Abstract
The ability to utilize the iron bound by high-affinity iron-binding proteins in the vertebrate host is an important virulence factor for the marine fish pathogen Vibrio anguillarum. Virulence in septicemic infections is due to the presence of a highly efficient plasmid-encoded iron transport system. AngR, a 110-kDa protein component of this system, appears to play a role in both regulation of the expression of the iron transport genes fatDCBA and the production of the siderophore anguibactin. Therefore, study of the expression of the angR gene and the properties of its product, the AngR protein, may contribute to the understanding of the mechanisms of virulence of this pathogen. In this work, we present genetic and molecular evidence from transposition mutagenesis experiments and RNA analysis that angR, which maps immediately downstream of the fatA gene, is part of a polycistronic transcript that also includes the iron transport genes fatDCBA and angT, a gene located downstream of angR which showed domain homology to certain thioesterases involved in nonribosomal peptide synthesis of siderophores and antibiotics. In order to dissect the specific domains of AngR associated with regulation of iron transport gene expression, anguibactin production, and virulence, we also generated a panel of site-directed angR mutants, as well as deletion derivatives. Both virulence and anguibactin production were dramatically affected by each one of the angR modifications. In contrast to the need for an intact AngR molecule for anguibactin production and virulence, the regulation of iron transport gene expression does not require the entire AngR molecule, since truncation of the carboxy terminus carrying the nonribosomal peptide synthetase cores, as well as the site-directed mutations, resulted in derivatives that retained their ability to regulate gene expression which was only abolished after truncation of amino-terminal sequences containing helix-turn-helix and leucine zipper motifs and a specialized heterocyclization and condensation domain found in certain nonribosomal peptide synthetases. The evidence, while not rigorously eliminating the possibility that a separate regulatory polypeptide exists and is encoded somewhere within the 5′-end region of the angR gene, strongly supports the idea that AngR is a bifunctional protein and that it plays an essential role in the virulence mechanisms of V. anguillarum. We also show in this study that the angT gene, found downstream of angR, intervenes in the mechanism of anguibactin production but is not essential for virulence or iron transport gene expression.
The bacterial fish pathogen Vibrio anguillarum, a gram-negative, polarly flagellated, comma-shaped rod, is responsible for both marine and freshwater fish epizootics throughout the world (1). V. anguillarum causes a highly fatal hemorrhagic septicemic disease in salmonids and other fish, including eels (1, 10). The disease caused by this bacterium has remarkable similarities to invasive septicemic disease in humans: the sequence of events immediately after infection mimics mammalian inflammation, except for obvious species-specific responses (1, 29). The molecular characteristics of the virulence determinants, the features of the infection process, which resemble very closely those found in human infections, and the fact that the bacterium is the actual pathogen of this vertebrate host, makes the V. anguillarum-fish system an ideal paradigm for studying eukaryotic host-bacterium interactions at the molecular level. The key feature which enables the pathogenic strains of V. anguillarum to survive within the vertebrate host is the possession of a 65-kb virulence plasmid, pJM1, which provides the bacteria with an iron-sequestering system that is crucial in overcoming the nonspecific defense mechanisms of the host (9, 16–22, 37, 39). This system centers upon the synthesis of the siderophore anguibactin, an iron-scavenging compound, and subsequent transport of the ferric-anguibactin complex into the cell cytosol via the cognate transport system proteins FatA, -B, -C, and -D (2–4, 33, 53). Anguibactin is produced by the virulent strains of this bacterium in the host and in any other environment in which the bacteria's sole source of iron is chelated by high-affinity iron binding compounds (17). The plasmid-encoded iron transport system and siderophore biosynthetic genes are controlled via the concentration of available iron through three plasmid-encoded regulators: two positive regulators, AngR (anguibactin system regulator) and TAF [transacting factor(s)], and a negative regulator, antisense RNAα. Repression also requires the chromosomally encoded Fur protein (11–14, 31, 44, 45, 55, 57, 59). The positive regulation of iron transport gene expression is enhanced by anguibactin (14). Synthesis of anguibactin per se requires expression of genes from the chromosome and the virulence plasmid pJM1 (12). One of these plasmid-harbored genes, angR, encodes the AngR protein, which possesses predicted regulatory domains such as helix-turn-helix and leucine zipper-like motifs at both the amino and the carboxy termini, a feature common to DNA-binding proteins involved in transcriptional control. Previous work showed that AngR, together with TAF, enhanced the expression of anguibactin biosynthetic genes (44). We also recently demonstrated that expression of the iron transport genes fatB and fatA under iron-limiting conditions is dramatically reduced in an AngR-deficient strain of V. anguillarum, suggesting another regulatory function for AngR in modulating expression of iron transport genes (5, 14).
However, in addition to its role as a regulator, AngR may also be a key enzyme in anguibactin biosynthesis since it possesses a characteristic domain found in nonribosomal peptide synthetases that catalyze the synthesis of siderophores, such as enterobactin, and antibiotics such as gramicidin S and bacitracin. These compounds are the products of condensation of hydroxy acids and amino acids via a multistep process catalyzed by nonribosomal peptide synthetases of adenylation, thioesterification, cyclization of heterocyclic compounds, and sometimes racemization or N-methylation of each amino acid or hydroxy acid, thereby creating various peptide structures (15, 18, 26, 32, 36, 38, 47–50). On the basis of the crystallographic structure and chemical analysis of anguibactin (3, 34), we predict that anguibactin might be synthesized from the nonribosomal enzymatic modification of 2,3-dihydroxybenzoic acid (DHBA), cysteine, and histamine. Indeed, recent investigations demonstrated that both DHBA and histamine are required for the biosynthesis of this siderophore (12, 51). Certain nonribosomal peptide synthetases, such as gramicidin S synthetase, the high-molecular-weight protein 2 (HMWP2) from Yersinia enterocolitica, and the EntF protein, which is involved in serine activation during biosynthesis of the siderophore enterobactin in Escherichia coli, possess a domain with six characteristic cores (27, 30, 36, 41, 42, 47, 51). These cores are required for many functions: cores 1 through 5 are required for adenylation; cores 2, 3, and 5 are involved in ATP binding; core 4 has an ATPase motif, and core 6 is the 4′-phosphopantetheine binding site involved in thioester formation. The AngR protein possesses all six cores; however, core 6 is defective and may not be functional (18). Because of the defective core 6, thioester formation may require another protein(s) to provide the essential core 6, possibly in a multienzyme complex with AngR. The siderophore anguibactin also shares a striking structural feature with the antibiotic bacitracin produced by Bacillus licheniformis and the siderophore yersiniobactin from Y. enterocolitica: a thiazoline ring which could be formed by interaction between cysteine and another amino acid or another compound, such as DHBA (32, 34). It was then of interest that AngR, in addition to the six cores found on the carboxy terminus of the molecule, also possesses sequences at the amino-terminal end that show significant homology with domains of the bacitracin synthetase BA1 protein from B. lichiniforme as well as of the protein HMWP2-2 from Y. enterocolitica which is involved in yersiniobactin biosynthesis (32). These sequences defined a new class of specialized condensation domain: the so-called heterocyclization domain, consisting of seven conserved regions (Cy1 to Cy7), which catalyzes both peptide bond and thiazoline ring formation in bacitracin synthesis (32) and may form a thiazoline ring between activated molecules of cysteine and DHBA in anguibactin biosynthesis. However, AngR could have another possible function in anguibactin biosynthesis, since it can complement E. coli mutants deficient in EntE, an AMP ligase only possessing the first four cores (51). This enzyme is involved in adenylating DHBA that is then incorporated into the enterobactin biosynthetic pathway (26, 41, 42). Since DHBA is also essential for anguibactin biosynthesis (12), AngR could play a role in V. anguillarum similar to that of EntE in E. coli, i.e., adenylation of DHBA during anguibactin biosynthesis. However, it is possible that in the V. anguillarum cytoplasm the AngR adenylation domain functions in the adenylation of cysteine prior to transpeptidation and thiazoline ring formation rather than in the adenylation of DHBA.
In experimental infections, the ability to synthesize anguibactin and the presence of the iron transport genes to allow for its uptake were demonstrated to be essential factors of virulence (60). Since AngR appears to play a role in both of these processes, we report in this study genetic and molecular experiments to characterize the expression of the angR gene at the transcriptional level and to dissect the functions associated with various domains of the AngR protein in relation to their possible role in the expression of the virulence phenotype. We also analyze in this work the contribution of angT, a gene found downstream from angR, to anguibactin production, iron transport gene regulation, and virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
V. anguillarum (pJHC-T2612, pJHC9-8) is a strain harboring two plasmids: the recombinant clone pJHC-T2612, which possesses genes required for synthesis of anguibactin and the iron transport complex, and pJHC9-8, which encodes the regulatory product(s) TAF that is essential for full expression of the iron uptake system (43, 44, 53). Figure 1 shows a map of pJHC-T2612 and some of its derivatives. Further details can also be found on Table 1. V. anguillarum (pJHC-T2612::TnangR4,pJHC9-8) harbors the mutant derivative pJHC-T2612#4 which has a Tn3Ho-Ho1 insertion (denoted with a “#” sign) within the angR gene of pJHC-T2612, while V. anguillarum strains harboring either pJHC-T2612::TnfatD20,pJHC9-8, pJHC-T2612fatC17,pJHC9-8, or pJHC-T2612fatB15,pJHC9-8 have a Tn3Ho-Ho1 insertion within the iron transport genes fatD, fatC, or fatB, respectively, carried on pJHC-T2612 (25; see also Fig. 1). These strains will be named TnangR4, TnfatD20, TnfatC17, and TnfatB15 respectively. Plasmid pBluescript SK(+) (Stratagene) was used for cloning of DNA fragments for site-directed mutagenesis and sequencing. Plasmids pKK223-3 and pJHC-S100 (Table 1) were used for subcloning the mutagenized DNA fragments in order to return them to the V. anguillarum cytoplasm, which was accomplished by mobilization with the plasmid pRK2073 (43, 44).
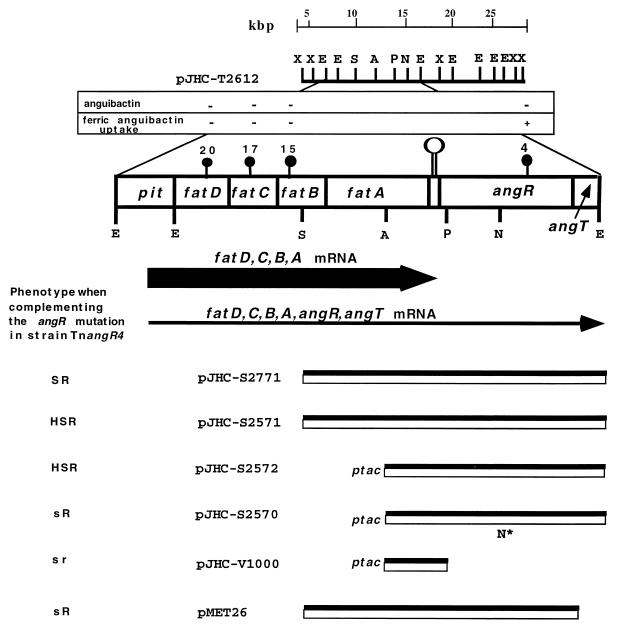
FIG. 1.
Genetic and physical map of plasmids used in this study. The dark circles symbolize the Tn3::HoHo1 transposon insertion mutants 20, 17, 15, and 4 in pJHC-T2612. The Tn3::HoHo1 transposon is approximately 14 kb and carries the ampicillin resistance marker in addition to a promoterless lacZ (52). Restriction sites are as follows: X, XhoI; E, EcoRI; S, SalI; A, AvaI; P, PstI; B, BamHI; N*, NcoI-site modified with the Klenow fragment of DNA polymerase I. The insert DNA from pJM1 is represented by the double black and white boxes. The vector DNA is not represented. pJHC-T2612 contains the iron uptake region of pJM1 cloned in the pVK102 vector. pJHC-S2771 and pMET26 are subclones carrying the angR gene from plasmid pJM1 cloned in pJHC-S100, a pBR325 derivative (see Table 1). pMET26 is an isogenic construct with pJHC-S2771 except that angT has been deleted. pJHC-2571 harbors the angR gene from pJHC1 also cloned in pJHC-S100. Plasmid pJHC-S2572 carries the angR gene from pJHC1 under the control of the ptac promoter by using the cloning vector pKK-223-3. Plasmid pJHC-2570 is an NcoI modification in the angR gene of pJHC-S2572, and pJHC-V1000 is a deletion at the PstI site within the angR gene, obtained from pJHC-2572. Pit is the promoter for the iron transport genes (11). The thick horizontal arrow represents the most abundant polycistronic transcript (fatDCBA mRNA) that terminates at or adjacent to the hairpin (open lollipop shape) located between the end of fatA and the beginning of angR. This hairpin has the characteristics of a rho-dependent transcription termination signal (2). The thin horizontal arrow represents a low-abundance polycistronic transcript (fatDCBA, angR, and angT mRNA). Phenotypes of the complementing plasmids in strain TnangR4 are as follows: HS, high siderophore production (531A-type); S, normal siderophore production (775-type); s, low or no siderophore production; R, positive for iron transport gene regulation; r, reduced or no iron transport gene regulation.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| XL1-Blue | recA1 endA1 gyrA46 thi hsdR17 supE44 relA lac F′[proAB+ lacIqlacZΔM15 Tn10(Tetr)] | Stratagene |
| HB101 | supE44 hsdS20 (rB, mB−) recA13 ara-14 proA2 lacyI galK2 rpsL20 xyl-5 mtl | 8 |
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1Δ (lac-proAB+) [F′ traD36 proAB lacIqlacZΔM15] | 46 |
| Vibrio anguillarum | ||
| 775(pJM1) | Wild type, Pacific Ocean prototype | 21 |
| 531A | Wild type, Atlantic Ocean prototype | |
| H775-3 | Plasmidless derivative of 775 | 22 |
| TnangR4 | H775-3 containing pJHC-T2612#4 and pJHC9-8 | 53 |
| TnfatD20 | H775-3 containing pJHC-T2612 #20 and pJHC9-8 | 53 |
| TnfatC17 | H775-3 containing pJHC-T2612 #17 and pJHC9-8 | 53 |
| TnfatB15 | H775-3 containing pJHC-T2612 #15 and pJHC9-8 | 53 |
| Plasmids | ||
| pJM1 | Indigenous plasmid in strain 775 | 16, 21 |
| pJHC1 | Indigenous plasmid in strain 531A | 44, 51, 54 |
| pJHC9-8 | pJM1 derivative carrying only the TAF region | 53, 58 |
| pJHC9-16 | pJM1 derivative anguibactin-deficient | 53, 58 |
| pJHC-S100 | Kmr cloning vector derived from pBR325 | 7, 44 |
| pBluescript SK | Cloning vector | Stratagene |
| pVK102 | Cloning vector derived from RP4 | 53 |
| pJHC-T2612 | Recombinant clone carrying the iron uptake region of pJM1 cloned in pVK102 | 53 |
| pJHC-T2612#4 | Iron uptake deficient, AngR-deficient recombinant clone pJHC-T2612 with Tn3::HoHo1 insertion in angR | 53 |
| pJHC-T2612#20 | Iron uptake-deficient recombinant clone derived from pJHC-T2612 by Tn3::-HoHo1 insertion in fatD | 53 |
| pJHC-T2612#17 | Iron uptake-deficient recombinant clone derived from pJHC-T2612 by Tn3::-HoHo1 insertion in fatC | 53 |
| pJHC-T2612#15 | Iron uptake-deficient recombinant clone derived from pJHC-T2612 by Tn3::-HoHo1 insertion in fatB | 53 |
| pJHC-S2771 | angR and angT genes from pJM1 cloned in pJHC-S100 | 44 |
| pJHC-S2571 | angR and angT genes from pJHC1 cloned in pJHC-S100 | 44 |
| pJHC-S2572 | angR and angT genes from pJHC1 cloned into pKK223-3 | 44 |
| pJHC-S2570 | angR and angT genes from pJHC1 with the NcoI modification cloned into pKK223-3 | 44 |
| pMET26 | Carries only angR gene; obtained by Klenow deletion of angT from pJHC2771 | This work |
| pMET13.1 | 92-bp ClaI-SalI fragment of the fatA gene cloned in pBluescript II SK(+) | 57 |
| pJHC-LW260 | 191-bp ThaI-RsaI fragment of the fatB region cloned in pBluescript II SK(+) | 57 |
| pQC3.5 | 420-bp SalI-ClaI fragment of aroC in pBluescript SK(+) | |
| pAWA1.1 | 100-bp fragment of the angR gene cloned in pBluescript SK(+) | 14 |
V. anguillarum was cultured in either trypticase soy broth or agar supplemented with 1% NaCl (TSBS and TSAS, respectively). For experiments determining iron uptake characteristics, the strains were first grown on TSAS supplemented with the appropriate antibiotics and passed on to M9 minimal agar with the same antibiotics. The resulting cultures were tested either on plates for bioassay or inoculated into M9 minimal medium supplemented with antibiotics and various concentrations of ethylenediamine-di-(o-hydroxyphenyl acetic acid) (EDDA) to achieve iron-limiting conditions and to determine the MIC for EDDA. Antibiotic concentrations used were as follows: ampicillin, 500 μg/ml, and tetracycline, 5 to 10 μg/ml.
General DNA procedures.
Plasmid DNA preparations were performed by using the alkaline lysis method of Birnboim and Doly (6, 35). Sequence-quality plasmid DNA was generated by using the appropriate Qiagen kits (Chatsworth, Calif.). Restriction endonuclease digestion of DNA was performed under the conditions recommended by the supplier (Life Technologies, Inc.). Transformations and other cloning strategies were carried out as described previously (34). Automated sequencing was performed by the Department of Molecular Microbiology and Immunology (MMI) Core Facility on the PE/ABI377 DNA sequencer with dye-terminator or dye-primer cycle sequencing chemistry or the A.L.F. Pharmacia fluorescent sequencer and either dye-primer sequencing with Sequenase or cycle sequencing with Taq polymerase. Manual sequencing was performed by the dideoxy chain-termination method (46) by using the Sequenase Kit (U.S. Biochemicals, Cleveland, Ohio) with the appropriate primers. Primers were all synthesized by the MMI Core Facility on a PE/ABD 394 automated synthesizer by using standard phosphoramidite chemistry on polystyrene solid supports. DNA and protein sequence analysis were carried out at the National Center for Biotechnology Information by using the BLAST network service and also by using the Sequence Analysis Software Package of the University of Wisconsin Genetics Computer Group (GCG). The GCG programs PILEUP and BESTFIT were used for comparisons of amino acid sequences.
Site-directed mutagenesis.
The SalI-EcoRI fragment containing the angR and angT genes from pJM1 was cloned into pBluescript SK(+) and then mutagenized by using the Muta-Gene Phagemid in vitro mutagenesis kit (Bio-Rad Laboratories, Richmond, Calif.) and synthetic mutagenic oligonucleotides. Site-specific mutations were confirmed by DNA sequencing with appropriate primers. In addition, the entire angR and angT genes were sequenced for most of the mutants to verify that no other regions of either angR or angT were affected during mutagenesis. Once mutagenized, the SalI-EcoRI fragments from each derivative were cleaved from the pBluescript vector and recloned into pJHC-S100 to generate the plasmids carrying the modified angR derivatives listed in Table 1 and Fig. 1 and 3. The modified angR derivatives cloned in the pJHC-S100 vector were then transferred by conjugation into V. anguillarum TnangR4 by using triparental matings as described previously (53). Each transconjugant was tested for regulation of gene expression by using a RNase protection assay with the iron transport gene-specific riboprobes for its MIC for the iron chelator EDDA, for anguibactin biosynthesis with bioassays, and for virulence in the trout model as described in the following corresponding sections.
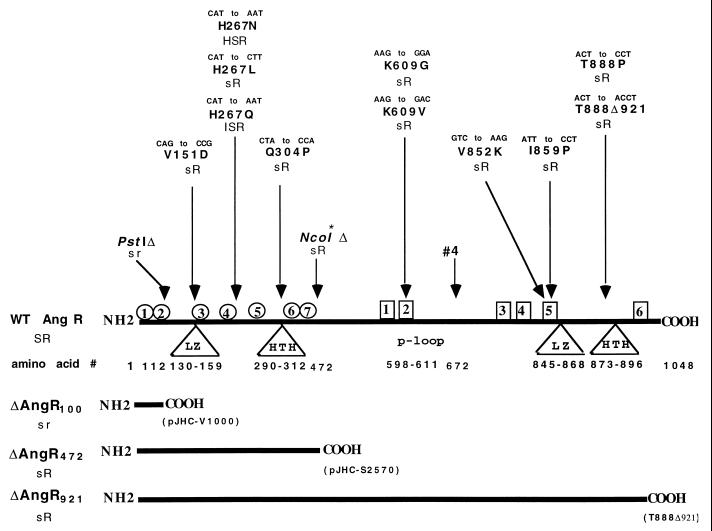
FIG. 3.
Location of mutations, modifications, and deletions on the angR gene and their phenotypes. Linear representation illustrates the regions of site-specific mutations, deletions, restriction site modification (NcoI*, with arrow), or transposon insertion (“#4”, with arrow). The mutations at the DNA level and the corresponding change in the amino acid at the protein level are indicated for each mutation. Triangles indicate that the region has structures that are predicted to be involved in regulation. The amino acids corresponding to the predicted domain are numbered below the triangle. LZ, predicted leucine zipper; HTH, predicted helix-turn-helix; p-loop, predicted ATP binding site. Circles represent the seven signature sequences (Cy1 to Cy7) corresponding to the highly conserved cyclization sequences identified in bacitracin synthetase BA1:1-2 from B. licheniformis, the proteins HMWP2-1 and HMWP2-2 from Y. enterocolitica, and the MTCY22H8.02 protein from M. tuberculosis. Amino acid numbers for each cyclization sequence are as follows: Cy1, 15 to 45; Cy2, 62 to 69; Cy3, 205 to 216; Cy4, 248 to 264; Cy5, 287 to 296; Cy6, 348 to 354; and Cy7, 365 to 391. Squares indicate regions where the core motifs found in nonribosomal peptide synthetases are located. Amino acid numbers for each core are as follows: core 1, 525 to 534; core 2, 598 to 611; core 3, 795 to 809; core 4, 838 to 842; core 5, 850 to 865; and core 6, 992 to 1002. The solid horizontal bars at the bottom show the protein molecules for the PstI deletion (ΔAngR100), the NcoI modification (ΔAngR472), and the truncation mutation T888Δ921 (ΔAngR921). Phenotypes are as follows: HS, high siderophore production (531A type); S, normal siderophore production (775 type); s, low or no siderophore production; IS, intermediate siderophore production; R, positive for iron transport gene regulation; r, reduced or no iron transport gene regulation.
Detection of the mutant AngR proteins.
We attempted to obtain AngR-specific antibodies by inoculating rabbits with overexpressed AngR protein obtained from a construct in which the angR gene was under the control of the ptac promoter. These putative antibodies reacted nonspecifically with many proteins even after absorption with proteins from the V. anguillarum plasmidless strain and from an E. coli strain (data not shown). These background problems, which would have likely obscured specific reactions, could be ascribed to the very low concentration of the AngR protein in the cell cytosol due to the low level of the angR transcript and the fact that the angR gene does not possess an optimal Shine-Dalgarno sequence (this work and reference 24). Therefore, we tried as an alternative approach to raise antibodies against specific oligopeptide sequences at the amino, carboxy, and middle regions of this protein. The oligopeptides were constructed by selecting regions with the highest probability to be antigenic and then used to generate antibodies in rabbits. The nonspecific reactions were considerably reduced; however, we could not detect any specific reaction with the AngR wild-type protein in V. anguillarum, although we could now detect AngR, if overexpressed, in the E. coli strains but not in V. anguillarum, which of course was not adequate (data not shown). Since neither of these immunological approaches worked in the V. anguillarum strains, we analyzed whether the AngR proteins in the mutants had the expected molecular weight by utilizing an in vitro-coupled transcription-translation system (Amersham Corp., Arlington Heights, Ill.). We used as templates the actual plasmids that were employed for the complementation experiments in V. anguillarum. The [35S]methionine-labeled proteins were electrophoresed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE); the gel was dried and exposed to X-ray film.
RNase protection assays.
Riboprobes were generated by in vitro transcription by using constructs pMET13.1 (linearized with ClaI for fatA), pJHC-LW260 (linearized with EcoRI for fatB), pAWA1.1 (linearized with AflII for angR), and pQC3.5 (linearized with RsaI for the internal marker aroC) (see Table 1 and references 12 to 14). After complete restriction endonuclease digestion, the DNA preparations were extracted with phenol and chloroform and then precipitated with ethanol. Riboprobes were then prepared by transcription of the above constructs with T3 RNA polymerase, [α-32P]UTP, and the appropriate unlabeled nucleotides, generating the fatA, fatB, angR, and aroC riboprobes. The values of the specific activities and quantities of the riboprobes were chosen according to their size and a standard hybridization curve by using 20 μg of total RNA from the wild-type strain and variable quantities of the labeled riboprobe. For each riboprobe we prepared a standard curve, and from this curve we were able to deduce the amount of the labeled riboprobe to be used. In a typical protection assay we would use ca. 105 cpm in 0.3 to 0.5 ng of a riboprobe. In the cases shown we used specific activities of 3.3 × 105 cpm/ng for the angR riboprobe, 3 × 105 cpm/ng for the fatB riboprobe, 2.7 × 105 cpm/ng for the fatA riboprobe, and 2.9 × 105 cpm/ng for the aroC riboprobe. Each probe was gel purified, resuspended in annealing buffer, and kept at −70°C until use. RNAs were prepared as follows: a 1:100 inoculum from an overnight culture grown in minimal medium plus the appropriate antibiotics was used. The inoculated cultures were grown with EDDA supplemented to just below the MIC to achieve maximal iron-limiting stress for each strain tested. Total RNA was prepared when the culture reached an optical density at 600 nm (OD600) of 0.4 to 0.6 by using the hot phenol method (56), and it was then stored at −70°C until analyzed. Between 10 and 20 μg of RNA was used for the RNase protections. The RNA was annealed for 8 to 16 h at 43 to 45°C with the fatA, fatB, or angR riboprobes, together with the aroC riboprobe (as an internal control for RNA concentrations) at the concentrations and radioactivity given above. The hybridized RNA was subsequently digested for 30 min at 30°C with RNases A and T1 (which were added to the hybridized samples but not to the tubes containing the control probes); proteinase K was then added and incubated an additional 30 min at 37°C. Samples were then treated with phenol and chloroform, followed by ethanol precipitation. The treated samples were next electrophoresed in a standard sequencing gel. Gels were exposed to X-ray film for 1 to 4 h with intensifying screens at −70°C.
Detection of anguibactin activity and determination of growth profiles under iron limitation.
Bioassays were performed to determine whether the mutant angR constructs would complement the siderophore-deficient phenotype of V. anguillarum TnangR4 after conjugation. Bioassays were carried out as described previously by using either culture supernatant of the transconjugants or by testing the strains grown on a minimal medium plate and then patched onto the bioassay lawn (53). The bioassay lawn is composed of an overnight culture of receptor-proficient, but anguibactin-deficient, V. anguillarum (pJHC9-16) (57). As a negative control the receptor-deficient and anguibactin-deficient V. anguillarum (pJHC9-8) was used (57). The following strains were each tested on the same bioassay plate: wild-type V. anguillarum 775 and strain TnangR4(pJHC-S2771), which serves as the positive control for complementation of the AngR-deficient strain, since pJHC-S2771 contains the entire angR and angT region cloned into pJHC-S100 (see Fig. 1), as the positive controls for anguibactin production; and the AngR-deficient strain TnangR4 as the negative control. Each mutant and deleted angR gene cloned in the same pJHC-S100 vector and conjugated into this AngR-deficient strain TnangR4 was tested on the bioassay plate. The plate was allowed to dry and subsequently was incubated at 20°C overnight. In addition, each mutant was tested for its ability to take up ferric anguibactin. This “reverse” bioassay uses the mutant strain as the lawn to assess whether it can be cross-fed with ferric anguibactin. The ferric complex was prepared by using purified anguibactin as previously described (3).
For each mutant we also determined the MIC for EDDA by using liquid cultures at increasing concentrations of EDDA (1, 2, 3, 4, and 5 μM) in M9 minimal medium at 20°C as previously described (53). The OD600 values at mid-log phase for each strain were plotted for each EDDA concentration, and the growth profiles were obtained. The experiments were performed in triplicate, and the standard deviations were calculated.
Fish infectivity assays.
Virulence tests were carried out on juvenile trouts (Salmo gairdnerii) weighing ca. 10 g which were anesthetized with tricaine methane sulfonate (0.1 g/liter). A total of 50 anesthetized fish were inoculated subcutaneously at the posterior base of the dorsal fin with 0.1 ml of each bacterial dilution, i.e., 50 fish per bacterial dilution. The dilutions were prepared with saline solution from 10-h cultures grown at 20°C in TSBS containing antibiotics for selection of the various plasmids harbored by the strains. The dilutions were prepared to test a range of cell concentrations from 102 to 108 cells/ml per strain. Therefore, 350 fish were tested per strain. After bacterial challenge, test fish were maintained in fresh water at 15°C for 1 month. Mortalities were checked daily, and kidney material was examined by bacteriological techniques. The isolated colonies were biotyped and serotyped, and their plasmid complement and antibiotic resistance levels were examined. Mortalities were considered to be due to a particular V. anguillarum strain when the bacterium could be reisolated in pure culture with the expected plasmid complement and antibiotic resistance. Virulence was quantified as the 50% lethal dose (LD50) (mean lethal dose: the number of microorganisms that will kill 50% of the animals tested) as determined by the method of Reed and Muench (40). Calculation of standard deviations for the LD50s indicate that, for a population of 50 fish per dilution tested, attenuations of virulence by a factor of 13 or more for high-virulence strains (LD50 <105) and a factor of 5 or more for low- and intermediate-virulence strains (LD50 >105) are significant with a 95% confidence limit (40).
RESULTS
Transposition mutagenesis of the iron transport region and its effect on angR expression.
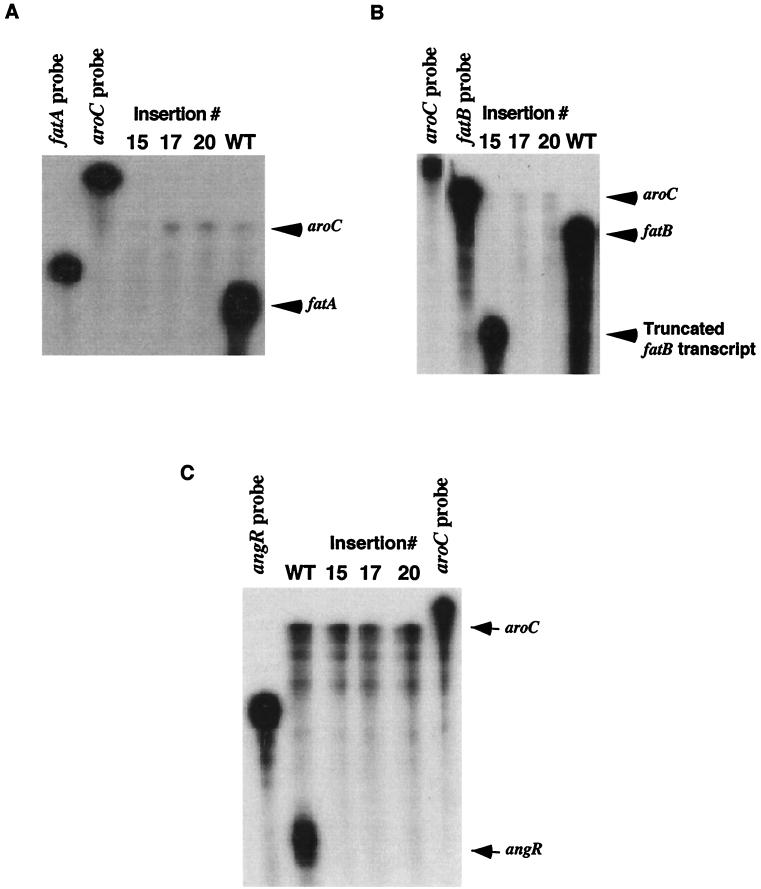
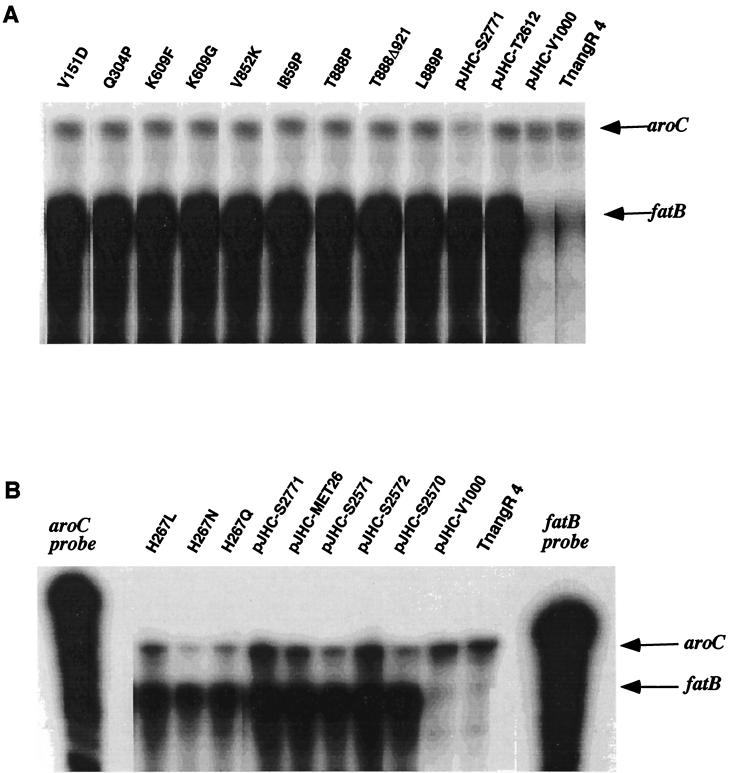
The diagram at the top of Fig. 1 illustrates that the iron transport genes fatD, fatC, fatB, and fatA and the angR gene are positioned in a head-to-tail fashion, which suggested that these genes could be part of an operon (53). Transposition mutations within each one of these genes all resulted in an iron transport-deficient and anguibactin-deficient phenotype (Fig. 1 and reference 53). However, we were not able to isolate any ca. 10-kb mRNA corresponding to the complete fatDCBA angR region by using Northern blots, possibly because of RNA processing or transcript instability (2, 57). To demonstrate that these genes are transcribed as an operon, we analyzed the expression of the fatA, fatB, and angR genes at the transcription level in the aforementioned transposition mutants. Riboprobes transcribed from constructs pMET13.1 and pJHC-LW260, by using T3 RNA polymerase, were used to detect specific fatA and fatB transcripts, respectively. Figure 2A and B illustrate that indeed transposition events in fatD (insertion 20, strain TnfatD20) and fatC (insertion 17, strain TnfatC17) lead to a significant attenuation of transcription for the fatA and fatB genes located downstream of the insertions compared to the internal aroC marker. In the case of insertion 15 (strain TnfatB15, panel B, lane 15), a truncated version of fatB mRNA can be detected, an expected result since the riboprobe used hybridizes to a region of fatB upstream and to another overlapping the actual site of insertion (14). Thus, the fatB mRNA which is protected by the riboprobe is smaller than the entire riboprobe. The polar effect of the transposon insertion mutations suggests that the fatDCBA genes must be contained within the same mRNA transcript. Figure 2C shows that, by using an angR-specific probe, expression of the angR gene was also affected by the upstream mutations in fatD, fatC, or fatB, suggesting that the angR gene must be part of the polycistronic message carrying fatD, fatC, fatB, and fatA. It was of interest that the signal band of angR-specific mRNA was always much lower in intensity than those for fatB or fatA compared to the aroC mRNA internal reference. We have determined that only a few transcriptional events go beyond a putative termination hairpin located between fatA and angR, resulting in a larger proportion of polycistronic transcripts that terminate at or around this hairpin, while a low level of transcriptional events traverse this hairpin and thus contain angR (2, 43). If we use a probe from the 3′ end of the angR gene and compare to the fatB probe we find the same result, i.e., angR transcripts are present at a lower level than fatB transcripts (43). These same results can be obtained if we compare the two angR probes with a fatA probe; abundance is also higher for the fatA-specific mRNA than the angR mRNA (43). Figure 1 shows schematically these two types of polycistronic transcripts and their relative abundance as determined from densitometer tracings of RNA gels (43). That these less-abundant transcripts are polycistronic messages containing angR together with fatDCBA was supported not only by the fact that the upstream insertion mutations in fatD, fatC, and fatB result in the disappearance of the angR signal but also because these insertions lead to a cessation of anguibactin biosynthesis (Fig. 1 and reference 53). The upstream transposition insertions also affected the expression of angT, a gene located downstream of angR which showed domain homology to certain thioesterases involved in nonribosomal peptide synthesis of siderophores, as determined by both RNase protection and reverse transcriptase PCR assays (unpublished data).
FIG. 2.
Effect of insertion mutations on transcription of the fatA, fatB, and angR genes as determined by RNase protection assays. Total RNA was harvested from V. anguillarum strains harboring various recombinant clones grown under iron-limiting conditions. Riboprobes were generated by using constructs pMET13.1, pJHC-LW260, and pQC3.5 (described in Table 1) and transcribed with T3 RNA polymerase, generating the fatA, fatB, and aroC probes, respectively. The aroC-specific riboprobe was included in the hybridization buffer as an internal control. For all three panels the specific transcripts were detected by RNase protection assays and are indicated by arrows, while the strains are indicated above each lane. Lanes marked aroC, fatB, fatA, or angR are the free riboprobes without RNase treatment. WT, TnangR4(pJHC-T2612); insertion 15, TnangR4(pJHC-T2612::TnfatB15); insertion 17, TnangR4(pJHC-T2612::TnfatC17); insertion 20, TnangR4(pJHC-T2612::TnfatD20). TnangR4 is V. anguillarum H775-3(pJHC-T2612::TnangR4, pJHC9-8). Tn is transposition sequence Tn3-HoHo1. Panels: A, detection of fatA-specific transcripts with the fatA probe; B, detection of fatB-specific transcripts with the fatB probe; C, detection of angR-specific transcripts with the angR probe.
Effect of modifications of the angR gene on anguibactin production and iron uptake.
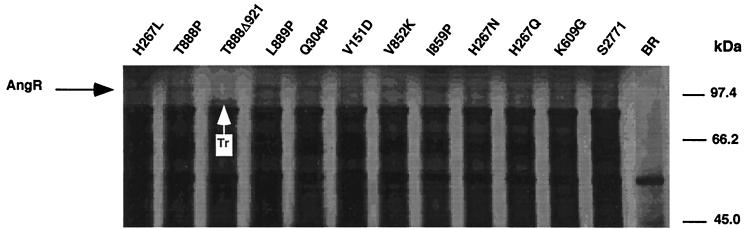
In order to dissect the functions of the angR gene, we performed site-directed mutagenesis and deletions at regions, shown in Fig. 3, containing helix-turn-helix, leucine zipper, or biosynthetic motifs, such as the heterocyclization domain and the synthetase cores, which could be involved in the properties of regulation of iron transport gene expression and anguibactin production ascribed to the AngR protein. The heterocyclization domain occurs on the amino-terminal end of the AngR molecule up to approximately amino acid 400, while the nonribosomal peptide synthetase motifs occur starting at amino acid 500 of AngR and continue towards the carboxy terminus up to amino acid 1002, just 46 amino acids before reaching the end of the molecule. The helix-turn-helix and leucine zipper motifs can be found at both the amino- and the carboxy terminus. Figure 3 identifies these characteristic sequences, as well as the location, nature, and phenotype of each mutation and deletion. In designing the mutations, we chose to change the amino acid at a particular site to amino acids that are predicted to alter the secondary structure of a given region. To verify that plasmids containing the site-specific mutations in angR were indeed synthesizing a stable AngR protein in V. anguillarum, we first tried to detect the modified AngR protein derivatives by immunological assays, which were unsuccessful (see Materials and Methods). Therefore, we analyzed whether the AngR proteins in the mutants had the expected molecular weight by utilizing an in vitro-coupled transcription-translation system. This method apparently leaves unanswered the question of the stability of the AngR proteins in the V. anguillarum cytosol; however, analysis of the effect of the mutations on the regulatory phenotype of AngR, which will be shown later in this work, suggests that the modified AngR proteins were stable in V. anguillarum. Figure 4 illustrates that all the mutants synthesized a protein of approximately 110 kDa or slightly less in the case of mutant T888Δ921, which is a frameshift mutant (see also Fig. 3). We have previously shown that transposon insertion 4 in the angR gene, as found in strain TnangR4, leads to a very unstable polycistronic mRNA and an AngR-deficient phenotype, which results in a dramatic decrease of both, expression of the iron transport genes, and of anguibactin production (reference 14 and Fig. 1). Therefore, the TnangR4 strain was used as the recipient for the plasmids containing the mutations and deletions of the angR gene cloned in plasmids pJHC-S100 or in pKK-223-3 (Table 1 and Fig. 3). The positive controls for these derivatives were the same strain TnangR4 carrying the wild-type angR gene from either pJM1 or pJHC1 (see Table 1) cloned in either of the aforementioned two vectors. Following the procedures diagrammed in Materials and Methods, we performed anguibactin bioassays and also determined the growth profiles of the strains under iron-limiting conditions for each of the site-specific angR mutants and truncations. In order to rule out a deficiency in the uptake of ferric siderophore resulting in a low MIC for EDDA, we also assayed the ability of each mutant to internalize ferric anguibactin. Each mutant was grown in minimal medium overnight at 20°C. The cultures were used to inoculate separate M9 plates supplemented with 10 μM EDDA to determine whether all of the modified AngR derivatives were able to be cross-fed when supplemented with a disc containing 5 μl of anguibactin.
FIG. 4.
Analysis of the synthesis of the AngR protein in the angR mutants. SDS-PAGE of polypeptides synthesized was done with coupled cell-free transcription-translation assays of recombinant clones harboring the mutated angR gene derivatives. BR represents the vector pJHC-S100 (see Table 1). The electrophoretic mobilities (in kilodaltons) of rabbit muscle phosphorylase b (97.4 kDa), bovine serum albumin (66.2 kDa), and hen egg white ovalbumin (45 kDa) used as markers are indicated in the right margin. Tr, truncated AngR. When the frameshift at amino acid 888 is introduced into mutant T888Δ921, a truncation is predicted. For the same amount of radioactivity, 5 × 104 cpm was loaded per lane.
Figure 5A shows that the strains harboring the angR site-directed mutants, except for H267N and H267Q, failed to produce measurable levels of anguibactin, while all of the control strains carrying an intact angR gene, either in the wild-type state or as a complementing plasmid, produced anguibactin. These results can also be found summarized in Fig. 3. Figure 5B shows that all of the mutants could utilize the ferric anguibactin provided. This cross-feeding experiment was carried out by first transferring the constructs to the plasmidless strain H775-3, since TnangR4, used as the recipient in all the crosses, is itself iron-uptake proficient when provided with ferric anguibactin (Fig. 1). Figure 5C demonstrates that the modified angR derivatives, except for H267N, pMET26, and H267Q (in order of proficiency), were also dramatically impaired in their ability to grow under iron-limited conditions. The negative control was the plasmidless strain H775-3, which did not produce anguibactin nor could utilize it.
FIG. 5.
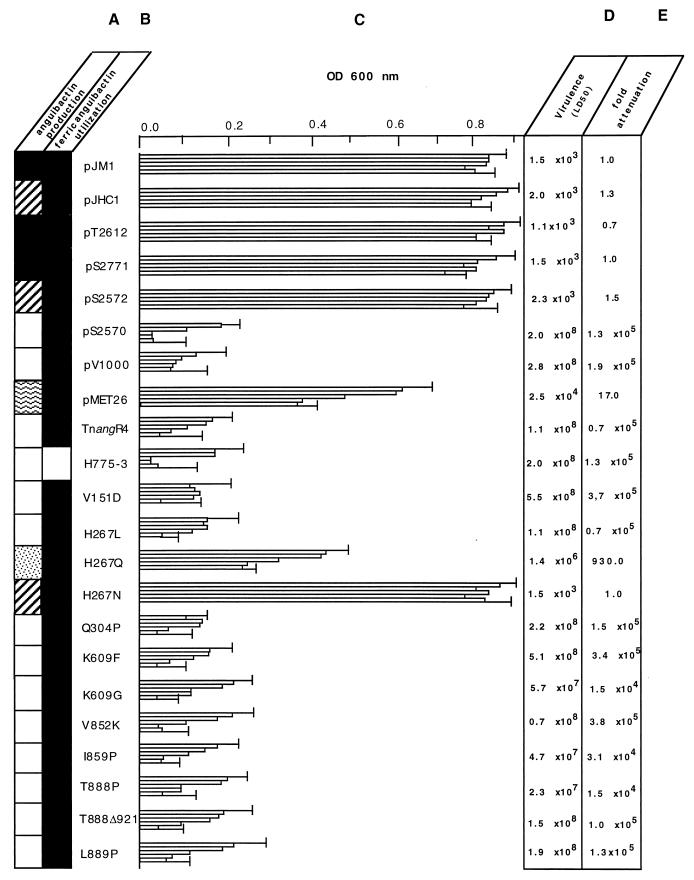
Phenotypes of the derivatives with the angR modifications. The strains are identified by the plasmid (either complementing or wild type) carrying the angR genes. Therefore, pJM1 corresponds to strain 775, pJHC1 to strain 531A, and pT-2612 to strain H775-3(pJHC-T2612). All of the other plasmid derivatives are in the TnangR4 strain background and are designated with a “p” before the number but omitting “JHC” for the sake of space. Mutants are designated by the actual transition in amino acids with the residue number in between the original amino acid and the mutated one. (A) Measurement of anguibactin production by bioassays. Values are obtained from the growth around disks inoculated with culture supernatant from the mutants and normalized to those obtained with the strains harboring the angR gene from pJM1, which was 12 ± 0.5 mm which is set to 100% (solid squares). Strains harboring angR from pJHC1, 300 to 400%, are indicated by striped squares. Strains with lower levels of anguibactin production are indicated as follows: 30 to 40%, wavy-line squares; 10 to 20%, dotted squares; no anguibactin produced, open squares. (B) Determination of ferric anguibactin uptake by reverse bioassay. Solid squares, positive ferric anguibactin utilization; open squares, unable to utilize ferric anguibactin. (C) Growth profiles of the strains at increasing concentrations of EDDA. Three cultures in M9 minimal medium for each strain were grown at 20°C for 6 h at each EDDA concentration. The OD600 values at mid-log phase for each strain were plotted for each EDDA concentration, and growth profiles were obtained. Each column for each strain corresponds to concentrations of EDDA, from top to bottom, of 1, 2, 3, 4, and 5 μM, respectively. Data shown are arithmetic means of the three cultures at each EDDA concentration. Error bars, shown only for the 1 and 5 μM EDDA concentrations, indicate standard deviations. (D) Quantification of virulence by the method of Reed and Muench (39) for the strains harboring plasmids with the angR modifications. Virulence tests were carried out on juvenile trouts (Salmo gairdnerii), weighing ca. 10 g, as described in Materials and Methods. (E) Relative values in fold attenuation normalized to the LD50 of strain 775(pJM1).
Mutation of residue 267 of AngR has been previously studied by us with respect to its influence on growth in the presence of EDDA (51). In this work we examined its correlation with anguibactin production and virulence. We determined that mutations in this position modulated anguibactin production in the following ways: when histidine (H) at the 267 location was replaced with asparagine (N), anguibactin levels increased. In this case it is of interest that the H267N mutation leads to the genotype of the angR gene found in 531A strains isolated in epizootics occurring in the Atlantic Ocean which produce a much higher level of anguibactin (54). Our results show that the high-siderophore production phenotype is independent of the cytoplasm of the V. anguillarum strain, since we performed the experiments in the 775 strain cytoplasmic background with the H267N mutation in the angR gene of pJM1 (i.e., strain 775) as part of plasmid pJHC-S2771. As expected, the angR gene from pJHC1 from strain 531A, cloned in plasmids pJHC-S2571 (vector pJHC-S100) and pJHC-S2572 (under the control of the ptac promoter in pKK223-3), also imparted the high-siderophore production phenotype. When H267 was replaced with glutamine (Q), anguibactin levels decreased below those of the wild-type derivative (H267), although glutamine is quite similar to asparagine, being simply one methylene group longer. Figure 3 shows a summary of these results. As demonstrated for the other single amino acid mutants, the ability to utilize ferric anguibactin was also unaffected in the mutants at position 267.
To assess whether siderophore production was affected by the complete elimination of ca. 60% of the carboxy-terminal end of the AngR molecule, which contains the specific synthetase cores 1 to 5 and defective core 6, we used construct pJHC-S2570 (Fig. 1 and Table 1). This plasmid harbors a NcoI modification by Klenow treatment of the pJHC1 angR gene cloned under the control of the ptac promoter, resulting in a truncated protein of 472 amino acids (Fig. 3 and reference 44). This modification was constructed in pJHC-S2572, with an asparagine at position 267, rather than pJHC-S2771, with a histidine at position 267, because of the higher siderophore production imparted by pJHC-2572, which would increase the sensitivity for detection of changes in the amount of siderophore production. As a further control, we also generated a DNA deletion at the PstI site in the angR gene of pJHC2572 that resulted in the truncation of ca. 90% of the AngR molecule, resulting in plasmid pJHC-V1000 (Fig. 1 and 3 and Table 1). Figure 5A through C shows that all three AngR truncations result in the complete loss of anguibactin production, affecting dramatically the growth profile of the strain under iron-limiting conditions with no effect on the ferric anguibactin uptake properties. Since most of the constructs possess the angT gene, which is found downstream of angR, we needed to prove whether or not angT is essential in anguibactin production and growth under iron limitation conditions. For this experiment we also used as a recipient of the various plasmids the strain TnangR4, in which the transposon insertion affects not only angR but also angT expression, since they are included in the same polycistronic transcript, resulting in an impairment of both anguibactin production and the ability to grow under iron-limiting conditions (see Fig. 5A and C). Figure 5A and C also show that complementation of the TnangR#4 transposon mutation with pJHC-S2771, a plasmid carrying both angR and angT, results in the restoration of anguibactin production and the ability to grow under iron-limiting conditions. If we complement with plasmid pMET26, in which angT is deleted, there is a decrease in anguibactin production and a concomitant impairment in the ability of V. anguillarum to grow under iron-limiting conditions; however, it is not quite as substantial as that found with the TnangR4 strain. The truncation mutations, i.e., the NcoI modification in pJHC-2570 and TΔ921 that cause premature termination of the AngR protein, because of their intrinsic nature, protein truncation, do not affect translation of angT. However, we have also assessed the existence of an active angT gene by complementation studies of these mutants with plasmid pMET26, a wild-type angR clone that does not harbor angT. As expected, the PstI DNA deletion does affect both angR and angT (unpublished results).
Contribution of the angR gene to multiplication and virulence of V. anguillarum in the vertebrate host.
Infectivity studies were performed to elucidate the contribution of angR to the pathogenicity of V. anguillarum and to determine the AngR domains that are essential for the spread of the bacterium in the host fish. We utilized the constructs and mutants that we have described in the previous section in experimental infections in the trout model system.
Figure 5D shows that the wild-type strains of V. anguillarum 775(pJM1) and 531A(pJHC1) had LD50s of 1.5 × 103 and 2 × 103, respectively. The strain TnangR4(pJHC-T2612) carrying the cloned iron uptake region also showed a high-virulence phenotype, with an LD50 of 1.1 × 103. The other two positive controls, which harbor plasmids with the cloned angR gene complementing the insertion mutation in angR in strain TnangR4, TnangR4(pJHC-S2771) and TnangR4(pJHC-S2572), showed LD50s of 1.5 × 103 and 2.3 × 103, respectively, being of the same order of magnitude as that of the V. anguillarum strain harboring pJHC-T2612 with an intact iron uptake region. Similar high virulence values were obtained with TnangR4(pJHC-S2571) carrying the angR gene from pJHC1 cloned in pJHC-S100 (data not shown). The negative control strains TnangR4, which carries a transposon insertion that causes a mutation in the angR gene, and the plasmidless H775-3 showed a dramatically reduced virulence with LD50s of 1.1 × 108 and 5.5 × 108, respectively. Therefore, our virulence analysis of the angR and angT modifications is very accurate since we are comparing LD50 values of isogenic strains containing the various constructs. Furthermore, calculation of standard deviations for the LD50s indicate that, for a population of 50 fish per dilution tested, attenuation of virulence by a factor of 13 or more for high-virulence strains (LD50 <105) and a factor of 5 or more for low- and intermediate-virulence strains (LD50 >105) are significant, with a 95% confidence limit (39).
Figure 5D and E demonstrate the following points: virulence is greatly reduced when AngR is mutated in the putative ATP-binding P loop, and attenuation is ca. 105 for both the K609G mutation and the K609D mutation. A similar decrease in virulence also occurs when either the first helix-turn-helix is changed by the helix breaker Q304P mutation or the second helix turn-helix is changed by the two helix-breaking mutations T888Δ921. Attenuation is ca. 105 fold for the T888P mutant, leading to a normal length AngR molecule, and ca. 104 fold for the T888Δ921 mutant, resulting in a truncated AngR. Also, mutations in the leucine zippers, V151D or I859P, result in a dramatic decrease in virulence. The 267 site, at which we reported modulation of anguibactin production, also shows a modulation of virulence. One of the mutations, H267L, has a very strong effect in reducing virulence by ca. 105 fold, while the mutation H267Q shows a less-dramatic decrease in virulence of 930 fold. The H267N mutation results in a high-virulence phenotype, with an LD50 of 1.5 × 103, the same order of magnitude as that of the isogenic wild types carrying pJHC-2771 (histidine in position 267, cloned from pJM1, LD50 of 1.5 × 103) or pJHC-S2572 (asparagine in position 267, cloned from pJHC1, LD50 of 2.3 × 103), although, as shown in the previous section, the H267N mutation resulted in a much higher production of anguibactin compared with that of the wild type. Obviously, there must be a threshold concentration of anguibactin at which maximum virulence is reached. As shown in Fig. 5D, both the NcoI modification, TnangR4(pJHC-S2570), and the PstI deletion of the angR gene, TnangR4(pJHC-V1000), also lead to a dramatic attenuation of virulence of ca. 105 fold. In summary, all of the modifications of AngR that lead to a cessation of anguibactin biosynthesis and a loss of the ability to grow under conditions of iron limitation also affect virulence dramatically. Since most of the constructs possess the angT gene found downstream of angR, we needed to prove whether angT influences the expression of the virulence phenotype. Therefore, we also tested the virulence of the strain in which the angT gene was deleted (see Fig. 1), TnangR4(pMET26). Figure 5D and E show that deletion of angT results in only a moderate attenuation of virulence (ca. 17 fold), correlating with the partial decrease in anguibactin production and consequent partial growth impairment of this strain.
Effect of the AngR modifications on the regulation of the expression of iron transport genes.
To assess the effect of each of the AngR mutations and deletions on the regulation of the expression of the iron transport genes, we determined the level of fatB-specific mRNA by using RNase protection assays since we have shown here that, as part of a polycistronic message, its expression is directly correlated with the expression of the other iron transport genes (14). The strains carrying the modified and wild-type angR genes were grown in minimal medium at the maximum iron limitation (based on the MIC of EDDA for the mutant), and total RNA was prepared as described in Materials and Methods. To perform the RNase protection experiments, the specific riboprobes for fatB and aroC (used as an internal control) were prepared by using T3 RNA polymerase as described in Materials and Methods. Figure 6 shows that none of the mutants caused a decrease in the fatB-specific transcript, suggesting that these AngR regions may not be required for the regulation of the iron transport genes. Even mutant T888Δ921, the frameshift mutant, retained full regulatory activity. As expected, the positive controls carrying an intact cloned angR gene introduced in strain TnangR4 showed full expression of fatB, while the negative control, strain TnangR4, which lacks active AngR, did not. Figure 6B shows that the mutations at amino acid position 267, which dramatically modulated anguibactin production, did not result in any major changes in expression of fatB relative to the aroC control (shown in the three lanes after the untreated aroC probe). Unexpectedly, Fig. 6A also shows that the mutation Q304P, which introduces a putative helix-breaking proline within the recognition helix of the predicted helix-turn-helix motif, does not affect appreciably the level of fatB-specific mRNA compared to the aroC control. This is also true for the mutation V151D in the leucine zipper, which affected anguibactin production and virulence (see previous sections) but did not affect the regulation of fatB expression by these modified AngR derivatives. Therefore, we tested the regulatory activity of the NcoI-modified angR gene which resulted in truncated AngR molecules that retained only the first predicted regulatory features and heterocyclization domains localized in the N-terminal 472 amino acids (in pJHC2570) or that lost most of this region (in pJHC-V1000). The NcoI modification truncation preserves the first helix-turn-helix and leucine zipper, as well as the cyclization sequences in the amino terminus of AngR, but no longer encodes the predicted synthetase cores, while the PstI deletion, in addition to having deleted the synthetase cores, also lost the helix-turn-helix-leucine zipper motifs and heterocyclization domain. Figure 6B shows that the NcoI modification truncation retained the ability to regulate the expression of fatB to levels similar to that of the wild type encoded by pJHC-S2771, pJHC-S2571, or pJHC-S2572, while the PstI deletion in pJHC-V1000 did not. It is also clear from Fig. 6B that AngR regulation of fatB expression can take place efficiently in the absence of the angT gene, as assessed when pMET26, which is AngR proficient but has a deletion of the angT gene, is the complementing plasmid in strain TnangR4.
FIG. 6.
Regulation of fatB gene expression by site-directed mutations and modifications or deletions of angR as determined by RNase protection assays. Total RNA was harvested from V. anguillarum strains harboring various recombinant clones grown under iron-limiting conditions. Strains were grown under their respective MICs for EDDA to achieve maximal iron stress as follows: bioassay-deficient mutants, 0.5 μM EDDA; bioassay-proficient mutants, 5 μM EDDA. Specific transcripts for fatB and aroC (their positions are indicated by arrows on the right) are detected by RNase protection by using the riboprobes for specific recognition of transcripts in the fatB or aroC region, respectively, as described in Materials and Methods. The aroC mRNA is constitutively expressed in V. anguillarum and is an internal control. All plasmids containing the mutagenized angR genes were constructed by cloning the SalI-EcoRI fragment harboring the mutant angR gene in pJHC-S100 in V. anguillarum TnangR4. The mutant strains are designated by the actual mutation. The wild-type angR genes are harbored by pJHC-T2612, pJHC-S2771, or pJHC-S2571. The negative control is strain TnangR4, which is AngR deficient. Lanes marked on top as aroC probe and fatB probe are the free riboprobes without RNase treatment. Specific RNase-protected RNAs are marked aroC and fatB in the right margin, with arrows pointing to their location in the gel. (A) Strain TnangR4 containing either plasmid pJHC-T2771 (wild-type angR cloned in pJHC-S100) or pJHC-T2612 (iron uptake region including the wild-type angR gene cloned in pVK102) served as positive controls, while the AngR-deficient strain, TnangR4, or the same strain containing the angRangT PstI deletion in plasmid pJHC-V1000, are the negative controls. (B) Mutations and plasmids are identified as in panel A. The clones are also carried on the TnangR4 strain and are designated above the lanes. Controls include the wild-type angR genes in pJHC-S100, pJHC-S2771, and pJHC-S2571 and, under the control of the ptac promoter, in pKK223-3, pJHC-S2572, pJHC-S2570, and pJHC-V1000.
DISCUSSION
AngR, a 110-kDa protein encoded on the pJM1 plasmid of V. anguillarum, appears to play a role in both regulation of the expression of the iron transport genes fatDCBA (5, 14) and the production of the siderophore anguibactin, either at the biosynthetic level (18, 51) or as a regulator of expression of siderophore biosynthetic genes (44). Early transposition mutagenesis studies revealed that a particular region of the pJM1 plasmid, a region, spanning nearly 25 kb, was required for siderophore production, iron acquisition, and virulence (53). A discrete subregion, spanning just under 10 kb, harbored the iron transport genes fatDCBA, the angR gene, and angT, a gene found downstream of angR, whose product may be a thioesterase acting in the release of the finished anguibactin from a pantothenate site (2, 18, 24, 44, 51, 53). The concerted regulation of these genetic determinants, as well as their consecutive locations, suggested that these genes could be transcribed as a single polycistronic transcript (18, 57). Earlier attempts to prove the polycistronic nature of this message utilized Northern blot experiments and led to the identification of a ∼2.5-kb transcript (2, 57) rather than the anticipated ∼10-kb that would harbor these six genes. Recent evidence suggests that the ∼10-kb mRNA containing fatDCBA, angR, and angT may be quite unstable and thus is found only at low concentrations, while processed products such as the 2.5-kb species accumulate (57). In the present study we have presented genetic and molecular evidence showing that expression of the angR and angT genes at the mRNA level decreases upon insertion of a transposon within any of the upstream fat genes. These insertions upstream of angR were also pleiotropic with respect to shutting off anguibactin production, thus underscoring the importance of AngR in anguibactin production and confirming the existence of an operon encoding a polycistronic mRNA with the fatDCBA, angR, and angT genes. We have recently identified the iron-regulated promoter element for this polycistronic message just upstream of fatD and were able to demonstrate not only that it binds Fur-Mn2+ complexes but also that expression from this promoter is repressed by Fur-iron complexes (11). Another important objective of the present study was to characterize the angR gene and its potential role as a virulence factor by using a panel of site-specific mutants and deletions to dissect domains of AngR that may be required for either regulation, such as helix-turn-helix and leucine zipper motifs, or for biosynthesis, such as the nonribosomal peptide synthetase cores and specialized heterocyclization domain, to assess their effect on virulence.
Examination of the properties of the angR modifications underscores the perfect correlation between virulence and anguibactin production: the dramatic decrease in virulence is prefaced by the complete shutoff of anguibactin production by these mutations. Therefore, does virulence depend on the amount of anguibactin produced and, if so, would a mutation in any other gene affecting siderophore production have the same negative effect on virulence as did the angR modifications? Our previous sequence analysis identified an open reading frame (ORF), angT, downstream from angR, which shows homology with certain thioesterases involved in nonribosomal peptide synthesis (24). The evidence shown in the present work indicates that this gene is indeed essential for anguibactin biosynthesis and growth under iron limitation. Deletion of the angT gene results in a decrease, but not in a complete shutoff, of anguibactin production and no change in the regulatory properties. However small, this amount of anguibactin was sufficient to allow for multiplication of the bacterium in the iron-limiting growth medium, as well as in the host fish, thus resulting in only a moderate reduction of virulence. The angT gene product may act in the release of the already-formed anguibactin from a panthotenate site (36, 48, 49). The strain harboring the plasmid pMET26 in which the angT gene was deleted, TnangR4(pMET26), may not be able to release anguibactin properly, although some anguibactin or anguibactin precursor molecules may be released by unknown mechanisms, which may include nonspecific hydrolysis. Therefore, the angT gene, although involved in anguibactin production, appears not to be as essential for virulence as is the case for the angR gene.
Another intriguing finding stemming from this work is the modulation of anguibactin production and virulence by mutations occurring at position 267 of AngR. Mutations from the wild-type histidine, found in the AngR protein of 775-type strains of the Pacific Northwest and Japan (24, 54), to asparagine leads to an AngR protein that has an enhanced capacity for anguibactin production. An AngR protein with an asparagine at position 267 is commonly found in 531A-type strains isolated in the East Coast of the United States and in Spain (53); these strains are high producers of anguibactin. We have shown here that the H267N mutant is virulent but not more so than the wild type carrying the histidine at position 267. This finding correlates with the fact that both Pacific Northwest and East Coast strains are highly virulent and have very similar LD50s. The amount of anguibactin produced by the strains carrying the AngR protein with histidine at position 267 may already be sufficient for the multiplication and spread in the host organism, and thus any extra anguibactin made by the AngR with asparagine at that position may be beyond the threshold level for virulence. An H267Q change leads to a decrease in anguibactin production and a concomitant reduction in virulence by 3 orders of magnitude, even though glutamine has the same uncharged polar group as asparagine with an extra methylene group. The H267L mutation, which results in a change to a nonpolar amino acid, leads to a dramatic reduction in virulence concomitant with the shutoff of anguibactin production. Neither of these changes affected the regulatory properties of AngR. Therefore, there is a tremendous capacity for modulation of anguibactin production with a concomitant change in virulence, depending on the amino acid present at this position. Ironically, no biosynthetic or regulatory motifs could be identified at this position, although, amino acid 267 is located within the amino-terminal end present in the truncated AngR obtained by the NcoI modification, 3 amino acids downstream of cyclization sequence 4 and 19 amino acids upstream of cyclization sequence 5. Scanning mutagenesis of neighboring sites to the 267 position, including the heterocyclization domain, is currently being carried out to pinpoint the actual region affected by the modulation phenomenon.
Truncation of the AngR molecule by modifying the NcoI restriction site resulted in a 472-amino-acid AngR protein derivative in which all the core biosynthetic motifs and one of the two helix-turn-helix and leucine zipper regions were missing. The strain harboring this derivative was unable to grow under iron limitation, resulting in a dramatic decrease in virulence; however, the truncated AngR molecule retained its ability to positively regulate expression of the iron transport genes. This result suggested that the first half of the AngR molecule contained the regulatory activity, which was then possibly associated with the heterocyclization domain, the first helix-turn-helix–leucine zipper region, or both. However, one mutant, Q304P, containing a proline, a known helix-breaker mutation in place of a glutamine in the predicted helix-turn-helix, could still regulate fatB gene expression. Mutations in the leucine zipper, such as V151D, also affected siderophore production, growth under iron limitation, and virulence but conserved regulatory capabilities. Therefore, these data suggest that the predicted helix-turn-helix or leucine zipper motifs may not be important for regulation of the expression of iron transport genes. Further deletions, such as that present in pJHCV1000, which left only about 10% of the AngR molecule, losing both the regulatory domains as well as the cyclization sequences, also lost the regulatory capability on the fatB gene expression, in addition to being affected in siderophore production, growth under iron limitation, and virulence. Our data are therefore consistent with the existence of a regulatory function within this amino-terminal end region of AngR. Single amino acid mutations in this region affect only siderophore production and virulence, with no effect on the regulation of fatB gene expression. In the whole AngR molecule, some of the single amino acid mutations occurred in predicted biosynthetic domains of angR, all of which had diminished anguibactin production, with no effect on the AngR regulatory activity. Therefore, we have to presume that these amino acid changes in either of the two portions of the AngR molecule may disturb the structure of the AngR protein, which is necessary for its synthetase function with no effect on the regulatory properties. Of course, while biosynthetic domains of many nonribosomal peptide synthetases are clearly recognized, it is also possible that all of these mutations are in unrecognized biosynthetic domains specific for anguibactin, in addition to the cyclization sequences and the biosynthetic cores. An alternative explanation, consistent with the phenotypes of the single-amino-acid mutations and the NcoI modification, is the existence of a separate small regulatory polypeptide encoded within the first half of the angR gene region that was not detected in our in vitro protein assays. In B. subtilis, the valine-activating nonribosomal peptide synthetase gene, srfAB1, is essential in the biosynthesis of the peptide antibiotic surfactin, as well as for competence during genetic transformation (25). These two activities, which had been thought to be two functions of the SrfAB1 peptide synthetase, were recently demonstrated to be the products of two different genes. A region encoded in the 5′ end of srfAB1 contains a small ORF, out-of-frame with srfAB1, which was actually responsible for the transformation competence activity (23, 25, 28). An out-of-frame ORF has not been identified within the angR gene, although an in-frame small ORF, expanding the site of the deletion in pJHC-V1000, could still exist. The evidence, therefore, while not rigorously eliminating the possibility that a separate, still unidentified, regulatory polypeptide exists and is encoded somewhere within the 5′-end region of the angR gene, strongly supports the idea that AngR is a bifunctional protein and that it plays an essential role in the virulence mechanisms of V. anguillarum. It is intriguing that the same stretch of amino acids that have the regulatory properties of AngR also carry the heterocyclization domain. Therefore, it is also possible that these sequences are involved in the biosynthesis of a precursor of anguibactin which might be the actual regulatory molecule.
In summary, our results suggest that, although the control of iron transport gene expression and siderophore production in V. anguillarum are intimately connected through AngR, the major effect of this protein on virulence is related to its central role in anguibactin production, either at the biosynthetic level or in the control of expression of anguibactin biosynthetic genes (44, 51). We are currently carrying out experiments to dissect the role of AngR in anguibactin production and gene regulation, as well as to identify the minimum AngR region required for the regulation of iron transport gene expression.
ACKNOWLEDGMENTS
This work was supported by Public Health grant AI19018 from the National Institutes of Health (NIH) to J.H.C. A.M.W. was the recipient of an NIH graduate training fellowship. A.M.W. and Q.C. were recipients of N. L. Tartar Research Fellowships from the Medical Foundation of Oregon. W.V. was a recipient of scholarships from the Hogeschool van Utrecht and the government of The Netherlands.
We thank Jo Ann Leong, Department of Microbiology, Oregon State University, for the use of the experimental infection facilities at the Salmon Disease Laboratory.
REFERENCES
- 1.Actis L A, Tolmasky M E, Crosa J H. Vibriosis. In: Woo P T K, Bruno D W, editors. Fish diseases and disorders, vol. 3. Viral, bacterial, and fungal infections. Wallingford, United Kingdom: Cab International Publishing; 1999. pp. 523–557. [Google Scholar]
- 2.Actis L A, Tolmasky M E, Farrell D H, Crosa J H. Genetic and molecular characterization of essential components of the Vibrio anguillarumplasmid-mediated iron-transport system. J Biol Chem. 1988;263:2853–2860. [PubMed] [Google Scholar]
- 3.Actis L A, Fish W, Crosa J H, Kellerman K, Ellenberger S R, Jauser F M, Sanders-Loehr J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum775(pJM1) J Bacteriol. 1986;167:57–65. doi: 10.1128/jb.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Actis L A, Potter S, Crosa J H. Iron-regulated outer membrane protein OM2 of Vibrio anguillarumis encoded by virulence plasmid pJM1. J Bacteriol. 1985;161:736–742. doi: 10.1128/jb.161.2.736-742.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Actis L A, Tolmasky M E, Crosa L M, Crosa J H. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol Microbiol. 1995;17:197–204. doi: 10.1111/j.1365-2958.1995.mmi_17010197.x. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene. 1978;4:121–126. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- 8.Boyer H, Roulland-Doussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 9.Bullen J J, Griffiths E. Iron binding proteins and host defense. In: Bullen J J, Griffiths E, editors. Iron and infection. 2nd ed. London, England: John Wiley & Sons, Ltd.; 1999. pp. 327–368. [Google Scholar]
- 10.Canestrini G. La malattia dominante delle anguille. Atti Ist Veneto Sci Lett Arti. 1893;7:809–814. [Google Scholar]
- 11.Chai S, Welch T, Crosa J H. Characterization of the interaction between Fur and the iron transport promoter of the virulence plasmid in Vibrio anguillarum. J Biol Chem. 1998;273:33841–33847. doi: 10.1074/jbc.273.50.33841. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, Actis L A, Tolmasky M E, Crosa J H. Chromosome-mediated 2,3-dihydroxybenzoic acid is a precursor in the biosynthesis of the plasmid-mediated siderophore anguibactin in Vibrio anguillarum. J Bacteriol. 1994;176:4226–4234. doi: 10.1128/jb.176.14.4226-4234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q, Crosa J H. Antisense RNA, Fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J Biol Chem. 1996;271:1885–1891. doi: 10.1074/jbc.271.31.18885. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Wertheimer A M, Tolmasky M E, Crosa J H. The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol Microbiol. 1996;22:127–134. doi: 10.1111/j.1365-2958.1996.tb02662.x. [DOI] [PubMed] [Google Scholar]
- 15.Crecy-Lagard V, Blanc V, Gil P, Naudin L, Lornzon S, Famechon A, Bamas-Jacques N, Crouzet J, Thibaut D. Pristinamycin I biosynthesis in Streptomyces pristinaespiralis: molecular characterization of the first two structural peptide synthetase genes. J Bacteriol. 1997;179:705–713. doi: 10.1128/jb.179.3.705-713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosa J H. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarumspecifies an iron-sequestering system. Nature. 1980;284:566–568. doi: 10.1038/284566a0. [DOI] [PubMed] [Google Scholar]
- 17.Crosa J H. Genetics and molecular biology of siderophore mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;67:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crosa J H. Molecular genetics of iron transport as a component of bacterial virulence. In: Bullen J J, Griffiths E, editors. Iron and infection. 2nd ed. London, England: John Wiley & Sons, Ltd.; 1999. pp. 255–288. [Google Scholar]
- 20.Crosa J H. The relationship of plasmid-mediated iron transport and bacterial virulence. Annu Rev Microbiol. 1984;38:69–89. doi: 10.1146/annurev.mi.38.100184.000441. [DOI] [PubMed] [Google Scholar]
- 21.Crosa J H, Schiewe M H, Falkow S. Evidence for plasmid contribution to the virulence of the fish pathogen Vibrio anguillarum. Infect Immun. 1977;27:509–513. doi: 10.1128/iai.18.2.509-513.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crosa J H, Hodges L, Schiewe M H. Curing of a plasmid is correlated with an attenuation of virulence in the marine fish pathogen Vibrio anguillarum. Infect Immun. 1980;27:897–902. doi: 10.1128/iai.27.3.897-902.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Souza C, Nakano M M, Zuber P. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:9397–9401. doi: 10.1073/pnas.91.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrell D H, Mikesell P, Actis L A, Crosa J H. A regulatory gene, angR, of the iron uptake system of Vibrio anguillarum: similarity with phage P22 croand regulation by iron. Gene. 1990;86:45–51. doi: 10.1016/0378-1119(90)90112-5. [DOI] [PubMed] [Google Scholar]
- 25.Fuma S, Fujishima Y, Corbell N, D'Souza C, Nakano M, Zuber P, Yamane K. Nucleotide sequence of the 5′ portion of sfrA that contains the region for competence establishment in Bacillus subtilis. Nucleic Acids Res. 1993;21:93–97. doi: 10.1093/nar/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehring A M, Bradley K A, Walsh C T. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 27.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley A, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarumand belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamoen L W, Eshuis H, Jongbloed J, Venema G, van Sinderen D. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol Microbiol. 1995;15:55–63. doi: 10.1111/j.1365-2958.1995.tb02220.x. [DOI] [PubMed] [Google Scholar]
- 29.Harbell S O, Hodgins H O, Schiewe M H. Studies on the pathology of vibriosis in coho salmon. J Fish Dis. 1979;2:527–535. [Google Scholar]
- 30.Heaton M P. Biosynthesis of d-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the d-alanine-activating enzyme. J Bacteriol. 1992;174:4707–4717. doi: 10.1128/jb.174.14.4707-4717.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunt M D, Pettis G S, McIntosh M A. Promoter and operator determinants for fur-mediated iron regulation in the bi-directional fepA-fes control region of the Escherichia colienterobactin gene system. J Bacteriol. 1994;176:3944–3955. doi: 10.1128/jb.176.13.3944-3955.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konz D, Klens A, Schorgendorfer K, Marahiel M A. The bacitracin biosynthesis operon of Bacillus licheniformisATCC 10176: molecular characterization of three multi-modular synthetases. Chem Biol. 1997;4:927–937. doi: 10.1016/s1074-5521(97)90301-x. [DOI] [PubMed] [Google Scholar]
- 33.Koster W L, Actis L A, Waldbeser L S, Tolmasky M E, Crosa J H. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum775. J Biol Chem. 1991;266:23829–23833. [PubMed] [Google Scholar]
- 34.Jalal M, Hossain D, van der Helm D, Sanders-Loehr J, Actis L A, Crosa J H. Structure of anguibactin, a unique plasmid-related bacterial siderophore from the fish pathogen Vibrio anguillarum. J Am Chem Soc. 1989;111:292–296. [Google Scholar]
- 35.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 36.Marahiel M A. Multidomain enzymes involved in peptide synthesis. FEBS Lett. 1992;307:40–43. doi: 10.1016/0014-5793(92)80898-q. [DOI] [PubMed] [Google Scholar]
- 37.Martinez J I, Herrero M, de Lorenzo V. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutAgenes. J Mol Biol. 1994;238:288–293. doi: 10.1006/jmbi.1994.1290. [DOI] [PubMed] [Google Scholar]
- 38.Merriman T R, Merriman M E, Lamont I L. Nucleotide sequence of pvdD, a pyoverdin biosynthesis gene from Pseudomonas aeruginosa: pvdDhas similarity to peptide synthetases. J Bacteriol. 1995;177:252–258. doi: 10.1128/jb.177.1.252-258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neilands J B. Mechanism and regulation of synthesis of aerobactin in Escherichia coliK12(pColV-K30) Can J Microbiol. 1992;38:728–733. doi: 10.1139/m92-119. [DOI] [PubMed] [Google Scholar]
- 40.Reed L J, Muench H. A simple method of estimating fifty percent end points. Am J Hyg. 1939;27:493–497. [Google Scholar]
- 41.Rusnak F, Faraci W S, Walsh C T. Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl) adenylate product. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 42.Rusnak F, Sakaitanis M, Drueckhammer D, Reichert J, Walsh C T. Biosynthesis of the Escherichia coli siderophore enterobactin: sequence of the entFgene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 43.Salinas P, Crosa J H. Regulation of angR, a gene with regulatory and biosynthetic functions in the pJM1 plasmid-mediated iron uptake system of Vibrio anguillarum. Gene. 1995;160:17–23. doi: 10.1016/0378-1119(95)00213-p. [DOI] [PubMed] [Google Scholar]
- 44.Salinas P C, Tolmasky M E, Crosa J H. Regulation of the iron uptake system in Vibrio anguillarum: evidence for a cooperative effect between two transcriptional activators. Proc Natl Acad Sci USA. 1989;86:3529–3533. doi: 10.1073/pnas.86.10.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salinas P, Waldbeser L S, Crosa J H. Regulation of the expression of bacterial iron transport genes: possible role of an antisense RNA as a repressor. Gene. 1993;123:33–38. doi: 10.1016/0378-1119(93)90535-b. [DOI] [PubMed] [Google Scholar]
- 46.Sanger F, Nicklen S, Coulson R A. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scholten J D, Chang K, Babbit P, Charest H, Sylvestre M, Dunaway-Mariano D. Novel enzymatic hydrolytic dehalogenation of a chlorinated aromatic. Science. 1991;253:182–185. doi: 10.1126/science.1853203. [DOI] [PubMed] [Google Scholar]
- 48.Stachelhaus T, Marahiel M A. Modular structure of peptide synthetases revealed by dissection of the multifunctional enzyme GrsA. J Biol Chem. 1995;270:6163–6169. doi: 10.1074/jbc.270.11.6163. [DOI] [PubMed] [Google Scholar]
- 49.Stachelhaus T, Marahiel M A. Modular structure of genes encoding multifunctional peptide synthetases required for nonribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 50.Toh H. Sequence analysis of firefly luciferase family reveals a conservative sequence motif. Protein Seq Data Anal. 1991;4:111–117. [PubMed] [Google Scholar]
- 51.Tolmasky M E, Actis L A, Crosa J H. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyperproduction of anguibactin. Infect Immun. 1993;61:3228–3233. doi: 10.1128/iai.61.8.3228-3233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tolmasky M E, Actis L A, Crosa J H. A histidine decarboxylase gene encoded by the Vibrio anguillarumplasmid pJM1: histamine is a precursor in the biosynthesis of anguibactin. Mol Microbiol. 1994;15:87–95. doi: 10.1111/j.1365-2958.1995.tb02223.x. [DOI] [PubMed] [Google Scholar]
- 53.Tolmasky M E, Actis L A, Crosa J H. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel transactivator of siderophore biosynthesis. J Bacteriol. 1988;160:860–866. doi: 10.1128/jb.170.4.1913-1919.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tolmasky M E, Salinas P C, Actis L A, Crosa J H. Increased production of the siderophore anguibactin mediated by pJM1-like plasmids in Vibrio anguillarum. Infect Immun. 1988;56:1608–1614. doi: 10.1128/iai.56.6.1608-1614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolmasky M, Wertheimer A M, Actis L A, Crosa J H. Characterization of the Vibrio anguillarum furgene: role in regulation of expression of the FatA outer membrane protein and catechols. J Bacteriol. 1994;176:213–220. doi: 10.1128/jb.176.1.213-220.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Gabain A, Belasco J, Schottel J, Chang A, Cohen S. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waldbeser L S, Tolmasky M E, Actis L A, Crosa J H. Mechanisms for negative regulation by iron of the FatA outer membrane protein gene expression in Vibrio anguillarum775. J Biol Chem. 1993;268:10433–10439. [PubMed] [Google Scholar]
- 58.Walter M, Potter S, Crosa J H. Iron uptake system mediated by Vibrio anguillarumplasmid pJM1. J Bacteriol. 1983;156:880–887. doi: 10.1128/jb.156.2.880-887.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wertheimer A M, Tolmasky M E, Actis L A, Crosa J H. Structural and functional analyses of mutant Fur proteins with impaired regulatory function. J Bacteriol. 1994;176:5116–5122. doi: 10.1128/jb.176.16.5116-5122.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf M, Crosa J H. Evidence for the role of a siderophore in promoting Vibrio anguillaruminfections. J Gen Microbiol. 1986;132:2949–2952. doi: 10.1099/00221287-132-10-2949. [DOI] [PubMed] [Google Scholar]