Abstract
The present study reports transcriptomic profiling of a Basidiomycota fungus, Podoscypha petalodes strain GGF6 belonging to the family Podoscyphaceae, isolated from the North-Western Himalayan ranges in Himachal Pradesh, India. Podoscypha petalodes strain GGF6 possesses significant biotechnological potential as it has been reported for endocellulase, laccase, and other lignocellulolytic enzymes under submerged fermentation conditions. The present study attempts to enhance our knowledge of its lignocellulolytic potential as no previous omics-based analysis is available for this white-rot fungus. The transcriptomic analysis of P. petalodes GGF6 reveals the presence of 280 CAZy proteins. Furthermore, bioprospecting transcriptome signatures in the fungi revealed a diverse array of proteins associated with cellulose, hemicellulose, pectin, and lignin degradation. Interestingly, two copper-dependent lytic polysaccharide monooxygenases (AA14) and one pyrroloquinolinequinone-dependent oxidoreductase (AA12) were also identified, which are known to help in the lignocellulosic plant biomass degradation. Overall, this transcriptome profiling-based study provides deeper molecular-level insights into this Basidiomycota fungi, P. petalodes, for its potential application in diverse biotechnological applications, not only in the biofuel industry but also in the environmental biodegradation of recalcitrant molecules.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12088-022-01037-6.
Keywords: Transcriptome, CAZyme, Lignocellulose, Ligninolytic, Fungal diversity, Laccase
Introduction
Lignocellulose biomass in its native form accounts for a potential feedstock for the biofuel industry; however, the crystalline nature of its components presents an array of technical obstacles to its valorization into valuable products. The cross-linking of cellulose with lignin restricts the biodegradation of lignocellulose biomass due to its complex recalcitrant characteristics [1]. The saccharification of these lignocellulose biomasses is further limited by the availability of the degradative enzymes known as carbohydrate-active enzymes (CAZymes), which are not readily available [2].
Several members of the white rot fungi are well known to degrade significant components of plant cell walls, including cellulose, hemicellulose, and lignin. Recent reports on white-rot fungi, especially from class Basidiomycota, suggest highly conserved core enzymatic systems associated with the breakdown of lignocellulose biomass [3, 4]. The fungal members of Basidiomycetes have a remarkable property of degrading a large proportion of lignin, thereby exposing the other structural polysaccharides to microbial attack and subsequent hydrolysis. The common enzymes reported in these Basidiomycetes species are diverse oxidative and hydrolytic enzymes such as phenol oxidases (which include manganese peroxidase, lignin peroxidase, versatile peroxidase, and laccase); auxiliary enzymes such as aryl-alcohol oxidase (AAO), methanol oxidase (MOX), pyranose 2-oxidase (P2O); and copper radical oxidases (such as glyoxal oxidase, GLX) [5, 6]. The synergistic action of all these enzymes is required for the complete degradation of lignocellulose biomass [7].
Lignocellulose biomass degradation by white-rot fungi has been previously studied in model organisms such as Phanerochaete and Trametes, emphasizing the growing interest in using white-rot fungi to convert recalcitrant plant residues to value-added products [8, 9]. In addition, recent studies on gene family expansions and transcriptome signatures have unearthed the regulation of conserved gene sets in white-rot fungi (Polyporales species) associated with lignocellulose decomposition [10].
Findings in the present study highlight the lignocellulosic degradation-associated transcriptomic signatures in Podoscypha petalodes strain GGF6, a white-rot fungus isolated from North-Western Himalayan ranges in Himachal Pradesh. The significance of the present study is further exemplified by the fact that this is the first attempt (to the best of our knowledge) to decipher transcriptome profiles of white-rot fungi in the family Podoscyphaceae, as no previous reports pertaining to omics-based analysis associated with lignocellulosic biomass degradation are available. Our research group has previously characterized the fungal culture for its lignocellulose enzyme potential in the biodegradation of wheat bran and dye decolorization [11–13] due to its endocellulase and laccase activities. The detailed molecular taxonomic analysis in the present study has allowed us to update the taxonomic classification of fungal culture to P. petalodes (NCBI Accession number KT008117.1). Furthermore, the transcriptomic analysis for functional characterization has revealed more profound insights into the evolutionary aspect of the lignocellulosic degradation machinery in white-rot fungi, especially in the family Podoscyphaceae. Findings presented in the manuscript have also led to identifying Podoscypha intra-genus genomic and functional diversity associated with proteins and enzymes associated with lignocellulose decomposition.
Materials and Methods
Culturing of the Fungus, DNA Extraction, and ITS Sequencing
P. petalodes cultures were maintained on Yeast Extract–Peptone–Dextrose (YPD) agar and stored at 4 °C. For DNA isolation, a fresh fungus culture was grown on Potato Dextrose Broth (PDB) for 72 h. The DNA was isolated from the mycelium collected from the broth culture. The mycelium was filtered and washed with sterilized water. The biomass was frozen and grounded into a fine powder with a pre-chilled mortar and pestle, followed by the genomic DNA extraction using standard protocols with minor modifications [14]. The ITS region was amplified with fungal-specific primer: ITS1 (5’TCCGTAGGTGAACCTGCGG3’) and ITS4 (5’TCCTCCGCTTATTGATATGC3’) [15]. The PCR amplification of the ITS region was carried out using PCR conditions (initial denaturation at 94 °C for 5 min; 30 cycles (95 °C for 45 s; 53 °C for 1 min; 72 °C for 2.5 min); followed by final extention of 10 min at 72 °C and final hold at 4 °C in a thermocycler (Bio-Rad T100). The PCR products were electrophoretically analyzed and visualized under the ultra violet light. The amplified PCR (ITS region) fragments were purified with a DNA purification kit (Real Biotech Corporation), as per the instructions from the manufacturer, and sequenced after that.
Phylogeny Analysis of the ITS Region of Fungus
To perform the taxonomy classification of the sequenced ITS region, we performed the Basic Local Alignment Search Tool (BLASTn) [16] against Fungi RefSeq ITS database using parameters [< 1e−5 E-value, > 35% query coverage and > 35% similarity], extracted top hits, and generated a maximum likelihood phylogenetic tree using MEGA [17]. For phylogenetic analysis, identified ITS sequence (KT008117.1) was subjected to Basic Local Alignment Search Tool (BLASTn) [18] against NCBI ITS sequence database with defined cutoff values: [< 1e−5 E-value, > 35% query coverage and > 35% similarity]. For taxonomy confirmation, the nucleotide sequences of the top 100 homologs were extracted, aligned using MUSCLE [19], and further subjected to RAxML version 8.2.12 using GRTGAMMA substitution model with 100 bootstraps to generate a maximum likelihood phylogeny [20]. The obtained ML tree was visualized in iTOL [21], and bootstrap information was depicted as a circle of size 1–10 for bootstrap values 1–100. We also used another marker, i.e., DNA-directed RNA polymerase II subunit (rpb1) protein, to generate a maximum-likelihood phylogeny. Protein rpb1 is present in one single copy in this organism. This protein sequence was used to find the top 100 close-related homolog sequences by subjecting it to BLASTp against the Non-redundant (NR) database. All homologs were aligned using MUSCLE, and the alignment was subjected to RAxML version 8.2.12 [22] using the GRTGAMMA substitution model with 100 bootstraps to generate a maximum likelihood phylogeny. The tree was visualized in iTOL, where color codes depict their taxonomy [21].
Illumina NextSeq PE Library Preparation
For RNA isolation, the fungus was grown in minimal agar media containing 2% wheat bran, 1% yeast extract, and 2% peptone incubated at 30 °C for 120 h. The growth was scraped from the plates, and the biomass was grounded in liquid nitrogen and used for RNA isolation using the TRIzol method and quantified using standard procedures on NanoDrop. The isolated RNA of the fungus was sequenced using Illumina paired-end sequencing technology from Eurofins Scientific, India. Total mRNA was enriched from total RNA using poly-T attached magnetic beads, followed by enzymatic fragmentation. The first strand of cDNA was synthesized to the second strand using a second strand mix and Act-D mix to facilitate RNA-dependent synthesis. Next, the ds-cDNA was purified using Ampure XP beads followed by A-tailing-based adapter ligation and further enriched using PCR amplification. RNA-Seq paired-end (PE) sequencing library was prepared from the RNA samples using Illumina TruSeq Stranded mRNA Sample Preparation Guide (15031047E). The PCR enriched libraries were analyzed in the 4200 Tape station system (Agilent Technologies, Germany) using High sensitivity D1000 screen tape as per the manufacturer’s instructions. Tap station profiling showed that the mean library fragment size was 315 bps. After confirming the Qubit concentration of the library and the mean peak size using the Agilent Tape Station profile, the PE Illumina library was loaded onto NextSeq 500 for cluster generation and sequencing using 2 × 75 bp chemistry.
Processing and Annotation of Transcriptomic Data
Quality confirmation of transcriptomic data was performed using FastQC [23]. Trimmomatic v0.35 [24] was used to clean the raw reads by removing adaptor sequences and retaining reads with a Phred score > 15 and a minimum read length of 36 bp. De novo transcriptome assembly of these quality reads was done by Trinity-v2.5.1 [25] by generating non-redundant contigs of transcripts using the default k-mer value. Then the resulting assembly is validated by keeping assembly as reference and mapping the raw reads back to it using Bowtie2. The most extended transcript within each assembled contig was classified as Unigene. Next, the TransDecoder.LongOrfs and TransDecoder.Predict algorithms from the TransDecoder [26] tool were used to find the Open Reading Frames (ORFs) and identify all coding sequences and the longest (–single_best_only) coding sequence. We also checked the completeness of assembled transcriptome using BUSCO v3.0.2 [27] against basidiomycota_odb10 (database consists of 133 species and 1764 BUSCOs in the database).
Functional Characterization Based on Transcriptomic Data
The identified protein-encoding sequences were subjected to BLASTp against the NCBI non-redundant protein database, PFAM, and TIGR to find close homologs with an E-value cutoff of 1e−5. To identify the potential biological pathways in which the predicted CDS might be associated, we mapped the CDS against the reference canonical pathways in KEGG. The KEGG analysis of P. petalodes transcriptome was based on the KEGG orthology (KO) assignments and corresponding enzyme commission (EC) numbers. The metabolic pathways of predicted CDS were determined using KEGG automated annotation server KAAS (https://www.genome.jp/kegg/kaas/) [28]. The P. petalodes transcriptome was also scanned using dbCAN2 metaserver [29] to identify all putative carbohydrate-active enzymes (CAZyme) [30] predicted by either one of the used methodologies. Based on these results, the distribution of proteins involved in lignocellulose degradation and ligninolytic function were identified.
Results and Discussion
The effectiveness of lignocellulose biodegradation significantly depends on the type and expression of enzymes involved in biodegradation [31]. As any reference genome is not available for P. petalodes or broadly for any member of the family Podoscyphaceae, lignocellulose biodegradation potential has not been studied before. Therefore, in this study, we explored the transcriptome of one of the family Podoscyphaceae members, i.e., P. petalodes, to provide insight into the proteins associated with lignocellulose degradation.
Morphology and Microscopic Examination
This fungal strain has already been reported for its potential to degrade lignocellulosic biomass to its monomers via saccharification via enzymatic machinery [11]. Moreover, it can also detoxify the azo dyes [12]. Based on external morphology (Fig. 1A), it depicts whitish mycelial growth on PDA media. After five days of incubation at 30 °C, a microscopic examination in a lactophenol cotton blue mount revealed ellipsoidal-shaped spores and aseptate hyphae (Fig. 1A). However, the morphology could not provide much information about its classification; therefore, a molecular identification strategy was performed using the internal transcribed spacer (ITS) region.
Fig. 1.
A Morphological characteristics of P. petalodes strain GGF6: (I) Fungal mycelial growth on PDA plate, (II) Microscopic observation of hyphae at 10x, (II) Microscopic observation of hyphae at 40x. B ITS sequence-based phylogenetic tree shows the position of P. petalodes strain GGF6 amongst its close relatives within the family Podoscyphaceae. Colour codes are according to the order-level fungus taxonomy shown on the left. Red-colored clade depicts the ITS sequences of those selected organisms from which we have identified the percent identity as in Table S1. C RPB1-protein-based maximum likelihood phylogenetic tree confirms the taxonomy of P. petalodes strain GGF6 amongst its close relatives within the family Podoscyphaceae. Colour codes are according to the order-level fungus taxonomy shown on the left. Both trees are drawn to scale, as shown on the top of the phylogenetic tree
Fungal Taxonomy
In our previous work [11–13], we classified this organism as Cotylidia pannosa belonging to the order Hymenochaetales. Here we have updated the taxonomic classification of this fungal strain using a well-known molecular marker, i.e., the ITS region. The sequenced ITS (656 bp) that has been updated in NCBI under the GenBank accession number KT008117.1 shows the closest homologs within P. Petalodes strains from order Polyporales. A phylogenetic tree (maximum-likelihood) was constructed after aligning the identified closest homolog sequences, confirming that this organism belongs to P. petalodes (Fig. 1B). As depicted in the tree, all ITS sequences belonging to each Podoscypha spp. are making their clades, suggesting that the phylogeny drawn here can demarcate each species group and most strains. It is also interesting to pinpoint a few cases where ITS sequences of a few strains were present along with a group of ITS sequences belonging to the same species, such as AB509635.1 Stereum sp. was present within the clade of P. yunnanensis and similarly, AJ537504.1 Stereopsis sp. was present nearby P. brasiliensis. Interpretations based on phylogenetic analysis suggest that these sequences should be examined further for their precise classification. It was also observed that percent identity between all closely related sequences within the clade, where P. petalodes strain GGF6 was located in the phylogeny, were significantly similar (Table S1). The phylogenetic analysis further revealed that the ITS sequence of this fungus shows 95.4% identity with P. petalodes voucher UOC BIB MB11, 93.24% with P. petalodes isolate DK09, and 91% identity with P. petalodes subsp. rosulate isolate AFTOL-ID 1931, suggesting a close relationship between P. petalodes strain GGF6 and other P. petalodes strains. Later we also used another phylogenetic marker protein, i.e., RPB1, to generate a maximum likelihood tree (Fig. 1C) and found that the RPB1 tree completely supports the ITS tree. Therefore, based on phylogenies and percent identity analysis, this fungal strain has been taxonomically classified as Podoscypha petalodes strain GGF6. The taxonomy of this ITS sequence has also been updated in the NCBI database under the GenBank accession number KT008117.1.
Transcriptomic Analysis of Podoscypha petalodes
The concentration of isolated RNA used for transcriptome sequencing was 450 ng/µl, and the 260/280 nm ratio was > 1.8, indicating high RNA purity and quality. Illumina NextSeq sequencer generated 16,919,336 pair-end raw reads equivalent to ~ 2.6 Gb data. Removal of adapter sequences, followed by ambiguous reads and low-quality sequences, we obtained 16,724,116 high-quality reads, i.e., 98.8% of total raw reads. Using Trinity-v2.5.1, these high-quality reads were assembled, resulting in 29,070,926 bp de novo assembly with an N50 value of 1,255 bp, 56.59% GC content, and 36,079 transcripts (Table 1). Furthermore, 94.25% of the raw reads were mapped back to the de novo assembly, showing that the generated assembly via Trinity was good quality. Using the TransDecoder Predict program, we found that P. petalodes encode 20,264 coding DNA sequences (CDS), including all possible transcripts such as full length, partial 3’ and partial 5’ open reading frames. Finally, 11,838 CDS, i.e., longest isoform per gene (called here as unigenes), were extracted from the assembly. Amongst all identified unigenes, the largest and smallest CDS were 5313 and 255 bp, and the average gene length was 736 bp (Fig. 2A). Thus, 6657 genes were more than 500 bps, whereas 383 were larger than 2000 bps. BUSCO analysis revealed that this transcriptome is 57.2% complete, whereas 20.2% is fragmented, and 22.6% is missing. Overall, this study was able to procure almost 77% transcriptome.
Table 1.
Transcriptome assembly statistics
| Attribute | Data |
|---|---|
| Assembly | |
| Organism Name | Podoscypha petalodes strain GGF6 |
| Sequencing Data (Reads; Bases) | 16,919,336 reads; 2,562,762,512 base |
| Quality Filtered Data (Reads; Bases) | 16,724,116 reads; 2,527,651,889 bases |
| Assembly Size | 29,070,926 bp |
| GC% | 56.59 |
| Identified Trinity Transcript count | 36,079 |
| Maximum Transcript length | 5831 bp |
| Minimum Transcript length | 200 bp |
| Average Transcript length | 805 bp |
| N50 Transcript length | 1255 bp |
| Unigene CDS | 11,838 |
| Percent GC | 56.81 |
| Maximum CDS length | 5,313 |
| Minimum CDS length | 255 |
| Average CDS length | 736 |
| CDS ≥ 500 | 6657 |
| Homologs in NR | 10,894 (92.02%) |
| Mapped PFAM Families | 5144 (43.45%) |
| Mapped TIGR families | 1386 (11.70%) |
| Mapped proteins in KEGG | 3791 (32.02%) |
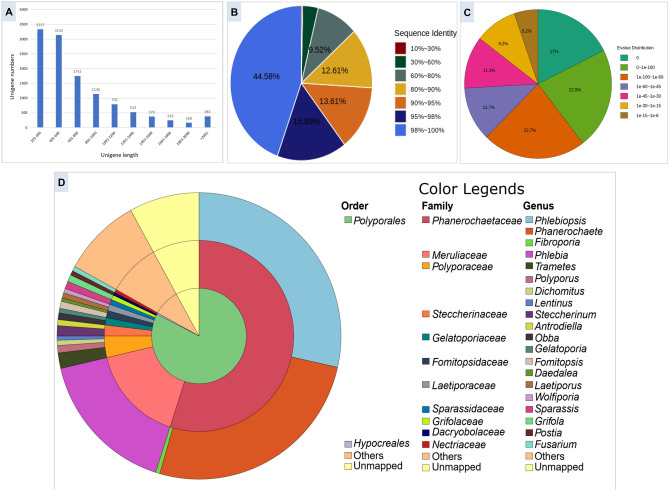
Fig. 2.
Homology information for the unigenes identified from P. petalodes strain GGF6 transcriptome: A Distribution of unigene length. B Sequence identity-based distribution of the top BLAST hits for each unigene-encoded protein against the NR database. C E‑value-based distribution as identified using the best BLASTp hit for each unigene-encoded protein against the non-redundant (NR) database. D Taxonomy classification (at order, family, and genus level) of best BLASTp homolog per each unigene-encoded protein against the non-redundant (NR) database and their relative distribution
Homology-Based Functional Characterization
All identified 11,838 unigenes were subjected to homology analysis against the NCBI NR database to annotate and characterize the functions of the P. petalodes transcriptome. A total of 10,894 unigenes showed a good homolog in the NR database representing 92.02% of total unigenes, suggesting that most unigenes are getting translated into valid proteins. Next, we looked into the sequence identity of the unigenes homology results against the NR database (Fig. 2B). Our results identified that ~ 61% of the mapped unigene encoded proteins have a 95–100% sequence identity, whereas only 13.31% of the mapped proteins showed less than 80% identity. Simultaneously, we also looked into the E-value distribution and found that almost 84.7% unigene encoded proteins show strong homology with < 1e−30 E-value, whereas only 15.3% showed higher E-value between 1e−30 and 1e−5 (Fig. 2C).
Most of the identified homologs (> 80%) were within order Polyporales members, as revealed by species distribution of the best-mapped homolog of each protein. At species level identity, we found that these unigenes are showing the maximum mapping with the homologs in Phlebiopsis gigantea 11061_1 CR5-6 (28.5%) and Phanerochaete carnosa HHB-10118-Sp (25.46%), which belongs to family Phanerochaetaceae, followed by Phlebia centrifuga (16.40%) in family Meruliaceae and Steccherinum ochraceum (1.16%) in family Steccherinaceae (Fig. 2D). As no genome/transcriptome sequence is available in the family Podoscyphaceae, the best hits (both at identity and quantity levels) of P. petalodes unigenes are found in the family Phanerochaetaceae, which suggests that at the sequence level, the family Phanerochaetaceae is closest to the family Podoscyphaceae. However, as family Podoscyphaceae and Phanerochaetaceae share only 55% homologs, it also depicts the diversity within the organisms of these two families. Increased genome sequencing of other members of the family Podoscyphaceae followed by the comparative genomic analysis of family Podoscyphaceae, and Phanerochaetaceae members will enable us to understand these relationships more precisely.
Family-Wise Functional Characterization Using PFAM and TIGR Databases
The present study explored the family-wise categorization and functional classification of identified unigenes using curated databases such as PFAM and TIGR. This study identified 5144 unigene-encoded proteins mapped to 2332 Pfam families (Fig. 3A, Table S2). Similarly, all unigene-encoded proteins were mapped to the TIGR database, and it was observed that 11.70% (1386) proteins were annotated for functional categorization and characterization (Table S3). This study further revealed that the maximum numbers (109 proteins) of unigene-encoded proteins in P. petalodes were involved in protein kinase function that helps in protein phosphorylation, a vital role in most cellular activities. Along with this, the significant representation of proteins is present in p450 (68 proteins), MFS_1 (54 proteins), Aldo_ket_red (48 proteins), adh_short (41 proteins), Fungal_trans (20 proteins), Hydrophobin (16 proteins), etc. Cytochrome P450s are known as heme-thiolate proteins and participate in the oxidative degradation of environmental toxins and mutagens [32]. MFS transporter proteins help in transporting small solutes during the chemiosmotic ion gradients. Proteins in the Pfam family, Aldo_ket_red, (function as aryl alcohol dehydrogenases), facilitate the degradation of β-aryl ethers within lignin [33, 34]. The abundance of these Aldo–keto reductase shows that P. petalodes could facilitate converting lignin-containing materials into fermentable products [35]. Previous findings reveal that the differential expression of genes linked to Aldo–keto reductase is associated with the wood decay process among all the brown-rot fungi [36].
Fig. 3.
Functional characterization of identified unigenes identified from P. petalodes strain GGF6 transcriptome: A The distribution of top 25 Pfam families B Distribution of CAZy families C Distribution of KEGG pathways
The present study also identified around 20 fungal-specific transcription factor domains (Fungal_trans), known to be peculiar in fungi [37]. These functions should be relevant for this fungus as all of these proteins are present in high representation. Our analysis also identified a family, Glyoxal_oxid_N, represented by three proteins. This family encodes plant and fungal-specific glyoxal oxidase enzymes (a copper metalloenzyme), an essential component of the extracellular lignin degradation pathways as observed in Phanerochaete chrysosporium [34]. Overall, these annotations provide a treasured resource for inspecting the precise function of a few proteins and associated pathways in P. petalodes.
Homology Studies Using the KEGG Pathway
To better understand the genes involved in diverse biological pathways of P. petalodes, all predicted unigenes were mapped against conical reference pathways in the KEGG database. As a result, 3791 unigenes (approximately 32% of the total identified unigenes) were mapped to 384 KEGG pathways. The top five pathways were ‘Signal transduction’ and ‘ Carbohydrate metabolism,’ ‘Amino acid metabolism,’ ‘Translation,’ and ‘Transport and catabolism.’ Thus, the unigene-encoded proteins have been classified into five major categories, i.e., metabolism, cellular processes, genetic information processing, environmental information processing, and organismal systems. This study revealed that 30.68% of genes are involved in metabolic pathways among all KEGG pathway mapped proteins, 20.25% CDS in genetic information processing, and 17.33% CDS in cellular processes (Fig. 3B).
Homology Studies Using Carbohydrate-Active Enzymes (CAZy) Database
The utilization of lignocellulosic biomass such as farming produce residues, agriculture crop leftovers, grass, wood, and other plants' dry matter depends on the secretion of an array of lignocellulolytic enzymes. These enzymes catalyze biochemical reactions such as hydrolysis, depolymerization, and biotransformation of aromatic compounds. Therefore, we identified all such proteins in P. petalodes, potentially degrading these materials. The carbohydrate-active enzyme (CAZy) database classified all biomass-degrading enzymes into six classes. CAZy database includes a total of 400 datasets that can be distributed; 153 glycoside hydrolase (GH, enzymes for the hydrolysis of glycosidic bond of polysaccharides), 105 glycosyltransferases (GT, transfer of saccharide moiety from donor to the acceptors and destruct glycosidic bonds in a variety of substrates), 15 auxiliary activity (AA, degradation, and mineralization of lignin), 16 carbohydrate esterase (CE, hydrolysis of carbohydrate substrates by removing the ester part), 28 polysaccharide lyases (PL, cleave non-hydrolytic cleavage of glycosidic bonds) and 83 carbohydrate-binding domains (adhesion to carbohydrates) (Fig. 3C). dbCAN HMMdb release 8.0 includes 641 CAZyme HMMs, 421 family HMMs, three cellulosome HMMs, and 217 subfamily HMMs. The transcriptomic analysis of P. petalodes resulted in 280 CAZy proteins. Among these proteins, 109, 70, 23, 12, 6, and 60 were identified as belonging to GH, GT, CE, CBM, PL, and AA families, respectively. As identified using omics studies, these enzymes can be explored for the biological pre-treatment processes of biomass substrates for bioenergy generation [31, 38–41].
Distribution of Proteins Participating in the Degradation of Cellulose/Hemicellulose/Pectin
The major constituents of lignocellulosic biomass are polysaccharides, viz. cellulose, hemicellulose, and pectin. Therefore, the essential proteins should degrade these polysaccharides into fermentable sugars. These sugars can be used for biofuel production or other compounds of value. For cellulose degradation, three main enzymes are required; (1) β-1,4-Endoglucanase (EC 3.2.1.4); (2) Exoglucanase or Exo-β-1,4-glucanase (EC 3.2.1.74); (3) Beta-glucosidase or cellobiase (EC 3.2.1.21). Hemicelluloses are a cumulative term for xylans, xyloglucans, arabinoxylans, and glucomannans, therefore amongst hemicellulase, endo-β-1,4-xylanases (EC 3.2.1.8), β-xylosidases (EC 3.2.1.37) degrade xylan backbone whereas other accessory protein, i.e., alpha-glucuronidase (EC 3.2.1), α-L-arabinofuranosidases (AFases; EC 3.2.1.55), acetyl xylan esterases (acetyl esterases; EC 3.1.1.6) help in the complete digestion of complex substituted xylans. Two enzyme categories, i.e., Homogalacturonases and Rhamnogalacturonases, are required to degrade major pectin components such as homogalacturonan (HG), apiogalacturonan, xylogalacturonan (XGA), rhamnogalacturonan I, and II (RGI and RGII). Using the CAZy database, we identified all CAZyme families known for degrading these constituents, as represented in Table 2.
Table 2.
Distribution of CAZy protein families involved in cellulose, hemicellulose, and pectin degradation
| Substrate | Known activity for respective CAZY category | CAZY category | Total unigene proteins | Total enzymes |
|---|---|---|---|---|
| Cellulose | Cellobiase | GH1 | 1 | 60 |
| Cellobiase | GH2 | 1 | ||
| Endoglucanase; Cellobiase | GH3 | 4 | ||
| Endoglucanase; Exoglucanase; Cellobiase | GH5 | 15 | ||
| Endoglucanase; Exoglucanase; | GH6 | 1 | ||
| Endoglucanase | GH7 | 1 | ||
| Endoglucanase; Exoglucanase; Cellobiase | GH9 | 1 | ||
| Endoglucanase | GH10 | 1 | ||
| Endoglucanase | GH12 | 1 | ||
| Endoglucanase | GH13 | 11 | ||
| Cellobiase | GH16 | 14 | ||
| Endoglucanase | GH27 | 2 | ||
| Endoglucanase; Cellobiase | GH30 | 1 | ||
| Endoglucanase | GH31 | 3 | ||
| Endoglucanase | GH74 | 1 | ||
| Endoglucanase | GH78 | 1 | ||
| Endoglucanase; Exoglucanase; | GH131 | 1 | ||
| Hemicellulose | Acetyl xylan esterase | CE1 | 2 | 51 |
| Acetyl xylan esterase | CE4 | 3 | ||
| Acetyl pectin esterase | CE12 | 1 | ||
| β-xylosidase | GH1 | 1 | ||
| β-mannosidase | GH2 | 1 | ||
| β-xylosidase | GH3 | 4 | ||
| endo-β-1,4-xylanase | GH5 | 15 | ||
| endo-β-1,4-xylanase | GH10 | 1 | ||
| Xyloglucanase | GH12 | 1 | ||
| Xyloglucanase | GH16 | 14 | ||
| α-galactosidase | GH27 | 2 | ||
| α-xylosidase | GH31 | 3 | ||
| β-xylosidase | GH43 | 2 | ||
| β‐glucanase | GH131 | 1 | ||
| Pectin | Pectin acetylesterase | CE12 | 1 | 32 |
| β-glucuronidase | GH1 | 1 | ||
| rhamnogalacturonyl hydrolase | GH105 | 1 | ||
| β-glucuronidase | GH2 | 1 | ||
| exo-β-1,4-glucanase | GH3 | 4 | ||
| α-L-arabinofuranosidase | GH43 | 2 | ||
| exo-β-1,4-glucanase | GH5 | 15 | ||
| α-L-rhamnosidase | GH78 | 1 | ||
| β-glucuronidase | GH79 | 4 | ||
| Pectate lyase | PL3 | 2 |
Bold fonts represent the unique nature of those families for that respective function. Bar plots represent the relative distribution of all unigene proteins
The present study identified 60 cellulases, 51 hemicellulases, and 32 pectinases distributed among different CAZyme categories. These identified proteins have multiple functions; however, a few (GH6, GH7, GH9, GH13, GH30, GH74, CE1, CE4, GH105, GH79, and PL3) are highly specific performing one particular function. GH5, GH16, and GH13 have the maximum representation; the GH5 category shares the properties of all three cellulases (Endoglucanase, Exoglucanase, Cellobiase), endo-β-1,4-xylanase, and exo-β-1,4-glucanase. A total of 14 GH16 CAZymes were identified that share cellobiase and Xyloglucanase functions involved in cellulose and hemicellulose degradation. Eleven GH13 proteins are identified as having endoglucanase activity. Several CAZyme families have multiple functions within cellulase categories; however, a few (i.e., GH1, GH2, GH7, GH74, etc.) have unique functionalities within cellulose degradation. In the present study, several unique cellulase CAZyme families such as GH6, GH7, GH9, GH30, and GH74 were identified, having one protein responsible for their respective functions. Several proteins involved in only endoglucanase and cellobiase functions were identified; however, exoglucanase function was not present alone. It was always along with endoglucanase and cellobiase functions, overall signifying that all three cellulose degradation functions are synergistic to each other. Among the hemicellulases, CE1 and CE4 CAZymes were unique in all identified categories. These categories are carboxylesterase or acetyl xylan esterase (EC 3.1. 1.72), which help the deacetylation of xylans and xylooligosaccharides. These two categories play a vital function in the complete enzymatic hydrolysis of lignocellulosic products [42]. Similarly, among the pectinases, GH105, GH79, and PL3 categories have 1, 4, and 2 proteins, respectively, and these categories have unique roles in pectin degradation, as they do not share any function with cellulases or hemicellulases. GH105 family proteins are rhamnogalacturonyl hydrolases, which act on unsaturated rhamnogalacturonan to produce unsaturated galacturonic acid [43]. Similarly, there were four proteins represented by GH79 known to be involved in β-glucuronidase activity. In the present study, two proteins with the PL3 CAZyme family were found, suggesting their role as pectate lyase, which is accountable for the cleavage of demethylated regions of pectin and yield oligosaccharide, i.e., Δ-4,5-D-galacturonate through a trans-elimination reaction. Overall, we identified many enzymes that allow this fungus to use cellulose/ hemicellulose/ pectin as a carbon source for their growth and efficiently degrade plant biomass in their adjacent ambiance.
Distribution of Proteins Involved in Ligninolytic Enzymes
Another focus of this study was identifying ligninolytic enzyme potential, which might allow this fungus to degrade lignin efficiently. Lignin degradation capacity has been reported in bacteria; however, the major contributors are representative fungal species, predominantly from the phylum Basidiomycota [44]. Based on the literature survey, we identified that laccase, manganese peroxidase (MnP), lignin peroxidase (LiP), and versatile peroxidase (VP) are the main fungal enzymes associated with the degradation and mineralization of lignin [45, 46]. The lignin-degrading enzymes may not directly act on carbohydrates. However, as lignin is customarily and well associated with carbohydrates in the plant cell wall, the ligninolytic enzymes are engaged with classical polysaccharide depolymerase [44].
This study observed that P. petalodes have 54 auxiliary enzymes, further categorized into two families (AA1 and AA2) of ligninolytic enzymes (Table 3). Fungal laccases are multi-copper phenol oxidases that play an essential role in diverse biological functions such as fruiting-body formation and lignin degradation in filamentous fungi. They are also one of the most studied fungal enzymes known for their capability to oxidize recalcitrant phenolic compounds, including polymeric lignin and humic organic substances [47]. Most ligninolytic fungal species in the soil environment encode at least one laccase enzyme [45]. In the present study, we also observed one AA1 CAZyme protein belonging to Laccase-like multicopper oxidase, reported in our previous report [12], portraying its essential role in decolorizing synthetic dyes during the treatment of wastewater from industrial effluents.
Table 3.
Distribution of auxiliary activity (AA) family proteins putatively involved in lignin degradation
| CAZy category | CAZy category function | Total unigene proteins |
|---|---|---|
| AA1 | Laccase-like multicopper oxidase | 1 |
| AA2 | Class II lignin-modifying peroxidases | 12 |
| AA3 | Glucose-methanol-choline (GMC) oxidoreductases family | 7 |
| AA4 | Vanillyl-alcohol oxidases (VAO) | 1 |
| AA5 | Copper radical oxidases | 3 |
| AA6 | 1,4-benzoquinone reductases | 3 |
| AA7 | Glucooligosaccharide oxidases (GOO) | 5 |
| AA8 | Iron reductase | 2 |
| AA9 | Copper-dependent lytic polysaccharide monooxygenases (LPMOs); lytic cellulose monooxygenase | 17 |
| AA12 | Pyrroloquinolinequinone-dependent oxidoreductase | 1 |
| AA14 | Copper-dependent lytic polysaccharide monooxygenases (LPMOs); lytic xylan monooxygenase | 2 |
Lignin-modifying peroxidases belong to class II peroxidases within the superfamily of plant and fungal peroxidases, also known as catalase-peroxidases [44]. Class II lignin-modifying peroxidases represented by auxiliary activities (AA2) CAZy family include all extracellular fungal heme peroxidases. We identified 12 homologs with AA2 family domains capable of attacking lignin polymers in a non-specific manner.
The findings in the present manuscript have revealed that P. petalodes has two families of lytic polysaccharide monooxygenases (LPMOs; AA9 and AA14). Furthermore, we identified 17 homologs of lytic cellulose monooxygenases and two lytic xylan monooxygenases. It has been reported that LPMOs are involved in depolymerizing inert parts of cellulose and other polysaccharides by removing the required electrons from the lignin part of plant biomasses [48].
The transcriptomic analysis of P. petalodes further revealed an enzyme, i.e., pyrroloquinoline quinone-dependent oxidoreductase, which belongs to the AA12 CAZy family. This CAZy family was first reported in a Basidiomycete mushroom, Coprinopsis cinerea, and is supposed to be directly or indirectly involved in lignin breakdown [49]. Thus, the presence of this protein in P. petalodes might help this fungus in better lignin degradation.
Lignin-Modifying Peroxidases
We identified the whole plethora of peroxidases in the present study by subjecting the predicted P. petalodes proteomes to standalone BLASTp against the Fungal Peroxidase Database available at http://peroxidase.riceblast.snu.ac.kr/class.php. The analysis revealed that this fungus encodes 82 peroxidases that can be grouped into two different categories (Table 4); heme peroxidases (73 proteins) and non-heme peroxidases (9 proteins). Heme peroxidases are a group of biocatalysts involved in the biodegradation of lignocelluloses and lignins, which are further categorized into different classes, such as Linoleate diol synthase (PGHS like) (33 proteins), DyP-type peroxidase D (11 proteins), Lignin peroxidase (Lip) (10 proteins), other class II peroxidase (7 proteins) and so on. All the peroxidases are supposed to possess lignin-degrading activities [50, 51], in which lignin peroxidase is the key lignin-degrading enzyme. However, we did not find any manganese peroxidase (MnP) and versatile peroxidase (VP) protein representative in this analysis.
Table 4.
Distribution and classification of peroxidase enzymes in P. petalodes strain GGF6
| Category | Class | Total proteins | Total proteins per category |
|---|---|---|---|
| Haem peroxidase | Catalase | 6 | 73 |
| Catalase-peroxidase | – | ||
| Cytochrome C peroxidase | 3 | ||
| Duox | – | ||
| DyP-type peroxidase D | 11 | ||
| Haloperoxidase (haem) | 3 | ||
| Hybrid Ascorbate-Cytochrome C peroxidase | – | ||
| Lignin peroxidase | 10 | ||
| Linoleate diol synthase (PGHS like) | 33 | ||
| Manganese peroxidase | – | ||
| NoxA | – | ||
| NoxB | – | ||
| NoxC | – | ||
| Other class II peroxidase | 7 | ||
| Prostaglandin H synthase (Cyclooxygenase) | – | ||
| Rbohs | – | ||
| Versatile peroxidase | – | ||
| Non haem peroxidase | 1-Cysteine peroxiredoxin | – | 9 |
| Atypical 2-Cysteine peroxiredoxin (type II, type V) | 2 | ||
| Atypical 2-Cysteine peroxiredoxin (type Q, BCP) | 3 | ||
| Carboxymuconolactone decarboxylase (no peroxidase activity) | – | ||
| Fungi-Bacteria glutathione peroxidase | 2 | ||
| No haem, Vanadium chloroperoxidase | – | ||
| Typical 2-Cysteine peroxiredoxin | 2 | ||
| Regulator | NoxR | – | |
| Total Peroxidases | 82 | ||
Conclusions
Several fungi are involved in breaking lignocellulose‐containing raw materials using a broad spectrum of enzymes which has been a significant research interest for all industry types. Earlier, we reported a fungal strain classified as Cotylidia pannosa as an efficient endocellulase-producer compared to Trichoderma reesei MTCC 164. Here in the present study, using the ITS gene and rpb1 protein sequence-based phylogenetic analysis, we have updated the taxonomy of this strain as P. petalodes strain GGF6 belonging to the family Podoscyphaceae. As no genome/transcriptome sequence information was available for the family Podoscyphaceae, in this study, we have performed the assembly, annotation, and functional characterization of the transcriptome of P. petalodes strain GGF6. This transcriptome-based data analysis revealed that this fungus has 280 CAZy proteins. In addition, we also identified many proteins with cellulase, hemicellulase, pectinase, laccase, and lignin peroxidase activities. These diverse arrays of proteins might be helping P. petalodes in the efficient degradation of lignocellulose biomass. Our study also revealed that P. petalodes has one AA12 family protein, i.e., pyrroloquinolinequinone-dependent oxidoreductase, well known to degrade lignin directly or indirectly. A total of 82 peroxidases were also identified within this fungus, including ten lignin peroxidases, which were heme‐containing enzymes responsible for catalyzing hydrogen peroxide‐dependent oxidative degradation of lignin. Overall, this study portrays P. petalodes strain GGF6 as an efficient lignocellulose biomass degrader based on the diversity of proteins involved in lignocellulose degradation and ligninolytic function as identified from its transcriptome assembly.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Table 1: Heatmap of %identity scores amongst the close relatives identified from its phylogenetic clade.Supplementary Table 2: Distribution of P. petalodes strain GGF6 unigenes among all Pfam families and their respective functions.Supplementary Table 3: Distribution of P. petalodes strain GGF6 unigenes among all TIGR families and their respective functions. (XLSX 130 KB)
Acknowledgements
The authors thank their respective Universities/ Institutes for providing essential facilities and environment for research. Furthermore, the authors sincerely acknowledge the help provided by Dr. R.S. Chauhan and Dr. B.M. Sharma regarding providing the fungal culture.
Abbreviations
- NCBI
National centre for biotechnology information
- BLAST
Basic local alignment search tool
- CAZyme
Carbohydrate active enzymes
- GH
Glycoside hydrolases
- GT
Glycosyl transferases
- CE
Carbohydrate esterases
- CBM
Carbohydrate binding-module
- PL
Polysaccharide lyases
- AA
Auxiliary activity
Authors' contributions
GG generated the idea. RM and DS cultured the fungi and isolated the RNA for transcriptome sequencing. SBH, VK, and GS performed the computational analysis. GS, RM, and GG wrote the manuscript. All authors edited and finalized the manuscript.
Funding
The research was funded by the Department of Science and Technology, Government of India for the Indo-Russian collaborative project “Elucidating the linkage between key limiting processes and microorganisms during anaerobic degradation of lignocellulosic waste” INT/RUS/RFBR/P-175 to GG. GS is supported by the DST-INSPIRE Faculty Award from DST, Government of India. In addition, this work was partially supported by the Department of Electronics, IT, BT, and S&T of the Government of Karnataka, India.
Data availability
Raw reads can be accessed using this link https://drive.google.com/drive/folders/1wnhbJvjAW3mYyJAFCpHYHAufwOY7PEvb?usp=sharing. The raw data can also be provided upon request.
Declarations
Conflict of interest
The authors declare no conflict of interest to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rishi Mahajan, B. Shenu Hudson, Deepak Sharma: Equal first authors.
Contributor Information
Gaurav Sharma, Email: gaurav.amit30@gmail.com.
Gunjan Goel, Email: gunjanmicro@gmail.com.
References
- 1.Mahajan R, Chandel S, Puniya AK, Goel G. Effect of pretreatments on cellulosic composition and morphology of pine needle for possible utilization as substrate for anaerobic digestion. Biomass Bioenergy. 2020;141:105705. doi: 10.1016/j.biombioe.2020.105705. [DOI] [Google Scholar]
- 2.van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;91:1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suryadi H, Judono JJ, Putri MR, Eclessia AD, Ulhaq JM, Agustina DN, Sumiati T. Biodelignification of lignocellulose using ligninolytic enzymes from white-rot fungi. Heliyon. 2022;8:e08865. doi: 10.1016/J.HELIYON.2022.E08865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyauchi S, Hage H, Drula E, Lesage-Meessen L, Berrin JG, Navarro D, Favel A, Chaduli D, Grisel S, Haon M, Piumi F, Levasseur A, Lomascolo A, Ahrendt S, Barry K, LaButti KM, Chevret D, Daum C, Mariette J, Klopp C, Cullen D, de Vries RP, Gathman AC, Hainaut M, Henrissat B, Hildén KS, Kües U, Lilly W, Lipzen A, Mäkelä MR, Martinez AT, Morel-Rouhier M, Morin E, Pangilinan J, Ram AFJ, Wösten HAB, Ruiz-Dueñas FJ, Riley R, Record E, Grigoriev IV, Rosso MN. Conservedwhite-rot enzymaticmechanismfor wood decay in the basidiomycota genus pycnoporus. DNA Res. 2020 doi: 10.1093/DNARES/DSAA011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monclaro AV, de O. Gorgulho Silva C, Gomes HAR, de S. Moreira LR, Filho EXF. The enzyme interactome concept in filamentous fungi linked to biomass valorization. Bioresour Technol. 2022;344:126200. doi: 10.1016/J.BIORTECH.2021.126200. [DOI] [PubMed] [Google Scholar]
- 6.Wu D, Wei Z, Mohamed TA, Zheng G, Qu F, Wang F, Zhao Y, Song C. Lignocellulose biomass bioconversion during composting: mechanism of action of lignocellulase, pretreatment methods and future perspectives. Chemosphere. 2022;286:131635. doi: 10.1016/J.CHEMOSPHERE.2021.131635. [DOI] [PubMed] [Google Scholar]
- 7.Singhania RR, Patel AK, Raj T, Chen CW, Ponnusamy VK, Tahir N, Kim SH, Di Dong C. Lignin valorisation via enzymes: a sustainable approach. Fuel. 2022;311:122608. doi: 10.1016/J.FUEL.2021.122608. [DOI] [Google Scholar]
- 8.Sena-Martins G, Almeida-Vara E, Duarte JC. Eco-friendly new products from enzymatically modified industrial lignins. Ind Crops Prod. 2008;27:189–195. doi: 10.1016/J.INDCROP.2007.07.016. [DOI] [Google Scholar]
- 9.Chaturvedi V, Verma P. An overview of key pretreatment processes employed for bioconversion of lignocellulosic biomass into biofuels and value added products. 3 Biotech. 2013;3:415–431. doi: 10.1007/s13205-013-0167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hage H, Miyauchi S, Virágh M, Drula E, Min B, Chaduli D, Navarro D, Favel A, Norest M, Lesage-Meessen L, Bálint B, Merényi Z, de Eugenio L, Morin E, Martínez AT, Baldrian P, Štursová M, Martínez MJ, Novotny C, Magnuson JK, Spatafora JW, Maurice S, Pangilinan J, Andreopoulos W, LaButti K, Hundley H, Na H, Kuo A, Barry K, Lipzen A, Henrissat B, Riley R, Ahrendt S, Nagy LG, Grigoriev IV, Martin F, Rosso MN. Gene family expansions and transcriptome signatures uncover fungal adaptations to wood decay. Environ Microbiol. 2021 doi: 10.1111/1462-2920.15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma D, Sud A, Bansal S, Mahajan R, Sharma BM, Chauhan RS, Goel G. Endocellulase production by Cotylidia pannosa and its application in saccharification of wheat bran to bioethanol. BioEnergy Res. 2018;11:219–227. doi: 10.1007/s12155-017-9890-z. [DOI] [Google Scholar]
- 12.Sharma D, Goel G, Sud A, Chauhan RS. A novel laccase from newly isolated Cotylidia pannosa and its application in decolorization of synthetic dyes. Biocatal Agric Biotechnol. 2015;4:661–666. doi: 10.1016/J.BCAB.2015.07.008. [DOI] [Google Scholar]
- 13.Sharma D, Garlapat VK, Goel G. Bioprocessing of wheat bran for the production of lignocellulolytic enzyme cocktail by Cotylidia pannosa under submerged conditions. Bioengineered. 2016;7:88–97. doi: 10.1080/21655979.2016.1160190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan R, Attri S, Sharma K, Singh N, Sharma D, Goel G. Statistical assessment of DNA extraction methodology for culture-independent analysis of microbial community associated with diverse environmental samples. Mol Biol Rep. 2018;45:297–308. doi: 10.1007/S11033-018-4162-3. [DOI] [PubMed] [Google Scholar]
- 15.White TJ, Bruns T, Lee SJ, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics—ScienceOpen. PCR Protoc Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babraham Bioinformatics—FastQC a quality control tool for high throughput sequence data, (n.d.). https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed July 13, 2022)
- 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Home·TransDecoder/TransDecoder Wiki·GitHub, (n.d.). https://github.com/TransDecoder/TransDecoder/wiki (accessed July 13, 2022)
- 27.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 28.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. DbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao L, Cao GL, Wang AJ, Ren HY, Dong D, Liu ZN, Guan XY, Xu CJ, Ren NQ. Fungal pretreatment of cornstalk with Phanerochaete chrysosporium for enhancing enzymatic saccharification and hydrogen production. Bioresour Technol. 2012;114:365–369. doi: 10.1016/j.biortech.2012.03.076. [DOI] [PubMed] [Google Scholar]
- 32.Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1:1–9. doi: 10.1186/GB-2000-1-6-REVIEWS3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tramontina R, Franco Cairo JPL, Liberato MV, Mandelli F, Sousa A, Santos S, Rabelo SC, Campos B, Ienczak J, Ruller R, Damásio ARL, Squina FM. The Coptotermes gestroi aldo–keto reductase: a multipurpose enzyme for biorefinery applications. Biotechnol Biofuels. 2017;10:4. doi: 10.1186/s13068-016-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whittaker MM, Kersten PJ, Cullen D, Whittaker JW. Identification of catalytic residues in glyoxal oxidase by targeted mutagenesis. J Biol Chem. 1999;274:36226–36232. doi: 10.1074/jbc.274.51.36226. [DOI] [PubMed] [Google Scholar]
- 35.Scharf M, Sethi A (2013) US patent application for novel lignases and aldo-keto reductases for conversion of lignin-containing materials to fermentable products patent. Google Patents, WO 2013126230 A1
- 36.Kameshwar AKS, Qin W. Systematic metadata analysis of brown rot fungi gene expression data reveals the genes involved in Fenton’s reaction and wood decay process. Mycology. 2019;11:22–37. doi: 10.1080/21501203.2019.1703052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shelest E. Transcription factors in fungi. FEMS Microbiol Lett. 2008;286:145–151. doi: 10.1111/j.1574-6968.2008.01293.x. [DOI] [PubMed] [Google Scholar]
- 38.Kuuskeri J, Hakkinen M, Laine P, Smolander OP, Tamene F, Miettinen S, Nousiainen P, Kemell M, Auvinen P, Lundell T. Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: growth on spruce wood and decay effect on lignocellulose. Biotechnol Biofuels. 2016;9:192. doi: 10.1186/s13068-016-0608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasco-Correa J, Li Y. Solid-state anaerobic digestion of fungal pretreated Miscanthus sinensis harvested in two different seasons. Bioresour Technol. 2015;185:211–217. doi: 10.1016/j.biortech.2015.02.099. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Wang ML, Tonnis B, Habteselassie M, Liao X, Huang Q. Fungal pretreatment of switchgrass for improved saccharification and simultaneous enzyme production. Bioresour Technol. 2013;135:39–45. doi: 10.1016/j.biortech.2012.10.095. [DOI] [PubMed] [Google Scholar]
- 41.Miyauchi S, Navarro D, Grisel S, Chevret D, Berrin JG, Rosso MN. The integrative omics of white-rot fungus Pycnoporus coccineus reveals co-regulated CAZymes for orchestrated lignocellulose breakdown. PLoS ONE. 2017;12:e0175528. doi: 10.1371/journal.pone.0175528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carvalho AFA, de O. Neto P, da Silva DF, Pastore GM. Xylo-oligosaccharides from lignocellulosic materials: chemical structure, health benefits and production by chemical and enzymatic hydrolysis. Food Res Int. 2013;51:75–85. doi: 10.1016/J.FOODRES.2012.11.021. [DOI] [Google Scholar]
- 43.Itoh T, Ochiai A, Mikami B, Hashimoto W, Murata K. Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate. Biochem Biophys Res Commun. 2006;347:1021–1029. doi: 10.1016/J.BBRC.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 44.Janusz G, Pawlik A, Sulej J, Świderska-Burek U, Jarosz-Wilkołazka A, Paszczyński A. Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol Rev. 2017;41:941–962. doi: 10.1093/FEMSRE/FUX049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldrian P. Fungal laccases—occurrence and properties. FEMS Microbiol Rev. 2006;30:215–242. doi: 10.1111/J.1574-4976.2005.00010.X. [DOI] [PubMed] [Google Scholar]
- 46.Min B, Park H, Jang Y, Kim JJ, Kim KH, Pangilinan J, Lipzen A, Riley R, Grigoriev IV, Spatafora JW, Choi IG. Genome sequence of a white rot fungus Schizopora paradoxa KUC8140 for wood decay and mycoremediation. J Biotechnol. 2015;211:42–43. doi: 10.1016/j.jbiotec.2015.06.426. [DOI] [PubMed] [Google Scholar]
- 47.Savinova OS, Moiseenko KV, Vavilova EA, Chulkin AM, Fedorova TV, Tyazhelova TV, Vasina DV. Evolutionary relationships between the laccase genes of polyporales: orthology-based classification of laccase isozymes and functional insight from Trametes hirsuta. Front Microbiol. 2019;10:152. doi: 10.3389/FMICB.2019.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westereng B, Cannella D, Wittrup Agger J, Jorgensen H, Larsen Andersen M, Eijsink VG, Felby C. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep. 2015;5:18561. doi: 10.1038/srep18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsumura H, Umezawa K, Takeda K, Sugimoto N, Ishida T, Samejima M, Ohno H, Yoshida M, Igarashi K, Nakamura N. Discovery of a eukaryotic pyrroloquinoline quinone-dependent oxidoreductase belonging to a new auxiliary activity family in the database of carbohydrate-active enzymes. PLoS ONE. 2014 doi: 10.1371/JOURNAL.PONE.0104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan Y, Zhang Z, Wang F, Li J, Hu K, Du Z. Lignin degradation in corn stover catalyzed by lignin peroxidase from Aspergillus oryzae broth: effects of conditions on the kinetics. Renew Energy. 2019;130:32–40. doi: 10.1016/j.renene.2018.06.037. [DOI] [Google Scholar]
- 51.Martínez AT, Camarero S, Ruiz-Dueñas FJ, Martínez MJ. Biological lignin degradation. In: Beckham GT, editor. Lignin valorization: emerging approaches. Royal Society of Chemistry; 2018. pp. 199–225. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Heatmap of %identity scores amongst the close relatives identified from its phylogenetic clade.Supplementary Table 2: Distribution of P. petalodes strain GGF6 unigenes among all Pfam families and their respective functions.Supplementary Table 3: Distribution of P. petalodes strain GGF6 unigenes among all TIGR families and their respective functions. (XLSX 130 KB)
Data Availability Statement
Raw reads can be accessed using this link https://drive.google.com/drive/folders/1wnhbJvjAW3mYyJAFCpHYHAufwOY7PEvb?usp=sharing. The raw data can also be provided upon request.