Abstract
Monocyte-derived macrophages (Mφs) are crucial regulators during muscularis inflammation. However, it is unclear which micro-environmental factors are responsible for monocyte recruitment and anti-inflammatory Mφ differentiation in this paradigm. Here, we investigate Mφ heterogeneity at different stages of muscularis inflammation and determine how environmental cues can attract and activate tissue-protective Mφs. Results showed that muscularis inflammation induced marked alterations in mononuclear phagocyte populations associated with a rapid infiltration of Ly6c+ monocytes that locally acquired unique transcriptional states. Trajectory inference analysis revealed two main pro-resolving Mφ subpopulations during the resolution of muscularis inflammation, i.e. Cd206+ MhcIIhi and Timp2+ MhcIIlo Mφs. Interestingly, we found that damage to the micro-environment upon muscularis inflammation resulted in EGC activation, which in turn stimulated monocyte infiltration and the consequent differentiation in anti-inflammatory CD206+ Mφs via CCL2 and CSF1, respectively. In addition, CSF1-CSF1R signaling was shown to be essential for the differentiation of monocytes into CD206+ Mφs and EGC proliferation during muscularis inflammation. Our study provides a comprehensive insight into pro-resolving Mφ differentiation and their regulators during muscularis inflammation. We deepened our understanding in the interaction between EGCs and Mφs, thereby highlighting pro-resolving Mφ differentiation as a potential novel therapeutic strategy for the treatment of intestinal inflammation.
Introduction
In recent years, significant advances have been made in our understanding of the phenotype of intestinal macrophage (Mφ) archeotypes, which perform a variety of niche-defined functions from preserving immune homeostasis to regulating motility and secretion via crosstalk with the enteric nervous system (ENS)1–3. Consequently, specific intestinal niches are populated by different Mφ subsets with a unique transcriptome to meet the diverse functional demands of the tissue micro-environment. For instance, lamina propria Mφs reside in close proximity to the epithelium and at the crypt base, where they are involved in the maintenance of gut homeostasis by contributing to epithelial barrier integrity and by phagocytosing luminal antigens2–5. In the deeper layers of the gut wall at the level of the muscularis externa, Mφs exert neurotrophic functions promoting enteric neuronal survival and supporting neuronal function1,6,7. However, during muscularis inflammation, activation of resident Mφs results in impairment of neuromuscular contractility due to the release of nitric oxide, prostaglandins and cytokines, simultaneously triggering the influx of inflammatory cells8–10. Although we have shown in a previous study that the influx of monocytes is essential for the resolution of muscularis inflammation due to their differentiation into pro-resolving monocyte-derived Mφs11, it remains to be determined which cell types are responsible for monocyte recruitment and which factors promote their differentiation towards anti-inflammatory Mφs during muscularis inflammation. The extensive crosstalk between muscularis Mφs and the ENS at homeostasis suggests a potential involvement also during inflammation6. The ENS is organized in a network of ganglia containing neurons and enteric glial cells (EGCs). Historically, EGCs were mainly considered as supporting cells of enteric neurons. However, recent evidence suggests that EGCs have a much broader function in gastro-intestinal (GI) physiology, contributing to motility and preserving epithelial barrier integrity by the interaction with innate lymphoid cells (ILCs), interstitial cells of Cajal, endothelial and epithelial cells12–17. Even more intriguing is the putative role of EGCs in immune regulation. In this regard, the interaction between astrocytes and microglia has been extensively studied in the brain during development, homeostasis and disease18. Yet, limited studies have investigated the interaction between Mφs and EGCs in the intestine19.
Here, we investigate Mφ heterogeneity and their role in tissue repair during muscularis inflammation. Using time-series single-cell transcriptomics, we observed that muscularis inflammation induced a prominent myeloid cell diversification, resulting in 7 myeloid subpopulations. Trajectory inference analysis indicated that incoming Ly6chi monocytes acquired diverse gene expression signatures in the injured muscularis, notably resulting in two main pro-resolving Mφ subpopulations characterized by Cd206+ MhcIIhi and Timp2+ MhcIIlo gene signatures. In addition, EGCs were identified as the main producers of the monocyte chemoattractant, CCL2, during the early phase of inflammation. Furthermore, these cells produced a variety of secreted factors that can potentially stimulate the differentiation of monocytes into pro-resolving Mφs. Specifically, we found that EGC-derived CSF1 is a critical signal for the generation of anti-inflammatory CD206+ Mφs in vitro and that blocking CSF1R in vivo diminishes CD206+ Mφ differentiation leading to damage to the EGCs. Overall, our findings imply that during intestinal inflammation, future therapies should be aimed at enhancing the pro-resolving and neurotrophic properties of Mφs to reduce possible damage to the ENS and promote functional GI recovery.
Results
Time-dependent recruitment of myeloid cells in the inflamed muscularis
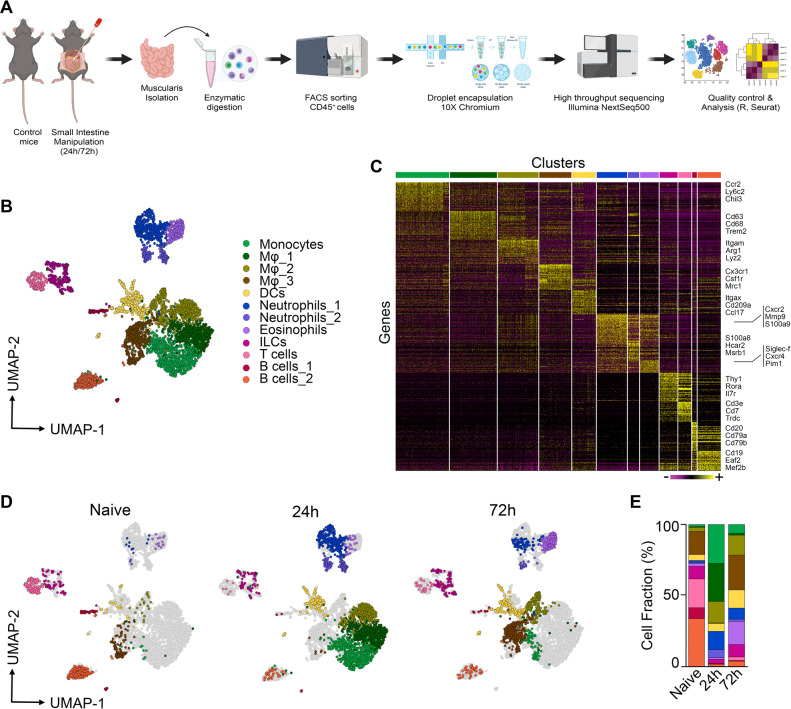
Surgery-induced damage to the muscularis leads to a transient impairment of GI motility associated with extensive recruitment of immune cells to the ENS. To characterize the immune cell infiltrate and their respective activation states during muscularis inflammation, droplet-based single cell RNA sequencing (scRNA-seq) was performed on sorted CD45+ immune cells from the muscularis of naïve mice, and during the acute inflammatory (24 h) and the recovery phase of muscularis inflammation (72 h) (10X Genomics Platform; Fig. 1a). Unsupervised clustering of 4102 cells and reference-based cell identification using Immgen (Fig. 1b, c and Supplementary Fig. 1a, b) revealed 12 independent immune cell populations including monocytes (Ccr2, Ly6c2, Chil3), 3 clusters of Mφs (1: Cd63, Cd68, Trem2; 2: Itgam, Arg1, Lyz2; 3: Cx3cr1, Csf1r, Mrc1), dendritic cells (DCs; Itgax, Cd209a, Ccl17), 2 clusters of neutrophils (1: Cxcr2, Mmp9, S100a9; 2:, S100a8, Hcar2, Msrb1), eosinophils (Siglec-f, Cxcr4, Pim1), ILCs (Thy1, Rora, Il7r), T cells (Cd3e, Cd7, Trdc) and 2 clusters of B cells (1: Cd79a, Cd20, CD79b; 2: Eaf2, Mef2b, CD19)20.
Fig. 1. Identification of CD45+ immune cell populations by unsupervised scRNA-seq clustering in the healthy and inflamed muscularis.
a Experimental pipeline of scRNA-seq experiment. b UMAP of sorted CD45+ immune cells from the healthy muscularis, 24 h and 72 h post-injury from WT mice. Each sample was pooled from 3–4 mice. c Heatmap of the 50 most differentially expressed genes in each cluster. d UMAPs of time-dependent infiltration of immune cells upon muscularis inflammation. Data of Fig. 1b has been split based on different time points. e Cell fraction of each cluster relative to the total number of CD45+ immune cells at different time points after muscularis inflammation.
To examine the role of different immune cell populations, we determined their gene expression signatures at different time points during muscularis inflammation. At homeostasis, the muscularis mainly consisted of a population of Cx3cr1+ Mφs (Mφ_3) together with T cells, B cells and ILCs (Fig. 1d, e). Consistent with previous observations11,21, a shift in the leukocyte populations was observed in the acute phase (24 h) after the induction of muscularis inflammation with a massive infiltration of monocytes in addition to Mφs (Mφ_1 and Mφ_2) and neutrophils (Neu_1 and Neu_2), pointing towards a pro-inflammatory micro-environment (Fig. 1d, e). During the resolution of muscularis inflammation (72 h), the muscularis was mainly populated by Cx3cr1+ Mφs (Mφ_3), DCs and eosinophils, indicating a return to homeostasis. Altogether, these results show that during muscularis inflammation a shift in the immune landscape favors the resolution of inflammation and a rapid return to homeostasis.
Identification of two distinct myeloid subpopulations during the resolution of muscularis inflammation
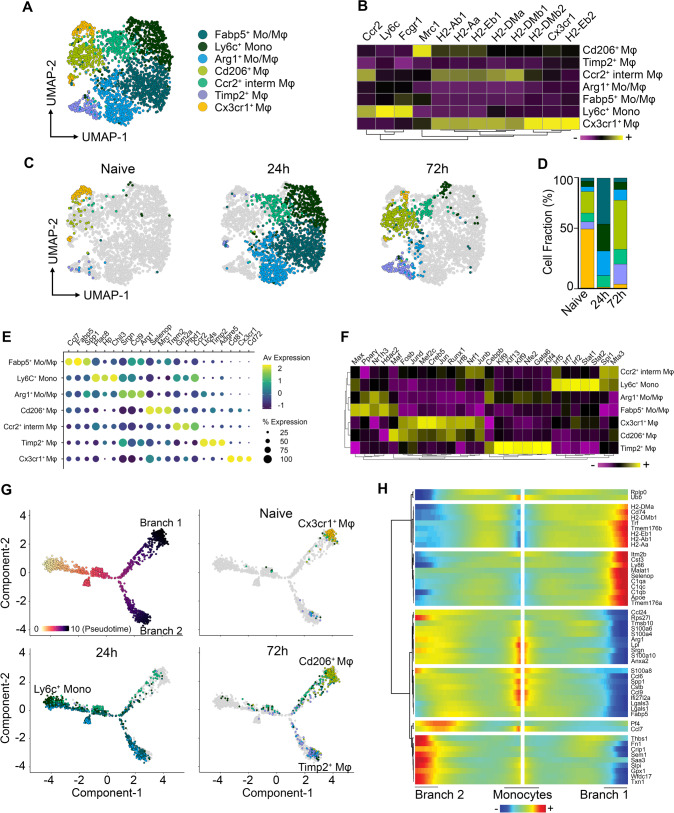
Monocyte-derived Mφs are essential for the recovery of GI motility during the resolution of muscularis inflammation11,21. However, their heterogeneity and differentiation trajectory towards tissue-protective Mφs have not yet been characterized during muscularis inflammation. To this end, subsets originally identified as monocyte/Mφ subpopulations in our scRNA-seq dataset (Fig. 1) were extracted and re-clustered to better define which types of myeloid cells might aid in the resolution of muscularis inflammation, thereby identifying 7 distinct subsets (Fig. 2a–d and Supplementary Fig. 2a). At homeostasis, the muscularis micro-environment mainly consisted of Cx3cr1+ Mφs with high expression of typical resident Mφ markers such as Cd81, Cd72 and H2-Eb1 (Fig. 2e). During acute muscularis inflammation, the most predominant subpopulation was the cluster of classical Ly6c+ monocytes, which was enriched for the expression of Plac8, Hp and Chil3 (Fig. 2b–e). Additionally, a subpopulation with a gene expression signature suggestive of an intermediate monocyte-to-Mφ differentiation state was observed (Ccr2+ int Mφs), which was underscored by their moderate MhcII expression as compared to Ly6c+ monocytes and homeostatic Cx3cr1+ Mφs (Fig. 2b). Besides Ly6c+ monocytes and Ccr2+ int Mφs, we observed a subcluster of Arg1+ Mo/Mφs with high expression of Ccl9 and Srgn alongside Fabp5+ Mo/Mφs with characteristic expression of Lgals1, Ccl7 and Flt1 (Fig. 2e). Of note, 72 h post-injury, during the resolution of muscularis inflammation, Ly6c+ monocytes and Ccr2+ int Mφs were present at a low percentage but two novel Mφ subclusters were identified: Cd206+ Mφs and Timp2+ Mφs (Fig. 2c, d and Supplementary Fig. 2b). The most abundant subcluster during the resolution of muscularis inflammation, Cd206+ Mφs, displayed high MhcII expression similar to homeostatic Cx3cr1+ Mφs, in addition to high expression of anti-inflammatory genes such as Selenop, Mrc1, Igf1, Trem2 and Stab1 (Fig. 2b and Supplementary Fig. 2b, c). In contrast, Timp2+ Mφs had lower MhcII expression compared to homeostatic Cx3cr1+ Mφs, but similarly expressed high levels of tissue reparative markers such as Ltc4s and Adgre5 (Fig. 2b, e and Supplementary Fig. 2b, c). To further investigate differences in the transcriptional regulation of Cd206+ and Timp2+ Mφs, single-cell regulatory network inference and clustering (SCENIC) was employed to assess specific regulon activities in different myeloid subclusters (Fig. 2f and Supplementary Fig. 2f). Cd206+ Mφs displayed high regulon activity of FosB, Runx1 and Irf8 similar to Cx3cr1+ Mφs, while Timp2+ Mφs had a distinctly altered transcription factor signature with high Cebpb and Gata6 regulon activity.
Fig. 2. Two main anti-inflammatory Mφ subpopulations with a unique transcriptional state are present during the resolution of muscularis inflammation.
a UMAP of reclustered monocyte/Mφ subpopulations from Fig. 1a from the healthy muscularis, 24 h and 72 h post-injury from WT mice. b Heatmap of typical monocyte and Mφ markers including MHCII genes. c UMAPs of time-dependent infiltration of monocyte and Mφ subsets upon muscularis inflammation. d Cell fraction of each subcluster relative to the total number of monocytes/Mφs at different time points after muscularis inflammation. e Dotplot showing expression of selected differentially expressed genes in each subcluster. f Heatmap of regulon activity per cluster according to SCENIC analysis. g Pseudotime analysis of monocytes/Mφs at different time points after muscularis inflammation. h Heatmap of gene expression showing the top 50 genes of different branches of the pseudotime trajectory tree.
To evaluate whether these subclusters can also be detected by flow cytometry, a comparative study was performed during muscularis inflammation. In the healthy muscularis, only 1 subset of MHCIIhi Mφs was observed similar to homeostatic Cx3cr1+ Mφs (Supplementary Fig. 2d, e). During acute muscularis inflammation, the muscularis micro-environment was mainly dominated by infiltrating Ly6Chi monocytes, which differentiated into Ly6C+ MHCII+ immature Mφs. However, at single cell level, we observed 4 different Mφ subclusters, indicating that Mφ heterogeneity was higher than originally considered. Interestingly, 72 h post-injury, flow cytometric analysis also revealed only two main Mφ subclusters (MHCIIhi and MHCIIlo Mφs), similar to our observations at single cell level (Supplementary Fig. 2d, e).
Identifying relevant gene expression changes in myeloid subclusters during muscularis inflammation could shed light on the molecular mechanisms regulating Mφ differentiation. In order to define these possible temporal transcriptional alterations leading to differentiation into Cd206+ and Timp2+ Mφs, Monocle-2 was used to superimpose subclusters on a trajectory placing Ly6c+ monocytes at the beginning of the pseudotime (Fig. 2g, h)22. Using this approach, we identified a major trajectory bifurcation leading to a branch of cells with high expression of genes found in homeostatic Mφs such as Cd74, Tmem176a/b, complement genes (C1qa, C1qb, C1qc) and MhcII genes (H2-Eb1, H2-Ab1, H2-Aa; Branch 1), while the cells in the second branch expressed less complement and MhcII genes but had increased expression of Fn1, Ltc4s and Saa3 (Branch 2) (Fig. 2h and Supplementary Fig. 2g, h). Interestingly, the Cd206+ and Timp2+ Mφ subsets were at two opposite ends of the trajectory suggesting different differentiation paths during the resolution of muscularis inflammation (Fig. 2g). Of note, homeostatic Cx3cr1+ Mφs were located in close proximity to Cd206+ Mφs in the pseudotime, underscoring the similarity between both Mφ subsets. Arg1+ and Fabp5+ Mo/Mφs were mainly present in Branch 2, while Ccr2+ int Mφs were exclusively present in Branch 1, indicating that these subpopulations might act as intermediates before attaining their terminal differentiation state. These findings support the hypothesis that there are two major monocyte-to-Mφ differentiation trajectories in the inflamed muscularis giving rise to Cd206+ and Timp2+ Mφs during the resolution of inflammation.
Pro-resolving Mφs originate from CCR2± monocytes during muscularis inflammation
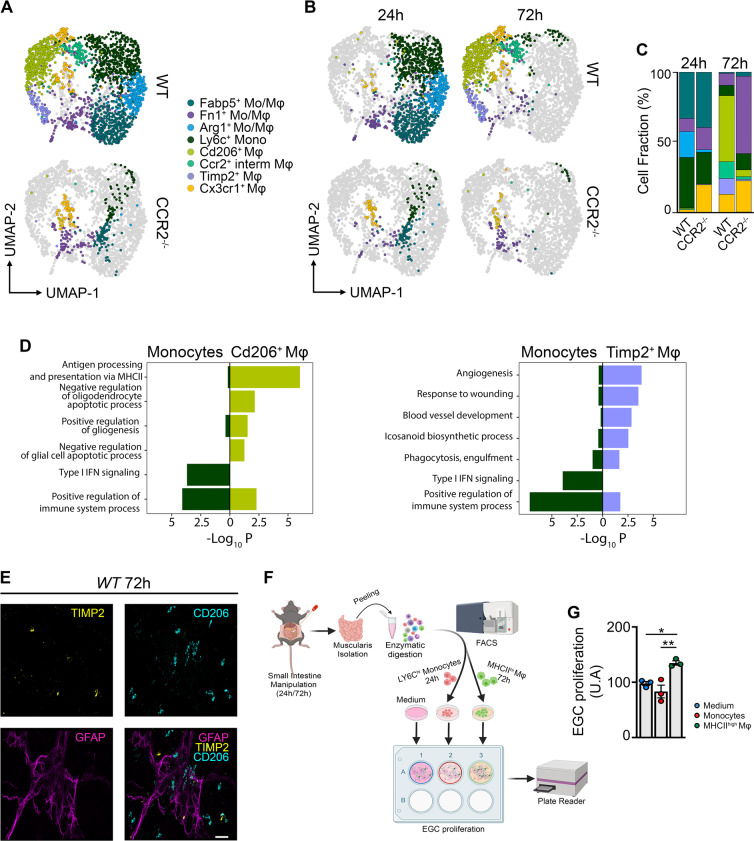
In a previous study, we have shown that the influx of CCR2+ monocytes is essential for resolution of muscularis inflammation, as blocking monocyte infiltration resulted in a delayed recovery of GI motility and damage to the ENS11. Thus, to investigate whether CCR2+ monocytes are the source of pro-resolving Mφs during muscularis inflammation, scRNA-seq was performed on sorted CD45+ immune cells from the muscularis of WT and CCR2−/− mice 24 h and 72 h post-injury (Fig. 3a–c and Supplementary Fig. 3a–d). After extraction and re-clustering of the monocyte and Mφ subsets (2,406 cells), the gene expression signatures from the myeloid subsets in Fig. 2 were crossmatched by singleR with this novel dataset to specifically annotate the subpopulations (Supplementary Fig. 3c), yielding only one additional cluster, i.e., Fn1+ Mo/Mφs. In CCR2−/− mice, there was a large alteration in the myeloid compartment after muscularis inflammation. As expected upon acute muscularis inflammation in CCR2−/− mice, there was an almost complete loss of Ly6c+ monocytes, and Arg1+ and Fabp5+ Mo/Mφs. In addition, we could only detect a small subpopulation of Cx3cr1+ Mφs representing the long-lived resident Mφs. During the resolution of muscularis inflammation, the Cd206+ and Timp2+ Mφ subsets were also completely absent in CCR2−/− mice, confirming that these subpopulations are derived from CCR2+ monocytes and that they possibly represent the pro-resolving Mφs essential for recovery of GI motility after tissue damage.
Fig. 3. Two distinct Mφ subpopulations during the resolution of muscularis inflammation are derived from CCR2+ monocytes.
a UMAP of monocyte and Mφ subclusters from the muscularis of WT and CCR2−/− mice at 24 h and 72 h after induction of muscularis inflammation. Each sample was pooled from 3–4 mice. b UMAPs of myeloid cells at different time points post-injury. c Cell fraction of each subcluster relative to the total number of myeloid cells at different time points after muscularis inflammation. d GO analysis of monocytes versus Cd206+ Mφs (left) or Timp2+ Mφs (right) showing negative Log10(p-value). e Immunofluorescent images of muscularis whole-mount preparations 3 days after the induction of muscularis inflammation stained for GFAP (purple), TIMP2 (yellow) and CD206 (light blue). Scale bar 15 µm. f Experimental outline of in vitro EGC proliferation by stimulation with supernatant of monocytes or Mφs from different time points post-injury. g Fold induction of EGCs stimulated with the supernatant of LY6Chi monocytes from 24 h post-injury or MHCIIhi Mφs from 72 h post-injury relative to control medium. Every data point is an independent sorting and culture experiment One-way ANOVA; test *p < 0.05; **p < 0.01; ns not significant.
To further investigate the phenotype of Cd206+ and Timp2+ Mφs during muscularis inflammation, gene ontology (GO) analysis was performed and revealed that both Mφ subsets seemed pro-resolving in nature (Fig. 3d). For instance, Timp2+ Mφs were implicated in pathways associated with tissue repair and regulation of the inflammatory response, such as angiogenesis, response to wounding, blood vessel development, icosanoid biosynthetic processes, and phagocytosis-engulfment. Conversely, Cd206+ Mφs were enriched for pathways associated with antigen processing and presentation via MHC class II, negative regulation of oligodendrocyte and glial cell apoptotic processes and positive regulation of gliogenesis. In contrast, monocytes were clearly enriched for pro-inflammatory pathways, such as type I interferon signaling and positive regulation of immune system processes. Interestingly, CD206+ Mφs were located in close proximity or even surrounded the ENS and outnumbered TIMP2+ Mφs, which were located further away from the ENS (Fig. 3e). To confirm that these Cd206+ MhcIIhi Mφs have a neuroprotective effect on the ENS, we nassessed their contribution to EGC proliferation in vitro (Fig. 3f). While Ly6c+ monocytes isolated 24 h post-injury did not alter the proliferation of EGCs, MHCIIhi Mφs from 72 h post-injury significantly induced EGC proliferation, underscoring their role in supporting EGC function (Fig. 3g). These results establish that CCR2+ monocytes differentiate into two distinct Mφ subpopulations with pro-resolving and/or neurotrophic functions.
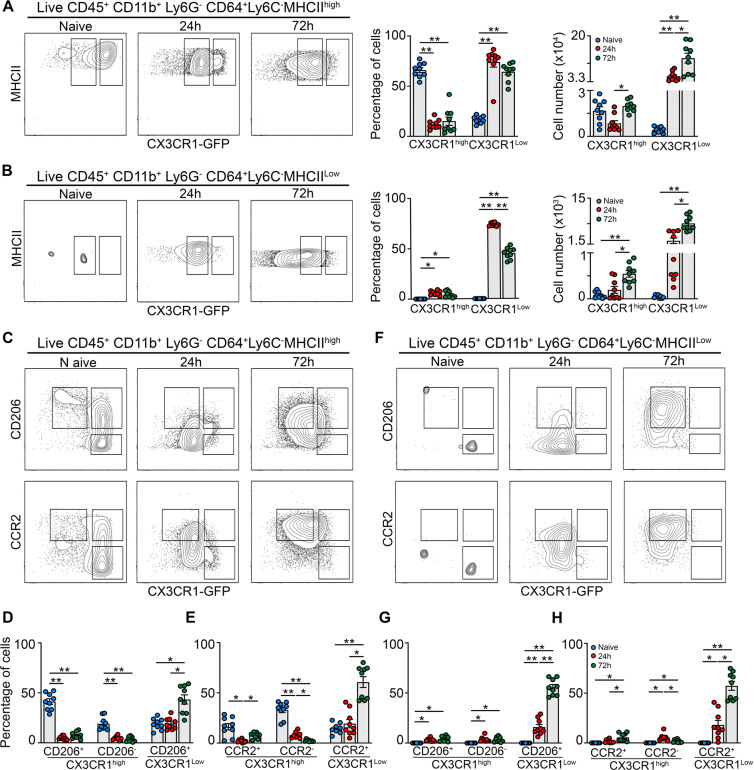
CX3CR1GFP-based mapping matches unique Mφ subsets observed in scRNA-seq during muscularis inflammation
To further define the phenotype of the Mφ subpopulations identified using scRNA-seq at the protein level, we induced muscularis inflammation in CX3CR1gfp/+ mice (Fig. 4a, b). Upon muscularis inflammation, the percentage of homeostatic CX3CR1hi Mφs was drastically reduced with a concomitant increase of CX3CR1lo MHCIIhi and CX3CR1lo MHCIIlo Mφs. As CCR2 and CD206 have been described as essential markers for recruited and anti-inflammatory Mφs respectively, their protein levels were determined in these CX3CR1hi/lo Mφ subpopulations (Fig. 4c–h and Supplementary Fig. 4a, b). In line with previous findings, CX3CR1hi MHCIIhi Mφs were mainly CD206+ and CCR2− at homeostasis (Fig. 4c–e). However, this subpopulation was gradually replaced by a combination of CX3CR1lo MHCIIlo and CX3CR1lo MHCIIhi Mφs, resembling Timp2+ and Cd206+ Mφs from our scRNA-seq dataset, respectively (Fig. 4c–h). Interestingly, CD206+ CX3CR1lo MHCIIhi Mφs 10-fold outnumbered CX3CR1lo MHCIIlo Mφs, further underscoring the importance of these anti-inflammatory CD206+ MHCIIhi Mφs (Supplementary Fig. 4a, b). Overall, our data validated at protein level the presence of two distinct Mφ subpopulations during the resolution of muscularis inflammation which are derived from CCR2+ monocytes and are anti-inflammatory in nature.
Fig. 4. Flow cytometry validates unique Mφ subpopulations during muscularis inflammation.
a–h from naïve CX3CR1GFP/+ mice, 24 h and 72 h after muscularis inflammation. a Contour plots representing CX3CR1 expression in Ly6C− MHCIIhi Mφs (left). Percentages and absolute numbers of CX3CR1hi and CX3CR1lo cells from Ly6C− MHCIIhi Mφs are shown as mean ± SEM (right). b Contour plots representing CX3CR1 expression in Ly6C− MHCIIlo Mφs (left). Percentages and absolute numbers of CX3CR1hi and CX3CR1lo cells from Ly6C− MHCIIlo Mφs are shown as mean ± SEM (right). c–h CD206 and CCR2 expression in Ly6C− MHCIIhi and MHCIIlo Mφs. Contour plots representing CD206 or CCR2 expression in Ly6C− MHCIIhi (c) and Ly6C− MHCIIlo Mφs (f). Percentages of CD206+ CX3CR1hi, CD206− CX3CR1hi and CD206+ CX3CR1lo cells in the LY6C− MHCIIhi Mφs (d) and MHCIIlo Mφs (g). Percentages of CCR2+ CX3CR1hi, CCR2− CX3CR1hi and CCR2+ CX3CR1lo cells in the LY6C− MHCIIhi Mφs (e) and MHCIIlo Mφs (h). One-way ANOVA; test *p < 0.05; **p < 0.01; ns not significant.
In line with our scRNA-seq datasets, we observed a drastic reduction in the percentage of homeostatic CX3CR1hi Mφs upon the induction of muscularis inflammation (Fig. 2c, d). To determine whether these CX3CR1hi Mφs are replaced by incoming monocyte-derived Mφs, CX3CR1+ resident Mφs were mapped by using a tamoxifen-inducible CX3CR1CreERT2 strain backcrossed with Rosa26-LSL-YFP mice. By comparing circulating unlabeled blood monocytes with resident YFP+ Mφs, we could show that resident Mφs did not diminish in number during muscularis inflammation as quantified by immunofluorescence and flow cytometry (Supplementary Fig. 4c–f). Taken together, these results indicate that CX3CR1hi Mφs were present alongside CX3CR1lo mature Mφs during muscularis inflammation.
Damage-activated EGCs initiate the recruitment and differentiation of monocytes upon muscularis inflammation
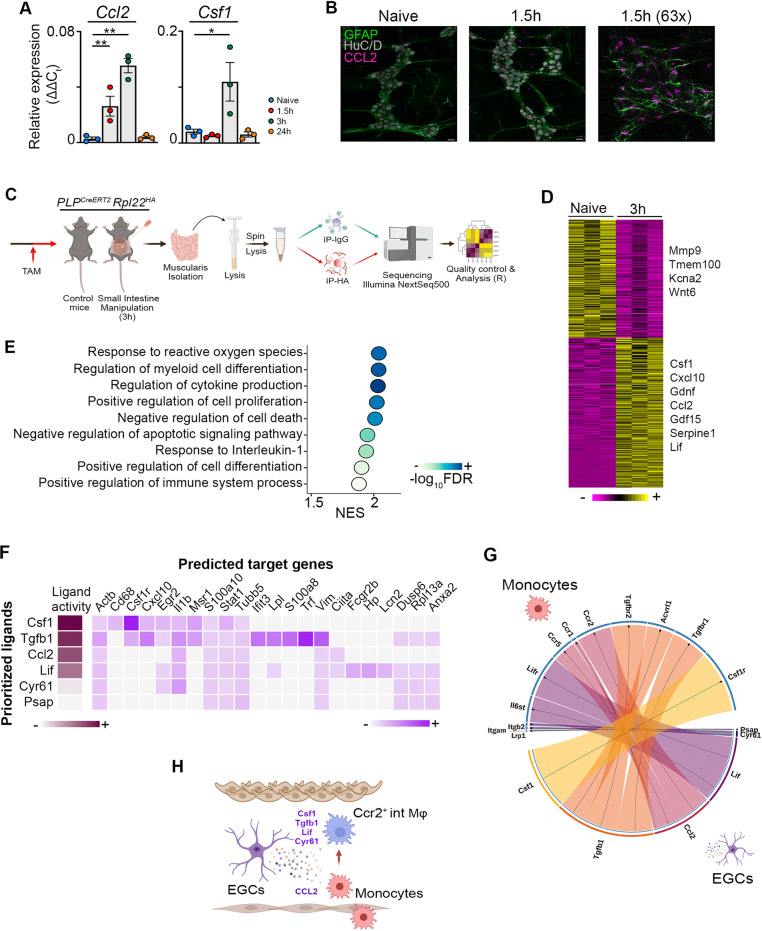
Monocytes originate from progenitors in the bone marrow and rely on local chemokine production such as MCP-1/CCL2 for their recruitment at the site of inflammation23. Considering the extensive interaction between CCR2+ monocytes and the ENS in the inflamed muscularis6,11, we examined the production of the major chemoattractant Ccl2 in the muscularis during the early stage of surgery-induced inflammation. Analysis of isolated enteric ganglia 1.5 h post-injury showed an increased gene expression of Ccl2 and Csf1 (Fig. 5a). Moreover, immunofluorescent images showed that CCL2 was specifically produced by GFAP+ EGCs (Fig. 5b), leading us to conclude that during the early stages of muscularis inflammation, EGCs likely initiate the recruitment of monocytes via CCL2 to stimulate tissue repair.
Fig. 5. EGCs produce factors essential for the recruitment and differentiation of monocytes during inflammation.
a Relative mRNA levels for Ccl2 and Csf1 normalized to the housekeeping gene rpl32 from ganglia isolated from the muscularis of the small intestine from naïve wild-type mice and 1.5 h, 3 h and 24 h after muscularis inflammation. One-way ANOVA; test *p < 0.05; **p < 0.01; ns not significant. b Immunofluorescent images of muscularis whole-mount preparations at homeostasis and 1.5 h after the induction of muscularis inflammation stained for GFAP (green), HuC/D (gray) and CCL2 (purple). Scale bar (25x) 25 µm, (63x) 15 µm. c Experimental overview of experiments in PLP-CreERT2 Rpl22HA mice. d Heatmap of HA-enriched differentially expressed genes between immunoprecipitated samples from naïve PLP-CreERT2 Rpl22HA mice and 3 h after intestinal manipulation. e Selected significant GO terms enriched (GSEA) in PLP1+ EGCs 3 h post-injury compared to naive PLP1+ EGCs. f Heat map of ligand-target pairs showing regulatory potential scores between top positively correlated prioritized ligands and their target genes among the differentially expressed genes between Ly6c+ monocytes and Ccr2+ int Mφs. g Circos plot showing top NicheNet ligand-receptor pairs between EGCs and Ly6c+ monocytes corresponding to the prioritized ligands in Fig. 5f. h Schematic overview of interactions between EGCs and infiltrating Ly6c+ monocytes.
To further explore the role of EGCs during the early phase of muscularis inflammation, PLPCreERT2 Rpl22HA mice were used, which allow immunoprecipitation (IP) of ribosome‐bound mRNAs from PLP1+ EGCs to study the transcriptome of EGCs at homeostasis and during early inflammation (3 h post-injury) (Fig. 5c and Supplementary Fig. 5a, b)24–26. Unlike pro-inflammatory reactive astrocytes during brain injury27, EGCs were activated early after muscularis inflammation and produced increased amounts of factors that are able to promote the differentiation of anti-inflammatory Mφs, such as Lif, Serpine-1, Gdf15, Gdnf, Cxcl10 and Csf1 (Fig. 5d and Supplementary Fig. 5c). To explore which pathways are activated in EGCs, gene set enrichment analysis was performed. Positive normalized enrichment scores were identified for response to ROS and cytokines including IL-1, indicating that EGCs are able to sense tissue damage in the muscularis micro-environment to initiate an appropriate immune response (Fig. 5e). These results were further underscored by showing that neurosphere-derived EGCs stimulated with IL1-α and/or IL1-β produced increased amounts of Ccl2 compared to unstimulated EGCs (Supplementary Fig. 5d). Interestingly, there was also an overrepresentation of genes involved in the regulation of myeloid differentiation during early muscularis inflammation in EGCs (Fig. 5e and Supplementary Fig. 5e). These results suggest that activation of EGCs by cytokines including IL-1 could lead to both the recruitment and differentiation of monocytes during the early stages of muscularis inflammation.
To determine whether EGCs are able to stimulate monocyte differentiation during muscularis inflammation, we analyzed the expression of ligand-receptor pairs based on differentially expressed genes in PLP1+ EGCs 3 h post-injury and Ly6c+ monocytes undergoing differentiation into Ccr2+ int Mφs. Their expression data was combined with prior knowledge of signaling and gene regulatory networks using Nichenet28. To identify genes regulated by the identified ligands, putative ligand-gene interactions were scored by NicheNet according to their “regulatory potential” (Fig. 5f). We next assessed whether the expression of the receptors for the putative ligand-receptor interaction were altered when Ly6c+ monocytes underwent differentiation into Ccr2+ int Mφs (Supplementary Fig. 5f–h). By determining a threshold for the receptor expression, we identified the following ligand-receptor interactions between EGCs and differentiating monocytes: Csf1-Csf1r, Tgfb1-Tgfbr2/Acvrl1/Tgfbr1, Ccl2-Ccr5/Ccr1/Ccr2, Lif-Lifr/Il6st, Cyr61-Itgam/Itgb2, and Psap-Lrp1 (Fig. 5g and Supplementary Fig. 5g). Taken together, damage to the muscularis micro-environment leads to activation of EGCs, that are able to recruit monocytes to the site of inflammation via CCL2 and that produce factors that have the potential to promote anti-inflammatory Mφ differentiation (Fig. 5h).
EGCs promote the differentiation of monocytes into pro-resolving CD206± Mφs in vitro
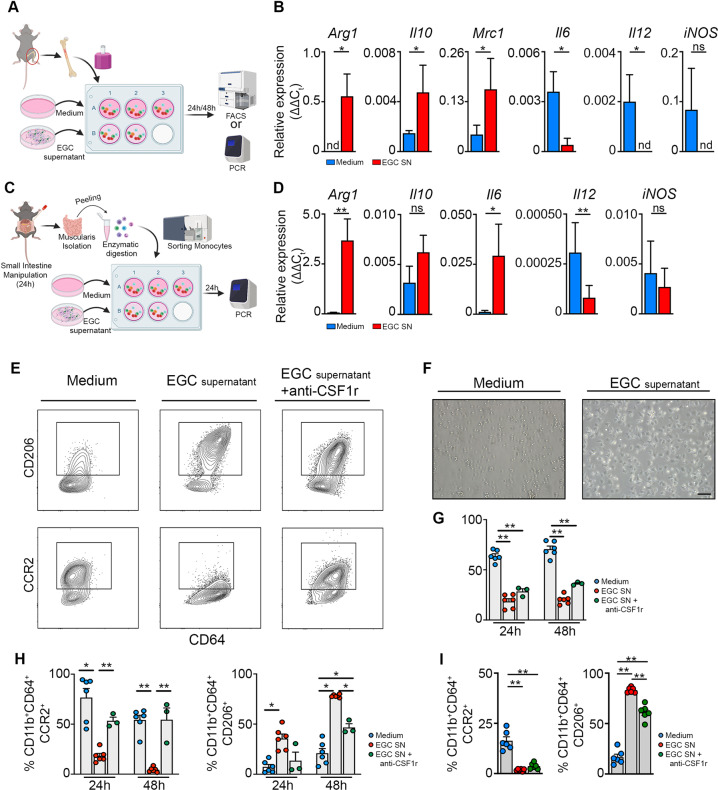
To directly assess the functional interaction between EGCs and monocytes, neurosphere-derived EGCs were analyzed by bulk RNAseq. The results revealed high expression of cytokines involved in the induction of anti-inflammatory Mφ differentiation (Supplementary Fig. 6a–c), similar to those observed in vivo in EGCs, including Lif, Csf1, Tgfb1 and Ccl2 (Fig. 5d, f). To demonstrate that EGC-derived factors are able to stimulate monocyte differentiation, an ex vivo model was set-up to co-culture bone marrow-derived monocytes with the supernatant of EGCs (Fig. 6a)29. Co-culture of bone marrow monocytes with EGC supernatant resulted in an upregulation of several anti-inflammatory markers such as Arg1, Il10 and Mrc1, while pro-inflammatory markers were downregulated, including Il6 and Il12 (Fig. 6b). In addition, when bone marrow monocytes were exposed to bacterial components (LPS), a similar anti-inflammatory effect of EGC supernatant on monocytes (EGCs+LPS) was observed compared to LPS alone on monocytes (Supplementary Fig. 6d). Finally, to confirm that muscularis monocytes responded to EGC supernatant in a similar manner as bone marrow monocytes given their different origin, ex vivo experiments were performed on sorted Ly6C+ monocytes from the muscularis 24 h post-injury (Fig. 6c). Also in this setting, monocytes acquired an anti-inflammatory phenotype upon co-culture with EGC supernatant with increased expression of Arg1 and decreased expression of il12 similar to that seen in bone marrow monocytes (Fig. 6d and Supplementary Fig. 6e). Furthermore, flow cytometric analysis of bone marrow monocytes stimulated with EGC supernatant confirmed the increased expression of CD206 at protein level, while CCR2 expression was reduced, along with increased survival compared to control monocytes (Fig. 6e–h). These results were further underscored by the ability of EGC secreted factors to induce the expression of CD206 on sorted Ly6C+ monocytes from the muscularis 24 h post-injury and decrease the expression of CCR2 (Fig. 6i). Taken together, our ex vivo data suggest a direct interaction between EGCs and monocytes that could support the resolution of inflammation.
Fig. 6. EGCs stimulate the differentiation of monocytes into anti-inflammatory CD206+ Mφs in part via CSF-1 in vitro.
a Experimental outline of in vitro primary bone marrow monocytes stimulated with supernatant of EGCs. b Bone marrow-derived monocytes were stimulated for 24 h with/without supernatant of EGCs. Relative mRNA levels for pro- and anti-inflammatory cytokines normalized to the housekeeping gene rpl32 in bone marrow monocytes cultured with/without EGC supernatant. c Experimental outline of in vitro experiment using sorted Ly6C+ MHCII− monocytes stimulated with/without EGC supernatant for 24 h. d Ly6C+ MHCII− monocytes were sorted from the muscularis of WT mice 24 h after the induction of muscularis inflammation and were stimulated for 24 h with/without supernatant of EGCs. Relative mRNA levels of pro- and anti-inflammatory mediators normalized to the housekeeping gene rpl32 in sorted Ly6C+ MHCII− monocytes stimulated with/without EGC supernatant. e–h Bone marrow monocytes were cultured for 24–48 h with/without EGC supernatant and supplemented with anti-CSF1r antibody. e Contour plots of bone marrow monocytes showing expression of CD206 (top) or CCR2 (bottom) upon culture for 48 h with/without EGC supernatant and supplemented with anti-CSF1r antibody. f Brightfield images of monocytes upon culture for 24 h with/without EGC supernatant. g Quantification of 7-AAD+ cells in bone marrow monocytes cultured for 24 h or 48 h with/without EGC supernatant and supplemented with anti-CSF1r antibody. h Percentages of CCR2+ and CD206+ cells in live CD45+ CD11b+ Ly6G− CD64+ population. i Ly6C+ MHCII− monocytes were sorted from the muscularis of WT mice 24 h after the induction of muscularis inflammation and were cultured with/without EGC supernatant and supplemented with anti-CSF1r antibody for 48 h. Percentages of CCR2+ and CD206+ cells in live CD45+ CD11b+ Ly6G− CD64+ population. a–d T-test. g–i one-way ANOVA. *p < 0.05; **p < 0.01.
As cellular interaction analysis by NicheNet identified Csf1-Csf1r as the top candidate for potential ligand-receptor interaction between EGCs and monocytes (Fig. 5f), we next aimed to determine whether EGC-derived CSF1 had the potential to induce anti-inflammatory CD206+ Mφs in our ex vivo co-culture model by antibody-mediated CSF1r blockade. Anti-CSF1r treatment in combination with EGC supernatant attenuated the differentiation of both bone marrow and muscularis monocytes into anti-inflammatory Mφs with reduced CD206 expression, while increased CCR2 expression was only observed for bone marrow monocytes compared to cells stimulated with EGC supernatant alone (Fig. 6e–i). No effect was observed on survival, CCR2 and CD206 expression when bone marrow monocytes were treated with anti-CSF1r compared to control monocytes, while stimulation of bone marrow monocytes with CSF-1 increased survival and reduced CCR2 expression with no effect on CD206 expression (Supplementary Fig. 6f–h). These results underscore that CSF-1 together with other factors are crucial for the differentiation of monocytes into anti-inflammatory CD206+ Mφs. Altogether, these data provide evidence for a direct interaction between monocytes and EGC-derived ligands ex vivo which stimulate the differentiation into anti-inflammatory CD206+ Mφs.
EGCs are involved in the differentiation of monocytes into CD206± Mφs, which are able to limit damage to EGCs through CSF-1 signaling during muscularis inflammation
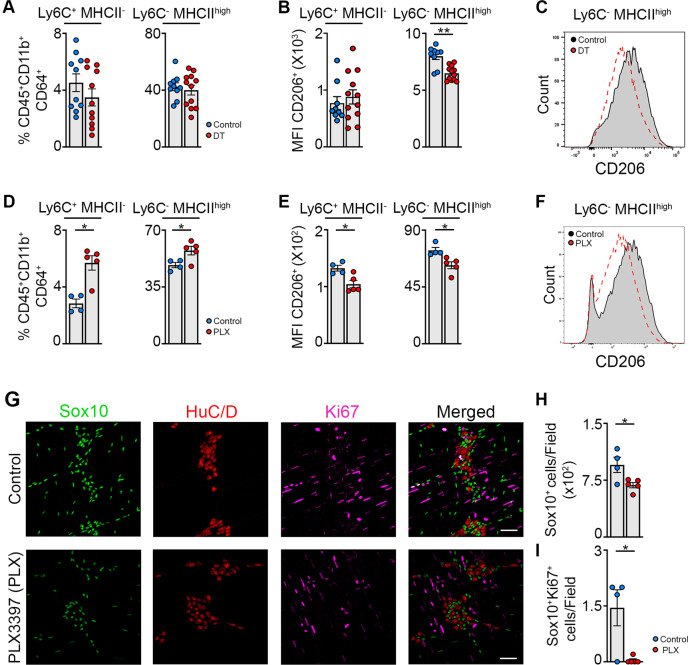
In order to definitively determine the contribution of EGCs in the differentiation of monocytes into anti-inflammatory CD206+ Mφs, 8-week-old PLPCreERT2/+ iDTR (PLP-iDTR) mice were injected with tamoxifen and a small part of the small intestine was exposed to saline or diphteria toxin (DT) at 12 weeks of age following the induction of muscularis inflammation. 72 h post-injury, there was a significant reduction of the volume and intensity of GFAP (Supplementary Fig. 7a, b) as well as in the number of Sox10+ EGCs (Supplementary Fig. 7a, c) in the DT treated PLP-iDTR mice compared to the control animals, thereby confirming EGC depletion upon DT administration. While no differences were observed in the percentage and cell numbers of the myeloid populations 72 h after the induction of muscularis inflammation (Fig. 7a and Supplementary Fig. 7d), there was a significant reduction of the MFI of CD206 in Ly6C− MHCIIhi Mφs of PLP-iDTR mice treated with DT compared with their control (Fig. 7b, c). These results are in line with our scRNA-seq data, where the highest CD206 expression was observed in the Mφ subpopulation that expresses high levels of MHCII genes. Therefore, we can conclude that EGCs are able to stimulate the differentiation of monocytes into CD206+ MHCIIhi Mφs in vivo during muscularis inflammation.
Fig. 7. EGCs are crucial for monocyte differentiation into anti-inflammatory CD206+ Mφs in part via CSF-1 in vivo.
a–c A small portion of the small intestine of PLPCreERT2 iDTR mice was exposed to saline or DT after intestinal manipulation and mice were sacrificed 72 h after the induction of muscularis inflammation. a Percentages of Ly6C+ MHCII− monocytes and Ly6C− MHCIIhi Mφs in live CD45+ CD11b+ Ly6G− CD64+ population. b Mean fluorescent intensity (MFI) of CD206 in Ly6C+ MHCII− monocytes and Ly6C− MHCIIhi Mφs. c Histogram of CD206 expression in Ly6C− MHCIIhi Mφs in control and DT exposed PLPCreERT2 iDTR mice 72 h after muscularis inflammation. d–h Wild-type mice were gavaged daily with vehicle or PLX-3397 (50 mg/kg) starting from the day of the manipulation and sacrificed 72 h after the induction of muscularis inflammation. D Percentages of Ly6C+ MHCII− monocytes and Ly6C− MHCIIhi Mφs in live CD45+ CD11b+ Ly6G− CD64+ population. e Mean fluorescent intensity (MFI) of CD206 in Ly6C+ MHCII− monocytes and Ly6C− MHCIIhi Mφs. f Histogram of CD206 expression in Ly6C− MHCIIhi Mφs in control and PLX treated mice 72 h after muscularis inflammation. g Immunofluorescent images of muscularis whole-mounts in control and PLX treated mice 72 h after muscularis inflammation stained for SOX10 (green), HuC/D (red) and Ki-67 (purple). Scale bar 25 µm. h Quantification of the number of SOX10+ cells per field in control and PLX treated mice 72 h after muscularis inflammation (average of 4–5 pictures/mouse). i Quantification of the number of Ki-67+ SOX10+ cells per field in control and PLX treated mice 72 h after muscularis inflammation (N = 4–5 pictures/mouse). a, b; d, e; h, i. T-test. *p < 0.05; **p < 0.01.
Based on the evidence that EGC-derived CSF-1 is able to stimulate the differentiation of monocytes into CD206+ Mφs with an anti-inflammatory phenotype in vitro, we hypothesized that blocking monocyte differentiation towards anti-inflammatory CD206+ Mφs in vivo via CSF1R inhibition might have neurodegenerative effects on EGCs. To test this hypothesis, WT mice were treated daily with pexidartinib (PLX-3397), an oral tyrosine kinase inhibitor of CSF1R, from the day of the induction of muscularis inflammation until the sacrifice at 72 h post-injury. Although long-term CSF1R inhibition can lead to Mφ depletion in vivo30,31, short-term PLX-3397 treatment did not result in an overall reduction of differentiated Mφs. Rather, we observed an increased percentage and cell number of neutrophils, monocytes and MHCIIhi Mφs compared to control treated animals 72 h post-injury (Fig. 7d and Supplementary Fig. 7e, f), showing normal development of the Mφ compartment with increased muscularis inflammation. These results can be explained by a reduced expression of CD206 in both monocytes and MHCIIhi Mφs in PLX treated mice compared to control mice (Fig. 7e, f), indicating a more pro-inflammatory phenotype of these myeloid subsets. As anti-inflammatory Mφs are essential to support the neuronal compartment in the muscularis externa7, we next sought to evaluate the effect of diminished CD206 expression on MHCIIhi Mφs on EGC growth and survival upon PLX treatment. Based on the immunofluorescent quantification of Sox10+ and Sox10+ Ki-67+ EGCs, we could observe that there was a reduction of Sox10+ EGCs in the PLX treated mice as compared to the control mice. In addition, in WT mice there was a small subset of EGCs that was Ki-67+ 72 h post-injury which was significantly reduced upon PLX treatment (Fig. 7g–i). A similar reduction in the number of Sox10+ EGCs was also observed in CCR2−/− mice as compared to WT mice 72 h after the induction of muscularis inflammation (Supplementary Fig. 7h, i). Based on these results, we conclude that EGCs are involved in the differentiation of monocytes into CD206+ Mφs in vivo, limiting damage to the EGCs during muscularis inflammation.
Discussion
Previous studies have shown that monocyte-derived Mφs are essential to resolve inflammation and restore gut motility following muscularis inflammation11,21. To date, it remains to be determined which Mφ subpopulations are responsible for tissue recovery following muscularis inflammation and which factors induce the pro-resolving Mφ phenotype. Our work provides a comprehensive overview of the differentiation of incoming monocytes into pro-resolving Mφs during muscularis inflammation, thereby characterizing gene expression dynamics during monocyte-to-Mφ transitions and uncovering transcriptional regulators of different Mφ subpopulations. Furthermore, we have identified a role for EGCs during muscularis injury, which via increased production of the monocyte chemoattractant CCL2 and elevated levels of multiple mediators could be involved in the differentiation of monocytes towards anti-inflammatory Mφs, thereby promoting tissue recovery.
Using time-resolved scRNA-seq of immune cells isolated from the inflamed muscularis, we revealed the heterogeneity and differentiation trajectory of different monocyte and Mφ subpopulations at homeostasis and during early and late inflammation. During acute muscularis inflammation, Ly6c+ monocytes were present along with other transient monocyte and Mφ subpopulations such as Fabp5+ and Arg1+ Mo/Mφs, showing substantial heterogeneity in monocytic cells during acute inflammation, rather than a single homogenous wave of Ly6c+ monocytes. Additionally, we identified Ccr2+ int Mφs as a transitional state between Ly6c+ monocytes and Cd206+ Mφs. These intermediate cells were mainly present near the branching point or in the branch of the trajectory diverging towards Cd206+ Mφs and resembled Ly6C+ MHCII+ immature Mφs characterized by flow cytometry. Furthermore, we identified two main pro-resolving Mφ subpopulations during the resolution of muscularis inflammation characterized by Cd206+ MhcIIhi and Timp2+ MhcIIlo gene signatures. Trajectory interference analysis during muscularis inflammation showed that these subpopulations are derived from infiltrating Ly6c+ monocytes. In line, scRNA-seq analysis showed that these two myeloid subpopulations are absent from the inflamed muscularis of CCR2 deficient mice, further supporting their monocytic origin. Accordingly, flow cytometric analysis of CX3CR1GFP/+ mice during muscularis inflammation showed that CX3CR1lo MHCIIhi and CX3CR1lo MHCIIlo Mφs are derived from CCR2+ monocytes and resemble Cd206+ and Timp2+ Mφs, respectively. Both phenotypically and functionally, Cd206+ MhcIIhi Mφs showed high resemblance to homeostatic Cx3cr1+ Mφs with a similar gene signature and activity of specific regulons (e.g., Junb, Irf8)32,33, while Timp2+ MhcIIlo Mφs displayed a different gene signature and regulon activity (e.g., Cebpb, Gata6)34,35. In this regard, Cd206+ Mφs were shown to have high regulon activity of Irf8, a transcription factor involved in the regulation of genes important for antigen-presentation (Cd86 and Cd40) and for lymphoid and myeloid chemotaxis (Cxcl9, Cxcl10, Ccl4, Ccl8, and Ccl12)33. In addition, these cells upregulated genes involved in efferocytosis (Axl and MerTK) and exhibited high MhcII gene expression. These data suggest that CD206+ Mφs might respond to apoptotic cells in their vicinity and undergo differentiation into an anti-inflammatory phenotype in response to efferocytosis. On the other hand, Timp2+ Mφs have high regulon activity of Cebpb, which typically controls the expression of anti-inflammatory genes such as Msr1, Il10, II13ra, and Arg1. Further studies on small and large peritoneal Mφs could be instrumental in understanding Mφ biology during muscularis inflammation, as these cells resemble Cd206+ and Timp2+ Mφs respectively, based on gene expression and regulon activity36,37. Furthermore, we also found other subpopulations that were characterized by highly specific gene signatures and regulon activities, such as Fabp5+ Mφs with enhanced Nr1h3 (LXRα) and Pparγ regulon activity, which may be involved in iron handling (Flt1, Fth1, Hmox1) and detoxification (Mt1, Akr1a1, Prdx1, Cd36). Further studies will be essential to unravel the functional roles of these different Mφ subsets during muscularis inflammation by defining their precise tissue localization and assessing their contribution to the pathophysiological response during local tissue damage.
The muscularis micro-environment has been shown to be highly regulated by a network of enteric neurons and EGCs constituting the ENS. Here, Mφs play an essential role to support neuronal function via the secretion of BMP-21,6, while neurons in turn produce CSF1, vital for Mφ maintenance6. In the current study and in line with Grubisic et al., we have shown that CSF1 can also be produced by EGCs19. This close neuro-immune interaction between the ENS and Mφs at homeostasis and during disease resembles the one found in other neuronal tissue niches. For instance, in the central nervous system, there is extensive crosstalk between astrocytes and microglia, critical for brain homeostasis38. Upon brain injury, microglia are the first cells to be recruited to the site of damage to phagocytose dead cells and debris39, similar to what is observed during muscularis inflammation11,21. Concurrently, reactive astrocytes are activated to release pro-inflammatory mediators that affect microglia and can recruit different leukocytes. Subsequently, astrocytes increase their expression of GFAP and undergo morphological changes leading to the formation of glial scars40. A recent report by Liddelow et al. proposed astrocyte polarization into A1 and A2 states as a response to various stimuli27. In line with our findings, they showed that IL-1β and IL-1α induce the expression of most pan-reactive marker genes and the A2 marker genes along with a minority of A1 markers including Gfap. Similarly, we have shown a comparable process taking place at the level of the ENS, in which EGCs are activated during the early stages of intestinal inflammation by damage to the micro-environment. In line with previous work from Stoffels et al. 41, our GSEA analysis confirmed that early activation of EGCs during muscularis inflammation is dependent upon IL-1 cytokines. Furthermore, these damage-activated EGCs closely interact with Mφs in the muscularis and act as safeguards of the micro-environment by initiating an inflammatory response via CCL2 to protect the tissue from further damage. As opposed to the detrimental effects of astrocytes in brain injury, EGCs induce monocyte differentiation towards anti-inflammatory CD206+ Mφs by the production of cytokines and growth factors, including CSF1. This process seems to be crucial for the resolution of ENS damage, as Mφs are able to stimulate EGC proliferation in vitro and depletion of EGCs in vivo leads to reduced expression of CD206 in MHCIIhi Mφs. Recently, it was also shown that during experimental autoimmune encephalomyelitis, astrocytes provide soluble signals to pro-inflammatory Mφs to induce an anti-inflammatory phenotype42. In line, blocking CSF1-CSF1R signaling (using an oral CSF1R inhibitor, PLX-3397) increased monocyte and neutrophil infiltration in the muscularis during the recovery of muscularis inflammation, and reduced CD206 expression in MHCIIhi Mφs associated with reduced number of EGCs as well as EGC proliferation. Overall, EGC-derived CSF1 represents a potent signal to initiate anti-inflammatory Mφ differentiation essential for tissue recovery and for avoiding excessive injury to the ENS during muscularis inflammation.
Overall, these insights may pave the way to novel therapeutic approaches aiming to prevent complications related to alterations of the ENS and reduced GI motility in patients affected by intestinal inflammatory disorders. Our findings on the impact of anti-inflammatory signals from EGCs on myeloid cells could be extrapolated beyond the context of the intestinal inflammation and may be expanded to several other acute and chronic inflammatory conditions and autoimmune diseases.
Material & methods
Mouse Model
Twelve week old female wild-type (WT; C57BL/6 J), CCR2 knockout (CCR2−/−)43, Cx3cr1GFP/+ 44, Cx3cr1CreERT2 45, Rosa26-LSL-YFP46, PLPCreERT2 47, Rpl22HA 24, PLPCreERT2 47, iDTR48 and their littermate controls were used. All lines were bred and maintained at the KU Leuven animal facility on a 12:12-h light-dark cycle and had ad libitum access to tap water and commercially available chow (ssniff® R/M-H, ssniff Spezialdiäten GmbH). All experimental procedures were approved by the Animal Care and Animal Experiments Ethical Committee of KU Leuven.
Experimental model of small intestinal inflammation
An established model of intestinal manipulation was used to induce small intestinal muscularis inflammation as previously reported49. In brief, animals were anesthetized by intraperitoneal injection of ketamine (Ketalar 100 mg/kg; Pfizer) and xylazine (Rompun 10 mg/kg; Bayer). After a midline laparotomy, the cecum and the small intestine were carefully externalized avoiding the pancreas and the ileocecal valve and stretching of the colon or the stomach. Next, the entire length of the exteriorized small intestine was manipulated three times back and forth using a purpose-designed device that enables the application of a constant pressure to the intestine by a cotton swab applicator attached to its end.
Statistical analysis
Results are shown as mean ± standard error of the mean (SEM). Significance between two mean groups was determined by an unpaired two-tailed Student’s t-test or a non-parametric Mann–Whitney test, while one-way analysis of variance (one-way ANOVA) followed by Dunnett’s Multiple comparison test was performed to compare the mean of multiple groups. GraphPad Prism V.9.1.0 software (GraphPad Inc) was used to generate graphs and to perform statistical analysis.
Supplementary information
Acknowledgements
The authors would like to thank Iris Appeltans, Naomi Fabre, Tine Gommers and Karlien Vranken (TARGID, KU Leuven) for their technical assistance, and Pier Andrée Penttila and Reena Chinnaraj (FACS Core, KU Leuven) for their assistance with flow cytometry and sorting, the GIGA-Genomics platform (University of Liège) for their assistance in our scRNA-seq experiments. Images were recorded at the Cell and Tissue Imaging Cluster (KU Leuven) using a Zeiss LSM 880 – Airyscan (supported by Hercules AKUL/15/37_GOH1816N and FWO G.0929.15 to Pieter Vanden Berghe) and a Zeiss LSM 780 – SP Mai Tai HP DS (supported by Hercules AKUL/11/37 and FWO G.0929.15 to Pieter Vanden Berghe). BioRender was used for making graphical images.
Author contributions
G.M. and S.I. conceptualized this study; S.I., S.A., and M.S. designed the experiments; M.S., S.A., V.D.S., G.G., N.S., L.V.B., Q.W., D.P., J.K., L.M., I.P. and S.T. performed the experiments; analyzed data; M.S., S.A., S.I. and G.M. interpreted data; L.B., J.V.G., J.T., S.J., E.P., P.V.S. and T.M. provided vital reagents; M.H., P.V., J.V.G., G.B., J.T., S.J. and T.M. gave technical support and conceptual advice, M.S., S.A., S.I. and G.M. contributed to manuscript writing. All authors edited and revised the manuscript.
Funding
M.S. was supported by a PhD fellowship from the FWO-Research Foundation – Flanders (1186317 N). V.D.S. was supported by a postdoctoral fellowship in Fundamental Research by the Stichting tegen Kanker. G.G. was supported by a postdoctoral research fellowship of FWO. S.I. was supported by a MSCA-IF (79756–GLIAMAC) and a fellowship from the European Crohn’s and Colitis Organization (ECCO). G.M.’s lab was supported by FWO grants G0D8317N, G0A7919N, G086721N and S008419N, a grant from the KU Leuven Internal Funds (C12/15/016 and C14/17/097 and from the International Organization for the Study of Inflammatory Bowel Diseases (IOIBD) as well as a research grant from ECCO.
Data availability
Single cell RNA sequencing and bulk RNA sequencing data generated for this study are available through Gene Expression Omnibus with accession number GSE167465.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Michelle Stakenborg, Saeed Abdurahiman.
Contributor Information
Sales Ibiza, Email: sales.IbizaMartinez@uantwerpen.be.
Gianluca Matteoli, Email: gianluca.matteoli@kuleuven.be.
Supplementary information
The online version contains supplementary material available at 10.1038/s41385-022-00563-2.
References
- 1.Gabanyi I, et al. Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell. 2016;164:378–391. doi: 10.1016/j.cell.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano K, et al. Intestinal CD169(+) macrophages initiate mucosal inflammation by secreting CCL8 that recruits inflammatory monocytes. Nat. Commun. 2015;6:7802. doi: 10.1038/ncomms8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadis U, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Mowat AM. Publisher Correction: To respond or not to respond - a personal perspective of intestinal tolerance. Nat. Rev. Immunol. 2018;18:536. doi: 10.1038/s41577-018-0033-3. [DOI] [PubMed] [Google Scholar]
- 5.Zigmond E, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40:720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Muller PA, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Schepper S, et al. Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell. 2018;175:400–415.e413. doi: 10.1016/j.cell.2018.07.048. [DOI] [PubMed] [Google Scholar]
- 8.Kalff JC, et al. Intra-abdominal activation of a local inflammatory response within the human muscularis externa during laparotomy. Ann. Surg. 2003;237:301–315. doi: 10.1097/01.SLA.0000055742.79045.7E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jonge WJ, et al. Postoperative ileus is maintained by intestinal immune infiltrates that activate inhibitory neural pathways in mice. Gastroenterology. 2003;125:1137–1147. doi: 10.1016/s0016-5085(03)01197-1. [DOI] [PubMed] [Google Scholar]
- 10.Wehner S, et al. Inhibition of macrophage function prevents intestinal inflammation and postoperative ileus in rodents. Gut. 2007;56:176–185. doi: 10.1136/gut.2005.089615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farro G, et al. CCR2-dependent monocyte-derived macrophages resolve inflammation and restore gut motility in postoperative ileus. Gut. 2017;66:2098–2109. doi: 10.1136/gutjnl-2016-313144. [DOI] [PubMed] [Google Scholar]
- 12.Savidge TC, Sofroniew MV, Neunlist M. Starring roles for astroglia in barrier pathologies of gut and brain. Lab Invest. 2007;87:731–736. doi: 10.1038/labinvest.3700600. [DOI] [PubMed] [Google Scholar]
- 13.Neunlist M, et al. Enteric glial cells: recent developments and future directions. Gastroenterology. 2014;147:1230–1237. doi: 10.1053/j.gastro.2014.09.040. [DOI] [PubMed] [Google Scholar]
- 14.Bach-Ngohou K, et al. Enteric glia modulate epithelial cell proliferation and differentiation through 15-deoxy-12,14-prostaglandin J2. J. Physiol. 2010;58814:2533–2544. doi: 10.1113/jphysiol.2010.188409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coelho-Aguiar Jde M, et al. The enteric glia: identity and functions. Glia. 2015;63:921–935. doi: 10.1002/glia.22795. [DOI] [PubMed] [Google Scholar]
- 16.Bassotti G, Villanacci V, Antonelli E, Morelli A, Salerni B. Enteric glial cells: new players in gastrointestinal motility? Lab Invest. 2007;87:628–632. doi: 10.1038/labinvest.3700564. [DOI] [PubMed] [Google Scholar]
- 17.Ibiza S, et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature. 2016;535:440–443. doi: 10.1038/nature18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vainchtein ID, Molofsky AV. Astrocytes and microglia: in sickness and in health. Trends Neurosci. 2020;43:144–154. doi: 10.1016/j.tins.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grubisic V, et al. Enteric glia modulate macrophage phenotype and visceral sensitivity following inflammation. Cell Rep. 2020;32:108100. doi: 10.1016/j.celrep.2020.108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desalegn G, Pabst O. Inflammation triggers immediate rather than progressive changes in monocyte differentiation in the small intestine. Nat. Commun. 2019;10:3229. doi: 10.1038/s41467-019-11148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl Acad. Sci. USA. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haimon Z, et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 2018;19:636–644. doi: 10.1038/s41590-018-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JS, et al. A binary cre transgenic approach dissects microglia and CNS border-associated macrophages. Immunity. 2021;54:176–190.e177. doi: 10.1016/j.immuni.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 29.Mich JK, et al. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. Elife. 2014;3:e02669. doi: 10.7554/eLife.02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald KP, et al. An antibody against the colony-stimulating factor 1 receptor depletes the resident subset of monocytes and tissue- and tumor-associated macrophages but does not inhibit inflammation. Blood. 2010;116:3955–3963. doi: 10.1182/blood-2010-02-266296. [DOI] [PubMed] [Google Scholar]
- 31.Patwardhan PP, et al. Sustained inhibition of receptor tyrosine kinases and macrophage depletion by PLX3397 and rapamycin as a potential new approach for the treatment of MPNSTs. Clin. Cancer Res. 2014;20:3146–3158. doi: 10.1158/1078-0432.CCR-13-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontana MF, et al. JUNB is a key transcriptional modulator of macrophage activation. J. Immunol. 2015;194:177–186. doi: 10.4049/jimmunol.1401595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langlais D, Barreiro LB, Gros P. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J. Exp. Med. 2016;213:585–603. doi: 10.1084/jem.20151764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruffell D, et al. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl Acad. Sci. USA. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosas M, et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosn EE, et al. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl Acad. Sci. USA. 2010;107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cain DW, et al. Identification of a tissue-specific, C/EBPbeta-dependent pathway of differentiation for murine peritoneal macrophages. J. Immunol. 2013;191:4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matejuk A, Ransohoff RM. Crosstalk between astrocytes and microglia: an overview. Front Immunol. 2020;11:1416. doi: 10.3389/fimmu.2020.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 40.Sofroniew MV. Astrogliosis. Cold Spring Harb. Perspect. Biol. 2014;7:a020420. doi: 10.1101/cshperspect.a020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoffels B, et al. Postoperative ileus involves interleukin-1 receptor signaling in enteric glia. Gastroenterology. 2014;146:176–187.e171. doi: 10.1053/j.gastro.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 42.Locatelli G, et al. Mononuclear phagocytes locally specify and adapt their phenotype in a multiple sclerosis model. Nat. Neurosci. 2018;21:1196–1208. doi: 10.1038/s41593-018-0212-3. [DOI] [PubMed] [Google Scholar]
- 43.Boring L, et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J. Clin. Invest. 1997;100:2552–2561. doi: 10.1172/JCI119798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 2000;20:4106–4114. doi: 10.1128/MCB.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- 48.Buch T, et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 49.van Bree SH, et al. Novel method for studying postoperative ileus in mice. Int J. Physiol. Pathophysiol. Pharm. 2012;4:219–227. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell RNA sequencing and bulk RNA sequencing data generated for this study are available through Gene Expression Omnibus with accession number GSE167465.