plus parA), the percentage of the population expressing the heterologous antigen remained at approximately 95% regardless of induction, while the mean fluorescence intensity increased from 52.1 (50 mM NaCl) to 89.2 (300 mM NaCl). It was noted that upon passage of these strains for an additional 24 h in the absence of antibiotic selection, less than 5% of bacteria continued to express functional GFPuv. Streaks of these cultures onto solid medium, prior to flow analysis, indicated that nonfluorescing bacteria remained viable; this was confirmed when nonfluorescing bacteria were sorted and shown by plating to be sensitive to antibiotic and nonfluorescing when irradiated with UV light, indicating loss of resident plasmids.
TABLE 4.
Stability within CVD 908-htrA of ori15A replicons, containing plasmid maintenance systems of increasing complexity, grown without selection and in the presence of increasing osmolaritya
| Strain or plasmid | Stability result with NaCl concn:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 50 mM
|
150 mM
|
300 mM
|
|||||||
| OD600b | % Fluorescing bacteria | Mean fluorescence intensityc | OD600 | % Fluorescing bacteria | Mean fluorescence intensityc | OD600 | % Fluorescing bacteria | Mean fluorescence intensityc | |
| Strain | |||||||||
| CVD 908-htrA | 0.98 | 100 | 0.6 | 1.11 | 100 | 0.6 | 1.12 | 100 | 0.6 |
| Plasmids | |||||||||
| pGEN91 | 1.00 | 13.2 | 28.6 | 1.17 | 11.4 | 42.9 | 1.26 | 10.9 | 65.5 |
| pGEN111 | 1.26 | 47.4 | 51.8 | 1.17 | 28.9 | 93.6 | 1.12 | 42.4 | 65.1 |
| pGEN121 | 1.01 | 80.5 | 53.3 | 1.20 | 73.8 | 74.0 | 1.15 | 56.7 | 105.3 |
| pGEN193 | 1.11 | 71.4 | 50.9 | 1.24 | 65.2 | 64.7 | 1.22 | 53.7 | 90.8 |
| pGEN222 | 1.01 | 96.8 | 52.1 | 1.28 | 93.3 | 67.8 | 1.13 | 95.3 | 89.2 |
These data are represented by histograms in Fig. 2.
OD600, optical density at 600 nm.
All standard error values are <0.6.
FIG. 2.
Flow cytometry histograms of GFPuv fluorescence for populations of attenuated S. typhi CVD 908-htrA containing expression plasmids with the ori15A origin of replication. Histograms are arranged in rows indicating analysis of bacterial populations grown under inducing conditions with either 50, 150, or 300 mM NaCl. The histogram columns indicate CVD 908-htrA carrying the expression plasmid listed at the bottom of the column, which contains the maintenance function(s) listed at the top of the column. Results are plotted as the log of GFPuv relative fluorescence intensity versus the number of fluorescing bacteria. The mean fluorescence intensity for a given subpopulation is indicated under each curve; these data are fully quantitated in Table 4.
Taken together, these data suggest that as copy number is reduced, the apparent stability of resident plasmids and proficiency of a live vector to synthesize a heterologous antigen such as GFPuv increase. However, as the induction of PompC1 and concomitant production of the heterologous antigen increase, the percentage of a growing population remaining capable of synthesizing antigen can be dramatically reduced; as plasmid maintenance functions accumulate within a given plasmid, apparent stability and antigen synthesis are restored.
DISCUSSION
The broad objective of the research presented here is to develop a plasmid maintenance system for the stabilization of multicopy expression plasmids encoding foreign antigens in an S. typhi live-vector vaccine strain, without additional modification of the chromosome. Attempts were made to enhance the maintenance of expression plasmids at two independent levels. First, dependence upon balanced-lethal maintenance systems that involve catalytic enzymes expressed from multicopy plasmids was removed; this was accomplished through incorporation into expression plasmids of a postsegregational killing system based on the noncatalytic hok-sok plasmid addiction system from the antibiotic resistance factor pR1. At least one naturally occurring plasmid partition function was also introduced into these expression plasmids, to potentially eliminate random segregation of such plasmids, thereby enhancing their inheritance and stability.
Although these expression plasmids are ultimately intended to express immunogenic and protective antigens for delivery to the human immune system, GFPuv was selected as a test reporter antigen, because quantitation of mean fluorescence in a population of growing live vectors could be used as a measure of the stability of resident plasmids within the live vector. All expression plasmids carried an identical antigen expression cassette, with a PompC1 allele controlling transcription and with translation optimized by incorporation of a consensus ribosome binding site. Because no catalytic activity is associated with the fluorescence of GFPuv, the level of fluorescence intensity measured by flow cytometry within individual bacteria could be correlated directly with gene dosage and copy number. In addition, use of an osmotically regulated ompC promoter allowed an assessment of plasmid stability and live-vector viability as increasing osmolarity induced higher levels of GFPuv synthesis and presumably higher levels of metabolic stress on the live vector. It was surprising and encouraging that although the PompC1 allele was engineered from the chromosomal locus of E. coli, it appeared to function more efficiently in S. typhi (Table 3).
The contributions of several plasmid maintenance functions to the stability of plasmids within CVD 908-htrA, growing in the absence of antibiotic selection, were examined. No combination of maintenance functions could stabilize plasmids containing oriE1 origins of replication; in fact, constructs containing maintenance functions were difficult to propagate even in the presence of antibiotic. These observations cast doubt upon the rationale for using higher-copy-number plasmids to optimize expression of heterologous antigens within the cytoplasm of S. typhi-based live vectors, a strategy that heretofore has been followed by other groups investigating salmonellae as live vectors (12).
Incorporation of plasmid maintenance functions into plasmids carrying an ori15A origin of replication was more encouraging. When live vectors carrying such plasmids were passaged without selection for 24 h at 37°C, the effects of various combinations of maintenance functions became apparent. In the absence of maintenance functions, the ori15A replicon pGEN91 was lost from greater than 85% of the population, regardless of the level of induction of PompC1 (Table 4 and Fig. 2). With incorporation of the hok-sok postsegregational killing locus in pGEN111, the percentage of bacteria expressing GFPuv tripled under all induction conditions, confirming the observations of others that the hok-sok locus enhances the stability of ori15A replicons (18, 20, 21). However, it was still noted that regardless of induction conditions, greater than 50% of the bacterial population no longer fluoresced. Since it was confirmed that at least a portion of this nonfluorescing population was still viable and lacked drug resistance, these data confirm previous reports (22, 39, 53) that the presence of a hok-sok postsegregational killing system is insufficient by itself to ensure that plasmidless viable bacteria will not arise in a growing population.
One possible mechanism that allows for escape from the influence of hok-sok involves spontaneous point mutations arising within the lethal Hok ORF, which could conformationally inactivate Hok and thereby allow plasmid loss to occur without lethality. This point emphasizes the requirement of multiple mechanisms for enhancing the stability of resident plasmids within growing bacteria; should one maintenance function become inactivated, the probability of other independent functions simultaneously becoming inactivated becomes vanishingly small. Indeed, such redundancy in maintenance functions is widespread within naturally occurring low-copy-number plasmids (36). For example, the E. coli sex factor F contains one active partitioning function (sop) and two killing systems (ccd and flm) (23, 29, 35, 51). Similarly, the drug resistance plasmid pR1 contains the active partitioning function parA, as well as the postsegregational killing system hok-sok; in addition, it carries yet another recently defined kis-kid killing system (7, 8, 41). We have demonstrated in the work reported here that insertion into multicopy ori15A replicons of a more complete maintenance system, composed of both a postsegregational system and two partition functions, dramatically improves the stability of these expression plasmids in the absence of selection, regardless of induction conditions for heterologous antigen expression. However, after passage without selection for 48 h, plasmids were eventually lost from the bacterial population, possibly due to escape from the lethality of Hok. This problem has recently been addressed by Pecota et al. (39), who reported that incorporation of dual killing systems significantly improved plasmid stability compared to the use of hok-sok alone; no partition functions were present in these plasmids. Perhaps inclusion of the kis-kid killing system, to more fully represent the complement of pR1 stability functions, may be required for optimal stability of higher-copy expression plasmids within S. typhi live vectors.
The efficiency of eliciting an immune response directed against a heterologous antigen will depend in part upon the ability of the live vector to present such antigens to the immune system. The ability of a live vector to present antigens will in turn depend upon the stability of multicopy expression plasmids that encode the heterologous antigens. However, we hypothesize that a significant metabolic burden is placed upon CVD 908-htrA carrying a multicopy expression plasmid; as copy number and/or level of gene expression increases, metabolic burden increases. Studies with E. coli have clearly established that plasmid-bearing bacteria grow more slowly than plasmidless bacteria (6, 30, 39, 46, 53). It has also been demonstrated that as copy number increases, the growth rate of such strains decreases; similarly, as induction of heterologous genes increases, growth rate decreases further (39, 53). Clearly, spontaneous plasmid loss would remove any metabolic burden and allow plasmidless bacteria to quickly outgrow the population of plasmid-bearing bacteria. Such a shift in antigen expression within a population of live-vector bacteria would be expected to reduce the efficiency of stimulating any immune response specific to the foreign antigen. Such reasoning suggests that the goal for an effective multivalent S. typhi-based live vector vaccine is to optimize viability by using stabilized lower-copy-number expression plasmids, capable of expressing high levels of heterologous antigen in response to an environmental signal likely to be encountered in vivo after the vaccine organisms have reached an appropriate ecological niche. We are currently testing this strategy by using the murine intranasal model to examine the immunogenicity of protein fusions involving fragment C of tetanus toxin fused at the carboxyl terminus to antigens from the malaria agent Plasmodium falciparum, expressed within CVD 908-htrA by using expression plasmids derived from pGEN222. It is hoped that such experiments will point the way toward development of single-dose, oral S. typhi-based live-vector vaccines capable of inducing protective immune responses against multiple unrelated human pathogens.
ACKNOWLEDGMENTS
This research was supported by grants 5 RO1 AI29471, RO1 AI40297, and Research contract NO1 AI45251 (M. M. Levine, principal investigator).
Footnotes
This work is dedicated to the memory of James F. Galen, Jr.
REFERENCES
- 1.Austin S, Friedman S, Ludtke D. Partition functions of unit-copy plasmids can stabilize the maintenance of plasmid pBR322 at low copy number. J Bacteriol. 1986;168:1010–1013. doi: 10.1128/jb.168.2.1010-1013.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balbas P, Soberon X, Merino E, Zurita M, Lomeli H, Valle F, Flores N, Bolivar F. Plasmid vector pBR322 and its special-purpose derivatives—a review. Gene. 1986;50:3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- 3.Barry E M, Gomez-Duarte O, Chatfield S, Rappuoli R, Pizza M, Losonsky G, Galen J, Levine M M. Expression and immunogenicity of pertussis toxin S1 subunit-tetanus toxin fragment C fusions in Salmonella typhivaccine strain CVD 908. Infect Immun. 1996;64:4172–4181. doi: 10.1128/iai.64.10.4172-4181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaucage S L, Miller C A, Cohen S N. Gyrase-dependent stabilization of pSC101 plasmid inheritance by transcriptionally active promoters. EMBO J. 1991;10:2583–2588. doi: 10.1002/j.1460-2075.1991.tb07799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomfield I C, Vaughn V, Rest R F, Eisenstein B I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacBgene and a temperature-sensitive pSC101 replicon. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 6.Boe L, Gerdes K, Molin S. Effects of genes exerting growth inhibition and plasmid stability on plasmid maintenance. J Bacteriol. 1987;169:4646–4650. doi: 10.1128/jb.169.10.4646-4650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bravo A, de Torrontegui G, Diaz R. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol Gen Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- 8.Bravo A, Ortega S, de Torrontegui G, Diaz R. Killing of Escherichia coli cells modulated by components of the stability system parDof plasmid R1. Mol Gen Genet. 1988;215:146–151. doi: 10.1007/BF00331316. [DOI] [PubMed] [Google Scholar]
- 9.Brosius J. Superpolylinkers in cloning and expression vectors. DNA. 1989;8:759–777. doi: 10.1089/dna.1989.8.759. [DOI] [PubMed] [Google Scholar]
- 10.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covarrubias L, Cervantes L, Covarrubias A, Soberon X, Vichido I, Blanco A, Kupersztoch-Portnoy Y M, Bolivar F. Construction and characterization of new cloning vehicles. V. Mobilization and coding properties of pBR322 and several deletion derivatives including pBR327 and pBR328. Gene. 1981;13:25–35. doi: 10.1016/0378-1119(81)90040-8. [DOI] [PubMed] [Google Scholar]
- 12.Covone M G, Brocchi M, Palla E, da Silveira W D, Rappuoli R, Galeotti C L. Levels of expression and immunogenicity of attenuated Salmonella enterica serovar Typhimurium strains expressing Escherichia colimutant heat-labile enterotoxin. Infect Immun. 1998;66:224–231. doi: 10.1128/iai.66.1.224-231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crameri A, Whitehorn E A, Tate E, Stemmer W P. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 14.Dam M, Gerdes K. Partitioning of plasmid R1: ten direct repeats flanking the parA promoter constitute a centromere-like partition site parC, that expresses incompatibility. J Mol Biol. 1994;236:1289–1298. doi: 10.1016/0022-2836(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 15.Galen J E, Nakayama K, Curtiss R., III Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonellavaccine strains. Gene. 1990;94:29–35. doi: 10.1016/0378-1119(90)90464-3. [DOI] [PubMed] [Google Scholar]
- 16.Galen J E, Gomez-Duarte O G, Losonsky G, Halpern J L, Lauderbaugh C S, Kaintuck S, Reymann M K, Levine M M. A murine model of intranasal immunization to assess the immunogenicity of attenuated Salmonella typhilive vector vaccines in stimulating serum antibody responses to expressed foreign antigens. Vaccine. 1997;15:700–708. doi: 10.1016/s0264-410x(96)00227-7. [DOI] [PubMed] [Google Scholar]
- 17.Galen J E, Vimr E R, Lawrisuk L, Kaper J B. Cloning, sequencing, and expression of the gene, nanH, for Vibrio cholerae neuraminidase. In: Sack R B, Zinnake Y, editors. Advances in research on cholera and related diarrheas. Tokyo, Japan: KTK Scientific Publishers; 1990. pp. 143–153. [Google Scholar]
- 18.Gerdes K. The parB (hok/sok) locus of plasmid R1: a general purpose plasmid stabilization system. Bio/Technology. 1988;6:1402–1405. [Google Scholar]
- 19.Gerdes K, Gultyaev A P, Franch T, Pedersen K, Mikkelsen N D. Antisense RNA-regulated programmed cell death. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Gerdes K, Jacobsen J S, Franch T. Plasmid stabilization by post-segregational killing. Genet Eng. 1997;19:49–61. doi: 10.1007/978-1-4615-5925-2_3. [DOI] [PubMed] [Google Scholar]
- 21.Gerdes K, Løve Larsen J E, Molin S. Stable inheritance of plasmid R1 requires two different loci. J Bacteriol. 1985;161:292–298. doi: 10.1128/jb.161.1.292-298.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerdes K, Rasmussen P B, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golub E I, Panzer H A. The F factor of Escherichia coli carries a locus of stable plasmid inheritance stm, similar to the parBlocus of plasmid R1. Mol Gen Genet. 1988;214:353–357. doi: 10.1007/BF00337735. [DOI] [PubMed] [Google Scholar]
- 24.Gultyaev A P, Franch T, Gerdes K. Programmed cell death by hok/sok of plasmid R1: coupled nucleotide covariations reveal a phylogenetically conserved folding pathway in the hokfamily of mRNAs. J Mol Biol. 1997;273:26–37. doi: 10.1006/jmbi.1997.1295. [DOI] [PubMed] [Google Scholar]
- 25.Hiszczynska-Sawicka E, Kur J. Effect of Escherichia coliIHF mutations on plasmid p15A copy number. Plasmid. 1997;38:174–179. doi: 10.1006/plas.1997.1307. [DOI] [PubMed] [Google Scholar]
- 26.Hone D M, Harris A M, Chatfield S, Dougan G, Levine M M. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine. 1991;9:810–816. doi: 10.1016/0264-410x(91)90218-u. [DOI] [PubMed] [Google Scholar]
- 27.Jensen R B, Gerdes K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parCcomplex. J Mol Biol. 1997;269:505–513. doi: 10.1006/jmbi.1997.1061. [DOI] [PubMed] [Google Scholar]
- 28.Karem K L, Chatfield S, Kuklin N, Rouse B T. Differential induction of carrier antigen-specific immunity by Salmonella typhimuriumlive-vaccine strains after single mucosal or intravenous immunization of BALB/c mice. Infect Immun. 1995;63:4557–4563. doi: 10.1128/iai.63.12.4557-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh S M, Cram D S, Skurray R A. Nucleotide sequence and transcriptional analysis of a third function (Flm) involved in F plasmid maintenance. Gene. 1988;66:259–268. doi: 10.1016/0378-1119(88)90362-9. [DOI] [PubMed] [Google Scholar]
- 30.McDermott P J, Gowland P, Gowland P C. Adaptation of Escherichia coligrowth rates to the presence of pBR322. Lett Appl Microbiol. 1993;17:139–143. doi: 10.1111/j.1472-765x.1993.tb01443.x. [DOI] [PubMed] [Google Scholar]
- 31.Meacock P A, Cohen S N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980;20:529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- 32.Miller C A, Beaucage S L, Cohen S N. Role of DNA superhelicity in partitioning of the pSC101 plasmid. Cell. 1990;62:127–133. doi: 10.1016/0092-8674(90)90246-b. [DOI] [PubMed] [Google Scholar]
- 33.Miller C A, Tucker W T, Meacock P A, Gustafsson P, Cohen S N. Nucleotide sequence of the partition locus of Escherichia coliplasmid pSC101. Gene. 1983;24:309–315. doi: 10.1016/0378-1119(83)90091-4. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama K, Kelley S M, Curtiss R., III Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonellavaccine strain. Bio/Technology. 1988;6:693–697. [Google Scholar]
- 35.Niki H, Hiraga S. Subcellular distribution of actively partitioning F plasmid during the cell division cycle of E. coli. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 36.Nordstrom K, Austin S J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 37.Norioka S, Ramakrishnan G, Ikenaka K, Inouye M. Interaction of a transcriptional activator, OmpR, with reciprocally osmoregulated genes, ompF and ompC, of Escherichia coli. J Biol Chem. 1986;261:17113–17119. [PubMed] [Google Scholar]
- 38.Oxer M D, Bentley C M, Doyle J G, Peakman T C, Charles I G, Makoff A J. High level heterologous expression in E. coli using the anaerobically-activated nirBpromoter. Nucleic Acids Res. 1991;19:2889–2892. doi: 10.1093/nar/19.11.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pecota D C, Kim C S, Wu K, Gerdes K, Wood T K. Combining the hok/sok, parDE, and pndpostsegregational killer loci to enhance plasmid stability. Appl Environ Microbiol. 1997;63:1917–1924. doi: 10.1128/aem.63.5.1917-1924.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puente J L, Verdugo-Rodriguez A, Calva E. Expression of Salmonella typhi and Escherichia coli OmpC is influenced differently by medium osmolarity; dependence on Escherichia coliOmpR. Mol Microbiol. 1991;5:1205–1210. doi: 10.1111/j.1365-2958.1991.tb01894.x. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Echevarria M J, Gimenez-Gallego G, Sabariegos-Jareno R, Diaz-Orejas R. Kid, a small protein of the parDstability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J Mol Biol. 1995;247:568–577. doi: 10.1006/jmbi.1995.0163. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Selzer G, Som T, Itoh T, Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983;32:119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- 44.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivasan J, Tinge S A, Wright R, Herr J C, Curtiss R., III Oral immunization with attenuated Salmonellaexpressing human sperm antigen induces antibodies in serum and the reproductive tract. Biol Reprod. 1995;53:462–471. doi: 10.1095/biolreprod53.2.462. [DOI] [PubMed] [Google Scholar]
- 46.Summers D K. Timing, self-control and sense of direction are the secrets of multicopy plasmid stability. Mol Microbiol. 1998;29:1137–1145. doi: 10.1046/j.1365-2958.1998.01012.x. [DOI] [PubMed] [Google Scholar]
- 47.Tacket C O, Kelley S M, Schödel F, Losonsky G, Nataro J P, Edelman R, Levine M M, Curtiss R., III Safety and immunogenicity in humans of an attenuated Salmonella typhivaccine vector strain expressing plasmid-encoded hepatitis B antigens stabilized by the Asd-balanced lethal vector system. Infect Immun. 1997;65:3381–3385. doi: 10.1128/iai.65.8.3381-3385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tacket C O, Sztein M, Losonsky G A, Wasserman S S, Nataro J P, Edelman R, Pickard D, Dougan G, Chatfield S N, Levine M M. Safety of live oral Salmonella typhi vaccine strains with deletions in htrA and aroC aroDand immune responses in humans. Infect Immun. 1997;65:452–456. doi: 10.1128/iai.65.2.452-456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tartera C, Metcalf E S. Osmolarity and growth phase overlap in regulation of Salmonella typhiadherence to and invasion of human intestinal cells. Infect Immun. 1993;61:3084–3089. doi: 10.1128/iai.61.7.3084-3089.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Twigg A J, Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980;283:216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- 51.Van Melderen L, Bernard P, Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol Microbiol. 1994;11:1151–1157. doi: 10.1111/j.1365-2958.1994.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 52.Wahle E, Kornberg A. The partition locus of plasmid pSC101 is a specific binding site for DNA gyrase. EMBO J. 1988;7:1889–1895. doi: 10.1002/j.1460-2075.1988.tb03022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu K, Wood T K. Evaluation of the hok/sokkiller locus for enhanced plasmid stability. Biotechnol Bioeng. 1994;44:912–921. doi: 10.1002/bit.260440807. [DOI] [PubMed] [Google Scholar]
- 54.Zurita M, Bolivar F, Soberon X. Construction and characterization of new cloning vehicles. VII. Construction of plasmid pBR327par, a completely sequenced, stable derivative of pBR327 containing the parlocus of pSC101. Gene. 1984;28:119–122. doi: 10.1016/0378-1119(84)90094-5. [DOI] [PubMed] [Google Scholar]