Abstract
Streptokinases secreted by nonhuman isolates of group C streptococci (Streptococcus equi, S. equisimilis, and S. zooepidemicus) have been shown to bind to different mammalian plasminogens but exhibit preferential plasminogen activity. The streptokinase genes from S. equisimilis strains which activated either equine or porcine plasminogen were cloned, sequenced, and expressed in Escherichia coli. The streptokinase secreted by the equine isolate had little similarity to any known streptokinases secreted by either human or porcine isolates. The streptokinase secreted by the porcine isolate had limited structural and functional similarities to streptokinases secreted by human isolates. Plasminogen activation studies with immobilized (His)6-tagged recombinant streptokinases indicated that these recombinant streptokinases interacted with plasminogen in a manner similar to that observed when streptokinase and plasminogen interact in the fluid phase. Analysis of the cleavage products of the streptokinase-plasminogen interaction indicated that human, equine, and porcine plasminogens were all cleaved at the same highly conserved site. The site at which streptokinase was cleaved to form altered streptokinase (Sk*) was also determined. This study confirmed not only the presence of streptokinases in nonhuman S. equisimilis isolates but also that these proteins belong to a family of plasminogen activators more diverse than previously thought.
Most group A, C, and G streptococci isolated from human hosts secrete a plasminogen activator known as streptokinase which catalyzes the conversion of the plasma zymogen, plasminogen, to the serine protease plasmin. Human plasminogen and streptokinase form a 1:1 stoichiometric complex that hydrolyzes other plasminogen molecules to generate plasmin, which subsequently can degrade fibrin, the primary protein component of blood clots. Introduced into clinical practice in the late 1950s, the intravenous infusion of streptokinase has become one of the treatments of choice in acute myocardial infarction. With regards to bacterial pathogenesis, plasmin may facilitate tissue invasion by dissolution of the fibrin barrier that forms at the site of infection, by hydrolysis of extracellular matrix proteins such as laminin or fibronectin, and by activation of latent collagenases and other zymogen forms of metalloproteinases (3, 21, 33). In addition, it has been hypothesized that certain isotypes of streptokinase are nephrotropic in nature and that the plasmin generated by the nephrostreptokinase-plasminogen complexes may be responsible for clinical and histopathological observations indicative of poststreptococcal glomerulonephritis (9, 13, 22, 30).
Given the clinical importance of this protein, a great deal of effort has been directed toward characterizing and understanding the molecular basis of the interaction of streptokinase with plasminogen. Most of this research has focused on the streptokinase secreted by a human isolate of the group C streptococcus S. equisimilis. These investigations, although increasing our understanding of the streptokinase-plasminogen interaction, have also created the impression that all streptokinases belong to a family of homologous proteins, with similar biophysical and biochemical properties. This has led to the failure to fully understand the importance of the concept of species specificity, with the result that the group C streptococci S. equi, S. zooepidemicus, and S. equisimilis, isolated from non-human hosts, have been regarded as non-streptokinase producers simply on the basis of the inability to activate human plasminogen (1, 6). However, McCoy et al. (26) and Nowicki et al. (31) demonstrated that group C streptococci isolated from nonhuman sources secreted streptokinases which preferentially activated plasminogen obtained from the host from which the isolate had been obtained. Although these streptokinases preferred to activate only the plasminogen derived from the host, they all bound plasminogen regardless of the host source. These observations suggested that there are two major events in the activation of plasminogen by streptokinase; a primary event (binding) which is not species specific and a secondary event (activation) which is species-specific. To better understand the basis of these observations, the streptokinase genes from two nonhuman group C S. equisimilis strains were cloned and sequenced; as a preliminary step in a more comprehensive investigation, these streptokinases, one from an S. equisimilis equine isolate (ESk) and one from an S. equisilimilis porcine isolate (PSk), were cloned and expressed in Escherichia coli as (His)6-tagged fusion proteins in order to study the interaction of these proteins with different mammalian plasminogens. These two streptokinases were compared to the streptokinase from an S. equisimilis human isolate (HSk).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
In this study the following strains of group C S. equisimilis streptococci were used: strain 87-542-W, isolated from an equine host; strain 89-272, isolated from a porcine host; and strain H46A, isolated from a human host (kindly provided by Anne Marie Bergholm, Astra Hässle, Mölndal, Sweden). Bacteria were grown at 37°C for 8 h in 500 ml of CDM medium (JRH Biosciences, Lenexa, Kans.) supplemented (10%, vol/vol) with a Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) ultrafiltrate (<10,000 kDa). The pH of the cultures was monitored and maintained at 7.0 by periodic addition of 10 N NaOH and sterile 10% (wt/vol) glucose.
Construction of streptococcal genomic library.
Bacterial cells were lysed and DNA was isolated by the procedure of Monsen et al. (27). Genomic DNA was purified by two rounds of CsCl gradient centrifugation in a Beckman Vti 80 rotor at 70,000 × g for 5.5 h. After extraction of ethidium bromide with water-saturated butanol, the DNA was dialyzed overnight against 6 liters of Tris-EDTA (TE) buffer. Purified DNA (100 to 200 μg) was partially digested with Sau3AI and subsequently size fractionated by centrifugation in a 10 to 40% sucrose-TE step gradient. Fractions were collected in 0.5-ml aliquots and examined by agarose gel electrophoresis; fractions that contained DNA fragments of 2 to 10 kb in size were combined, diluted with water, and ethanol precipitated overnight at −20°C. DNA fragments were resuspended in 50 μl of TE buffer; 0.25, 0.5, and 1.0 μg of DNA were ligated to 0.5 μg of a predigested (BamHI/CIAP) Lambda ZAP Express vector (Stratagene, La Jolla, Calif.) in a total ligation volume of 5 μl. One microliter from each ligation was packaged by using Gigapak II Gold packaging extract (Stratagene), and the resulting phage were infected into the host strain E. coli XL1-Blue MRF′ and subsequently applied to the top agar layer of NZY agar plates containing IPTG (1 mM) and X-Gal. Ligations with the vector-to-insert ratio that yielded the most plaques and the best blue-white color selection were packaged and set aside for further screening. E. coli XL1-Blue MRF′ cells were infected with the recombinant phage and grown on 150-mm NZY plates for 8 h. Each plate was then tested with a modification of the casein overlay procedure of Malke and Ferretti (23). Briefly, 18 ml of a warm (50°C) buffer-agar solution (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, and 1% agar) containing 2 ml of skim milk (Difco Laboratories), prepared according to the manufacturer's instructions, and 200 μg of equine or porcine plasminogen were carefully layered on top of each plate and incubated for a further 8 to 12 h. Plaques showing evidence of caseinolysis were isolated. Positive recombinant phagemids were excised from the parent phage by infection into E. coli XLOLR in the presence of helper phage. One colony from each clone was grown overnight in 5 ml of LB medium supplemented with 50 μg of kanamycin per ml, and the supernatant was acidified by addition of a one-fifth volume of 60% (wt/vol) trichloroacetic acid. After centrifugation at 15,000 × g for 15 min, pellets were washed with 95% (vol/vol) ethanol containing 5% (vol/vol) saturated sodium acetate and dried. Pellets were resuspended in 50 μl of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) solubilization buffer, subjected to SDS-PAGE, and subsequently transferred to nitrocellulose prior to testing for the presence of streptokinase in a solid phase plasminogen activation assay using the chromogenic substrate H-d-valyl-leucyl-lysine-para-nitroanilide (12, 16).
PCR analysis of cloned DNA.
As a preliminary step to sequencing, the size of the various inserts in the streptokinase secreting clones was determined by PCR analysis. Clones were grown in 5 ml of LB supplemented with kanamycin (50 μg/ml), and the double-stranded phagemids were isolated by using a commercially available plasmid purification kit (QIAprep; Qiagen, Chatsworth, Calif.). DNA (25 ng) from each clone was amplified by using 25 pmol of forward (5′-ATTAACCCTCACTAAAGGGA-3′) and reverse (5′-AATACGACTCACTATAGGGC-3′) primers complementary to the DNA sequences flanking the multiple cloning site of the vector. PCR was performed by using a programmable thermal control unit (MJ Research, Cambridge, Mass.) programmed for 30 cycles to denature the template at 94°C for 1 min, anneal primers to the template at 55°C for 1 min, and extend polymerization at 72°C for 5 min.
DNA sequencing.
Clone pSTKE22, containing a 2.9-kb insert of 87-542-W genomic DNA, and clone pSTKP8, containing a 3.0-kb insert of 89-272 genomic DNA, were sequenced in both directions according to Sanger et al. (34) by using a Sequenase, version 2.0, kit (Amersham, Arlington Heights, Ill.). Radiolabeled ([α-35S]dATP) samples were heated to 85°C for 3 min prior to addition to a 6% 19:1 acrylamide:bis-acrylamide–7 M urea wedge gel prepared in Tris-borate-EDTA (TBE) buffer and cast in a Macrophor sequencing apparatus (Pharmacia, Piscataway, N.J.). Oligonucleotides were resolved at 33 W for 2 to 4 h, after which the gel was washed twice with 10% acetic acid, air dried, and developed by autoradiography with Biomax-MR imaging film (Eastman Kodak, Rochester, N.Y.).
Cloning and expression of the streptokinase genes into the prokaryotic expression vector pQE-30.
The streptokinase genes were cloned into the pQE-30 vector (Qiagen) to produce recombinant streptokinases appended at the N terminus with a polyhistidine (His)6 domain to facilitate the binding of recombinant streptokinase molecules to an IMAC (Talon) affinity column (Clontech, Palo Alto, Calif.). This would not only facilitate purification of the recombinant streptokinase molecules from the bacterial cytosol but also permit the study of the interaction of mammalian plasminogens with matrix-bound streptokinases. Genomic DNA from the parent strains was used as template for PCR amplification of the streptokinase genes. The forward primers used in these amplifications were 5′-CGCGGATCCAATAATTACGCCAAGCCT-3′ for 87-542-W, 5′-CGCGGATCCATTGGTGGCAGAGAC-3′ for 89-272, and 5′-CGCGGATCCATTGCTGGACCTGAG-3′ for H46A streptokinase genes. The sequences chosen were complementary to the DNA sequences (underlined) coding for the start of the mature proteins, with additional codons to facilitate directional cloning into the BamHI and SmaI restriction endonuclease sites of the pQE30 vector. The reverse primers, 5′-CTTATTTGTTTGATTCGTTGTACC-3′ for 87-542-W, 5′-TCCCCCGGGTTATTTGATCATATG-3′ for 89-272, and 5′-TCCCCCGGGTTATTTGTCGTTAGGGTT-3′ for H46A streptokinase genes, were complementary to the sequences coding for the carboxyl termini of the mature proteins. This cloning strategy resulted in recombinant constructs in which a (His)6GlySer domain was appended to the N terminus of the native molecule. Amplified DNA was purified with a commercially available PCR purification kit (Boehringer Mannheim, Indianapolis, Ind.). Amplified 87-542-W DNA was treated with Pfu polymerase (Stratagene) to polish the 3′ end, as it did not have an internal SmaI site. Purified PCR products were incubated with BamHI overnight, phenol extracted, ethanol precipitated, and either incubated with SmaI (89-272 and H46A) or phosphorylated with T4 kinase (87-542-W) prior to ligation into pQE-30 (Qiagen), which had been previously restricted with BamHI and SmaI. Competent Top 10 E. coli cells (Invitrogen, Carlsbad, Calif.) were transformed with 1 μl of the ligation mix, and the resulting clones were lysed with chloroform fumes and screened for expression of streptokinase with the casein overlay procedure using the appropriate plasminogen. Transformed Top 10/pQE30 cells were grown at 30°C to mid-log phase, and expression of the streptokinase gene was induced by addition of IPTG (1 mM). After 4 h of growth at 30°C, cells were harvested by centrifugation at 5,000 × g, and the bacterial pellet was suspended in 15 ml of lysis buffer (20 mM Tris-Cl [pH 8.0]–100 mM NaCl–8 M urea) per 50 ml of original culture volume, sonicated briefly, and clarified by centrifugation at 14,000 × g for 30 min. The clarified supernatant was applied to a 1-ml IMAC (Talon) agarose column (capacity, 2 to 3 mg/ml) equilibrated in lysis buffer. The column was washed twice with lysis buffer, followed by four column bed volumes of 20 mM Tris-Cl, pH 8.0, containing 100 mM NaCl. The bound protein was eluted from the resin with 100 mM EDTA and dialyzed overnight against 10 mM Tris-Cl, pH 8.0. Eluted protein was assayed for purity by SDS-PAGE and for activity by the solid-phase plasminogen activation assay (16).
Cloning of streptokinase genes from strains 87-542-W and 89-272 into the procaryotic expression vector pQ60.
The N-terminal amino acid sequence of both 87-542-W (ESk) and 89-272 (PSk) mature streptokinases was confirmed by cloning the respective genes into the expression vector pQE-60 (Qiagen). This vector permits the expression of cloned genes containing the initial ATG codon of the signal sequence and is expressed with an appended (His)6 tail at the C terminus to facilitate purification. Genomic DNA from both the 89-272 and 87-542-W streptococcal strains was amplified by PCR using nucleotide primers complementary to the 5′ and 3′ termini of the genome representing the immature protein. Nucleotide primers complementary to the genome coding for the N terminus of the streptokinase molecules were appended with nucleotides containing an NcoI restriction endonuclease cleavage site, and nucleotide primers complementary to the DNA sequence representing the C terminus of the streptokinase molecules were appended with a nucleotide sequence containing a BglII restriction endonuclease cleavage site to facilitate directional cloning into the pQE-60 vector. After cloning, expression, and purification by metal affinity chromatography, the proteins were sequenced and the N-terminal amino acid sequences were determined.
Plasminogen activation studies with recombinant streptokinases.
Immobilized plasminogen activation studies were performed by a modification of the procedure of Lizano and Johnston (20). (His)6GlySer streptokinases (100 μg) in 10 mM Tris-Cl (pH 8.0) and 100 mM NaCl were added to 100 μl of metal-chelating IMAC affinity matrix. After incubation at 22°C for 5 min, the slurry was applied to a Spin-X microcentrifuge filter (Costar, Cambridge, Mass.) fitted with a 0.45-μm cellulose acetate filter. The matrix was pelleted by centrifugation at 2,000 × g for 3 min and subsequently washed several times with 20 mM Tris-Cl, pH 7.4. The matrix was removed from the Spin-X unit, placed in a microcentrifuge tube, and resuspended in 200 μl of 50 mM Tris-Cl buffer, pH 7.4. To a series of microcentrifuge tubes, 25-μl aliquots of the matrix were added to an equimolar amount of plasminogen in 50 mM Tris-Cl buffer, pH 7.4. Samples were incubated at 22°C and placed on a rotating platform to keep the matrix in suspension. At different intervals (0 to 120 min), a sample was selected and the reaction was terminated by the addition of 0.1 volumes of 10× stop buffer (1.0 M NaHCO3, 1.0 M ɛ-aminocaproic acid [pH 9.4]). The sample was transferred to a Spin-X microcentrifuge tube and pelleted by centrifugation at 2,000 × g for 3 min. Immobilized reactants were eluted by addition of 25 μl of 100 mM EDTA, followed by centrifugation at 5,000 × g for 10 min. Samples were prepared for SDS-PAGE analysis by addition of 25 μl of 2× SDS buffer containing β-mercaptoethanol, boiled for 5 min, and applied to an SDS–10% polyacrylamide gel.
PAGE and protein blotting.
SDS-PAGE was performed according to Laemmli (17). For sequencing of electrotransferred proteins, the upper buffer reservoir of the electrophoretic chamber contained 1 mM reduced glutathione to act as a scavenger of potential acrylamide polymerization by-products. Gels were either stained with Coomassie blue or equilibrated in carbonate buffer (10 mM NaHCO3–3 mM Na2CO3 [pH 9.9]–20% [vol/vol] methanol) for 10 min and electrophoretically transferred to 0.45-μm nitrocellulose membranes (Bio-Rad, Hercules, Calif.) at 900 mA and 4°C for 30 min, according to Dunn (7), in a Hoefer miniblotting transfer cell (Hoefer Scientific, San Francisco, Calif.) or to Immobilon-CD-PVDF transfer membranes (Millipore, Bedford, Mass.) for subsequent N-terminal amino acid sequence analysis.
N-terminal amino acid analysis.
Proteins transferred after SDS-PAGE to PVDF membranes were visualized by staining with 0.1% (wt/vol) Ponceau S in 1% (vol/vol) acetic acid. Those protein bands which represented either cleaved plasminogen or streptokinase molecules in the plasminogen activation studies were excised and subjected to amino acid sequencing on an Applied BioSystems model 4767A protein sequencer by the LSUMC Core Laboratories staff. To determine which band represented which cleavage product, amino acid sequence data were compared to published sequences of human (8), porcine (25), and equine (35) plasminogen and H46A streptokinase (24) and to the sequence data for ESk and PSk described in this communication.
Purification of equine and human plasminogen.
Human and equine plasminogen were purified from fresh frozen plasma by lysine-Sepharose chromatography according to Deutsch and Mertz (5). l-Lysine was coupled to Sepharose 4B activated with CnBr according to Kohn and Wilchek (15). Bound plasminogen was eluted by addition of 0.2 M ɛ-aminocaproic acid in 0.1 M phosphate buffer, pH 7.2, containing 5 μg of aprotinin per ml. Plasminogen-containing fractions were concentrated by ultrafiltration, washed with 50 mM Tris-Cl buffer, pH 7.5, and stored at −80°C.
Isolation of native 89-272 streptokinase (PSk) by equine plasminogen affinity chromatography.
Equine plasminogen was covalently coupled to agarose by the method described by Nowicki et al. (31). The equine plasminogen affinity column was equilibrated with 40 mM Tris-Cl, pH 7.5. The filtered supernatant of a 89-272 bacterial culture was passed over the column at a rate of 1 ml/min at 4°C. Bound PSk streptokinase was eluted by addition of 8 M urea in 40 mM Tris-Cl, pH 7.5. Fractions containing the streptokinase were subjected to SDS-PAGE, electroblotted, and subjected to N-terminal amino acid analysis.
MALDI-TOF mass spectral analysis of recombinant streptokinases.
Mass analysis of the recombinant streptokinases was done by using a Perceptive Biosystems Voyager-DE MALDI-TOF (matrix-assisted laser desorption ionization–time of flight) mass spectrometer. A matrix solution of 10 mg of sinapinic acid per ml in acetonitrile-water (1:1) was prepared, mixed with an equal amount (vol/vol) of dialyzed protein solution, and spotted on a sample plate. The sample was run under positive ion mode according to the following parameters: method, LDE1008A; mode, linear; accelerating voltage, 20,000; laser, 2,000; low mass gate, 400; grid voltage, 94%: scan average, 222; guide wire voltage, 0.050%; pressure, 1.09 × 10−6.
Analysis of sequence data.
Manipulations of the DNA and protein sequence data were done by PC/Gene (Intelligenetics, Geneva, Switzerland). The DNA sequences were translated into protein by using the PC/Gene TRANSL program. One-on-one alignments were done by Myers and Miller's (28) method with a Dayhoff MDM-78 Comparison Matrix open gap cost of 50 and a unit gap cost of 100. Protein comparisons were done by the method of Needleman and Wunsch (29), with a bias of 60 and a gap penalty of 60.
Reagents and methods not described in text.
All restriction endonucleases and DNA-modifying enzymes were purchased from New England Biolabs (Beverly, Mass.), unless otherwise noted. All chemical reagents, including bovine, porcine, and rabbit plasminogen, were purchased from Sigma (St. Louis, Mo.), unless otherwise noted. Total protein concentrations were determined by a microassay adaptation of the bicinchoninic acid (BCA) method of Smith et al. (36). When streptokinases were assayed, a standard curve using streptokinase isolated from strain H46A (14) was used; for plasminogen, purified human plasminogen was used. Methods not otherwise described were obtained from Ausubel et al. (2).
Nucleotide sequence accession numbers.
The nucleotide sequences of the streptococcal streptokinase (sk) genes reported here have been deposited in the GenBank nucleotide sequence database. The sk gene from strain 87-542-W has been assigned accession no. AF104301, and the sk gene from strain 89-272 has been assigned accession no. AF104300. The GenBank accession number of the sk gene from strain H46A used in this study is K02986.
RESULTS
Comparison of cloned and native streptokinases.
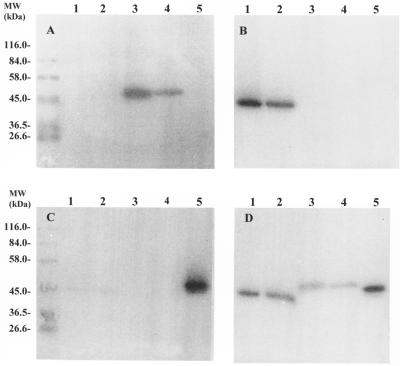
Streptococcal DNA purified from S. equisimilis strains isolated from either an equine host (87-542-W) or a porcine host (89-272) and partially digested with Sau3AI and ligated into a Lambda ZAP Express vector produced numerous clones which had streptokinase activity, as observed by the casein-plasminogen overlay assay. One clone (pSTKE22) contained a 2.9-kb fragment of 87-542-W streptococcal DNA, and another clone (pSTKP8) contained a 3.0-kb fragment of 89-272 streptococcal DNA. Transfer of the phagemids to E. coli DH5α and reanalysis by the casein-plasminogen overlay assay indicated that the E. coli transcriptional and translational machinery recognized the streptococcal promoters as well as extracellular transport signals. Cloned streptokinases were compared to their native counterparts in terms of apparent molecular size and activation properties in the presence of equine, human, and porcine plasminogen by using the solid-phase plasminogen activation assay. Clone pSTKE22 secreted a streptokinase which activated equine plasminogen and was indistinguishable from the native streptokinase secreted by strain 87-542-W (Fig. 1A, lanes 3 and 4); both cloned and wild-type streptokinases failed to activate porcine plasminogen (Fig. 1B, lanes 3 and 4) and human plasminogen (Fig. 1C, lanes 3 and 4). Clone pSTKP8 secreted a streptokinase that activated porcine plasminogen and was indistinguishable from the wild-type streptokinase (Fig. 1B, lanes 1 and 2); both cloned and native streptokinases failed to activate equine plasminogen (Fig. 1A, lanes 1 and 2) but could weakly activate human plasminogen (Fig. 1C, lanes 1 and 2). Figure 1D illustrates the differences in apparent molecular size of both cloned and native streptokinases from porcine (lanes 1 and 2), equine (lanes 3 and 4), and human (lane 5) streptococcal isolates in a solid-phase plasminogen activation assay in which human, equine, and porcine plasminogens were all present. Cloned streptokinase from the porcine isolate (89-272) had an apparent molecular mass of ∼44 kDa, cloned streptokinase from the equine isolate (87-542-W) had an apparent molecular mass of ∼49 kDa, and cloned streptokinase from a human isolate (H46A) had an apparent molecular mass of ∼47 kDa. Each cloned streptokinase has the same apparent molecular size as the corresponding native streptokinases.
FIG. 1.
Solid-phase plasminogen activation assay of native and cloned streptokinases in the presence of equine plasminogen (A), porcine plasminogen (B), human plasminogen (C), and a mixture of equine, human, and porcine plasminogens (D). Lanes 1, native streptokinase from a porcine isolate of S. equisimilis, strain 89-272; lanes 2, recombinant streptokinase from a porcine isolate of S. equisimilis, strain 89-272, expressed by clone DH5α/pSTKP8; lanes 3, native streptokinase from an equine isolate of S. equisimilis, strain 87-542-W; lanes 4, recombinant streptokinase from an equine isolate of S. equisimilis, strain 87-542-W, expressed by clone DH5α/pSTKE22; lanes 5, native streptokinase from a human isolate of S. equisimilis, strain H46A.
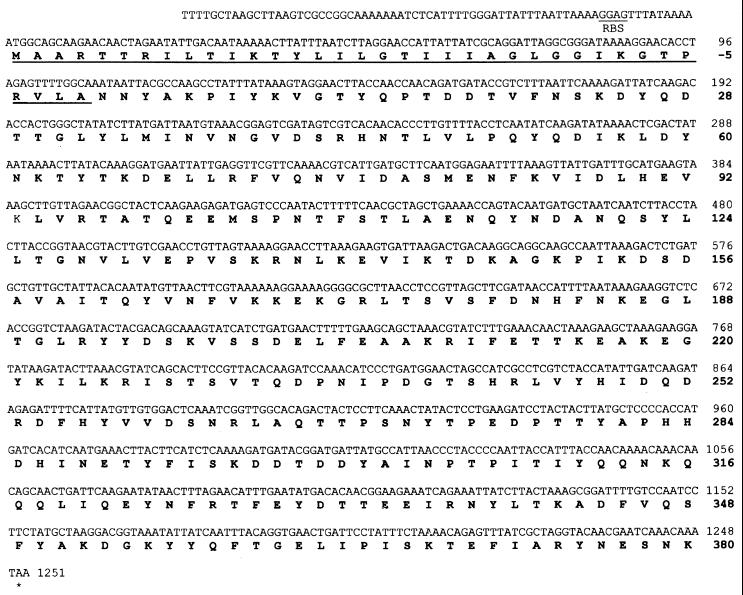
Nucleotide and deduced amino acid sequence of the cloned streptokinase gene from S. equisimilis 87-542-W (ESk) and MALDI-TOF mass spectral analysis of the expressed gene product.
The streptokinase gene from strain 87-542-W was found to be 1,251 bp in size and to code for an immature protein of 416 amino acids (Fig. 2). Posttranslational cleavage of the first 36 amino acids would result in a mature protein having 380 amino acids and a molecular mass of 44.12 kDa. From the N-terminal amino acid data (NNYAKPIYKVGTYQPTDDTVFNSKDAYQDT-30GLYLT35 [31]) of wild-type 87-542-W streptokinase (ESk), it was deduced that Asn37 was the N-terminal residue of the mature protein. This was confirmed by sequence analysis of the pQ60-expressed ESk. The first 34 amino acids of the cloned sequence agreed with the native N-terminal sequence except that Thr35 was replaced by a Met residue in the cloned sequence. The protein sequence data did not resolve a residue at position 30, but in the cloned sequence a Thr residue was indicated. Although SDS-PAGE analysis indicated a molecular mass of ∼49 kDa and genetic analysis indicated a molecular size of 44.12 kDa, the electrophoretic mobility of both the native and cloned ESk indicated that the entire gene had been cloned. MALDI-TOF mass spectral analysis of the pQE30-expressed (His)6 GlySer-ESk showed it to be a 45.192-kDa protein, which agreed closely with the calculated molecular mass (44.120 kDa) from the sequence data plus the mass of the appended (His)6GlySer residues (0.985 kDa). Further confirmation that the entire gene had been cloned came from MALDI-TOF mass spectral analysis of the ESk-(His)6 expressed in pQE60; this protein had a mass of 44.82 kDa, which agreed with the calculated molecular mass obtained from the sequence data plus the mass of the carboxyl terminus-appended (His)6 tag (0.702 kDa).
FIG. 2.
Nucleotide and deduced amino acid sequence of the streptokinase gene from the equine isolate S. equisimilis 87-542-W. The signal sequence is underlined, and the deduced amino acid sequence is in boldface type.
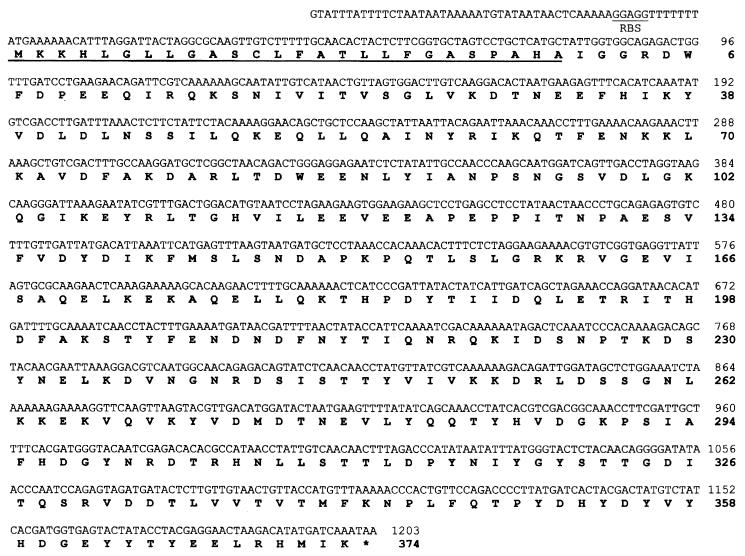
Nucleotide and deduced amino acid sequence data of the cloned streptokinase gene from S. equisimilis 89-272 (PSk) and MALDI-TOF mass spectral analysis of the expressed gene product.
The sequence data indicated that the 89-272 streptokinase gene had 1,203 bp (Fig. 3) and coded for an immature protein of 400 amino acids. The mature protein was deduced to begin at amino acid 27, resulting in a protein of 374 amino acids; the first 28 amino acids of the mature protein agreed with the N-terminal analysis (IGGRDWFDPEEQIRQKSNI VITVSGLVKK29 T) of the native protein purified from 89-272 culture supernatants by equine plasminogen-Sepharose affinity chromatography. This was confirmed by sequence analysis of PSk cloned and expressed in pQE60. The only exception was Lys29, which was replaced by an Asp residue in the cloned sequence. The apparent molecular mass of the mature protein determined by SDS-PAGE (∼44 kDa) agreed closely with the molecular mass calculated from the sequence data (43.406 kDa). MALDI-TOF mass spectral analysis indicated the pQE30-expressed (His6)GlySer-PSk streptokinase to be a 44.350-kDa protein, which agreed closely with the sequence data-calculated mass (43.406 kDa) plus the contribution from the (His)6GlySer appended residues resulting from the cloning strategy. In addition, MALDI-TOF mass spectral analysis of the PSk-(His)6 expressed in pQE60 indicated that this protein had a mass of 45.112 kDa, which also agreed with the calculated molecular mass from the sequence data plus the mass of the carboxyl terminus-appended (His)6 tag.
FIG. 3.
Nucleotide and deduced amino acid sequence of the streptokinase gene from the porcine isolate S. equisimilis 89-272. The signal sequence is underlined, and the deduced amino acid sequence is in boldface type.
Amino acid homology.
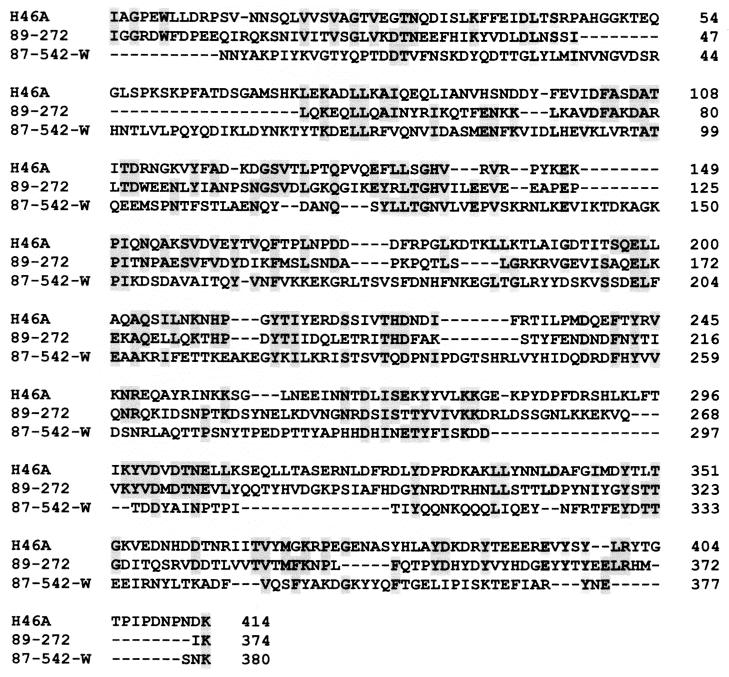
The two streptokinases described in this study were quite dissimilar from each other and from the streptokinase secreted by the human isolate. ESk had only 21.4% identity with PSk and 25.4% identity with HSk at the amino acid level. PSk had 35.3% amino acid identity with HSk. In addition, analysis of the primary sequence did not indicate any significant regions of homology (Fig. 4) between ESk, PSk, and HSk.
FIG. 4.
Multiple alignment of deduced amino acid sequences of streptokinases from S. equisimilis H46A (GenBank accession no. K02986), S. equisimilis 89-272 (GenBank accession no. AF104300), and S. equisimilis 87-542-W (GenBank accession no. AF104301). Shaded boxes indicate identical amino acid residues.
Interaction of immobilized (His)6GlySer-ESk with different mammalian plasminogens.
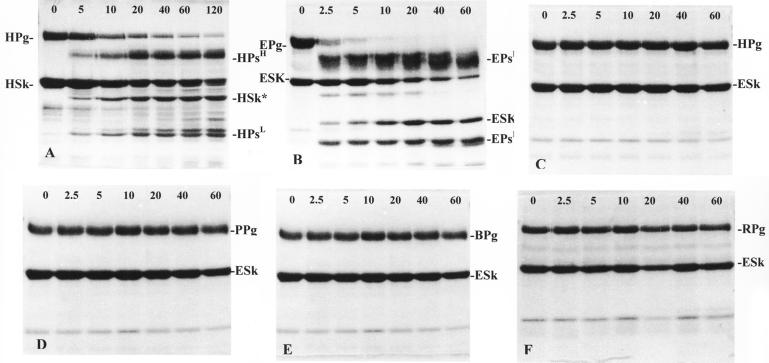
To study the interaction of the recombinant streptokinases with different mammalian plasminogens, (His)6GlySer-streptokinase fusion constructs were immobilized on a metal-chelating IMAC affinity matrix. Figure 5 illustrates the interaction of equine (Fig. 5B), human (Fig. 5C), porcine (Fig. 5D), bovine (Fig. 5E), and rabbit (Fig. 5F) plasminogen with immobilized (His)6GlySer-ESk. Samples taken over a period of 5 to 120 min after application of plasminogen to the immobilized streptokinase indicated that this immobilized streptokinase fusion protein was capable of binding and activating equine plasminogen to plasmin. Although human, porcine, bovine, and rabbit plasminogens were bound by this streptokinase, none were activated. These results were consistent with the solid-phase assay results (Fig. 1). N-terminal amino acid analysis of all of the breakdown products resulting from the streptokinase-plasminogen interaction (Fig. 5B) indicated that a 27-kDa protein band had the N-terminal amino acid sequence AGKPI. From the sequence data (Fig. 2), it was apparent that this band represented a cleaved product of ESk. Immobilized (His)6GlySer-ESk was cleaved between amino acids Lys147 and Ala148 to yield an altered 27.173-kDa streptokinase (ESk*). Generation of ESk* occurred within 5 min after addition of equine plasminogen in a manner similar to that observed in the interaction of human plasminogen with immobilized (His)6GlySer-HSk (Fig. 5A).
FIG. 5.
SDS-PAGE analysis of the interaction of immobilized recombinant (His)6GlySer-H46A streptokinase with human plasminogen (A) and of recombinant (His)6GlySer-87-542-W streptokinase with equine (B), human (C), porcine (D), bovine (E), and rabbit (F) plasminogen over time (in minutes). (A) HPg, human plasminogen; HSk, H46A streptokinase; HPsH, plasmin heavy chain; HPsL, plasmin light chain; HSk*, altered H46A streptokinase (40.998 kDa). (B) EPg, equine plasminogen; ESk, 87-542-W streptokinase; EPsH, equine plasmin heavy chain; EPsL, equine plasmin light chain; ESk*, altered 87-542-W streptokinase (27.173 kDa). (C) HPg, human plasminogen; ESk, 87-542-W streptokinase. (D) PPg, porcine plasminogen; ESk, 87-542-W streptokinase. (E) BPg, bovine plasminogen; ESk, 87-542-W streptokinase. (F) RPg, rabbit plasminogen; ESk, 87-542-W streptokinase.
Interaction of immobilized (His)6GlySer-PSk with different mammalian plasminogens.
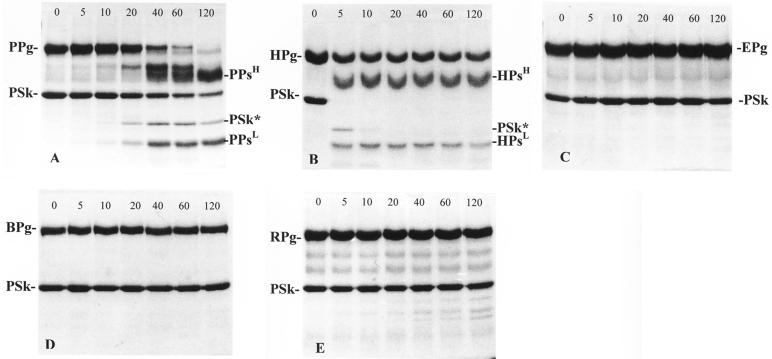
The interaction of immobilized (His)6GlySer-PSk streptokinase with human, porcine, equine, bovine, and rabbit plasminogen resulted in activation of porcine plasminogen (Fig. 6A), partial activation of human plasminogen (Fig. 6B), and no activation of equine (Fig. 6C), bovine (Fig. 6D), or rabbit plasminogen (Fig. 6E). These observations agreed with the results obtained in the solid-phase assay with native and cloned PSk (Fig. 1). Activation of porcine plasminogen by immobilized (His)6GlySer-PSk streptokinase resulted in the formation of several breakdown products (Fig. 6A). N-terminal amino acid analysis of all of the breakdown products resulting from this streptokinase-plasminogen interaction indicated that a 31-kDa protein band had the N-terminal sequence LTGHV. From the sequence data (Fig. 3), it was apparent that this band represented a cleaved product of PSk. Immobilized (His)6GlySer-PSk was cleaved between Arg109 and Leu110 upon incubation with porcine plasminogen, which resulted in an altered streptokinase (PSk*) 30.784-kDa molecule. However, generation of PSk* by the interaction of porcine plasminogen with immobilized (His)6GlySer-PSk proceeded at a much lower rate. Cleavage of the Arg109-Leu110 bond in the immobilized PSk did not occur until after 20 min of incubation with porcine plasminogen. This was very different from the activation profiles observed in the interaction of human plasminogen with (His)6GlySer-HSk (Fig. 5A) or of equine plasminogen with (His)6GlySer-ESk (Fig. 5B), in which cleavage of the immobilized streptokinases occurred within 5 min after addition of homologous plasminogen. Activation of human plasminogen by immobilized (His)6GlySer-PSk streptokinase occurred within 5 min but then stopped, with no further conversion of plasminogen to plasmin (Fig. 6B). N-terminal amino acid analysis of the breakdown products resulting from the interaction of (His)6GlySer-PSk with human plasminogen indicated that PSk was cleaved at the same peptide bond (Arg109-Leu110) that was cleaved in the interaction with porcine plasminogen. However, the protein band identified as PSk* (Fig. 6B) disappeared between the 5- and 10-min incubation time points, indicating extensive degradation of this molecule.
FIG. 6.
SDS-PAGE analysis of the interaction of immobilized recombinant (His)6GlySer-89-272 streptokinase with human (A), porcine (B), equine (C), bovine (D), and rabbit (E) plasminogen over time (in minutes). (A) PPg, porcine plasminogen; PSk, 89-272 streptokinase; PPsH, porcine plasmin light chain; PPsL, porcine plasmin light chain; PSk*, altered 89-272 streptokinase (30.784 kDa). (B) HPg, human plasminogen; PSk, 89-272 streptokinase; HPsH, plasmin heavy chain; HPsL, plasmin light chain; PSk*, altered 89-272 streptokinase (30.784 kDa). (C) EPg, equine plasminogen; PSk, 89-272 streptokinase. (D) BPg, bovine plasminogen; PSk, 89-272 streptokinase. (E) RPg, rabbit plasminogen; PSk, 89-272 streptokinase.
Interaction of immobilized (His)6GlySer-HSk streptokinase with different mammalian plasminogens.
As a control, the interaction of immobilized (His)6GlySer-HSk with human plasminogen was assessed. Incubation of immobilized HSk with human plasminogen over a 120-min period indicated that this immobilized streptokinase was capable of activating human plasminogen (Fig. 5A). Activation occurred within 5 min after incubation with human plasminogen, resulting in the generation of several breakdown products. N-terminal amino acid analysis of these breakdown products indicated that a 41-kDa protein corresponded to altered streptokinase (HSk*). This altered streptokinase had the N-terminal sequence SKPFA. This indicated that HSk had been cleaved between Lys59 and Ser60, which resulted in the generation of a 40.998-kDa HSk*. Although HSk bound human, equine, porcine, bovine, and rabbit plasminogens, it did not activate equine, porcine, bovine, or rabbit plasminogens. The activation profiles were similar to those observed in Fig. 5C to F, which illustrate the interaction of immobilized (His)6GlySer-ESk streptokinase with human (Fig. 5C), porcine (Fig. 5D), bovine (Fig 5E), and rabbit (Fig. 5F) plasminogens.
Analysis of plasminogen cleavage products.
The interaction of immobilized recombinant streptokinases with their corresponding host plasminogens resulted in the generation of plasmin heavy and light chains. N-terminal amino acid analysis of the breakdown products in Fig. 5A indicated that immobilized (His)6GlySer-HSk cleaved human plasminogen to generate human plasmin heavy (HPsH) and light (HPsL) chains; the plasmin light chain (∼25 kDa) had the N-terminal amino acid sequence VVGGC. When immobilized (His)6GlySer-PSk was incubated with human plasminogen (Fig. 6B), HPsH and HPsL chains were also generated, each a having the same apparent molecular size as the chains produced by the human plasminogen-HSk interaction; the HPsL chain generated by the interaction with PSk had an N-terminal amino acid sequence (VVGGC) identical to that of HPsL generated by the interaction of human plasminogen with HSk. Immobilized (His)6GlySer-PSk cleaved porcine plasminogen (Fig. 6A) to release HPsL, which had the same size (∼25 kDa) and N-terminal amino acid sequence (VVGGC) as the HPsL released by the activation of human plasminogen by immobilized (His)6GlySer-HSk. When immobilized (His)6GlySer-ESk was incubated with EPg (Fig. 5B), equine plasmin heavy (EPsH) and light (EPsL) chains were generated; equine plasminogen was cleaved to yield EPsL with the N-terminal sequence IVGGC and a size (∼25 kDa) similar to both the PsL and HPsL chains generated by the interaction of these streptokinases with their homologous plasminogens. These observations indicated that cleavage of the plasminogen molecules occurred at the same cleavage site (Arg561-Val562) in human plasminogen, a site which is highly conserved in a number of mammalian plasminogens (8, 25, 35).
DISCUSSION
All streptokinases sequenced to date, namely, those secreted by group A S. pyogenes strains NZ131 (10), SF13013 (37), and A374 (32), group C S. equisimilis H46A (24), and group G Streptococcus sp. strain G19909 (38), were originally isolated from streptococci which had infected human hosts. These streptokinases have been shown to be remarkably similar to one another, both functionally and structurally, with greater than 85% homology at the amino acid level. In addition, these streptokinases have the same number of amino acid residues, namely, 414. In contrast, the streptokinase secreted by a streptococcus (S. equisimilis 87-542-W) isolated from an equine host was 380 amino acids in length and exhibited at the amino acid level only 25.4% identity with streptokinase secreted by a streptococcus (S. equisimilis H46A) isolated from a human host. The second streptokinase studied, a streptokinase secreted by an S. equisimilis porcine isolate (strain 89-272), was 374 amino acids in length and had only 35.3% identity with the streptokinase from the S. equisimilis human isolate H46A. The identity between the two non-human-associated streptokinases was only 21.4%. Obviously, these two streptokinases are distinctly different from each other, not only at the primary structural level but also from human-associated streptokinases. Recently, streptokinases from Streptococcus uberis (18) and Streptococcus dysgalactiae (19), which preferentially activate bovine plasminogen, have been described. Johnsen et al. (11) have determined the primary structure of a streptokinase from S. uberis (NCTC 3858; accession no. AJ006413). This molecule exhibited 19.2% amino acid identity with streptokinase from the porcine isolate (strain 89-272) and 18.0% amino acid identity with streptokinase from the equine isolate (strain 87-542-W).
According to the current “activator complex” model of plasminogen activation by streptokinase, an initial binding event is followed by a conformational change imposed by the streptokinase molecule on the plasminogen moiety. This event uncovers an active site in the latter which leads to a series of catalytic and autocatalytic events that result in the generation of a streptokinase-plasmin complex with the cleavage of streptokinase to form an altered streptokinase still associated with the complex (4). This study demonstrated that immobilized streptokinases behave in a manner very similar to that observed in fluid-phase assays (4). The immobilized H46A streptokinase was cleaved at the same trypsin-sensitive peptide bond as that observed in fluid-phase studies (4). The streptokinase secreted by the human isolate is cleaved between amino acids Lys59 and Ser60, while the streptokinases from the equine and porcine isolates are cleaved between amino acids Lys147 and Ala148 and between Arg109 and Leu110, respectively. The observation that streptokinases secreted by streptococci from different hosts were able to differentially activate only the plasminogen derived from the same host but were able to bind human, porcine, equine, bovine, and rabbit plasminogens strongly suggests that these streptokinases must share a common plasminogen binding domain; however, analysis of the primary sequences of these streptokinases did not indicate any major contiguous regions of similarity. In addition, these three streptokinases did not share the amino acids demonstrated to bind to the light chain of human plasminogen, as described by Wang et al. (39). However, activation of plasminogen by streptokinase is a species-specific event. Logically, primary structural differences in those regions involved in activation among streptokinases from different species should be reflected in differences in secondary structure and finally in three-dimensional conformation; otherwise, activation would be nonspecific. Although the region required for activation has not been fully elucidated, the streptokinases from the porcine and equine isolates certainly contain enough differences in primary structure to account for potentially different conformations which would affect activation. As the streptokinase from the equine isolate is unable to activate either human or porcine plasminogen and the streptokinase from the porcine isolate can weakly activate human plasminogen, one would expect the primary structural differences between the equine, human, and porcine streptokinases to be greater than between the porcine and human streptokinases. This, in fact, is the case, as shown by the alignment studies and homology indexes.
The results from the plasminogen activation studies performed with the various recombinant His-tagged streptokinases were of particular interest because they demonstrated that these constructs were capable of forming a streptokinase-plasminogen complex that could progress to an altered streptokinase-plasmin complex while immobilized on an affinity matrix. The plasminogen domain where cleavage by the activator complex generates the heavy and light chains of plasmin seems to be highly conserved among plasminogen from different species. The actual cleavage site occurs at the same site (Arg561-Val562) for both human and porcine plasminogen and at a corresponding site (Arg-Ile) for equine plasminogen. It should be noted that Val and Ile are both hydrophobic amino acids with an aliphatic side chain bearing no net charge and are thus considered to be similar by Myers and Miller's method (28) of protein comparison.
The data presented in this paper nevertheless conclusively support the view that streptokinase belongs to a family of plasminogen activators whose members display greater diversity in primary structure than previously suspected. The observation that streptokinases demonstrate species-specific activation of plasminogen, reflecting the origin of the streptococcal isolate, implies a role for streptokinase in the pathogenesis of streptococcal infections. The data presented in this paper reveal distinct structural differences in streptokinase molecules to account for selective plasminogen activation. Detailed structure-function relationships can now be better defined for these plasminogen activators.
ACKNOWLEDGMENTS
This investigation was supported in part by a grant-in-aid from the American Heart Association, Florida Affiliate, to R.L. and by a grant from NIH-NIDDK (R01-DK45014) to K.H.J.
The MALDI-TOF mass spectral analyses performed by Anthony Haag are gratefully appreciated.
REFERENCES
- 1.Arditi M, Shulman S T, Davis T, Yoger R. Group C β hemolytic streptococcal infections in children: nine pediatric cases and review. Rev Infect Dis. 1989;11:34–45. doi: 10.1093/clinids/11.1.34. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1993. [Google Scholar]
- 3.Boyle M D P, Lottenberg R. Plasminogen activation by invasive human pathogens. Thromb Haemost. 1997;77:1–10. [PubMed] [Google Scholar]
- 4.Brockway W J, Castellino F J. A characterization of native streptokinase and altered streptokinase isolated from a human plasminogen activator complex. Biochemistry. 1974;10:2063–2070. doi: 10.1021/bi00707a010. [DOI] [PubMed] [Google Scholar]
- 5.Deutsch D G, Mertz E T. Plasminogen. Purification from human plasmin by affinity chromatography. Science. 1970;170:1095–1096. doi: 10.1126/science.170.3962.1095. [DOI] [PubMed] [Google Scholar]
- 6.Diebel R H, Steeley W H. Streptococcacea. In: Buchanan R E, Gibbons N E, editors. Bergey's manual of determinative bacteriology. Baltimore, Md: Williams and Wilkins; 1974. pp. 490–517. [Google Scholar]
- 7.Dunn S D. Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal Biochem. 1986;157:144–153. doi: 10.1016/0003-2697(86)90207-1. [DOI] [PubMed] [Google Scholar]
- 8.Forsgren M, Raden B, Israelsson M, Larsson K, Heden L-O. Molecular cloning and characterization of a full-length cDNA clone for human plasminogen. FEBS Lett. 1987;213:254–260. doi: 10.1016/0014-5793(87)81501-6. [DOI] [PubMed] [Google Scholar]
- 9.Holm S E, Bergholm A-M, Johnston K H. A streptococcal plasminogen activator in the focus of infection and in the kidneys during the initial phase of experimental streptococcal glomerulonephritis. Acta Pathol Microbiol Scand. 1988;96:1097–1108. doi: 10.1111/j.1699-0463.1988.tb00987.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang T T, Malke H, Ferretti J J. The streptokinase gene of group A streptococci: cloning, expression in E. coli and sequence analysis. Mol Microbiol. 1989;3:197–205. doi: 10.1111/j.1365-2958.1989.tb01808.x. [DOI] [PubMed] [Google Scholar]
- 11.Johnsen L B, Poulsen K, Killian M, Petersen T E. Purification and cloning of a streptokinase from Streptococcus uberis. Infect Immun. 1999;67:1073–1078. doi: 10.1128/iai.67.3.1072-1078.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston K H. Solid and fluid phase assays for bacterial plasminogen activators. J Microbiol Methods. 1993;18:267–274. [Google Scholar]
- 13.Johnston K H, Chaiban J E, Wheeler R C. Analysis of the variable domain of the streptokinase gene from streptococci associated with post-streptococcal glomerulonephritis. Zentbl Bakteriol Suppl. 1992;22:339–342. [Google Scholar]
- 14.Johnston K H, Zabriskie J B. Purification and partial purification of the nephritis-strain associated protein from Streptococcus pyogenes. J Exp Med. 1986;163:697–712. doi: 10.1084/jem.163.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohn J, Wilchek J B. A new approach (cyano-transfer) for cyanogen bromide activation of Sepharose at neutral pH, which yields activated resins, free of interfering nitrogen derivatives. Biochem Biophys Res Commun. 1982;107:878–884. doi: 10.1016/0006-291x(82)90604-0. [DOI] [PubMed] [Google Scholar]
- 16.Kulisek E S, Holm S E, Johnston K H. A chromogenic assay for the detection of plasmin generated by plasminogen activator immobilized on nitrocellulose using a para-nitroanilide synthetic peptide substrate. Anal Biochem. 1989;177:78–84. doi: 10.1016/0003-2697(89)90017-1. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Leigh J A. Purification of a plasminogen activator from Streptococcus uberis. FEMS Microbiol Lett. 1994;118:153–158. doi: 10.1111/j.1574-6968.1994.tb06818.x. [DOI] [PubMed] [Google Scholar]
- 19.Leigh J A, Hodgkinson S M, Lincoln R A. The interaction of Streptococcus dysgalactiaewith plasmin and plasminogen. Vet Microbiol. 1998;61:121–135. doi: 10.1016/s0378-1135(98)00179-5. [DOI] [PubMed] [Google Scholar]
- 20.Lizano S, Johnston K H. Streptokinase-mediated plasminogen activation using a recombinant dual fusion protein construct. A novel approach to study bacterial-host protein interactions. J Microbiol Methods. 1995;23:261–280. [Google Scholar]
- 21.Lottenberg R, Broder C C, Boyle M D, Kain S J, Schroeder B L, Curtiss R., III Cloning, sequence analysis, and expression in Escherichia coliof a streptococcal plasmin receptor. J Bacteriol. 1992;174:5204–5210. doi: 10.1128/jb.174.16.5204-5210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malke H. Polymorphism of the streptokinase gene: implications for the pathogenesis of post-streptococcal glomerulonephritis. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;278:246–257. doi: 10.1016/s0934-8840(11)80842-x. [DOI] [PubMed] [Google Scholar]
- 23.Malke H, Ferretti J J. Streptokinase: cloning, expression and excretion by E. coli. Proc Natl Acad Sci USA. 1984;81:3557–3561. doi: 10.1073/pnas.81.11.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malke H, Roe B, Ferretti J J. Nucleotide sequence of the streptokinase gene from Streptococcus equisimilisH46-A. Gene. 1985;34:357–362. doi: 10.1016/0378-1119(85)90145-3. [DOI] [PubMed] [Google Scholar]
- 25.Marti T, Schaller J, Rickli E E. Determination of the complete amino-acid sequence of porcine miniplasminogen. Eur J Biochem. 1985;149:279–285. doi: 10.1111/j.1432-1033.1985.tb08923.x. [DOI] [PubMed] [Google Scholar]
- 26.McCoy H E, Broder C C, Lottenberg R. Streptokinases produced by pathogenic group C streptococci demonstrate species specificity plasminogen activation. J Infect Dis. 1991;164:515–521. doi: 10.1093/infdis/164.3.515. [DOI] [PubMed] [Google Scholar]
- 27.Monsen J J, Holm S E, Burman L G. A general method for cell lysis and preparation of DNA from streptococci. FEMS Microbiol Lett. 1983;16:19–24. [Google Scholar]
- 28.Myers E W, Miller W. Optical alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 29.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 30.Nordstrand A, Norgren M, Ferretti J J, Holm S E. Streptokinase as a mediator of post-streptococcal glomerulonephritis in an experimental animal model. Infect Immun. 1998;66:315–321. doi: 10.1128/iai.66.1.315-321.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowicki S T, Minning-Wenz D, Johnston K H, Lottenberg R. Characterization of a novel streptokinase produced by Streptococcus equisimilisof non-human origin. Thromb Haemost. 1994;72:595–603. [PubMed] [Google Scholar]
- 32.Ohkuni H, Todome Y, Suzuki H, Mizuse M, Kotanl N, Horiuchi K, Shikama N, Tsugita A, Johnston K H. Immunochemical studies and complete amino acid sequence of the streptokinase from Streptococcus pyogenes(group A) M type 12 strain A374. Infect Immun. 1992;60:278–283. doi: 10.1128/iai.60.1.278-283.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy K N N. Streptokinase—biochemistry and clinical applications. Enzyme. 1988;40:79–89. doi: 10.1159/000469149. [DOI] [PubMed] [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaller J, Straub C, Kampfer U, Rickli E E. Complete amino acid sequence of equine miniplasminogen. Protein Seq Data Anal. 1991;4:69–74. [PubMed] [Google Scholar]
- 36.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 37.Walter F, Siegel M, Malke H. Nucleotide sequence of a streptokinase gene from S. pyogenestype 1 strain. Nucleic Acids Res. 1989;17:1261. doi: 10.1093/nar/17.3.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter F, Siegel M, Malke H. Nucleotide sequence of the streptokinase from group G streptococci. Nucleic Acids Res. 1989;17:1262. doi: 10.1093/nar/17.3.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Lin X, Loy J A, Tang J, Zhang X C. Crystal structure of the catalytic domain of human plasmin complexed with streptokinase. Science. 1998;282:1662–1665. doi: 10.1126/science.281.5383.1662. [DOI] [PubMed] [Google Scholar]