Abstract
Sugarcane accounts for a large portion of the worlds sugar production. Modern commercial cultivars are complex hybrids of S. officinarum, S. spontaneum, and several other Saccharum species, resulting in an auto-allopolyploid with 8–12 copies of each chromosome. The current genome assembly gold standard is to generate a long read assembly followed by chromatin conformation capture sequencing to scaffold. We used the PacBio RSII and chromatin conformation capture sequencing to sequence and assemble the genome of a South East Asian commercial sugarcane cultivar, known as Khon Kaen 3. The Khon Kaen 3 genome assembled into 104,477 contigs totalling 7 Gb, which scaffolded into 56 pseudochromosomes containing 5.2 Gb of sequence. Genome annotation produced 242,406 genes from 30,927 orthogroups. Aligning the Khon Kaen 3 genome sequence to S. officinarum and S. spontaneum revealed a high level of apparent recombination, indicating a chimeric assembly. This assembly error is explained by high nucleotide identity between S. officinarum and S. spontaneum, where 91.8% of S. spontaneum aligns to S. officinarum at 94% identity. Thus, the subgenomes of commercial sugarcane are so similar that using short reads to correct long PacBio reads produced chimeric long reads. Future attempts to sequence sugarcane must take this information into account.
Subject terms: Agricultural genetics, Evolutionary biology, Genome
Introduction
Sugarcane is an important crop species and is the major source of processed sugar in the world. The name sugarcane does not refer to a single species, but rather refers to any of several species in the genus Saccharum. Taxonomic classification of these species is difficult, confounded by many years of cultivation and cross breeding, which makes phenotypic classification unreliable, and has been a topic of much debate. It was originally considered that six species exist: S. spontaneum, S. robustum, S. officinarum, S. sinense, S. edule and S. barberi1. Although, now it is common for S. edule to be excluded from this genus because in situ hybridisation revealed it to be a likely hybrid between S. officinarum and S. robustum, leaving only five species1. Only two of these species, S. robustum and S. spontaneum, are considered wild species, and the remaining species are all cultivated2,3.
Modern commercial cultivars of sugarcane are complex hybrids of S. officinarum as the maternal donor crossed with S. spontaneum and, to a lesser extent, some other species and hybrids (for review see1,4,5). Sugarcane is believed to have originated in the South Pacific, but was widely dispersed by early explorers making it difficult to pinpoint the exact origin. It is believed that S. spontaneum originates from India, but can be found growing wild from eastern and northern Africa, through the Middle East, to India, China, South East Asia, and through the Pacific to New Guinea. The other wild species, S. robustum, is indigenous to New Guinea and can be found along river banks. It is considered that S. officinarum also most likely originated in New Guinea and was likely derived from S. robustum. Modern commercial cultivars have complex polyploid genomes as a result of many generations of hybridisation.
The genome of modern commercial sugarcane consists of ten unique chromosomes with 8–12 copies of each, giving a variable chromosome count of 100–1206–9. The genome of S. officinarum consists of ten unique chromosomes and a copy number of eight, 2n = 8x = 80 (GCA_020631745.1). The genome of S. spontaneum consists of eight unique chromosomes and a copy number of eight, 2n = 8x = 6410. Some variability exists for S. spontaneum chromosome number with cases of 9 and 10 chromosomes identified11,12. When S. officinarum is crossed with S. spontaneum the offspring have two copies of the S. officinarum chromosomes and one copy of the S. spontaneum chromosomes9. Both the S. officinarum and S. spontaneum genomes have a high degree of gene collinearity to S.orghum bicolor6,10,13,14. A one haploid S. spontaneum, AP85-441 1n = 4x = 32, genome was sequenced and assembled into the full set of 32 chromosomes with each homoeologue separated into a distinct chromosome10. This S. spontaneum has strong gene collinearity to S. bicolor, despite having two fewer chromosomes, because the reduction in chromosome count was the result of fission in two chromosomes and translocation to the ends of four other chromosomes with little to no rearrangements10. The strong similarity between sugarcane and S. bicolor led many to suggest that that S. bicolor can be used as an aid in assembling the sugarcane genome and S. bicolor was often used as a reference for sugarcane transcriptome sequencing.
A monoploid mosaic sugarcane genome was assembled by generating and sequencing a series of bacterial artificial chromosomes that contained inserts of sugarcane R570 variety15. These sequences were then assembled into a single set of ten chromosomes aided by gene collinearity with S. bicolor to scaffold contigs. The result of such an approach was that each of the ten chromosomes were assembled, but the sequence of any particular locus can come from any of the homoeologous chromosomes, including potential mixing of chromosomes from the ancestral species S. officinarum and S. spontaneum15. This genome assembly is a valuable resource, but lacks much of the genetic variation that exists in the whole genome. Sequencing and comparing the nuclear and mitochondrial genomes of multiple sugarcane cultivars may yield some insight into the history of sugarcane and provide a valuable resource for genetic improvement.
The work presented here began as a collaboration effort to sequence and assemble S. spontaneum, S. officinarum, and a commercial cultivar, widely used in Thailand, known as Khon Kaen 3 (KK3). The intent was to use the genomes of the progenitor species, S. spontaneum and S. officinarum, to aid in assembly and annotation of the commercial variety KK3. All three were sequenced simultaneously using PacBio and the assembly, annotation and comparative analysis of KK3 is presented here.
Materials and methods
Sample and DNA/RNA extraction
The sugarcane we used is a popular commercial hybrid that has been developed in Thailand known as Khon Kaen 3 (KK3). This cultivar was generated by crossing K84-200 (ROC1 × CP63-588) with 85-2-352 (SP70-1143 × Q76) and is a cultivar that is commonly used in Southeast Asia. Leaf tissue was collected, with all relevant permissions obtained and in accordance with all relevant guidelines and legislation, from a single plant and used for DNA extraction with the standard CTAB method followed by clean-up using a DNeasy Mini spin column from Qiagen. Total RNA was extracted using the standard LiCl method from young leaf, mature leaf, meristem, young stem, and mature stem.
Genome size estimation
The 2C DNA content was estimated by flow cytometry according to the protocol in Dolezel and Bartos16. Two reference species, Zea mays and Pisum sativum, were used for comparison.
Genome sequencing and assembly
DNA was used to prepare libraries for the PacBio RSII following the Pacific Biosciences ‘Procedure and Checklist—20 Kb Template Preparation Using BluePippin Size-Selection System’ protocol. DNA (10 µg) was sheared with a Covaris gTube, 4500 rpm for 2 min and the BluePippin cassette used was ‘0.75% DF Marker S1 high-pass 15–20 Kb’ with selection of 12–50 Kb. Sequencing was performed for 700 cells on the PacBio RSII.
Raw reads longer than 10 Kb (27,848,780 reads totalling 428.2 Gb) were used as seed reads and reads shorter than 10 Kb were used to correct them using CANU v1.317. The corrected long reads were then assembled using CANU v1.3 with an estimated genome size of 10 Gb. The assembly was then corrected using quiver. Default settings were used for each step as they are optimised for PacBio type data. Four Hi-C sequencing libraries were constructed following the steps described in Zhang et al.10 and sequenced on an Illumina HiSeq X Ten to generate 407,313,208 paired-end 150 bp reads. Hi-C was used to scaffold the assembled contigs into pseudo-chromosomes using the program ALLHiC, which is a version of HiRise software that was adapted for highly polyploid genomes18 and successfully used to assemble the S. spontaneum genome10. The ALLHiC parameters were derived from the data as described described in Zhang et al.10, with the exception that the k values were set to the expected number of haplotypes for KK3, even integers between 50 and 60 were tested, and the optimal k found to be 56.
RNA-seq and annotation
Total RNA from stem (cane stalk), from the 9th internode, 3rd internode, 1st internode, mature leaf, and young leaf was collected from KK3 and immediately frozen in liquid nitrogen. Sequencing libraries were prepared from each collected tissue according to Ion Total RNA-Seq Kit ptocols and sequenced on an Ion S5 XL System.
Annotation of KK3 was performed by EvidenceModeler v1.1.1 r2015-07-03 using both transcriptome based evidence and ab initio prediction19. The transcriptome data consisted of EST data from the sugarcane SUCEST project20,21 and RNA-seq from KK3, this data was mapped to the genome assembly using PASA2 v2.0.119. In addition, seven PacBio cells of RNA-seq was mapped to the genome assembly using GMAP22. The ab initio prediction program SNAP23 was trained using the PASA2 alignment assembly. All gene predictions were then combined by EvidenceModeler to generate consensus gene models using the following weights for each evidence type: PASA2 = 1; GMAP = 0.5; SNAP = 0.3.
Repeat sequence annotation
RepeatModeler version 1.0.8 was used to identify and classify de novo repeat families on the assembled genome24. The repeat sequences identified in the genome were aligned to Genbank’s non-redundant protein database (using BLASTX with an E-value cutoff of 1 × 10−6) to exclude repeat sequences that contain large families of protein-coding genes. Repeat masking was performed on the assembled genome by RepeatMasker version 4.0.6 (http://repeatmasker.org) using repetitive sequences in the RepeastMasker consensus library (20150807; http://www.girinst.org) and the custom repeat library generated by RepeatModeler.
Comparative genomics
The genome of S. spontaneum was aligned to the genome of S. officinarum using the NUCmer script of MUMmer325. The coords output of NUCmer was analysed to determine the total amount of mapped sequence and the percent identity of the two genomes. Multiple overlapping alignments of individual sequences were reduce to a single range using the GenomicRanges package in R to calculate the total percent of S. spontaneum that mapped to S. officinarum. The same method was applied to the individual wheat subgenomes26 to determine identity statistics. See supplementary File 1 for a step-by-step description.
The KK3 genome assembly was compared to the genome assemblies of S. officinarum and S. spontaneum by mapping the KK3 contigs separately using the two progenitor assemblies as a reference. The mapping was performed to each reference using BWA-mem27 with default settings and the minimum match score (-T) set to 1000. The CIGAR string from the resulting SAM file was parsed to extract the ranges of each KK3 contig that could be mapped to each of the genomes. Cases where overlapping parts of the KK3 contigs mapped to multiple locations in the reference were reduce to a single range using the GenomicRanges package in R to calculate the percent of the KK3 assembly that mapped to each reference. Similarly, to calculate the percent of each reference that matched KK3, reference contigs with overlapping ranges of KK3 match were reduced to a single range. Mapping results using BWA-mem with default settings (more stringent) was used to differentiate between KK3 contigs that mapped equally to both references allowing contigs to be more accurately grouped. See supplementary File 1 for a step-by-step description.
Results and discussion
Sugarcane genome assembly
The KK3 genome was estimated to be 10.8 Gb (2C) based on flow cytometry. We assembled the KK3 genome using long PacBio reads into 104,477 contigs totalling 7 Gb. The contigs ranged in size from 10 kb to 1.8 Mb and had an N50 of 83 kb. Raw reads were mapped back to the contigs revealing a median read depth of 13. Based on the read depth of each contig compared to the median read depth, we estimated that approximately 4.6 Gb of sequence was single copy and 2.4 Gb was multiple copy that represent collapsed sequence. The KK3 contigs were compared to the assembled genomes of S. officinarum and S. spontaneum to identify which progenitor species they most resembled. This identified 4.9 Gb of the genome to have S. officinarum origins and 2.1 Gb with S. spontaneum origins. In addition, this comparison suggested that 6819 contigs showed recombination between the two ancestral genomes, which is a substantially larger than expected number of recombinations. We performed the same mapping using the R570 monoploid sugarcane genome15, and found 407 recombinant contigs. Accounting for differences in chromosome number, the KK3 assembly had three times the number of recombinant contigs than the R570 monoploid genome. The R570 monoploid genome is chimeric by design and this fact, coupled with the following results and discussion, led us to conclude that the KK3 genome assembly is artifactually chimeric.
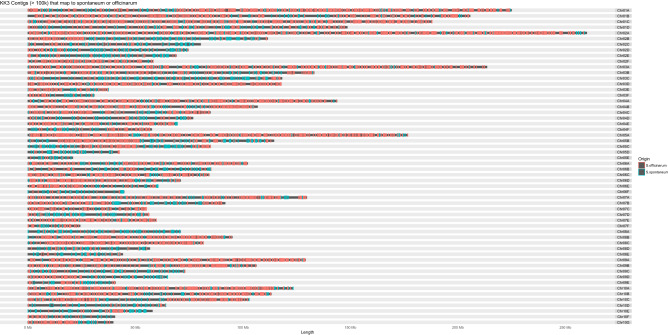
Scaffolding with Hi-C data produced 56 pseudo chromosomes containing 5.2 Gb, consistent with the 2C genome size estimation. There was 1.8 Gb of unplaced sequence, which consisted largely of alleleic sequence. Approximately 1.36 Gb (75%) of the unplaced sequence aligns to the pseudo chromosome scaffolds, leaving approximately 450 Mb of unplaced unique sequence. The unplaced sequence is mostly connected to allelic sequence, suggesting that it consists of prescence/absence alleles. Comparison of gene order and location in the 56 pseudo chromosomes to Sorghum bicolor was consistent with prior reports showing a high degree of gene colinearity (Fig. 1). However, all pseudo chromosomes contained sequence from both ancestral species (Fig. 2), which would require a large amount of recombination between the ancestral chromosomes. When only repeat masked sequence was analysed and plotted, the result looks the same (supplementary Fig. 1), meaning that the apparent recombination is not just repeat sequence or active transposon sequence. This was unexpected since Flourescence In Situ Hybridisation (FISH) and Genome In Situ Hybridisation (GISH) have shown that the genomes of each ancestral species remain largely isolated7,9. Genome in situ hybridisation has identified that 10–23% of the chromosomes of commercial sugarcane varieties are completely S. spontaneum7,9. However, when Cuadrado et al.7 used whole genome DNA from S. officinarum as one probe and whole genome DNA from S. spontaneum as another probe, they found that all chromosomes were labelled with the same probes because of high homology between the two species.
Figure 1.
Gene synteny between the KK3 pseudo chromosomes and Sorghum bicolor. Dot plot showing the relative location of each homologous gene between the KK3 chromosomes and Sorghum bicolor chromosomes. Chromosomes are scaled to maintain relative distance between genes, but remove size differences between chromosomes. The chromosomes of KK3 are numbered according to which S. bicolor chromosome they represent and assigned a letter to differentiate between homoeologs.
Figure 2.
Chromosomes of KK3 showing length with similarity to each ancestral genome indicated by color. Each KK3 chromosome is represented by a horizontal bar with the length indicated on the x-axis. The ancestral origin of each contig larger than 100 kb, based on sequence alignment results, is indicated by colored rectangles on each chromosome. The size of colored rectangles is proportionate to the relative size of the contig, but not proportionate to the actual bp size. Pink represents contigs that had more sequence overlap with S. officinarum and teal represents S. spontaneum.
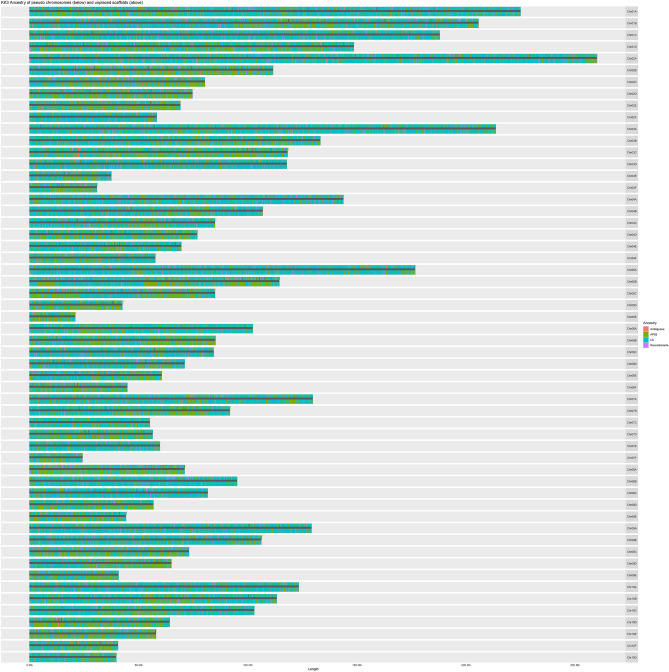
We first considered that sequence similarity between the homoeologous chromosomes may have resulted in paired reads linking allelic sequence from separate homoeologous chromosomes. This could cause artefactual mixing of homoeologous sequence in the scaffolding step while maintaining the correct gene order. If this was the case, the unplaced allelic contigs would likely be from the opposite ancestral species as the corresponding pseudo chromosome sequence. However, this was largely not the case as 59% of the aligned unplaced contig sequence was identified as originating from the same ancestral species as the pseudo chromosome sequence to which it aligns (Fig. 3). We next partitioned the contigs based on ancestral origin and performed Hi-C scaffolding on each partition separately. We expected this would reconstruct each ancestral set of full chromosomes and partial segments of any recombinant chromosomes if that was the true sequence. The scaffolding of contigs identified as having S. spontaneum ancestry was attempted first, with the expected chromosome number set to 15. This produced 10 large scaffolds that ranged in size from 122 to 264 Mb and five small scaffolds that ranged in size from 24 to 38 Mb.
Figure 3.
Chromosomes of KK3 showing two tracks per chromosome with the bottom track representing ancestry of scaffolded contigs and the top track showing ancestry of aligned unplaced contigs. Each KK3 chromosome is represented by a horizontal bar with the length indicated on the x-axis. The ancestral origin of each scaffolded contig, based on sequence alignment results, is indicated by colored rectangles on the bottom of each chromosome. The ancestral origin of each unplaced contig, based on sequence alignment results, is indicated by colored rectangles on the top of each chromosome. The size of colored rectangles is proportionate to the relative size of the contig, but not proportionate to the actual bp size. Teal represents contigs that had more sequence overlap with S. officinarum (LA), green represents S. spontaneum (AP85), pink represents contigs that mapped almost equally to both ancestors, and purple represents contigs that mapped half to one ancestor and half to the other.
The S. spontaneum genome (n = 8) has gene synteny to S. bicolor (n = 10) with two of the S. bicolor chromosomes each split in half and fused onto the ends of other chromosomes giving rise to the chromosome count of 810. Sequence data for S. spontaneum with n = 9 or n = 10 is not available, but similarly high gene synteny to S. bicolor would be expected as synteny is also found in S. officinarum and commercial cultivars. The obtained 10 large scaffolds could be consistent with ancestry of an S. spontaneum with n = 10. However, the gene order for this assembly relative to sorghum was substantially less colinear than the full assembly was, which means that partitioning the contigs in this way resulted in a lower quality assembly (Supplementary Fig. 2).
The amount of recombination observed in the KK3 assembly is much larger than would be expected, suggesting a problem with the assembly. The genome was assembled using only PacBio reads, which have a large error rate that is corrected by aligning short reads to the longest reads and correcting the long reads to generate consensus long reads28. If the progenitor genomes are similar enough, this error correction stage could result in reads from both subgenomes correcting long reads from both subgenomes and produce corrected long reads that are artificial chimeras of the two subgenomes. In such a situation, only the most divergent sequence between the two progenitors would produce accurate corrected long reads, while regions that are most conserved between the two would generate chimeric sequence skewing toward S. officinarum sequence. This process would explain the large amount of apparent recombination observed in the assembly and why there were so few S. spontaneum chromosomes found. This means that the homoeologous chromosomes are likely all chimeric and cannot be reliably used to dissect progenitor genome contribution. To investigate this possibility, the S. spontaneum genome was aligned to the recently assembled S. officinarum genome using NUCmer and it was found that 91.8% of the S. spontaneum genome aligned to the S. officinarum genome with an average nucleotide identity of 94.3%. This means that the error rate of the uncorrected PacBio reads (10–15% error rate)28 is higher than the percentage of difference between the two progenitor genomes and thus, the error correction step is probably producing chimeric reads rendering the entire assembly chimeric. The high amount of repeat sequence combined with the high ploidy also caused problems with the Hi-C paired sequence mapping (supplementary file 2) and may have generated scaffolds that contain homologous and homoeologous sequences, which would explain the large size variation within chromosome groups (Fig. 2).
The methods we used to assemble KK3 are essentially the methods that were used to successfully assemble the hexaploid wheat genome26. To investigate why it worked for wheat and not for sugarcane, we compared the individual subgenomes of hexaploid wheat to each other to add some perspective to the sugarcane progenitor genome similarity results. The wheat subgenomes are designated A, B and D26. We mapped the A genome against the B and D genomes and the B genome against the D genome. This showed that between 83.7 and 86.7% of each subgenome mapped to the other subgenomes at 91.2 to 91.4% identity, which is lower than the similarity between sugarcane progenitors, but still quite high. The wheat assembly project used short read sequencing to assemble contigs and PacBio reads to scaffold these contigs prior to using Hi-C to generate chromosome level scaffolds. Optical mapping of the wheat genome later identified the presence of some chimeric sequence and some sequence assigned to incorrect locations, which is likely to be, at least in part, caused by the high level of similarity between the subgenomes29. If a short read assembly method was used for KK3 sugarcane we would likely have a much larger number of correct shorter contigs. However, scaffolding using long reads would likely still produce chimeric scaffolds, similar to our current assembly. So the sugarcane genome may require an assembly approach where chromosomes are isolated and individually sequenced and assembled. Alternatively, updates to PacBio sequencing chemistry, known as HiFi reads (12–22 kb), occurred after the sequencing for this project was completed that sacrifice some read length for greatly increased accuracy, above 99%, and would avoid the chimeric long reads experienced in this work. However, the trade-off of shorter read length is considerably costly for a genome with such a high ploidy and repeat level and would result in reads not being able to span conserved allelic or repeat sequence, resulting in a more fragmented assembly. This means the high similarity of homoeologous chromosomes would likely still result in problematic allele mixing at repeat regions and regions of high sequence conservation. This would likely cause the generation of chimeric Hi-C scaffolded chromosomes, similar to a monoploid genome.
Sugarcane genome annotation
The genome was annotated by combining protein data from the progenitor species, EST data from the SUCEST database30, RNA-seq data, and ab initio gene prediction methods. This resulted in 242,406 genes, which is a large number of genes, but this number is high because the assembly produced 4–7 copies of each chromosome with 6 copies being the most common. Grouping this gene set using Orthofinder produced 30,927 groups of orthologous genes. While most chromosomes had five or six homoeologous copies, very few orthogroups had genes from each homoeologous chromosome and the most common distribution was for each othogroup to include genes from two to four homoeologous chromosomes. Most orthogroups had multiple genes that occurred within a single homoeologous chromosome, which likely reflects the problems of the assembly. The genome was annotated to consist of 56.13% repeat sequence with the most prevalent repeat type being LTR Gypsy at 27.8%, followed by LTR Copia at 15.4% (Table 1). RepeatMasker masked 60.13% of the genome (stretches less than 10 bp between repeats are also masked) and 4238 of these masked regions were larger than 20 kb. This is likely an under-representation considering that the sequenced genome was 7 Gb and the measured genome was 10.8 Gb, meaning over 3 Gb of sequence is likely to be collapsed allele or repeat sequence.
Table 1.
Repeat sequence annotated in KK3 genome.
| Repeat class | No. of elements | Total bp masked | % genome masked |
|---|---|---|---|
| DNA/CMC-EnSpm | 519,278 | 255,462,349 | 3.64 |
| DNA/MULE-MuDR | 240,450 | 77,168,806 | 1.10 |
| DNA/PIF-Harbinger | 721,693 | 153,386,043 | 2.19 |
| LINE/L1 | 236,740 | 133,767,239 | 1.91 |
| LTR/Copia | 685,340 | 1,083,387,117 | 15.43 |
| LTR/Gypsy | 1,357,965 | 1,951,756,200 | 27.80 |
| Unknown | 743,186 | 286,426,659 | 4.08 |
| Total | 4,504,652 | 3,941,354,413 | 56.15 |
Of particular interest to sugarcane farming is the identification of genes that affect sugar production or provide resistance. There were 17,316 resistance genes that were annotated in the KK3 genome and 1077 genes that are involved in the starch and sucrose metabolism KEGG pathway. There were 294 sugar transporter genes identified in KK3, which is only slightly more than the 286 that were found in S. spontaneum. The number of genes for most transporter families was similar or reduced compared to S. spontaneum, with the polyol transporter (PLT) and tonoplast sugar transporter (TST) families showing a larger number of genes (Table 2). Considering the chimeric nature of the assembly it cannot be determined which progenitor genome provided which gene.
Table 2.
Sugar transporter genes from S. spontaneum that were identified in KK3.
| Transporter group | S. spontaneum | KK3 |
|---|---|---|
| INT | 15 | 12 |
| pGlcT | 8 | 6 |
| PLT | 66 | 102 |
| SFP | 22 | 18 |
| STP | 83 | 78 |
| SUT | 19 | 8 |
| SWEET | 53 | 38 |
| TST | 13 | 32 |
| VGT | 7 | 0 |
| Total | 286 | 294 |
Conclusions
Assembling genomes from long read sequence data with scaffolding from Hi-C is the current gold standard in de-novo genome assembly and has produced chromosome level assemblies for a large number of species, including the hexaploid wheat genome. Application of this method to the auto-allopolyploid commercial sugarcane, KK3, appears to have produced a chimeric assembly because of a combination of consensus long read sequencing chemistry, the high ploidy level, and the high level of nucleotide identity between the genomes of the two progenitor species. Although the assembly produced from this work is unlikely to be a true reflection of what the actual genome should look like, it does at least capture much of the alleleic variation that exists in this commercial sugarcane and may be of some use when combined with one of the published monoploid mosaic sugarcane genomes or a future assembly that can overcome these problems. A good approach for sugarcane could be to isolate, sequence and assemble individual chromosomes. This work will hopefully benefit any future attempts to assemble difficult polyploid genomes, such as sugarcane.
Supplementary Information
Acknowledgements
The authors would like to acknowledge funding from the National Science and Technology Development Agency, Thailand, and Mitr Phol Sugarcane Research Center.
Author contributions
S.Tr., R.M., S.Ta., S.To., and J.R.S. conceived of the study. J.R.S., C.S., C.Na., C.Ng., J.Z., X.Z. performed bioinformatics work and prepared figures. W.P., T.Y., D.S., N.J., J.P., N.W., K.U., P.P., P.K., and K.S. performed sample collection, laboratory work and library construction. J.R.S. wrote the manuscript and all authors reviewed it.
Data availability
All raw read data was deposited in the NCBI SRA data archive under the bioproject: PRJNA820955. The genome assemble can be found in the NCBI genome archive: JALQSO000000000.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jeremy R. Shearman, Email: jeremy.she@biotec.or.th
Sithichoke Tangphatsornruang, Email: sithichoke.tan@biotec.or.th.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-24823-0.
References
- 1.Daniels J, Roach BT. Chapter 2—taxonomy and evolution. In: Heinz DJ, editor. Developments in Crop Science. Elsevier; 1987. pp. 7–84. [Google Scholar]
- 2.Irvine JE. Saccharum species as horticultural classes. Theor. Appl. Genet. 1999;98:186–194. doi: 10.1007/s001220051057. [DOI] [Google Scholar]
- 3.D’Hont A, Paulet F, Glaszmann JC. Oligoclonal interspecific origin of ‘North Indian’ and ‘Chinese’ sugarcanes. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2002;10:253–262. doi: 10.1023/A:1015204424287. [DOI] [PubMed] [Google Scholar]
- 4.Ming R, et al. Sugarcane improvement through breeding and biotechnology. In: Janick J, et al., editors. Plant Breeding Reviews. Wiley; 2005. pp. 15–118. [Google Scholar]
- 5.Purseglove JW. Tropical Crops: Monocotyledons. Longman Group Ltd.; 1979. [Google Scholar]
- 6.Aitken KS, et al. A comprehensive genetic map of sugarcane that provides enhanced map coverage and integrates high-throughput Diversity Array Technology (DArT) markers. BMC Genomics. 2014;15:152. doi: 10.1186/1471-2164-15-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuadrado A, Acevedo R, Moreno Díaz de la Espina S, Jouve N, de la Torre C. Genome remodelling in three modern S. officinarum × S. spontaneum sugarcane cultivars. J. Exp. Bot. 2004;55:847–854. doi: 10.1093/jxb/erh093. [DOI] [PubMed] [Google Scholar]
- 8.Jannoo N, Grivet L, David J, D’Hont A, Glaszmann J-C. Differential chromosome pairing affinities at meiosis in polyploid sugarcane revealed by molecular markers. Heredity. 2004;93:460–467. doi: 10.1038/sj.hdy.6800524. [DOI] [PubMed] [Google Scholar]
- 9.Piperidis G, Piperidis N, D’Hont A. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol. Genet. Genomics. 2010;284:65–73. doi: 10.1007/s00438-010-0546-3. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018;50:1565–1573. doi: 10.1038/s41588-018-0237-2. [DOI] [PubMed] [Google Scholar]
- 11.Meng Z, et al. Characterization of a Saccharum spontaneum with a basic chromosome number of x = 10 provides new insights on genome evolution in genus Saccharum. Theor. Appl. Genet. 2020;133:187–199. doi: 10.1007/s00122-019-03450-w. [DOI] [PubMed] [Google Scholar]
- 12.Meng Z, et al. Chromosome painting provides insights into the genome structure and evolution of sugarcane. Front. Plant Sci. 2021;12:731664. doi: 10.3389/fpls.2021.731664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guimarães CT, Sills GR, Sobral BWS. Comparative mapping of Andropogoneae: Saccharum L. (sugarcane) and its relation to sorghum and maize. Proc. Natl. Acad. Sci. 1997;94:14261–14266. doi: 10.1073/pnas.94.26.14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, et al. Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics. 2010;11:261. doi: 10.1186/1471-2164-11-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garsmeur O, et al. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 2018;9:2638. doi: 10.1038/s41467-018-05051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolezel J, Bartos J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koren S, et al. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017 doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Zhang S, Zhao Q, Ming R, Tang H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants. 2019;5:833–845. doi: 10.1038/s41477-019-0487-8. [DOI] [PubMed] [Google Scholar]
- 19.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telles GP, Silva FRD. Trimming and clustering sugarcane ESTs. Genet. Mol. Biol. 2001;24:17–23. doi: 10.1590/S1415-47572001000100004. [DOI] [Google Scholar]
- 21.Vettore AL, da Silva FR, Kemper EL, Arruda P. The libraries that made SUCEST. Genet. Mol. Biol. 2001;24:1–7. doi: 10.1590/S1415-47572001000100002. [DOI] [Google Scholar]
- 22.Wu TD, Watanabe CK. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 23.Zaharia, M. et al.Faster and More Accurate Sequence Alignment with SNAP. ArXiv11115572 Cs Q-Bio (2011).
- 24.Flynn JM, et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimin AV, et al. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. GigaScience. 2017;6:gix097. doi: 10.1093/gigascience/gix097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinform. Oxf. Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dohm JC, Peters P, Stralis-Pavese N, Himmelbauer H. Benchmarking of long-read correction methods. NAR Genomics Bioinform. 2020;2:lqaa037. doi: 10.1093/nargab/lqaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu T, et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. Cell Mol. Biol. 2021;107:303–314. doi: 10.1111/tpj.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Telles GP, et al. Bioinformatics of the sugarcane EST project. Genet. Mol. Biol. 2001;24:9–15. doi: 10.1590/S1415-47572001000100003. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw read data was deposited in the NCBI SRA data archive under the bioproject: PRJNA820955. The genome assemble can be found in the NCBI genome archive: JALQSO000000000.