Abstract
Wastewater-based epidemiology is an economical and effective tool for monitoring the COVID-19 pandemic. In this study we proposed sampling campaigns that addressed spatial-temporal trends within a metropolitan area. This is a local study of detection and quantification of SARS-CoV-2 in wastewater during the onset, rise, and decline of COVID-19 cases in Salta city (Argentina) over the course of a twenty-one-week period (13 Aug to 30 Dec) in 2020. Wastewater samples were gathered from 13 sewer manholes specific to each sewershed catchment, prior to convergence or mixing with other sewer lines, resulting in samples specific to individual catchments with defined areas. The 13 sewershed catchments selected comprise 118,832 connections to the network throughout the city, representing 84.7 % (534,747 individuals) of the total population. The number of COVID19-related exposure and symptoms cases in each area were registered using an application developed for smartphones by the provincial government. Geographical coordinates provided by the devices were recorded, and consequently, it was possible to geolocalise all app-cases and track them down to which of the 13 sampling catchments belonged. RNA fragments of SARS-CoV-2 were detected in every site since the beginning of the monitoring, anticipating viral circulation in the population. Over the course of the 21-week study, the concentrations of SARS-CoV-2 ranged between 1.77 × 104 and 4.35 × 107 genome copies/L. There was a correspondence with the highest viral load in wastewater and the peak number of cases reported by the app for each catchment. The associations were evaluated with correlation analysis. The viral loads of SARS-CoV-2 in wastewater were a feasible means to describe the trends of COVID-19 infections. Surveillance at sewershed scale, provided reliable and strategic information that could be used by local health stakeholders to manage the COVID-19 pandemic.
Keywords: SARS-CoV-2, Wastewater, Monitoring, COVID-19, Wastewater-based epidemiology, SALTA COVID app
Graphical abstract
1. Introduction
The current pandemic coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) started in December 2019 in Wuhan, China. In few months, facilitated by global travel, the disease expanded to all the continents and almost all countries reported COVID-19 cases. The city of Salta, main city in the province of Salta, located in the northwest of Argentina, was no exception. On March 18, 2020, the first COVID-19 case was reported in a person returning from Spain. Virus spread remained under control with sporadic cases; however, on 26th August community transmission of the virus was declared. By the end of the year 2020, according to official reports, the virus had infected >22 thousand and killed 1024 people in Salta province (https://www.argentina.gob.ar/coronavirus/informe-diario). A high percentage of cases (47.6 %) occurred in Salta city where >631,000 citizens are settled.
From the beginning of the pandemic the medical and the scientific communities joined efforts to understand better the symptoms that would help to an early diagnose of the disease. Many symptoms were identified including diarrhoea, which led to the search of SARS-CoV-2 in stool samples. According to a review, where >73 hospitalised patients were studied, SARS-CoV-2 was detectable in stool in up to half of COVID-19 patients independently of gender and age (Xiao et al., 2020) as well as in stool of asymptomatic individuals (Jiang et al., 2020; Rampelli et al., 2020; Park et al., 2021).
Asymptomatic people represented (still does) a challenge for the health system around the world as they were silent virus spreaders and their number remained unknown, unless massive testing was performed. Some developed countries or small communities were able to control the viral dissemination; massive testing and strict tracking of contacts were the key to success. However, generalized massive testing is unpractical and impossible in most of the places due to the limitation of resources and to the speed of viral transmission. In this context, wastewater surveillance raised as a tool to assess the viral circulation in a community as faeces were carrying the virus excreted by symptomatic as well as by asymptomatic individuals (Medema et al., 2020; Thompson et al., 2020; Wannigama et al., 2021).
Several research groups around the world have reported the presence of SARS-CoV-2 RNA fragments in untreated wastewaters in the Netherlands (Izquierdo-Lara et al., 2021), Australia (Ahmed et al., 2020a), Italy (Rimoldi et al., 2020; Maida et al., 2022), France (Wurtzer et al., 2020), United States (Wu et al., 2020a; Gonzalez et al., 2020), Canada (Acosta et al., 2022), Mexico (Carrillo-Reyes et al., 2021) and elsewhere in South America it was reported in Brazil (Claro et al., 2021; Fongaro et al., 2021; Prado et al., 2021; Prado et al., 2020), Argentina (Barrios et al., 2021; Giraud-Billoud et al., 2021; Iglesias et al., 2021), and Chile (Ampuero et al., 2020).
Comparing or trying to correlate the concentration of SARS-CoV-2 in wastewater to the number of COVID-19 cases tested in the community (Mallapaty, 2020) is challenging due mainly to two limitations. The first one is that the viral concentration excreted in faeces is highly variable, especially in the first wave of COVID-19 when population have not yet developed immunity, as the value varied among patients and changed over time. Miura and collaborators estimated the median concentration of SARS-CoV-2 in faeces as 3.4 (95 % CI: 0.24–6.5) log copies per gram-faeces (Miura et al., 2021), and it was independent of the severity (or absence) of symptoms (Chen et al., 2020). The second important limitation was the variability of the timeframe for the persistence of SAR-CoV-2 in stools samples, with report of faeces remaining positive for 7 days (Chen et al., 2020), 26 days (95 % CrI: 21.7–34.9) (Miura et al., 2021), 33 days and as much as five weeks (Wu et al., 2020b). Despite those difficulties, wastewater monitoring proved useful to identify COVID-19 hotspots, trends, and to deliver early warnings of SARS-CoV-2 outbreaks (Ahmed et al., 2020a).
Most frequently, wastewater samples are collected at the entrance of wastewater treatment plant (WWTP) facilities, as they represent a main sampling point for the total population served. Nevertheless, wastewater sampling performed at different points of the sewage network (sewershed), is better to detect COVID-19 hotspots within an urban settlement and to provide more useful information to control the dissemination of the disease (Hoar et al., 2022; Mao et al., 2020; Nghiem et al., 2020).
Here we present a local study of detection and quantification of SARS-CoV-2 in wastewater during the onset, rise, and decline of COVID-19 cases in the city of Salta over the course of a twenty-one-week period (13 Aug to 30 Dec) in 2020 at the sewershed scale. In this study we proposed sampling campaigns that addressed spatial-temporal trends within a metropolitan area, and assessed their usefulness to detect and localise outbreaks, providing strategic information for public health policy.
2. Materials and methods
2.1. Sampling design
Salta city is located in a subtropical region in north-western Argentina. The estimated population in Salta city for 2020, according to previous census, was 631,058 inhabitants. Sanitation services cover the whole city, with >140,000 households connected to the sewage system. There are two wastewater treatment plants in Salta city. About 88 % of the collected sewage is treated in an aerobic sewage treatment plant located in the south-east of the city (WWTP-S), while the remaining 12 % (approx.) is treated in a system of waste stabilization ponds (WSP) in the north of the city (WWTP-N).
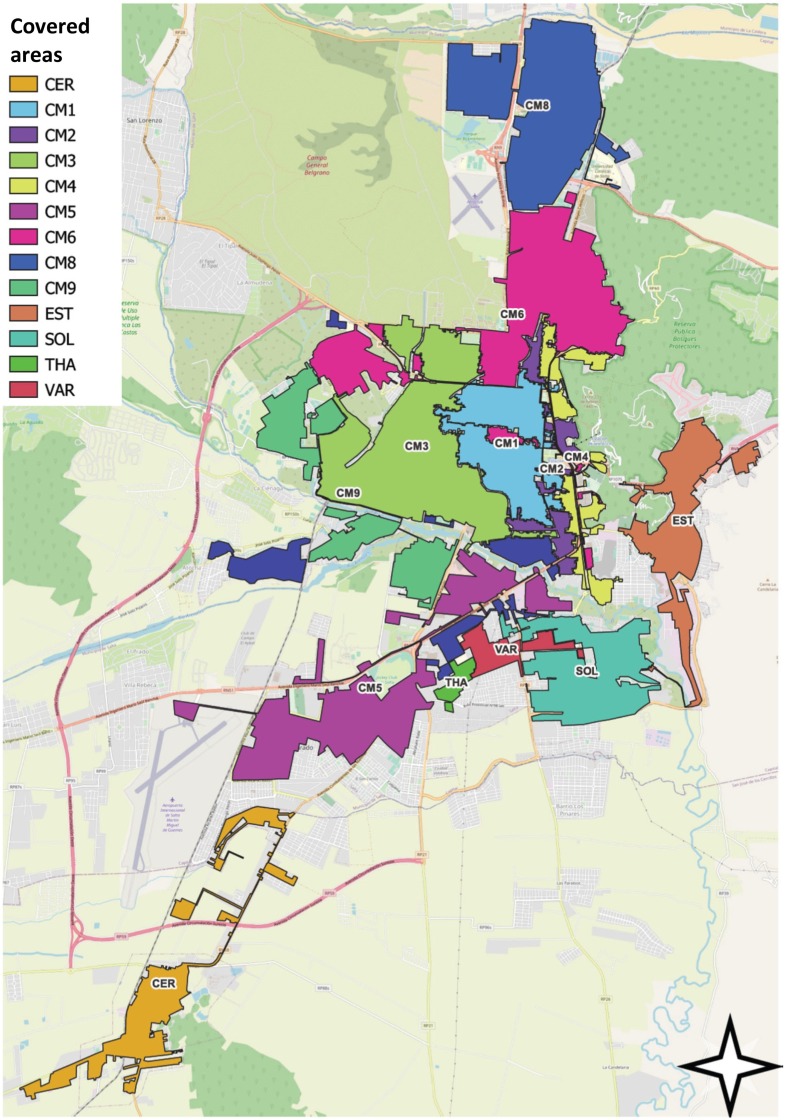
Wastewater samples were gathered (in a joint effort with the company in charge of sanitation in Salta City, Aguas del Norte) initially from 11 sites (CM1, CM2, CM3, CM4, CM5, CM6, CM8, CM9, EST, CER, SOL), and on 23 September two more (THA and VAR) were added to the sampling scheme. Thus, the monitoring involved a total of 13 sewer manholes specific to each sewershed catchment, prior to convergence or mixing with other sewer lines, resulting in samples specific to individual sub-catchments with defined communities (Fig. 1 ).
Fig. 1.
Map of Salta city. Thirteen areas, indicated by different colours, were identified as represented by wastewater collected at manholes: CM1, CM2, CM3, CM4, CM5, CM6, CM8, CM9, CER, SOL, EST, THA, VAR. Light grey areas indicate areas that were not covered by the design.
The 13 sewershed catchments selected contemplated 118,832 connected users to the network throughout the city, accounting for the 84.7 % (534,747 individuals) of the total population (Table 1 ). Therefore, only a 15 % of the population was not reflected in this study.
Table 1.
Selected sewershed catchments in Salta City. Connections and estimated population served considering a factor of 4.5 persons per household.
| WWTP discharge | Sewershed catchment ID | Connections | Population (inhabitants) | Estimated load (m−3 day−1) |
|---|---|---|---|---|
| North | CM8 | 18,214 | 81,963 | 23,605 |
| South | CM1 | 13,760 | 61,920 | 17,833 |
| CM2 | 4102 | 18,459 | 5316 | |
| CM3 | 21,299 | 95,846 | 27,604 | |
| CM4 | 5048 | 22,716 | 6542 | |
| CM5 | 14,433 | 64,949 | 18,700 | |
| CM6 | 17,502 | 78,759 | 22,683 | |
| CM9 | 9096 | 40,932 | 11,788 | |
| EST | 5788 | 26,046 | 7501 | |
| CER | 4425 | 19,913 | 5735 | |
| SOL | 1587 | 7142 | 2057 | |
| THA | 1203 | 5414 | 1559 | |
| VAR | 2375 | 10,688 | 3078 | |
| Total South | 100,618 | 452,784 | 130,396 | |
| Total city | 118,832 | 534,747 |
The sewershed catchment CM3 was the service area with the greatest number of houses connected (21,299) accounting for 18 % of the population included in this study (95,846 inhabitants). The second largest service area was CM8 with 15.3 % of users (81,963 inhabitants) connected. Meanwhile, the smallest catchment was THA with 5414 inhabitants (1 % of population) distributed in 1203 households (Table 1).
Wastewater was systematically monitored between 8 and 11 am biweekly from 13th of August to 30th of December 2020, during the first wave of COVID-19 in Salta. During that period, a total of 137 grab samples (500 ml) were collected from manholes using a pole and submerging a bucket into the flowing wastewater and later transferred into sterile bottles that were brought to the laboratory on ice. They were straightaway inactivated upon receipt in a water bath at 60 °C for 90 min following the procedure recommended by Wu and collaborators (2020a). After that, the concentration was performed.
2.2. Sample concentration, RNA extraction and quantification
Sample concentration was performed by adding 4 g of PEG 8000 (Sigma-Aldrich) and 0.9 g of NaCl (Biopack, Argentina) to 40 ml of inactivated wastewater, contained in 50-ml sterile tubes, and stirred for 5 min for total dissolution of PEG. Tubes were incubated overnight at 4 °C. After incubation, the suspension was centrifuged at 10,000 ×g until total dissolution of PEG (approximately 60 min), at 4 °C. Supernatant was discarded and the pellet resuspended in 450 μl of Trizol (Sigma-Aldrich). The final concentrate samples were immediately stored at −70 °C until further use.
RNA extractions were conducted as described in the manual by the manufacturer of Puro Virus kit (PBL, Productos Biológicos, Argentina). Briefly, 140 μl of concentrate sample in Trizol was used for RNA extraction with fast-spin columns holding silica membranes that bind specifically viral nucleic acids, and after three washing steps, nucleic acids were eluted in 60 μl of nuclease free water.
RT-qPCR was performed targeting the N gene of SARS-CoV-2 using primers and probe for N1 (F: GACCCCAAAATCAGCGAAAT; R: TCTGGTTACTGCCAGTTGAATCTG; P: FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1) from 2019-nCoV validated by the US Centers for Disease Control and Prevention CDC (Lu et al., 2020). Reaction mixtures (20 μl) contained 5 μl of 4× Taqman Fast Virus 1-Step Master Mix (Applied Biosystems), 0.16 μl of each primer, 0.5 μl of probe with final concentrations of 400 and 250 nM, respectively, and nuclease-free water plus 5 μl of extract sample. Each RT-qPCR run included a positive (nCoV-All plasmid, Eurofins) and a negative no template control and a control of nuclease free water (MilliQ water). All the amplification reactions were run in duplicate using the Step One Plus real time PCR system (Applied Biosystems, Inc. Foster city, CA). The thermal cycling protocol for RT-qPCR was as follows: initial reverse transcription of 5 min at 50 °C and 20 s at 95 °C, then 45 cycles at 95 °C for 15 s and 59 °C for 60 s.
The standard curve for SARS-CoV-2 quantification (−3.379x + 42.276; R2 = 0.993, dynamic range: 7) was generated using ten-fold serial dilutions of a SARS-CoV-2 linear plasmid control (nCoV-All) provided by Eurofins containing the complete nucleocapsid (N) gene. Limit of detection (LOD) was calculated by running serial dilutions over eight orders of magnitude in triplicate (LOD = 4 cg/reaction).
2.3. Prevalence of reported COVID-19 cases within WWTP sub-catchment areas
Under strict lockdown and government encouraging people to stay home, it is difficult to concur to health care facilities for COVID tests. Furthermore, qPCR swabs tests availability was restrained. Therefore, a remote COVID Symptom tracker app SALTA COVID (https://appcovid.salta.gob.ar/#/ui/home) was developed by the Modernization Secretary, a governmental institution, to give support to the Ministry of Health. This app was widely used to track COVID-related exposure and symptoms. Registered people were classified as at low, medium, and high risk according to their symptoms and their close contacts. Simultaneously, geographical coordinates of their location provided by the smartphone device was recorded (geolocation). Consequently, it was possible to localise each App-case and track them down to which of the 13 sampling catchments belonged. As SARS-CoV-2 can be released in faeces during 8 to 33 days (Miura et al., 2021; Wu et al., 2020b) we took an average of 14 days (active cases) and computed total app-cases within two-weeks periods before the sampling day.
2.4. Statistical analysis
All figures and analysis were performed using R Statistical software version 4.0.2 (R Core Team, 2020). Specifically, packages ggplot2 (Wickham, 2016) and ggpubr (Kassambara, 2020) were used for plotting, and dplyr (Wickham et al., 2020) for data manipulation and correlation analysis. Nonparametric Kendall's tau was used to test hypotheses regarding associations and trends as data were neither normally nor log normally distributed based on Shapiro-Wilk tests. Kendall correlation coefficient was calculated for each sewershed using corr function from stats R package.
Clinical testing data (confirmed-cases) with qPCR swab tests were collected from Argentinean Sanitary Integrated System (SISA) through the Ministry of Health daily reports (www.datos.salud.gob.ar). These data were not geo-localised and were reported as total for the whole city. App-cases were gathered as described above.
Maps were drawn up with the free and open-source software Qgis 3.2 (https://www.qgis.org/en/site/).
3. Results and discussion
3.1. SISA (qPCR confirmed) vs App cases in Salta city
The first confirmed case of COVID-19 in Salta was reported on 18 March 2020 when a passenger returned from Spain and was kept in isolation. Strict restrictions were imposed the following months in all the country and the total accumulated confirmed cases since March to 12 August 2020, when wastewater monitoring started, were 168 for Salta city according to SISA (www.sisa.msal.gov.ar). It is important to note that from March to July, confirmed lab tested cases were sporadic and the average per week was below ten cases.
As it is well known, the number of confirmed cases depends on the positive rate (total amount of cases/number of tests performed). During the peak of cases the national government reported a positivity rate above 50 % (https://www.argentina.gob.ar/coronavirus/informes-diarios/sala-de-situacion) (Fig. S1). Therefore, it was expected that the true number of cases was underestimated. A high positive rate indicates that the number of confirmed cases is likely to represent only a small fraction of the true number of infections. According to criteria published by WHO, a positive rate of <5 % is one indicator that the epidemic is under control (https://ourworldindata.org/coronavirus-testing) nevertheless a rising positive rate suggests the virus is actually spreading faster than the growth seen in confirmed cases. Consequently, the reported cases by SALTA COVID app were considered to reflect more accurately the actual epidemiological situation at that moment.
From 13 August to 30 December the total number of confirmed cases in Salta city was 10,375, meanwhile, the app reported cases were 28,729. Under the assumption that App cases reflected the actual number of cases, this means the confirmed cases were underestimated 2.7 times on average and only 36.1 % of SARS-CoV-2 infected individuals have sought medical care in Salta City. This is slightly higher than reported by other authors in the US where it was predicted that only 32 % of patients were tested (Silverman et al., 2020).
The difference was clearly higher during the peak weeks (9 to 30 Sep). According to SISA there were 1371 and 1670 confirmed cases for weeks of 16 and 23-Sept, respectively. However, the app reported 5092 and 5610 cases during the same weeks, indicating that clinically confirmed cases were respectively 3.7 and 3.4 times lower (Fig. 2 ) and positivity rate was between 46 and 53 % (Fig. S1).
Fig. 2.
Recorded cases of COVID-19 in Salta city by week for the study period. Cases confirmed by laboratory testing were recorded in the Argentinean Sanitary Integrated System (SISA), while symptomatic individuals were registered in the tracker app SALTA COVID (App).
Nevertheless, it is important to mention that passive surveillance of COVID-19 based on monitoring only clinical confirmed cases, always underestimated the actual number of cases since it depended on symptomatic infections. Thus, these rates were strongly biased since lab testing was only carried out on people who recognized their symptoms or had close contact with infected people and attended to the hospital or testing centres for swab examinations (Omori et al., 2021). Also, it is well-known that symptomatic infections depended on age. Even a greater bias was introduced because passive surveillance does not take into account asymptomatic infected people; indeed, there were estimations that there were four to five asymptomatic people per symptomatic one. Therefore, actual number of cases was difficult to be estimated.
3.2. App-cases by sewershed catchments
As app-reported cases were geo-localised, it was possible to follow the trend of cases precisely in each sewershed catchment. The number of viral particles shed in faeces depends on several variables. Among others, the severity of symptoms and the age affect the virus shedding kinetics (Omori et al., 2021). The virus can be released from body fluids from 8 to 33 days (Wu et al., 2020b) and according to viral RNA degradation rates, nucleic acids persist in wastewater during 8.04 to 27.8 days (Ahmed et al., 2020b). Therefore, the cumulative number of cases within the 14 days before the sampling date were considered as active cases that contribute to the viral load in the wastewater.
During the first two weeks of study, at least one case was reported in each sewershed catchment. The spike of number of cases was reported on the 23-Sep week in all catchments (Fig. S2) when 11 out 13 areas were above 100 cases. The highest number of cases was 1218 in sewershed CM3 as expected, since its service area covered for the greatest amount of people (95,845 individuals). However, normalised cases by catchment population (Fig. S3) showed the highest prevalence of 9.1 % in sewershed SOL and it was above 1.0 % until mid-November. It is important to consider that the major regional hospital, where most of the COVID-19 cases were treated is located within this zone. Therefore, it was expected that several cases were geo-localised in this area, increasing the figure number in this catchment. During that period the incidence was from 0.3 to 1.3 % in the other catchments. In six out of 13 areas, the incidence was above 0.8 % (Fig. S3) during the peak. The lowest number of cases in most of areas were found in the first two weeks of December; the occurrences (normalised by population) were around 0 (in three areas) to 0.008 %. By the end of the year, during the festive season, a slight increment in prevalence values between 0.01 and 0.04 % was observed in most of the serviced area, besides SOL where it was 0.13 %.
3.3. Spatial-temporal SARS-CoV-2 trends in sewershed catchments
Over the course of the 21-weeks study, the concentrations of CoV-2 were between 1.77 × 104 and 4.35 × 107 gc/L. These values are in line with the concentrations reported in Spain (Randazzo et al., 2020) and France (Wurtzer et al., 2020). Only in one sewershed (CM9) SARS-CoV-2 was not detected in two occasions. After the second detection event, further investigations established that an industrial effluent discharge was disturbing the measurements. Therefore, the sampling site was set 300 m upstream the original manhole, without impacting the residential users covered by the service area.
RNA fragments of SARS-CoV-2 were detected in every site since the beginning of the monitoring on 13 August 2020 (Fig. 4). Thus, these results revealed that there was viral community circulation in the city even though, the government confirmed such situation on 26 August, two weeks after the wastewater monitoring had started. During the first sampling dates, the highest viral load (9.62 × 105 gc/L) was found in sewershed CM8 where 26 app-cases were reported, and it was also the highest number of cases during that week. The lowest concentration was detected in sewershed CM6 (4.14 × 104 gc/L) where 6 app-cases were cumulative within two weeks before sampling date, in a total population of 78,759 inhabitants. That meant there was a high likelihood that if one case was actively shedding SARS-CoV-2 RNA, it could be detected in a catchment of 13,127 people. This demonstrate the platform is reliable and sensitive for detecting infections at the community scale, even when case prevalence is low (Hewitt et al., 2022). Interestingly, even though no cases were registered in sewershed CER, the viral load in wastewater was already 1.06 × 105 gc/L. Overall, during the same period, the total number of confirmed cases were 61 and 98 cases according to the app for the whole city.
Fig. 4.
Viral SAR-CoV-2 loading for each of the 13 sewershed (CER, CM1, CM2, CM3, CM4, CM5, CM6, CM8, CM9, EST, SOL, THA, VAR) with LOESS smoothing over the 21-weeks study period.
The viral concentration trends for each site showed rising and declining loads over the study period (Fig. 4). Within 42 days of monitoring a sharp increase in detections, with three orders of magnitude higher, was observed in each sewershed. Even though, the peak of cases reported was on 23rd September in each catchment, high viral loads in raw wastewater were determined two weeks before (9-Sep) in some of the sewershed (CER, CM1, CM2, CM4, CM5, and EST). Meanwhile, for the rest of the sites (CM3, CM6, CM8, and SOL) there was a correspondence between the highest viral load in wastewater and the peak number of cases reported by the app. Thus, the highest viral load 4.35 × 107 gc/L was detected in CM3 on 23rd September, this is expected since this sewershed served the greatest amount of people.
Nonetheless, when population normalised viral load (gene copies per capita) was considered it allowed assessing for catchment outbreaks and more precise trends at a finest scale were perceived (Fig. S4). A critical situation was detected in sewershed SOL where for one month (9-Sept to 7-Oct) viral loads per capita were above 103 gene copies/person. As mentioned before, the reference hospital of the city, where most of the COVID-19 cases were hospitalised, was located in this area. Therefore, more SARS-CoV-2 particles shedding by patients were expected to impact in the sewage. This fact proved the sensitivity of the methodology. Other zones with high viral loads per capita (>103) were CER, THA, VAR, CM2, and EST during September. These trends are easy to visualize and would help decision-makers to adopt specific measures in these areas to control the virus transmission (Tiwari et al., 2022).
Following the peak there was a sustained decrease in the viral concentrations in the raw wastewater analysed. It is important to note that from 21-September to 8-November, the government increased the lockdown restrictions that were lifted the previous months for winter holidays and religious festivities. Probably, those relaxed measures allowed the viral transmission in the community. The containment measures had a positive effect since the viral load in wastewater and COVID-19 cases were in gradual decline.
At the beginning of December, within the first two weeks, the lowest concentrations were detected in most of the sites. Indeed, concentrations were even one order of magnitude below the first detection performed in August in several sewershed.
During the festive season, on 30th December detections of SARS-CoV-2 particles increased in all the sewershed catchments. These results were in agreement with the number of cases reported by the app (Fig. 3 ).
Fig. 3.
Fourteen-days cumulative app-cases reported by sewershed catchment (CER, CM1, CM2, CM3, CM4, CM5, CM6, CM8, CM9, EST, SOL, THA, VAR) during the study period.
3.4. Correlation of SARS-CoV-2 RNA viral load in wastewater with COVID-19 disease burden at sewershed scale
Trends of SARS-CoV-2 RNA viral load in wastewater mirrored the number of cases reported by the Salta COVID-app in most of the sewershed (except CM9) during the 21-week time frame of the study (Fig. 3, Fig. 4 ). Wastewater testing captured inputs from all the individual (including those with mild or asymptomatic infections) in the local community and thus, had the potential for estimating the true prevalence of COVID-19 (Nemudryi et al., 2020), independently of lab confirmed cases and of if infected people had symptoms or not.
Therefore, we analysed the strength of association between viral concentration in wastewater and cases reported by the app using Kendall rank correlation analysis to test if monitoring sewershed for SARS-CoV-2 provides a useful epidemiological metric that could alert public health officials about emerging outbreaks at different areas of the city (Fig. 5 ).
Fig. 5.
Kendall rank correlation analysis for each sewershed with LOESS smoothing confidence intervals. Dots represent experimental data from: CM1, CM2, CM3, CM4, CM6 (n = 11); CER, CM5, CM8, CM9, EST, SOL (n = 10); THA and VAR (n = 7). Blue lines indicate the trend-lines (indicated by the equation), τ: Kendall's rank correlation coefficient, p: denotes the p-value of τ under the null hypothesis of no association (when p < 0.05, reject null hypothesis).
Statistically significant and positive correlations were measured in every sampled sewershed (besides CM9) with different association strengths. According to the correlation coefficients values, strong associations (τ > 0.85 and p < 0.001) were found in five sewershed: CM2, CM3, CM6, EST, and SOL. Four sampling sites showed moderate associations (0.85 < τ < 0.70) and were measured in CER (p = 0.004), CM4 (p = 0.005), CM8 (p = 0.004) and THA (p = 0.01). Weaker associations (0.70 < τ < 0.55) were calculated in CM1 (p = 0.016), CM5 (p = 0.017) and VAR (p = 0.034). Some authors have reported that bigger catchments show better association since small sewershed did no exhibit clear tendencies (Barrios et al., 2021; Gonzalez et al., 2020). In our sampling sites the catchments that covered the lesser number of users showed different behaviours. THA was the smallest sewershed (5414 inhabitants) and showed a moderate association (τ = 0.8), meanwhile VAR with double the people (10,688 persons) showed weak correlation (τ = 0.62). A high coefficient value (τ = 0.87) was observed in SOL with only 7142 people in the catchment. It is important to note that SOL had the highest median incidence rate due to the location of the reference hospital. Larger sewersheds as CM3 and CM6 and a small one as CM2, with similar median virus loads and incidence rates, showed strong association with τ values of 0.89, 0.93, and 0.88, respectively. Rusiñol et al. (2021) observed that small wastewater treatment plants (<24,000 inhabitants) had lower median loads of SARS-CoV-2 despite similar incidence of infection.
Even though correlations were measured in each study sewershed, we cannot directly relate to the total number of cases since not all the COVID-19 patients shed SARS-CoV-2 viral particles in their stools. However, this tool provides a more reliable view of the actual situation of prevalence of the disease across the city. Passive surveillance of COVID-19 relies only on tracking clinical test results, but this involves intrinsic delays, and is highly dependent of the capacity of clinical testing.
3.5. Spatial-temporal viral load evolution across Salta city during the first COVID-19 wave in 2020
To maximize the utility of wastewater monitoring, heat-maps of the viral concentration data from the 13 sewershed catchments by week sampled were represented (Fig. 6 ). These maps were helpful to rapidly visualize and identify “hot-spots” of the city where the disease was spreading faster, and measurements needed to be implemented. This finer resolution (at sewershed level) of a large geographic region (city with >600 thousand inhabitants) allowed for localised public health interventions. For example, lockdowns could be implemented by neighbourhoods rather than the whole city.
Fig. 6.
Spatial-temporal viral concentration (VC) in the city of Salta along the monitoring campaigns in 2020. Green: 104 gc/L ≤ VC < 105 gc/L; yellow: 105 gc/L ≤ VC < 106 gc/L; orange: 106 gc/L ≤ VC < 107 gc/L; red: 107 gc/L ≤ VC < 108 gc/L. Grey areas (crossed lines) indicate areas that were not covered by the design.
Even more, considering that the positive rate was a good metric for how adequately countries were testing when there was scarcity of resources (for testing), these heatmaps could help health-care staff to identify areas where more tests need to be applied. Thus, in low-income countries where the positive rate was high during outbreaks, this tool was suitable to select areas to increase testing and therefore, to properly monitor and control the spread of the virus.
Even though over the period of study, the average viral loading trend mirrored the total number of cases (Fig. 7 ), it is important to highlight the usefulness of identifying specific spots to take actions and to efficiently manage the pandemic. It is important to note that this approach may be useful in modelling future outbreaks of SARS-CoV-2 or any other pathogen (enteroviruses and bacteria) shed in faeces.
Fig. 7.
Weighted average loading by population data from all 13 sewershed catchments and daily number of cases registered for Salta city.
3.6. Relevance of sewershed surveillance in threshold countries
The usefulness and advantages of using wastewater-based epidemiology (WBE) as a surveillance tool to track severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cases in a city were demonstrated during the COVID-19 pandemic. Moreover, as Safford et al. (2022) highlighted, it is a cost-effective way to track disease trends, while replacing some clinical testing could save billions of dollars without compromising surveillance accuracy. Therefore, this tool should be helpful for many low- and middle-income countries with scarce resources to clinically test their communities. Indeed, for such countries WBE may represent the only viable means of effective population surveillance (Hart and Halden, 2020; Usman et al., 2020). Paradoxically, according to Naughton and collaborators, WBE monitoring is primarily carried on in high-income countries (65 %) and only 35 % of low- and middle-income countries have access to this valuable tool (Naughton et al., 2021).
Some limitations of WBE include the potential to trace amounts of viral RNA shed from an infected individual going undetected in wastewater, to pinpoint the specific location, and the numbers of inhabitants in each monitored area (Kitajima et al., 2022). The uncertainty is even higher when samples are collected from inlets of wastewaters treatment plants at the city level. A way to decrease it is monitoring at the sewershed level, that means wastewater samples are collected from the area of lands where all the sewers of a neighbourhood flow to a single end point. Thus, hotspots could be easily identified and understanding the spatial changes of the disease trend could help public health officials to focus the mitigations efforts locally (Safford et al., 2022).
Most of the sewershed surveillance studies have been applied in developed countries; meanwhile, many Latin-American countries that used this tool, have predominantly sampled the inlets of wastewater treatment plants, sometimes combined with sewage from manholes or open sewage in marginal neighbourhoods (Barrios et al., 2021; Carrillo-Reyes et al., 2021; Fongaro et al., 2021; Giraud-Billoud et al., 2021; Iglesias et al., 2021; Mota et al., 2021; Prado et al., 2020; Prado et al., 2021). To the best of our knowledge, this is the only report in Latin-America executed at the sewershed level, where the whole city was segmented in 13 areas covering almost 85 % of the population connected to the network. In many Latin-American countries clinical testing of the surveyed population is scarce, deficient, and very expensive. In addition, considering that clinical testing programs only provide information on the subset of individuals who consent to testing, the estimation of the disease prevalence is biased and not useful to manage a pandemic. Furthermore, the use of technology could provide a way to overcome the lack of testing and help to pinpoint case location. In the province of Salta, the government developed a smartphone application that allowed individuals to record their symptoms and thus, geo-localise that case. Throughout collaborative multidisciplinary work data was accessible to our research group to further inform public health actions, perform meta-analysis, better coordinate, and determine equitable distribution of monitoring sites.
Finally, a great advantage of our approach is that after a period of collecting and analysing information and trends, it is possible to determine sentinel sewersheds that can be used for the efficient surveillance of the evolution of infectious diseases caused by pathogens following the oral-faecal route (i.e. poliovirus). In that case, resource-constrained WBE practitioners should also consider monitoring fewer sites more frequently, sacrificing some spatial granularity to achieve greater sampling frequency. As an outcome of this successful experience in the city of Salta, a surveillance program has been carried out up to date monitoring six manholes in a collaboration effort between the company in charge of wastewater treatment, the government (Department of Epidemiology), and our research group.
4. Conclusions
-
•
The app was a good estimator of the actual number of COVID-19 cases in Salta city and allowed to geo-localise cases across the metropolitan area, facilitating the identification of hot spots where virus spreading was occurring. This was highly useful when level of testing was low and positive rate was high therefore, confirmed lab cases were underestimated and did not represent the actual epidemiological situation.
-
•
Detection of SARS-COV-2 in wastewater was a good tool for surveillance at the sewershed scale thus, confined neighbourhood decision could be made.
-
•
The tool developed showed correlation with the number of cases reported during the study period.
-
•
We observed that viral loads of SARS- CoV-2 in wastewater were a feasible means to describe the trends of COVID-19 infections. Our results indicate that a SARS-CoV-2 wastewater surveillance at sewershed scale will provide reliable and useful information that can be used by local health stakeholders and policy makers to manage responses to the COVID-19 pandemic.
CRediT authorship contribution statement
Mercedes Cecilia Cruz: Conceptualization, methodology, data curation, formal analysis, data analysis, discussion of results, visualization, investigation, writing - review & editing.
Diego Sanguino-Jorquera: Sample collection, nucleic acid extraction, discussion of results, review and editing.
Monica Aparicio-Gonzalez: Samples concentration, discussion of results, review and editing.
Veronica Patricia Irazusta: Samples concentration, discussion of results, review and editing.
Hugo Ramiro Poma: Methodology, discussion of results, review and editing.
Héctor Antonio Cristóbal: Methodology, discussion of results, review and editing.
Veronica Rajal: Conceptualization, data analysis, discussion of results, resources, funding acquisition and administration, writing - review & editing.
Funding
This research was funded by Project COVID-19 233-785, from Fondo para la Investigación Científica y Tecnológica (FONCyT), Argentina. Diego Gastón Sanguino-Jorquera is recipient of doctoral fellowship from CONICET.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank all members of the network “Detection of Coronavirus in the Environment” coordinated by Dr. Carolina Vera from the Argentinean Ministry of Science and Technology (MINCyT) for helpful discussions. The authorities of Universidad Nacional de Salta supported this research allowing us to use the university vehicles for sampling; we are very thankful of all the driver's patience. Aguas del Norte personnel, especially Javier Jurado and Ana Cardozo, are gratefully acknowledged for selecting the monitoring points that would best cover the city and Gerardo Tarcaya (deceased) and the rest of the staff for the collection of samples. We would also like to thank the personnel from Epidemiology in the Public Health Ministry of the Salta Province, to Luis Miño and Jairo Martínez in particular for preparing the maps, from the Modernization Secretary, the Ministry of Education, and from the Secretary of Science and Technology of the Salta Province, for sharing the information and helping with the visualization of the app-cases. We also acknowledge Sarita Reyes for helping with the sample collection.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.160573.
Appendix A. Supplementary data
Supplementary figures
Data availability
Data will be made available on request.
References
- Acosta N., Bautista M.A., Waddell B.J., McCalder J., Beaudet A.B., Man L., Pradhan P., Sedaghat N., Papparis C., Bacanu A., Hollman J. Longitudinal SARS-CoV-2 RNA wastewater monitoring across a range of scales correlates with Total and regional COVID-19 burden in a well-defined urban population. Water Res. 2022;220 doi: 10.1016/j.watres.2022.118611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampuero M., Valenzuela S., Valiente-Echeverría F., Soto-Rifo R., Barriga G.P., Chnaiderman J., Rojas C., Guajardo-Leiva S., Díez B., Gaggero A. Infectious Diseases (Except HIV/AIDS) 2020. SARS-CoV-2 detection in sewage in Santiago, Chile - preliminary results (preprint) [DOI] [Google Scholar]
- Barrios M.E., Díaz S.M., Torres C., Costamagna D.M., Blanco Fernández M.D., Mbayed V.A. Dynamics of SARS-CoV-2 in wastewater in three districts of the Buenos Aires metropolitan region, Argentina, throughout nine months of surveillance: a pilot study. Sci. Total Environ. 2021;800 doi: 10.1016/j.scitotenv.2021.149578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Reyes J., Barragán-Trinidad M., Buitrón G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process Eng. 2021;40 doi: 10.1016/j.jwpe.2020.101815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Qi T., Liu L., Ling Y., Qian Z., Li T., Li F., Xu Q., Zhang Y., Xu S., Song Z., Zeng Y., Shen Y., Shi Y., Zhu T., Lu H. Clinical progression of patients with COVID-19 in Shanghai, China. J. Infect. 2020;80:e1–e6. doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claro I.C.M., Cabral A.D., Augusto M.R., Duran A.F.A., Graciosa M.C.P., Fonseca F.L.A., Speranca M.A., Bueno R.de F. Long-term monitoring of SARS-COV-2 RNA in wastewater in Brazil: a more responsive and economical approach. Water Res. 2021;203 doi: 10.1016/j.watres.2021.117534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fongaro G., Rogovski P., Pereira Savi B., Dorighello Cadamuro R., Faria Pereira J.V., Sant Anna I.H., Rodrigues I.H., Marques Souza D.S., Torres Saravia E.G., Rodríguez-Lázaro D., da Silva Lanna M.C. SARS-CoV-2 in human sewage and river water from a remote and vulnerable area as surveillance tool in Brazil. Food Environ. Virol. 2021 doi: 10.1007/s12560-021-09487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Billoud M., Cuervo P., Altamirano J.C., Pizarro M., Aranibar J.N., Catapano A., Cuello H., Masachessi G., Vega I.A. Monitoring of SARS-CoV-2 RNA in wastewater as an epidemiological surveillance tool in Mendoza, Argentina. Sci. Total Environ. 2021;796 doi: 10.1016/j.scitotenv.2021.148887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt J., Trowsdale S., Armstrong B.A., Chapman J.R., Carter K.M., Croucher D.M., Trent C.R., Sim R.E., Gilpin B.J. Sensitivity of wastewater-based epidemiology for detection of SARS-CoV-2 RNA in a low prevalence setting. Water Res. 2022;211 doi: 10.1016/j.watres.2021.118032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoar C., Chauvin F., Clare A., McGibbon H., Castro E., Patinella S., Katehis D., Dennehy J.J., Trujillo M., Smyth D.S., Silverman A.I. Monitoring SARS-CoV-2 in wastewater during New York City's second wave of COVID-19: sewershed-level trends and relationships to publicly available clinical testing data. Environ. Sci. Water Res. Technol. 2022;8(5):1021–1035. [Google Scholar]
- Iglesias N.G., Gebhard L.G., Carballeda J.M., Aiello I., Recalde E., Terny G., Ambrosolio S., L’Arco G., Konfino J., Brardinelli J.I. SARS-CoV-2 surveillance in untreated wastewater: detection of viral RNA in a low-resource community in Buenos Aires,Argentina. Rev. Panam. Salud Pública. 2021;45:1. doi: 10.26633/RPSP.2021.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Lara R., Elsinga G., Heijnen L., Munnink B.B.O., Schapendonk C.M.E., Nieuwenhuijse D., Kon M., Lu L., Aarestrup F.M., Lycett S., Medema G., Koopmans M.P.G., de Graaf M. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 2021;27:1405–1415. doi: 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Luo M., Zou Z., Wang X., Chen C., Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J. Med. Virol. 2020;92:1807–1809. doi: 10.1002/jmv.25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. ggpubr: 'ggplot2' based publication ready plots. R package version 0.4.0. 2020. https://CRAN.R-project.org/package=ggpubr
- Kitajima M., Murakami M., Kadoya S.S., Ando H., Kuroita T., Katayama H., Imoto S. Association of SARS-CoV-2 load in wastewater with reported COVID-19 cases in the Tokyo 2020 Olympic and Paralympic Village from July to September 2021. JAMA Netw. Open. 2022;5(8) doi: 10.1001/jamanetworkopen.2022.26822. e2226822-e2226822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida C.M., Amodio E., Mazzucco W., La Rosa G., Lucentini L., Suffredini E., Palermo M., Andolina G., Iaia F.R., Merlo F., Chiarelli M.G., Siragusa A., Vitale F., Tramuto F., Segreto D., Schembri P., Cuffari G., Conti A., Casamassima G., Polizzi A., Ferrara M., Gullo G., Lo Verde A., Russo A., Casuccio A., Costantino C., Restivo V., Immordino P., Graziano G. Wastewater-based epidemiology for early warning of SARS-COV-2 circulation: a pilot study conducted in Sicily, Italy. Int. J. Hyg. Environ. Health. 2022;242 doi: 10.1016/j.ijheh.2022.113948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580(7802):176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Mao K., Zhang K., Du W., Ali W., Feng X., Zhang H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opin. Environ. Sci. Health. 2020;17:1–7. doi: 10.1016/j.coesh.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., Kitajima M., Omori R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: re-analysis of patient data using a shedding dynamics model. Sci. Total Environ. 2021;769 doi: 10.1016/j.scitotenv.2020.144549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota C.R., Bressani-Ribeiro T., Araújo J.C., Leal C.D., Leroy-Freitas D., Machado E.C., Espinosa M.F., Fernandes L., Leao T.L., Chamhum-Silva L., Azevedo L., Morandi T., Tadeu G., Freitas O., Costa M.S., Carvalho B.O., Tulius M., Reis P., Melo M.C., Ayrimoraes S.R., Chernicharo C.A.L. Assessing spatial distribution of COVID-19 prevalence in Brazil using decentralised sewage monitoring. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C.C., Roman F.A., Jr., Alvarado A.G.F., Jr., Tariqi A.Q., Jr., Deeming M.A., Jr., Bibby K., Jr., Bivins A., Jr., Rose J.B., Jr., Medema G., Jr., Ahmed W., Jr., Katsivelis P., Jr., Allan V., Jr., Sinclair R., Jr., Zhang Y., Jr., Kinyua M.N., Jr. 2021. Show us the data: global COVID-19 wastewater monitoring efforts, equity, and gaps. MedRXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem L.D., Morgan B., Donner E., Short M.D. The COVID-19 pandemic: considerations for the waste and wastewater services sector. Case Stud. Chem. Environ. Eng. 2020;1 doi: 10.1016/j.cscee.2020.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori R., Miura F., Kitajima M. Age-dependent association between SARS-CoV-2 cases reported by passive surveillance and viral load in wastewater. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C.-W., Park D.-I., Woo H.-Y., Cheong H.S., Shin H.C., Ahn K., Kwon M.-J., Joo E.-J. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2021;19:1387–1394. doi: 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Maranhão A.G., Siqueira M.M., Miagostovich M.P. Preliminary results of SARS-CoV-2 detection in sewerage system in Niterói municipality, Rio de Janeiro,Brazil. Mem. Inst. Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., do Vale V.H., Braz R.M.S., de Andrade J.S.R., Maranhão A.G., Miagostovich M.P. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191 doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S., Biagi E., Turroni S., Candela M. 2020. Retrospective Search for SARS-CoV-2 in Human Faecal Metagenomes. Available at SSRN 3557962. [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language And Environment for Statistical Computing. https://www.R-project.org/ [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewater and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Zammit I., Itarte M., Forés E., Martínez-Puchol S., Girones R., Borrego C., Corominas Ll, Bofill-Mas S. Monitoring waves of the COVID-19 pandemic: inferences from WWTPs of different sizes. Sci. Total Environ. 2021;787 doi: 10.1016/j.scitotenv.2021.147463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford H.R., Shapiro K., Bischel H.N. Opinion: wastewater analysis can be a powerful public health tool—if it's done sensibly. Proc. Natl. Acad. Sci. 2022;119(6) doi: 10.1073/pnas.2119600119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J.D., Hupert N., Washburne A.D. Using influenza surveillance networks to estimate state-specific prevalence of SARS-CoV-2 in the United States. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lin Lee W., Rajal V.B., Haines M.B., Girones R., Ching Ng L., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A., Lipponen A., Hokajärvi A.-M., Luomala O., Sarekoski A., Rytkönen A., Österlund P., Al-Hello H., Juutinen A., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. Detection and quantification of SARS-CoV-2 RNA in wastewater influent in relation to reported COVID-19 incidence in Finland. Water Res. 2022;215 doi: 10.1016/j.watres.2022.118220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman M., Farooq M., Hanna K. Existence of SARS-CoV-2 in wastewater: implications for its environmental transmission in developing countries. Environ. Sci. Technol. 2020;54(13):7758–7759. doi: 10.1021/acs.est.0c02777. [DOI] [PubMed] [Google Scholar]
- Wannigama D.L., Amarasiri M., Hurst C., Phattharapornjaroen P., Abe S., Hongsing P., Rad S.M.A.H., Pearson L., Saethang T., Luk-in S., Kueakulpattana N., Storer R.J., Ounjai P., Jacquet A., Leelahavanichkul A., Chatsuwan T. Tracking COVID-19 with wastewater to understand asymptomatic transmission. Int. J. Infect. Dis. 2021;108:296–299. doi: 10.1016/j.ijid.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wickham H., François R., Henry L., Müller K. Dplyr: a grammar of data manipulation. R package version 1.0.2. 2020. https://CRAN.R-project.org/package=dplyr
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Vol. 5. 2020. p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J., Maday Y., Teyssou R., Richard E., Almayrac J., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020;25(50) doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Data Availability Statement
Data will be made available on request.