Abstract
The gut microbiome lies at the intersection between the environment and the host, with the ability to modify host responses to disease-relevant exposures and stimuli. This is evident in how enteric microbes interact with the immune system, e.g., supporting immune maturation in early life, affecting drug efficacy via modulation of immune responses, or influencing development of immune cell populations and their mediators. Many factors modulate gut ecosystem dynamics during daily life and we are just beginning to realise the therapeutic and prophylactic potential of microbiome-based interventions. These approaches vary in application, goal, and mechanisms of action. Some modify the entire community, such as nutritional approaches or faecal microbiota transplantation, while others, such as phage therapy, probiotics, and prebiotics, target specific taxa or strains. In this review, we assessed the experimental evidence for microbiome-based interventions, with a particular focus on their clinical relevance, ecological effects, and modulation of the immune system.
Introduction
To fully appreciate the stakes and challenges of microbiome-based interventions, it is important to detail a few important features of gut microbiomes, which is done in this introductory section.
The gut microbiome is a highly complex ecosystem
The human body is colonised by rich communities of microorganisms at various body sites, with the gastrointestinal tract being home to a cell-rich and diverse community. The intestinal microbiome consists of various types of eukaryotic (fungi, helmiths and protozoa)1 and prokaryotic (bacteria and archaea)2 microbes, as well as viruses3. While protozoa and helminths are frequent colonisers of the gut within developing nations, their prevalence is significantly reduced in developed countries, leading to them being understudied4. Both groups have generally been viewed as pathogenic in nature within the human gut, however they have been shown to interact with the gut microbiome and influence the enteric immune system5, as well as wider allergic inflammation6. Bacteria within the gut have been extensively studied and will be the major topic of this review. Due to the complexity and highly dynamic nature of enteric microbial populations, study of their interactions with the host is made difficult.
With the emergence of large population-based studies from different continents, our understanding of the diverse landscape within the human gut continues to expand7. While the human gut may represent a total diversity of ~2000 bacterial species8 and almost 200,000 phages3, an individual’s gut is likely home to a few hundred bacterial species, although sequencing methods can only detect 100–200 within an individual sample9,10. The core human gut microbiome, which are species found in the majority of individuals, consists of less than one hundred out of the thousands of bacterial species found in the intestine11. Although these dominant species constitute 99% of the mapped reads in sequencing studies of the human microbiome11, each of these core species may be represented by different strains with functional differences, further contributing to the complexity of gut microbiomes12,13. Currently it remains unclear how strain-level diversity detected by sequencing translates into physiological differences that are of relevance for functional variations of the entire community between individuals. These issues are further confounded due to much of this variation occurring within species and genes of unknown function14,15.
The complex, yet stable gut microbial ecosystem in adults contrasts dramatically with the rapid seeding of the sterile intestinal tract of new-borns. This rapid process of colonisation makes early life an important period during which both the ecosystem trajectory and immune responses are shaped.
The perinatal period is crucial for establishment of microbe-host interactions
The perinatal period is defined as pregnancy and up to one year post birth and is a time of immense physiological programming that influences health. Although previous studies have suggested an in utero (fetal) and placental microbiome during healthy pregnancies, the detected bacterial ‘communities’ are likely the result of contamination, either during DNA extraction (through contamination of reagents and purification columns), tissue collection, and/or during the sample processing and sequencing16,17. Those microbes that have been definitely identified in these otherwise sterile sites include those associated with negative birth outcomes, such as Streptococcus alginate18. Gut microbes do play a key role in foetal development; however, this is via the maternal gut microbiome, with microbial mediators and metabolites entering the circulation and impacting particular developmental and immune pathways in utero (see sections below)19. Interestingly, the maternal gut microbiome alters during pregnancy, including reduction in diversity over the course of gestation and increasing levels of Bifidobacterium in women and mice which appears to be partly driven by progesterone20. The direct ecological interactions between the gut microbiome and the infant host starts immediately post birth, as this is when initial seeding, from a range of environments and sources, occurs.
Vertical transmission of microbes from mother to baby during vaginal child birth provides a source of Bacteroides and Bifidobacterium (from the maternal gut) that shapes the initial ecosystem and longer term profiles21. Caesarean section delivery (CSD) can interrupt this microbial transfer, and these infants are often characterised by a delay in the acquisition of these commensals, and increased levels of hospital acquired bacteria that may also be multi-drug resistant22. As gut bacteria are dependent on the host diet, a diet solely comprising maternal milk drives a Bifidobacterium dominant profile (between 50–90% of the total community). This is due to the enzymatic machinery encoded by specific strains of Bifidobacterium that are able to break down complex human milk sugars (i.e., human milk oligosaccharides [HMOs]), that are not actively metabolised by the infant23. In contrast to the gut microbiome of breast-fed infants, formula-fed infants are characterised by higher microbial diversity and elevated levels of potentially pathogenic organisms, such as Escherichia coli and Clostridioides difficile (formerly Clostridium), and reduced levels of beneficial Bifidobacterium24.
The introduction of solid food, paralleled with reduced intake of human milk, and then cessation of breast-feeding during weaning results in another highly dynamic phase of ecosystem maturation and structuring. The ever-expanding nutritional content and oral intake of environmental matter drives the introduction, transient and then longer-term colonisation of horizontally acquired microbes. Genera such as Veillonella, Roseburia, Bacteroides, and members of the Lachnospiraceae family begin to establish from month 6 onwards, and although Bifidobacterium abundance reduces, there is replacement of strains able to utilise HMOs, with those that are able to use complex plant carbohydrates and starches25, leading to the replacement of infant-associated taxa (e.g. Bifidobacterium longum subsp. infantis) with adult-associated species (such as Bifidobacterium adolescentis).
Throughout this perinatal period, the infant gut microbiome is highly dynamic, and therefore more susceptible to microbiome perturbations, which is linked to a plethora of acute and chronic (including immune-mediated) conditions. This is why understanding the ecological principles that govern microbiome assembly during this developmental window is important and a prime-time period for beneficially manipulating the gut microbiome to promote appropriate immune development.
Equilibrium and imbalance within the gut
The diversity of microbial taxa within an individual’s gut microbiome leads to a large range of microbial communities between individuals. It has been suggested that these communities can be grouped into three categories, originally termed ‘enterotypes’, based on the dominance of species within the genera Ruminococcus, Bacteroides, or Prevotella26,27. Since the proposition of enterotypes in 2011, there has been great debate as to if they constitute real ‘clusters’ of the microbial communities or are driven by artificial clustering. While this matter has been reviewed in greater depth elsewhere28, enterotypes do not appear to be stable as an individual’s microbiome can swap enterotypes during the course of a year29.
While the exact taxa of high importance within the gut are still debated, it is generally agreed that a diverse gut microbiome is beneficial. However, this is a simplistic approach to the microbiome as it is not the diversity of the microbiome per se which is beneficial, but the functions those microbes contribute. In part the association between taxonomic diversity and healthy states is because a diverse community will cover a greater number of functional niches. Furthermore, the interactions between commensal species provide additional benefits that no single species provides on its own.
Despite the apparent simplicity of the gut microbiome that establishes postnatally, important cross-feeding interactions already establish and contribute to the health of the host30. Bifidobacteria and lactobacilli produce the necessary enzymes to break down the complex milk oligosaccharides into simpler saccharides like lactose or glucose31. These simpler sugars are either absorbed by the host, or alternatively further used as an energy source by, for example, fast growing lactic acid bacteria. These produce lactic acid (or lactate in conjugate form), which itself can be metabolised into short-chain fatty acids (SCFA) such as butyrate and propionate32. Thus, even in this early phase of dietary homogeneity a trophic food chain of cross-feeding mechanisms between gut bacteria begins to establish33.
Disruption of these metabolic interactions between different gut bacteria can result in disease. For example, the production of hydrogen gas by specific types of lactate-utilising bacteria has been postulated to contribute to infant colic34. As the host matures and their diet transitions to solid food, the gut microbiome matures and gains in complexity. This introduces a plethora of novel bacteria that each take on different functional roles, termed niches, within a larger metabolic network that converts primary dietary components (mostly complex fibres, see sections below) into resources for growth or end metabolites35.
The occupation of these functional niches, as well as their integration within greater metabolic interactions prevent opportunistic pathogens from being able to colonise. This is termed niche exclusion and occurs if a current occupant of the environment already utilises the same substrates as the pathogen, preventing the pathogen accessing the nutrients it requires. This has been shown to occur with Klebsiella oxytoca preventing colonisation of Klebsiella pneumoniae36, and E. coli preventing the colonisation of Salmonella enterica37. In both cases it was the shared utilisation of specific substrates by the resident microbe which prevented colonisation by the invading pathogen. These examples highlight the benefits of a diverse microbiome, as the more diverse the microbiome is, the more functional niches are occupied and the harder it is for pathogens to colonise.
Biogeography of the gut is important
The gastrointestinal tract itself is not a homogenous ecosystem but instead consists of a diverse range of environments for microbial species to reside. The small intestine is generally viewed as being sparsely colonised with a limited number of microbial species, which increases towards the distal gastrointestinal tract38,39. The limited number of studies that have looked at small intestinal communities suggest the presence of a few dominant taxa such as Streptococcus, Escherichia, Gemella, and Veillonella spp.40,41. Distally, the colon is home to a more diverse community of microbes that also varies depending on the segment studied42.
While it is the luminal content which is generally studied, the mucosal surface is also home to a distinct community of microbes compared to that of the lumen43. A major component of the mucus layer is mucin, a glycosylated protein which can be utilised by specialised species44. While mucus in the small intestine and cecum/proximal colon is unstructured and easily detachable, the distal colon is covered in a double layer of dense mucus, limiting direct microbiome-host interaction45. This difference in proximity of the host and microbiome is immunologically relevant, with the small intestine acting as the major site of immune priming46,47. The mucus also acts as a nutrient source used by the microbiome, facilitating a symbiotic relationship between the host and its microbes which is discussed in more detail later within this review. The mucosal-associated microbiome is enriched with members of Bacteroidaceae, Lachnospiraceae48, Akkermansia muciniphila49, and Enterococcus gallinarum50. Similar to the lumen, the mucosal microbiome varies greatly across gut locations, sharing <60% of species across its length51.
These differences in composition along the gastrointestinal tract are driven by a range of factors, including the availability of substrates40, pH of the environment52, the flow of the environment53, and partial pressure of oxygen54. Simple sugars are absorbed quickly from the fast-flowing content within the small intestine, and oligosaccharides are utilised quickly by the dominant taxa present including lactobacilli55, while longer-chain dietary fibres are fermented within the slow-flowing content of the colon56. Colonic fibre fermentation is conducted by a diverse community of species, each containing a unique repertoire of carbohydrate utilisation enzymes which work in tandem to release substrates, forming trophic chains57,58. Most bacterial species within the human gut are strict anaerobes59. Small amounts of oxygen diffuse into the gut lumen from the epithelial cells, leading the mucosal microbiome to contain a greater number of aerotolerant species than the lumen60. Disruption of the homeostatic release of oxygen by the host, such as during inflammation, leads to a disturbed microbiota, driven by the growth of facultative anaerobes61. For readers interested in the other factors that differ along the biogeography of the human gut, we direct them towards reviews that have covered this subject in greater depth39.
Current methods for directly sampling the gut are highly invasive, hence most human microbiome studies are limited to the study of faeces. While the faecal microbiome can provide insights into general shifts within the microbiome, it represents only the end-point of a dynamic process occurring along the gastrointestinal tract. Comparison of its profile to that of other gut locations has shown many species which are dominant throughout the gut are not detected within the faeces by sequencing, however may still be present51. Ingestible devices are currently under development that facilitate sampling from sites along the gastrointestinal tract, although further development and testing is required before wide-spread application62.
In summary, this introduction aimed to provide key information about the diversity and development of the gut microbiome, as understanding the innate complexity of the system is crucial for the interpretation of how microbiome-based interventions can modify microbial communities for the benefit of the host. The various layers of complexity mentioned above (different microbes; strain-level diversity; many unknown ecosystem members; biogeography of the gut) complicate the understanding of interactions between enteric microbes and the host. Nonetheless, progress has been made in all of these aspects of research, facilitating the implementation of microbiome-based applications in health and disease.
Methods of microbiome-based interventions, their rationales, and effect on gut ecology
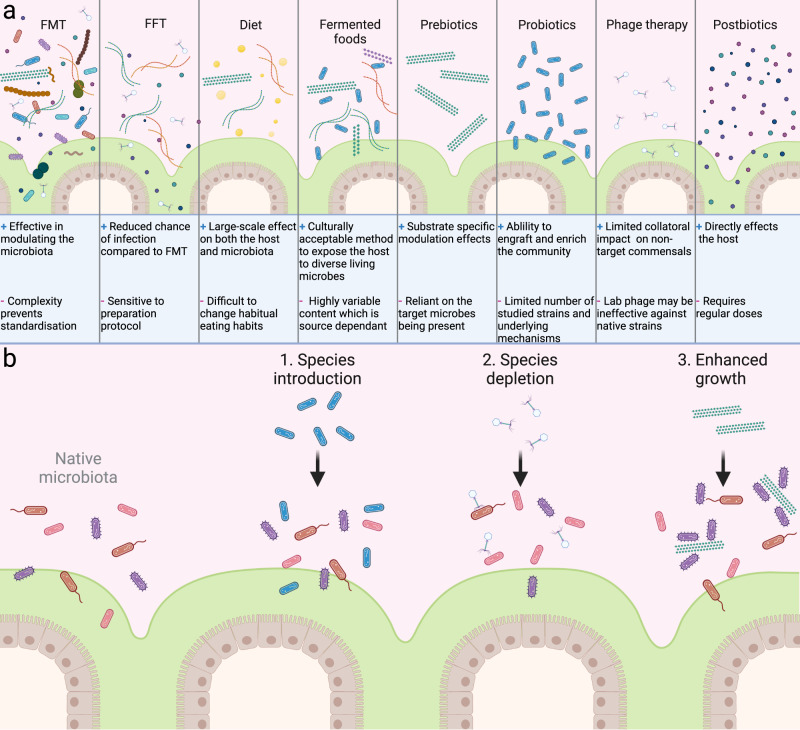
The rationales for microbiome-based interventions are diverse, ranging from the prevention of acute infections to the improvement of life-long health (Table 1). Depending on the intervention, the mechanism of action by which the microbiome-based intervention works also vary, from restructuring of the entire community, to supplementation of specific molecules that directly affect the host (Fig. 1a). Based on this, the ecological impact of each intervention can be conceptually defined as belonging to at least one of three categories of ecological impacts: species introduction, species depletion, and enhanced growth (Fig. 1b). For example, phage therapy aims to deplete a species within the ecosystem, hence it can be assigned to the ‘species depletion’ category, while prebiotics are selected based on their specific utilisation by a subset of bacteria, hence they belong to the ‘enhanced growth’ category. Grouping microbiome-based interventions into three categories is a simplistic and reductionist approach as many interventions belong to two or all three categories and any modification to the gut microbiota is likely to have secondary effects.
Table 1.
Overview of the reasons for microbiome-based interventions.
| Rationale | Brief concept | Examples |
|---|---|---|
| Prevention of enteric infections | Microbiome-based interventions can help prevent infections via multiple mechanisms. One such mechanism is niche exclusion, which involves commensal species utilising nutrients that the pathogen is specialised into using. Another mechanism is the improvement of barrier functions, which plays a particularly important role in early life. Bacterial production of antagonistic substances such as antimicrobial peptides, or the production of secondary bile acids, have also been shown to be effective in altering susceptibility to infection. |
• Reduced epithelial apoptosis during treatment with probiotic strains of Lacticaseibacillus spp. (previously Lactobacillus) has been proposed as a mechanism for the reduction of necrotising enterocolitis (NEC)278. • Colonisation with a closely related strain can lead to competitive exclusion of pathogenic strains. This has been shown for Klebsiella oxytoca which prevents colonisation of K. pneumoniae colonisation via utilisation of carbohydrates in collaboration with other commensals36. |
| Treatment of infections | While microbiome-therapies can prevent infection, they can also be utilised to treat infections, either by active depletion of the pathogenic species, or by competing for the niche the pathogen occupies. |
• Faecal microbiota transplants (FMT) have been utilised globally for treatment of Clostridioides difficile infections64 with a success rate between 80–94% depending on the method used and cohort65. • Phage therapy for the treatment of gastrointestinal infections have been highly effective in animal models279, although there have been few clinical trials and those that have been conducted are of limited success176. |
| Gut restoration | Repeated antibiotic use or poor diet over long periods of time can lead to the gut remaining in a perturbed state. Replacement of lost or underrepresented species into the ecosystem can provide the opportunity for the gut ecosystem to recover. |
• Meta-analysis of 82 studies confirmed that probiotics can reduce antibiotic-associated diarrhoea280 • FMT has proven effective for improved microbiome recovery from antibiotic treatment78 |
| Prevention of chronic diseases | Chronic diseases can develop over a long period of time, with multiple factors influencing their chance to occur. By modulating the contribution by gut microbes, we can reduce the chance of chronic diseases developing. | • Long-term (26 years) intake of dietary fibre has been associated with decreased risk of both ulcerative colitis and Crohn’s diseases281. Consistent high fibre intake is essential for this association as other studies that show no association between dietary fibre and inflammatory bowel disease (IBD) incidence utilised a single baseline questionnaire to determine dietary intake rather than regular questionnaires282. |
| Disease treatment | The gut microbiome has been observed to exacerbate certain inflammatory and metabolic conditions. Modification of the microbiome in patients suffering from such conditions may be a route to reduce symptoms and improve patient health. |
• The probiotic bacterium, Hafnia alvei HA4597, has been shown to reduce feed intake by patients, improving weight loss during dieting283. This was linked to the expression of ClpB, a protein similar to α-melanocyte stimulating hormone, which is involved in the regulation of energy balance284. • FMT has been studied for inducing remission in patients with ulcerative colitis285. Success was achieved in combination with pre-treatment using antibiotics286. However, the choice of antibiotics seems important and must be selected with care based on the profiling of antibiotic resistant strains already present within the recipient’s microbiome287. |
| Early modulation of the immune system | The early life gut microbiome coincides with mucosal and systemic immune development—and therefore represents an ideal’window of opportunity’ for targeted modulation of the gut to direct immune programming. This is particularly important in neonates/infants that have ‘perturbed’ microbial ecosystems, such as preterm babies, those born via C-section or who receive antibiotics. | • Human milk oligosaccharides (HMOs) can be deemed prebiotic181 due to their bifidogenic effect288. Bifidobacterium spp. have in turn been shown to be better able to enhance barrier function within the gut when grown on HMOs289. Specific probiotic interventions can substantially affect immune responses early in life. For instance, feeding of B. infantis EVC001 was linked to upregulation of IFNβ and silencing of intestinal Th2 and Th17 responses211. |
| Improved nutrition | Enteric microbes directly interact with both food components and the host. Some foods have been observed to alter the gut ecosystem in a way to improve the nutritional status of the host. This can be done as part of a holistic diet or via targeted intake of specific nutrients. |
• Food specifically designed to modify the gut microbiome for the purpose of enhancing host nutritional status have been developed290. The successful application of MDCF-2 to improve growth of undernourished children as well as modification of the gut microbiome confirms the usefulness of such microbiome-targeted approaches291. • Variability in glycaemic response to the same diet is linked to a range of microbial functions. Incorporating these variables in a model allowed personally tailored diets to be designed that altered glycaemic response of the individual. The personalised diets had consistent effects on the composition of the gut microbiome, suggesting this may be partly how they influence the host292. |
| Improved vaccine efficacy | The presence of a microbiome has also been shown to influence vaccine efficiency. This can be due to shared sequence similarity between microbes and the sequences targeted by vaccines, priming the immune system and enhancing the immune-stimulatory effect of these vaccines. Another method is that the microbiota can enhance vaccine response via activation of convergent pathways, leading to a greater response than the vaccine would cause alone. |
• The application of probiotic bifidobacteria has been shown to enhance the immune response to a neoepitope-based cancer vaccine293. This was suggested to be caused by the probiotic leading to an increase in taxa with high sequence similarity to the neoepitope, leading to further immune stimulation. • Activation of TLR5 by the gut microbiota, specifically flagellated bacteria, significantly increases antibody production against influenza vaccination, in mice294. |
| Modulation of drug therapy | The gut microbiome can modify the ability of an individual to respond to medication. This can occur via direct interaction between the microbes and the drugs, e.g., storage or biochemical modification, or by priming the host to be responsive to antibody therapies. | • Bioaccumulation of common drugs has been shown to be a common feature of commensal gut microbes295. This may in turn modify the drug dosage received by a patient, preventing clinical improvement296. Microbiome modulation to delete species that impact specific drugs may enhance patient response. |
For each rationale for microbiome-based intervention we provide details about the concept and published examples.
Fig. 1. Constituents and mechanisms of microbiome-based interventions.
a Common constituents of microbiome-based intervention methods are broadly illustrated. b Schematic of the three major categories by which interventions influence the gut microbiome. The simplified native microbiome members are coloured shades of red to purple, while introduced species are shades of blue and prebiotic fibres are green. FMT faecal microbiota transplant, FFT faecal filtrate transplant.
The impact of each intervention method on the ecology of the gut microbiome are detailed below, along with mechanistic insight into how they can influence the immune system. Each section, based on a single intervention, have been ordered on the complexity of their interactions with both the resident microbes and immune system. As such, FMT is the most complex, consisting of live microbes, remnant dietary components, phage and metabolites. On the other end of the spectrum, postbiotics are purified molecules derived from a microbe, which have a direct effect on either the ecosystem or the host, leading to a desired outcome.
Faecal microbiota transplant (FMT)
FMT is the administration of stool preparation obtained from a healthy donor to a diseased patient (recipient) with the goal to treat or alleviate the pathology. Such transplants have been done since the 4th century in China63, but the last decade has seen an increase in its clinical application, enhancing our understanding. Modern interest in FMT began with the successful application of FMT to treat Clostridioides difficile infections64. A recent meta-analysis of 45 studies (n = 3768 patients) on the efficacy of FMT to treat C. difficile showed that although highly effective (average success rate between 50.2–96.4%), its success depends on the frequency of transplants, as well as the method of transplantation65. The most successful method of transplantation to date is lower gastrointestinal endoscopy (96.4%), with enema being the least effective (50.2%). Repeat administration significantly improved the effectiveness of FMT, independent of the transplantation method.
FMT can lead to community-wide changes in the ecology of the recipient’s microbiome66, including the engraftment of species or strains not previously present within the recipient’s microbiome67. The effectiveness of FMT to maintain remission in patients with Crohn’s disease is reliant on the engraftment of the donor microbiome68. Long-term engraftment of the donor’s microbiome can be difficult to predict as it is dependent on multiple factors including xenobiotics, donor-recipient similarity, and taxonomic profile of the recipient69. It has also been suggested that the engraftment of bacteria is not determinative of successful treatment for inflammatory bowel disease (IBD)70, which may suggest that, in these cases, the non-living elements (e.g. phage and metabolites) are the beneficial elements (see section on faecal filtrate transplants).
Donor-recipient mapping seems to be a key factor in predicting and increasing the success of FMT. The variability in FMT success has been identified to be partly due to the microbiome of the donor; improved success rate is associated with higher richness as well as the relative abundance of specific taxa, such as A. muciniphila and Ruminococcus spp.67, although such parameters are not universal. It may also be the recipients microbiome which is key, with a depleted recipient microbiome giving the highest chance for engraftment70,71. A lower diversity of eukaryotic viruses within the recipients’ faeces has also been suggested to be a marker of FMT success, highlighting the need to screen both donors and recipients prior to transplant72. Preliminary findings suggested that the donor’s microbiome profile was the main indicator of success, leading to the concept of ‘super donors’, individuals with a highly diverse microbiome that were most effective for FMT73. This concept has been criticised due to lacking consistent experimental support74 and ignoring other factors, such as metabolite production by the donor’s microbiome75.
To circumvent the issue of relying on donors, autologous FMT (aFMT) has been explored. aFMT refers to patients receiving their own stool, biobanked before undergoing treatment, rather than receiving faeces from separate donors. For instance, when using samples obtained after completing a dietary intervention (Mediterranean or green Mediterranean) and then taken regularly over the course of 6 months afterwards, aFMT improved retention of weight loss and glycaemic control conferred by the diet76. aFMT has also been shown to be effective in enhancing recovery of a patient’s microbiome after antibiotic treatment77. Further experiments have confirmed that aFMT enhances the recovery of both the luminal and mucosal microbiome after antibiotic treatment78.
While the majority of FMT trials have focused on adults, its application during early life has also been explored. Children born via CSD have been identified to have a disturbed microbiome during early-life79. This altered microbiome has also been linked to reduced immune priming during the first days of life, which may result in the observed increase in occurrence of immune conditions in CSD children in later life80. To avoid this, FMT using faecal samples from the mother has been tested. During an initial trial applying FMT to babies to restore a conventional microbiome, the faeces from 29% of mothers were positive for pathogens, highlighting the need for in depth donor screening81. Colonisation of a microbiome deprived environment such as the neonatal gut with pathogens may in turn lead to increased incidence of necrotising enterocolitis (NEC) and other infections. An alternative approach to faecal transfers is the transfer of the vaginal microbiome either via a “swabbing” approach82 or oral administration83. Potentially due to this variation in approaches, or due to the vagina being a poor source of gut microbes, results have so far been inconsistent.
Ethical concerns have been raised based on the undefined nature of the material during FMT84. This has led to increasing interest in the identification of the key species, consortia of species85, or molecules that confer the health benefits86.
Faecal filtrate transplant (FFT)
FMT success is generally attributed to the dominant bacterial populations in stool, with a large focus placed on engraftment of donor strains and species87. However, the lack of standardisation and use of complex material has been raised as a concern for the wide-spread implementation of FMT. Faecal filtrates aim at removing microbial cells, but retain the viral and small molecules in the donor sample. Faecal filtrate transfer (FFT) was shown to be effective in treating C. difficile which has been proposed to be due to the transfer of bacteriophages88. FFT has also been shown to be more effective than FMT when treating NEC in piglets89. Comparison of the effectiveness of faecal sediment vs. supernatant for treatment of C. difficile infection has shown a 27% increase in effectiveness when using the supernatant, confirming that active agents other than the bacterial component are important90. However, FFT has been tested in few studies that have relied on a small number of patients. Systematic comparisons between FMT and FFT are required using optimised protocols for each approach (application route for FMT and method of filtration for FFT) before solid conclusions can be made. While FFT has been shown to significantly alter the composition of viruses within the gut91, the impact on the entire ecosystem is less understood.
Diet
There is a large overlap between the chronic diseases linked to diet and those that have been linked to the gut microbiome92. These pathologies, such as allergies, obesity (plus metabolic comorbidities), autoimmune, neurological, and oncological disorders, are complex diseases that are to some degree immune mediated with inflammation as a common determinant93. Decreased microbial diversity in the gut microbiome is observed both in chronic diseases but also population-wide in Western societies, in which the rates of these diseases are higher94. Western diets (WD) are high in processed foods, animal products, and refined grains, and low in whole plant foods such as vegetables, fruits, nuts, and legumes92. The impact of a lard-based WD on mice has been shown to be reliant on the presence of a microbiota95, as germ-free mice are resistant to obesity from this diet96. This interaction between WDs and the microbiota has been confirmed by microbiota transfer experiments into germ-free mice96. These showed that microbiota of obese mice significantly increased body-fat gained compared to transfer of a lean mouses microbiota97.
Different food components and dietary patterns shape the gut microbiome with associated immune modulating effects. Whole plant foods (fruits, vegetables, wholegrains, legumes, and nuts) are the main source of naturally occurring dietary fibres (see section on prebiotics)92. Fermentable dietary fibers are converted into metabolites such as SCFAs which are important mediators for the interaction between the gut microbiome and immune system98 and affect the balance between pro- and anti-inflammatory mechanisms. For example, the SCFA butyrate increased the generation of extrathymic Treg cells in mice, additionally, periphery de novo Treg cell generation can be stimulated by the SCFA propionate99. Fibre-rich diets also promote a mature intestinal mucus layer and barrier, decreasing pathogen infection and risk of colitis100. In contrast, fibre deficient diets are low in available nutrients for the gut microbiome and can promote microbial degradation of mucus, which contributes to the erosion of the colonic mucus barrier, increasing pathogen susceptibility100.
The refined food components of processed foods, which dominate WDs, are easily fermented in the small intestine to promote bacterial overgrowth and an undesirable microbial composition and metabolic profile that may negtively influence immune functions101 while not being available to colonic microbes102. Additionally, the high fat content of WDs may promote the growth of the pathobiont Bilophilia wadsworthia which has been associated with inflammation, intestinal barrier dysfunction, glucose dysmetabolism and hepatic steatosis in mice103. Emulsifiers present in processed foods such as carboxymethylcellulose have been shown to alter gut microbiota and enhance chronic intestinal inflammation in mice by inducing the expression of gene clusters mediating Crohn’s-disease- associated adherent-invasive E.coli104. The combined effects WDs negatively influence the composition of the gut microbiome and may weaken mucosal barriers to promote an inflammatory response.
Fruits and vegetables contain a variety of fibres with varying physicochemical properties. Human intervention studies have noted an inhibitory role of fruit and vegetables intake against the growth of pathogenic clostridia (Clostridium histolyticum/perfringens)105 among other positive effects on the microbiome106. Whole grains contain a diverse array of hemicellulose fibres, such as xylans and β-glucans, in addition to cellulose, resistant starches, and oligosaccharides92. Increasing consumption of wholegrains (whole-grain-barely and/or brown rice) to 60 g/day has been shown to increase microbial diversity and reduce both, plasma interleukin-6 (IL-6) ratio and peak postprandial glucose107. The response to increased whole-grain consumption may be highly individualised. Improvements in glucose metabolism in response to consuming a high fibre barley-kernel bread (over 3-days) was associated with an increase in Prevotella27 in responders (who had a higher ratio of Prevotella/Bacteroides)27,108.
Dietary patterns such as the Mediterranean diet (MD) combine many universally accepted food-based dietary guidelines and are recommended for their health promoting properties76. The MD is characterised by a high intake of plant-based foods, moderate intakes of olive oil, fish and poultry, and low intakes of dairy products, and red meat109. This dietary pattern is inversely associated with a reduced risk of cancer and cardiovascular disease109. A meta-analysis of 16S rRNA amplicon data (1931 human faecal samples) noted that the gut microbiome associated with the MD was enriched in bacteria with potential anti- Vs. pro-inflammatory properties (e.g. Akkermansia Vs. Fusobacterium)110. Additionally, high level of adherence to a MD was associated with an increase in faecal SCFAs whilst low levels of adherence were associated with higher urinary trimethylamine oxide levels (linked to atherosclerosis and cardiovascular disease)111. Due to individual variability and low adherence to prescribed diets, altering their impact, strategies are needed to maintain the impact of diets while being tailored to increase individual adherence112.
Ketogenic diets (KDs), restrict carbohydrate intake and provide high levels of fat and adequate protein. KDs have been associated with a reduced incidence of seizures in children with therapy resistant epilepsy113 the effects of which may be mediated by the gut microbiome (via bacterial cross-feeding and increases in Akkermansia and Parabacteroides) through their modulation of hippocampal GABA/glutamate ratios114. The restriction of carbohydrate intake, has been shown to be beneficial to patients with non-alcoholic fatty liver disease (NAFLD)115. Over 14 days on this diet, community-wide shifts caused significant decrease in SCFA production, but increase in folate production which was associated with decreased liver fat. When compared to restriction of fat intake, patients undergoing carbohydrate restriction for 24 weeks lost significantly more weight116. However, compositional shifts observed on KDs may have a less positive influence on gut and overall health, noted by decreases in health promoting (and fibre fermenting) bacteria such as bifidobacteria and Eubacterium rectale and their metabolites113. Though evidence highlights short-term beneficial effects of KDs in certain population groups, its restriction of fermentable fibre and high fat content may detrimentally impact the gut microbiome and immune response in the long term.
Personalised nutrition focuses on the use of person-specific factors (genetic, phenotypic, medical, nutritional or other relevant information) to develop nutritional recommendations tailored for each individual with the goal to preserve, or increase health117. Given the pronounced inter-individual variation of the gut microbiome118,119, the microbiome could constitute a key determinant of personalised nutrition. By identifying key microbiome features that predict the response to particular food components personalised nutrition can inform the design of a diet offering favourable health outcomes120.
For example, there is emerging evidence that an individual’s microbial composition may influence their ability to lose weight when following specific diets. Stratifying individual’s according to their microbial community may help predict their responses to certain diets121. Individuals with a gut microbiome dominated by Prevotella27 may achieve optimal weight loss by adhering to a high fibre diet122,123 which is not observed among individuals with a gut microbiome dominated by Bacteroides121. The value of a personalized use of diet informed through microbiome information is also supported by the Prevotella-dependent27 effects of barley-kernel bread on glucose discussed above108. However, increasing bifidobacteria in individuals with a gut microbiome dominated by Bacteroides has been shown to improve metabolic parameters which could be used as a weight loss strategy121. Although the concept of personalised nutritional advice incorporating microbiome analysis is promising, this area of research is still in its early stages with noteworthy limitations including cost and a lack of causal data from clinical trials underpinning recommendations.
Fermented food
The International Scientific Association for Probiotics and Prebiotics (ISAPP) has defined fermented foods and beverages as “foods made through desirable microbial growth and enzymatic conversions of food components”124. Fermented foods in which live organisms are present include yoghurt, sour cream, kefir, most cheeses, sauerkraut, kimchi, miso, natto, kombucha, some beers, and non-heat-treated (raw) fermented sausages (e.g. salami). If served uncooked, fermented foods often contain a high number of live microbes and have a long history of safe consumption124.
Lactic acid bacteria (LAB) are essential in the production of fermented foods and large quantities of live LAB may be consumed via fermented foods e.g., yoghurt and cheeses. A large-scale genome-wide analysis (analysing 9445 metagenomes from human faecal samples) noted that the occurrence of LAB species was typically low and associated with age, lifestyle, and geographical location125. However, it was noted that similar LAB strains occur in both food and gut microbiomes indicating that fermented foods may be the direct source of LAB found in the microbiome. It is important to consider that although live microbes from fermented foods reach the gut microbiome alive126, they are not adapted to the gut and thus unlikely to persist and have a major effect on the overall microbial community127. Due to this, it is likely that the metabolites produced by these species, which are found in high concentrations within the fermented foods, are the active agents which confer any beneficial effect128.
Although the microbes present in fermented foods are unlikely to exert a major impact on the gut microbiome structure, evidence is emerging that they can influence the host and its immune system. In a 17-week randomised prospective study (n = 18) during which participants increased their baseline average intake of fermented foods from 0.4 ± 0.6 to 6.3 ± 2.9 servings per day, it was found that a diet high in fermented foods steadily increased gut microbiome diversity and decreased inflammatory markers in the blood129. Yoghurt consumption has been associated with a higher relative abundance of species used as yoghurt starters (Streptococcus thermophilus and Bifidobacterium animalis subsp. lactis) in participants who also showed improved metabolic health characterised by reduced visceral fat130. Yoghurt contains concentrated milk fermentation metabolites such as branched chain hydroxy acids (BCHA)131. Recent studies in mice have noted that feeding the equivalent of 2 servings per day of yoghurt prevented insulin resistance and hepatic steatosis in diet-induced obesity. This was partly driven by changes in gut microbiome composition and maintaining BCHA levels which are typically reduced in diet-induced obesity131.
Although there is limited and conflicting evidence that fermented foods contribute to the repopulation and densification of the gut microbiome, they are culturally acceptable and safe vehicles by which live microorganisms can temporarily reside and pass through the human alimentary canal, influencing the immune system. Such foods they may confer health benefits attributed to their low pH and unique preparation methods which expose individuals to microbial species not otherwise present in the food chain.
Prebiotics
A prebiotic is currently defined as a substrate that is selectively utilised by host microorganisms conferring a health benefit132. Although current definitions of prebiotics have been criticised for not allowing a clear distinction between dietary fibres that are prebiotic and those that are not133–135, non-digestible carbohydrates (NDCs) can be used to modulate both composition and function of the gut microbiome.
The administration of fibre supplements, no matter if they are considered traditional prebiotics or not, result in specific changes in the gut microbiome136,137. These changes are fibre-type specific, meaning, different fibres enrich for different bacterial types. Several studies have established dose-response relationships between the administration of NDCs and compositional shifts138,139. At higher doses (10–30 g), the magnitude of the effects of NDCs can be significant, enriching some species by several thousand percent to reach relative proportions of >20% of the gut microbiome136,138,139. Using a dose-response trial with three type-IV resistant starches, crystalline and phosphate cross-linked starch structures were shown to induce divergent and highly specific effects on microbiome composition that are linked to directed shifts in the output of either propionate or butyrate139, These findings support the concept of precise gut microbiome composition modification through discrete fibre chemical structures139. However, these responses are for the most part individualised and reversible when the host is no longer receiving the substrate104.
Ecology can be used to explain the effect of dietary fibres and prebiotics. Fibres provide substrates (resources) that select for microbes that are the most adapted to utilising these fibres under the competitive conditions present. The colon consists of a complex ecosystem of microbes in which primary degraders, secondary degraders, and cross-feeders are capable of growing on nondigestible fermentable carbohydrates (NDFCs) and utilising its by-products140. Varying microbial species are required to degrade specific fibre types and certain bacteria are highly specialised and significantly contribute to fibre degradation, often referred to as keystone species104. Intervention trials with resistant starch, a microbiome accessible NDFC136,141, noted significant increases in Ruminococcus bromii and E. rectale with supplementation. R. bromii utilisation of NDFCs can benefit other microbial species by releasing sugars and acetate and can therefore be considered a keystone species142. Interestingly, the more complex fibre structures become, the more specific the response. For example, while the effects of high molecular arabinoxylan is highly specific to the species B. longum and Prevotella copri27,143, yet arabinoxylan oligosaccharides seem to have lower specificity and promote multiple species of Bifidobacterium and Prevotella27, as well as several additional genera (e.g., Eubacterium and Roseburia144). Specificity is not only determined by the ability of bacteria to utilise the fibre but also by selective colonisation (attachment) to the substrates145.
In addition to impacting the composition of the microbiome, fibre supplements can influence the production of microbial-derived metabolites that influence immune responses (see section on diet). SCFA production is dependent on the fibre type and species present in the host microbiome104, providing a framework for directing SCFA production by utilising different fibres139 (for a recent review see Vinelli et al.146).
Despite the many potential positive effects of fibre supplementation, rodent studies have indicated that high-dose supplementation with certain soluble fibres (such as inulin and pectin) can induce cholestasis, hepatic inflammation and icteric hepatocellular carcinoma (HCC) in mice147. Singh and colleagues suggested that the production of excessive amounts of butyrate (beyond levels tolerable to the host) in combination with changes to the microbiota caused by a high fat diet, and inflammation may accelerate tumour formation. In addition, fermentable fibers have been shown to exacerbate severity of DSS-induced colitis in mice148. These findings show potential detrimental impact when using certain soluble fibers as supplements in high doses.
The capacity to identify the mechanisms by which specific fibre types regulate host microbial populations and metabolism may offer the possibility for targeted therapeutic dietary fibre interventions to regulate disease states associated with altered metabolism and inflammation104. However, well controlled human clinical trials that establish the effects of fibre supplements or prebiotics on the gut microbiome and host health parameters are scarce and effects are highly inconsistent149.
Probiotics
Probiotics are live microorganisms, and thus directly add to the diversity of the native gut ecosystem. The most recent definition of a probiotic is “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host”150. This definition is purposely broad and inclusive, but the difficulties lie in the demonstration of a health benefit. Probiotics come in a range of complexities, from single strains151 to complex microbial assemblages of multiple strains from different species152 that are either administered in foods or as supplements.
Traditionally, probiotics have consisted of single strains which may or may not have been isolated from the environment they are applied to. One of the most studied probiotic strains is Lacticaseibacillus rhamnosus GG (LGG)153. LGG has been used to treat C. difficile colitis in humans154 and has been shown to restore leptin responsiveness in mice fed a high-fat diet, suggesting it may improve weight loss155. When given to mice fed a high-fat diet, LGG was observed to decrease the proportion of Pseudomonadota (formerly Proteobacteria), without impacting overall microbiome diversity. Until recently, probiotics have been limited to a few known taxa (mainly lactobacilli and bifidobacteria), for which multiple strains have been identified and characterised.
Increased activity in cultivating previously unknown taxa from the human gut has provided access to a wider array of strains that may be used as probiotics2,156–158. Advances in sequencing technology have also facilitated greater insight into the association of these species and strains with health conditions159,160. By utilising the results from sequencing to identify which strains may be beneficial for specific conditions, we can generate probiotics which were not possible before. Probiotics selected via this format have been termed ‘next generation probiotics’ (NGPs)161. Due to these strains being selected to treat or prevent specific diseases or conditions, they are likely to fall under the broader category of ‘live biotherapeutic products’ (LBPs) (discussed in Box 1), which would impose a more stringent regulatory framework than those required to be deemed safe as a food supplement and given ‘generally considered as safe’ (GRAS) status. One of the most studied NGPs is A. muciniphila, of which one strain (Muc) has been approved, in pasturised form, as a food supplement and categorised as a novel food by the European Food Safety Authority (EFSA)162. Ingestion of either live or pasteurized A. muciniphila have been shown, both in human163 and mouse trials164, not to cause community wide changes in the microbiome while improving host metabolic parameters including insulin sensitivity and plasma cholesterol levels163. In addition to A. muciniphilia, strains of Faecalibacterium prausnitzii165 and Bacteroides fragilis166 have also been studied as potential NGPs, although human trials have not yet been reported. So far, human studies failed to show a significant effect of probiotics on the gut microbiota167. This may be in part due to many of these NGP species being strict anaerobes, meaning the higher partial pressure of oxygen within the small intestine could reduce their viability within the distal gut. Engraftment of the probiotic strain into the already present and complex ecosystem is not guaranteed. However, if autochthonous strains are used, permanent engraftment has been reported168.
Multi-strain consortia can apply a holistic approach towards probiotics rather than the traditional single strain approach. Including multiple bacterial strains into a single probiotic can have benefits that leverage different approaches. The likelihood of achieving a specific functional target, for example, butyrate production, could be improved by including a number of different strains that have different requirements or interactions with a resident microbiome. The presence of trophic chains within the gut microbiome is one such opportunity, whereby interactions can be manipulated to increase the production of a wanted end-product, such as butyrate. Lactate is produced by many lactobacilli and bifidobacteria as an end-product of carbohydrate fermentation, however, it can then be utilised by butyrate producing species. Supplementation of faecal samples with a multi-strain lactate-producing consortia was observed to cause increased butyrate concentrations after a week169. While multi-strain consortia offer a unique opportunity to utilise our knowledge of the microbiome, producing multi-strain probiotics at scale poses technological challenges (see Box 1).
Box 1 Biotechnological trials and tribulations of microbiome therapies.
Traditional probiotics, i.e. lactate producing bacteria, which have been ‘generally considered as safe’ (GRAS) by the US Food and Drug Administration (FDA) and European Food Safety Agency (EFSA), have been broadly used as food supplements. The promise of novel intestinal microorganisms without GRAS status to treat disease has prompted the FDA to define a new regulatory category called ‘live biotherapeutic products’ (LBPs) that fall into the broad definition of probiotics, but are used to treat health conditions. This new category of LBPs are regulated as a drug and are hence subject to the stringent regulations in terms of safety, clinical efficacy, and current good manufacturing practices297,298.
The production of novel single strain probiotics that are classified as LBPs can directly benefit from the experience of producing probiotics at scale, though many strict anaerobic gut bacteria are more fastidious than lactobacilli or bifidobacteria. This requires extra investment to optimise growth conditions during production—even more so for the intestinal microbes that depend on close interactions with other microorganisms for optimal growth.
The production of multi-strain consortia, however, poses additional biotechnological challenges. Optimal growth conditions will vary between strains, complicating the overall process. When produced as individual mono-cultures that are subsequently mixed in a final product, production complexity scales linearly with the number of strains.
Alternatively, multi-strain consortia can be directly produced as cocultures. While this addresses the complexity problem of scaling, determining growth conditions that apply to all strains simultaneously is far from trivial. Furthermore, because each strain will typically have different growth rates, guaranteeing reproducibility and consistency of the final product is hard. These challenges can be overcome by incorporating ecological interactions into consortium design299.
The challenge of multi-strain consortium LBPs expands beyond the production process into clinical application, where consistent activity is a requirement for regulators. The control over pharmacokinetics and pharmacodynamics, i.e. growth and exertion of the desired activity of this new modality is currently the biggest challenge that remains to be shown in humans and will be the make or break of the efforts to mimic FMT in a standardised fashion using defined bacterial consortia300.
Phage therapy
Bacteriophages (or phage) are viruses that target bacteria. Most have a narrow host range, meaning they infect closely related strains within a species or related species, limiting their ability to infect other species and reducing collateral damage to the resident microbiome170. One example is the development of phage cocktails to treat C. difficile infections, which have, in fermenter experiments, been shown to eliminate C. difficile, while also avoiding major impact to the commensal groups (bifidobacteria, enterococci, enterobacteria, and lactobacilli)171. Phage isolated against disease-associated bacteria, such as Adherent invasive Escherichia coli (AIEC), have also been applied to reduce the severity of inflammation in DSS treated mice172. Another species targeted to reduce inflammation is Klebsiella pneumoniae. A consortium of five phage that target K. pneumoniae has been developed and shown to reduce the severity of DSS induced inflammation, and an initial human study has shown this consortium is safe for human use, and viable173.
However, the interdependency between gut microbes due to cross-species interactions mediated by metabolite production/utilisation can nonetheless lead to collateral effects. Phage treatment against E. coli and Clostridium sporogenes has been observed to have knock-on effects on non-target species, reducing the abundance of B. fragilis, while significantly enhancing the abundance of Phocaeicola vulgatus174. This in turn modified the metabolome of the gut, which can affect the bioavailability of molecules to the host175. While this was observed in mice colonised with a minimal microbial consortium, the impact to a complex community is likely lesser.
The specific nature of phage is also proposed to be a limitation to the application of phage therapy to treat gastrointestinal infections as the phage maintained in culture in laboratories may no-longer be able to replicate in the native pathogenic strains176. Phage resistance has also been observed to occur within gut bacterial species, leading to a coexistence between phage and their host, rather than depletion of the latter177. Due to their narrow host range and potential coexistence, the ability of phages to modulate the gut microbiome is thought to be limited173.
The abundance of phage within the gut has also been linked to increased inflammation caused by IFN-γ activation through a TLR9-dependent pathway178. Stimulation of IFN-γ production from dendritic cells incubated with phage DNA, but not empty phage capsids, confirms that phage DNA is immunostimulatory. The immunogenicity of phage proteins is likely specific to each phage due to the diversity of phage that exist3. For example, the T4 phage contains multiple capsid proteins that stimulate an immune response in humans179 although safety testing of T4 has confirmed it causes no adverse effect after oral treatment180.
While thousands of phage have been identified to exist within the human gut, few have been isolated directly from this environment and fewer still have had their impact on the community studied3. Further research is required in this area to understand the direct and indirect impact of phage therapy on both the microbial community and the host.
Postbiotics
Postbiotics are functionally bioactive molecules produced by microbes, formally defined as a “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host”181. It is key within this definition that while intact microbial cells may be deemed a postbiotic, the preparation is not alive or viable. This prevents the chance for colonisation and means health benefits conferred by postbiotics are reliant on regular intake to maintain the presence of the bioactive molecules.
An example of an intact cellular postbiotic is the use of pasteurised A. muciniphila cells. A trial comparing pasteurised A. muciniphila cells to a living culture showed that the pasteurised cells, but not the living cells, improved insulin sensitivity and reduced plasma cholesterol levels163. The active molecule responsible for the beneficial effects of pasteurised A. muciniphila cells has been identified to be a membrane protein, Amuc_1100182.
p40 is another protein which has been identified to be the element conferring the beneficial effects of the common probiotic, LGGs, as it secretes p40 into the environment183. While this protein is discussed in greater detail in the next section, in brief it has been shown to improve barrier function184. It is possible that this protein influences the ecology of the gut, as administration of LGG significantly increased the richness and evenness of the microbiome when administered during the first week of life185.
Direct supplementation of SCFAs as postbiotics for improving mucosal development has been investigated since the 1950’s186. As important microbial metabolic molecules, it is not surprising that supplementation of SCFAs can alter the gut microbiome. Butyrate supplementation (300 mg/Kg daily) increased the relative abundance of multiple genera in mice, including Bacteroides and Rikenella187. In humans, 4 g daily butyrate supplementation for a month across 30 individuals was observed to cause a community wide shift, led by Lachnospiraceae, including Dorea formicigenerans188. Similarly, propionate supplementation (1% within drinking water) reduced the impact of high-fat diet in mice, recovering the microbiome richness to control levels and promoting the occurrence of Bacteroides and reducing that of Lactobacillus189.
Secondary bile acids (SBAs) represent another main category of microbial molecules proposed for use as postbiotics, created by modification of primary bile acids released by the host into the gut lumen. The gut microbiome therefore acts as a key modulator of the bile acid pool, which directly regulates certain intestinal immune cell populations190. Ursodeoxycholic acid (UDCA) is a SBA produced from chenodeoxycholic acid (CDCA) by a two-step reaction involving 7α- and 7β-hydroxysteroid dehydrogenases (HSDHs)191. It contributes a minor fraction of the human bile pool, but is a major component of black bear bile192. Traditionally, black bear bile, also called “Yutan”, has been used to treat hepatobiliary disorders193, but UDCA has been confirmed as the bioactive molecule conferring the beneficial effects. Based on this, UDCA has been widely used as a postbiotic to treat different disorders. UDCA has also been suggested to reduce C. difficile associated inflammation via modulation of the host bile pool and via inhibition of C. difficile spore germination194,195. However, unlike antibiotics, UDCA intake caused minor changes to the gut microbiome of human males but not females, even after three years of daily intake196. This is in contrast to its impact on the bile acid pool, which after two years of daily treatment with UDCA, a study of 55 patients observed the dominance of cholic acid (CA) in the bile acid pool was replaced with UDCA197.
Tauroursodeoxycholic acid (TUDCA) is the conjugated form of UDCA, produced after re-uptake and conjugation of UDCA by the host, and has been shown to reduce inflammation in mice, potentially by preventing the downregulation of nuclear receptors observed under inflammatory conditions198. TUDCA has also been observed to affect gut microbiome structure, increasing the relative abundance of Bacillota (formerly Firmicutes) and decreasing Bacteroidota (formerly Bacteroidetes) in mice199. However, these findings are in contrast to observations of increased SBA-producing genes in the gut of patients with colorectal cancer200. The detrimental impact of SBAs is supported by studies in mice showing that DCA can induce colonic tumour formation201.
Currently only a few microbial metabolites and proteins have had their therapeutic potential studied, yet the gut microbiome produces thousands of metabolites and proteins202 with more discovered each year203. Detailed description of the molecular effector landscape of the gut microbiome will be a major endeavour for coming years. For those molecules that have been studied, standardisation of dosages will facilitate greater understanding of their impact on the ecology of the microbiome as current variation in treatment between studies prevents direct comparison of results.
Immune modulation by microbiome-based interventions
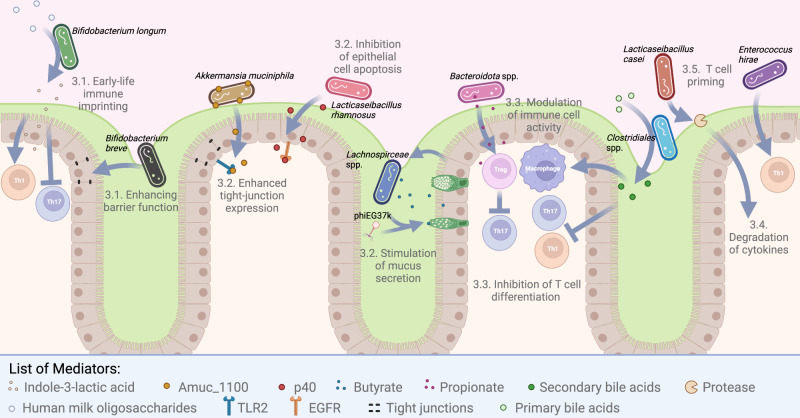
In this section, we detail some of the known effects of microbiome-based interventions on the immune system. As this is a broad field of research, we have focused this section on interactions for which mechanisms have been studied and those with therapeutic application. Each subsection focuses on a mechanism by which microbial effectors (e.g., FMT) impact immune targets (e.g., cytokine production). Many studies which identify an association between a change in the microbiome, such as one caused by a microbiome-based intervention, and a change in host phenotype fail to provide a mechanisms204. Due to the difficulty of conducting mechanistic experiments in humans, we also consider mouse and cell lines experiments to provide greater insights into the pathways and cells involved. However, mechanisms identified in mouse experiments may not transfer to humans, hence their validation is still required in humans205. An overview of the discussed mechanisms is shown in Fig. 2.
Fig. 2. Visualisation of some discussed mechanisms of how microbiome-based interventions can modulate the immune system.
Each interaction is numbered based on the sub-section they are detailed in. The key mediators and molecules involved are detailed below the scheme. Species known to conduct these interactions are given as examples in black. The mucus layer is depicted in green. As a schematic, the sizes and location of mediators and microbes have been modified to enhance visualisation.
Modulation of early-life priming of the immune system
Establishment of the gut microbiome in early life overlaps with the immune priming window. Epidemiological and more recent mechanistic studies indicate that a perturbed perinatal gut microbiome is associated with heightened risk of developing immune-mediated disorders, including atopic dermatitis, asthma, food allergies, chronic intestinal diseases, and autoimmune conditions206,207.
Immune priming begins in utero with the mother and can be linked to increased Bifidobacterium over the course of pregnancy. Supplementation of germ-free dams with Bifidobacterium breve UCC2003 is associated with transcriptional and morphological changes in the placenta and foetal compartments, including alterations of foetal liver immune pathways208. Other studies in humans have also indicated that maternal diet, such as fibre, and microbiome shape immune factors in cord blood, being associated with cord blood IgA209.
As discussed above, there is a strong evolutionary link between diet, especially human milk, the gut microbiome, and infant health. Components of human milk are known to beneficially drive immune programming and maturation (for a recent review see Singh et al.210). Several recent studies have suggested a number of mechanisms by which human milk-Bifidobacterium interactions directly influence early life immune responses. A lack of Bifidobacterium and associated HMO-metabolism genes is associated with a heightened inflammatory state in infants, including higher levels of neutrophils and basophils (and plasma TNF-α and IL-17A), while infants with higher endogenous Bifidobacterium were observed to have had a higher frequency of anti-inflammatory monocytes and regulatory T cells, and elevated levels of circulating IL-10. Direct supplementation with B. longum subsp. infantis EVC001, which encodes a range of HMO metabolism genes, was associated with modulation of intestinal T cell responses; Th2 and Th1-associated cytokines were reduced to undetectable limits, whilst interferon β (IFNβ) increased in faeces. Moreover, supplemented infants had higher levels of a key metabolite, indole-3-lactic acid (ILA), which exerted direct regulatory effects on Th2 and Th17 cells in vitro, via the induction of regulatory galectin-1, which is known to limit T cell activation (Fig. 2)211. Linking to ILA, microbial-derived components and metabolites signal via the aryl hydrocarbon receptor (AhR), a transcriptional factor that regulates the host immune system212. Endogenous ligands of AhR are contained in foods such as brassicas and are converted into AhR ligands such as indole and indole derivatives by the gut microbiome. During early life, certain Bifidobacterium species in the infant gut can metabolise human milk derived components, i.e., aromatic amino acids (tryptophan, phenylalanine and tyrosine), into their respective aromatic lactic acids (ILA, phenyllactic acid and 4-hydroxyphenyllactic acid) via aromatic lactate dehydrogenases. ILA from Bifidobacterium activates AhR in a dose-dependent manner in vitro, and modulated CD4 + T cells and monocytes responses ex vivo213. These studies have begun to shed light on how key microbial-derived metabolites modulate developing immune responses in early life, and may provide a key link as to why formula-fed infants (that have reduced levels of Bifidobacterium, and HMOs) are, in some cases, at a heightened risk of later life immune-mediated conditions like asthma214 given their potentially skewed initial immune priming.
Strengthening of the epithelium barrier during early life is critical to limit unregulated stimulation of the mucosal and systemic immune system that may lead to inflammatory associated conditions215. A particular condition of concern during early life is NEC, which impacts between 5–15% of preterm infants. Previous studies in mice have indicated that Bifidobacterium plays an oversized role in guiding epithelial barrier development. Specifically, supplementation with B. breve UCC203 during the neonatal window was associated with whole scale transcriptional responses in the small intestinal epithelial compartment, with stem cell marker genes being selectively targeted, indicating an increased regenerative potential of the epithelial layer (Fig. 2)216. This is important in the context of NEC, as numerous studies have shown that supplementation with Bifidobacterium (and Lactobacillus) probiotics significantly reduces NEC incidence in preterm infants217, including strain EVC001218. This links to previous findings in various animal models and ex vivo studies also showing how addition of specific strains of Bifidobacterium can reduce intestinal epithelial cell apoptosis and NEC pathology in vivo219.
Modulation of innate immune function
As discussed in the previous section, enhancing barrier function in early life is a key mechanism by which probiotics can help reduce the incidence of NEC, but barrier function is also important in adults. The term ‘leaky gut’ has been used to describe when reduced epithelial barrier function leads to increased occurrence of microbial products within the body220. Both probiotics and prebiotics have previously been found to improve barrier function in patients with obesity221.
Intestinal permeability is also impacted by SCFAs. In experiments using CaCo-2 cells, Bifidobacterium bifidum was shown to reduce the impact of TNF-α induced barrier permeability via production of acetate222. In mice, reduced barrier permeability cause by oral acetate supplementation (300 mM in the drinking water beginning two days prior to the experiment and continuing for the duration) reduced susceptibility to DSS colitis223. Acetate is not the only SCFA to influence barrier function; butyrate has been shown to improve barrier integrity, but increase permeability at high doses224. When administered to mice at 5% of total food weight, given ad libitum, butyrate significantly increased colonic expression of occludin, claudin-5, and zonula occludens protein-1 (ZO-1), leading to a significant decrease in intestinal permeability225. Cell line experiments have shown that butyrate also upregulates expression of IL-10 receptor α subunit, a molecule shown to be essential for barrier formation226.
The probiotic strain LGG secretes two small soluble proteins, named p75 and p40, both of which inhibit cytokine-induced apoptosis in cultured epithelial cells227. By activating the epidermal growth factor receptor (EGFR), and its downstream target, Akt, p40 has been identified to prevent apoptosis and maintain barrier integrity in mice (Fig. 2)228. This provides a mechanism by which LGG improves intestinal development and tight-junction formation during the first week of life in mice185. Production of p40 by LGG can be modulated by the release of extracellular vesicles containing heat-shock protein 90 (HSP90) from host epithelial cells229. It has been shown in mice that p40 can be provided as a postbiotic and confer the same benefits, without host factors influencing its expression184.
The protein, Amuc_1100, from A. muciniphila Muc, has also been shown to improve barrier function when applied to CaCo2 cells230 via interacting with toll-like receptor 2 (TLR2)231. Cell line experiments have also shown that activation of TLR2 can increase the expression of tight junction proteins232, which may be the mechanism by which A. muciniphila improves barrier function (Fig. 2)233. In human trials, A. muciniphila Muc can improve barrier function, as shown by reduced plasma LPS levels, however this effect was only significant when pasteurised cells and not live cells were ingested163.
Before microbes can reach the intestinal epithelium, they must pass the mucus layer which forms a physical barrier between the microbes and the host. Decreased secretion of structural components of the mucus layer, such as MUC2, has been associated with the development of inflammatory conditions within humans234. The mucin layer can be enhanced in different ways. Butyrate has been shown to increase MUC gene expression when applied directly to goblet cells (Fig. 2)235. In mice, increased release of mucin after administration of butyrate causes the mucus-associated microbiome to expand and reduces the impact of infection-induced inflammation236. Exposure of mice to bacterial products such as LPS and peptidoglycan enhances mucin production and recovery of the mucin layer after inflammatory intervention237, although this is likely to be dependent on the species present. For example, A. muciniphila is known to degrade mucin238, however it also increases the number of goblet cells within the ileum of treated mice239. It has been suggested that A. muciniphila enhances expression of the Wnt/B-catenin pathway, increasing intestinal epithelial cell proliferation, however this has only been shown in chickens240. The prebiotic berberine has also been shown to enhance MUC2 expression in mice, leading to increased mucin production and an associated increase in A. muciniphila241. While A. muciniphila is one of the best studied mucin-associated bacteria, Bifidobacterium spp. have been shown to enhance goblet cell activity in mice, reportedly via production and secretion of acetate242. Stimulation of mucin production is not limited to bacteria either; the phiEG37k phage has also been shown to stimulate MUC2 expression, reducing the severity of gastrointestinal inflammation in treated mice243.
Mononuclear phagocytes, macrophages and dendritic cells (DCs), within the gastrointestinal tract have also been observed to be modulated by microbiota-based interventions. Butyrate can stimulate differentiation of monocytes to macrophages via inhibition of histone deacetylase 3 (HDAC3) and alter activity by increasing antimicrobial peptide production via reducing activity of mTOR kinase244. This was confirmed in vivo by giving mice butyrate in their drinking water (150 mM for seven days), after which their colonic macrophages had higher antimicrobial activity than those given a saline solution. In vitro experiments have also shown that conditioned media from probiotic strains, such as LGG, can enhance the rate at which macrophages kill ingested bacteria. This is done via increasing reactive oxygen species (ROS) generation by enhancing the expression of NADPH oxidase245. Some LAB strains have been shown to modify the activity of DCs via the induction of tolerogenic properties. When bone marrow-derived dendritic cells, treated with the probiotic strain L. rhamnosus LR-32 were transferred into mice, they conferred a protective phenotype on the recipients to 2, 4, 6-trinitrobenzenesulfonic acid (TNBS)-induced colitis due to the higher abundance of tolerogenic DCs246. LAB strains found in fermented foods, particularly sauerkraut, produce D-phenyllactic acid, leading to increased serum levels, which is recognised by the HCA3 receptor on monocytes, altering their migration247.
Modulation of T cell populations
In addition to modulation of innate immunity, microbiome-based interventions can have profound effects on the adaptive immune system. For example, in patients with end-stage renal disease, daily intake of propionate (1000 mg) was shown to expand the Treg population within the peripheral blood248 and has consistently been shown to decrease the levels of C-reactive protein, a marker of inflammation, in peripheral blood by half, although its influence on other inflammation markers (IL-2, IL-17) are inconsistent249. Propionate has also been shown to modulate the activity of immune cells, including increased production of IL-10 by Tregs, which leads to inhibition of Th17 cells (Fig. 2). In the context of Multiple sclerosis, patients receiving propionate daily (1000 mg) for 14 days reported alleviation of clinical symptoms and a reduced risk of disease progression if taken continuously over a year250.
Modification of bile acids by the gut microbiome produces a wide range of SBAs, many of which impact the immune system and some of which have been applied therapeutically. The SBA lithocholic acid (LCA) is a potent inhibitor of IL-1β production in macrophages via inhibiting activation of the NLRP3 inflammasome. Dominant human gut bacteria, including Gordonibacter pamelaeae, Eggerthella lenta, and B. fragilis, were identified to modify LCA into 3-oxolithocholic acid (3-oxoLCA) and isolithocholic acid (isoLCA)251. Both of these metabolites reduced the differentiation of Th17 cells but had no impact on Treg or Th1 cells in human cell lines and mouse models. However, LCA itself has been shown to inhibit differentiation and activation of Th1 cells (Fig. 2)252. Taken together, these results suggest that bacterial modification of bile acids, particularly the production of LCA and its derivatives, have a substantial impact on T cell populations and their activity.
Some microbial metabolites have been identified to act as AhR ligands, detailed above, and modulate the T cell population of the gut253. Metabolism of tryptophan by the gut microbiome produces indole-3-acetic acid (IAA)254, leading to activation of AhR via IL-22 production, improving intestinal recovery in colitis mouse models255. IAA-induced IL-22 production is key for colonisation resistance to gastrointestinal fungal infections in mice256. While postbiotic treatment with IAA has not been studied in humans, many lactobacilli used as probiotics are IAA producers.
Many other immune cells, other than T cells which have been the focus of this section, are impacted by the gut microbiome. As such, they can be part of the mechanism by which microbiome-based interventions impact the immune system. Recently, probiotic bacterial consortia have been developed to modulate immune populations. GUT-108 is one such consortia which reduces the CD4 + T cell population within the gut, reducing the abundance of multiple inflammatory cytokines152. While effective in mice, clinical trials are required to confirm its application in humans and better understand the mechanisms by which it affects the immune system.
Modulation of cytokine levels
Cytokines are host molecules used for communication between immune cells. Altering the abundance of these molecules can change the migration and activity of specific immune cell types. Gut microbes have evolved multiple methods to modulate the ability of cells to produce and secrete these molecules. Lactocepin is a protease produced by Lacticaseibacillus casei which is able to degrade IP-10 (CXCL10), a proinflammatory molecule (Fig. 2)257. Based on this, lactocepin-producing probiotic species have been proposed to be of therapeutic use for IBD patients258, while no clinical trials studied their application, probiotic use of L. casei DN-114 001 has proven effective against gastrointestinal inflammatory conditions259.
Degradation of inhibitory cytokines, leading to increased activation of the immune system, is another method of interaction. Cgr2 is an enzyme produced by E. lenta that can convert digoxin, a cardiac medication, into an inactive molecule260. Cgr2 also degrades RORγt inhibitors increasing levels of IL-17a (CTLA-8) and activating Th17 cells within mice, although the specific inhibitor degraded is currently unknown261. This proinflammatory process can itself be inhibited via dietary arginine which appears to inactivate Cgr2262. Therefore, ingestion of arginine may be a method to modulate Cgr2 activity and reduce gastrointestinal inflammation, as studies in mice have shown263. In addition to cytokine degradation, microbes can alter their secretion. For instance, a probiotic consortium altered the ratio of inflammatory to anti-inflammatory cytokines released by Caco2 cells169 and when applied to patients with IBS, reduced symptom severity264.
Ursodeoxycholic acid (UDCA), commercially called ursodiol, is a SBA which has been used clinically for decades197 and inhibits cultured epithelial cells releasing proinflammatory chemokines, therefore reducing the severity of inflammatory responses265. However, trials have so far failed to prove their effectiveness to treat conditions such as IBD266. As the impact of bile acids is dose-dependent267, it may be that an insufficient dose was applied. Additionally, high-dosage treatment with UDCA has also proven detrimental, being linked to increased risk of colorectal neoplasia in patients with ulcerative colitis and primary sclerosing cholangitis268.
Stimulation of immune cells to enhance response to check-point inhibitors
The gut microbiome has been identified as a factor influencing host response to medication, such as check-point inhibitor (CPI) therapy269. CPIs are a form of immunotherapy for treatment of cancer which target the immune escape mechanisms used by cancerous cells. There are three main target molecules that are inhibited; CTLA-4, PD-1, and PD-L1. The interaction between the microbiome and CPIs has been shown to be complex and is not limited to a single species, instead showing cohort-specific microbiome signatures270.
Based on this, FMT has been used to modulate the microbiome of non-responders and improve CPI therapy. This was successful in 30% of patients (n = 10) treated with anti-PD-1 CPI therapy271 and further improved to 40% when using faecal material from responders272. While the exact mechanism by which FMT improves anti-PD-1 response is undefined, the microbiome-dependent nature of CTLA-4 CPI therapy has been better characterised. B. fragilis and Bacteroides thetaiotaomicron were identified to be key for T cell priming, without which CTLA-4 blockade is ineffective (Fig. 2). Specifically, the zwitterionic polysaccharides found in the capsule of these species stimulates the immune response. Responders have also been confirmed to have significantly greater abundance of these taxa within their gut microbiome273. Increasing the abundance of these species via application of microbiome-based interventions may improve response to CTLA-4 CPI, however, such methods should be targeted as other members of this genus (Bacteroides) are not beneficial to the host274. Bacteroides intestinalis has been linked to toxicity during combined CPI therapy275. This suggests the interactions between this genus of bacteria and the immune system are complex and require further study to ensure the best outcome for the patient.
Other members of the gut microbiome associated with clinical response to PD-1, and PD-L1 treatment are A. muciniphila and Enterococcus hirae. Germ-free mice gavaged with these species were observed to become responsive to PD-1 treatment. This may be in part due to stimulating the secretion of IL-12 by DCs, Th1 cell reactivity to A. muciniphila, and Tc cell reactivity to A. muciniphila and E. hirae276, however the exact mechanism remains unclear.
Conclusions
While microbiome-based interventions influence the host immune system using multiple ways, our understanding is still in its infancy and further work is needed to understand these mechanisms and utilise this information to design better intervention strategies. However, implementation of this information has already led to great advancements over a relatively short period of time and future advancements are likely to occur at a fast rate. In Box 2 we detail a few of the most exciting developments in recent and coming years. Additionally, while this review has focused on the bacterial and viral fractions of the microbiome, the fungal residents of the gut have also been identified as being important to host health and their potential use should be studied further277.
Box 2 Future development of microbiome-based interventions.
Genetic modification of commensal strains to produce additional molecules such as IL-10301, butyrate302, as well as enzymes such as bile salt hydrolase, allowing for the modification of bile acid metabolism, may enhance the strains beneficial properties as a probiotic. However, such modified strains have not yet been tested in humans161. Further analysis of factors influencing strain engraftment or sensitivity to drugs may also improve probiotic design.
Advances in bioinformatic approaches combined with cultivation efforts may enable the selection of microbial consortia usable as personalised probiotic mixtures, designed to treat the condition of interest303.
Using our knowledge of each species metabolic requirements, generation of open niches has been proposed as a method to achieve long-term engraftment. By supplementing nutrients that few species can utilise, engraftment of specific strains can be achieved304.
Modification of how probiotics are prepared and administered has also advanced in recent years. One such advancement is nanocoating of probiotics in a mixture of tannic acid and ferric ions confers some resistance to antibiotics. In mice, this allows the probiotic to colonise during antibiotic treatment, reducing adverse effects of antibiotic treatment such as weight loss and diarrhoea305.
The evolutionary trajectory of gut microbes can also be directed via modification of the hosts immune response, such as stimulation of specific IgA responses306. While this has only been applied to pathogens to reduce colonisation, a similar approach could be applied to enhance colonisation of beneficial commensals.
By designing food, not only for the host, but for the microbiome, host-microbe interactions can be utilised to enhance host health290. Microbiota-directed complementary food (MDCFs) applied this principle and have been shown to modify the microbiome and lead to improve the growth of undernourished children291.
Acknowledgements
Figures were created in BioRender.
Author contributions
T.C.A.H. and T.C. initiated the review, detailing manuscript structure and defined its content. All authors contributed to drafting text sections and provided comments on the structure and overall content. T.C.A.H. coordinated the writing process. All authors reviewed the final version and agreed with its content.
Funding
Open Access funding enabled and organized by Projekt DEAL. T.C.A.H. and T.C. received funding from the DFG (German Research Foundation): project-ID 403224013 (SFB 1382).
Competing interests
T.C.A.H. and T.C. have ongoing scientific collaborations with HiPP GmbH. T.C. is also a member of the scientific advisory board of Savanna Ingredients GmbH.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li XV, et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature. 2022;603:672–678. doi: 10.1038/s41586-022-04502-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forster SC, et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat. Biotechnol. 2019;37:186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nayfach S, et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 2021;6:960–970. doi: 10.1038/s41564-021-00928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beghini F, et al. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017;11:2848–2863. doi: 10.1038/ismej.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karo-Atar, D. et al. Helminth-induced reprogramming of the stem cell compartment inhibits type 2 immunity. J. Exp. Med. 219, e20212311 (2022). [DOI] [PMC free article] [PubMed]
- 6.Zaiss MM, et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity. 2015;43:998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groussin M, et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell. 2021;184:2053–2067.e18. doi: 10.1016/j.cell.2021.02.052. [DOI] [PubMed] [Google Scholar]
- 8.Almeida A, et al. A new genomic blueprint of the human gut microbiota. Nature. 2019;1:1. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faith, J. J. et al. The long-term stability of the human gut microbiota. Science341, 1237439 (2013). [DOI] [PMC free article] [PubMed]
- 10.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schloissnig S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2012;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podlesny D, et al. Metagenomic strain detection with SameStr: identification of a persisting core gut microbiota transferable by fecal transplantation. Microbiome. 2022;10:1–15. doi: 10.1186/s40168-022-01251-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Filippis F, et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe. 2019;25:444–453.e3. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Thomas, A. M. & Segata, N. Multiple levels of the unknown in microbiome research. BMC Biol. 17–20 (2019). [DOI] [PMC free article] [PubMed]
- 15.Hitch, T. C. A. et al. Recent advances in culture-based gut microbiome research. Int. J. Med. Microbiol. 311, 151485 (2021). [DOI] [PubMed]
- 16.Blaser MJ, et al. Lessons learned from the prenatal microbiome controversy. Microbiome. 2021;9:1–7. doi: 10.1186/s40168-020-00946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter J, Hornef MW. A philosophical perspective on the prenatal in utero microbiome debate. Microbiome. 2021;9:1–9. doi: 10.1186/s40168-020-00979-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Goffau MC, et al. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;572:329–334. doi: 10.1038/s41586-019-1451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Agüero MG, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 20.Nuriel-Ohayon M, et al. Progesterone Increases Bifidobacterium Relative Abundance during Late Pregnancy. Cell Rep. 2019;27:730–736.e3. doi: 10.1016/j.celrep.2019.03.075. [DOI] [PubMed] [Google Scholar]
- 21.Shao Y, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell CM, et al. Delivery Mode Affects Stability of Early Infant Gut Microbiota. Cell Rep. Med. 2020;1:100156. doi: 10.1016/j.xcrm.2020.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson, M. A. E. et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 10.1038/s41396-019-0553-2 (2019). [DOI] [PMC free article] [PubMed]
- 24.Stewart CJ, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laursen MF, Bahl MI, Michaelsen KF, Licht TR. First foods and gut microbes. Front. Microbiol. 2017;8:1–8. doi: 10.3389/fmicb.2017.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costea PI, et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2017;3:8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hitch, T. C. A. et al. A taxonomic note on the genus Prevotella: Description of four novel genera and emended description of the genera Hallella and Xylanibacter. Syst. Appl. Microbiol.45, 126354 (2022). [DOI] [PubMed]
- 28.Cheng M, Ning K. Stereotypes About Enterotype: the Old and New Ideas. Genomics, Proteom. Bioinforma. 2019;17:4–12. doi: 10.1016/j.gpb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knights D, et al. Rethinking enterotypes. Cell Host Microbe. 2014;16:433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunesova V, Lacroix C, Schwab C. Mucin Cross-Feeding of Infant Bifidobacteria and Eubacterium hallii. Microb. Ecol. 2018;75:228–238. doi: 10.1007/s00248-017-1037-4. [DOI] [PubMed] [Google Scholar]
- 31.Laursen MF, Bahl MI, Licht TR. Settlers of our inner surface-factors shaping the gut microbiota from birth to toddlerhood. FEMS Microbiol. Rev. 2021;45:1–14. doi: 10.1093/femsre/fuab001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourriaud C, et al. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J. Appl. Microbiol. 2005;99:201–212. doi: 10.1111/j.1365-2672.2005.02605.x. [DOI] [PubMed] [Google Scholar]
- 33.Robertson RC, Manges AR, Finlay BB, Prendergast AJ. The Human Microbiome and Child Growth – First 1000 Days and Beyond. Trends Microbiol. 2019;27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Pham VT, Lacroix C, Braegger CP, Chassard C. Lactate-utilizing community is associated with gut microbiota dysbiosis in colicky infants. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-11509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischbach MA, Sonnenburg JL. Eating for two: How metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osbelt L, et al. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe. 2021;29:1663–1679.e7. doi: 10.1016/j.chom.2021.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Eberl C, et al. E. coli enhance colonization resistance against Salmonella Typhimurium by competing for galactitol, a context-dependent limiting carbon source. Cell Host Microbe. 2021;29:1680–1692.e7. doi: 10.1016/j.chom.2021.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Stearns JC, et al. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011;1:1–9. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoetendal EG, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barlow JT, et al. Quantitative sequencing clarifies the role of disruptor taxa, oral microbiota, and strict anaerobes in the human small-intestine microbiome. Microbiome. 2021;9:1–17. doi: 10.1186/s40168-021-01162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan FJ, et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang M, et al. Mucosal-Associated Microbiota Other Than Luminal Microbiota Has a Close Relationship With Diarrhea-Predominant Irritable Bowel Syndrome. Front. Cell. Infect. Microbiol. 2020;10:1–12. doi: 10.3389/fcimb.2020.515614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeder BO. Fight them or feed them: How the intestinal mucus layer manages the gut microbiota. Gastroenterol. Rep. 2019;7:3–12. doi: 10.1093/gastro/goy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson MEV, Sjövall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kayama H, Okumura R, Takeda K. Interaction between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu. Rev. Immunol. 2020;38:23–48. doi: 10.1146/annurev-immunol-070119-115104. [DOI] [PubMed] [Google Scholar]
- 47.Liu HY, et al. Distinct B cell subsets in Peyer’s patches convey probiotic effects by Limosilactobacillus reuteri. Microbiome. 2021;9:198. doi: 10.1186/s40168-021-01128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyauchi E, et al. Analysis of colonic mucosa-associated microbiota using endoscopically collected lavage. Sci. Rep. 2022;12:1–8. doi: 10.1038/s41598-022-05936-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collado MC, Derrien M, Isolauri E, De Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, Y. et al. Within-host evolution of a gut pathobiont facilitates liver translocation. Nature607, 563–570 (2022). [DOI] [PMC free article] [PubMed]
- 51.Vaga S, et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-71939-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurer JM, et al. Gastrointestinal pH and transit time profiling in healthy volunteers using the IntelliCap system confirms ileo-colonic release of ColoPulse tablets. PLoS One. 2015;10:1–17. doi: 10.1371/journal.pone.0129076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cremer J, Arnoldini M, Hwa T. Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc. Natl Acad. Sci. USA. 2017;114:6438–6443. doi: 10.1073/pnas.1619598114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, et al. Primary Human Colonic Mucosal Barrier Crosstalk with Super Oxygen-Sensitive Faecalibacterium prausnitzii in Continuous. Cult. Med. 2021;2:74–98.e9. doi: 10.1016/j.medj.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watson D, et al. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J. Appl. Microbiol. 2013;114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 56.Clausen MR, Bonnén H, Mortensen PB. Colonic fermentation of dietary fibre to short chain fatty acids in patients with adenomatous polyps and colonic cancer. Gut. 1991;32:923–928. doi: 10.1136/gut.32.8.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubino F, et al. Divergent functional isoforms drive niche specialisation for nutrient acquisition and use in rumen microbiome. ISME J. 2017;11:932–944. doi: 10.1038/ismej.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tasse L, et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010;20:1605–1612. doi: 10.1101/gr.108332.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jalili-Firoozinezhad S, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019;3:520–531. doi: 10.1038/s41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albenberg L, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee, J.-Y., Tsolis, R. M. & Bäumler, A. J. The microbiome and gut homeostasis. Science. 377, eabp9960 (2022). [DOI] [PubMed]
- 62.Tang, Q. et al. Current Sampling Methods for Gut Microbiota: A Call for More Precise Devices. Front. Cell. Infect. Microbiol. 10, 1–10 (2020). [DOI] [PMC free article] [PubMed]
- 63.Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation. Am. J. Gastroenterol. 2012;107:1755. doi: 10.1038/ajg.2012.251. [DOI] [PubMed] [Google Scholar]
- 64.Nood Evan, et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 65.Baunwall SMD, et al. Faecal microbiota transplantation for recurrent Clostridioides difficile infection: An updated systematic review and meta-analysis. EClinicalMedicine. 2020;29–30:100642. doi: 10.1016/j.eclinm.2020.100642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okahara, K. et al. Matching between Donors and Ulcerative Colitis Patients Is Important for Long-Term Maintenance after Fecal Microbiota Transplantation. J. Clin. Med. 9, 1650 (2020). [DOI] [PMC free article] [PubMed]
- 67.Kump P, et al. The taxonomic composition of the donor intestinal microbiota is a major factor influencing the efficacy of faecal microbiota transplantation in therapy refractory ulcerative colitis. Aliment. Pharmacol. Ther. 2018;47:67–77. doi: 10.1111/apt.14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sokol H, et al. Fecal microbiota transplantation to maintain remission in Crohn’s disease: A pilot randomized controlled study. Microbiome. 2020;8:1–14. doi: 10.1186/s40168-020-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staley C, et al. Durable Long-Term Bacterial Engraftment following Encapsulated Fecal Microbiota Transplantation To Treat Clostridium difficile Infection. MBio. 2019;10:1–12. doi: 10.1128/mBio.01586-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Podlesny, D. et al. Identification of clinical and ecological determinants of strain engraftment after fecal microbiota transplantation using metagenomics. Cell Rep. Med. 100711 10.1016/j.xcrm.2022.100711 (2022). [DOI] [PMC free article] [PubMed]
- 71.Schmidt, T. S. et al. Drivers and Determinants of Strain Dynamics Following Faecal Microbiota Transplantation. bioRxiv 2021.09.30.462010 (2021). [DOI] [PMC free article] [PubMed]
- 72.Conceição-Neto N, et al. Low eukaryotic viral richness is associated with faecal microbiota transplantation success in patients with UC. Gut. 2018;67:1558–1559. doi: 10.1136/gutjnl-2017-315281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson BC, et al. Strain engraftment competition and functional augmentation in a multi-donor fecal microbiota transplantation trial for obesity. Microbiome. 2021;9:1–16. doi: 10.1186/s40168-021-01060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olesen SW, Gerardin Y. Re-Evaluating the Evidence for Faecal Microbiota Transplantation ‘Super-Donors’ in Inflammatory Bowel Disease. J. Crohn’s Colitis. 2021;15:453–461. doi: 10.1093/ecco-jcc/jjaa170. [DOI] [PubMed] [Google Scholar]
- 75.Duvallet C, et al. Framework for rational donor selection in fecal microbiota transplant clinical trials. PLoS One. 2019;14:1–18. doi: 10.1371/journal.pone.0222881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rinott E, et al. Effects of Diet-Modulated Autologous Fecal Microbiota Transplantation on Weight Regain. Gastroenterology. 2021;160:158–173.e10. doi: 10.1053/j.gastro.2020.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taur, Y. et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci. Transl. Med. 10, eaap9489 (2018). [DOI] [PMC free article] [PubMed]
- 78.Suez J, et al. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell. 2018;174:1406–1423.e16. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 79.Long G, et al. The Influence of Cesarean Section on the Composition and Development of Gut Microbiota During the First 3 Months of Life. Front. Microbiol. 2021;12:1–13. doi: 10.3389/fmicb.2021.691312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Busi SB, et al. Persistence of birth mode-dependent effects on gut microbiome composition, immune system stimulation and antimicrobial resistance during the first year of life. ISME Commun. 2021;1:1–12. doi: 10.1038/s43705-021-00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korpela K, et al. Maternal Fecal Microbiota Transplantation in Cesarean-Born Infants Rapidly Restores Normal Gut Microbial Development: A Proof-of-Concept Study. Cell. 2020;183:324–334.e5. doi: 10.1016/j.cell.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 82.Dominguez-Bello MG, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilson, B. C. et al. Oral administration of maternal vaginal microbes at birth to restore gut microbiome development in infants born by caesarean section: A pilot randomised placebo-controlled trial. EBioMedicine69, 103443 (2021). [DOI] [PMC free article] [PubMed]
- 84.Ma Y, Liu J, Rhodes C, Nie Y, Zhang F. Ethical Issues in Fecal Microbiota Transplantation in Practice. Am. J. Bioeth. 2017;17:34–45. doi: 10.1080/15265161.2017.1299240. [DOI] [PubMed] [Google Scholar]
- 85.Xiao Y, Angulo MT, Lao S, Weiss ST, Liu YY. An ecological framework to understand the efficacy of fecal microbiota transplantation. Nat. Commun. 2020;11:1–17. doi: 10.1038/s41467-020-17180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clark RL, et al. Design of synthetic human gut microbiome assembly and butyrate production. Nat. Commun. 2021;12:1–16. doi: 10.1038/s41467-021-22938-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smillie CS, et al. Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplantation. Cell Host Microbe. 2018;23:229–240.e5. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ott SJ, et al. Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients With Clostridium difficile Infection. Gastroenterology. 2017;152:799–811.e7. doi: 10.1053/j.gastro.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 89.Brunse A, et al. Fecal filtrate transplantation protects against necrotizing enterocolitis. ISME J. 2022;16:686–694. doi: 10.1038/s41396-021-01107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Varga A, et al. How to Apply FMT More Effectively, Conveniently and Flexible – A Comparison of FMT Methods. Front. Cell. Infect. Microbiol. 2021;11:1–11. doi: 10.3389/fcimb.2021.657320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zuo T, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2018;67:634–643. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Armet AM, et al. Rethinking healthy eating in light of the gut microbiome. Cell Host Microbe. 2022;30:764–785. doi: 10.1016/j.chom.2022.04.016. [DOI] [PubMed] [Google Scholar]
- 93.Furman D, et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019;25:1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:36–44. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kübeck R, et al. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol. Metab. 2016;5:1162–1174. doi: 10.1016/j.molmet.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl Acad. Sci. USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 98.Ranjbar R, Vahdati SN, Tavakoli S, Khodaie R, Behboudi H. Immunomodulatory roles of microbiota-derived short-chain fatty acids in bacterial infections. Biomed. Pharmacother. 2021;141:111817. doi: 10.1016/j.biopha.2021.111817. [DOI] [PubMed] [Google Scholar]
- 99.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Desai MS, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zinöcker MK, Lindseth IA. The western diet–microbiome-host interaction and its role in metabolic disease. Nutrients. 2018;10:1–15. doi: 10.3390/nu10030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spreadbury I. Comparison with ancestral diet suggests dense acellular carbohydrates promote an inflammatory microbiota, and may be the primary dietary cause of leptin resistance and obesity. Diabetes, Metab. Syndr. Obes. Targets Ther. 2012;5:175–189. doi: 10.2147/DMSO.S33473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Natividad JM, et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 2018;9:1–15. doi: 10.1038/s41467-018-05249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cronin P, Joyce SA, O’toole PW, O’connor EM. Dietary fibre modulates the gut microbiota. Nutrients. 2021;13:1–22. doi: 10.3390/nu13051655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klinder A, et al. Impact of increasing fruit and vegetables and flavonoid intake on the human gut microbiota. Food Funct. 2016;7:1788–1796. doi: 10.1039/C5FO01096A. [DOI] [PubMed] [Google Scholar]
- 106.Cui J, et al. Dietary Fibers from Fruits and Vegetables and Their Health Benefits via Modulation of Gut Microbiota. Compr. Rev. Food Sci. Food Saf. 2019;18:1514–1532. doi: 10.1111/1541-4337.12489. [DOI] [PubMed] [Google Scholar]
- 107.Martínez I, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kovatcheva-Datchary P, et al. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 109.Renzella J, et al. What national and subnational interventions and policies based on Mediterranean and Nordic diets are recommended or implemented in the WHO European Region, and is there evidence of effectiveness in reducing noncommunicable diseases? World Heal. Organ. Reg. Eur. 2018;1:74. [PubMed] [Google Scholar]
- 110.Illescas O, Rodríguez-Sosa M, Gariboldi M. Mediterranean diet to prevent the development of colon diseases: A meta-analysis of gut microbiota studies. Nutrients. 2021;13:1–21. doi: 10.3390/nu13072234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.De Filippis F, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 112.Gibson, A. A. & Sainsbury, A. Strategies to improve adherence to dietaryweight loss interventions in research and real-world settings. Behav. Sci.7, 44 (2017). [DOI] [PMC free article] [PubMed]
- 113.Lindefeldt M, et al. The ketogenic diet influences taxonomic and functional composition of the gut microbiota in children with severe epilepsy. npj Biofilms Microbiomes. 2019;5:5. doi: 10.1038/s41522-018-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Olson CA, et al. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell. 2018;173:1–14. doi: 10.1016/j.cell.2018.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mardinoglu A, et al. An Integrated Understanding of the Rapid Metabolic Benefits of a Carbohydrate-Restricted Diet on Hepatic Steatosis in Humans. Cell Metab. 2018;27:559–571.e5. doi: 10.1016/j.cmet.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yancy WSW, Olsen MMK, Guyton JJR, Bakst RRP, Westman EEC. A low-carbohydrate, ketogenic diet versus a low-fat diet to treat obesity and hyperlipidemiaA randomized, controlled trial. Ann. Intern. Med. 2004;140:769–777. doi: 10.7326/0003-4819-140-10-200405180-00006. [DOI] [PubMed] [Google Scholar]
- 117.Ordovas JM, Ferguson LR, Tai ES, Mathers JC. Personalised nutrition and health. BMJ. 2018;361:1–7. doi: 10.1136/bmj.k2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 119.Rothschild D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 120.Kolodziejczyk AA, Zheng D, Elinav E. Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019;17:742–753. doi: 10.1038/s41579-019-0256-8. [DOI] [PubMed] [Google Scholar]
- 121.Christensen L, Roager HM, Astrup A, Hjorth MF. Microbial enterotypes in personalized nutrition and obesity management. Am. J. Clin. Nutr. 2018;108:645–651. doi: 10.1093/ajcn/nqy175. [DOI] [PubMed] [Google Scholar]
- 122.Hjorth MF, et al. Pre-treatment microbial Prevotella-to-Bacteroides ratio, determines body fat loss success during a 6-month randomized controlled diet intervention. Int. J. Obes. 2018;42:580–583. doi: 10.1038/ijo.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hjorth MF, et al. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: results from a post-hoc analysis. Int. J. Obes. 2019;43:149–157. doi: 10.1038/s41366-018-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marco, M. L. et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat. Rev. Gastroenterol. Hepatol. 10.1038/s41575-020-00390-5 (2021). [DOI] [PMC free article] [PubMed]
- 125.Pasolli E, et al. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-16438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dal Bello F, Walter J, Hammes WP, Hertel C. Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb. Ecol. 2003;45:455–463. doi: 10.1007/s00248-003-2001-z. [DOI] [PubMed] [Google Scholar]
- 127.Walter J, Maldonado-Gómez MX, Martínez I. To engraft or not to engraft: an ecological framework for gut microbiome modulation with live microbes. Curr. Opin. Biotechnol. 2018;49:129–139. doi: 10.1016/j.copbio.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Taylor, B. C. et al. Consumption of Fermented Foods Is Associated with Systematic Differences in the Gut Microbiome and Metabolome. mSystems5, e00901–19 (2020). [DOI] [PMC free article] [PubMed]
- 129.Wastyk HC, et al. Gut-microbiota-targeted diets modulate human immune status. Cell. 2021;184:4137–4153.e14. doi: 10.1016/j.cell.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roy CILe, et al. Yoghurt consumption is associated with changes in the composition of the human gut microbiome and metabolome. BMC Microbiol. 2022;22:1–12. doi: 10.1186/s12866-021-02364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Daniel N, et al. Gut microbiota and fermentation-derived branched chain hydroxy acids mediate health benefits of yogurt consumption in obese mice. Nat. Commun. 2022;13:1–18. doi: 10.1038/s41467-022-29005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gibson GR, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 133.Katsnelson A. Prebiotics gain prominence but remain poorly defined. Proc. Natl Acad. Sci. USA. 2016;113:14168–14169. doi: 10.1073/pnas.1618366113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Verspreet J, et al. A Critical Look at Prebiotics Within the Dietary Fiber Concept. Annu. Rev. Food Sci. Technol. 2016;7:167–190. doi: 10.1146/annurev-food-081315-032749. [DOI] [PubMed] [Google Scholar]
- 135.Bindels LB, Delzenne NM, Cani PD, Walter J. Opinion: Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2015;12:303–310. doi: 10.1038/nrgastro.2015.47. [DOI] [PubMed] [Google Scholar]
- 136.Martínez, I., Kim, J., Duffy, P. R., Schlegel, V. L. & Walter, J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One5, e15046 (2010). [DOI] [PMC free article] [PubMed]
- 137.Vandeputte D, et al. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut. 2017;66:1968–1974. doi: 10.1136/gutjnl-2016-313271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Davis LMG, Martínez I, Walter J, Hutkins R. A dose dependent impact of prebiotic galactooligosaccharides on the intestinal microbiota of healthy adults. Int. J. Food Microbiol. 2010;144:285–292. doi: 10.1016/j.ijfoodmicro.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 139.Deehan EC, et al. Precision Microbiome Modulation with Discrete Dietary Fiber Structures Directs Short-Chain Fatty Acid Production. Cell Host Microbe. 2020;27:389–404.e6. doi: 10.1016/j.chom.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 140.Dobranowski, P. A. & Stintzi, A. Resistant starch, microbiome, and precision modulation. Gut Microbes13, 1926842 (2021). [DOI] [PMC free article] [PubMed]
- 141.Bendiks ZA, Knudsen KEB, Keenan MJ, Marco ML. Conserved and variable responses of the gut microbiome to resistant starch type 2. Nutr. Res. 2020;77:12–28. doi: 10.1016/j.nutres.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nguyen NK, et al. Gut microbiota modulation with long-chain corn bran arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate. Microbiome. 2020;8:1–21. doi: 10.1186/s40168-020-00887-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Benítez-Páez A, et al. A Multi-omics Approach to Unraveling the Microbiome-Mediated Effects of Arabinoxylan Oligosaccharides in Overweight Humans. mSystems. 2019;4:1–16. doi: 10.1128/mSystems.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Leitch ECMW, Walker AW, Duncan SH, Holtrop G, Flint HJ. Selective colonization of insoluble substrates by human faecal bacteria. Environ. Microbiol. 2007;9:667–679. doi: 10.1111/j.1462-2920.2006.01186.x. [DOI] [PubMed] [Google Scholar]
- 146.Vinelli V, et al. Effects of Dietary Fibers on Short-Chain Fatty Acids and Gut Microbiota Composition in Healthy Adults: A Systematic Review. Nutrients. 2022;14:2559. doi: 10.3390/nu14132559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Singh V, et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell. 2018;175:679–694.e22. doi: 10.1016/j.cell.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Miles JP, et al. Supplementation of Low- and High-fat Diets with Fermentable Fiber Exacerbates Severity of DSS-induced Acute Colitis. Inflamm. Bowel Dis. 2017;23:1133–1143. doi: 10.1097/MIB.0000000000001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Armet AM, Deehan EC, Thöne JV, Hewko SJ, Walter J. The Effect of Isolated and Synthetic Dietary Fibers on Markers of Metabolic Diseases in Human Intervention Studies: A Systematic Review. Adv. Nutr. 2020;11:420–438. doi: 10.1093/advances/nmz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hill C, et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 151.Hui, Y. et al. The effect of early probiotic exposure on the preterm infant gut microbiome development. Gut Microbes13, 1951113 (2021). [DOI] [PMC free article] [PubMed]
- 152.Lelie Dvander, et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat. Commun. 2021;12:1–17. doi: 10.1038/s41467-021-23460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rohani M, Papizadeh M, Nahrevanian H, Hosseini SN, Shojaosadati SA. Lactobacillus rhamnosus Gorbach-Goldin (GG): A Top Well-Researched Probiotic Strain. J. Med Bacteriol. 2016;5:46–59. [Google Scholar]
- 154.Biller JA, Katz AJ, Flores AF, Buie TM, Gorbach SL. Treatment of recurrent Clostridium difficile colitis with lactobacillus GG. J. Pediatr. Gastroenterol. Nutr. 1995;21:224–226. doi: 10.1097/00005176-199508000-00016. [DOI] [PubMed] [Google Scholar]
- 155.Cheng YC, Liu JR. Effect of lactobacillus rhamnosus gg on energy metabolism, leptin resistance, and gut microbiota in mice with diet-induced obesity. Nutrients. 2020;12:1–13. doi: 10.3390/nu12092557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hitch, T. C. A. et al. Automated analysis of genomic sequences facilitates high-throughput and comprehensive description of bacteria. ISME Commun. 1, 1–16 (2021). [DOI] [PMC free article] [PubMed]
- 157.Poyet M, et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 2019;25:1442–1452. doi: 10.1038/s41591-019-0559-3. [DOI] [PubMed] [Google Scholar]
- 158.Afrizal, A. et al. Anaerobic single-cell dispensing facilitates the cultivation of human gut bacteria. Environ. Microbiol. 10.1111/1462-2920.15935 (2022). [DOI] [PubMed]
- 159.Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 160.Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. In press, (2019). [DOI] [PMC free article] [PubMed]
- 161.O’Toole PW, Marchesi JR, Hill C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017;2:1–6. doi: 10.1038/nmicrobiol.2017.57. [DOI] [PubMed] [Google Scholar]
- 162.Turck, D. et al. Safety of pasteurised Akkermansia muciniphila as a novel food pursuant to Regulation (EU) 2015/2283. EFSA J. 19, 6780 (2021). [DOI] [PMC free article] [PubMed]
- 163.Depommier C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Everard A, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl Acad. Sci. USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ueda A, et al. Identification of Faecalibacterium prausnitzii strains for gut microbiome-based intervention in Alzheimer’s-type dementia. Cell Rep. Med. 2021;2:100398. doi: 10.1016/j.xcrm.2021.100398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang Y, et al. Safety evaluation of a novel strain of Bacteroides fragilis. Front. Microbiol. 2017;8:1–10. doi: 10.3389/fmicb.2017.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kristensen, N. B. et al. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: A systematic review of randomized controlled trials. Genome Med. 8, 1–11 (2016). [DOI] [PMC free article] [PubMed]
- 168.Maldonado-Gómez MX, et al. Stable Engraftment of Bifidobacterium longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 169.Moens F, et al. A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro. Int. J. Pharm. 2019;555:1–10. doi: 10.1016/j.ijpharm.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 170.Montso PK, Mlambo V, Ateba CN. Characterization of Lytic Bacteriophages Infecting Multidrug-Resistant Shiga Toxigenic Atypical Escherichia coli O177 Strains Isolated From Cattle Feces. Front. Public Heal. 2019;7:1–13. doi: 10.3389/fpubh.2019.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nale JY, Redgwell TA, Millard A, Clokie MRJ. Efficacy of an optimised bacteriophage cocktail to clear Clostridium difficile in a batch fermentation model. Antibiotics. 2018;7:1–15. doi: 10.3390/antibiotics7010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Galtier M, et al. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J. Crohn’s Colitis. 2017;11:840–847. doi: 10.1093/ecco-jcc/jjw224. [DOI] [PubMed] [Google Scholar]
- 173.Federici S, et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell. 2022;185:2879–2898.e24. doi: 10.1016/j.cell.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 174.Hsu BB, et al. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe. 2019;25:803–814.e5. doi: 10.1016/j.chom.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014;20:769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sarker SA, et al. Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine. 2016;4:124–137. doi: 10.1016/j.ebiom.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lourenço M, et al. The Spatial Heterogeneity of the Gut Limits Predation and Fosters Coexistence of Bacteria and Bacteriophages. Cell Host Microbe. 2020;28:390–401.e5. doi: 10.1016/j.chom.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 178.Gogokhia L, et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe. 2019;25:285–299.e8. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.D browska K, et al. Immunogenicity Studies of Proteins Forming the T4 Phage Head Surface. J. Virol. 2014;88:12551–12557. doi: 10.1128/JVI.02043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bruttin A, Brüssow H, Bru H. Human Volunteers Receiving Escherichia coli Phage T4 Orally: a Safety Test of Phage Therapy These include: Human Volunteers Receiving Escherichia coli Phage T4 Orally: a Safety Test of Phage Therapy. Antimicrob. Agents Chemother. 2005;49:2874–2878. doi: 10.1128/AAC.49.7.2874-2878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Salminen, S. et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 10.1126/science.32.809.20 (2021). [DOI] [PMC free article] [PubMed]
- 182.Plovier H, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 183.Bäuerl C, Pérez-Martínez G, Yan F, Polk DB, Monedero V. Functional analysis of the p40 and p75 proteins from lactobacillus casei BL23. J. Mol. Microbiol. Biotechnol. 2011;19:231–241. doi: 10.1159/000322233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shen X, et al. Supplementation of p40, a Lactobacillus rhamnosus GG-derived protein, in early life promotes epidermal growth factor receptor-dependent intestinal development and long-term health outcomes. Mucosal Immunol. 2018;11:1316–1328. doi: 10.1038/s41385-018-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yan F, et al. Neonatal colonization of mice with LGG promotes intestinal development and decreases susceptibility to colitis in adulthood. Mucosal Immunol. 2017;10:117–127. doi: 10.1038/mi.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sander EG, Warner RG, Harrison HN, Loosli JK. The Stimulatory Effect of Sodium Butyrate and Sodium Propionate on the Development of Rumen Mucosa in the Young Calf. J. Dairy Sci. 1959;42:1600–1605. doi: 10.3168/jds.S0022-0302(59)90772-6. [DOI] [Google Scholar]
- 187.Yu C, et al. Effect of exercise and butyrate supplementation on microbiota composition and lipid metabolism. J. Endocrinol. 2019;243:125–135. doi: 10.1530/JOE-19-0122. [DOI] [PubMed] [Google Scholar]
- 188.de Groot PF, et al. Oral butyrate does not affect innate immunity and islet autoimmunity in individuals with longstanding type 1 diabetes: a randomised controlled trial. Diabetologia. 2020;63:597–610. doi: 10.1007/s00125-019-05073-8. [DOI] [PubMed] [Google Scholar]
- 189.Song B, et al. Propionate alleviates high-fat diet-induced lipid dysmetabolism by modulating gut microbiota in mice. J. Appl. Microbiol. 2019;127:1546–1555. doi: 10.1111/jam.14389. [DOI] [PubMed] [Google Scholar]
- 190.Song X, et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zheng MM, Wang RF, Li CX, Xu JH. Two-step enzymatic synthesis of ursodeoxycholic acid with a new 7β-hydroxysteroid dehydrogenase from Ruminococcus torques. Process Biochem. 2015;50:598–604. doi: 10.1016/j.procbio.2014.12.026. [DOI] [Google Scholar]
- 192.Hagey LR, et al. Ursodeoxycholic acid in the Ursidae: Biliary bile acids of bears, pandas, and related carnivores. J. Lipid Res. 1993;34:1911–1917. doi: 10.1016/S0022-2275(20)35109-9. [DOI] [PubMed] [Google Scholar]
- 193.Ishizaki K, Imada T, Tsurufuji M. Hepatoprotective bile acid ‘ursodeoxycholic acid (UDCA)’: Property and difference as bile acids. Hepatol. Res. 2005;33:174–177. doi: 10.1016/j.hepres.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 194.Thanissery R, Winston JA, Theriot CM. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe. 2017;45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Winston, J. A. et al. Ursodeoxycholic acid (udca) mitigates the host inflammatory response during clostridioides difficile infection by altering gut bile acids. Infect. Immun. 88, e00045-20 (2020). [DOI] [PMC free article] [PubMed]
- 196.Pearson T, et al. Effects of ursodeoxycholic acid on the gut microbiome and colorectal adenoma development. Cancer Med. 2019;8:617–628. doi: 10.1002/cam4.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Combes B, et al. Biliary bile acids in primary biliary cirrhosis: Effect of ursodeoxycholic acid. Hepatology. 1999;29:1649–1654. doi: 10.1002/hep.510290618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bossche LVanDen, et al. Tauroursodeoxycholic acid protects bile acid homeostasis under inflammatory conditions and dampens Crohn’s disease-like ileitis. Lab. Investig. 2017;97:519–529. doi: 10.1038/labinvest.2017.6. [DOI] [PubMed] [Google Scholar]
- 199.Lu Q, Jiang Z, Wang Q, Hu H, Zhao G. The effect of Tauroursodeoxycholic acid (TUDCA) and gut microbiota on murine gallbladder stone formation. Ann. Hepatol. 2021;23:100289. doi: 10.1016/j.aohep.2020.100289. [DOI] [PubMed] [Google Scholar]
- 200.Wirbel J, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bernstein C, et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch. Toxicol. 2011;85:863–871. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lai Y, et al. High-coverage metabolomics uncovers microbiota-driven biochemical landscape of interorgan transport and gut-brain communication in mice. Nat. Commun. 2021;12:1–16. doi: 10.1038/s41467-021-26209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Quinn RA, et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature. 2020;579:123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fischbach MA. Microbiome: Focus on Causation and Mechanism. Cell. 2018;174:785–790. doi: 10.1016/j.cell.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Walter J, Armet AM, Finlay BB, Shanahan F. Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell. 2020;180:221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 206.Jain N. The early life education of the immune system: Moms, microbes and (missed) opportunities. Gut Microbes. 2020;12:1–20. doi: 10.1080/19490976.2020.1824564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ege MJ, et al. Exposure to Environmental Microorganisms and Childhood Asthma. N. Engl. J. Med. 2011;364:687–696. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 208.Lopez-Tello, J. et al. Maternal microbiota Bifidobacterium promotes placental vascularization, nutrient transport and fetal growth in mice. Cellular and Molecular Life Sciences79, 386 (2022). [DOI] [PMC free article] [PubMed]
- 209.Rio-Aige K, et al. Association of maternal microbiota and diet in cord blood cytokine and immunoglobulin profiles. Int. J. Mol. Sci. 2021;22:1–23. doi: 10.3390/ijms22041778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Singh RP, Niharika J, Kondepudi KK, Bishnoi M, Tingirikari JMR. Recent understanding of human milk oligosaccharides in establishing infant gut microbiome and roles in immune system. Food Res. Int. 2022;151:110884. doi: 10.1016/j.foodres.2021.110884. [DOI] [PubMed] [Google Scholar]
- 211.Henrick BM, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884–3898.e11. doi: 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 212.Lamas B, Natividad JM, Sokol H. Aryl hydrocarbon receptor and intestinal immunity review-article. Mucosal Immunol. 2018;11:1024–1038. doi: 10.1038/s41385-018-0019-2. [DOI] [PubMed] [Google Scholar]
- 213.Laursen MF, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021;6:1367–1382. doi: 10.1038/s41564-021-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Azad, M. B. et al. Breastfeeding, maternal asthma and wheezing in the first year of life: A longitudinal birth cohort study. Eur. Respir. J. 49, 1602019 (2017). [DOI] [PubMed]
- 215.Ley D, et al. Early life nutrition influences susceptibility to chronic inflammatory colitis in later life. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-019-54308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kiu R, et al. Bifidobacterium breve UCC2003 Induces a Distinct Global Transcriptomic Program in Neonatal Murine Intestinal Epithelial Cells. iScience. 2020;23:101336. doi: 10.1016/j.isci.2020.101336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Robertson C, et al. Incidence of necrotising enterocolitis before and after introducing routine prophylactic Lactobacillus and Bifidobacterium probiotics. Arch. Dis. Child. Fetal Neonatal Ed. 2020;105:380–386. doi: 10.1136/archdischild-2019-317346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tobias J, et al. Bifidobacterium longum subsp. infantis EVC001 Administration Is Associated with a Significant Reduction in the Incidence of Necrotizing Enterocolitis in Very Low Birth Weight Infants. J. Pediatr. 2022;244:64–71.e2. doi: 10.1016/j.jpeds.2021.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ganguli K, et al. Probiotics prevent necrotizing enterocolitis by modulating enterocyte genes that regulate innate immune-mediated inflammation. Am. J. Physiol. - Gastrointest. Liver Physiol. 2013;304:132–141. doi: 10.1152/ajpgi.00142.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hollander D. Intestinal permeability, leaky gut, and intestinal disorders. Curr. Gastroenterol. Rep. 1999;1:410–416. doi: 10.1007/s11894-999-0023-5. [DOI] [PubMed] [Google Scholar]
- 221.Krumbeck JA, et al. Probiotic Bifidobacterium strains and galactooligosaccharides improve intestinal barrier function in obese adults but show no synergism when used together as synbiotics. Microbiome. 2018;6:121. doi: 10.1186/s40168-018-0494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hsieh CY, et al. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol. Rep. 2015;3:1–17. doi: 10.14814/phy2.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Laffin M, et al. A high-sugar diet rapidly enhances susceptibility to colitis via depletion of luminal short-chain fatty acids in mice. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-48749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Peng L, He Z, Chen W, Holzman IR, Lin J. Effects of butyrate on intestinal barrier function in a caco-2 cell monolayer model of intestinal barrier. Pediatr. Res. 2007;61:37–41. doi: 10.1203/01.pdr.0000250014.92242.f3. [DOI] [PubMed] [Google Scholar]
- 225.Beisner J, et al. Prebiotic Inulin and Sodium Butyrate Attenuate Obesity-Induced Intestinal Barrier Dysfunction by Induction of Antimicrobial Peptides. Front. Immunol. 2021;12:1–18. doi: 10.3389/fimmu.2021.678360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zheng L, et al. Microbial-Derived Butyrate Promotes Epithelial Barrier Function through IL-10 Receptor–Dependent Repression of Claudin-2. J. Immunol. 2017;199:2976–2984. doi: 10.4049/jimmunol.1700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yan F, et al. Soluble Proteins Produced by Probiotic Bacteria Regulate Intestinal Epithelial Cell Survival and Growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yan F, et al. A lactobacillus rhamnosus GG-derived soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J. Biol. Chem. 2013;288:30742–30751. doi: 10.1074/jbc.M113.492397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yang L, et al. Production of a Functional Factor, p40, by Lactobacillus rhamnosus GG Is Promoted by Intestinal Epithelial Cell-Secreted Extracellular Vesicles. Infect. Immun. 2019;87:1–17. doi: 10.1128/IAI.00113-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ottman N, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 2017;12:1–18. doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wang J, et al. The outer membrane protein Amuc_1100 of: Akkermansia muciniphila promotes intestinal 5-HT biosynthesis and extracellular availability through TLR2 signalling. Food Funct. 2021;12:3597–3610. doi: 10.1039/D1FO00115A. [DOI] [PubMed] [Google Scholar]
- 232.Gu MJ, et al. Barrier protection via Toll-like receptor 2 signaling in porcine intestinal epithelial cells damaged by deoxynivalnol. Vet. Res. 2016;47:1–11. doi: 10.1186/s13567-016-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhang FL, et al. Surface-Displayed Amuc_1100 From Akkermansia muciniphila on Lactococcus lactis ZHY1 Improves Hepatic Steatosis and Intestinal Health in High-Fat-Fed Zebrafish. Front. Nutr. 2021;8:1–13. doi: 10.3389/fnut.2021.726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Post SVanDer, et al. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut. 2019;68:2142–2151. doi: 10.1136/gutjnl-2018-317571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gaudier E, et al. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. - Gastrointest. Liver Physiol. 2004;287:1168–1174. doi: 10.1152/ajpgi.00219.2004. [DOI] [PubMed] [Google Scholar]
- 236.Jiminez, J. A., Uwiera, T. C., Abbott, D. W., Uwiera, R. R. E. & Inglis, G. D. Butyrate Supplementation at High Concentrations Alters Enteric Bacterial Communities and Reduces Intestinal Inflammation in Mice Infected with Citrobacter rodentium. mSphere2, e00243-17 (2017). [DOI] [PMC free article] [PubMed]
- 237.Petersson J, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. - Gastrointest. Liver Physiol. 2011;300:327–333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Shin J, et al. Elucidation of akkermansia muciniphila probiotic traits driven by mucin depletion. Front. Microbiol. 2019;10:1–12. doi: 10.3389/fmicb.2019.01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Shin NR, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 240.Zhu L, et al. Akkermansia muciniphila protects intestinal mucosa from damage caused by S. pullorum by initiating proliferation of intestinal epithelium. Vet. Res. 2020;51:1–9. doi: 10.1186/s13567-020-00755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Dong C, et al. Berberine, a potential prebiotic to indirectly promote Akkermansia growth through stimulating gut mucin secretion. Biomed. Pharmacother. 2021;139:111595. doi: 10.1016/j.biopha.2021.111595. [DOI] [PubMed] [Google Scholar]
- 242.Engevik MA, et al. Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways. MBio. 2019;10:e01087–19. doi: 10.1128/mBio.01087-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Nishio J, et al. Identification and characterization of a novel Enterococcus bacteriophage with potential to ameliorate murine colitis. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-021-99602-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Schulthess J, et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity. 2019;50:432–445.e7. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Nanjundaiah YS, et al. Modulation of Macrophage Function by Lactobacillus-Conditioned Medium. Front. Cell Dev. Biol. 2020;8:1–13. doi: 10.3389/fcell.2020.00723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Foligne, B. et al. A key role of dentric cells in probiotic functionality. PLoS One2, e313 (2007). [DOI] [PMC free article] [PubMed]
- 247.Peters A, et al. Metabolites of lactic acid bacteria present in fermented foods are highly potent agonists of human hydroxycarboxylic acid receptor 3. PLOS Genet. 2019;15:e1008145. doi: 10.1371/journal.pgen.1008145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Meyer F, et al. Propionate supplementation promotes the expansion of peripheral regulatory T-Cells in patients with end-stage renal disease. J. Nephrol. 2020;33:817–827. doi: 10.1007/s40620-019-00694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Marzocco, S. et al. Supplementation of short-chain fatty acid, sodium propionate, in patients on maintenance—“beneficial effects on inflammatory parameters and gut-derived uremic toxins”—a pilot study (Plan study). J. Clin. Med. 7, 315 (2018). [DOI] [PMC free article] [PubMed]
- 250.Duscha A, et al. Propionic Acid Shapes the Multiple Sclerosis Disease Course by an Immunomodulatory Mechanism. Cell. 2020;180:1067–1080.e16. doi: 10.1016/j.cell.2020.02.035. [DOI] [PubMed] [Google Scholar]
- 251.Guo C, et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity. 2016;45:802–816. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 252.Pols TWH, et al. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the Vitamin D receptor. PLoS One. 2017;12:1–16. doi: 10.1371/journal.pone.0176715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Monteleone I, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248.e1. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 254.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 2012;13:144–152. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 257.Schillde MAVon, et al. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe. 2012;11:387–396. doi: 10.1016/j.chom.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 258.Hörmannsperger G, Schillde MAvon, Haller D. Lactocepin as a protective microbial structure in the context of IBD. Gut Microbes. 2013;4:152–157. doi: 10.4161/gmic.23444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Llopis M, et al. Lactobacillus casei downregulates commensals’ inflammatory signals in Crohn’s disease mucosa. Inflamm. Bowel Dis. 2009;15:275–283. doi: 10.1002/ibd.20736. [DOI] [PubMed] [Google Scholar]
- 260.Koppel N, Bisanz JE, Pandelia M, Turnbaugh PJ, Balskus EP. Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. Elife. 2018;7:1–32. doi: 10.7554/eLife.33953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Alexander M, et al. Human gut bacterial metabolism drives Th17 activation and colitis. Cell Host Microbe. 2022;30:17–30.e9. doi: 10.1016/j.chom.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Haiser HJ, et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Singh K, et al. Dietary arginine regulates severity of experimental colitis and affects the colonic microbiome. Front. Cell. Infect. Microbiol. 2019;9:1–15. doi: 10.3389/fcimb.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sisson G, Ayis S, Sherwood RA, Bjarnason I. Randomised clinical trial: A liquid multi-strain probiotic vs. Placebo in the irritable bowel syndrome - A 12 week double-blind study. Aliment. Pharmacol. Ther. 2014;40:51–62. doi: 10.1111/apt.12787. [DOI] [PubMed] [Google Scholar]
- 265.Ward JBJ, et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am. J. Physiol. - Gastrointest. Liver Physiol. 2017;312:G550–G558. doi: 10.1152/ajpgi.00256.2016. [DOI] [PubMed] [Google Scholar]
- 266.Sjöqvist U, et al. Ursodeoxycholic acid treatment in IBD-patients with colorectal dysplasia and/or DNA-aneuploidy: A prospective, double-blind, randomized controlled pilot study. Anticancer Res. 2004;24:3121–3127. [PubMed] [Google Scholar]
- 267.Paik, D. et al. Human gut bacteria produce Τ Η 17-modulating bile acid metabolites. Nature10.1038/s41586-022-04480-z (2022). [DOI] [PMC free article] [PubMed]
- 268.Eaton JE, et al. High-dose ursodeoxycholic acid is associated with the development of colorectal neoplasia in patients with ulcerative colitis and primary sclerosing cholangitis. Am. J. Gastroenterol. 2011;106:1638–1645. doi: 10.1038/ajg.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Katayama Y, et al. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl. Lung Cancer Res. 2019;8:847–853. doi: 10.21037/tlcr.2019.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lee KA, et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. 2022;28:535–544. doi: 10.1038/s41591-022-01695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Baruch EN, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2020;5920:1–16. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 272.Davar D, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Vétizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mills, R. H. et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat. Microbiol. 2022 1–15 10.1038/s41564-021-01050-3 (2022). [DOI] [PMC free article] [PubMed]
- 275.Andrews MC, et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat. Med. 2021;27:1432–1441. doi: 10.1038/s41591-021-01406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Routy B, et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 277.Doron I, et al. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in Crohn’s disease. Nat. Microbiol. 2021;6:1493–1504. doi: 10.1038/s41564-021-00983-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lin PW, Nasr TR, Berardinelli AJ, Kumar A, Neish AS. The probiotic Lactobacillus GG may augment intestinal host defense by regulating apoptosis and promoting cytoprotective responses in the developing murine gut. Pediatr. Res. 2008;64:511–516. doi: 10.1203/PDR.0b013e3181827c0f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tanji Y, et al. Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J. Biosci. Bioeng. 2005;100:280–287. doi: 10.1263/jbb.100.280. [DOI] [PubMed] [Google Scholar]
- 280.Hempel S, et al. Probiotics for the Prevention and Treatment of Antibiotic-Associated Diarrhea. Jama. 2012;307:1959–1969. doi: 10.1001/jama.2012.3507. [DOI] [PubMed] [Google Scholar]
- 281.Ananthakrishnan AN, et al. A prospective study of long-term intake of dietary fiber and risk of Crohn’s disease and ulcerative colitis. Gastroenterology. 2013;145:970–977. doi: 10.1053/j.gastro.2013.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Andersen V, et al. Fibre intake and the development of inflammatory bowel disease: A European prospective multi-centre cohort study (EPIC-IBD) J. Crohn’s Colitis. 2018;12:129–136. doi: 10.1093/ecco-jcc/jjx136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Déchelotte P, et al. The probiotic strain h. Alvei ha4597® improves weight loss in overweight subjects under moderate hypocaloric diet: A proof-of-concept, multicenter randomized, double-blind placebo-controlled study. Nutrients. 2021;13:1–14. doi: 10.3390/nu13061902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Kravchychyn ACP, et al. The Long-Term Impact of High Levels of Alpha-Melanocyte-Stimulating Hormone in Energy Balance among Obese Adolescents. Ann. Nutr. Metab. 2018;72:279–286. doi: 10.1159/000488005. [DOI] [PubMed] [Google Scholar]
- 285.Rossen NG, et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology. 2015;149:110–118.e4. doi: 10.1053/j.gastro.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 286.Haifer C, et al. Lyophilised oral faecal microbiota transplantation for ulcerative colitis (LOTUS): a randomised, double-blind, placebo-controlled trial. lancet Gastroenterol. Hepatol. 2022;7:141–151. doi: 10.1016/S2468-1253(21)00400-3. [DOI] [PubMed] [Google Scholar]
- 287.Oliphant, K. et al. Effects of Antibiotic Pretreatment of an Ulcerative Colitis-Derived Fecal Microbial Community on the Integration of Therapeutic Bacteria In Vitro. mSystems5, e00404-19 (2020). [DOI] [PMC free article] [PubMed]
- 288.Ma J, et al. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-72635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chichlowski M, De Lartigue G, Bruce German J, Raybould HE, Mills DA. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J. Pediatr. Gastroenterol. Nutr. 2012;55:321–327. doi: 10.1097/MPG.0b013e31824fb899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Gehrig, J. L. et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science365, eaau4732 (2019). [DOI] [PMC free article] [PubMed]
- 291.Chen RY, et al. A Microbiota-Directed Food Intervention for Undernourished Children. N. Engl. J. Med. 2021;384:1517–1528. doi: 10.1056/NEJMoa2023294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zeevi D, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell. 2015;163:1079–1095. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 293.Tomasi M, et al. Commensal bifidobacterium strains enhance the efficacy of neo‐epitope based cancer vaccines. Vaccines. 2021;9:1–19. doi: 10.3390/vaccines9111356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Klünemann M, et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature. 2021;597:533–538. doi: 10.1038/s41586-021-03891-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Steiner HE, et al. Role of the gut microbiome in cardiovascular drug response: The potential for clinical application. Pharmacotherapy. 2022;42:165–176. doi: 10.1002/phar.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Rouanet A, et al. Live Biotherapeutic Products, A Road Map for Safety Assessment. Front. Med. 2020;7:1–13. doi: 10.3389/fmed.2020.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Olle B. Medicines from microbiota. Nat. Biotechnol. 2013;31:309–315. doi: 10.1038/nbt.2548. [DOI] [PubMed] [Google Scholar]
- 299.Fabienne Kurt et al. Co-cultivation is a powerful approach to produce a robust functionally designed synthetic consortium as a live biotherapeutic product (LBP). bioRxiv 6 10.1101/2021.10.13.464188v1 (2021). [DOI] [PMC free article] [PubMed]
- 300.Ducarmon QR, Kuijper EJ, Olle B. Opportunities and challenges in development of live biotherapeutic products to fight infections. J. Infect. Dis. 2021;223:S283–S289. doi: 10.1093/infdis/jiaa779. [DOI] [PubMed] [Google Scholar]
- 301.Steidler L, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 302.Kim K, et al. Engineering Bacteroides thetaiotaomicron to produce non-native butyrate based on a genome-scale metabolic model-guided design. Metab. Eng. 2021;68:174–186. doi: 10.1016/j.ymben.2021.10.005. [DOI] [PubMed] [Google Scholar]
- 303.Kumar, N., Hitch, T. C. A., Haller, D., Lagkouvardos, I. & Clavel, T. MiMiC: a bioinformatic approach for generation of synthetic communities from metagenomes. Microb. Biotechnol. 10.1111/1751-7915.13845 (2021). [DOI] [PMC free article] [PubMed]
- 304.Shepherd, E. S., Deloache, W. C., Pruss, K. M., Whitaker, W. R. & Sonnenburg, J. L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature10.1038/s41586-018-0092-4 (2018).. [DOI] [PMC free article] [PubMed]
- 305.Pan J, et al. A single-cell nanocoating of probiotics for enhanced amelioration of antibiotic-associated diarrhea. Nat. Commun. 2022;13:1–11. doi: 10.1038/s41467-022-29672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Diard M, et al. A rationally designed oral vaccine induces immunoglobulin A in the murine gut that directs the evolution of attenuated Salmonella variants. Nat. Microbiol. 2021;6:830–841. doi: 10.1038/s41564-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]