Abstract
Obesity and related metabolic diseases are major problems worldwide. Some probiotics are currently considered potential therapeutic strategies for obesity. We aimed to investigate the antiobesity efficacy of Latilactobacillus sakei WIKIM31 in obese mice induced by a high fat diet. The administration of a high-fat diet with L. sakei WIKIM31 reduced body weight gain, epididymal fat mass, triglyceride and total cholesterol levels in the blood, and remarkably decreased the expression of lipogenesis-related genes in the epididymal adipose tissue and liver. Interestingly, intake of L. sakei WIKIM31 improved gut barrier function by increasing the gene expression of tight junction proteins and suppressing the inflammatory responses. Additionally, L. sakei WIKIM31 enhanced the production of short-chain fatty acids, such as butyrate and propionate, in the intestinal tract. These results showed that L. sakei WIKIM31 can be used as a potential therapeutic probiotic for obesity.
Keywords: Latilactobacillus sakei WIKIM31, probiotics, obesity, inflammation
Introduction
Obesity is a serious disease worldwide and is associated with many metabolic diseases, such as diabetes, fatty liver, and cardiovascular diseases [1]. Recent studies have shown that the gut microbiome, which manifested by changes in bacterial compositions and function, is closely related to obesity [2]. Changes in specific intestinal microbiota, particularly, the ratio of the phyla Firmicutes and Bacteroidetes, which are representative dominant bacteria, is the main cause of obesity [3]. These changes in dominant bacteria affect the energy metabolism of the host, which can cause metabolic problems [4]. In addition, emerging research indicates that chronic inflammation is highly associated with the development of obesity [5]. Thus, obese individuals have increased circulating levels of interleukin (IL)-6, tumor necrosis factor (TNF)-α, and monocyte chemoattractant protein-1 (MCP-1) [6]. Obesity induced inflammation is related with leakage of microbial, gut-derived molecules, such as lipopolysaccharide (LPS) [7]. Alterations in gut microbiota induced by a high-fat diet cause intestinal inflammation, which leads to the leakage of gut microbes and contributes to inflammation associated with obesity [8]. Therefore, modulation of the gut microbiota is very important as a potential therapeutic strategy for metabolic disorders.
Probiotics, as living bacteria, not only contribute to the health of the gastrointestinal tract, but also have a positive effect on health when administered in appropriate amounts to the host [9]. Other beneficial effects of probiotics include improving blood glucose, cholesterol, hypertension, immune response, and energy metabolism [10]. Several studies have shown that probiotic bacteria inhibit the lipid metabolism and differentiation of adipocytes [11, 12]. Although a number of studies have been reported on the beneficial effects of probiotics on host metabolism, the mechanisms by which probiotics modulate host metabolism remain to be elucidated.
L. sakei WIKIM31 was isolated from kimchi. Kimchi is fermented by various lactic acid bacteria, of which L. sakei is one of the representative dominant bacteria [13]. Previous studies have shown that L. sakei inhibits the growth of harmful bacteria and has the immunomodulatory effects by inhibiting of allergic Th2 responses and increasing the production anti-inflammatory cytokines [14, 15]. Also, L. sakei has anti-adipogenesis and anti-obesity effects [16, 17]. In this study, we investigated whether oral administration of WIKIM31 could suppress lipid accumulation in adipocytes by regulating the expression of lipogenesis genes, inflammatory cytokines, and tight junction genes in vivo.
Materials and Methods
Isolation and Preparation of L. sakei WIKIM31
L. sakei WIKIM31 was isolated from homemade kimchi (Gangwon province, Republic of Korea). The kimchi was homogenized and filtered by stomacher filter bag. The homogenate was spread onto Man-Rogosa-Sharpe (MRS; BD Difco, USA) agar. The plate were incubated to at 30°C for 48 h. Isolated strains were obtained from subculture. After pre-screening for anti-obesity effects in vitro, selected strain was identified by 16S rRNA gene sequence. WIKIM31 was deposited at Korean Federation of Culture Collection as KFCC 11654P. To prepare the strain for use in animal experiments. WIKIM31 was cultured at 30°C for 18 h in MRS broth. The strain was harvested by centrifugation and washed twice with PBS. Next, the strain was resuspended in PBS (1 × 109 CFU) for oral administration to mice.
Cell Culture and Oil Red O Staining
3T3-L1 (ATCC-CL-173) preadipocyte cells were maintained in Dulbecco Modified Eagle Medium (DMEM; Gibco, USA) supplemented with 10% newborn calf serum (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin. For differentiation to adipocytes, preadipocytes were cultured to full confluency, and the medium was changed with differentiation media (DMEM supplemented with 10% fetal bovine serum [FBS]) with 1 μg/ml insulin, 0.1 μM dexamethasone, and 0.5 mM isobutyl methylxanthine. After 2 days, the cells were cultured in DMEM containing 10% FBS for additional 4 days. For Oil Red O staining, cells were washed with PBS, fixed with 10% formalin for 1 h, and rinsed with 60 % isopropyl alcohol. Cells were stained with Oil Red O solution for 10 min, and washed with distilled water.
Animals and Experimental Design
The care and study of the laboratory mice followed the protocols of the Institute Animal Care and Use Committee for the World Institute of Kimchi (WIKIM IACUC 201608). 6-week-old male C57BL/6 mice were purchased from OrientBio (Korea) and acclimated to the environment for 1 week. All mice were cared in individually ventilated cages where temperature and humidity were controlled on a 12 h light/dark cycle. Food and water were supplied ad libitum. Each group of mice were fed a normal diet (ND mice, 10% kcal as fat, D10001, Research Diet Inc., USA), high-fat diet (HFD mice, 45% kcal fat, D12451, Research Diet), or HFD with WIKIM31 (HFD-WIKIM31 mice) for 12 weeks. WIKIM31 was orally administered at a daily (10 am) concentration of 1 × 109 CFU per 200 μl. The well-being of the animals, body weight, and food intake were measured weekly. The body composition of the mice was determined by MRI relaxometry (EchoMRI-500, Echo Medical System, USA). After 12 weeks, the mice were sacrificed using CO2 asphyxiation. Serum separated from blood was stored at -80°C until analysis.
Histological Analysis
Histological analysis was performed as described previously [18]. Histological images of the liver and adipose tissue were examined under a microscope (Olympus DP73, Japan).
Measurements of Triglyceride, Cholesterol, Glucose and Adipokines Levels
Serum triglyceride, cholesterol, and glucose levels were determined by a FUGI DRI-CHEM 7000 (Fujifilm, Japan) according to the manufacturer’s protocol. The concentrations of leptin, adiponectin, and resistin were measured using a Bio-Plex Pro Mouse Diabetes-Plex Assay (Bio-Rad, USA).
Isolation of Stromal Vascular Fraction and Measurements of Cytokine Levels
Isolation of stromal vascular fraction (SVF) was performed as previously described [18]. Briefly, epididymal fat (EF) tissues were digested using 2 mg/ml collagenase (Sigma-Aldrich, USA) at 37°C for 45 min, filtered using 100 μm cell strainers (BD Falcon), and centrifugated at 300 ×g for 10 min at room temperature. The adipocyte layer and the supernatant were separated, and SVF pellets were collected. For the measurement of pro-inflammatory cytokine secretion, the isolated SVFs (5 × 105 cells/well) were seeded in a 96-well plate, added to LPS (100 ng/ml), and cultured for 24 h at 37°C. Levels of IL-6, TNF-α, and MCP-1 in the cultured supernatant were measured using a cytometric bead array kit (BD Biosciences, USA).
RNA Isolation and Real-Time PCR Analysis
Total RNA was isolated from frozen tissues and cDNA was synthesized using a TOPscript cDNA synthesis kit (Enzynomics, Korea). Real-time PCR was performed using a CFX9600 (Bio-Rad). PCR experiments were performed under the same cycling conditions: 95°C for 5 min, followed by 39 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s. The PCR primer sequences are listed in Table 1. β-Actin RNA levels were used to normalize each gene.
Table 1.
List of oligonucleotide primer sequences for real time RT-PCR.
| Gene | Sense | Antisense |
|---|---|---|
| PPARγ | 5′-GCCCTTTGGTGACTTTATGGA-3′ | 5′-GCAGCAGGTTGTCTTGGATG-3′ |
| C/EBPα | 5′-TGCTCTGATTCTTGCCAAA-3′ | 5′-ACCCAAAATCCCTAAACC-3′ |
| FAS | 5′-TGTTCCTTGTGCACCCCATT-3′ | 5′-GTAGGTGTGTGAGCCGTCAA-3′ |
| SREBP-1c | 5′-AGATCCAGGTTTGAGGTGGG-3′ | 5′-AGATCCAGGTTTGAGGTGGG-3′ |
| SCD1 | 5′-CCTCCGGAAATGAACGAGAGAA-3′ | 5′-CCTGATAGGTGGGGTCGTGA-3′ |
| CD36 | 5′-CTTCACAGTTCTGAATCTGGCTGT-3′ | 5′-GAGGCTGCGTCTGTGCCATTAATCATG-3′ |
| UCP2 | 5′-AGAACGAGACACCTTTAGAGA-3′ | 5′-GAAGATGGAGAGAAATTGGAGAA-3′ |
| CTP-1α | 5′-ACCCTGAGGCATCTATTGACAG-3′ | 5′-ATGACATACTCCCACAGATGGC-3′ |
| TNF-α | 5′-ACGGCATGGATCTCAAAGAC-3′ | 5′-AGATAGCAAATCGGCTGACG-3′ |
| IL-6 | 5′-CCTTCCTACCCCAATTTCCAA-3′ | 5′-AGATGAATTGGATGGTCTTGGTC-3′ |
| MCP-1 | 5′-GTGACTCGGACTGTGATG-3′ | 5′-CATTGAAAGTGTTGAATCTG-3′ |
| Claudin-2 | 5′-TATGTTGGTGCCAGCATTGT-3′ | 5′-TCATGCCCACCACAGATATA-3′ |
| Claudin-5 | 5′-GCAAGGTGTATGAATCTGTGCT-3′ | 5′-GCAAGGTGTATGAATCTGTGCT-3′ |
| Occludin | 5′-TTGAAAGTCCACCTCCTTACAGA-3′ | 5′-CCGGATAAAAAGAGTACGCTGG-3′ |
| ZO-1 | 5′-GCCGCTAAGAGCACAGCAA-3′ | 5′-TCCCCACTCTGAAAATGAGGA-3′ |
| GPR40 | 5′-CCCACGCTAAACTGCGACT-3′ | 5′-CGCTGAGAGCAGCTAGGAAG-3′ |
| GPR41 | 5′ -CTAAACCTGACCATTTCGGACC-3′ | 5′-GATAGGCCACGCTCAGAAAAC-3′ |
| GPR43 | 5′-ATCCTCCTGCTTAATCTGACCC-3′ | 5′-CGCACACGATCTTTGGTAGG-3′ |
| GPR120 | 5′-TGCCCCTCTGCATCTTGTTC-3′ | 5′-CGCGATGCTTTCGTGATCTG-3′ |
| β-actin | 5′-GTTACCACTGGGACGAC-3′ | 5′-CTCAAACATGATCTGGGTCA-3′ |
Statistical Analysis
All data are presented as the mean ± standard error (S.E.). Student’s t-test was used for the statistical analysis of data. Analysis of variance (ANOVA) was used to determine the level of significance.
Results
Selection and Identification of a Candidate Strain with Anti-Obesity Efficacy
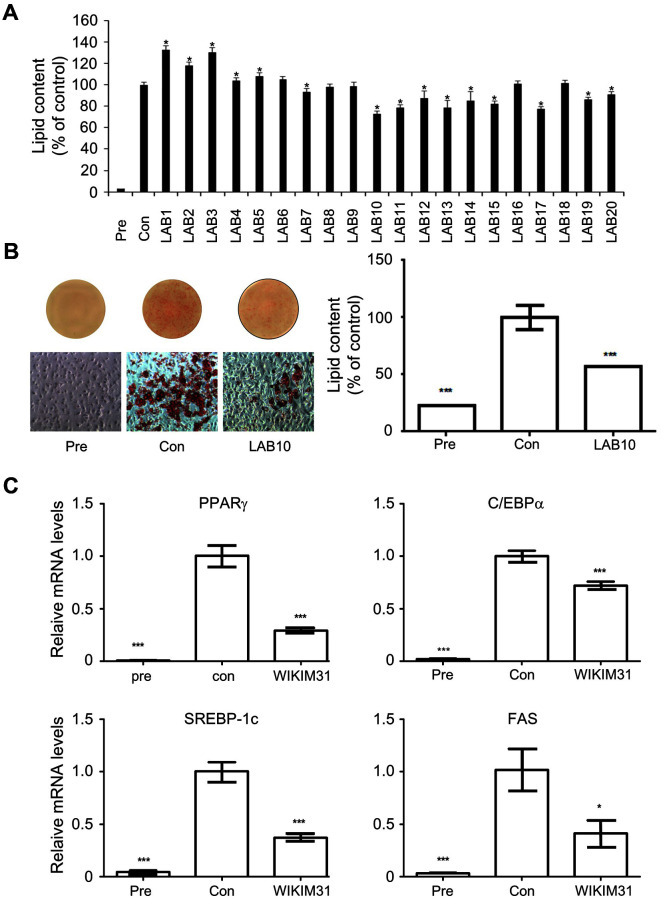
To screen lactic acid bacteria (LAB) with high anti-obesity efficacy in vitro, we measured the level of lipid accumulation by LAB treatment in 3T3-L1 adipocytes. Among 20 isolates, we selected a LAB10 strain for further study as it showed the highest inhibition of lipid accumulation in 3T3-L1 adipocytes (Fig. 1A). Even in a repeated assay, LAB10 reduced by about 44% compared to the control (Fig. 1B). In addition, we monitored the expression of genes related to adipogenes to assess whether WIKIM31 could suppress adipogenesis. The expression of PPARγ, C/EBPα, SREBP-1c, and FAS remarkably decreased compared with the control (Fig. 1C). Analysis of the 16S rRNA gene sequences of LAB10 showed 99.9% similarity with Latilactobacillus sakei (data not shown). Therefore, we identified a LAB10 strain as L. sakei. Based on these results, L. sakei LAB10 (= WIKIM31) was selected as a candidate with anti-obesity efficacy.
Fig. 1. The inhibitory effect of L. sakei WIKIM31 on lipid accumulation.
(A, B) 3T3-L1 cells were treated with 10% bacterial extracts for 10 days, and then intracellular lipid were stained with Oil Red O (Pre, no differentiated cells; Con, fully differentiated cells) (C) Gene expression of lipogenesis were determined by real-time RT-PCR. LAB10 was renamed as WIKIM31. Data are presented as the mean ± SD of triplicate tests. *p < 0.05, ***p < 0.001 vs. Con.
L. sakei WIKIM31 Administration Suppresses Weight Gain in HFD-Induced Obese Mice
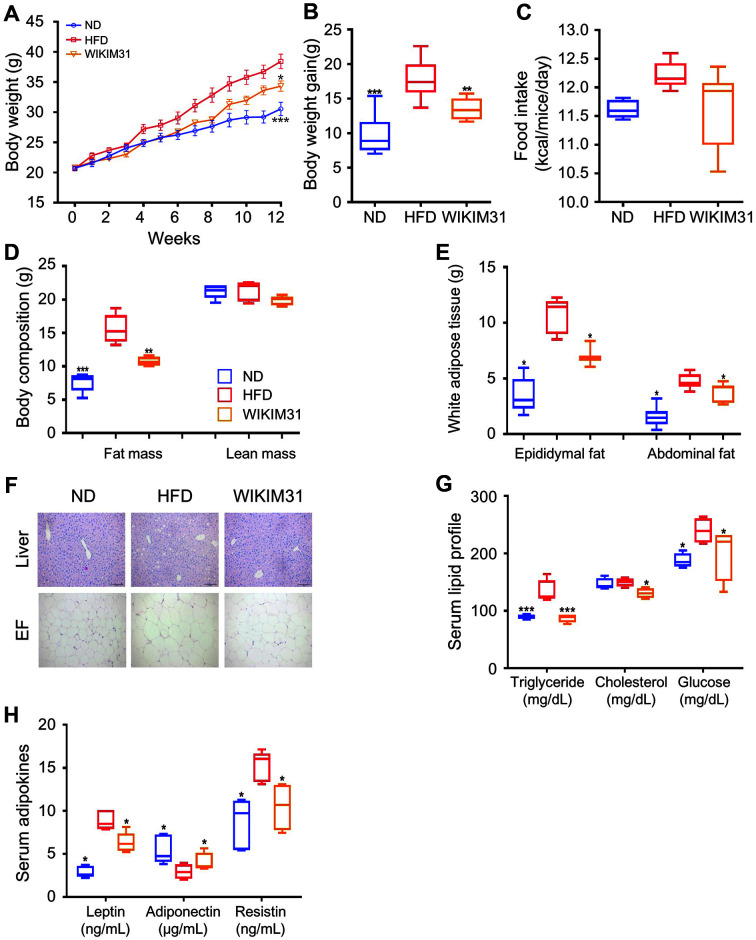
To determine whether WIKIM31 can improve HFD-induced obesity, WIKIM31 was orally administered to 7 week old male mice for 12 weeks. Mice fed both HFD and WIKIM31 showed reduced weight gain compared to mice fed a HFD (Fig. 2A, p = 0.0148; B, p = 0.0022), although food intake was not different (Fig. 2C). WIKIM31 led to a significant reduction in fat mass gain (29%), without suppressing lean mass gain, compared to HFD mice (p = 0.0049) (Fig. 2D). In addition, the weights of epididymal (p = 0.01) and abdominal adipose tissue (p = 0.0113) in HFD-WIKIM31 mice were significantly lower than those in HFD mice (Fig. 2E). Consistently, adipocyte size in EF and lipid droplet deposition in the liver were reduced by WIKIM31 administration (Fig. 2F). Furthermore, the serum levels of triglycerides, cholesterol, and glucose in HFD-WIKIM31 mice were reduced by 35% (p = 0.0006), 13% (p = 0.0155), and 16% (p = 0.0195), respectively (Fig. 2G). Alterations in serum adipocytokine levels are closely associated with obesity. For example, serum leptin and resistin levels increased, whereas adiponectin levels decreased in HFD mice. Therefore, we investigated whether WIKIM31 administration to mice during HFD feeding affected the serum adipocytokine levels. Leptin (p = 0.0072) and resistin (p = 0.0098) serum levels decreased, and adiponectin (p = 0.0177) levels in HFD-WIKIM31 mice increased (Fig. 2H). Together, these results indicate that WIKIM31 administration suppresses HFD-induced obesity.
Fig. 2. The effect of L. sakei WIKIM31 administration on HFD-induced obesity.
WIKIM31 or PBS was administered daily to C57BL/6 mice for 12 weeks during feeding with an ND or HFD. (A) Body weight. (B) Gain in body weight. (C) Food intake. (D) Total white fat mass. (E) Partial white fat mass. (F) Representative images of liver and epididymal fat (EF) staining. (G, H) Serum levels of triglycerides, cholesterol, glucose, leptin, adiponectin, and resistin. Data are presented as the mean ± SE. *p < 0.05, **p < 0.01 vs. HFD.
L. sakei WIKIM31 Administration Alters Metabolic Gene Expression in HFD-Induced Obese Mice
To identify the mechanism underlying the phenotype of HFD-WIKIM31 mice, the expression of lipid metabolic genes in the liver and EF was determined by real-time RT-PCR. The expression levels of hepatic or EF of lipogenesis-related genes, such as PPARγ (liver, p = 0.023; EF, p = 0.0043) , C/EBPα (liver, p = 0.0088; EF, p = 0.034), FAS (liver, p = 0.024; EF, p = 0.0009), SREBP-1c (liver, p = 0.0042; EF, p = 0.0044), SCD1 (EF, p = 0.0456), and CD36 (liver, p = 0.0103; EF, p = 0.0117) were substantially lower in HFD-WIKIM31 mice than in HFD mice (Figs. 3A and 3B). However, the expression of UCP2 (liver, p = 0.0204; EF, p = 0.0005) and CPT-1α (liver, p = 0.0019; EF, p = 0.0001) genes involved in β-oxidation, was remarkably increased in HFD-WIKIM31 mice (Figs. 3C and 3D). These results indicate that WIKIM31 administration improves lipid and energy metabolism in the liver and adipose tissue by modulating the expression of lipogenesis -and β-oxidation-related genes.
Fig. 3. The effect of L. sakei WIKIM31 treatment on lipogenesis and energy metabolism.
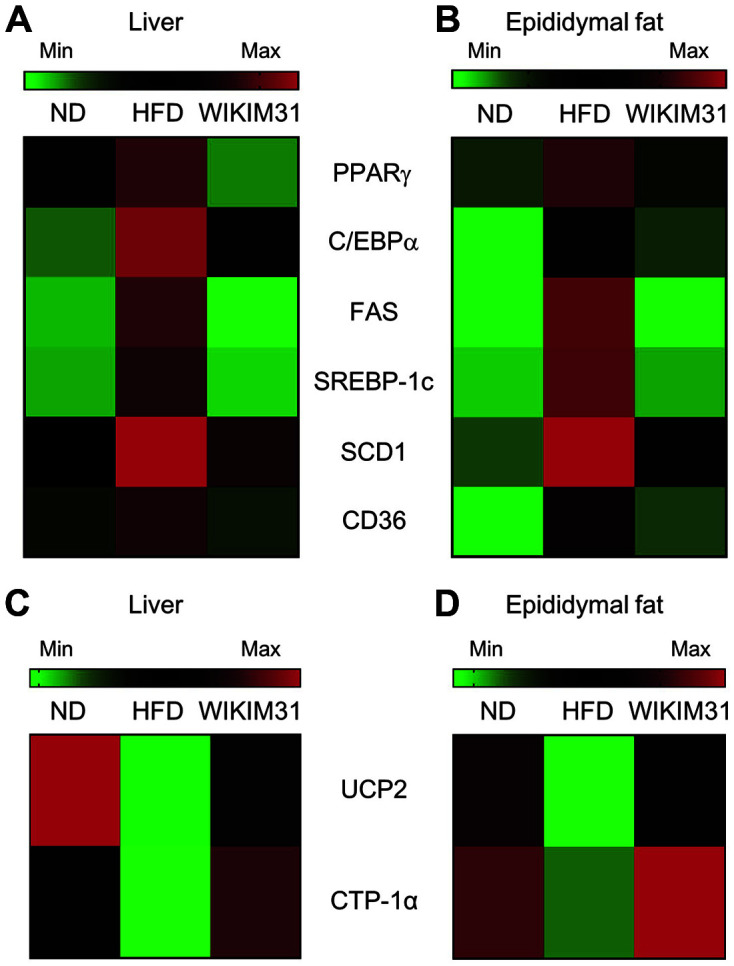
(A, B) The mRNA levels of lipogenesis and (C, D) energy metabolic genes were determined by real-time RT-PCR in liver and epididymal fat. Data are presented as the mean ± SE. *p < 0.05 vs. HFD.
L. sakei WIKIM31 Treatment Reduces Inflammatory Response and Ameliorates Gut Barrier Function in HFD-Induced Obese Mice
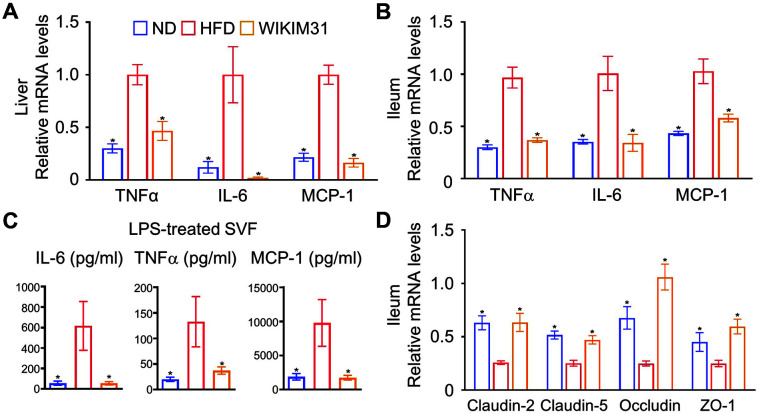
It is known that the secretion of pro-inflammatory cytokines and gene expression increases in HFD-induced obesity [5]. Therefore, we examined the expression levels of pro-inflammatory cytokine genes to determine whether WIKIM31 administration could reduce the inflammatory response. The expression of TNF-α, IL-6, and MCP-1, pro-inflammatory gene, were remarkably increased in the liver and ileum of HFD mice compared to those in ND mice. However, WIKIM31 administration strongly reduced the expression of these genes (TNF-α, p = 0.0048 in liver; p = 0.0003 in ileum, IL-6; liver: p = 0.0042, ileum: p = 0.0058, and MCP-1; liver: p = 0.0001, ileum: p = 0.0052) (Figs. 4A and 4B). Obesity is associated with chronic inflammation characterized by macrophages and monocytes infiltration, and pro-inflammatory cytokines production [5]. To address whether oral administration of WIKIM31 regulates the suppression of inflammatory response in the adipose tissue, SVF isolated from EF were re-stimulated with LPS for 24 h, and the levels of pro-inflammatory cytokines in the supernatant were measured. The increased levels of IL-6 (p = 0.047), TNF-α (p = 0.041), and MCP-1 (p = 0.0489) production in the HFD mice were remarkably decreased in the WIKIM31 mice (Fig. 4C). Intestinal inflammation is associated with intestinal barrier dysfunction [19, 20]. The effect of oral administration of WIKIM31 on the expression of tight junction proteins in the ileum of mice was further explored. Compared to HFD mice, WIKIM31 administration upregulated the expression levels of tight junction protein genes, such as claudin-2 (p = 0.0019), claudin-5 (p = 0.0017), occluding (p = 0.001), and zonula occludens-1 (ZO-1) (p = 0.0018) (Fig. 4D). Taken together, these results indicate that WIKIM31 treatment suppresses the immune responses related to obesity and further improves intestinal barrier function.
Fig. 4. The effect of L. sakei WIKIM31 administration on gut barrier function and inflammation response.
(A, B) Gene expression of pro-inflammatory cytokines was determined by real-time RT-PCR. (C) After isolated SVF were treated with LPS for 24 h, the levels of IL-6, TNF-α, and MCP-1 in the supernatant were detected using the Cytometric Bead Array kit. (D) Gene expression of tight junction proteins was determined by real-time RT-PCR. Data are presented as the mean ± SE. *p < 0.05 vs. HFD.
L. sakei WIKIM31 Administration Increases SCFA Content in HFD-Induced Obese Mice
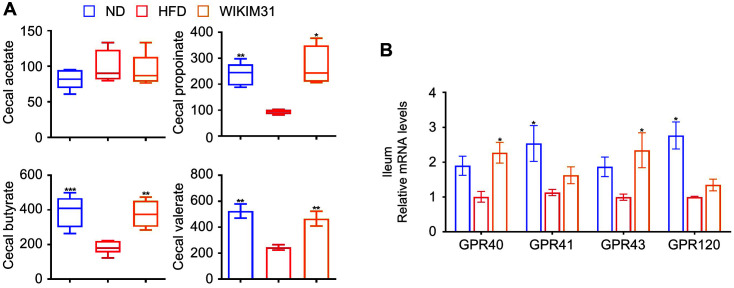
SCFAs are metabolites of the intestinal microbiota and are beneficial to the host body [21]. The contents of propionate, butyrate, and valerate in HFD mice were distinctly lower than those in ND mice (Fig. 5A). After WIKIM31 administration, these SCFAs increased in comparison with HD mice (propionate, p = 0.0014; butyrate, p = 0.0006; valerate, p = 0.0035). However, acetate content was not significantly different. Consistent with these results, the expression of G protein-coupled receptor (GPR) 40 (p = 0.0102) and GPR43 (p = 0.038), known as receptors for SCFAs, was remarkably increased (Fig. 5B).
Fig. 5. Concentration of short-chain fatty acids (SCFAs) from cecal contents and the gene expression of G protein receptors (GPRs) by L. sakei WIKIM31.
(A) Content of SCFAs (total SCFAs, acetate, propionate, butyrate, and valerate) in cecum. (B) Gene expression of GPRs in ileum was determined by real-time RT-PCR. Data are presented as mean ± SE. *p < 0.05, **p < 0.01, ***p < 0.001 vs. HFD.
Discussion
The pharmacological approach to the treatment of obesity can cause serious side effects, and probiotic administration has been considered an alternative [22]. Several clinical trials and studies have demonstrated the usefulness of probiotics in the treatment of metabolic diseases [23]. Probiotics improve obesity status by regulating the host’s energy metabolism and gut microbiota [24, 25].
In this study, to obtain LAB with anti-obesity from kimchi, we isolated 20 LAB strains from kimchi and examined their ability to inhibit lipid accumulation in vitro. L. sakei WIKIM31 was selected as it showed the highest inhibitory effect. Our in vivo results showed that the oral administration of L. sakei WIKIM31 in HFD-induced obese mice remarkably reduced body weight and fat mass gain without reducing food intake (Fig. 2). The liver and white adipose tissues are central organs with metabolic and lipogenic functions in the body [1, 26]. Therefore, dysregulation of energy metabolism in these organs is one of potential factors for metabolic disorders, such as obesity and diabetes [27]. It has been previously reported that L. sakei strains OK67 and ADM14 isolated from kimchi induce anti-obesity effects by modulating lipid metabolism in adipose tissue [16, 17]. Similarly, L. sakei WIKIM31 administration effectively reduced serum triglyceride contents by inhibiting the expression of lipogenesis-related genes (PPARγ, C/EBPα, FAS, SREBP-1c, SCD1, and CD36) and promotes energy expenditure by increasing the expression of β-oxidation-related genes, UCP2 and CPT-1α in liver and white adipose tissues.
In obese individuals, as tight junction protein levels decrease, intestinal permeability increases, resulting in the infiltration of pro-inflammatory molecules that lead to inflammatory responses [28, 29]. Tight junction proteins such as occludin and claudins, play important roles in the intestinal barrier [30]. Previous studies have reported that ingestion of probiotic strains increases the expression of tight junction proteins in the intestine [16-18]. In the present study, L. sakei WIKIM31 not only increased the expression of tight junction protein genes to the level of the normal group, but also reduced the expression of pro-inflammatory cytokines in the ileum. GPRs, receptors for SCFAs, have been known to modulate the expression of tight junction proteins upon activation, as well as to improve intestinal inflammation and host energy metabolism [21, 31, 32]. However, it is unclear whether L. sakei can regulate the expression of GPRs in the intestine. In this study, we clearly demonstrated that L. sakei WIKIM31 administration increases the level of propionate and butyrate, resulting in the enhanced expression of their receptors, GPR41 and GPR43.
In conclusion, oral administration of L. sakei WIKIM31 ameliorated obesity by suppressing lipogenesis and inflammatory responses via improvement of gut barrier functions and the production of SCFAs. These results suggest that L. sakei WIKIM31 can be used as a therapeutic agent for obesity.
Acknowledgments
This research was supported by a grant from the World Institute of Kimchi (KE2101-1) funded by the Ministry of Science and ICT, Republic of Korea.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Arner P, Bernard S, Salehpour M, Possnert G, Liebl J, Steier P, et al. Dynamics of human adipose lipid turnover in health and metabolic disease. Nature. 2011;478:110–113. doi: 10.1038/nature10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 2010;51:1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellulu MS, Patimah I, Khaza'ai H, Rahmat A, Abed Y. Obesity and inflammation: the linking mechanism and the complications. Arch. Med. Sci. 2017;13:851–863. doi: 10.5114/aoms.2016.58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makki K, Froguel P, Wolowczuk I. Adipose tissue in obesity-related inflammation and insulin resistance: cells, cytokines, and chemokines. ISRN Inflamm. 2013;2013:139239. doi: 10.1155/2013/139239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3:279–288. doi: 10.4161/gmic.19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George Kerry R, Patra JK, Gouda S, Park Y, Shin HS, Das G. Benefaction of probiotics for human health: a review. J. Food Drug Anal. 2018;26:927–939. doi: 10.1016/j.jfda.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He MQ, Shi BY. Gut microbiota as a potential target of metabolic syndrome: the role of probiotics and prebiotics. Cell. Biosci. 2017;7:54. doi: 10.1186/s13578-017-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo SR, Kim YJ, Park DY, Jung UJ, Jeon SM, Ahn YT, et al. Probiotics L. plantarum and L. curvatus in combination alter hepatic lipid metabolism and suppress diet-induced obesity. Obesity (Silver Spring) 2013;21:2571–2578. doi: 10.1002/oby.20428. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Liu F, Lu J, Shi J, Guan J, Yan F, Li B, Huo G. Probiotic mixture of Lactobacillus plantarum strains improves lipid metabolism and gut microbiota structure in high fat diet-fed mice. Front. Microbiol. 2020;11:512. doi: 10.3389/fmicb.2020.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong SH, Lee SH, Jung JY, Choi EJ, Jeon CO. Microbial succession and metabolite changes during long-term storage of Kimchi. J. Food Sci. 2013;78:M763–M769. doi: 10.1111/1750-3841.12095. [DOI] [PubMed] [Google Scholar]
- 14.Kwon MS, Lim SK, Jang JY, Lee J, Park HK, Kim N, et al. Lactobacillus sakei WIKIM30 ameliorates atopic dermatitis-like skin lesions by inducing regulatory T cells and altering gut microbiota structure in mice. Front. Immunol. 2018;9:1905. doi: 10.3389/fimmu.2018.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong YF, Kim H, Kim HR, Gim MG, Chung DK. Different immune regulatory potential of Lactobacillus plantarum and Lactobacillus sakei isolated from Kimchi. J. Microbiol. Biotechnol. 2014;24:1629–1635. doi: 10.4014/jmb.1406.06062. [DOI] [PubMed] [Google Scholar]
- 16.Lim SM, Jeong JJ, Woo KH, Han MJ, Kim DH. Lactobacillus sakei OK67 ameliorates high-fat diet-induced blood glucose intolerance and obesity in mice by inhibiting gut microbiota lipopolysaccharide production and inducing colon tight junction protein expression. Nutr. Res. 2016;36:337–348. doi: 10.1016/j.nutres.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Won SM, Chen S, Park KW, Yoon JH. Isolation of lactic acid bacteria from kimchi and screening of Lactobacillus sakei ADM14 with anti-adipogenic effect and potential probiotic properties. LWT Food Sci. Technol. 2020;126:109296. doi: 10.1016/j.lwt.2020.109296. [DOI] [Google Scholar]
- 18.Lee J, Jang JY, Kwon MS, Lim SK, Kim N, Lee J, et al. Mixture of two Lactobacillus plantarum strains modulates the gut microbiota structure and regulatory T cell response in diet-induced obese mice. Mol. Nutr. Food Res. 2018;62:e1800329. doi: 10.1002/mnfr.201800329. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez de Medina F, Romero-Calvo I, Mascaraque C, Martinez-Augustin O. Intestinal inflammation and mucosal barrier function. Inflamm. Bowel Dis. 2014;20:2394–2404. doi: 10.1097/MIB.0000000000000204. [DOI] [PubMed] [Google Scholar]
- 20.Meddings J. The significance of the gut barrier in disease. Gut. 2008;57:438–440. doi: 10.1136/gut.2007.143172. [DOI] [PubMed] [Google Scholar]
- 21.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bessesen DH, Van Gaal LF. Progress and challenges in anti-obesity pharmacotherapy. Lancet Diabetes. Endocrinol. 2018;6:237–248. doi: 10.1016/S2213-8587(17)30236-X. [DOI] [PubMed] [Google Scholar]
- 23.Kadooka Y, Sato M, Imaizumi K, Ogawa A, Ikuyama K, Akai Y, et al. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur. J. Clin. Nutr. 2010;64:636–643. doi: 10.1038/ejcn.2010.19. [DOI] [PubMed] [Google Scholar]
- 24.Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr. Pharm. Des. 2009;15:1546–1558. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- 25.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Invest. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy JK, Rao MS. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G852–G858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- 27.Choe SS, Huh JY, Hwang IJ, Kim JI, Kim JB. Adipose tissue remodeling: its role in energy metabolism and metabolic disorders. Front. Endocrinol. 2016;7:30. doi: 10.3389/fendo.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad R, Rah B, Bastola D, Dhawan P, Singh AB. Obesity-induces organ and tissue specific tight junction restructuring and barrier deregulation by claudin switching. Sci. Rep. 2017;7:5125. doi: 10.1038/s41598-017-04989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018;50:1–9. doi: 10.1038/s12276-018-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Priyadarshini M, Kotlo KU, Dudeja PK, Layden BT. Role of short chain fatty acid receptors in intestinal physiology and pathophysiology. Compr. Physiol. 2018;8:1091–1115. doi: 10.1002/cphy.c170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, Chen Y, Jiang H, Robbins GT, Nie D. G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int. J. Cancer. 2011;128:847–856. doi: 10.1002/ijc.25638. [DOI] [PubMed] [Google Scholar]