Abstract
Acinetobacter strains are widely present in the environment. Some antimicrobial-resistant strains of this genus have been implicated in infections acquired in hospitals. Genetic similarities have been reported between Acinetobacter strains in nosocomial infections and those isolated from foods. However, the antimicrobial resistance of Acinetobacter strains in foods, such as meat, remains unclear. This study initially aimed to isolate Campylobacter strains; instead, strains of the genus Acinetobacter were isolated from meat products, and their antimicrobial resistance was investigated. In total, 58 Acinetobacter strains were isolated from 381 meat samples. Of these, 32 strains (38.6%) were from beef, 22 (26.5%) from pork, and 4 (4.8%) from duck meat. Antimicrobial susceptibility tests revealed that 12 strains were resistant to more than one antimicrobial agent, whereas two strains were multidrug-resistant; both strains were resistant to colistin. Cephalosporin antimicrobials showed high minimal inhibitory concentration against Acinetobacter strains. Resfinder analysis showed that one colistin-resistant strain carried mcr-4.3; this plasmid type was not confirmed, even when analyzed with PlasmidFinder. Analysis of the contig harboring mcr-4.3 using BLAST confirmed that this contig was related to mcr-4.3 of Acinetobacter baumannii. The increase in antimicrobial resistance in food production environments increases the resistance rate of Acinetobacter strains present in meat, inhibits the isolation of Campylobacter strains, and acts as a medium for the transmission of antimicrobial resistance in the environment. Therefore, further investigations are warranted to prevent the spread of antimicrobial resistance in food products.
Keywords: Acinetobacter, meat, antimicrobial resistance, food surveillance, colistin resistance
Introduction
Acinetobacter baumannii, a gram-negative opportunistic pathogen that causes multidrug-resistant infections, can grow under conditions selective for Campylobacter spp. [1]. Members of the genus Acinetobacter are typically found in the soil, where they play an important role in mineralization and organic compound decomposition for plant use [2]. However, in recent years, the genus Acinetobacter, particularly multidrug-resistant A. baumannii, has become a major causative agent of opportunistic infections. These bacteria, found in emergency rooms and intensive care units, are responsible for various infectious diseases such as pneumonia, wound tissue infection, sepsis, and urinary tract infection [2].
Recently, genetic similarities have been reported between the strains responsible for human infection in hospitals and A. baumannii isolated from food [3]. In addition, infections caused by colistin-resistant Acinetobacter have been reported [4]. In gram-negative pathogenic bacteria, such as Acinetobacter, the emergence of a strain carrying mobile colistin resistance (mcr) gene, which induces colistin resistance, is a major global health challenge [5]. Therefore, the spread of the colistin-resistant gene in various environments must be monitored and restricted [6]. Specifically, understanding the overall status of antimicrobial resistance in the food chain is important for identifying transmission pathways for resistant bacteria and blocking their transmission [7].
In this regard, the antimicrobial resistance of food-derived A. baumannii and its molecular dynamics requires special attention. Several studies performed in Korea have shown that A. baumannii causes infections in hospitals; however, data regarding the presence and antimicrobial resistance of A. baumannii on foods, such as meat distributed in the market, remain limited. This study initially aimed to isolate and characterize Campylobacter, including antimicrobial resistance; however, we instead isolated Acinetobacter strains since they were more abundant. Despite this change in study aim, our results addressed several insufficiencies reported by previous studies.
In the present study, we isolated the strains of the genus Acinetobacter grown in a Campylobacter-specific environment and identified them using molecular methods to confirm their distribution rate and antimicrobial resistance status. We also selected a colistin-resistant strain and performed next-generation sequencing (NGS) analysis to compare the sequence similarities with those of strains isolated from the clinical environment.
Materials and Methods
Sample Collection
In the present study, we collected five types of meat and meat products. The sample types were selected based on their consumption rates. The samples comprised domestic and imported meat products, including beef, pork, chicken, duck, and aquatic products. These samples (number of samples indicated in square brackets, number of domestic and imported products are separated with a comma, respectively) included beef [n = 59, 60], pork [n = 70, 37], chicken meat [n = 83 (only domestic)], duck meat [n = 42 (only domestic), and aquatic products [n = 10, 20]. In total, 381 samples (domestic meat [n = 264] and imported meat [n = 117]) were collected from 13 hypermarkets in Korea between February and October 2019. Samples were transported under refrigerated conditions within 24 h of collection to the laboratory for analysis. The sample distribution is shown in Table 1.
Table 1.
Distribution and identification of Acinetobacter spp. isolated from meat samples using Campylobacterselective conditions.
| Species | Beef | Pork | Chicken meat | Duck meat | Aquatic product | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Number of samples | 119 | 107 | 83 | 42 | 30 | ||
|
| |||||||
| Sample origin | Domestic (n = 59) | Imported (n = 60) | Domestic (n = 70) | Imported (n = 37) | Domestic (n = 83) | Domestic (n = 42) | Domestic & imported (n = 30) |
| A. baumannii | 7 | 11 | 8 | 4 | 3 | ||
| A. nosocomialis | 2 | 7 | 3 | 4 | 1 | ||
| A. pittii | 1 | ||||||
| A. seifertii | 5 | 2 | |||||
| Total | 9 | 23 | 12 | 10 | 0 | 4 | 0 |
Isolation of Acinetobacter spp.
Acinetobacter spp. were isolated as previously described in the Korean Food Code 2020 for Campylobacter spp. isolation [8]. Twenty-five grams of each meat sample was inoculated into 225ml of Bolton medium (Oxoid, UK) and cultured at 42°C under microaerophilic conditions (5% O2, 85% N2, and 10% CO2) for 48h. Next, the cultures were spread on modified Charcoal-Cefoperazone-Deoxycholate Agar (Oxoid) and incubated at 42°C under microaerophilic conditions for 24h. Subsequently, three or more colonies with a circular or irregular shape and translucent or transparent in appearance in grayish-white tones were selected and identified using the VITEK-MS system (BioMérieux Vitek, France). The isolates, identified as belonging to “A. baumannii complex,” were selected for further molecular characterization.
Identification of A. baumannii
The collected strains were identified using molecular and microbiological methods. First, collected strains were cultured in trypticase soy agar (Oxoid) for 24 h. The cultured colonies were suspended in 200 μl of sterile distilled water to obtain genomic DNA, followed by amplification and sequencing of two segments of the 16S rRNA (27F: 5'-AGA GTT TGA TCC TGG CTC AG-3', 1492R: 5'-TAC GGY TAC CTT GTT ACG ACT T-3'), rpoB zone 1 (Ac696F: 5'-TAY CGY AAA GAY TTG AAA GAA G-3', Ac1093R: 5'-MAC ACC YTT GTT MCC RTG A-3') and rpoB zone 2 (Ac1055F: 5'-GTG ATA ARA TGG CBG GTC GT-3', Ac1598R: 5'-CGB GCR TGC ATY TTG TCR T-3'). The 16S rRNA gene was compared to the Eztaxon database of ChunLab (https://www.ezbiocloud.net/) to confirm the high agreement (sequence identity of > 98.7%), whereas, for rpoB, zones 1 and 2 (two variable areas bordered by highly conserved regions in rpoB. For zone 1, between positions 2,900 and 3,250, while for zone 2, between positions 3,250 and 3,700) were sequenced and compared via BLAST analysis [9].
Antimicrobial Susceptibility Tests
Antimicrobial susceptibility tests were performed using Sensititre KRNVF5F panels (Trek Diagnostic Systems, USA) to measure the MIC value by the broth microdilution method. The following 16 antimicrobials were used to evaluate the antimicrobial susceptibility of the isolated strains: amoxicillin/clavulanic acid (2:1), cefoxitin, chloramphenicol, ciprofloxacin, gentamicin, tetracycline, nalidixic acid, meropenem, cefepime, colistin, trimethoprim/sulfamethoxazole, ampicillin, ceftazidime, sulfisoxazole, ceftiofur, and streptomycin. The breakpoint of the antimicrobials was decided based on the CLSI guidelines [10]. However, no resistance criteria have been provided in the CLSI guidelines for the following antimicrobials: amoxicillin/clavulanic acid (2:1), cefoxitin, chloramphenicol, nalidixic acid, ampicillin, sulfisoxazole, ceftiofur, and streptomycin. For these agents, the degree of increase in resistance was determined by referring to the resistance level of the standard strain, ATCC 17978 [6].
NGS and Bioinformatics Analyses
Colistin-resistant Acinetobacter spp. isolates were subjected to NGS (Macrogen, Korea), and the genome sequences of these strains were obtained using the Illumina HiSeqXten platform (USA). The acquired antimicrobial resistant gene and plasmid replicon typing were identified in silico using ResFinder 3.2 and the PlasmidFinder 2.1 webserver (https://cge.cbs.dtu.dk, accessed June 24, 2020), respectively [11, 12]. The genome was annotated using RAST (http://rast.theseed.org/) to analyze the genetic environment of mcr [13]. BLAST was used to align the genetic sequences flanking mcr (www.ncbi.nlm.nih.gov/BLASTih.gov/BL). The results were visualized using SnapGene (GSL Biotech; available at snapgene.com). ISfinder was used to check the presence and type of the insertion sequence in a contig (database URL, http://www-is.biotoul.fr) [14].
Results
Isolation and Identification of Acinetobacter spp.
Overall, 58 strains (15.2%) of Acinetobacter spp. were isolated from 381 samples using the Campylobacter growth environment, identified with Vitek-MS IVD mode. A total of 32 strains (55.2%) were isolated from beef, 22 (37.9%) from pork, and 4 (6.9%) from duck meat; no strains were isolated from chicken meat and aquatic products. Sequence analysis of 16S rRNA and rpoB from the isolated strains showed that 33 of the isolated strains exhibited sequence homology with A. baumannii. In contrast, 17 strains showed sequence homology to Acinetobacter nosocomialis, 7 strains to Acinetobacter seifertii, and one strain to Acinetobacter pittii (Table 1). This result was used as a reference for species identification.
Antimicrobial Resistance Profile of Acinetobacter Isolates
The antimicrobial susceptibility test results of the 58 isolates are summarized in Tables 2 and 3. Of the 16 antimicrobials tested, the isolates were resistant to only five types of antimicrobials. The isolates were the most resistant to trimethoprim/sulfamethoxazole, with 10 isolates being resistant to this drug combination. Three isolates were resistant to gentamicin and tetracycline; only two isolates were resistant to colistin, and one isolate was resistant to cefepime. The MICs of the two isolates resistant to colistin were 16 μg/ml (A. nosocomialis, KUFSE-ACN036) and 4 μg/ml (A. seifertii KUFSE-ACS055), respectively.
Table 2.
Minimum inhibitory concentration (MIC) distribution of Acinetobacter spp. isolates.
| Antimicrobial | Test range | Distribution of MICs (μg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| ≤0.12 | ≤0.25 | ≤0.5 | ≤1 | ≤2 | 4≤ | 8≤ | 16≤ | 32≤ | 64≤ | 128≤ | 256≤ | ||
| Amoxicillin/clavulanic acid 2:1* | 2–32 | 2 | 15 | 37 | 4 | ||||||||
| Cefepime | 0.25–16 | 4 | 19 | 20 | 6 | 5 | 3 | |1 | |||||
| Cefoxitin* | 1–32 | 2 | 1 | 10 | 45 | ||||||||
| Trimethoprim/sulfamethoxazole | 0.25–4 | 22 | 26 | 2 | 4 | |4 | |||||||
| Chloramphenicol* | 2–64 | 1 | 2 | 1 | 5 | 16 | 30 | 3 | |||||
| Ampicillin* | 2–64 | 1 | 10 | 25 | 12 | 7 | 3 | ||||||
| Ciprofloxacin | 0.12–16 | 43 | 12 | 2 | |1 | ||||||||
| Streptomycin* | 16–128 | 28 | 18 | 7 | 4 | 1 | |||||||
| Gentamycin | 1–64 | 49 | 1 | 5 | 2 | | | 1 | ||||||
| Ceftazidime | 1–16 | 5 | 7 | 31 | 13 | 2 | | | ||||||
| Tetracycline | 2–128 | 49 | 5 | 1 | |1 | 1 | 1 | ||||||
| Sulfisoxazole* | 16–256 | 38 | 12 | 8 | |||||||||
| Nalidixic acid* | 2–128 | 11 | 31 | 13 | 2 | 1 | |||||||
| Ceftiofur* | 0.5–8 | 28 | 30 | ||||||||||
| Meropenem | 0.25-4 | 41 | 15 | 1 | 1 | | | |||||||
| Colistin | 2–16 | 56 | |1 | 1 | |||||||||
Vertical lines in each row indicate the breakpoint of each antimicrobial.
*Indicates no breakpoint in the CLSI guidelines.
Table 3.
Antimicrobial resistance profile of the strains.
| Strain | Sample | Sampling date | Sample origin | Antimicrobial resistance |
|---|---|---|---|---|
| Acinetobacter seifertii KUFSE-ACS004 | Imported Beef | 2019-02-18 | Australia | GEN |
| Acinetobacter seifertii KUFSE-ACS007 | Imported Pork | 2019-02-18 | Spain | TET |
| Acinetobacter nosocomialis KUFSE-ACN017 | Domestic Beef | 2019-06-17 | Chungcheong, Korea | SXT |
| Acinetobacter baumannii KUFSE-ACB021 | Domestic Beef | 2019-06-17 | Chungcheong, Korea | SXT |
| Acinetobacter nosocomialis KUFSE-ACN022 | Imported Beef | 2019-06-17 | Australia | SXT |
| Acinetobacter nosocomialis KUFSE-ACN025 | Domestic Pork | 2019-06-17 | Chungcheong, Korea | FEP, SXT, TET |
| Acinetobacter nosocomialis KUFSE-ACN026 | Domestic Pork | 2019-06-17 | Chungcheong, Korea | SXT, GEN, TET |
| Acinetobacter baumannii KUFSE-ACB029 | Imported Beef | 2019-07-08 | Australia | SXT |
| Acinetobacter nosocomialis KUFSE-ACN033 | Domestic Pork | 2019-07-08 | Gyeonggi, Korea | SXT |
| Acinetobacter baumannii KUFSE-ACB035 | Imported Pork | 2019-07-08 | Spain | SXT |
| Acinetobacter nosocomialis KUFSE-ACN036 | Imported Pork | 2019-07-08 | Netherlands | COL |
| Acinetobacter seifertii KUFSE-ACS055 | Imported Beef | 2019-09-17 | Australia | GEN, COL |
*SXT, trimethoprim/sulfamethoxazole; FEP, Cefepime; TET, tetracycline; COL, colistin; GEN, gentamicin
Two isolates were resistant to three antimicrobials, one isolate was resistant to two antimicrobials, while the remaining nine isolates were resistant to only one antimicrobial, as shown in Table 3. The resistant isolates included A. nosocomialis (n = 6), A. baumannii (n = 3), and A. seifertii (n = 3). The results of antimicrobial susceptibility analysis are summarized in Table 3.
Detection of mcr-4.3 and in silico Molecular Characteristic Analysis
Table 4 shows the ResFinder and PlasmidFinder analysis results for the colistin-resistant strain (KUFSE-ACN036, MIC: 16 μg/ml). This colistin-resistant strain (A. nosocomialis KUFSE-ACN036) harbored mcr-4.3, while the other colistin-resistant strain (KUFSE-ACS055) did not harbor any mcr genes. However, these strains did not carry any other antimicrobial resistance genes.
Table 4.
Results of Resfinder analysis of KUFSE-ACN036.
| Colistin | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Resistance gene | Identity | Query/Template length | Contig | Position in contig | Predicted phenotype | Accession number |
| mcr-4.3 | 100 | 1626/1626 | ac36_first_contig_40 length 22852 coverage 1846.3 normalized_cov 3.55 | 12743..14368 | Colistin resistance | MG026621 |
Genetic Characterization of Colistin Resistance Strains
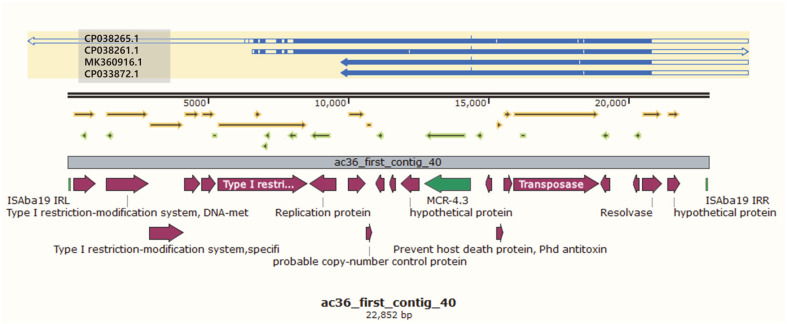
The results of contig 40 analysis of KUFSE-ACN036 harboring mcr-4.3 using BLAST showed that the top 4 matches based on the maximum score carried plasmids derived from A. baumannii (Table 5). Among them, the strain with accession number CP038265.1 showed the highest maximum score of 26,502. This strain carried the A. baumannii plasmid, which was previously reported to possess mcr-4.3. The remaining top three strains carried plasmids with mcr-4.3 (CP038261.1, MK360916.1, and CP033872.1); however, their query covers were only 63–80%; therefore, no plasmids in GenBank matched all contig 40. Analysis using ISfinder showed that contig 40 was related to ISAba19. Although not all these sequences were available, the sequences of ISAba19 IRL (5′-TGA ACC GTA CCG GGT TTG TCG GAG AGT CAA TAT TCT GAG AGA CTA TCC CG-3′) and IRR (5′-TGA ACC GTA CCG GGT TTG TCG GAG ACT TTT TTA TTT AAG TTA AGC CAC CT-3′) were included in contig 40. Contig 40 has a total length of 21,130 base pairs, in which CP03261.1 matched over a specific part (6702..21,130) in the middle (Fig. 1). As BLAST analysis was performed on both ends (1..6701 and 21,131..22,852) of contig 40 (except for the middle portion), the match with other bacteria included in the Acinetobacter spp. was confirmed. Interestingly, the top matched gene sequences of each two-part BLAST results were the same (CP028138.1, CP044018.1, CP046595.1, and CP044445.1) (Table 6).
Table 5.
Top 4 BLAST search results in full part of contig 40 of KUFSE-ACN036.
| Description | Max Score | Total Score | Query Cover | E value | Per. Ident | Accession number |
|---|---|---|---|---|---|---|
|
Acinetobacter baumannii strain EC plasmid pEC_mcr4.3, complete sequence |
26,502 | 27,081 | 0.65 | 0 | 0.99 | CP038265.1 |
|
Acinetobacter baumannii strain EH plasmid pEH_mcr4.3, complete sequence |
26,020 | 26,599 | 0.63 | 0 | 0.9922 | CP038261.1 |
|
Acinetobacter baumannii plasmid pAB18PR065-MCR-4.3, complete sequence |
20,740 | 31,213 | 0.79 | 0 | 0.9953 | MK360916.1 |
|
Acinetobacter baumannii strain MRSN15313 plasmid pAb-MCR4.3, complete sequence |
20,740 | 31,768 | 0.8 | 0 | 0.9953 | CP033872.1 |
Fig. 1. Sequence alignment of the top four BLAST search results in the full part of contig 40 of KUFSEACN036.
Blue arrows indicate the degree and orientation of the reference genes (CP038265.1, CP038261.1, MK360916.1, and CP033872.1). The light green square indicates the insertion sequence ISAba19. The green arrow indicates mcr-4.3. Red arrows indicate other genes. Arrows highlighted in yellow and light green indicate open reading frames. Sequence comparison, alignment, and drawing were performed using SnapGene 5.0.8.
Table 6.
Top 4 BLAST search results on both ends part of contig 40 of KUFSE-ACN036.
| Description | Max Score | Total Score | Query Cover | E value | Per. Ident | Accession number |
|---|---|---|---|---|---|---|
| 1..6701 of contig 40 | ||||||
|
Acinetobacter baumannii strain NCIMB 8209 chromosome, complete genome |
4303 | 7087 | 0.68 | 0 | 0.9416 | CP028138.1 |
|
Acinetobacter indicus strain HY20 chromosome, complete genome |
4146 | 7027 | 0.75 | 0 | 0.9321 | CP044018.1 |
|
Acinetobacter indicus strain FS42-2 chromosome, complete genome |
4095 | 6954 | 0.76 | 0 | 0.9283 | CP046595.1 |
|
Acinetobacter indicus strain CMG3-2 chromosome, complete genome |
2614 | 6985 | 0.74 | 0 | 0.9182 | CP044445.1 |
| 21,131..22,852 of contig 40 | ||||||
|
Acinetobacter baumannii strain NCIMB 8209 chromosome, complete genome |
4303 | 7087 | 0.68 | 0 | 0.9416 | CP028138.1 |
|
Acinetobacter indicus strain HY20 chromosome, complete genome |
4146 | 7027 | 0.75 | 0 | 0.9321 | CP044018.1 |
|
Acinetobacter indicus strain FS42-2 chromosome, complete genome |
4095 | 6954 | 0.76 | 0 | 0.9283 | CP046595.1 |
|
Acinetobacter indicus strain CMG3-2 chromosome, complete genome |
2614 | 6985 | 0.74 | 0 | 0.9182 | CP044445.1 |
Discussion
We successfully isolated Acinetobacter genus strains from commercial meat products using conditions selective for Campylobacter isolation. Microbiological and molecular methods were used to characterize the isolates to the species level; the characterized strains included A. nosocomialis, A. pittii, and A. seifertii, in addition to A. baumannii. To the best of our knowledge, no previous study has reported the isolation of strains belonging to the genus Acinetobacter from food using the microbiological and molecular methods discussed in the present study. Although a previous study by Fernando et al. [1] reported the isolation of A. baumannii from environmental reservoirs, its presence in meat products openly sold in retail markets has not been studied. Due to the availability and large-scale consumption of meat products, combined with the environmental transmission of mcr, it is important to identify bacterial isolates present in meat products.
This study used mCCDA as a selection medium, while the modified Karmali agar was used by Fernando et al.[1]. However, both media contained antimicrobials that increased the specificity of isolating Campylobacter (modified Karmali agar: cefoperazone-32 mg/l, vancomycin-20 mg/l, and amphotericin B-10 mg/l; mCCDA: cefoperazone-32 mg/l, and amphotericin B-10 mg/l) [1]. Since Campylobacter is naturally resistant to these antimicrobials, they were added to inhibit the growth of other strains. However, in this study, Acinetobacter spp. could grow in media containing these antimicrobials, indicating that the resistance of Acinetobacter spp., which can be isolated from the environment, has increased [15, 16]. Thus, it is presumed that these resistance factors were disseminated to Acinetobacter strains in the environment. In this study, no resistant strains were found in poultry meat, while 6 resistant strains were isolated from pork and beef. There is a difference in antimicrobials used depending on the type of livestock. Further studies on Acinetobacter strains in livestock are warranted to better understand the migration pattern of this resistance factor.
Antimicrobial resistance genes spread in livestock environment can be continuously transmitted between other bacteria [17]. Originally, this study aimed to isolate only Campylobacter bacteria under selective conditions; however, only two C. jejuni and three C. coli strains were isolated from chicken and duck meat (data not shown here). This result suggests that using a selective media that can inhibit the growth of certain bacteria while allowing the growth of others is no longer sufficient due to the increasing number of resistant strains. For instance, the growth of Acinetobacter bacteria, resistant to these antimicrobials, can suppress the growth or selection of Campylobacter under Campylobacter-specific culture conditions.
A high rate of Acinetobacter isolation can be achieved from various raw meat and agricultural products using CHROMagar and Dijkshoorn's media [18, 19], which can be used to isolate Acinetobacter. Additionally, Acinetobacter is intrinsically resistant to cephalosporin-based antimicrobials. Therefore, to effectively isolate Campylobacter, it is necessary to consider the differential biochemical properties of target strains apart from antimicrobial resistance profile.
Acinetobacter is commonly present in the environment, and the transmission of antimicrobial resistance can increase the number of antimicrobial-resistant strains in our environment and the food we consume. Particularly, an increase in antimicrobial resistance hampers clinical treatment, and consumption of food items infested by these resistant strains poses serious risks to immunocompromised individuals [20]. Therefore, studies on the presence and prevalence of Acinetobacter strains in food products are urgently needed.
The colistin-resistant gene can spread horizontally, and the genus Acinetobacter plays a key role in carrying and disseminating mobile colistin resistance genes in the environment. These are known as mcr-gene-containing bacteria, which have been propagated by horizontal/lateral transfer to various ecosystems, including wild animals or livestock; environments such as soil, stream water, plant or agricultural systems; and human communities [21]. In the present study, mcr-4.3 was identified in only one strain. To the best of our knowledge, this is the first study to report the isolation of a strain belonging to the genus Acinetobacter from meats in Korean food hypermarkets. Moreover, the strain (KUFSE-ACN036) was isolated from pork imported from the Netherlands. Although no Acinetobacter strain isolated from domestic meat showed resistance to colistin, further research is needed to investigate the distribution of mcr genes in Acinetobacter strains from domestic livestock farms. mcr-4 was first reportedly isolated in Italy, while the mcr-4.3 genotype has been widely reported in Europe, which is consistent with our study results [22-24]. However, recent studies have reported this gene in countries outside Europe, including China and Brazil [5, 25]. The path of dissemination of antimicrobial resistance warrants further investigation.
BLAST analysis results showed that mcr-4.3 identified in this study is related to A. baumannii. However, there is no complete alignment in BLAST results with contig 40. The aligned part containing mcr-4.3 showed results consistent with the plasmids, while the other parts (both ends) showed results consistent with chromosomes. No plasmid type was identified through PlasmidFinder, suggesting that the plasmid containing mcr-4.3 identified in the present study is novel. The entire plasmid remains to be confirmed because NGS analysis was based on the short-read method, which is a limitation of the present study. Therefore, further studies are warranted to identify the complete sequence of the plasmid using long-read sequencing; epidemiological studies based on molecular biological methods can provide deeper insights into antimicrobial resistance transmission.
In conclusion, we isolated Acinetobacter bacteria from culture conditions specific for Campylobacter isolation. Further, we confirmed the antimicrobial resistance properties and molecular epidemiological characteristics of the isolated strains, in addition to the presence of the colistin resistance gene. This study suggests the transmission of antimicrobial resistance to microbes commonly present in our environments and food products and highlights the imminent threat posed by the increased number of infections caused by these microbes. Moreover, their multidrug resistance profile may limit therapeutic options. Therefore, continuous efforts are needed to prevent the spread of antimicrobial resistance by judicious use of antimicrobials and identifying novel microbial agents to treat multidrug-resistant strains.
Acknowledgments
This work was supported by a grant from the Ministry of Food and Drug Safety (nos. 15161MFDS645), the Korea National Institute of Health (grant number 2017-NI41004 and 2017ER540601), and the Korea Rural Development Administration (grant number PJ010500). The findings and conclusions of this article are ours and do not necessarily reflect the views of MFDS, NIH, and KRDA.
Footnotes
Conflict of interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Fernando DM, Khan IUH, Patidar R, Lapen DR, Talbot G, Topp E, et al. Isolation and characterization of Acinetobacter baumannii recovered from Campylobacter selective medium. Front. Microbiol. 2016;7:1871–1871. doi: 10.3389/fmicb.2016.01871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visca P, Seifert H, Towner KJ. Acinetobacter infection-an emerging threat to human health. IUBMB Life. 2011;63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 3.Maboni G, Seguel M, Lorton A, Sanchez S. Antimicrobial resistance patterns of Acinetobacter spp. of animal origin reveal high rate of multidrug resistance. Vet. Microbiol. 2020;245:108702. doi: 10.1016/j.vetmic.2020.108702. [DOI] [PubMed] [Google Scholar]
- 4.Leite GC, Stabler RA, Neves P, Perdigão Neto LV, Ruedas Martins RC, Rizek C, et al. Genetic and virulence characterization of colistin-resistant and colistin-sensitive A. baumannii clinical isolates. Diagn. Microbiol. Infect. Dis. 2019;95:99–101. doi: 10.1016/j.diagmicrobio.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Ma F, Shen C, Zheng X, Liu Y, Chen H, Zhong L, et al. Identification of a novel plasmid carrying mcr-4.3 in an Acinetobacter baumannii strain in China. Antimicrob. Agents Chemother. 2019;63:e00133–19. doi: 10.1128/AAC.00133-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mustapha MM, Li B, Pacey MP, Mettus RT, McElheny CL, Marshall CW, et al. Phylogenomics of colistin-susceptible and resistant XDR Acinetobacter baumannii. J. Antimicrob. Chemother. 2018;73:2952–2959. doi: 10.1093/jac/dky290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha M-H, Ryu J-G, Chi Y-M, Woo G-J. Characterization of extended-spectrum β-lactamase-producing Escherichia coli isolated from fresh produce and agricultural environments in Korea. J. Food Protect. 2020;83:1115–1124. doi: 10.4315/JFP-19-483. [DOI] [PubMed] [Google Scholar]
- 8.Ministry of Food and Drug Safety, author. Korean Food Code. Cheongju, Korea: 2020. pp. 224–225. [Google Scholar]
- 9.La Scola B, Gundi VAKB, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 2006;44:827–832. doi: 10.1128/JCM.44.3.827-832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute, author. CLSI Document M100. Wayne, PA. USA: 2017. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 11.Carattoli A, Zankari E, Garcìa-Fernandez A, Larsen MV, Lund O, Villa L, et al. PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, et al. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013;68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 13.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siguier P, Pérochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark NM, Zhanel GG, Lynch III JP. Emergence of antimicrobial resistance among Acinetobacter species: a global threat. Curr. Opin Crit. Care. 2016;22:491–499. doi: 10.1097/MCC.0000000000000337. [DOI] [PubMed] [Google Scholar]
- 16.Choi J-Y, Kim Y, Ko EA, Park YK, Jheong W-H, Ko G, et al. Acinetobacter species isolates from a range of environments: species survey and observations of antimicrobial resistance. Diagn. Microbiol. Infect. Dis. 2012;74:177–180. doi: 10.1016/j.diagmicrobio.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 17.Woolhouse M, Ward M, van Bunnik B, Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. Lond., B, Biol. Sci. Philos. T R Soc. B. 2015;370:20140083. doi: 10.1098/rstb.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalheira A, Casquete R, Silva J, Teixeira P. Prevalence and antimicrobial susceptibility of Acinetobacter spp. isolated from meat. Int. J. Food Microbiol. 2017;243:58–63. doi: 10.1016/j.ijfoodmicro.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Carvalheira A, Ferreira V, Silva J, Teixeira P. Enrichment of Acinetobacter spp. from food samples. Food Microbiol. 2016;55:123–127. doi: 10.1016/j.fm.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 20.de Amorim AMB, dos Santos Nascimento J. Acinetobacter: an underrated foodborne pathogen? J. Infect. Dev. Countr. 2017;11:111–114. doi: 10.3855/jidc.8418. [DOI] [PubMed] [Google Scholar]
- 21.Anyanwu MU, Jaja IF, Nwobi OC. Occurrence and characteristics of mobile colistin resistance (Mcr) gene-containing isolates from the environment: a review. Int. J. Environ. Res. Public Health. 2020;17:1028. doi: 10.3390/ijerph17031028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bitar I, Medvecky M, Gelbicova T, Jakubu V, Hrabak J, Zemlickova H, et al. Complete nucleotide sequences of mcr-4.3-carrying plasmids in Acinetobacter baumannii sequence type 345 of human and food origin from the Czech republic, the first case in Europe. Antimicrob. Agents Chemother. 2019;63:e01166–19. doi: 10.1128/AAC.01166-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro. Surveill. 2017;22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavda B, Lv J, Hou M, Chavda KD, Kreiswirth BN, Feng Y, et al. Coidentification of mcr-4.3 and blaNDM-1 in a clinical Enterobacter cloacae isolate from China. Antimicrob. Agents Chemother. 2018;62:e00649–18. doi: 10.1128/AAC.00649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins-Sorenson N, Snesrud E, Xavier DE, Cacci LC, Iavarone AT, McGann P, et al. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 2020;75:60–64. doi: 10.1093/jac/dkz413. [DOI] [PMC free article] [PubMed] [Google Scholar]